Abstract
We investigate the hypothesis that, in Escherichia coli, while the concentration of RNA polymerases differs in different growth conditions, the fraction of RNA polymerases free for transcription remains approximately constant within a certain range of these conditions. After establishing this, we apply a standard model-fitting procedure to fully characterize the in vivo kinetics of the rate-limiting steps in transcription initiation of the Plac/ara-1 promoter from distributions of intervals between transcription events in cells with different RNA polymerase concentrations. We find that, under full induction, the closed complex lasts ∼788 s while subsequent steps last ∼193 s, on average. We then establish that the closed complex formation usually occurs multiple times prior to each successful initiation event. Furthermore, the promoter intermittently switches to an inactive state that, on average, lasts ∼87 s. This is shown to arise from the intermittent repression of the promoter by LacI. The methods employed here should be of use to resolve the rate-limiting steps governing the in vivo dynamics of initiation of prokaryotic promoters, similar to established steady-state assays to resolve the in vitro dynamics.
Keywords: free RNA polymerase, in vivo transcription dynamics, rate-limiting steps, reversible closed complex formation, repressor binding dynamics
1. Introduction
Gene expression has been intensively studied with the relatively new tools provided by fluorescent proteins and microscopy techniques with single-molecule resolution, in both prokaryotic1–5 and eukaryotic6,7 systems. These studies have established that this process cannot be fully characterized by the mean protein production rate,8–12 since cells exhibit fluctuations (i.e. noise) over time and diversity in numbers across populations,13 which, among other things, generates phenotypic diversity.8 The noise has generally been investigated through indirect means, such as by observing the diversity in RNA and protein numbers in cell populations.2,3,10,11,14 Other, more direct means consist of observing the distribution of intervals between RNA productions2,4,5 and between protein bursts in individual cells.3,15
From these observations, a wide range of gene expression behaviours have been reported and, therefore, significantly different probabilistic models of transcription have been proposed.2,4,16–18 In general, higher-than-Poissonian variability in RNA numbers has been explained by models in which the promoter intermittently switched into an inactive state, resulting in bursty RNA production dynamics.2,16,19 Meanwhile, lower-than-Poissonian variability appears to be more consistent with models assuming multiple rate-limiting steps.4,5,16,20,21
There is direct experimental evidence for the existence of both mechanisms. Recently, Chong et al.19 showed that bursts of RNA production can emerge due to positive supercoiling build-up on a DNA segment, which eventually stops transcription initiation for a short period until the release of the supercoiling by gyrase. On the other hand, the existence of rate-limiting steps was established by studies using steady-state assays.22–24 Also, more recently, by fitting a monotone piecewise-constant function to the fluorescence signal from MS2-GFP tagged RNAs in individual cells, it was shown that in vivo RNA production can be a sub-Poissonian process.4,5,20,21
Recent studies have considered the possibility that both mechanisms can be present in a single promoter.16,25 In ref. 25, a model including both mechanisms was proposed, and statistical methods were developed to select the relevant components and estimate the kinetics of the intermediate steps in initiation based on empirical data. However, this method cannot distinguish the order of the steps which occur after the start of transcription initiation, nor can it determine their reversibility, which recent evidence suggests may play a significant role in the dynamics of RNA production.26
A complete model for transcription in prokaryotes must account, apart from the genome-wide variability in noise levels,17,27,28 for the well-established genome-wide variability in mean transcription rate2,3,8 and in fold change (ratio of production rate between zero and full induction)29 in response to induction found, e.g. in Escherichia coli promoters. For example, in vitro measurements on fully induced variants of the lar promoter showed that the mean interval between transcription events of these variants differs by hundreds of seconds.29 Promoters also differ widely in range of induction, even when differing only by a couple of nucleotides.29,30 For example, while PlarS17 has an induction range of 500 fold, PlarconS17 has an induction range of 4.5-fold, even though it only differs by 3 point mutations.29 This wide behavioural diversity is likely made possible by the sequence dependence of each step in transcription initiation.29
Thus far, the strategies used in vitro to characterize the kinetics of the steps involved in transcription initiation22,26 have not been applied in vivo since they rely on measuring transcription for different RNA polymerase (RNAp) concentrations. Such a change in cells is expected to have a multitude of unforeseen effects31 (in addition to the side effects of the means used to alter RNAp concentrations), which hampers the assessment of its consequences to the duration of the closed complex formation of a specific promoter. However, it is reasonable to hypothesize that, for certain small ranges of RNAp concentrations, these side effects will be negligible and thus, in such ranges, the inverse of the rate of transcription will be linear with respect to the inverse of the free RNAp concentration.
Importantly, in E. coli, RNAp concentrations have been shown to vary widely with differing growth conditions.32 As such, here we make use of different media richness to achieve different RNAp concentrations and test whether within this range of conditions, the RNA production rate changes hyperbolically with the RNAp concentrations (i.e. if the inverse of this rate changes linearly with the inverse of the RNAp concentration). Having established this relationship, we make use of it to study the in vivo kinetics of transcription initiation of Plac/ara-1. In particular, we perform measurements of the time intervals between RNA productions at the single molecule level in different intracellular RNAp and inducer concentration conditions, which we use to derive a more detailed model of transcription initiation of Plac/ara-1. For this, we first extrapolate the mean interval between production events to the limit of infinite RNAp concentration, so as to estimate the in vivo durations of the open and closed complex formations of this promoter. Next, we examine the significance of an intermittent inactive promoter state, and the role of LacI in the emergence of this state. Finally, for the first time in vivo, we determine the reversibility of the closed complex formation.
2. Materials and methods
2.1. Cells and plasmids
For single-cell RNAp fluorescence measurements, we used E. coli W3110 and RL1314,33 generously provided by Robert Landick, University of Wisconsin-Madison. For single-cell transcription interval measurements, we used E. coli DH5α-PRO (generously provided by Ido Golding, Baylor College of Medicine, Houston). The strain information is: deoR, endA1, gyrA96, hsdR17(rK- mK+), recA1, relA1, supE44, thi-1, Δ(lacZYA-argF)U169, Φ80δlacZΔM15, F-, λ-, PN25/tetR, PlacIq/lacI and SpR. This strain contains two constructs: a high-copy reporter plasmid vector PROTET-K133 (carrying MS2d-GFP under the control of PLtetO-1) and a single-copy plasmid vector pIG-BAC carrying the target transcript (mRFP1 followed by 96 MS2-binding sites) under the control of Plac/ara-1.2 This promoter is located approximately 2 and 9 kb from the origin of replication (Ori2) and the plasmid size is 11.5 kb.2 This system has been used to measure the distribution of time intervals between RNA production events due to its ability to detect individual target RNA molecules consisting of numerous MS2 coat protein binding sites, which are rapidly bound by fluorescently tagged MS2 coat proteins. These can be seen as they are produced under a fluorescence microscope as fluorescent foci.2,4,5,20,21 Finally, we used the plasmid pAB332 carrying hupA-mCherry to visualize nucleoids (generously provided by Nancy Kleckner, Harvard University, Cambridge, MA, USA). For our measurements, we inserted this plasmid into DH5α-PRO cells so as to detect nucleoids in individual cells during the live cell microscopy sessions. HupA is a major nucleoid associated protein (NAP) that participates in its structural organization.34
2.2. Chemicals
The components of Lysogeny Broth (LB) were purchased from LabM (UK), and antibiotics from Sigma-Aldrich (USA). For RT-PCR, cells were fixed with RNAprotect bacteria reagent (Qiagen, USA). Tris and EDTA for lysis buffer were purchased from Sigma-Aldrich and lysozyme from Fermentas (USA). The total RNA extraction was done with RNeasy RNA purification kit (Qiagen). DNase I, RNasefree for RNA purification, was purchased from Promega (USA). iScript Reverse Transcription Supermix for cDNA synthesis and iQ SYBR Green supermix for RT-PCR were purchased from Biorad (USA). Agarose, isopropyl β-d-1-thiogalactopyranoside (IPTG), arabinose, and anhydrotetracycline (aTc) are from Sigma-Aldrich.
2.3. Growth media
To achieve different RNAp concentrations in cells, we altered their growth conditions as in.35 For this, we used modified LB media which differed in the concentrations of some of their components. The media used are denoted as m×, where the composition per 100 ml are: m grams of tryptone, m/2 gram of yeast extract and 1 g of NaCl (pH = 7.0). For example, 0.25× media has 0.25 g of tryptone and 0.125 g of yeast extract per 100 ml.
2.4. Relative RNAp quantification
We measured relative RNAp concentrations in cells using four different methods. First, relative RNAp concentrations in the strains W3110 and DH5α-PRO were measured from the relative rpoC transcript levels obtained using RT-PCR. Cells containing the target plasmid with Plac/ara-1-mRFP1-96BS and the reporter plasmids were grown overnight in respective media. Cells were diluted into fresh media to an OD600 of 0.05. After 110 min, cells were re-diluted to an OD600 of 0.05 into respective media containing IPTG (1 mM) and arabinose (1%). After 70 min, RNA protect reagent was added to fix the cells, followed by enzymatic lysis with Tris–EDTA lysozyme buffer (pH 8.3). RNA was isolated from cells using RNeasy mini-kit (Qiagen). One microgram of RNA was used as the starting material. The RNA samples were treated with DNase free of RNase to remove residual DNA. Next, RNA was reverse transcribed into cDNA using iSCRIPT reverse transcription super mix (Biorad). RT-PCR was performed using Power SYBR-green master mix (Life Technologies) with primers for the amplification of the target gene at a concentration of 200 nM. Reactions were carried out in triplicate with 500 nM per primer with a total reaction volume 20 µl. The following primers were used for quantification: RpoC-F: CGTCAGATGCTGCGTAAAGC, RpoC-R: GCGATCTTGACGCGAGAGTA, mRFP1-F: TACGACGCCGAGGTCAAG, mRFP1-R: TTGTGGGAGGTGATGTCCA. Estimated relative RNAp concentrations in each condition m, and their standard uncertainties , were calculated according to the method.36
Second, E. coli RL1314 cells with fluorescently tagged β′ subunits were grown overnight in respective media. A pre-culture was prepared by diluting cells to an OD600 of 0.1 with fresh specific medium, and grown to an OD600 of 0.5 at 37°C at 250 rpm. Cells were pelleted by centrifugation and re-suspended in saline. Fluorescence from the cell population was measured using a fluorescent plate-reader (Thermo Scientific Fluoroskan Ascent Microplate Fluorometer).
Third, relative RNAp concentrations were also estimated based on the growth rates of DH5α-PRO cells in Supplementary Fig. S1. First, we fit a power law function to the ‘RNA polymerase molecules per cell’ row of Table 3 from ref. 32, which we found to be R=106µ−1.426, where µ is the cell doubling time. Relative RNAp concentrations were then estimated from the measured cell doubling times.
Table 3.
Fit parameters of the transcription initiation model in Reaction (3) to the measured intervals in the 1× media with and without induction by IPTG
| Joint parameters | Condition | (s) | (s) | (s) | (s) | ΔBIC | |
|---|---|---|---|---|---|---|---|
| Independent | IPTG+ | 110 | Fast | 0.11 | Fast | 5 | 14.3 |
| IPTG− | 48 | Fast | 0.01 | Fast | Fast | ||
| IPTG+ | 4,444 | Fast | 11.50 | Fast | 964 | 120.3 | |
| IPTG− | Fast | ∞ | Fast | 2,919 | |||
| IPTG+ | 7 | Fast | ∞ | Fast | 964 | 152.9 | |
| IPTG− | 320 | 1.86 | Fast | 2,919 | |||
| IPTG+ | 326 | Fast | ∞ | Fast | 964 | 145.7 | |
| IPTG− | 1.94 | Fast | 2,918 | ||||
| IPTG+ | 106 | Fast | 0.11 | Fast | Fast | 0.0 | |
| IPTG− | 48 | Fast | 0.01 |
The relationship between these parameters and the parameters in Table 2 are discussed in the Materials and methods and Supplementary Material. Five models are considered, differing in which parameters are assumed to be the same between the two induction conditions. Parameters denoted ‘fast’ are too fast to present on the timescale of seconds. As and were found to be fast in all models, the ratio is also shown. ΔBIC values are given as the difference of the model's BIC from the BIC of the best-fitting model (the one with ΔBIC = 0). Models with lower ΔBIC are favoured over models with higher ΔBIC.58
Lastly, we measured the relative RNAp concentrations in RL1314 cells under the microscope using fluorescently tagged RpoC (described in the next section).
2.5. Microscopy
DH5α-PRO cells containing the target and the reporter plasmids were grown as described previously. Briefly, cells were grown overnight in respective media, diluted into fresh media to an OD600 of 0.1, and allowed to grow to an OD600 of ∼0.3. For the reporter plasmid induction, aTc (100 ng/ml) was added 1 h before the start of the measurements. For the target plasmid, arabinose (1%) was added at the same time as aTc (following the protocol in ref. 2), and IPTG (1 mM) was added 10 min before the start of the measurements. Cells were pelleted and resuspended to fresh medium. A few microliters of cells were placed between a coverslip and an agarose gel pad (2%), which contains the respective inducers, in a thermal imaging chamber (FCS2, Bioptechs), heated to 37°C. The cells were visualized using a Nikon Eclipse (Ti-E, Nikon, Japan) inverted microscope with a C2+ confocal laser-scanning system using a 100× Apo TIRF objective. Images were acquired using the Nikon Nis-Elements software. GFP fluorescence was measured using a 488 nm argon ion laser (Melles-Griot) and 514/30 nm emission filter. Phase-contrast images were acquired with the external phase contrast system and a Nikon DS-Fi2 camera. Fluorescence images were acquired every 1 min for a total duration of 2 h. Phase-contrast images were acquired simultaneously every 5 min during the measurements.
We tested for phototoxicity due to the fluorescence and the phase-contrast imaging in these measurements. Supplementary Results suggest that there is no significant phototoxicity. Additionally, we verified that the relative RNAp concentrations under the microscope are similar to those measured in the previous section by repeating the above procedure with RL1314 cells and imaging RpoC::GFP fluorescence, 1 h after being placed in the thermal imaging chamber (see Supplementary Fig. S4). The relative RNAp concentration was estimated from the mean fluorescence concentrations of cells growing in each media.
2.6. Image analysis
Cells were detected from the phase contrast images as described in ref. 37. First, the images were temporally aligned using cross-correlation. Next, an automatic segmentation of the cells was performed by MAMLE,38 which was checked and corrected manually. Next, cell lineages were constructed by CellAging.39 Alignment of the phase-contrast images with the confocal images was done by manually selecting 5–7 landmarks in both images, and using thin-plate spline interpolation for the registration transform. Fluorescent spots and their intensities were detected from the confocal images using the Gaussian surface-fitting algorithm from.40
Jumps were detected in each cell's spot intensity timeseries using a least-deviation jump-detection method.41 Given the level of noise in the timeseries, jump sizes, i.e. the intensity of ‘one RNA’, were selected by manual inspection of the timeseries of total foreground spot intensities within cells of a given timeseries, and cross-referencing these values with the observed numbers of spots in the cells. After performing the jump detection process making use of the complete timeseries, jumps occurring within 5 min of the beginning or end of a cell's lifetime were disregarded due to our observation that the jump detection method tends to produce spurious jumps in these regions due to insufficient data. The remaining jumps were interpreted as RNA production times, from which intervals between transcription events were calculated. Finally, censored intervals were calculated as the time from the last RNA production in a cell until the last time at which a jump could have been observed (i.e. until 5 min prior to cell division or the end of the timeseries). This removes the possibility of false positives while not affecting the distribution of intervals.
This method, when first proposed, made two assumptions on the fluorescence of MS2-GFP tagged RNAs (named ‘spots’). Importantly, both assumptions were recently shown to be valid.42 First, an individual spot is bound sufficiently rapidly by MS2-GFPs such that its fluorescence intensity, when first detected, is already within the range of fluorescence of fully formed MS2-GFP-RNA spots (when taking one image per minute). In other words, the spot intensity of a newly transcribed RNA jumps from 0 to ‘full’ in <1 min, rather than slowly ramping up. Namely, since the transcription elongation rate of mRNA in E. coli is ∼50 nt/s32 and the target gene is ∼3,200 bp long,1 the time to elongate the MS2-binding site region of the target RNA is ∼60 s. Provided that MS2-GFP binding to its RNA-binding sites is fast, there will therefore be a maximum of one timepoint at which the fully transcribed target RNA may have reduced fluorescence. Since MS2-GFP is produced in excess in the cell and its binding affinity is strong (dissociation constant of ∼0.04 nM43), most binding sites will be saturated very shortly after being produced. In agreement with ref. 42, no gradual increase in spot fluorescence was observed around the time of the first appearance of a spot.
Second, once formed, MS2-GFP-RNA spots, as well as their fluorescence, are resistant to degradation for the duration of our measurements (2 h). This was shown by measurements of the dissociation rate of MS2 coat proteins from their RNA binding sites (on the order of several hours43), and by measurements of the lifetimes of the fluorescence of MS2-GFP tagged RNAs kept under observation for more than 2 h.1,2,5,42,44 Relevantly, no detectable decrease in fluorescence was observed during this time.42
2.7. Model of transcription initiation
We first consider a model that allows for RNA production dynamics to range from sub-Poissonian to super-Poissonian, given the results from genome-wide studies of the variability in RNA numbers27,45 and from studies of the transcription dynamics of individual genes.2,4,5,17,20 The features of the model that allow it to reproduce these numbers are based on processes known to occur during transcription initiation in E. coli (e.g. the open complex formation16,22,23 and an ON/OFF mechanism16,19). Then, based on our novel empirical data and methodology, we aim to obtain the most parsimonious version of the model that fits the data for a given promoter. We expect this procedure to be applicable to any promoter, and to result in slightly different models due to their differing dynamics and regulatory mechanisms.
The full model of transcription initiation considered here consists of the following set of reactions:
| (1) |
| (2) |
Reaction (1) represents the multi-step process of transcription initiation of an active promoter in prokaryotes.23,24,46,47 It begins with the formation of the closed complex (RPc), i.e. the binding of the RNA polymerase (R) to a free promoter (PON). Once at the start site, the polymerase must open the DNA double helix, a process that includes several long-lived intermediate states,23,26,46,48 resulting in the open complex (RPo). Finally, the polymerase begins RNA elongation, though before clearing the promoter, it may engage in abortive RNA synthesis in which short RNA transcripts (<10 nt) are produced.47,49 The reactions in (1) should not be interpreted as elementary transitions. Rather, they represent the effective rates of the rate-limiting steps in the process, thus defining the promoter strength, and have been shown to be sequence-dependent.50
Specifically, k1 represents the rate at which polymerases find and bind to the promoter region, which is the overall result of the promoter search process which includes non-specific binding of the polymerases to the DNA, followed by a 1D diffusive search,51,52 collectively referred to here as the closed complex formation. Subsequently, several rapid, possibly reversible isomerization reactions occur until the polymerase melts the DNA and forms the transcription ‘bubble’.51 In Reaction (1), the RPc state represents all substates until the first irreversible reaction in this chain. Consequently, k2 and k−1 should be interpreted as the product of the rates of the elementary reactions which exit from this group of substates, and the steady-state probability of being in the appropriate substates for these reactions to occur.
Similarly, the RPo state may represent numerous substates between the first state after which the complex is committed to initiation, and successful initiation. However, after this point, we cannot distinguish the reversibility of any of the following steps, since the time-interval distribution of a sequence of elementary reversible reactions of arbitrary rates is observationally equivalent to a sequence of irreversible reactions.25 The remaining steps (here, only k3) therefore represent the rates of the slowest of these irreversible reactions. Such steps may include additional isomerization reactions, abortive RNA synthesis and promoter escape and clearance.35
Reaction (2) represents the promoter intermittently transitioning to a transcriptionally inactive state (POFF). Experimentally verified mechanisms by which this can occur are the binding and unbinding of repressors and activators,29 the accumulation of positive supercoiling in the DNA.19 Additional mechanisms have also been hypothesized, such as transcriptional pausing53,54 and others.55
For a given concentration of R, the interval distribution between transcription events described by Reactions (1) and (2) (i.e. the first-passage time distribution to reach the final state, starting in the PON state) is observationally equivalent to the interval distribution described by a model of the form:
| (3) |
where the system starts in state S0. The relationship between the parameters of these two models is described in Supplementary Table S1. Note that the states Si do not correspond to the promoter states in Reactions (1) and (2). For details on how to derive and evaluate the distribution function for this model, see Supplementary Material and.25
It is noted that this model assumes that only one copy of the promoter is present in each cell at any given time. In the experiments performed here, in all conditions tested, the bacteria divided sufficiently slowly such that they spent most lifetime with only one chromosome. Specifically, cells spent no more than 11.4 ± 1.0% of their lifetime with two copies of the target promoter (Supplementary Material).
Finally, it is noted that the present model does not consider the influence of σ factors' numbers on the dynamics of transcription initiation, focussing instead solely on the concentration of RNA polymerases (in particular, on the concentration of holoenzymes containing a σ70, i.e. Eσ70, since our promoter of interest can only be transcribed by Eσ70). This is based on the fact that, in all conditions tested, most RNA polymerases are occupied by σ factors.56,57 Further, this occupation is made largely by σ70 since, first, when altering media richness, only σ32's concentration is significantly altered56 and, second, the binding affinity of σ70 to E is much higher than that of any other σ factor (e.g. it is approximately 9 times higher than that of σ32).57
2.8. Parameter estimation
Parameter estimates in Tables 1–3 were obtained by a maximum likelihood fit using the samples of the distribution of time intervals between production events obtained above (the intervals and censored intervals), as in.25 The complete model-fitting procedure is detailed in the Supplementary Material. The uncertainty of the fit of the model parameters was estimated using the negative of the Hessian of the log-likelihood surface, evaluated at the maximum likelihood estimate.
Table 1.
Statistics of the measured distributions of intervals between transcription events from lac/ara-1 promoters
| Condition | Number of cells | Number of intervals | Number of censored intervals | Inferred interval mean and uncertainty (s) | Inferred CV2 |
|---|---|---|---|---|---|
| 0.25× | 196 | 371 | 323 | 1,899 ± 105 | 1.08 |
| 0.5× | 302 | 1,027 | 605 | 1,553 ± 50 | 1.06 |
| 0.75× | 146 | 620 | 345 | 1,205 ± 51 | 1.09 |
| 1× | 206 | 1,202 | 573 | 1,005 ± 112 | 1.21 |
Shown are the condition, the number of cells (which is the cell count at the start of the measurements), the numbers of whole and censored intervals extracted, and finally the inferred mean (and its standard uncertainty) and CV2 of the interval distribution.
The mean of the time interval distribution between transcription initiation events, I(R), predicted by Reactions (1) and (2) is, for a given RNAp concentration R:
| (4) |
where is the mean time taken by the initial binding of RNAp for a given RNAp concentration, and is the mean time taken by the steps occurring after the polymerase has committed to transcription until the clearance of the promoter region (due to the initiation of elongation). As such, we expect the majority of the duration of to consist of the open complex formation as defined in.46 The remaining of its duration we attribute to failures in promoter escape.59
Estimates of and , denoted and , were obtained from the best-fit parameters of the most parsimonious model, as given in Table 3. The standard uncertainties of the estimators and , denoted and , were obtained using the Delta Method60 from the uncertainties of the model parameters.
Finally, mean durations of intervals between transcription events for each media condition , were estimated by fitting the model in Reaction (3) to the data from only that condition, and taking the mean of the distribution. This procedure was followed to include the censored intervals in the estimate of to avoid underestimating the mean interval duration due to the limited observation times. The standard uncertainty was estimated using the Delta Method.60
2.9. Validation of the τ-plot slope
We verified the slope of the τ-plot in Fig. 4 using the RT-PCR measurements from Fig. 3. These measurements are both linear with respect to , but differ by an unknown scaling factor. We denote the estimated production rate as measured by RT-PCR in media condition m as , with standard uncertainty . We found this scaling factor by fitting the parameter c in by weighted total least squares61 (WTLS), with the measurements weighted by the inverse of their uncertainty (i.e. and ). This method was chosen since it accounts for the uncertainty in both of the measurements. It results in the estimate . The dashed line in Fig. 4 was obtained by fitting the scaled points against by WTLS. The uncertainty shown includes both the uncertainty in the WTLS fit of this line, as well as the uncertainty in .
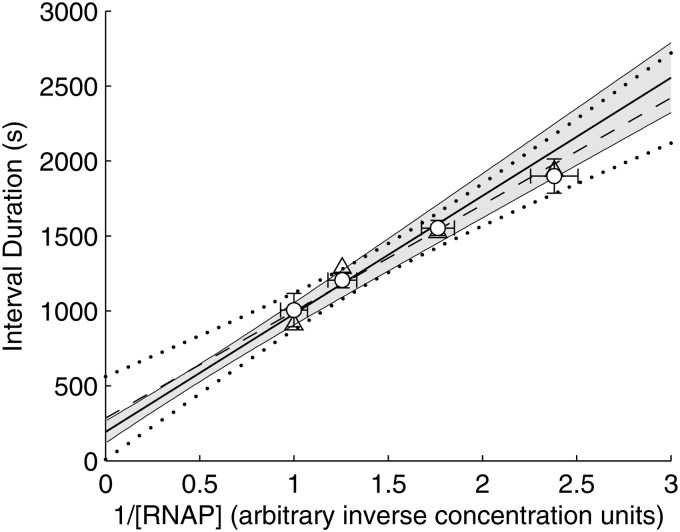
Figure 4.
τ-plot for Plac/ara-1, showing the mean interval between transcription events in individual cells for each media condition (white circles), with their standard uncertainties (vertical error bars) and the standard uncertainties of the relative RNAp concentrations (horizontal error bars). Also shown is the best-fit line (solid line), as determined by the intercept and slope obtained from the best-fitting model (Table 3), with one standard uncertainty estimated by Scheffé's method69 combined with the Delta Method60 (grey area). In addition, the figure shows the data from Fig. 3 (triangles), and the best-fitting line (dashed line, see Materials and methods) with one standard uncertainty estimated by Scheffé's method69 (dotted black curves).
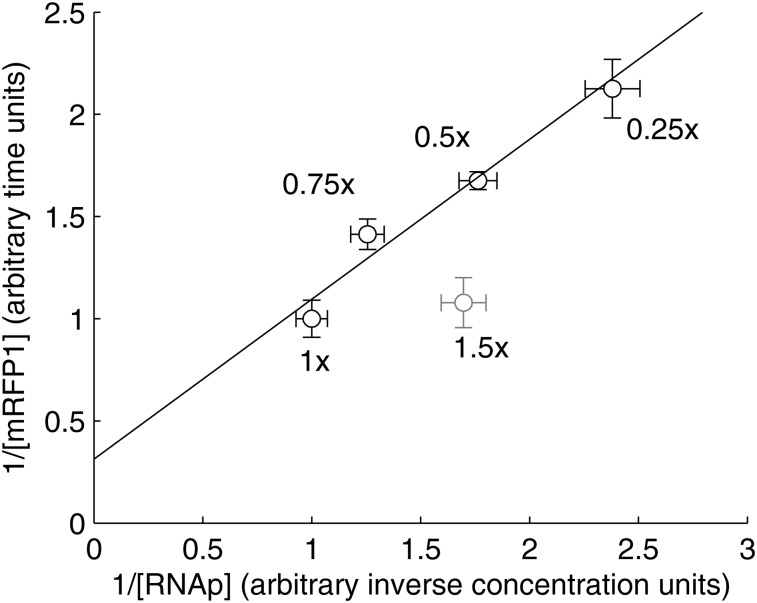
Figure 3.
Lineweaver–Burk plot of the inverse of the production rate of mRFP1 from the Plac/ara-1 promoter against the inverse of the total RNAp concentrations for the same growth conditions as in Fig. 2 (black points), and for 1.50× media (grey point). Standard uncertainties are shown for both quantities (horizontal and vertical error bars). Relative production rates were measured by RT-PCR with two biological replicates with three technical replicates each.
2.10. Method to infer the duration of the closed complex of a promoter
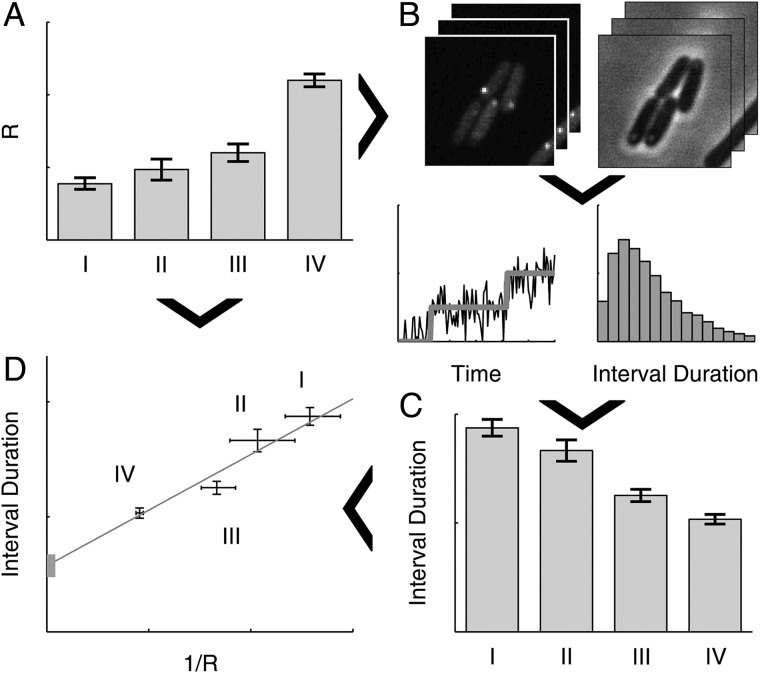
The method to infer the kinetics of transcription initiation in vivo is illustrated in Fig. 1. First, conditions are selected that differ widely in free intracellular RNAp concentrations (step A in Fig. 1). Next, an in vivo single-molecule detection technique is used to sample the time interval distribution between consecutive transcription events in individual cells in each of the conditions (step C in Fig. 1). To obtain these intervals, here we used the MS2d-GFP single RNA detection system4 (step B in Fig. 1). Then, we fit a general model of transcription initiation to the empirical data (see above), which includes both the multi-step nature of transcription initiation as well as the possibility of an intermittently inactive promoter state25 (Reactions (1) and (2)). From this fit, we obtain an estimate of the in vivo mean duration of the open complex formation by extrapolating the duration of intervals between transcription events to infinite RNAp concentrations, similar to the in vitro extrapolation presented in ref. 22 (step D in Fig. 1). The model fit will also assess the importance of an intermittent inactive promoter state and the reversibility and kinetics of the closed complex formation.
Figure 1.
Schematic representation of the in vivo measurement of the initiation kinetics, using simulated data. (A) First, several conditions are selected, labelled I–IV, differing in intracellular RNAp concentration, R. (B) Next, we obtain timeseries of fluorescence and phase contrast (for cell segmentation purposes) images of cells expressing MS2d-GFP and target RNA under the control of the promoter of interest in each condition, from which time intervals between individual transcription events are determined. This is done by jump detection in the total RNA spot intensity of each cell (lower-left in B), from which the interval distribution is obtained (lower-right in B). (C) Mean interval durations are then estimated from these interval distributions for each condition. (D) Finally, the mean interval durations and measurements of R are combined into a τ-plot,22 from which estimates of the mean times taken by the closed complex and open complex formation are obtained for each condition. Arrows depict the flow of information in the measurement procedure.
3. Results
3.1. Changing free RNA polymerase concentrations
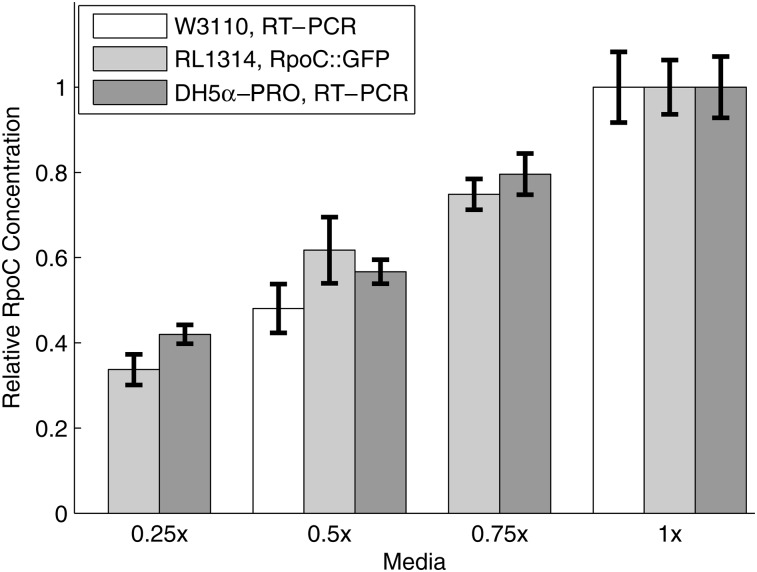
We first verified that it is possible to change intracellular RNAp concentration by a wide range by changing the growth conditions of the cells.32,35,62 As such, we grew cells in four media (described in the Materials and methods), labelled 1×, 0.75×, 0.5×, and 0.25×, which solely differ in richness of two components (tryptone and yeast extract). We then measured the relative RNAp concentrations in cells grown in these four media using RT-PCR of the rpoC gene, i.e. the gene coding for the β′ subunit, which is the limiting factor in the assembly of the RNAp holoenzyme.48,57,62 Results in Fig. 2 (dark grey bars) show that, in the range tested, the RNAp concentration in the cells increases significantly with increasing media richness.
Figure 2.
Measurements of the relative intracellular RNAp concentrations for cells growing in the four different media. Bars show the standard uncertainties of the measurements. Data is from two replicates with 3 technical replicates each (DH5α-PRO, RT-PCR, and W3110, RT-PCR), and three replicates with three technical replicates each (RL1314, RpoC::GFP). All data are presented relative to the RNAp concentration at 1×. The media used are denoted as m×, where the composition per 100 ml is: m grams of tryptone, m/2 grams of yeast extract and 1 g of NaCl (pH = 7.0).For example, 0.25× media has 0.25 g of tryptone and 0.125 g of yeast extract per 100 ml.
To validate this result, we measured the relative RNAp concentrations by plate reader in cells expressing fluorescently tagged RpoC in the strain RL1314 (derived from W3110),33 in the same four media. In addition, we also measured the levels of the rpoC transcripts in the strain W3110 by RT-PCR in the 0.5× and 1× conditions. Results (Fig. 2) show that the relative changes in the protein and mRNA levels of rpoC match the measurements by RT-PCR of the rpoC gene in DH5α-PRO.
Note that, even though the experimental procedures and strains differ, our measurements are in agreement with the relative changes in RNAp concentrations reported in ref. 32, for the difference in growth rates observed here between the 0.25× and 1× conditions (Supplementary Fig. S1), which we estimate to be ∼0.48 (Materials and methods). In this regard, given that the same result applies to (at least) three different strains, we expect it to be significantly strain-independent.
Finally, to verify that the relative RNAp concentrations measured in Fig. 2 are maintained under the microscope, we measured the relative RNAp concentration in the RL1314 cells expressing fluorescently tagged RpoC under the microscope between the two extreme conditions (0.25× and 1×), after 1 h in the thermal imaging chamber (Materials and methods). The relative RNAp concentration between the conditions was measured to be 0.367 ± 0.012, which is consistent with the measurements in Fig. 2. Lastly, from these images, we did not observe significant cell-to-cell variability in the RNAp concentrations (Supplementary Fig. S4), indicating that the mean concentrations reported in Fig. 2 are representative of the populations.
These measurements show that the relative RNAp concentration changes widely between the selected growth conditions. However, the variable affecting transcription kinetics is the relative free RNAp concentration. As such, we must verify whether the relative total RNAp concentration can be used as a proxy for the relative free RNAp concentrations. If this holds true and there are no other factors affecting the production rate of the promoter of interest in these conditions, then the RNA production rate should be hyperbolic with respect to the RNAp concentration. That is, the reciprocal of the RNA production rate from this promoter should be linear when plotted against the reciprocal of the measured relative RNAp concentrations, and one should obtain a line on a Lineweaver–Burk plot.
There are several reasons why this plot may not be linear. If, for example, the ratio of free RNAp to total RNAp is not constant in this range of growth conditions, with a higher fraction of free RNAp in the poorer growth conditions due to increased ppGpp,31 then we expect a curve with positive curvature on this plot. Meanwhile, a negative curvature would be obtained if the promoter of interest could be induced by increased cAMP in the poorer growth conditions, or if the cells spent, on average, a significantly increased amount of time with multiple copies of the plasmid in the richer growth conditions, among other possibilities. In these cases, to dissect the transcription initiation kinetics of such promoters, another method of modifying the free RNAp concentration will be required.
Given the above, we interpret a straight line on the Lineweaver–Burk plot as evidence that, for the conditions tested, (i) the relative free RNAp concentrations can be assessed from the total RNAp concentrations, and (ii) no factors other than the changes in the free RNAp concentration affect the target promoter.
Here, we tested this by measuring the RNA production rate from Plac/ara-1 in E. coli DH5α-PRO by RT-PCR in the same four media conditions as in Fig. 2. We selected this promoter, since its dynamics has been extensively characterized2,21,29,63–67 and because it has the same logical structure as the lac promoter, with an activator and a repressor.63 The resulting Lineweaver–Burk plot is shown in Fig. 3 where a linear relationship is clearly observed between these points (black points). To determine whether the small deviations from linearity are statistically significant, we performed a likelihood ratio test between a linear fit by WTLS61 (shown as a line in Fig. 3), and fits with higher order polynomials (also by WTLS by minimizing χ2 as in61). No test rejected the linear model (all P > 0.25). As noted earlier, this relationship is only expected to occur in a limited range of growth conditions. To illustrate this, we repeated the same measurements in 1.5× media (grey point in Fig. 3). The result shows that this hyperbolic relationship is lost in very rich media (including this point causes the likelihood ratio test to reject the linear model, P = 0.0014). We conclude that, for the growth conditions in Fig. 2, the relative free RNAp concentrations are well-approximated by the total RNAp concentrations, and there are no significant other factors affecting the initiation dynamics of Plac/ara-1.
3.2. Interval distributions between consecutive RNA productions
Given this, it is possible to apply a standard model-fitting procedure to fully characterize the in vivo kinetics of the rate-limiting steps in transcription initiation of the Plac/ara-1 promoter from distributions of intervals between transcription events in cells with different RNA polymerase concentrations.
We measured the distribution of time intervals between transcription events (hereafter referred to as ‘intervals’) for Plac/ara-1 in each cell growth condition using the MS2d-GFP single-RNA detection system,1 with a least-deviation jump-detection procedure41 (Materials and methods). This measurement results in samples from the interval distribution as well as ‘censored’ intervals, i.e. intervals for which we only observe the beginning due to cell division or the end of the time series. Both censored and uncensored intervals were accounted for in all parameter estimates to avoid biasing the estimates. For example, note that taking the mean of the uncensored intervals alone would underestimate the mean of the true interval distribution since long unobservable intervals would be absent from the estimate. Including the censored intervals balances this by considering long intervals that are at least as long as the censored interval length.25
From these distributions, we estimated the true mean and the squared coefficient of variation (CV2, defined as the variance over the squared mean) of the interval distributions (Materials and methods). We chose CV2 for quantifying the noise in the interval distribution since, to a good approximation, this quantity reflects the level of noise in the protein levels regardless of the actual shape of the transcription interval distribution.68 Further, this variable equals 1 for the interval distribution of a Poisson process (i.e. an exponential distribution), regardless of the mean rate. These results, along with the amount of empirical data used, are shown in Table 1.
From Table 1, the mean interval decreases significantly with increasing media richness, as expected from the increased RNAp concentrations. Meanwhile, the CV2 does not exhibit the same dependence on the media richness, and remains slightly >1 in all conditions tested.
3.3. Decomposition of the in vivo kinetics
From the data in Table 1, we next recreate the Lineweaver–Burk plot in Fig. 3 (white circles in Fig. 4), using the mean interval durations between RNA productions, as this quantity is an absolute measure of the inverse rate of RNA production (this plot is called a τ-plot).
Previously, using in vitro techniques, it has only been possible to extract from a τ-plot the mean duration of the open complex formation (the y-intercept of the plot, here denoted ), because the plot is based on the steady-state assay which only measures the mean rate of abortive transcription initiations. However, the distributions of time intervals between RNA productions contain information about the stochasticity of the process (i.e. the variability between intervals). As such, it is possible to extract a more complete model of the process of transcription. Namely, aside from the open complex formation, as mentioned in Materials and methods, it is possible to extract information on the closed complex and on an intermittent state prior to the closed complex formation.
In particular, we consider the detailed model of transcription initiation presented in Materials and methods (Reactions (1) and (2)), along with simplified models that can be considered if certain steps of the more detailed model do not influence the distribution of intervals. This model assumes that only one copy of the promoter is present in each cell at any given time, since in all conditions, the bacteria divided slowly, which suggests that they spent most lifetime with only one chromosome. We then consider three simplified models. First, if the time spent in the OFF state is very small, or if the system switches between OFF and ON very rapidly when compared with the forward reaction, then Reaction (2) will not affect the RNA production dynamics. A sufficient condition for both of these situations is that kON >> k1. The other two simplifications are two limits of the closed complex formation, first considered in22: (i) k−1 >> k2, i.e. it is reversible (Limiting Mechanism I), and (ii) k2 >> k−1, i.e. irreversible (Limiting Mechanism II). Limiting Mechanism I was found to be more likely in several in vitro measurements of various promoters.22,23,26
While all three simplifications are consistent with a line on a τ-plot, they produce significantly different distributions of intervals between RNA production events. For example, a significant ON/OFF mechanism will result in a more noisy distribution (a higher CV2).25 Similarly, Limiting Mechanism I effectively eliminates one limiting step, which also results in higher noise when compared with Limiting Mechanism II (Supplementary Fig. S2).
We fit the full and simplified models of transcription initiation to the observed dynamics of Plac/ara-1 from all media conditions (Materials and methods). We used the Bayesian Information Criterion70 (BIC) to compare the fits. The BIC is a model selection criterion which balances goodness-of-fit with the number of parameters to determine which model is most likely the ‘truth’. The difference between BIC values (ΔBIC) can be interpreted as evidence against the model with higher BIC, with a ΔBIC > 5 being interpreted as strong evidence.58 Results are shown in Table 2. Since, for several of the models, the optimal fit was for , we also considered models that do not include another rate-limiting step after the open complex formation.
Table 2.
Fit parameters of the transcription initiation model in Reactions (1) and (2), and the models derived by applying the listed simplifying assumptions
| Limiting mechanisms | Simplifications | (s) | (s) | (R−1) | (Rs) | (R−1) | (s) | (s) | (s) | ΔBIC | ΔBICC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model | 87 | Fast | 8,313 | Fast | 2,247 | Fast | 177 | Fast | 14.8 | 15.7 | |
| I | , | 87 | Fasta | 7,446 | 192 | Fast | 8.1 | 8.5 | |||
| I, k3 = ∞ | , , k3= ∞ | 87 | Fasta | 6,469 | 192 | 0.0 | 0.0 | ||||
| II | 90 | Fast | 0.10 | Fast | 7 | 7 | 18.3 | 18.8 | |||
| II, k3= ∞ | , k3= ∞ | 86 | Fast | 0.09 | Fast | 10 | 10.7 | 10.7 | |||
| No ON/OFF | Fast | 0.49 | Fast | 326 | Fast | 188.1 | 188.1 | ||||
| No ON/OFF, I | , | 0.50 | 328 | Fast | 180.1 | 179.6 | |||||
| No ON/OFF, I, k3= ∞ | , , k3= ∞ | 0.50 | 328 | 172.0 | 171.1 | ||||||
| No ON/OFF, II | , | 910 | Fast | Fast | 201.0 | 200.6 | |||||
| No ON/OFF, II, k3= ∞ | , , k3= ∞ | 910 | Fast | 192.9 | 192.0 |
Parameters denoted ‘fast’ are too fast to present on the timescale of seconds. When competing fast reactions occur, relevant ratios are given. ΔBIC values are given as the difference of the model's BIC from the BIC of the best-fitting model (the one with ΔBIC = 0). Models with lower ΔBIC are favoured over models with higher ΔBIC.58 Censored intervals were included in ΔBICC, but not in ΔBIC. The best-fitting model is shaded. Rates (and ratios) involving are given relative to the intracellular RNAp concentration in the 1× media.
a.
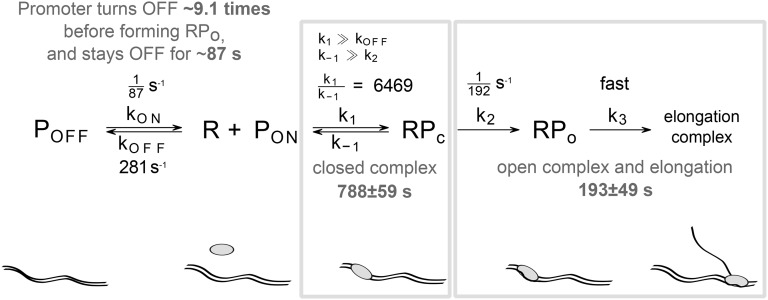
From Table 2, the initiation kinetics of Plac/ara-1 is best-fit by Limiting Mechanism I (i.e. a reversible closed complex), with very high certainty (ΔBIC of all other models >8). We also find evidence for a significant ON/OFF mechanism. Though the time spent in each OFF state is short (∼87 s), it will turn OFF, on average, ∼9.1 times before committing to transcription in the 1× case (see Supplementary Material). This results in an interval distribution which is only slightly more noisy than what would be expected if the production process were Poissonian (i.e. a CV2 of the interval distribution of 1; see the CV2 values in Table 1). Interestingly, this implies that the noise in transcription of this promoter is representative of the behaviour of the majority of promoters in E. coli.27 Finally, the steps after the commitment to transcription are fast, indicating that abortive initiation events do not play a significant role in the dynamics of RNA production by Plac/ara-1. This model is depicted graphically in Fig. 5.
Figure 5.
Best fitting model of transcription initiation (with ON/OFF mechanism and reversible close complex formation). The model parameters are specified in black and estimated durations of the transcription initiation steps for 1× LB media are shown in grey.
In addition, from Table 2, we find that is 193 ± 49 s. Meanwhile, the slope of the line on the τ-plot, here denoted , is 788 ± 59 R·s (R is the polymerase concentration such that R = 1 is the polymerase concentration in 1× media). The line given by these values is shown in Fig. 4 (solid line). As a side note, the uncertainties of these estimates exaggerate the uncertainty of the inference, since the estimates are highly correlated (correlation coefficient of −0.6). This correlation is responsible for the hyperbolic shape of the confidence bounds (grey region in Fig. 4).
We verified the slope of the solid line in Fig. 4 using the RT-PCR measurements presented in Fig. 3, scaled to match the timescale of the intervals (Materials and methods). The resulting line is shown in Fig. 4 (dashed line), and is in good agreement with both the line given by our estimates of and (solid line), and the inferred interval means (white circles).
Lastly, we note that the BIC depends on the number of samples used to calculate the likelihood. Thus, BIC values calculated assuming that each censored interval is ‘one sample’ will over-penalize models with more parameters, while removing them will under-penalize them. Both sets of ΔBIC values are presented in Table 2 and, in our case, both result in the same conclusion, and thus the distinction does not affect the results for Plac/ara-1. If, for another promoter, this turns out to be the case, additional measurements will be required to distinguish between the models.
Our results are in agreement with previous measurements of the kinetics of this and similar promoters. For example, a previous study reported that, under full induction in LB media (1× media here), Plac/ara-1 expresses ∼4 RNA/h2 (i.e. 1 RNA every ∼900 s), while we inferred the time between transcription events to be ∼980 s. Using the steady-state assay, was measured to be ∼330 s for Plac71 (with or without CRP-cAMP), while we obtained ∼193 s.
3.4. Determining the source of the intermittent inactive state for Plac/ara-1
We identified the presence of an ON/OFF mechanism in the dynamics of Plac/ara-1. It is worth noting that this ON/OFF phenomenon differs from the one reported in refs 2 and 19 since, first, we only observe OFF periods on the order of ∼87 s, while in ref. 2 the OFF periods reported for Plac/ara-1were on the order of 37 min. In addition, both here and in ref. 2, the promoter of interest is integrated in a single-copy plasmid, and thus the OFF periods cannot be explained by the buildup of positive supercoiling, since the plasmid is not topologically constrained.19 We therefore hypothesized that the OFF periods observed here more likely result from the intermittent formation of a DNA loop, due to the transient binding of LacI, which exists in high concentration in DH5α-PRO (∼3,000 copies vs. ∼20 in wild type63).
If LacI is responsible for the ON/OFF behaviour, then reducing the concentration of IPTG should affect the ON/OFF dynamics, and not change the dynamics following the closed complex formation.29 To test this prediction, and demonstrate the utility of the model-fitting approach, besides considering the interval measurements in 1× in Table 1, we also measured the interval distribution of Plac/ara-1 using MS2d-GFP in the 1× media without induction by IPTG. From 130 cells, we extracted 57 intervals and 117 censored intervals between transcription events. From these, we inferred a mean interval of 3,374 ± 462 s, and a CV2 of 1.03. This mean is significantly greater than the mean measured in the fully induced condition (1,005 ± 112 s), consistent with the much stronger repression of the promoter by LacI in this condition.
Given the wide difference in dynamics of RNA production between the induced and non-induced cases, we used the model fitting procedure to determine which steps are significantly affected by LacI. For this, we performed independent fits of a reduced model of initiation to the induced and the non-induced conditions. This model is observationally equivalent to the full model of initiation (Reactions (1) and (2)) for a single value of R, and is presented in Reaction (3). This reduced model is necessary since we do not have measurements of the uninduced case at multiple values of R with which to fit all parameters of the full model. The reduced model's parameters are denoted by λx, which are related to, but are not equal to the values of kx. Their relationship is presented in Supplementary Table S1. The fitting results are shown in Table 3 (labelled ‘Independent’). We also considered joint models where parameters were fixed between conditions, and used the BIC to select the most likely model.
The first three models with joint parameters test for whether or not the parameters controlling the ON/OFF mechanism change with induction strength. Consistent with this hypothesis, the models with joint are strongly rejected (ΔBIC much higher than that of the Independent model). Surprisingly, the model with only joint was also rejected, implying that the mean OFF times might also vary with induction strength. Additional studies are needed to elucidate why such OFF times depend on the induction strength.
Having established that and differ between conditions, we next assessed whether only these parameters differ. For that, we fixed and , and verified that this model is the most parsimonious model (ΔBIC relative to the Independent model of −14.3). We conclude that only and differ between conditions, confirming the prediction that LacI is responsible for the ON/OFF mechanism affecting the RNA production dynamics.
Finally, other models were considered, e.g. the hypothesis that , , and/or do not differ between conditions. These models were also strongly rejected in favour of the parsimonious model, and are not shown for brevity.
3.5. Precision of the estimates
We define the precision of the estimates of and as the ratio between the timescale of the intervals (i.e. the mean interval in the condition with greatest R) and the standard uncertainties of and , respectively. Specifically, the precision of 's estimate is , and the precision of 's estimate is . Given this, here, with the volume of data in Table 1, we achieved and PCC = 17.0, corresponding to errors of ∼5 and ∼6%, respectively.
In addition, we found that this precision is highly dependent on the dynamic range of RNAp concentrations. For example, for a small dynamic range of 1.5 (our measurements in Fig. 2 have a range of ∼2.4), the precisions (in ) and PCC(in ) would have been reduced to ∼11.2 and ∼6.7, respectively. Losses in precision due to reduced dynamic ranges can, however, to some extent, be offset by collecting more samples for the interval distributions (see estimation of precision in Supplementary Material).
4. Discussion
We established that, in E. coli, the concentration of free RNA polymerases differs significantly within a certain range of growth conditions, and that the inverse of the target RNA production rate under the control of Plac/ara-1 varies linearly with the inverse of the free RNAp concentration (which are the conditions imposed in the in vitro measurements the open complex formation by steady state assays22,24,72). Thus, we were able to apply a standard model-fitting procedure to fully characterize the in vivo kinetics of the rate-limiting steps in transcription initiation of the Plac/ara-1 promoter from distributions of intervals between transcription events in cells with different RNA polymerase concentrations. This revealed that this promoter has two rate-limiting steps: a reversible closed complex formation and a significant open complex formation. Further, it also intermittently switches to a short-lived inactive state. Based on the inferred timescale of this inactive state, we predicted that this state is the result of the intermittent binding of the repressor LacI, which we verified by measuring the interval distribution when the promoter is not induced by IPTG. We believe that the complexity of this process is the reason why it has not been reported before. Namely, previous studies only considered either multiple rate-limiting steps,4,5,22,23,66 or an ON/OFF process,2,17,19,73,74 while this promoter exhibits both.
We note that, provided that the promoter has a reversible closed complex formation, the model fitting procedure proposed here allows the duration and order of two steps following the closed complex to be obtained (specifically, the ratio between k2 and k3 can be determined from how the CV2 of the interval distribution changes with R; see Supplementary Fig. S2). Here, this additional step was not found. However, we expect that, for other promoters, or in different conditions (e.g. low temperatures72), this step may be significant. Meanwhile, if Limiting Mechanism II is found to be the best-fitting model, the order of the last two steps will remain ambiguous due to the lack of reversibility.
Finally, it is worth noting that in previous works, we have not found evidence for an ON/OFF mechanism for Plac/ara-1, due to the low levels of noise detected in the time intervals between transcription events.4,21,66 This can be explained by, first, we did not consider censored intervals, which contribute significantly to the increase of the tail of the distribution of intervals.25 Second, the OFF period is quite short, and thus its detection requires a large volume of data and a sensitive inference methodology.25 Our results show that, by solving these two issues (by applying the methods in refs 41 and 25), our methodology can identify and characterize many relevant steps in transcription initiation, including those with lesser influence.
In the future, it would be of interest to extend the model to consider what occurs when more than one copy of a promoter is present in the cell. We expect that variations in the promoter copy numbers would, in that case, explain some of the variance of the data, instead of this variance being solely determined by the ON/OFF mechanism and the sequential steps.
We expect the methodology employed here to be applicable to promoters, native or synthetic, whose changes in the inverse of the transcription rate are linear with the inverse of the free RNAp concentrations. Also, it should be applicable to promoters evolved to interact with multiple transcription factors (TF), provided their fast binding and unbinding (compared with competing events), as they could be accounted for by tuning the rate constants of some of the reactions of the model. Further, multiple slow TFs, including activators, can be accounted for by adding appropriate TF-bound states, with differing production rates, in a similar manner to the ON/OFF model. As such, the methodology should be applicable at a genome wide scale. It should also be applicable to eukaryotes, provided suitable means to alter polymerase concentrations. Lastly, it should be useful in detecting differences in transcription initiation kinetics of a promoter subject to different intra- or extra-cellular conditions.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Academy of Finland (257603 to A.S.R.); Centre of International Mobility (13.1.2014/TM-14-91361/CIMO to A.S.R.); Jenny and Antti Wihuri Foundation (to A.H.); and the Tampere University of Technology President's Graduate Programme (to J.L.-P. and S.S.). Funding to pay the Open Access publication charges for this article was provided by Academy of Finland (257603 to A.S.R.).
Supplementary Material
Acknowledgements
We thank Axel Oikari, Abhishekh Gupta, and Antti Martikainen for valuable advice.
References
- 1.Golding I., Cox E.C.. 2004, RNA dynamics in live Escherichia coli cells, Proc. Natl Acad. Sci. USA, 101, 11310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golding I., Paulsson J., Zawilski S.M., Cox E.C.. 2005, Real-time kinetics of gene activity in individual bacteria, Cell, 123, 1025–36. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Xiao J., Ren X., Lao K., Xie X.S.. 2006, Probing gene expression in live cells, one protein molecule at a time, Science, 311, 1600–3. [DOI] [PubMed] [Google Scholar]
- 4.Kandhavelu M., Mannerström H., Gupta A. et al. 2011, In vivo kinetics of transcription initiation of the lar promoter in Escherichia coli. Evidence for a sequential mechanism with two rate-limiting steps, BMC Syst. Biol., 5, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muthukrishnan A.-B., Kandhavelu M., Lloyd-Price J. et al. 2012, Dynamics of transcription driven by the tetA promoter, one event at a time, in live Escherichia coli cells, Nucleic Acids Res., 40, 8472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fusco D., Accornero N., Lavoie B. et al. 2003, Single mRNA Molecules Demonstrate Probabilistic Movement in Living Mammalian Cells, Curr. Biol., 13, 161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S.. 2006, Stochastic mRNA synthesis in mammalian cells, PLoS Biol., 4, 1707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaern M., Elston T.C., Blake W.J., Collins J.J.. 2005, Stochasticity in gene expression: from theories to phenotypes, Nat. Rev. Genet., 6, 451–64. [DOI] [PubMed] [Google Scholar]
- 9.Arkin A.P., Ross J., McAdams H.H.. 1998, Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells, Genetics, 149, 1633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S.. 2002, Stochastic gene expression in a single cell, Science, 297, 1183–6. [DOI] [PubMed] [Google Scholar]
- 11.Raser J.M., O'Shea E.K.. 2005, Noise in gene expression: origins, consequences, and control, Science, 309, 2010–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozbudak E.M., Thattai M., Kurtser I., Grossman A.D., van Oudenaarden A.. 2002, Regulation of noise in the expression of a single gene, Nat. Genet., 31, 69–73. [DOI] [PubMed] [Google Scholar]
- 13.McAdams H.H., Arkin A.P.. 1999, It's a noisy business! Genetic regulation at the nanomolar scale, Trends Genet., 15, 65–9. [DOI] [PubMed] [Google Scholar]
- 14.Süel G.M., Garcia-Ojalvo J., Liberman L.M., Elowitz M.B.. 2006, An excitable gene regulatory circuit induces transient cellular differentiation, Nature, 440, 545–50. [DOI] [PubMed] [Google Scholar]
- 15.Cai L., Friedman N., Xie X.S.. 2006, Stochastic protein expression in individual cells at the single molecule level, Nature, 440, 358–62. [DOI] [PubMed] [Google Scholar]
- 16.Mitarai N., Dodd I.B., Crooks M.T., Sneppen K.. 2008, The generation of promoter-mediated transcriptional noise in bacteria, PLoS Comput. Biol., 4, e1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So L.-H., Ghosh A., Zong C., Sepúlveda L.A., Segev R., Golding I.. 2011, General properties of transcriptional time series in Escherichia coli, Nat. Genet., 43, 554–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhdanov V.P. 2011, Kinetic models of gene expression including non-coding RNAs, Phys. Rep., 500, 1–42. [Google Scholar]
- 19.Chong S., Chen C., Ge H., Xie X.S.. 2014, Mechanism of transcriptional bursting in bacteria, Cell, 158, 314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandhavelu M., Häkkinen A., Yli-Harja O., Ribeiro A.S.. 2012, Single-molecule dynamics of transcription of the lar promoter, Phys. Biol., 9, 026004. [DOI] [PubMed] [Google Scholar]
- 21.Kandhavelu M., Lloyd-Price J., Gupta A., Muthukrishnan A.-B., Yli-Harja O., Ribeiro A.S.. 2012, Regulation of mean and noise of the in vivo kinetics of transcription under the control of the lac/ara-1 promoter, FEBS Lett., 586, 3870–5. [DOI] [PubMed] [Google Scholar]
- 22.McClure W.R. 1980, Rate-limiting steps in RNA chain initiation, Proc. Natl Acad. Sci. USA, 77, 5634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClure W.R. 1985, Mechanism and control of transcription initiation in prokaryotes, Annu. Rev. Biochem., 54, 171–204. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand-Burggraf E., Lefèvre J.F., Daune M.. 1984, A new experimental approach for studying the association between RNA polymeras and the tet promoter of pBR322, Nucleic Acids Res., 12, 1697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häkkinen A., Ribeiro A.S.. 2016, Characterizing rate limiting steps in transcription from RNA production times in live cells, Bioinformatics, http://bioinformatics.oxfordjournals.org/content/early/2016/01/28/bioinformatics.btv744.abstract. [DOI] [PubMed] [Google Scholar]
- 26.Friedman L.J., Gelles J.. 2012, Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation, Cell, 148, 679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi Y., Choi P.J., Li G.-W. et al. 2010, Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells, Science, 329, 533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez A., Garcia H.G., Jones D., Phillips R., Kondev J.. 2011, Effect of promoter architecture on the cell-to-cell variability in gene expression, PLoS Comput. Biol., 7, e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz R., Lozinski T., Ellinger T., Bujard H.. 2001, Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator, Nucleic Acids Res., 29, 3873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia H.G., Sanchez A., Boedicker J.Q. et al. 2012, Operator sequence alters gene expression independently of transcription factor occupancy in bacteria, Cell Rep., 2, 150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gummesson B., Magnusson L.U., Lovmar M. et al. 2009, Increased RNA polymerase availability directs resources towards growth at the expense of maintenance, EMBO J., 28, 2209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bremer H., Dennis P.P.. 1996, Modulation of Chemical Composition and Other Parameters of the Cell by Growth Rate. In: Neidhardt F.C., (ed.), Escherichia Coli and Salmonella, 2nd ed ASM Press, Washington, DC, pp. 1553–69. [Google Scholar]
- 33.Bratton B.P., Mooney R.A., Weisshaar J.C.. 2011, Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli., J. Bacteriol., 193, 5138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dillon S.C., Dorman C.J.. 2010, Bacterial nucleoid-associated proteins, nucleoid structure and gene expression, Nat. Rev. Microbiol., 8, 185–95. [DOI] [PubMed] [Google Scholar]
- 35.Liang S.-T., Bipatnath M., Xu Y.-C. et al. 1999, Activities of constitutive promoters in Escherichia coli., J. Mol. Biol., 292, 19–37. [DOI] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D.. 2001, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method, Methods, 25, 402–8. [DOI] [PubMed] [Google Scholar]
- 37.Gupta A., Lloyd-Price J., Oliveira S.M.D., Yli-Harja O., Muthukrishnan A.-B., Ribeiro A.S.. 2014, Robustness of the division symmetry in Escherichia coli and functional consequences of symmetry breaking, Phys. Biol., 11, 066005. [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury S., Kandhavelu M., Yli-Harja O., Ribeiro A.S.. 2013, Cell segmentation by multi-resolution analysis and maximum likelihood estimation (MAMLE), BMC Bioinformatics, 14, S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Häkkinen A., Muthukrishnan A.-B., Mora A., Fonseca J.M., Ribeiro A.S.. 2013, CellAging: A tool to study segregation and partitioning in division in cell lineages of Escherichia coli, Bioinformatics, 29, 1708–9. [DOI] [PubMed] [Google Scholar]
- 40.Häkkinen A., Kandhavelu M., Garasto S., Ribeiro A.S.. 2014, Estimation of fluorescence-tagged RNA numbers from spot intensities, Bioinformatics, 30, 1146–53. [DOI] [PubMed] [Google Scholar]
- 41.Häkkinen A., Ribeiro A.S.. 2015, Estimation of GFP-tagged RNA numbers from temporal fluorescence intensity data, Bioinformatics, 31, 69–75. [DOI] [PubMed] [Google Scholar]
- 42.Tran H., Oliveira S.M.D., Goncalves N., Ribeiro A.S.. 2015, Kinetics of the cellular intake of a gene expression inducer at high concentrations, Mol. Biosyst., 11, 2579–87. [DOI] [PubMed] [Google Scholar]
- 43.Johansson H.E., Dertinger D., LeCuyer K.A., Behlen L.S., Greef C.H., Uhlenbeck O.C.. 1998, A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein, Proc. Natl Acad. Sci. USA, 95, 9244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golding I., Cox E.C.. 2006, Physical Nature of Bacterial Cytoplasm, Phys. Rev. Lett., 96, 98102. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein J.A., Khodursky A.B., Pei-Hsun L., Lin-Chao S., Cohen S.N.. 2002, Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays, Proc. Natl Acad. Sci. USA, 99, 9697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saecker R.M., Record M.T., Dehaseth P.L.. 2011, Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis, J. Mol. Biol., 412, 754–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeHaseth P.L., Zupancic M.L., Record M.T.. 1998, RNA polymerase-promoter interactions: The comings and goings of RNA polymerase, J. Bacteriol., 180, 3019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chamberlin M.J. 1974, The selectivity of transcription, Annu. Rev. Biochem., 43, 721–75. [DOI] [PubMed] [Google Scholar]
- 49.Hsu L.M. 2009, Monitoring abortive initiation, Methods, 47, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulligan M.E., Hawley D.K., Entriken R., McClure W.R.. 1984, Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity, Nucleic Acids Res., 12, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai L., Santangelo T.J., Wang M.D.. 2006, Single-molecule analysis of RNA polymerase transcription, Annu. Rev. Biophys. Biomol. Struct., 35, 343–60. [DOI] [PubMed] [Google Scholar]
- 52.Wang F., Greene E.C.. 2011, Single-molecule studies of transcription: From one RNA polymerase at a time to the gene expression profile of a cell, J. Mol. Biol., 412, 814–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Artsimovitch I., Landick R.. 2000, Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals, Proc. Natl Acad. Sci. USA, 97, 7090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajala T., Häkkinen A., Healy S., Yli-Harja O., Ribeiro A.S.. 2010, Effects of transcriptional pausing on gene expression dynamics, PLoS Comput. Biol., 6, e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Nahum G., Nudler E.. 2001, Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli, Cell, 106, 443–51. [DOI] [PubMed] [Google Scholar]
- 56.Grigorova I.L., Phleger N.J., Mutalik V.K., Gross C.a.. 2006, Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA, Proc. Natl Acad. Sci. USA, 103, 5332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda H., Fujita N., Ishihama A.. 2000, Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase, Nucleic Acids Res., 28, 3497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kass R.E., Raftery A.E.. 1995, Bayes Factors, J. Am. Stat. Assoc., 90, 773–95. [Google Scholar]
- 59.Hsu L.M. 2002, Promoter clearance and escape in prokaryotes, Biochim. Biophys. Acta, 15,77, 191–207. [DOI] [PubMed] [Google Scholar]
- 60.Casella G., Berger R.L.. 2001, The Delta Method. Statistical Inference, 2nd ed Duxbury Press, Pacific Grove, CA, pp. 240–5. [Google Scholar]
- 61.Krystek M., Anton M.. 2008, A weighted total least-squares algorithm for fitting a straight line, Meas. Sci. Technol., 19, 79801. [Google Scholar]
- 62.Klumpp S., Hwa T.. 2008, Growth-rate-dependent partitioning of RNA polymerases in bacteria, Proc. Natl Acad. Sci. USA, 105, 20245–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lutz R., Bujard H.. 1997, Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements, Nucleic Acids Res., 25, 1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J.. 2008, A fast, robust and tunable synthetic gene oscillator, Nature, 456, 516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martins L., Mäkelä J., Häkkinen A. et al. 2012, Dynamics of transcription of closely spaced promoters in Escherichia coli, one event at a time, J. Theor. Biol., 301, 83–94. [DOI] [PubMed] [Google Scholar]
- 66.Mäkelä J., Kandhavelu M., Oliveira S.M.D. et al. 2013, In vivo single-molecule kinetics of activation and subsequent activity of the arabinose promoter, Nucleic Acids Res., 41, 6544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kandhavelu M., Lihavainen E., Muthukrishnan A.B., Yli-Harja O., Ribeiro A.S.. 2012, Effects of Mg2+ on in vivo transcriptional dynamics of the lar promoter, BioSystems, 107, 129–34. [DOI] [PubMed] [Google Scholar]
- 68.Pedraza J.M., Paulsson J.. 2008, Effects of molecular memory and bursting on fluctuations in gene expression, Science, 319, 339–43. [DOI] [PubMed] [Google Scholar]
- 69.Casella G., Berger R.L.. 2001, Simultaneous Estimation and Confidence Bands. Statistical Inference, 2nd ed Duxbury Press, Pacific Grove, CA, USA, pp. 559–63. [Google Scholar]
- 70.Schwarz G. 1978, Estimating the dimension of a model, Ann. Stat., 6, 461–4. [Google Scholar]
- 71.Malan T.P., Kolb A., Buc H., McClure W.R.. 1984, Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter, J. Mol. Biol., 180, 881–909. [DOI] [PubMed] [Google Scholar]
- 72.Buc H., McClure W.R.. 1985, Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps, Biochemistry, 24, 2712–23. [DOI] [PubMed] [Google Scholar]
- 73.Sanchez A., Choubey S., Kondev J.. 2013, Stochastic models of transcription: From single molecules to single cells, Methods, 62, 13–25. [DOI] [PubMed] [Google Scholar]
- 74.Schwabe A., Rybakova K.N., Bruggeman F.J.. 2012, Transcription stochasticity of complex gene regulation models, Biophys. J., 103, 1152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.