Abstract
Background
The transmembrane subunit of the HIV envelope protein, gp41 is a vulnerable target to inhibit HIV entry. There is one fusion inhibitor T20 (brand name: Fuzeon, generic name: enfuvirtide) available by prescription. However, it has several drawbacks such as a high level of development of drug resistance, a short-half life in vivo, rapid renal clearance, low oral bioavailability, and it is only used as a salvage therapy. Therefore, investigators have been studying a variety of different modalities to attempt to overcome these limitations.
Methods
Comprehensive literature searches were performed on HIV gp41, inhibition mechanisms, and inhibitors. The latest structural information was collected, and multiple inhibition strategies targeting gp41 were reviewed.
Results
Many of the recent advances in inhibitors were peptide-based. Several creative modification strategies have also been performed to improve inhibitory efficacy of peptides and to overcome the drawbacks of T20 treatment. Small compounds have also been an area of intense research. There is a wide variety in development from those identified by virtual screens targeting specific regions of the protein to natural products. Finally, broadly neutralizing antibodies have also been important area of research. The inaccessible nature of the target regions for antibodies is a challenge, however, extensive efforts to develop better neutralizing antibodies are ongoing.
Conclusion
The fusogenic protein, gp41 has been extensively studied as a promising target to inhibit membrane fusion between the virus and target cells. At the same time, it is a challenging target because the vulnerable conformations of the protein are exposed only transiently. However, advances in biochemical, biophysical, structural, and immunological studies are coming together to move the field closer to an understanding of gp41 structure and function that will lead to the development of novel drugs and vaccines.
Keywords: HIV, envelope protein, gp41, neutralizing antibodies, peptide inhibitors, viral entry, membrane fusion
INTRODUCTION
Structure of gp41 in the Env Complex
HIV-1 is an enveloped virus with 8-10 envelope ‘spike’ complexes per virion [1]. The envelope spike is made up of two protein subunits, the surface subunit, gp120, and the transmembrane subunit, gp41. These two subunits form a trimer of heterodimers that are non-covalently associated on the virus or on infected cells and are attached to the membranes via the transmembrane region of gp41.
The envelope complex is expressed as a precursor protein encoded by the envelope gene of HIV-1. It is expressed into the endoplasmic reticulum and transits the Golgi apparatus during which 25-30 N-linked glycans are added and trimmed [2-4].
The surface subunit, gp120, has a globular structure composed of five conserved domains (C1-C5) and five variable loops (V1-V5) [5, 6]. Gp120 has 18 cysteine residues, which form a loop structure connecting V1 to V4 by disulfide bonds [7]. These highly glycosylated variable loops shield the conserved regions of gp120 and protect the virus from antibodies. This is a protective barrier that the virus utilizes to evade the immune system, which is often referred to as the glycan shield [8].
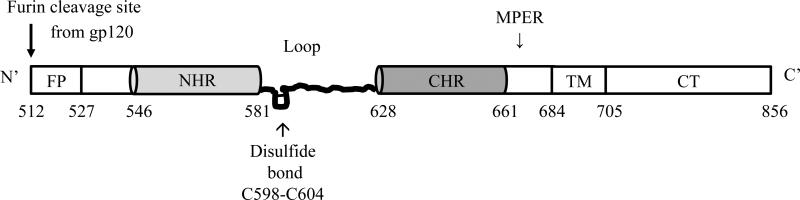
Gp41 is divided into multiple functional domains (Fig. 1). Beginning at the N-terminus, there is a fusion peptide, which is necessary for membrane fusion. Moving toward the C-terminus there are two helical heptad repeat (HR) regions, which are designated N-terminal heptad repeat (NHR) and C-terminal heptad repeat (CHR). These two regions are connected to a loop region that is more mobile than the helical heptad repeat regions and also contains an important disulfide bond [9-12]. The CHR is followed in sequence by a membrane proximal external region (MPER). This region has been a very promising target for drug and immunogen development as it contains epitopes that bind some of the neutralizing antibodies that have been identified such as 2F5, 4E10, Z13, and 10E8 [13-20] (see below). Next in sequence is a highly conserved transmembrane domain (TM) of 22 amino acids followed by a C-terminal cytoplasmic region (Fig. 1).
Fig. (1). The primary structure of gp41.
Functional domains of gp41 from the N-terminus to C-terminus are: the fusion peptide (FP), N-terminal heptad repeat (NHR), a disulfide-bonded immunodominant loop region, C-terminal heptad repeat (CHR), a membrane proximal external region (MPER), and a transmembrane domain (TM) followed by a C-terminal cytoplasmic tail (CT). (Amino acids numbers are noted based on conventional numbering of the HIV-1 HXB2 strain).
Atomic level structures of portions of HIV gp41 larger than single domain studies were limited for many years to the ecotodomain in a six-helical bundle, hairpin-like conformation, which researchers in the field consider to be the post-fusion structure. Of these, there were several x-ray crystallographic structures made up of the core sequences of the gp41 NHR/CHR regions of the gp41 ectodomain either incubated together as individual peptides, and allowed to form the 6HB, or tethered covalently, and there was one NMR structure that included the NHR, the loop region, and the CHR [21-27]. The 6HB conformation is made up of three NHR regions, which bind together in parallel forming a three helical bundle. Three CHR regions wrap around in an antiparallel manner, each CHR coming into contact with two of the NHR helices due to the oblique angle of the CHR regions. This results in the disulfide-bonded loop region of gp41 forming the top of a hairpin-like structure.
In 2010, a crystal structure was reported that included sequences further toward the fusion peptide and further toward the viral membrane including the MPER [28]. While most of the structure showed a coiled-coil conformation, terminal sections near the fusion peptide and the viral membrane were not in a canonical coiled-coil, and several residues were situated so that their aromatic side chains would be oriented toward what would be the viral membrane. Interestingly, prior computational work [29] predicted the importance of peptide inhibitor-lipid interactions in what would be an MPER-like bound state.
A construct known as the BG505 SOSIP.664 gp140 trimer was crystallized in complex with a broadly neutralizing antibody (PGT122) and the structure was solved to 4.7 Å [30]. Very briefly, this is a construct that includes gp120 and terminates before the transmembrane region of gp41. There is a disulfide bond inserted between gp120 and gp41 and some of the residues from MPER have been deleted. Interesting findings include a similarity in structure between the internal three helix bundle made up of gp41 NHR and the same portion of the trimer in previous atomic level structures of the 6HB. Also, the authors note the presence of a “hole” in the electron density that they mention is consistent with that observed for the influenza and ebola fusion proteins. The 3HB section (NHR) is stated to be the location of stabilizing contacts between gp41 and gp120 in this structure.
Crystal structures were solved to 3.5 Å in 2014 in complex with two neutralizing antibodies (PGT122 and 35O22) again using the envelope complex mentioned above, BG505SOSIP.664 [31]. The addition of the second antibody (35O22) helped researchers to obtain crystals that diffracted to the higher resolution. The higher resolution allowed the authors to detail very interesting portions of gp41 such as a 4 helix structure termed a “collar” that appears to hold the N- and C- termini of gp120 in a clasp or as the authors call it, a “tryptophan sandwich.” This work also allowed for valuable modeling and detailed comparisons of glycan shield integrity and immune evasion between what the authors identify as their “pre-fusion mature closed state” structure to the previous structures widely considered to be post-fusion structures.
It has been suspected for many years that HIV Env functions by undergoing large, transient conformational changes. The underlying structural changes are propagated over long distances as compared to better-studied enzymatic function. Indeed, this is what has made the study of the conformational changes responsible for HIV entry, especially with regards to the membrane protein gp41, extremely challenging and also what helps to protect the virus from the immune system. HIV Env is also very sensitive to the addition of fluorescent proteins or tags that have been widely utilized to tease out the details of conformational changes in various other systems. Researchers are making advances, however, and single molecule FRET studies have revealed some very interesting clues to the details of the Env conformational changes [32, 33]. Observations of envelope from smFRET with labeled gp120 showed three distinct states representing dynamic prefusion conformations. The occupancy of these states was reported to shift upon addition of CD4 and the co-receptor mimic, antibody 17b. By studying the sampling of the three states and the effect of addition of CD4 and 17b in isolation and in combination, the authors were able to produce data that verify the two-state nature of the activation mechanism of Env. Interestingly it was also apparent that the conformational shift from a low FRET or “closed ground” state occurred less frequently with a primary HIV-1 strain (JR-FL) than with a lab-adapted strain (NL4-3). These data correlate well with the capacity these strains have for neutralization with the lab-adapted strains being much more susceptible to neutralization.
Hydrogen-deuterium exchange coupled to mass spectrometry has also been utilized to study the conformational sampling of Env [34]. Utilizing the soluble SOSIP.664 construct briefly described above and broadly neutralizing antibodies, these results also suggest that there is transient sampling of an ‘open state’ conformation and that the kinetics of this sampling is important to the capacity of antibodies to neutralize the virus. Both smFRET and hydrogen-deuterium exchange studies suggest that broadly neutralizing antibodies are recognizing a ‘closed’ or ‘ground’ state conformation of envelope and are functioning to neutralize the virus by stabilizing this state of envelope [33, 34].
Recently, the first x-ray crystallographic study with an envelope construct that is unliganded was solved [35]. Prior to this publication, structures were liganded with CD4 or a variety of antibodies. This report presents a combination of analyses that are illustrative of how structural knowledge and antigen studies can be combined to inform immunogen design. The authors use “antigenicity-guided conformational fixation” to search for a construct that will be fixed in a conformation that binds only to the most effective neutralizing antibodies and not to those that are poorly neutralizing. One of the most interesting findings of this study was evidence that the entry pathway may be asymmetric with an initial step in which a single CD4 molecule binds followed by the binding of additional CD4 molecules. The initial CD4 binding did not initiate the conformational changes considered the classical changes seen upon CD4-binding (i.e., the formation of a bridging sheet in gp120), however, subsequent CD4 binding did produce these changes [35]. Although there is not a lot of information regarding the structure or structural changes specific to regions of gp41, the identification and isolation of an intact ground state trimer is a very important advancement to potential future structural and mechanistic studies involving gp41.
A key set of findings related to structural aspects of Env and specifically important for gp41 has come from an intricate set of biochemical experiments. Crooks et al. utilized enzyme digests followed by detailed gel analysis as a way to analyze for and eventually eliminate aberrantly folded or “junk” envelope [36]. These include a gp160 that is retained in the endoplasmic reticulum and not fully matured, a mature gp160 that is not fully cleaved at the furin cleavage site between gp120 and gp41, and trimeric and monomeric gp41 stumps along with variations in glycosylation. These aberrant forms of Env are hypothesized to act as decoys for the virus eliciting non-neutralizing antibodies. Tong et al. were able to show that enzyme digests could be utilized to produce virus-like particles bearing native Env trimers, which were only able to be recognized by neutralizing antibodies [37]. The varied population of variants of Env is an important aspect of consideration in the design of targeted inhibitors and also in the design of potential immunogens.
HIV Entry Mechanism
HIV-1 entry is initiated by binding of gp120 to CD4 which is a primary receptor for HIV-1 infection on the target cell surface [38-42]. Association of the conserved domains of gp120 to the CD4 receptor causes rearrangements of the V1 and V2 loops, and then the V3 loop is able to bind to the co-receptor, either chemokine receptor 5, CCR5 (CD195 [43, 44]) and C-X-C chemokine receptor 4, CXCR4 (fusin or CD184 [45, 46]). Receptor binding leads to exposure of hydrophobic regions of gp41 and insertion of the fusion peptide into the host cell membrane.
Upon insertion of the fusion peptide of gp41 into the host cell membrane, the NHR and CHR regions of gp41 come together in an antiparallel fashion forming the hairpin-like six-helix bundle (6HB) conformation of gp41 which is exceptionally hyperthermostable [22]. 6HB formation is thought to be the driving force that brings the viral and cellular membranes in close proximity to facilitate fusion (reviewed in [47, 48]). Initial contact is of the outer leaflets of the viral and cellular membrane, which undergo lipid mixing followed by small pore formation. As merging of the two membranes progresses, the fusion pore enlarges and finally, the viral core enters into the cell cytoplasm [49-54]. It has not been definitively shown whether one or multiple trimers are needed for membrane fusion and this is an area of ongoing debate in the field [48, 49].
NHR- and CHR-Based Peptides
Peptide inhibitors made up of sequences from NHR or CHR (Table 1) when added exogenously bind to form heterogeneous 6HB molecules preventing a collapse of gp41 to the stable 6HB conformation and thereby, inhibiting membrane fusion and viral entry or cell-cell fusion [55-58] (for a recent review see [59]). Various fusion inhibitors targeting pre-hairpin intermediate structures have been under development [55, 56, 58-61]. Entry inhibitor studies also have been very important in helping to shed light on how gp41 mediates membrane fusion [62-71].
Table 1.
Peptide sequences targeting HIV-1 gp41.
| Type | Name | Sequence | gp41 Location | Ref. |
|---|---|---|---|---|
| NHR Peptides | DP-107 (T21) | NNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQ | 553-590 | [55] |
| N36 | SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | 546-581 | [22] | |
| N42 | STMGAASMTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLT | 528-569 | [72] | |
| N36F10 | TLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLT | 536-569 | [73] | |
| CHR peptides | SJ-2176 | EWDREINNYTSLIHSLIEESQNQQEKNEQEGGC | 637-666 | [56] |
| T20 | YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF | 638-673 | [55, 57] | |
| C34 | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL | 628-661 | [22] [27] | |
| T1249 | WQEWEQKITALLEQAQIQQEKNEYELQKLDKWASLWEWF | Chimera | [78, 79] | |
| T1144 | TTWEAWDRAIAEYAARIEALLRALQEQQEKNEAALREL | Chimera | [80] | |
| TLT35 | a chimeric peptide of T20 and T1144 | Chimera | [81] | |
| 2DLT | a chimeric peptide of T1144 and CD4 D1D2 domains | Chimera | [82, 83] | |
| Sifuvirtide | SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE | Chimera | [84] |
Although there are NHR-based peptides such as N36, N42, and N36F10, investigators have placed a greater focus on the development of CHR-based peptides [27, 72, 73]. The primary reason is that CHR-peptide inhibitors are monomeric in solution and show a higher potency, whereas, NHR-peptides tend to aggregate in solution and exhibit lower potency [74].
The pre-hairpin intermediate structure is the target of the only fusion inhibitor in the clinic. The inhibitory peptide T20 (brand name: Fuzeon, generic name: enfuvirtide) is a peptide sequence from the CHR/MPER regions (residues 638-673) of gp41 [55, 57, 63], which was approved by the US FDA as an injectable HIV-1 fusion inhibitor in 2003. Since that time, many other peptides have shown great promise and are in continuing development (reviewed in [59, 75]).
There is a need to develop novel fusion inhibitors that have higher potency than T20, are active against T20 resistant viruses, and exhibit a higher genetic barrier to resistance. To fulfill this mission, many researchers have focused on the highly conserved hydrophobic pocket of gp41. The notion behind this is that the pocket is at the c-terminus of the NHR trimer and is bound to the pocket-binding domain (PBD) in the six helix bundle, made up of residues W628, W631, and I635 [22, 76, 77]. However, the T20 sequence does not include the PBD so that this inhibitor does not take advantage of the interaction between the NHR and PBD. Examples of next generation fusion peptide inhibitors that include the PBD are T1249 [78, 79], T1144 [80], TLT35- a chimera protein of T20 and T1144 [81], and 2DLT-a bivalent recombinant protein of T1144 and the CD4 D1D2 domains [82, 83].
T1249 and T1144 were designed to overcome T-20 resistance. T1249 is composed of gp41 sequences derived from HIV-1, HIV-2, and SIV and this construct exhibited anti-HIV potency to T20 resistant isolates [78, 79]. T1144 was designed to have enhanced helical structure with multiple substitutions of amino acid residues from T2410, which is a peptide that begins with Met and Thr followed by the C34 sequence [80]. This peptide was active against T20- and T1249-resistant HIV strains [80], however, rapid proteolysis of the peptide inhibitor in vivo and high cost of the peptide synthesis remain issues.
To solve these problems, two chimeric proteins were created. TLT35 is a chimeric protein based upon T20 and T1144, which is large enough to be produced in E. coli. This can be produced with lower cost than synthetic peptides. TLT35 exhibited anti-HIV activity in the low nM range against lab-adapted strains, T20 resistant strains, and primary isolates of a variety of clades [81]. There was ~10-100 fold increased antiviral potency compared to T20 and T1144 alone. TLT35 exhibited notably increased stability in PBMC culture (the half-life was increased ~18 fold over that of T20) and in the presence of proteinase K and pepsin (the half-life was increased 8-35 fold over that of T20) [81].
Another bivalent chimeric protein called 2DLT was made up of T144 and the D1D2 domains of CD4. 2DLT was also produced in E. coli and showed promising antiviral potency [82]. However, the mechanism by which this construct inhibits is unique compared to most peptide inhibitors. It is able to trigger pre-exposure of vulnerable gp41 regions before viral fusion occurs. The D1D3 fragment binds to gp120 of HIV, it triggers conformational changes and exposure of gp41 and then another fragment, T1144 binds the exposed gp41 region and results in rapid inactivation of cell-free virus [82]. Importantly, 2DLT resulted in synergistic effects in combination with other antiretroviral drugs [83].
Sifuvirtide (SFT) is the most thoroughly studied peptide inhibitor after T20 [84-88]. It was designed to have a unique 36 amino acid sequence different from the previously studied peptide inhibitors (C34, T20, and T-1249). It has 16 residues that are different than in C34 (underlined below in the SFT sequence: SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE), 22 residues different from T20 (underlined in SFT sequence: SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE), and 24 residues different from T-1249 (underlined in SFT sequence: SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE) [84]. The peptide maintains the gp41 pocket binding sequence that is included in C34, but that T20 is lacking. SFT is missing, however, the lipid binding domain that is present in T20. As such, SFT shares more characteristics in common with C34 than with T20 such as a random coil conformation pattern, secondary α-helix formation in the presence of N36, inability to interact with the lipid membrane, and formation of a 6HB with the N36 peptide [84]. However, the additional residues over the base sequence of C34 that are contained in SFT seem to provide the capacity for SFT to form a tighter 6HB with N36 than the 6HB formed by N36 and C34 (Tm of N36/SFT = 72°C > Tm of N36/C34 = 62°C), which will provide stronger activity against NHR [84]. This peptide produced promising results both in vitro and in clinical studies [84, 86]. It exhibited high potency against a wide range of HIV strains including T20-resistant strains [86, 88] and showed improved clinical pharmacokinetics characteristics when compared to T20 in a clinical trial reported in 2014 [89].
Modified Peptides
CHR peptides have exhibited great promise as inhibitors, however, there are still many problems in using peptide inhibitors as therapeutic agents. Peptides have short half-lives due to degradation by cellular enzymes and rapid renal clearance. There is also limited oral bioavailability so injections are necessary and compliance becomes an issue especially when there are injection site reactions as is the case with T20. Peptides are also very expensive to produce and require specialized storage methods such as either refrigeration or reconstitution with solvent. Thus, investigators have been trying to overcome these problems by modifying peptide inhibitors with a variety of design strategies.
Covalent Peptides
Covalent inhibitors are designed specifically to bind permanently to their targets of inhibition (for a general review on covalent drugs, see [90]). Our group applied this concept to gp41 fusion peptide inhibitors. We designed covalent inhibitors to trap the vulnerable gp41 fusion intermediate permanently [71, 91] using a base C34 peptide. We tested covalent CHR peptide inhibitors made up of residues 628-661 of gp41 [22, 26, 27]. Placement of the maleimide reactive group was based upon the atomic level structures of the HIV gp41 ectodomain, which are considered to be the postfusion structure of gp41 [10, 92, 93]. The maleimide modification of the peptide inhibited gp41 mediated cell-cell fusion [71] and virus-cell fusion [91] after extensive washing by making the association between the peptide and gp41 permanent as shown by extensive washing followed by functional assays.
Albuvirtide (ABT, also known as FB006) is another C34-based covalently reactive peptide, which is chemically modified with 3-maleimidopropionic acid on the Ser640 residue after it had been mutated to a Lys [94]. This construct was able to form secondary α-helical structures with N36 with slightly higher thermostability (Tm of ABT/N36=56°C) than C34/N36 (Tm=54°C). It also blocked formation of the 6HB between C34 and N36 in a dose-dependent manner and inhibited gp41 mediated cell-cell fusion as well as virus entry with more anti-HIV potency than T20 [94]. However, it is not yet known which residues of NHR react with the maleimidopropionic acid group of ABT [94].
Recently, an alternative reactive group has been utilized to create a covalent linkage. A thioester was placed onto C34 in order to initiate an acyl transfer reaction. This is projected to be a better mimic of what is happening under physiological conditions when polypeptides are synthesized [95, 96]. In the process of nonribosomal peptide synthesis, a peptide bond is formed between a carboxy-thioester of the upstream substrate and a free amine on a downstream amino acid by 4’ phosphopantetheinyl cofactor (reviewed in [97]). This acyl transfer reaction is able to form a covalent bond between the modified C34 and the NHR resulting in high potency anti-HIV activity, which correlated to increased thermostability of 6HB when studied in vitro [95, 96].
The benefits of covalent peptides are not limited to improving efficacy of inhibition against gp41. This design methodology can also be used in a variety of areas such as improving drug delivery by attaching molecules to prolong the half-life of the peptides. One such strategy was utilized by attaching human serum albumin [98, 99] to the peptide inhibitor C34. Surprisingly, this did not result in a loss of potency as was expected because the conformational space available between the cellular and viral membranes was thought to be restrictive to something as big as Albumin-C34 (~MW 72 kDa). Inhibition potency was the same as the peptide alone and animal studies showed that the inhibitor efficacy was three fold greater with the albumin attachment.
Constructs that entrap the fusogenic intermediate are also promising tools for mechanistic studies of viral entry with the potential to allow for localization, kinetic, and biophysical studies of the intermediate. A covalently trapped gp41 intermediate also has the potential to elicit novel immune responses. It will be important to carefully consider the increasing amount of structural, biophysical, and immunological information regarding envelope conformational changes when designing these strategies for gp41.
D-Peptides
Investigators designed D-peptide inhibitors, which are protease-resistant to overcome the sensitivity of L-peptide inhibitors to proteolytic degradation. These were designed to increase the half-life in blood and to increase the potential for oral bioavailability. The first D-peptide, D10-p5-2K identified by mirror-image phage display inhibited both gp41-mediated cell-cell membrane fusion and viral entry [77]. However, the potency of D10-p5-2K was only in the micromolar range.
The next generation of D-peptides were discovered by modified mirror-image phage display screening. The D-peptide, PIE7, was reported to have high anti-HIV potency (picomolar range) in its trimeric version against many HIV strains [100]. However, the PIE7 trimer was only moderately potent for the neutralization-resistant clinical isolate JR-FL (IC50=220 nM) compared to HXB2 (IC50=250 pM) or BaL (IC50=650 pM).
The third generation of D-peptide PIE12 trimer showed improved potency against diverse pseudotyped viruses (including clades A-D, circulating recombinant forms, and T20-resistant strains) and replication-competent viruses [101, 102]. Importantly, PIE12 trimer exhibited a better potency for JR-FL (about 4 times better for a pseudotyped virus and 2 times better for a replication-competent virus) than PIE7 trimer [102]. It also displayed better inhibition than T20 in all virus strains that were tested. This improved efficacy of the PIE12 trimer was attributed to a high affinity for the gp41 pocket as the Tm value of PIE12 trimer from its in vitro binding target IZN17 was measured to be 81°C (compared to the Tm of IZN17+PIE7 trimer = 73°C) [102].
Cholesterol-Conjugated Peptides
Another valuable modification that has been made is the covalent addition of cholesterol to a peptide inhibitor. The cholesterol group was chosen as a lipid anchor for the C34 peptide inhibitor based on the importance of its role in HIV fusion [103]. Cholesterol is highly enriched in the lipid membranes of HIV [104-106] and CD4 receptor is enriched in lipid rafts where cholesterol is prevalent [107]. The membranotropic nature of the cholesterol-conjugated C34 (C34-Chol, also known as DS007 or L'644) enhances the interaction between the conjugated peptide and the biological membrane where viral membrane fusion occurs [108].
Indeed, this type of modification provided C34-Chol with ~25-100 fold more potency than C34, ~50-400 fold more potency than T20, and ~15-300 fold more potency than T1249 in multiple isolates in vitro [103]. Further investigation of cholesterol conjugation of a C34 dimer was analyzed and this construct showed improved anti-HIV potency (IC50 = 210±370 pM for HIV-1IIIB and IC50 = 10±10 pM for HIV-1BaL) over C34 (IC50 = 31.12±36 nM for HIV-1IIIB and IC50 = 60.14±84 nM for HIV-1BaL), but it exhibited comparable potency with monomeric C34-Chol (IC50 = 80±70 pM for HIV-1IIIB and IC50 = 360±330 nM for HIV-1BaL) [109].
Importantly, the cholesterol modification allowed the peptide inhibitor to retain antiviral potency even after washing [103] and it was more resistant to proteinase K than T20 and C34 [110]. It also exhibited promising anti-HIV potency in ex vivo genital and colorectal tissue explant models [111]. Therefore, C34-Chol seems to be an attractive topical microbicide candidate as well [111].
The conjugation of cholesterol has been shown to improve the antiviral potency of HR-derived peptide inhibitors of the viral membrane fusion machinery in many other virus systems (reviewed in [112]) such as influenza [113] and Ebola virus [114]. Interestingly, this cross-virus effectiveness could make this strategy a potential rapid response alternative to combat emerging viral infections if combined with rapidly acquired sequence information from advanced bioinformatics (for more perspective on this see review [112]).
Small Molecule Inhibitors
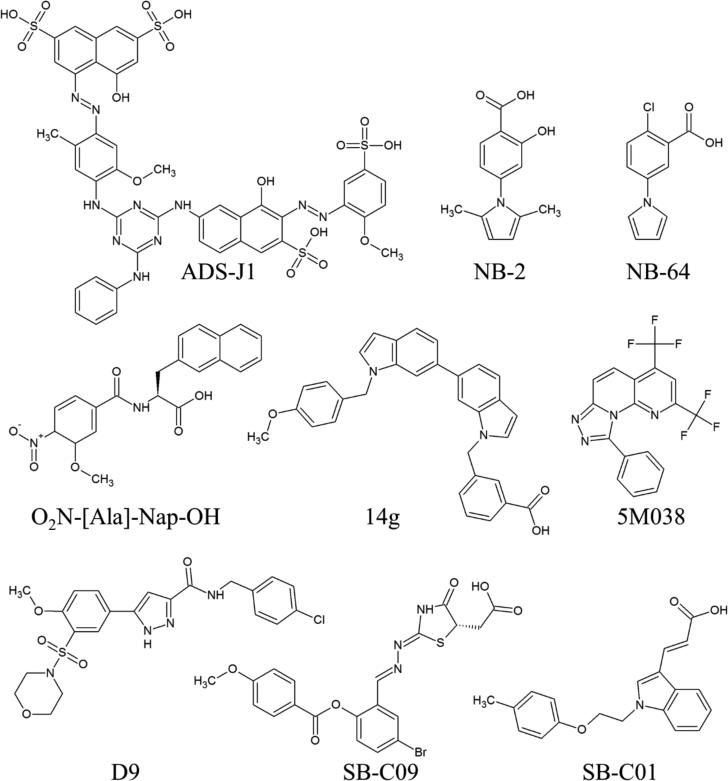
In addition to peptides, considerable efforts to identify small molecule inhibitors of fusion have also been reported. Recent reviews include Cai and Jiang [75], Gochin and Zhou [115], and Allen and Rizzo [116]. Representative examples of small molecule inhibitors are shown in Fig. (2). In general, small molecules have been designed to disrupt 6HB formation by targeting a known hydrophobic pocket on the N-helical interface [76]. Jiang, Debnath and coworkers reported the first such inhibitors, ADS-J1, which was discovered in 1999 through computational docking [117]. Later experiments by the same group found low molecular weight compounds NB-2 and NB-64 using an experimental high throughput ELISA assay [118]. These compounds required a carboxylic acid for activity that is believed to interact with K574, a residue that forms a key salt bridge with the CHR during 6HB formation [119]. This salt bridge is usually recreated by later generations of compounds. Follow-up studies by the group designed several additional inhibitor series [120-122].
Fig. (2).
Representative small molecule inhibitors of HIV gp41. See text for references.
Developing a novel 5-helix bundle protein construct, presenting a solvent accessible N-helix groove, allowed Frey et al. [123] to perform a high throughput screen. Looking for ligands that could displace a fluorescent probe from the N-helix groove, the authors report several inhibitors including 5M038 and 5M041. The authors note that because the 5-helix bundle construct presents only one binding groove per unit instead of the three native ones, this construct represents a more stringent test of molecular activity. Stable folding and a single exposed binding site makes this construct a promising target in ongoing attempts to crystallize a small molecule inhibitor with gp41.
Knowledge-based scoring functions have been used to select active compounds from virtual screens. Holden et al. [124] performed a virtual screen and selected compounds using both traditional energy-based and knowledge-based scoring functions, specifically measuring the ability of a small molecule to recreate key interactions seen between the NHR and CHR. Three of the seven hits identified, including SB-C01 would not have been tested if the authors had used only traditional scoring functions. Compound SB-C09 was well ranked by both traditional and knowledge based scoring functions.
Peptidomimetic small molecules have also shown promise against gp41. Gochin et al. screened a library of 400 molecules using a competitive inhibition fluorescence intensity assay [125] and showed the activity for some molecules was correlated with the presence of aromatic groups at two customizable regions of the peptidomimetic scaffold (analogous to sidechains at i and i+4 of an alpha helix; e.g. O2N-[Ala]-Nap-OH, Fig. 2). NMR based constraints in conjunction with computational docking were used to identify binding poses for two of the compounds. The same group also had success using structure-based design to develop a series of indole inhibitors, the most active compound, 14g shows sub-micromolar activity [126].
Wang et al. [127] designed a series of inhibitors by covalently linking a series of small molecules based off of NB-2 and A12 to a truncated version of peptide C34 termed P26. Although the constructs were unable to obtain the full potency of C34 the constructs performed one-two orders of magnitude better in cell-cell fusion assays compared to P26. Ferrer et al. [128] employed a similar strategy to the discovery of small molecule inhibitors by covalently linking molecules from a combinatorial chemical library to a CHR peptide.
Work has not been limited to only the hydrophobic pocket. NMR-based fragment screening studies have identified an alternative pocket proximate to the hydrophobic region [129]. The authors posit that small molecule potency could be improved through a strategy of tethering binders of each pocket together. More recently, Allen et al. [130] used computational screening to identify inhibitors predicted to bind to theoretical pockets on the inner groove of two N helices thereby preventing N-helical trimer formation. Two molecules, which were selected based on their predicted ability to recreate interactions of the third N helix, were shown to have dose-dependent anti-fusion activity in the micromolar range. Although there is no direct experimental evidence the inhibitors act via the proposed NHR-trimer obstruction mechanism, companion experiments suggest the inhibitors do not work through binding to the familiar hydrophobic pocket. Follow up molecular dynamics simulations showed compound D9 was particularly structurally and energetically stable in terms of the predicted binding pose.
Work has also been done to test the ability of natural products to inhibit HIV, some of which have been found to inhibit 6HB formation. Interested readers should consult reviews by Cai and Jiang [75] and Teixeira et al. [131]. Natural products include triterpenoids [132] [133] [134], tannin [135], theaflavin and catechin derivatives [136], and Chinese medicinal herbal extracts [137].
Neutralizing Antibodies
The MPER of gp41 located between the CHR and the viral membrane is highly conserved and has long been known as an important region for the development of neutralizing antibodies [138-147]. Human antibodies, 2F5, 4E10, Z13, and 10E8 bind to MPER and neutralize a broad range of HIV-1 strains [13-20]. 2F5 and 4E10 were the first to be identified followed by Z13, which was identified from a phage display library using a peptide containing epitope sequences from both 2F5 and 4E10 [19, 148-150]. 10E8 was identified by analysis of healthy HIV-infected patient sera [17]. It was found to be targeted to MPER but to not bind phospholipids or cause auto-reactivity. These antibodies in general are known to disrupt MPER function during the membrane fusion step of the entry mechanism [151, 152]. However, MPER has also been shown to be occluded in the native structure before contact with the cellular receptors and is exposed only transiently at a relatively late stage in the entry mechanism [153-156]. MPER is widely considered to be a membrane interacting domain and inaccessibility to MPER has been one of the obstacles in developing vaccines and therapeutic intervention methods targeted to this region. Structural studies of MPER have produced diverse details depending upon the sequence, the experimental method utilized, and binding partners that were present (for a more extensive review of MPER structure and immunogenicity, see reference [138]).
Recent strategies to target Env have included more detailed studies of B cell maturation, antigen-specific single B-cell sorting, and the use of mimotopes in phage display (for a recent review, see [157]). Studies have been done with sequences from both the immunodominant loop (603-609) and from MPER [13, 143, 158, 159].
An alternative strategy was reported that utilized the isolation and cloning of switch memory B cells from peripheral blood in order to identify human monoclonal antibodies without prior information regarding target sequences [160]. This strategy revealed a broadly neutralizing and potent antibody that binds to the non-covalent interface between gp41 and gp120 [161]. This epitope was found to be cleavage dependent and structural details have been reported [162].
CONCLUSION
The trimer of noncovalently bound heterodimers that is the HIV envelope spike is responsible for specificity to the cell surface, bringing two phospholipid bilayers into close proximity, forming a hemifusion diaphragm and finally opening of a fusion pore to allow for entry of the viral genome into the cytoplasm. This process requires a complex set of large conformational changes that are most likely propagated over long distances and may involve oligomerization state changes or asymmetries yet to be delineated. Because this process is not synchronized between the very sparse number of envelope spikes that are present on the virus surface, it has been exceedingly challenging to solve structures for the transient intermediate states that are the most vulnerable to inhibition. However, due to the elegant use of classic biochemical techniques and advances in a combination of biophysical, structural, and antigenic studies, the field is moving closer to an understanding of the complex conformational changes that the HIV-1 envelope spike undergoes during attachment and membrane fusion. These are key advancements to identifying the optimal target to elicit effective neutralizing antibodies and model and screen for potential small molecule inhibitors. It remains to be stated, however, that studies of the details of the conformational changes, especially at the atomic level, in the case of the transmembrane subunit gp41 still lag behind those of the surface subunit gp120.
ACKNOWLEDGEMENTS
This work was funded by NIH grants R21AI102796 (to A.J.) and R01GM083669 (to R.C.R.).
Biography
 Amy Jocobs
Amy Jocobs
REFERENCES
- 1.Zhu P, Chertova E, Bess J, Jr., et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100(26):15812–7. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinter A, Honnen WJ, Tilley SA, et al. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63(6):2674–9. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Earl PL, Moss B, Doms RW. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65(4):2047–55. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozarsky K, Penman M, Basiripour L, Haseltine W, Sodroski J, Krieger M. Glycosylation and processing of the human immunodeficiency virus type 1 envelope protein. J Acquir Immune Defic Syndr. 1989;2(2):163–9. [PubMed] [Google Scholar]
- 5.Starcich BR, Hahn BH, Shaw GM, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45(5):637–48. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 6.Willey RL, Rutledge RA, Dias S, et al. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc Natl Acad Sci U S A. 1986;83(14):5038–42. doi: 10.1073/pnas.83.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265(18):10373–82. [PubMed] [Google Scholar]
- 8.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4(6):679–84. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 9.Caffrey M, Cai M, Kaufman J, et al. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17(16):4572–84. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffrey M. Model for the structure of the HIV gp41 ectodomain: insight into the intermolecular interactions of the gp41 loop. Biochim Biophys Acta. 2001;1536(2-3):116–22. doi: 10.1016/s0925-4439(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 11.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Gronenborn AM, et al. Determination of the secondary structure and global topology of the 44 kDa ectodomain of gp41 of the simian immunodeficiency virus by multidimensional nuclear magnetic resonance spectroscopy. J Mol Biol. 1997;271(5):819–26. doi: 10.1006/jmbi.1997.1217. [DOI] [PubMed] [Google Scholar]
- 12.Sen J, Jacobs A, Jiang H, Rong L, Caffrey M. The disulfide loop of gp41 is critical to the furin recognition site of HIV gp160. Protein Sci. 2007;16(6):1236–41. doi: 10.1110/ps.072771407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson JD, Brunel FM, Jensen R, Crooks ET, Cardoso RM, Wang M, et al. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol. 2007;81(8):4033–43. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73(5):4009–18. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17(18):1757–65. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 19.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10(4):359–69. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 21.Caffrey M, Kaufman J, Stahl SJ, Wingfield PT, Gronenborn AM, Clore GM. 3D NMR experiments for measuring 15N relaxation data of large proteins: application to the 44 kDa ectodomain of SIV gp41. J Magn Reson. 1998;135(2):368–72. doi: 10.1006/jmre.1998.1583. [DOI] [PubMed] [Google Scholar]
- 22.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 23.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci U S A. 1997;94(23):12303–8. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–30. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 25.Weissenhorn W, Wharton SA, Calder LJ, et al. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 1996;15(7):1507–14. [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2(12):1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 27.Lu M, Kim PS. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J Biomol Struct Dyn. 1997;15(3):465–71. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 28.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6(5):e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGillick BE, Balius TE, Mukherjee S, Rizzo RC. Origins of resistance to the HIVgp41 viral entry inhibitor T20. Biochemistry. 2010;49(17):3575–92. doi: 10.1021/bi901915g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julien JP, Cupo A, Sok D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancera M, Zhou T, Druz A, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–61. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro JB, Gorman J, Ma X, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346(6210):759–63. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munro JB, Mothes W. Structure and Dynamics of the Native HIV-1 Env Trimer. J Virol. 2015;89(11):5752–5. doi: 10.1128/JVI.03187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttman M, Cupo A, Julien JP, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Do Kwon Y, Pancera M, Acharya P, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22(7):522–31. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85(12):5825–39. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong T, Crooks ET, Osawa K, Binley JM. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86(7):3574–87. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–7. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 39.Klatzmann D, Champagne E, Chamaret S, et al. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312(5996):767–8. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 40.McDougal JS, Mawle A, Cort SP, et al. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol. 1985;135(5):3151–62. [PubMed] [Google Scholar]
- 41.McDougal JS, Kennedy MS, Sligh JM, Cort SP, Mawle A, Nicholson JK. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231(4736):382–5. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 42.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47(3):333–48. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 43.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272(5270):1955–8. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 44.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 45.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–7. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 46.Berson JF, Long D, Doranz BJ, Rucker J, Jirik FR, Doms RW. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70(9):6288–95. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melikyan GB. Membrane fusion mediated by human immunodeficiency virus envelope glycoprotein. Current topics in membranes. 2011;68:81–106. doi: 10.1016/B978-0-12-385891-7.00004-0. [DOI] [PubMed] [Google Scholar]
- 48.Wilen CB, Tilton JC, Doms RW. HIV: Cell Binding and Entry. Cold Spring Harb Perspect Med. 2012;2(8) doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caffrey M. HIV envelope: challenges and opportunities for development of entry inhibitors. Trends Microbiol. 2011 doi: 10.1016/j.tim.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan C, Liu S, Jiang S. HIV-1 gp41 Fusion Intermediate: A Target for HIV Therapeutics. J Formos Med Assoc. 2010;109(2):94–105. doi: 10.1016/S0929-6646(10)60029-0. [DOI] [PubMed] [Google Scholar]
- 51.Tilton JC, Doms RW. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res. 2010;85(1):91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Permanyer M, Ballana E, Este JA. Endocytosis of HIV: anything goes. Trends Microbiol. 2010;18(12):543–51. doi: 10.1016/j.tim.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Blumenthal R, Durell S, Viard M. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem. 2012;287(49):40841–9. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci U S A. 1992;89(21):10537–41. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang S, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365(6442):113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 57.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9(11):1051–3. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 58.Jiang S, Lin K, Strick N, Neurath AR. Inhibition of HIV-1 infection by a fusion domain binding peptide from the HIV-1 envelope glycoprotein GP41. Biochem Biophys Res Commun. 1993;195(2):533–8. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 59.Zhang D, Li W, Jiang S. Peptide fusion inhibitors targeting the HIV-1 gp41: a patent review (2009 - 2014). Expert opinion on therapeutic patents. 2015;25(2):159–73. doi: 10.1517/13543776.2014.987752. [DOI] [PubMed] [Google Scholar]
- 60.Wild C, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res Hum Retroviruses. 1995;11(3):323–5. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 61.Neurath AR, Strick N, Jiang S. Synthetic peptides and anti-peptide antibodies as probes to study interdomain interactions involved in virus assembly: the envelope of the human immunodeficiency virus (HIV-1). Virology. 1992;188(1):1–13. doi: 10.1016/0042-6822(92)90729-9. [DOI] [PubMed] [Google Scholar]
- 62.Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5(4):276–9. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 63.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91(21):9770–4. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shai Y. Functional domains within fusion proteins: prospectives for development of peptide inhibitors of viral cell fusion. Biosci Rep. 2000;20(6):535–55. doi: 10.1023/a:1010411021326. [DOI] [PubMed] [Google Scholar]
- 65.Kliger Y, Shai Y. Inhibition of HIV-1 entry before gp41 folds into its fusion-active conformation. J Mol Biol. 2000;295(2):163–8. doi: 10.1006/jmbi.1999.3368. [DOI] [PubMed] [Google Scholar]
- 66.Kliger Y, Gallo SA, Peisajovich SG, et al. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J Biol Chem. 2001;276(2):1391–7. doi: 10.1074/jbc.M004113200. [DOI] [PubMed] [Google Scholar]
- 67.Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci USA. 2001;98(20):11187–92. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derdeyn CA, Decker JM, Sfakianos JN, et al. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J Virol. 2001;75(18):8605–14. doi: 10.1128/JVI.75.18.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bewley CA, Louis JM, Ghirlando R, Clore GM. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J Biol Chem. 2002;277(16):14238–45. doi: 10.1074/jbc.M201453200. [DOI] [PubMed] [Google Scholar]
- 70.Gallo SA, Sackett K, Rawat SS, Shai Y, Blumenthal R. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J Mol Biol. 2004;340(1):9–14. doi: 10.1016/j.jmb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 71.Jacobs A, Quraishi O, Huang X, et al. A covalent inhibitor targeting an intermediate conformation of the fusogenic subunit of the HIV-1 envelope complex. J Biol Chem. 2007;282(44):32406–13. doi: 10.1074/jbc.M705577200. [DOI] [PubMed] [Google Scholar]
- 72.Noah E, Biron Z, Naider F, Arshava B, Anglister J. The membrane proximal external region of the HIV-1 envelope glycoprotein gp41 contributes to the stabilization of the six-helix bundle formed with a matching N' peptide. Biochemistry. 2008;47(26):6782–92. doi: 10.1021/bi7023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S, Lu H, Niu J, Xu Y, Wu S, Jiang S. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J Biol Chem. 2005;280(12):11259–73. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 74.Liu S, Wu S, Jiang S. HIV Entry Inhibitors Targeting gp41: From Polypeptides to Small-Molecule Compounds. Curr Pharm Des. 2007;13(2):143–62. doi: 10.2174/138161207779313722. [DOI] [PubMed] [Google Scholar]
- 75.Cai L, Jiang S. Development of peptide and small-molecule HIV-1 fusion inhibitors that target gp41. ChemMedChem. 2010;5(11):1813–24. doi: 10.1002/cmdc.201000289. [DOI] [PubMed] [Google Scholar]
- 76.Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci U S A. 1998;95(26):15613–7. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–15. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 78.Eron JJ, Gulick RM, Bartlett JA, et al. Short-Term Safety and Antiretroviral Activity of T-1249, a Second-Generation Fusion Inhibitor of HIV. Journal of Infectious Diseases. 2004;189(6):1075–83. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 79.Miralles GD LJ, Bellos N, et al. Program and abstracts of the 10th Conference on Retroviruses and Opportunistic Infections. Foundation for retrovirology and human health; Boston Alexandria, VA: 2003. T-1249 demonstrates potent antiviral activity over 10-day dosing in most patients who have failed a regimen containing enfuvirtide: planned interim analysis of T1249-102, a phase I/II study. p. 63. abstract 14Ib. [Google Scholar]
- 80.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, et al. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc Natl Acad Sci USA. 2007;104(31):12772–7. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan C, Cai L, Lu H, Lu L, Jiang S. A novel chimeric protein-based HIV-1 fusion inhibitor targeting gp41 glycoprotein with high potency and stability. J Biol Chem. 2011;286(32):28425–34. doi: 10.1074/jbc.M111.241992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu L, Pan C, Li Y, Lu H, He W, Jiang S. A bivalent recombinant protein inactivates HIV-1 by targeting the gp41 prehairpin fusion intermediate induced by CD4 D1D2 domains. Retrovirology. 2012;9:104. doi: 10.1186/1742-4690-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu W, Wang Q, Yu F, Lu L, Jiang S. Synergistic effect resulting from combinations of a bifunctional HIV-1 antagonist with antiretroviral drugs. J Acquir Immune Defic Syndr. 2014;67(1):1–6. doi: 10.1097/QAI.0000000000000265. [DOI] [PubMed] [Google Scholar]
- 84.He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, et al. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem. 2008;283(17):11126–34. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 85.Franquelim HG, Loura LM, Santos NC, Castanho MA. Sifuvirtide screens rigid membrane surfaces. establishment of a correlation between efficacy and membrane domain selectivity among HIV fusion inhibitor peptides. Journal of the American Chemical Society. 2008;130(19):6215–23. doi: 10.1021/ja711247n. [DOI] [PubMed] [Google Scholar]
- 86.Wang RR, Yang LM, Wang YH, et al. Sifuvirtide, a potent HIV fusion inhibitor peptide. Biochem Biophys Res Commun. 2009;382(3):540–4. doi: 10.1016/j.bbrc.2009.03.057. [DOI] [PubMed] [Google Scholar]
- 87.Franquelim HG, Veiga AS, Weissmuller G, Santos NC, Castanho MA. Unravelling the molecular basis of the selectivity of the HIV-1 fusion inhibitor sifuvirtide towards phosphatidylcholine-rich rigid membranes. Biochim Biophys Acta. 2010;1798(6):1234–43. doi: 10.1016/j.bbamem.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 88.Yao X, Chong H, Zhang C, et al. Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J Biol Chem. 2012;287(9):6788–96. doi: 10.1074/jbc.M111.317883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meng Q, Dong T, Chen X, et al. Pharmacokinetics of sifuvirtide in treatment-naive and treatment-experienced HIV-infected patients. J Pharm Sci. 2014;103(12):4038–47. doi: 10.1002/jps.24174. [DOI] [PubMed] [Google Scholar]
- 90.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10(4):307–17. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 91.Yi HA, Diaz-Aguilar B, Bridon D, Quraishi O, Jacobs A. Permanent inhibition of viral entry by covalent entrapment of HIV gp41 on the virus surface. Biochemistry. 2011;50(32):6966–72. doi: 10.1021/bi201014b. [DOI] [PubMed] [Google Scholar]
- 92.Jacobs A, Simon C, Caffrey M. Thermostability of the HIV gp41 wild-type and loop mutations. Protein Pept Lett. 2006;13(5):477–80. doi: 10.2174/092986606776819510. [DOI] [PubMed] [Google Scholar]
- 93.Krell T, Greco F, Engel O, et al. HIV-1 gp41 and gp160 are hyperthermostable proteins in a mesophilic environment. Characterization of gp41 mutants. Eur J Biochem. 2004;271(8):1566–79. doi: 10.1111/j.1432-1033.2004.04068.x. [DOI] [PubMed] [Google Scholar]
- 94.Chong H, Yao X, Zhang C, et al. Biophysical property and broad anti-HIV activity of albuvirtide, a 3-maleimimidopropionic acid-modified peptide fusion inhibitor. PLoS One. 2012;7(3):e32599. doi: 10.1371/journal.pone.0032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bai Y, Xue H, Wang K, et al. Covalent fusion inhibitors targeting HIV-1 gp41 deep pocket. Amino acids. 2013;44(2):701–13. doi: 10.1007/s00726-012-1394-8. [DOI] [PubMed] [Google Scholar]
- 96.Bai Y, Xue H, Ling Y, Cheng M, Cai L, Liu K. Inter-chain acyl transfer reaction in a peptide six-helical bundle: a chemical method for regulating the interaction between peptides or proteins. Chem Commun (Camb) 2012;48(36):4320–2. doi: 10.1039/c2cc17428f. [DOI] [PubMed] [Google Scholar]
- 97.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chemical reviews. 2005;105(2):715–38. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 98.Stoddart CA, Nault G, Galkina SA, et al. Albumin-conjugated C34 peptide HIV-1 fusion inhibitor: equipotent to C34 and T-20 in vitro with sustained activity in SCID-hu Thy/Liv mice. J Biol Chem. 2008;283(49):34045–52. doi: 10.1074/jbc.M805536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xie D, Yao C, Wang L, et al. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob Agents Chemother. 2010;54(1):191–6. doi: 10.1128/AAC.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Potent D-peptide inhibitors of HIV-1 entry. Proc Natl Acad Sci U S A. 2007;104(43):16828–33. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Francis JN, Redman JS, Eckert DM, Kay MS. Design of a modular tetrameric scaffold for the synthesis of membrane-localized D-peptide inhibitors of HIV-1 entry. Bioconjug Chem. 2012;23(6):1252–8. doi: 10.1021/bc300076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Welch BD, Francis JN, Redman JS, et al. Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J Virol. 2010;84(21):11235–44. doi: 10.1128/JVI.01339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ingallinella P, Bianchi E, Ladwa NA, et al. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci U S A. 2009;106(14):5801–6. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103(8):2641–6. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90(11):5181–5. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aloia RC, Jensen FC, Curtain CC, Mobley PW, Gordon LM. Lipid composition and fluidity of the human immunodeficiency virus. Proc Natl Acad Sci U S A. 1988;85(3):900–4. doi: 10.1073/pnas.85.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Popik W, Alce TM, Au W-C. Human Immunodeficiency Virus Type 1 Uses Lipid Raft-Colocalized CD4 and Chemokine Receptors for Productive Entry into CD4+ T Cells. Journal of Virology. 2002;76(10):4709–22. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hollmann A, Matos PM, Augusto MT, Castanho MA, Santos NC. Conjugation of cholesterol to HIV-1 fusion inhibitor C34 increases peptide-membrane interactions potentiating its action. PLoS One. 2013;8(4):e60302. doi: 10.1371/journal.pone.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pessi A, Langella A, Capito E, et al. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PLoS One. 2012;7(5):e36833. doi: 10.1371/journal.pone.0036833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao L, Tong P, Chen YX, et al. A multi-functional peptide as an HIV-1 entry inhibitor based on self-concentration, recognition, and covalent attachment. Organic & biomolecular chemistry. 2012;10(32):6512–20. doi: 10.1039/c2ob25853f. [DOI] [PubMed] [Google Scholar]
- 111.Harman S, Herrera C, Armanasco N, Nuttall J, Shattock RJ. Preclinical evaluation of the HIV-1 fusion inhibitor L'644 as a potential candidate microbicide. Antimicrob Agents Chemother. 2012;56(5):2347–56. doi: 10.1128/AAC.06108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pessi A. Cholesterol-conjugated peptide antivirals: a path to a rapid response to emerging viral diseases. Journal of peptide science : an official publication of the European Peptide Society. 2015;21(5):379–86. doi: 10.1002/psc.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee KK, Pessi A, Gui L, Santoprete A, Talekar A, Moscona A, et al. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem. 2011;286(49):42141–9. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Higgins CD, Koellhoffer JF, Chandran K, Lai JR. C-peptide inhibitors of Ebola virus glycoprotein-mediated cell entry: effects of conjugation to cholesterol and side chain-side chain crosslinking. Bioorg Med Chem Lett. 2013;23(19):5356–60. doi: 10.1016/j.bmcl.2013.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gochin M, Zhou G. Amphipathic properties of HIV-1 gp41 fusion inhibitors. Current topics in medicinal chemistry. 2011;11(24):3022–32. doi: 10.2174/156802611798808488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Allen WJ, Rizzo RC. Computer-Aided Approaches for Targeting HIVgp41. Biology. 2012;1(2):311–38. doi: 10.3390/biology1020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Debnath AK, Radigan L, Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. Journal of medicinal chemistry. 1999;42(17):3203–9. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- 118.Jiang S, Lu H, Liu S, Zhao Q, He Y, Debnath AK. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrobial agents and chemotherapy. 2004;48(11):4349–59. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He Y, Cheng J, Li J, et al. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. Journal of virology. 2008;82(13):6349–58. doi: 10.1128/JVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jiang S, Tala SR, Lu H, et al. Design, synthesis, and biological activity of novel 5-((arylfuran/1H-pyrrol-2-yl)methylene)-2-thioxo-3-(3-(trifluoromethyl)phenyl)thi azolidin-4-ones as HIV-1 fusion inhibitors targeting gp41. Journal of medicinal chemistry. 2011;54(2):572–9. doi: 10.1021/jm101014v. [DOI] [PubMed] [Google Scholar]
- 121.Jiang S, Tala SR, Lu H, et al. Design, synthesis, and biological activity of a novel series of 2,5-disubstituted furans/pyrroles as HIV-1 fusion inhibitors targeting gp41. Bioorganic & medicinal chemistry letters. 2011;21(22):6895–8. doi: 10.1016/j.bmcl.2011.08.081. [DOI] [PubMed] [Google Scholar]
- 122.Katritzky AR, Tala SR, Lu H, et al. Design, synthesis, and structure-activity relationship of a novel series of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl)furans as HIV-1 entry inhibitors. Journal of medicinal chemistry. 2009;52(23):7631–9. doi: 10.1021/jm900450n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(38):13938–43. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holden PM, Kaur H, Goyal R, Gochin M, Rizzo RC. Footprint-based identification of viral entry inhibitors targeting HIVgp41. Bioorg Med Chem Lett. 2012;22(8):3011–6. doi: 10.1016/j.bmcl.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gochin M, Whitby LR, Phillips AH, Boger DL. NMR-assisted computational studies of peptidomimetic inhibitors bound in the hydrophobic pocket of HIV-1 glycoprotein 41. Journal of computer-aided molecular design. 2013;27(7):569–82. doi: 10.1007/s10822-013-9662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou G, Wu D, Snyder B, Ptak RG, Kaur H, Gochin M. Development of indole compounds as small molecule fusion inhibitors targeting HIV-1 glycoprotein-41. Journal of medicinal chemistry. 2011;54(20):7220–31. doi: 10.1021/jm200791z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang C, Shi W, Cai L, Lu L, Wang Q, Zhang T, et al. Design, synthesis, and biological evaluation of highly potent small molecule-peptide conjugates as new HIV-1 fusion inhibitors. Journal of medicinal chemistry. 2013;56(6):2527–39. doi: 10.1021/jm3018964. [DOI] [PubMed] [Google Scholar]
- 128.Ferrer M, Kapoor TM, Strassmaier T, et al. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nature structural biology. 1999;6(10):953–60. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 129.Chu S, Gochin M. Identification of fragments targeting an alternative pocket on HIV-1 gp41 by NMR screening and similarity searching. Bioorganic & medicinal chemistry letters. 2013;23(18):5114–8. doi: 10.1016/j.bmcl.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Allen WJ, Yi HA, Gochin M, Jacobs A, Rizzo RC. Small molecule inhibitors of HIVgp41 N-heptad repeat trimer formation. Bioorg Med Chem Lett. 2015;25(14):2853–9. doi: 10.1016/j.bmcl.2015.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Teixeira C, Gomes JR, Gomes P, Maurel F, Barbault F. Viral surface glycoproteins, gp120 and gp41, as potential drug targets against HIV-1: brief overview one quarter of a century past the approval of zidovudine, the first anti-retroviral drug. European journal of medicinal chemistry. 2011;46(4):979–92. doi: 10.1016/j.ejmech.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 132.Fujioka T, Kashiwada Y, Kilkuskie RE, et al. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. Journal of natural products. 1994;57(2):243–7. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
- 133.Evers M, Poujade C, Soler F, et al. Betulinic acid derivatives: a new class of human immunodeficiency virus type 1 specific inhibitors with a new mode of action. Journal of medicinal chemistry. 1996;39(5):1056–68. doi: 10.1021/jm950670t. [DOI] [PubMed] [Google Scholar]
- 134.Soler F, Poujade C, Evers M, Carry JC, Henin Y, Bousseau A, et al. Betulinic acid derivatives: a new class of specific inhibitors of human immunodeficiency virus type 1 entry. Journal of medicinal chemistry. 1996;39(5):1069–83. doi: 10.1021/jm950669u. [DOI] [PubMed] [Google Scholar]
- 135.Lu L, Liu SW, Jiang SB, Wu SG. Tannin inhibits HIV-1 entry by targeting gp41. Acta pharmacologica Sinica. 2004;25(2):213–8. [PubMed] [Google Scholar]
- 136.Liu S, Lu H, Zhao Q, et al. Theaflavin derivatives in black tea and catechin derivatives in green tea inhibit HIV-1 entry by targeting gp41. Biochimica et biophysica acta. 2005;1723(1-3):270–81. doi: 10.1016/j.bbagen.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 137.Liu S, Jiang S, Wu Z, et al. Identification of inhibitors of the HIV-1 gp41 six-helix bundle formation from extracts of Chinese medicinal herbs Prunella vulgaris and Rhizoma cibotte. Life sciences. 2002;71(15):1779–91. doi: 10.1016/s0024-3205(02)01939-2. [DOI] [PubMed] [Google Scholar]
- 138.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiology and molecular biology reviews : MMBR. 2008;72(1):54–84. doi: 10.1128/MMBR.00020-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6(2):143–55. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 140.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 141.Kim M, Song L, Moon J, et al. Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J Biol Chem. 2013;288(44):31888–901. doi: 10.1074/jbc.M113.494609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou T, Zhu J, Yang Y, et al. Transplanting supersites of HIV-1 vulnerability. PLoS One. 2014;9(7):e99881. doi: 10.1371/journal.pone.0099881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morris L, Chen X, Alam M, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One. 2011;6(9):e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim M, Sun ZY, Rand KD, et al. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat Struct Mol Biol. 2011;18(11):1235–43. doi: 10.1038/nsmb.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ofek G, Guenaga FJ, Schief WR, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107(42):17880–7. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blish CA, Nguyen MA, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5(1):e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Muster T, Steindl F, Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–7. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Moore JP, Cao Y, Leu J, Qin L, Korber B, Ho DD. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70(1):427–44. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parren PW, Wang M, Trkola A, et al. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72(12):10270–4. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vujcic LK, Quinnan GV., Jr. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retroviruses. 1995;11(7):783–7. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- 151.Song L, Sun ZY, Coleman KE, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci U S A. 2009;106(22):9057–62. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sun ZY, Oh KJ, Kim M, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28(1):52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 153.Chakrabarti BK, Walker LM, Guenaga JF, et al. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol. 2011;85(16):8217–26. doi: 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ofek G, Tang M, Sambor A, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–37. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46(5):1398–401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- 156.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105(10):3739–44. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Geiss Y, Dietrich U. Catch Me If You Can--The Race Between HIV and Neutralizing Antibodies. AIDS Rev. 2015;17(2):107–13. [PubMed] [Google Scholar]
- 158.Enshell-Seijffers D, Smelyanski L, Vardinon N, Yust I, Gershoni JM. Dissection of the humoral immune response toward an immunodominant epitope of HIV: a model for the analysis of antibody diversity in HIV+ individuals. FASEB J. 2001;15(12):2112–20. doi: 10.1096/fj.00-0898com. [DOI] [PubMed] [Google Scholar]
- 159.Zhou M, Meyer T, Koch S, et al. Identification of a new epitope for HIV-neutralizing antibodies in the gp41 membrane proximal external region by an Env-tailored phage display library. Eur J Immunol. 2013;43(2):499–509. doi: 10.1002/eji.201242974. [DOI] [PubMed] [Google Scholar]
- 160.Huang J, Doria-Rose NA, Longo NS, et al. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc. 2013;8(10):1907–15. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang J, Kang BH, Pancera M, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515(7525):138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Blattner C, Lee JH, Sliepen K, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–80. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]