Abstract
Background
The detection rate for identifying the underlying mutation in neurocutaneous syndromes is affected by the sensitivity of the mutation test and the heterogeneity of the disease based on the diagnostic criteria. Neurofibromatosis type (NF1) has been defined for 29 years by the National Institutes for Health (NIH) criteria which include ≥ 6 Café au Lait macules (CAL) as a defining criterion. The discovery of SPRED1 as a cause of Legius syndrome which is manifested by CAL, freckling and learning difficulties has introduced substantial heterogeneity to the NIH criteria.
Methods
We have defined the sensitivity of comprehensive RNA analysis on blood of presumed NF1 patients meeting NIH criteria with at least one nonpigmentary criterion and determined the proportion of children with ≥ 6 CAL and no family history that has an NF1 or SPRED1 genetic variant. RNA analysis was carried out from 04/2009–12/2015 on 361 NF1 patients.
Findings
A presumed causative NF1 mutation was found in 166/171 (97.08%–95% CI 94.56–99.6%) of familial cases and 182/190 (95.8%–95% CI 92.93–98.65%) sporadic de novo cases. Two of thirteen (15%) mutation negative individuals had dysembryoplastic neuroepithelial tumour (DNET) compared to 2/348 (0.6%) with an NF1 variant (p = 0.007). No SPRED1 variants were found in the thirteen individuals with no NF1 variant. Of seventy-one individuals with ≥ 6 CAL and no non-pigmentary criterion aged 0–20 years, 47 (66.2%) had an NF1 variant six (8.5%) a SPRED1 variant and 18 (25.3%) no disease causing variant. Using the 95.8% detection rate the likelihood of a child with ≥ 6 CAL having constitutional NF1 drops from 2/3 to 1/9 after negative RNA analysis.
Interpretation
RNA analysis in individuals with presumed NF1 has high sensitivity and includes a small subset with DNET without an NF1 variant. Furthermore negative analysis for NF1/SPRED1 provides strong reassurance to children with ≥ 6 CAL that they are unlikely to have NF1.
Keywords: RNA, NF1, Splicing mutation, Deep intronic, Legius, SPRED1, Café au lait
Highlights
-
•
RNA testing for NF1 mutations has very high sensitivity (c.96%) and is significantly more sensitive than DNA testing.
-
•
Mosaicism is not a major feature in those with classical NF1.
-
•
Around two thirds of children with just 6 or more café au lait spots have NF1, 8% Legius syndrome.
-
•
Patients with normal RNA testing who meet NF1 criteria but also have a DNET may have a fault in a yet to be identified gene.
Identifying the underlying genetic mutation is of benefit to patients and their families as it clarifies the diagnosis, can give information on the likely disease course and allow predictive testing in pregnancy and early childhood. The present study has shown that testing of blood RNA has very high sensitivity (96%) and allows substantial reassurance to parents whose children have multiple Café au lait birthmarks that they are unlikely to have the poorer outcomes of NF1 if they test negative. Furthermore the study suggests that a different mechanism may underlie the association of NF1 features and a rare benign brain tumour called DNET.
1. Introduction
Neurofibromatosis type 1 (NF1) is a multisystem autosomal dominantly inherited tumour predisposition neurocutaneous syndrome characterised by pigmentary skin changes and benign nerve sheath tumors (neurofibromas) (Anderson and Gutmann, 2015, Huson et al., 1988). NF1 affects around 1 in 1/1900–2500 live births (Huson et al., 1989, Evans et al., 2010, Uusitalo et al., 2015) and significantly reduces life expectancy primarily due to the development of Malignant Peripheral Nerve Sheath Tumors (MPNST) in around 10% of affected people, but also gliomas and other malignancies including breast cancer (Evans et al., 2011). The National Institutes of Health (NIH) defined diagnostic criteria in 1987 (National Institutes of Health Consensus Development Conference, 1987) and these have largely remained unchanged also including bone dysplasia and Lisch nodules as criteria (Table 1) (Anderson and Gutmann, 2015, Gutmann et al., 1997). For such a large gene containing 58 coding exons (Shen et al., 1996) little is still known about domains outside the GTPase-activating protein-related domain (GRD) domain which is thought to be the main region associated with tumour suppression through down-regulation of the oncogene ras (Anderson and Gutmann, 2015, Ferner, 2007). There are substantial risks of neurological deficits including cognitive impairment, epilepsy, spinal cord compression, cerebrovascular disease, and multiple sclerosis (Ferner, 2007).
Table 1.
NIH criteria for NF1.
| NIH diagnostic criteria for Neurofibromatosis type 1 |
|---|
| Two or more of the following: 6 or more café au lait macules (0.5 cm in children or 1.5 cm in adults) 2 or more cutaneous/subcutaneous neurofibromas or one plexiform neurofibroma Skin fold freckling Optic pathway glioma 2 of more Lisch nodules (iris hamartomas seen on slit lamp examination) Bony dysplasia (sphenoid wing dysplasia, bowing of long bone, pseudarthrosis) First degree relative with NF1 |
The gene was cloned in 1990 and initial studies of the large complex gene identified only between 30 and 65% of germline mutations in clearly affected individuals (Shen et al., 1996). Furthermore, phenotype analysis in pedigrees suggested that there would be little significant genotype–phenotype correlation (Easton et al., 1993) and apart from the NF1 microdeletions first reported in 1994 (Kayes et al., 1992), there was little interest in this field until relatively recently. It was not until 10 years after the gene was identified that a study using an exhaustive approach including RNA analysis identified 64/67 (95%) of mutations in clearly affected NF1 individuals (Messiaen et al., 2000). Since then mutations in the SPRED1 gene have been identified as the cause of Legius syndrome characterised by multiple (CAL) patches and macrocephaly, but without the tumour features of NF1. Among a cohort of 42 SPRED1 mutated individuals 48% fulfilled NIH NF1 diagnostic criteria based on the presence of > 5 CAL with or without freckling or an NF1-compatible family history (Messiaen et al., 2009). Of 94 probands with familial CAL with or without freckling and no other NF1 features, 69 (73%; 95% CI, 63%–80%) had an NF1 mutation and 18 (19%; 95% CI, 12%–29%) had a pathogenic SPRED1 mutation (Messiaen et al., 2009). Variants have not been identified in any further gene producing resulting in an NF1-like syndrome. However, since the finding that certain NF1 mutations give rise to a CAL and freckling only phenotype (Upadhyaya et al., 2007, Rojnueangnit et al., 2015 (Nov) and that patients with ‘spinal phenotype’ have an excess of splicing and missense mutations there has been a resurgence of interest in genotype–phenotype correlation. Although post zygotic mutations in embryogenesis leading to mosaicism have been shown to be the cause at least 33% of de novo Neurofibromatosis type 2 (NF2) affected individuals with classical disease fulfilling NIH criteria with bilateral vestibular schwannoma (National Institutes of Health Consensus Development Conferen, Evans et al., 2007), it has rarely been reported as the cause of classical non segmental NF1.
As the reference laboratory for the nationally funded highly specialised complex NF1 service in England, from 2009 we have applied comprehensive RNA analysis of the NF1 gene coupled with MLPA based copy number analysis using the approach developed by Messiaen et al. (Messiaen et al., 2000) The current study aimed to determine the sensitivity of this strategy to detect mutations in a large cohort of well characterised individuals with NF1 who met NIH diagnostic with more than just pigmentary criteria. This would potentially identify whether other genes may still cause features compatible with NF1. Furthermore the study aimed to assess the likely contribution of mosaicism to classically affected de novo cases.
2. Materials and Methods
Individuals referred to the Manchester service who fulfilled NIH criteria (Table 1), which was not confined to a body segment and who had at least one non-pigmentary criteria for NF1 in them or their affected relative were included. NF1 affected individuals were divided into familial where there was at least one affected first degree relative who also met NIH criteria and sporadic de novo cases. All the families tested were unrelated and did not contain known multiple branches of the same family.
In addition, a separate analysis was carried out on consecutive children with at least 6 CAL with or without freckling, but no other NF1 diagnostic criterion who also had no parent with an NF1 criterion and were aged < 20 years of age at assessment. This was for a 5-year period from November 2010–November 2015.
NF1 mutation analysis was carried out in the Genomic Diagnostic Laboratory at the Manchester Centre for Genomic Medicine in St Mary's Hospital, Manchester. This is a clinically accredited medical testing laboratory. RNA and genomic DNA were prepared from peripheral blood samples. RNA was isolated from short term PHA stimulated cultures pre-treated before RNA extraction with Puromycin to inhibit nonsense mediated decay. RNA was reverse transcribed to cDNA using standard procedures, and the cDNA was PCR amplified in 5 overlapping fragments of approx. 2 kb in size. Each fragment was then Sanger sequenced with between 8 and 11 primers to give overlapping sequence data of the whole fragment thus highlighting abnormalities in NF1 splicing or mutations within the coding sequence. Short term culture followed by Puromycin treatment was used in preference to RNA stabilising blood collection tubes e.g. PAXgene, due to more consistent and robust RNA quality achieved in preliminary tests. Mutation status was confirmed in genomic DNA. Multiplex ligation dependent probe amplification (MLPA) for dosage analysis was additionally performed in samples without a clearly pathogenic mutation identified on cDNA analysis using the MRC-Holland P081 and P082 probe sets. In samples where no clearly pathogenic NF1 mutation was identified the SPRED1 gene was screened for mutations by bidirectional Sanger sequencing of the whole coding region and flanking splice donor and acceptor sites to ± 15 bp plus MLPA dosage analysis using the MRC-Holland P295 probe set.
Statistical analysis was carried out using chi square testing with two tailed Fisher's exact test.
3. Results
RNA analysis was carried out on 361 NF1 affected individuals who fulfilled NIH criteria with at least one non pigmentary criterion. Twenty one familial cases with just pigmentary NIH criteria were excluded (14 NF1 mutations (12 missense mutations); 7 SPRED1 mutations). One hundred and seventy one individuals were from families with multiple affected individuals and 190 were sporadic de novo (no evidence of NF1 family history) affected people. Potentially causative variants in the NF1 gene were identified in 166/171 (97.08%–95% CI 94.56–99.6%) of familial cases compared to 182/190 (95.8%–95% CI 92.93–98.65%) of sporadic samples (p = 0.58-Table 2). This dropped to 154/171 (90.06%) and 175/190 (92.10%) respectively if class 3 variants were excluded. Although truncating mutations (nonsense and frameshift) were seen slightly more frequently in sporadic cases and non-truncating mutations in familial affected individuals these differences were not significant (Table 2). Whole gene deletions were present in 6/166 (3.6%) of familial samples with identified mutations and 15/182 (8.2%) of sporadic samples but this difference did not quite reach significance (p = 0.08). However, when whole gene deletions found before Nov 2010 were included the 21 found in sporadic de novo cases was significantly higher than the 9 in familial cases (p = 0.05). Two sporadic cases had a point mutation detected at low level in their lymphocyte RNA and were assumed to be mosaic for the mutation. One patient with the c.2530C > T (p.Lys844Phe) missense mutation detected at low level in blood lymphocytes (we estimate that the mutation is present in approx. 40% of lymphocytes in this sample, this equates to a ~ 20% mutant allele fraction) has had an affected child who has the same mutation. On reassessment of her skin although she clearly met NIH criteria with > 5 CAL, freckling and neurofibromas her pattern of skin involvement meant various body segments were completely unaffected.
Table 2.
Mutation detection in familial and de novo NF1 patients.
| Type mutation | Familial number | Familial %+ | De novo number | De novo % | p value | Sporadic ≥ 6 CAL group | ≥ 6 CAL % |
|---|---|---|---|---|---|---|---|
| WGD on MLPA (type 1;2;3) |
6 (5;1;0) |
3.61% | 15 (13;1;1) |
8.24% | 0.076 | 1 | 2.13% |
| MLPA deletion | 3 | 1.81% | 4 | 2.20% | Ns | 1 | 2.13% |
| Frameshift | 53 | 31.93% | 61 | 33.52% | ns | 10 | 21.28% |
| IFD | 7 | 4.22% | 6 | 3.30% | Ns | 1 | 2.13% |
| Nonsense | 35 | 21.08% | 41 | 22.53% | Ns | 15 | 31.91% |
| Missense class 4/5 | 7 | 4.21% | 7⁎ | 3.85% | 6⁎ | 12.77% | |
| Missense class 3 | 11 | 6.63% | 6⁎ | 3.30% | 0.26 | 3⁎ | 6.38% |
| Splice site | 40 | 24.10% | 40 | 21.98% | Ns | 10 | 21.28% |
| 5′ UTR class 4/5 | 3 | 1.81% | 1 | 0.55% | ns | 0 | 0.00% |
| 5′UTR class 3 | 1 | 0.60% | 1 | 0.55% | ns | 0 | 0.00% |
| Total variant found | 166 | 97.08% | 182 | 95.79% | 0.58 | 47 | 66.2% |
| Total pathogenic variant found | 154 | 90.06% | 175 | 92.10% | ns | 44 | 62.0% |
| No mutation | 5 | 8 | 24 | ||||
| All tested | 171 | 190 | 71 | ||||
| Only identified through RNA | 21 | 12.65% | 20 | 10.99% | ns | 3 | 6.40% |
| Truncating | 88 | 53.01% | 102 | 56.04% | 0.39 | 25 | 53.19% |
| Non truncating | 29 | 17.47% | 21 | 11.54% | 0.29 | 10 | 21.28% |
+ Mutation proportions are of total where mutation was found.
Ns —not significant; WGD —whole gene deletion; IFD in frame deletion; 5′ UTR —untranslated region.
+ This is percentage of found mutations.
difference between de novo NIH criteria with at least one non-pigmentary criterion and de novo ≥ 6 CAL group significant for missense variants (p = 0.023).
A similar proportion of inherited and sporadic samples had mutations identified that were only classifiable by RNA analysis (Table 3). A total of 41/348 (11.8%) of samples with proven mutations met this criteria with ten (2.9%) variants occurring > 10 nucleotides from the consensus splice site of which six would have been well outside the boundaries of intron exon regions screened in standard DNA analysis. Seven mutations which would have been classified as a variant of uncertain significance (VUS) (6 missense mutations, one synonymous change) and eight patients with six nonsense mutations on DNA analysis alone were also found to affect the normal splicing of NF1 RNA.
Table 3.
Mutations outside the canonical splicing domains in the NF1 gene classified as causative due to effects on in vitro NF1 splicing.
| Inherited | Number found | RNA mutation | DNA mutation | Original classification | Deep intronic | Reported previously |
|---|---|---|---|---|---|---|
| No | 1 | r.100_204del | c.204 + 3_204 + 6delGAGT | splice site | ||
| Yes | 1 | r.288_289ins288 + 1917_288 + 2024 | c.288 + 2025T > G | splice site | Yes | |
| No × 2 | 2 | r.289_479del | c.479 + 5G > A | splice site | ||
| Yes | 1 | r.480_586del | c.586 + 3_586 + 4delinsGG | splice site | ||
| Yes | 1 | r.480_586del | c.586 + 5G > A | splice site | ||
| No | 1 | r.587_654del | c.587-12_587-10delinsGGG | splice site | ||
| Yes | 1 | r.888_889ins888 + 710_888 + 784 | c.888 + 789A > G | splice site | Yes | |
| Yes | 1 | r.1063_1185del | c.1185 + 5G > C | splice site | Yes | |
| Yes × 1 No × 1 |
2 | r.1260_1261ins1260 + 1605_1260 + 1646 | c.1260 + 1604 A > G | splice site | Yes | Yes but missed on DNA |
| Yes × 2 | 2 | r.1393_1527del | c.1527 + 4_1527 + 7delAGTA | splice site | ||
| No | 1 | r.2252_2325del | c.2325 + 3A > G | splice site | ||
| No | 1 | r.2409_2410ins2410–15_2410–1 | c.2410–16A > G | splice site | Yes | |
| Yes | 1 | r.2851_2990del | c.2851–6-2851-3del | splice site | ||
| No | 1 | r.3494_3496dupuag p.(Ile1165_Gly1166insVal) | c.3497-4T > G | splice site | ||
| No | 1 | r.4110_4111 ins4110 + 836_4110 + 940 | c.4110 + 945A > G | splice site | Yes | Yes but missed on DNA |
| No | 1 | r.4514_4515ins17 | c.4515-14T > G | splice site | ||
| Yes | 1 | r.5152_5205del p.(Phe1719_Val1736del) | c.5205 + 3A > T | splice site | ||
| Yes | 2 | r.[5206_5546del; 5206_5749del] | c.5206-38A > G | splice site | Yes | |
| Yes | 1 | r.5749_5750ins5749 + 155_5749 + 331 | c.5749 + 332A > G | splice site | Yes | Yes but missed on DNA |
| No | 1 | r.6085_6364del | c.6364 + 4A > G | splice site | ||
| Yes | 1 | r.[6756_6757ins6757-2_6757–1; 6757_6858del] | c.6757-3A > G | splice site | ||
| No | 1 | r.7907_7908ins7908-391_7908–322 | c.7908-321C > G | splice site | Yes | Yes but missed on DNA |
| No | 1 | r.2952_2990del | c.2953C > T p.(Gln985Ter) | Nonsense | ||
| Yes | 1 | r.5940_5943delgcag | c.5941C > T p.(Gln1981Ter) | Nonsense | ||
| No | 1 | r.6642_6756del | c.6754A > T p.(Lys2252Ter) | Nonsense | ||
| Yes x 1 No x 2 |
3 | r.6757_6858del | c.6792C > A p.(Tyr2264Ter) | Nonsense | Yes | |
| No | 1 | r.6756_6858del | c.6792C > A p.(Tyr2264Ter) | Nonsense | Yes | |
| Yes | 1 | r.7647_7675del29 | c.7648A > T p.(Arg2550Ter) | Nonsense | ||
| Yes | 2 | r.[1229u > a; 1260_1261ins1260 + 1_1260 + 13] | c.1229T > A p.(Leu410Gln) | VUS missense | ||
| No | 1 | r.1722_1748del | c.1748A > G p.(Lys583Arg) | VUS missense | Yes | |
| No | 1 | r.2252_2325del | c.2325G > T p.(Glu775Asp) | VUS missense |
||
| Yes | 1 | r.[5206_5546del; 5206_5749del] | c.5546G > A (p.Arg1849Gln) | VUS missense | ||
| Yes | 1 | r.[5206_5546del; 5206_5749del] | c.5546G > T p.(Arg1849Leu) | VUS Missense |
||
| No | 1 | r.5940_5943delgcag | c.5943G > A p.(Gln1981Gln) | VUS Synonymous | ||
| Total | 41 | 26 Splice 7 VUS 8 Nonsense |
||||
| Additional samples only classifiable by RNA without study criteria or information to determine this | Deep intronic | ||||
|---|---|---|---|---|---|
| 1 | r.655_730del | c.655-17_655-5del13 | Splice site | ||
| 1 | r.[1260 + 1_1260 + 13ins;1260 + 4a > c] | c.1260 + 4A > C | Splice site | ||
| 1 | r.1393_1527del | c.1393-13_1393-3del11 | Splice site | ||
| 1 | r.3494_3496dupuag p.(Ile1165_Gly1166insVal) | c.3497-4T > G | Splice site | ||
| 1 | r.[6642_6858del, 6757_6858del] | c.6757–14T > G | Splice site | ||
| 1 | r.8050_8051ins8050 + 1_8050 + 85 | c.8050 + 86A > G | Splice site | Yes | |
| 1 | r.289_479del | c.479G > T p.(Arg160Met) | VUS missense | ||
| Subtotal | 7 | 6 splice 1 missense | |||
| Previously reported variants outside study criteria and outside canonical domain | |||||
| 1 | r.1642_1721del | c.1721 + 3A > G | Splice site | ||
| 1 | r.4110_4111ins4110 + 4160_4110 + 4239 | c.4110 + 4159A > G | Splice site | Yes | |
| 1 | r.1466_1527del | c.1466 A > G p.(Tyr489Cys) | VUS missense | ||
| 2 | r.1846_1886del41 | c.1885G > A p.(Gly629Arg) | VUS missense | ||
| 1 | r.2252_2325del | c.2325G > T p.(Glu775Asp) | VUS missense | ||
| Total | 13 | 5 missense 8 splice |
|||
Table 4 shows the variants which were not truncating and did not affect the splicing at RNA level. These were predominantly missense mutations some of which have previous published evidence for pathogenicity (Griffiths et al., 2007, Upadhyaya et al., 1997, Ahmadian et al., 2003, Valero et al., 2011, Fahsold et al., 2000). However, six samples (4 familial) occurred in an evolutionary conserved region of the 5′ untranslated region (UTR). Although we did confirm bi-allelic NF1 expression in two samples that were heterozygous at the DNA level for transcribed SNPs (c.-272G > A and c.-273A > C), the first variant segregated with disease in the family. Furthermore c.-273A > C was shown to have arisen de novo as it was not carried by the unaffected parents of a sporadic case. Classification of pathogenicity in Table 4 is as per accepted guidelines (Plon et al., 2008, Wallis et al., 2013, Richards et al., 2015).
Table 4.
Missense and 5′UTR variants and their likely pathogenicity in main study cohort. (Reference sequence used for mutation names NM_000267.3).
| Inherited | Times seen in study cohort | Variant | Type | Exon | Segregation analysis | In silico analysis | Classification of pathogenicity1 |
|---|---|---|---|---|---|---|---|
| Yes | 1 | c.139T > C p.(Ser47Pro) | missense | 2 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| No | 1 | c.412G > C p.(Ala138Pro) | missense | 4 | Seen in 1 individual- not detected in parents | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4- Likely pathogenic |
| No | 1 | c.581T > C p.(Leu194Pro) | missense | 5 | Seen in 1 individual- not detected in parents | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic |
| No | 1 | c.988G > C p.(Ala330Pro) | missense | 9 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.1658A > G p.(His553Arg) | missense | 15 | Seen in 3 individuals- appeared to segregate with NF1 features in 1st family & not detected in parents in 2nd family, no segregation studies done in 3rd family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4- Likely pathogenic |
| Yes | 1 | c.1808T > G p.(IIe603Arg) | Missense | 16 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| No | 1 | c.2329T > A p.(Trp777Arg) | Missense | 20 | Seen in 2 individuals- no segregation studies done in 1st family, not detected in parents in 2nd study family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4- Likely pathogenic |
| Yes | 1 | c.2339C > A p.(Thr780Lys) | Missense | 20 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.2530C > T p.(Lys844Phe) | Missense | 21 | Mosaic with heterozygous mutation in daughter so likely pathogenic | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4 Likely pathogenic |
| No | 1 | c.2540T > C p.(Leu847Pro) | 21 | Seen in 2 individuals- no segregation studies done in 1st family, not detected in 1 parent in 2nd study family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic (Also reported in an NF1 patient inFahsold et al. (2000) | |
| Yes | 1 | c.2543G > A p.(Gly848Glu) | Missense | 21 | Seen in 1 individual- appeared to segregate with NF1 features in the family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.2870A > T p.(p.Asn957Ile) | Missense | 22 | Seen in 1 individual- appeared to segregate with NF1 features in the family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.3044T > C p.(Leu1015Pro) | Missense | 23 | Seen in 1 individual- Did NOT appear to segregate with NF1 symptoms in the family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.3047G > A p.(Cys1016Tyr) | Missense | 23 | Seen in 1 individual- appeared to segregate with NF1 symptoms in the family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.3251C > A p.(Pro1084His) | Missense | 25 | Seen in 1 individual on a DNA screen- appeared to segregate with NF1 symptoms in the family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.3447G > A p.(Met1149Ile) | Missense | 26 | Seen in 2 individuals- appeared to segregate with NF1 symptoms in 1st family, no segregation studies done in 2nd family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4- Likely pathogenic de novo in Griffiths et al. (2007) |
| No | 1 | c.3610C > G p.(Arg1204GIy) | Missense | 27 | Seen in 2 individuals- not detected in parents in 1st study family, no segregation studies done in 2nd family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4 Likely pathogenic |
| No | 1 | c.3649G > T p.(Asp1217Tyr) | Missense | 27 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.3827G > A p.(Arg1276Gln) | Missense | 28 | Seen in 2 individuals- no segregation studies done in 1st family, appeared to segregate with Watson syndrome symptoms in 2nd family | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4- Likely pathogenic |
| No | 1 | c.4016T > G p.(Leu1339Arg) | Missense | 30 | Seen in 1 individual- not detected in parents | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic |
| No | 1 | c.4172G > C p.(Arg1391Thr) | Missense | 32 | Seen in 2 individuals- not detected in 1 parent in 1st family, no segregation studies done in 2nd family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3- Unknown pathogenicity |
| No | 1 | c.4173A > T p.(Arg1391Ser) | Missense | 32 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic (Functional studies in Upadhyaya et al. (1997) |
| No | 1 | c.4288A > G p.(Asn1430Asp) | Missense | 33 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic (located within the NF1 RAS GAP domain. shown to be deleterious to RAS interaction in vitro) Ahmadian et al. (2003) |
| Yes | 1 | c.4306A > G p.(Lys1436Glu) | Missense | 33 | Seen in 2 individuals- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4- Likely pathogenic (de novo in Valero et al. (2011) |
| No | 1 | c.4715T > C p.(Phe1572Ser) | Missense | 36 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3- Unknown pathogenicity |
| Yes | 1 | c.4805T > C p.(Leu1602Pro) | Missense | 37 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3-Unknown pathogenicity |
| Yes | 1 | c.4986C > G p.(Asn1662Lys) | Missense | 37 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3-Unknown pathogenicity |
| No | 1 | c.5450C > G p.(Ser1817Cys) | Missense | 38 | Seen in 1 individual- no segregation studies done | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3-Unknown pathogenicity |
| Yes | 1 | c.5681T > G p.(Leu1894Arg) | Missense | 39 | Seen in 1 individual- appeared to segregate with NF1 features in the family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 3-Unknown pathogenicity |
| No | 1 | c.6947T > C p.(Leu2316Pro) | Missense | 47 | Seen in 1 individual- not detected in parents | Affects an evolutionarily conserved amino-acid residue, gave conflicting results on whether it is likely to be disruptive to normal function | Class 4-Likely pathogenic |
| Yes | 1 | c.6950T > C p.(Leu2317Pro) | Missense | 47 | Seen in 2 individuals- not detected in parents in 1st family, no segregation studies done in 2nd family | Affects an evolutionarily conserved amino-acid residue and is predicted to disrupt normal function | Class 4-Likely pathogenic |
| Yes × 2 | 2 | c.-272G > A | 5′ UTR | Seen in 2 individuals- appeared to segregate with NF1 features in both families, | Class 4-Likely pathogenic | ||
| Yes | 1 | c.-272G > C, NF1 RNA NORMAL | 5′ UTR | Seen in 1 individual- appeared to segregate with NF1 symptoms in the family | Class 3-Unknown pathogenicity | ||
| No × 1 Yes × 1 |
2 | c.-273A > C | 5′ UTR | Seen in 2 individuals- not detected in parents or unaffected sister in 1st family, no segregation studies done in 2nd family | Class 4-Likely pathogenic | ||
| No | 1 | c.-280C > T | 5′ UTR | Seen in 1 individual- no segregation studies done | Class 3-Unknown pathogenicity |
Classification of pathogenicity follows those proposed by Plon et al. (2008),Wallis et al. (2013) and Richards et al. (2015).
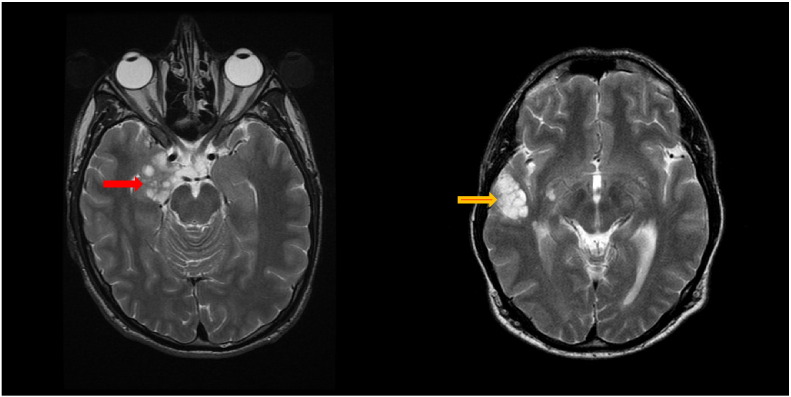
Of the thirteen cases with no identified mutation two had a dysembryoplastic neuroepithelial tumour (DNET) identified on MRI of the brain (Fig. 1a and b) both associated males with epilepsy. One had inherited NF1 from a mother affected with classical features of NF1. The other was sporadic but did appear to have major nerve root nerve sheath tumors on whole body MRI and had tumour resection because of his intractable epilepsy. Histopathology of tissue removed at surgery to relieve epilepsy in the sporadic case showed sections of cortical grey matter revealing indistinct glioneuronal nodules, the familial case histopathology showed oligodendroglia-like cells forming bundles around axonal profiles with occasional floating neurons, both consistent with the radiological diagnosis of DNET. Both patients tested negative for SPRED1 mutations. The only two other NF1 affected individuals with a DNET had identified NF1 nonsense mutations (c.6763G > T p.(Glu2255Ter; c.4084C > T p.(Arg1362Ter). They both had classical NF1 inherited from a clearly affected parent with > 6 CAL and neurofibromas, but did not have epilepsy and diagnosis was based on imaging appearances alone. The presence of DNET in 2/13 (15%) negative screens versus 2/348 (0.6%-Relative Risk of 0.004) with mutations is highly significant (p = 0.007).
Fig. 1.
Axial T2-weighted images showing multi-lobulated mass lesions in right mesial temporal and amygdala (red arrow) in case 1 and right temporal lobe, including superior temporal and middle temporal gyral cortex (yellow arrow) in case 2. Lesions were T1-hypointense and failed to enhance with contrast.
Seventy one individuals aged < 20 with 6 + CAL and no non-pigmentary NF1 criterion had an RNA sample assessed (Table 5). 44/71 (62%) had a clearly deleterious NF1 mutation, this included six with missense variants that were shown to have arisen de novo (c.2329T > C p.(Trp777Arg); c.3610C > G p.(Arg1204GIy); c.4016T > G (p.Leu1339Arg); c.4267A > G p.(Lys1423Glu); c.5425C > T p.(Arg1809Cys); c.6950T > C p.(Leu2317Pro); and also an in frame deletion not present in the parents lymphocyte DNAs (c.3327_3329del p.(Leu1109del)). All of these variants were also predicted to be deleterious on in silico analysis and were in conserved amino-acid residues across species and the latter four missense variants also excluded in both parents. There were also: fifteen nonsense, ten splice site/splicing variants (including c.1260 + 1604A > G also seen in Table 3) with three only classified by RNA, ten frameshift and one deletion of exon2 and one whole gene deletion (type 1). One nonsense mutation was detected as a mosaic (estimated 18% mutant cellular fraction) in lymphocyte RNA in an individual aged 19 years at assessment who had 6 faint CAL (c.5839C > T p.(Arg1947Ter)). A further three individuals had missense mutations that had in silico evidence and conservation across species to predict they were disease causing (c.1658A > G p.(His553Arg); c.5425C > T p.(Arg1809Cys); c.6554C > G p.(Thr2185Arg)) the former two also had evidence from a previous patients that they had arisen de novo.
Table 5.
Identification of NF1 and Legius syndrome based on testing sporadic childhood cases with > 5 CAL with no other features of NF1 and extrapolation to reassurance of negative testing with both RNA and DNA based testing.
| Clear pathogenic NF1 mutation (definite NF1) |
Missense or other vus probably disease causing (probable NF1) |
SPRED1 mutation (Legius syndrome) | No mutation (probable sporadic CAL or other disorder) | Likelihood of a missed mutation assuming sensitivity of (RNA) and (DNA) | Likelihood that no full germline NF1 mutation exists | Likelihood has NF1 if NF1 testing negative | |
|---|---|---|---|---|---|---|---|
| Actual testing of 71 samples | 62% (n = 44)a | 4.2% (n = 3) | 8.4% (n = 6) | 25.4% (n = 18) | – | ||
| Predicted likelihood based on 95.8% sensitivity from RNA testing | 64.9% | 4.2% | 8.4% | 22.5% | 2.90% (95.79%) | 2.9/25.4 = 11.4% | 11.4% 1 in 9 |
| Predicted identification rate from DNA testing | 55.7%b | 8.6%+ | 8.4% | 27.3% | 4.8% (93.05%) |
4.8/27.3 = 17.6 | 17.6% 1 in 6 |
Includes 4 missense mutations shown to have arisen de novo by exclusion in parents.
The reduction from RNA testing assumes 2.9% with deep intronic splicing mutations (overall 1.9% of samples) will be missed and that a further 6.6% with splicing variants outside consensus splice site or missense mutations causing splicing will not be classifiable and moved to the VUS column.
There were six patients from this cohort with predicted loss of function SPRED1 mutations (8.4%) one was proven de novo (c.190C > T p.(Arg64Ter)) and four further mutations were assumed to be de novo as the parents had no features (c.177dupT p.(Ile60TyrfsTer18); c.360delA p.(Ile120MetfsTer32); c.1048_1060del13 p.(Gly350MetfsTer52); c.304dupA p.(Thr102AsnfsTer7)) and a further mutation was found to have been inherited from a mother who on closer inspection was found to have 4 CAL (c.482_483delCA p.(Thr161SerfsTer5)). Seventeen individuals (24%) had no mutation identified in either NF1 or SPRED1 and one had a missense variant c.4768C > T (p.Arg1590Trp) in NF1 that was inherited from his father without CAL and assumed to be unrelated to his CAL.
Since 2013 we have screened one hundred and thirty two samples from patients that did not meet NIH criteria where no NF1 mutation or variant was identified following comprehensive RNA level and MLPA copy number analysis.
4. Discussion
The present study has shown a very high detection rate for NF1 mutations in classically affected individuals meeting NIH criteria. There was no difference in detection rates between the familial and de novo group although two patients with de novo NF1 including one with an affected child had mosaicism detected in lymphocyte RNA. This study would therefore suggest that levels of mosaicism undetectable in RNA/DNA are unlikely to cause classical NF1. Mosaicism is nonetheless well documented as the cause of segmental NF1 where identical NF1 mutations can be detected in melanocyte cultures from anatomically separate CAL (Maertens et al., 2007). Nonetheless mosaic large deletions that are harder to detect by MLPA (as this will usually only detect down to around 20% level) have been shown to cause an occasional generalised case (Messiaen et al., 2011). However, both cases reported in the series with large deletions were present at 50% or greater level (25% mutant allele fraction) and should therefore have been identified in our current study. It is nonetheless possible that the more severe phenotype associated with whole gene deletions may cause generalised disease with even lower levels of mosaicism, whereas this would be less likely for point mutations at below 10% mutant allele fraction. A previous RNA analysis identified 64/67 (95%) of mutations in clearly affected NF1 individuals (Messiaen et al., 2000). This study was based on much smaller numbers: 29/29 with familial and 35/38 sporadic had mutations identified, but was not able to differentiate statistically between inherited and sporadic cases (p = 0.25).
A substantial proportion of variants identified (12.5% familial; 11% sporadic) were not classifiable based solely on RNA analysis as functionally relevant. Nonetheless several of those reported here have been shown to be disease causing by functional analysis and other evidence such as being proven to have arisen de novo. Most occur at residues that are highly conserved across species. Using the ExAC database (http://exac.broadinstitute.org/gene/) from exome sequencing of over 120,000 alleles in humans the prevalence of NF1 missense variants was only 2.2% per allele with 489 variants being seen 2426 times with 28 variants being seen in a homozygous state. The homozygous variants are shown in Table 6 and are very unlikely to be functional variants as NF1 is thought to be lethal when homozygous (Brannan et al., 1994). Overall, excluding these variants observed in a homozygous state only 1.2% of NF1 alleles on ExAC contained missense variants which approximates to 2.4% of people. This compares to 28/33 (84%) of familial NF1 cases and 19/27 (70%) sporadic de novo cases in our cohort where no other NF1 sequence change was present. It is therefore highly likely that most of these variants are disease causing.
Table 6.
Missense variants that appear on the ExAC database as occurring in homozygous state.
| Missense variant | Type | Number seen | Number of alleles tested | Number homozygous |
|---|---|---|---|---|
| p.Pro678Leu | Missense | 36 | 120726 | 1 |
| p.Met895Ile | Missense | 10 | 121340 | 1 |
| p.Asn974Ser | Missense | 28 | 120726 | 1 |
| p.Thr1324Ala | Missense | 5 | 121372 | 1 |
| p.Ile2127Val | Missense | 40 | 121386 | 1 |
| p.Met645Val | Missense | 139 | 121230 | 2 |
| p.Ser665Phe | Missense | 89 | 121022 | 2 |
| p.Asp176Glu | Missense | 397 | 120814 | 3 |
| p.Ile1679Val | Missense | 449 | 121404 | 16 |
Five of the six variants identified in the NF1 5′ UTR region involve just two nucleotides at position c.-272 and c.-273. These nucleotides are highly conserved through species and the c.-273A > C variant was shown to occur de novo in a sporadic case in our series. Furthermore, these variants were not seen on the other allele when a pathogenic NF1 mutation has been found in over 500 full screens. Whilst we have shown that these variants are still associated with bi-allelic RNA expression in vitro, as transcribed polymorphisms in these patients did not show any perceptible skewing on Sanger analysis of cDNA, we hypothesise that they could still act via a reduction in production of mature mRNA in vivo from the affected allele. For instance these variants may activate a cryptic translation initiation site which could in turn suppress the use of the normal NF1 translation initiation site leading to an aberrant NF1 protein or nonsense mediated decay if a premature stop codon is incorporated upstream of the normal initiation site. This mechanism has been described in a 5UTR mutation in BMPR2 in patients with pulmonary hypertension (Aldred et al., 2007).Further experimentation is however needed to determine the mode of action.
Two families, one sporadic de novo and one familial presenting with a DNET did not have an identifiable mutation despite the individuals meeting NIH criteria. DNETs have been reported as histologically proven in 5/25 (20%) NF1 patients who underwent epilepsy surgery, but NF1 mutational status was not reported (Barba et al., 2013). DNETs are slow growing low grade brain tumors with excellent prognosis following surgery the vast majority present with seizures. We hypothesise that DNETs may define an NF1 like syndrome caused by heterozygous variants in a gene other than NF1 given the highly significant association of DNETs with absence of identifiable mutation. Thus another as yet unidentified gene could account for the approximately 4% of patients/families meeting NIH criteria or some may possibly be caused by an extragenic deletion or promoter methylation affecting RNA expression of one copy of the NF1 gene.
A number of other researchers have employed an RNA based analysis to identify NF1 mutations which emphasise the importance of identifying splicing variants. Sensitivity against NIH criteria could be ascertained in 53/56 (95%) in a Spanish study (Valero et al., 2011) and 546/565 (97%) in French study (Sabbagh et al., 2013). Another study from Barcelona identified splicing mutations in 173/374 (46%) independent samples with pathogenic variants of which nine (2.4%) were deep intronic (Pros et al., 2008). The large French study (Sabbagh et al., 2013, Pasmant et al., 2010) found a rate of patients with deletion involving the entire NF1 locus of 4.3% with 3.9% having other MLPA rearrangements and 13/565 (2.3%) having deep intronic splicing mutations. These results are similar to the present study although comparison of rates of whole gene deletions between sporadic and familial cases was not possible to discern from the data in the manuscripts. It is unclear whether next generation sequencing testing will add any advantages over comprehensive RNA analysis (Balla et al., 2014, Pasmant et al., 2015). Whole gene deletions of NF1 have been associated with a more severe phenotype including significantly higher incidence of learning disabilities and facial dysmorphism (Pros et al., 2008) as well as malignancy (De Raedt et al., 2003). This may well explain a lower genetic fitness due to the severity of the phenotype caused to account for the higher rate of these in de novo cases (Smith et al., 2016). The predominance of the larger 1.4 MB type 1 whole gene deletion in our UK study (18/21–86%) is similar to other studies at 78% (54/70) (Pasmant et al., 2010).
The present study quantifies the level of reassurance a parent may have on diagnostic testing that their child does not have NF1 if there is no sequence variant in NF1 or SPRED1 when their child has 6 or more CAL. Around 25% of children tested did not have an identifiable mutation. Using a Bayesian calculation and assuming 95.8% sensitivity for testing in an isolated case fulfilling NIH criteria there would still be a 1 in 9 chance that their child had NF1 (Table 5). However, this rises to 1 in 6 if only DNA testing was carried out. DNA testing would also increase the likelihood that the parents would be given an uncertain result with the VUS rate rising from 4.2% to 8.6% because of failure to be able to classify missense and splicing variants outside the consensus splicing region as functional. We also found a significant rate of 8.5% (1 in 12) for SPRED1 mutations. These were previously shown to be more frequent in familial CAL than in sporadic cases with 19% of familial CAL due to SPRED1 (Messiaen et al., 2009).
The present study has defined a very high sensitivity for a mutation screening approach incorporating RNA testing in identifying a mutation in NF1 affected individuals meeting NIH criteria with at least one non-pigmentary criterion. This enables a great deal of reassurance to unaffected parents of children with > 5 CAL as the likelihood of their child having constitutional NF1 drops from over 60% to as little as 11%. DNA based testing alone classifies fewer cases as having definite NF1 and a negative screen leaves a 1 in 6 chance the child will still have NF1. Finally, among the very small number without an identifiable mutation who do meet NIH criteria it remains possible that a rare further second gene may be responsible for a condition which also predisposes to DNET.
Author Contributions
Conception, study design: DGE, SMH, AJW; patient data contribution: DGE, SMH, MP-S, GV, AD, EB-W, SG, EH, JE; RNA analysis and MLPA: EM, NB, AJW; data analysis DGE, AJW, WN, VS-K; Interpretation: DGE, AJW, WN, SMH; manuscript drafting all; approval of final version: all.
Acknowledgements
The complex NF1 service is funded by the NHS England programme for highly specialised services. We are grateful to Professor Ludwine Messiaen for her assistance in the initial stages of setting up this service and for continued valued discussion of difficult cases. We would like to thank Kirsty Henshaw for her help in obtaining notes and supporting the service.
References
- Ahmadian M.R., Kiel C., Stege P., Scheffzek K. Structural fingerprints of the Ras-GTPase activating proteins neurofibromin and p120GAP. J. Mol. Biol. 2003;329(4):699–710. doi: 10.1016/s0022-2836(03)00514-x. [DOI] [PubMed] [Google Scholar]
- Aldred M.A., Machado R.D., James V., Morrell N.W., Trembath R.C. Characterization of the BMPR2 5′-untranslated region and a novel mutation in pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2007;176(8):819–824. doi: 10.1164/rccm.200701-164OC. [DOI] [PubMed] [Google Scholar]
- Anderson J.L., Gutmann D.H. Neurofibromatosis type 1. Handb. Clin. Neurol. 2015;132:75–86. doi: 10.1016/B978-0-444-62702-5.00004-4. [DOI] [PubMed] [Google Scholar]
- Balla B., Árvai K., Horváth P. Fast and robust next-generation sequencing technique using ion torrent personal genome machine for the screening of neurofibromatosis type 1 (NF1) gene. J. Mol. Neurosci. 2014;53(2):204–210. doi: 10.1007/s12031-014-0286-7. [DOI] [PubMed] [Google Scholar]
- Barba C., Jacques T., Kahane P. Epilepsy surgery in neurofibromatosis type 1. Epilepsy Res. 2013;105(3):384–395. doi: 10.1016/j.eplepsyres.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Brannan C.I., Perkins A.S., Vogel K.S. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8(9):1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- De Raedt T., Brems H., Wolkenstein P. Elevated risk for MPNST in NF1 microdeletion patients. Am. J. Hum. Genet. 2003;72(5):1288–1292. doi: 10.1086/374821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D.F., Ponder M.A., Huson S.M., Ponder B.A.J. An analysis of variation in expression of NF1: evidence for modifying genes. Am. J. Hum. Genet. 1993;53:305–315. [PMC free article] [PubMed] [Google Scholar]
- Evans D.G., Ramsden R.T., Shenton A. Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. J. Med. Genet. 2007;44(7):424–428. doi: 10.1136/jmg.2006.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.G., Howard E., Giblin C. Birth incidence and prevalence of tumour prone syndromes: estimates from a UK genetic family register service. Am. J. Med. Genet. 2010;152A(2):327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- Evans D.G., Howard E., Wilding A. Mortality in neurofibromatosis 1. Eur. J. Hum. Genet. 2011;19(11):1187–1191. doi: 10.1038/ejhg.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsold R., Hoffmeyer S., Mischung C. Minor lesional mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner R.E. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6(4):340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- Griffiths S., Thompson P., Frayling I., Upadhyaya M. Molecular diagnosis of neurofibromatosis type 1: 2 years experience. Familial Cancer. 2007;6(1):21–34. doi: 10.1007/s10689-006-9001-3. [DOI] [PubMed] [Google Scholar]
- Gutmann D.H., Aylesworth A., Carey J.C. The diagnostic and multi-disciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997;278:51–57. [PubMed] [Google Scholar]
- Huson S.M., Harper P.S., Compston D.A. Von Recklinghausen neurofibromatosis. A clinical and population study in south-east wales. Brain. 1988;111:1355–1381. doi: 10.1093/brain/111.6.1355. [DOI] [PubMed] [Google Scholar]
- Huson S.M., Compston D.A.S., Harper P.S. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. I. Prevalence, fitness, mutation rate, and effect of parental transmission on severity. J. Med. Genet. 1989;26:704–711. doi: 10.1136/jmg.26.11.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes L.M., Riccardi V.M., Burke W., Bennett R.L., Stephens K. Large de novo DNA deletion in a patient with sporadic neurofibromatosis 1, mental retardation, and dysmorphism. J. Med. Genet. 1992 Oct;29(10):686–690. doi: 10.1136/jmg.29.10.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O., De Schepper S., Vandesompele J. Molecular dissection of isolated disease features in mosaic neurofibromatosis type 1. Am. J. Hum. Genet. 2007;81(2):243–251. doi: 10.1086/519562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L.M., Callens T., Mortier G. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 2000;15(6):541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Messiaen L., Yao S., Brems H. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA. 2009;302(19):2111–2118. doi: 10.1001/jama.2009.1663. [DOI] [PubMed] [Google Scholar]
- Messiaen L., Vogt J., Bengesser K. Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1) Hum. Mutat. 2011;32(2):213–219. doi: 10.1002/humu.21418. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Conference Statement on Neurofibromatosis. Arch. Neurol. 1987;45:575–579. [Google Scholar]
- Pasmant E., Sabbagh A., Spurlock G. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum. Mutat. 2010;31(6):E1506–E1518. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- Pasmant E., Parfait B., Luscan A. Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations? Eur. J. Hum. Genet. 2015;23(5):596–601. doi: 10.1038/ejhg.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S.E., Eccles D.M., Easton D., Foulkes W.D. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29(11):1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pros E., Gómez C., Martín T., Fábregas P., Serra E., Lázaro C. Nature and mRNA effect of 282 different NF1 point mutations: focus on splicing alterations. Hum. Mutat. 2008;29(9):E173–E193. doi: 10.1002/humu.20826. [DOI] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;10:294–300. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojnueangnit K., Xie J., Gomes A. High incidence of Noonan syndrome features including short stature and pulmonic stenosis in patients carrying NF1 missense mutations affecting p.Arg1809: genotype–phenotype correlation. Hum. Mutat. 2015 (Nov);36(11):1052–1063. doi: 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh A., Pasmant E., Imbard A. NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum. Mutat. 2013;34(11):1510–1518. doi: 10.1002/humu.22392. [DOI] [PubMed] [Google Scholar]
- Shen M.H., Harper P.S., Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1) J. Med. Genet. 1996 Jan;33(1):2–17. doi: 10.1136/jmg.33.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.J., Urquhart J.E., Harkness E.F. The contribution of whole Gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum. Mutat. 2016;37(3):250–256. doi: 10.1002/humu.22938. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M., Osborn M.J., Maynard J., Kim M.R., Tamanoi F., Cooper D.N. Mutational and functional analysis of the neurofibromatosis type 1 (NF1) gene. Hum. Genet. 1997;99(1):88–92. doi: 10.1007/s004390050317. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M., Huson S.M., Davies M. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): evidence of a clinically significant NF1 genotype–phenotype correlation. Am. J. Hum. Genet. 2007;80(1):140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo E., Leppävirta J., Koffert A. Incidence and mortality of neurofibromatosis: a total population study in Finland. J. Investig. Dermatol. 2015;135(3):904–906. doi: 10.1038/jid.2014.465. [DOI] [PubMed] [Google Scholar]
- Valero M.C., Martín Y., Hernández-Imaz E. A highly sensitive genetic protocol to detect NF1 mutations. J. Mol. Diagn. 2011;13(2):113. doi: 10.1016/j.jmoldx.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis Y., Payne S., McAnulty C. Practice guidelines for the evaluation of pathogenicity and the reporting of sequence variants in clinical molecular genetics. ACGS epub. 2013. http://www.acgs.uk.com/