Abstract
Nutrition plays a significant role in the increasing prevalence of metabolic and brain disorders. Here we employ systems nutrigenomics to scrutinize the genomic bases of nutrient–host interaction underlying disease predisposition or therapeutic potential. We conducted transcriptome and epigenome sequencing of hypothalamus (metabolic control) and hippocampus (cognitive processing) from a rodent model of fructose consumption, and identified significant reprogramming of DNA methylation, transcript abundance, alternative splicing, and gene networks governing cell metabolism, cell communication, inflammation, and neuronal signaling. These signals converged with genetic causal risks of metabolic, neurological, and psychiatric disorders revealed in humans. Gene network modeling uncovered the extracellular matrix genes Bgn and Fmod as main orchestrators of the effects of fructose, as validated using two knockout mouse models. We further demonstrate that an omega-3 fatty acid, DHA, reverses the genomic and network perturbations elicited by fructose, providing molecular support for nutritional interventions to counteract diet-induced metabolic and brain disorders. Our integrative approach complementing rodent and human studies supports the applicability of nutrigenomics principles to predict disease susceptibility and to guide personalized medicine.
Keywords: Systems nutrigenomics, Fructose, Omega-3 fatty acid, DHA, Epigenome, Transcriptome, Brain networks, Metabolic diseases, Brain disorders, Extracellular matrix
Highlights
-
•
Fructose promotes transcriptomic and epigenomic reprogramming to perturb brain networks linking metabolism and brain function.
-
•
The extracellular matrix genes Bgn and Fmod emerge as key regulators of gene networks responsive to fructose.
-
•
The omega-3 fatty acid DHA reverses fructose-induced genomic and network reprogramming.
Meng et al. report fructose as a powerful inducer of genomic and epigenomic variability with the capacity to reorganize gene networks critical for central metabolic regulation and neuronal processes in the brain; conversely, an omega-3 fatty acid, DHA, has the potential to normalize the genomic impact of fructose. Our findings help explain the pathogenic actions of fructose on prevalent metabolic and brain disorders and provide proof-of-concept for nutritional remedies supported by nutrigenomics evidence. Our integrative approach complementing rodent and human studies supports the applicability of nutrigenomics principles to predict disease susceptibility and to guide personalized medicine.
1. Introduction
Metabolic disorders (MetDs) such as metabolic syndrome, obesity, and type 2 diabetes (T2D) have become a pressing health apprehension worldwide due to their increasing prevalence and high mortality rate, and even more recently to their ability to escalate the pathology of neurological and psychiatric disorders (Bomfim et al., 2012, Newcomer, 2007, Farooqui et al., 2012, Lowette et al., 2015). Among the potential culprits for the rising epidemic of metabolic and brain disorders are dietary components introduced through industrialization (Chassaing et al., 2015, Suez et al., 2014). In particular, fructose, which has been widely used as a “safe and healthy” sweetener in soft drinks and processed foods in the past decades, is emerging as a significant contributor to MetDs in humans (Lyssiotis and Cantley, 2013, Lustig et al., 2012). Fructose-induced MeDs has been shown to reduce hippocampal-dependent memory (Agrawal and Gomez-Pinilla, 2012) and to worsen the pathology of brain disorders in rodents (Agrawal et al., 2015). Conversely, the omega-3 fatty acid docosahexaenoic acid (DHA) has been shown to attenuate MetDs (Steffen et al., 2015, Virtanen et al., 2014, De Caterina, 2011), and to counteract the deleterious effects of fructose on brain function and plasticity (Bremer et al., 2014, Agrawal and Gomez-Pinilla, 2012). Our understanding of the molecular mechanisms underlying the actions of fructose and DHA on MetDs and brain disorders has been limited by conventional approaches focusing on isolated molecular events. This limitation has delayed major advances in the utilization of nutrient-based strategies for the prevention and treatment of common complex disorders.
As fundamental aspects of gene regulation, disruptions in epigenomic reprogramming, transcript abundance, alternative splicing, and gene–gene interactions are increasingly recognized as core aspects of wide-ranging pathogenesis (Chen et al., 2008, Zhang et al., 2013, Rhinn et al., 2013, Narayanan et al., 2014, Makinen et al., 2014, Yang et al., 2009). Systems nutrigenomics is emerging as a powerful approach to reveal the hidden aspects of pathogenesis under dietary modulation (Zhao et al., 2015, De Graaf et al., 2009, Panagiotou and Nielsen, 2009). Here we apply systems nutrigenomics to unveil the multidimensional molecular interactions driven by fructose and DHA that regulate pathogenesis and recovery, and to provide proof-of-principle on the potential of systems nutrigenomics to guide personalized medicine. The comparative account of nutrigenomics signals between fructose and DHA is crucial to understand how select diets impact the molecular substrates governing the balance between normal brain function and disease, and holds potential for guiding effective preventative and therapeutic strategies to mitigate common human diseases.
2. Materials and Methods
We describe essential methods in the main text and detailed experimental procedures are available in the Supplemental Materials.
2.1. Overall Study Design
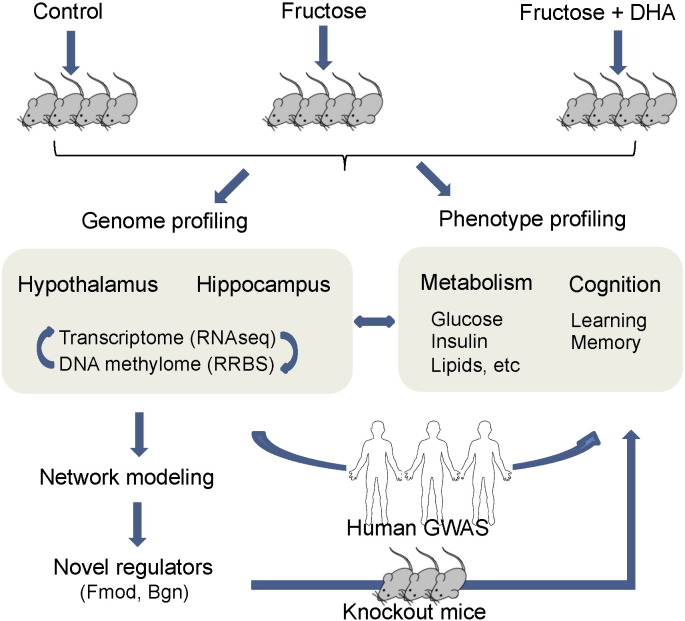
As depicted in the analysis flow in Fig. 1, we focus our study on two key regions in the rodent brain that are important for the regulation of metabolism (hypothalamus) and cognition (hippocampus), and therefore can play a major role in fructose-induced metabolic and brain dysregulation as well as DHA-mediated recovery. We analyzed the transcriptome and DNA methylome using next generation sequencing, followed by investigation of the regulatory relationship between the methylome and transcriptome. Network approaches were then applied to model gene–gene interactions and to predict essential perturbation or regulatory points. Knockout mouse models were subsequently used to validate the predicted regulatory genes with regard to their ability to modulate metabolic and behavior phenotypes. To infer translatability to human pathophysiology, we assessed the intersection of the molecular signals from our rodent models with human genome-wide association studies (GWAS) of metabolic and brain disorders.
Fig. 1.
Overall study design and analysis flow.
2.2. Rat model of fructose consumption and DHA supplementation
Male Sprague–Dawley rats (Charles River Laboratories, Inc., MA, USA) of 2 months old weighing 200-220 g were randomly assigned to 15% fructose treatment (n = 8, 15% w/v fructose in the drinking water), 15% fructose plus an omega-3 fatty acid diet rich in DHA (n = 8; 0.5% of flaxseed oil supplying ALA and 1.2% of DHA capsule oil, Nordic Naturals, Inc., CA, USA), or a control group (n = 8, without fructose in drinking water or DHA supplement) for six weeks. Sample size was chosen to yield > 80% statistical power to detect 30% between-group difference with 10% within-group difference in a phenotype using two-sided Student's t-test. The rats were singly housed in polyacrylic cages with free access to water and their respective diets, and maintained under standard housing conditions (room temperature 22–24 °C) with 12 h light/dark cycle. Daily food and drink intake were monitored. The fructose intake level is approximately equivalent to long-term daily consumption of 130 g sugar in 1–2 l soda drinks in a 60 kg human. The total fat content in the control and DHA diets was 10 g per 100 g of diet. The rats were then examined for changes in MetD-related phenotypes (serum levels of insulin, glucose, and triglycerides, and insulin resistance index (fasting glucose [mg/dl] × fasting insulin [ng/ml] / 16.31)). Rats were trained in the Barnes maze test for 5 days to learn the task prior to diet treatment and then tested for memory retention in the Barnes Maze after 6 weeks of treatment as previously described (Agrawal and Gomez-Pinilla, 2012). Mice were sacrificed, and hypothalamus and hippocampus were dissected out, flash frozen, and stored at − 70 °C for transcriptome and DNA methylome sequencing experiments.
2.3. RNA Sequencing (RNA-Seq) and Data Analyses
RNA-Seq libraries were prepared for 24 RNA samples (n = 4 per treatment group per brain region) and sequenced in paired-end mode by HiSeq 2000 (Illumina Inc., CA, USA) as detailed in Supplemental Materials. The short reads data were analyzed for different transcription between treatment and control groups using the Tuxedo tool package (Trapnell et al., 2012) and false discovery rate (FDR) was estimated using the q-value approach. One hypothalamus sample from the fructose group failed standard quality control and was removed from analysis. Genes and transcripts showing differential expression or alternative splicing at p < 0.01 in each brain region were defined as a gene “signature” for further integrative analyses. The reliability of transcriptome signals at p < 0.01 from RNA-Se1 was confirmed using quantitative real-time PCR (qPCR) for 12 selected genes (Supplemental Materials). Relative gene expression was represented by delta Ct = Ctgene − CtGapdh. The RNA-Seq data was deposited to Gene Expression Ominbus (GEO) under accession numbers GSE59918 (control and fructose groups) and GSE 64815 (DHA).
2.4. Reduced Representation Bisulfite Sequencing (RRBS) of DNA Methylome
RRBS libraries were constructed for 24 DNA samples (n = 4 per treatment group per brain region) and sequenced using an Illumina HiSeq 2500 System (Illumina Inc., CA, USA) as described in Supplemental Materials. Bisulfite-converted reads were processed using the bisulfite aligner BS Seeker2 (Guo et al., 2013). One hypothalamus sample from the DHA group failed standard quality control and was removed from analysis. Differential methylation between treatment and control groups was calculated using the R package methylKit (Akalin et al., 2012) and FDR was estimated using the q-value approach. Loci with methylation levels > 25% between groups and FDR < 0.05 were defined as differentially methylated loci (DMLs). The potential cis- and trans-regulatory effects of DNA methylome on transcriptome were further examined (Supplemental Materials). RRBS data was deposited to GEO under accession numbers GSE59893 (control and fructose groups) and GSE64816 (DHA).
2.5. Assessing Overlap Between Fructose Signatures and Candidate Causal Genes From Human GWAS of MetDs and Brain Disorders
We tested the consistency between the genes identified from fructose treatment in our study and those from human GWAS related to MetDs and brain functions in two ways. First, we cross-checked genes identified in our study with top GWAS candidate genes reported in the GWAS catalog (http://www.genome.gov/gwastudies/) (Hindorff et al., 2009) for direct overlap. Second, we used a SNP Set Enrichment Analysis (SSEA) (Makinen et al., 2014), which goes beyond the top GWAS hits and tests the overall enrichment of the genes affected in our fructose animal model for SNPs that demonstrated disease association in humans using Kolmogorov–Smirnov (KS) test and Fisher's exact test. Bonferroni-corrected p < 0.05 by either KS test or Fisher's exact test was considered significant. The detailed methods and GWAS datasets involved are described in Supplemental Materials.
2.6. Network Modeling and Identification of Key Drivers (KD) of Fructose and DHA Signatures
We obtained both protein–protein interaction (PPI) networks from the HPRD database (http://www.hprd.org) (Keshava Prasad et al., 2009) and tissue-specific Bayesian networks (BNs) (Zhu et al., 2004, Zhu et al., 2008) for hypothalamus and hippocampus constructed based on genetic and transcriptomic data from multiple large-scale mouse studies (Supplemental Materials). We used the fructose or DHA signature genes identified from the RNA-Seq analysis as seeds to extract the top most connected subnetworks as described previously (Yang et al., 2009). The network components including nodes and edges as well as topological structures were visualized using Cytoscape (Smoot et al., 2011). A Key Driver Analysis (KDA; detailed in Supplemental Materials) (Yang et al., 2010, Zhang et al., 2013) was used to identify potential key regulators for the fructose and DHA signatures based on the topology of BNs and PPI networks. Briefly, the neighboring subnetwork of each gene in a given network was first extracted and then compared with the tissue-matched fructose or DHA signature to assess the enrichment of genes in the latter using Fisher's exact test. Network genes that reach Bonferroni-adjusted p < 0.05 were reported as KDs. KDs identified from multiple networks were ranked according to a composite score that favors KDs that are highly significant in each KDA and are consistent across network models used for KDA (Supplemental Materials).
2.7. KD Validation Using Bgn and Fmod Knockout (KO) Models
Bgn KO mice (Xu et al., 1998) and Fmod KO mice (Svensson et al., 1999) were given as gifts from National Institute of Dental and Craniofacial Research and re-derived at the University of California, Los Angeles. Mice were housed in standard polyethylene cages (3–4 mice per cage) in an environmentally controlled room (22–24 °C) with a 12 h light/dark cycle and free access to food and water. Homozygous male KO mice (Bgn: n = 16, Fmod: n = 11) of 9-week age were tested against male wild type (WT) controls (n = 8) for phenotypic changes in metabolic (glucose tolerance, plasma lipid panel) and learning and memory traits (Barnes maze) as described in the Supplemental Materials.
2.8. Statistics
For metabolic and behavior phenotypes, two-sided Student's t-test was used to test statistical differences in between fructose-treated mice and control mice, or between the fructose and fructose + DHA group, or between KO mice (Bgn KO or Fmod KO) and wild-type mice. Two-way ANOVA with Holm-Sidak post hoc analysis for multiple comparisons was performed for the learning curves. For RNA-Seq, RRBS, KDA, and SSEA analyses, the statistical tests and multiple testing correction methods were described in detail under the respective method sections. All statistical and bioinformatics analyses were performed in R (http://www.R-project.org/).
2.9. Study Approval
All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Chancellor's Animal Research Committee of the University of California, Los Angeles.
3. Results
3.1. Induction of Metabolic and Behavioral Abnormalities by Fructose and Normalization by DHA
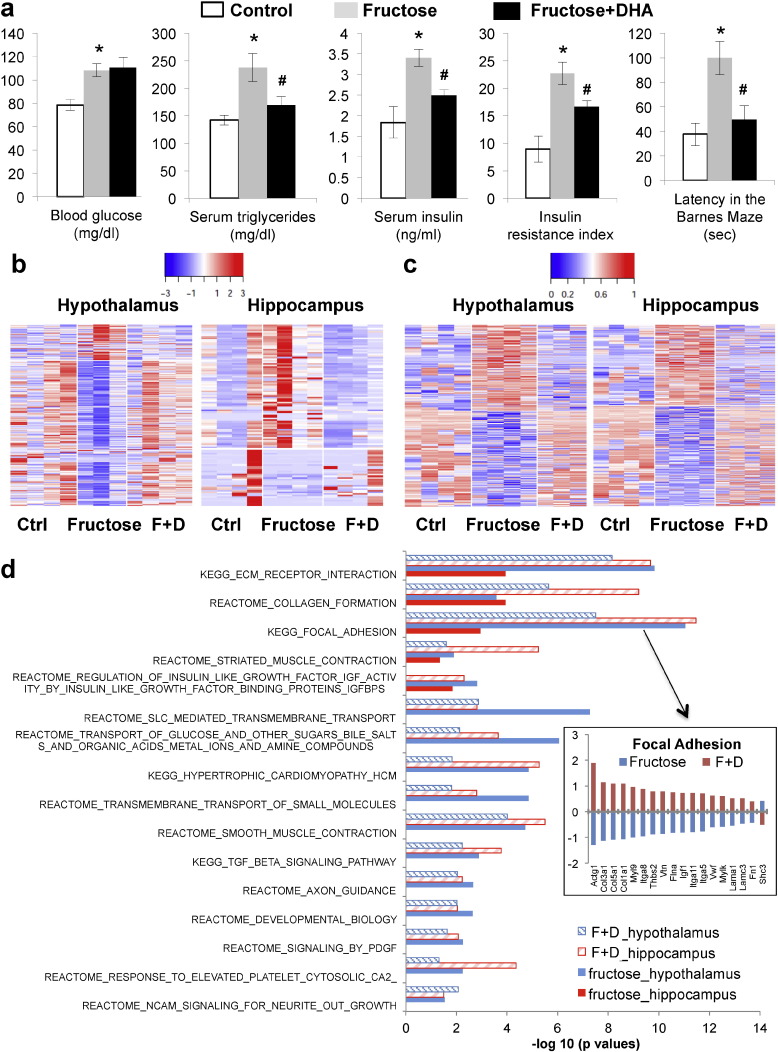
Compared to the control group (n = 8), adult rats consuming fructose (n = 8) had MetD characteristics including significantly increased blood glucose, triglycerides, insulin, and insulin resistance index (Fig. 2a). The animals on fructose also displayed impaired memory as demonstrated by prolonged latency in the Barnes Maze test (Fig. 2a). On the other hand, fructose-treated animals with DHA supplementation (DHA + fructose) showed significantly improved metabolic parameters including lower serum triglycerides, insulin, insulin resistance index, as well as improved memory as reflected by the reduced latency time in the Barnes Maze test (Fig. 2a). The measurements in the DHA + fructose group were not significantly different from the control group with no fructose treatment for triglycerides, insulin, and latency time, indicating that DHA supplementation counteracted the fructose effects. Of note, total caloric intake between groups was similar between groups.
Fig. 2.
Changes in metabolic and behavior phenotypes, transcriptome, DNA methylome, and biological pathways in response to fructose treatment and DHA supplementation. (a) Metabolic and behavior phenotypes. From left to right: blood glucose, serum triglycerides, serum insulin, insulin resistance index, and latency time in the memory retention probe in the Barnes Maze test. Fructose group was compared with the control group, and the fructose + DHA group was compared to the fructose only group. ⁎p < 0.01 and #p < 0.05 by 2-sided Student's t-test. Error bars in the plots are standard errors. N = 8/group. (b) Heatmap of gene expression changes in hypothalamus and hippocampus. Blue to red colors indicate low to high expression values. (c) Heatmap of DNA methylation changes in hypothalamus and hippocampus. Blue to red colors indicate low to high methylation levels. (d) Select biological pathways affected by fructose and DHA in hypothalamus and hippocampus. Bars are the –log10 enrichment p values of the pathways. The insert plot on the right shows the opposite direction of changes in genes in the “focal adhesion” pathway in the hypothalamus between fructose and DHA: fructose mostly inhibits whereas DHA reverses the expression levels. In the insert plot Y-axis indicates log10 fold changes. The fructose + DHA group is labeled as F + D in panels b–d.
3.2. Transcriptomic Changes in Hypothalamus and Hippocampus Induced by Fructose and Normalization by DHA
Using RNA-Seq, we identified 581 and 146 differentially expressed genes, 352 and 79 differentially expressed transcripts, and 99 and 53 genes showing alternative exon usage in hypothalamus and hippocampus, respectively, at p < 0.01 (Dataset S1). These results support large-scale alterations in transcriptional activities affecting both differential expression and alternative splicing, and define 734 unique hypothalamic genes (312 passed FDR < 5%) and 206 hippocampal genes (56 passed FDR < 5%) as “fructose gene signatures”. The reliability of the differentially signals at p < 0.01 was supported by consistent qPCR experiments on 12 selected genes (Table S1). Among the signatures were transcription factors (e.g., Aff3, Junb, Zbtb16, Tcf7l2), epigenetic regulators (e.g., Trmu, Suv420h2, Rgs9bp), and alternative splicing regulators (e.g., Bicc1, Prpf31, Rbpms) (Dataset S1), which may partially explain the large-scale transcriptional alterations observed.
The fructose signatures showed strong tissue-specificity, with 664 genes unique to the hypothalamus, 136 unique to the hippocampus, and 70 overlapping between tissues. In particular, only in hypothalamus we observed upregulation of Slc2a5 (encoding the major fructose transporter GLUT5) and downregulation of Slc2a4 (encoding the glucose transporter protein GLUT4). The signatures also differ by direction of change between the two brain compartments: 68% of hypothalamic signatures showed down-regulation compared to 42% in hippocampus. Among the overlapping genes between the two brain regions (Dataset S1), only 6 showed consistent upregulation (Atp5f1, Pnpla1, Pomc, Sepw1, Slc39a12, and Ubc) and 5 showed consistent downregulation (Alox15, Dstn, Rpl30, Slc39a4, and Slc5a7). The consistent genes are involved in mitochondria function, energy balance, antioxidation, inflammation, and cognition.
Compared to the fructose group, the DHA + fructose group showed differential expression of 651 and 462 genes, 449 and 277 transcripts, and alternative exon usage for 155 and 81 genes in hypothalamus and hippocampus at p < 0.01, respectively (Dataset S1), which together define “DHA gene signatures” of 910 unique genes (382 passed FDR < 5%) in hypothalamus and 569 unique genes (246 passed FDR < 5%) in hippocampus. Strikingly, these DHA signatures significantly overlapped with the fructose signatures (Table 1), with DHA reversing the expression of the common signatures perturbed by fructose in both brain regions (Fig. 2b; Dataset S1).
Table 1.
Overlap between fructose and DHA signatures. Numbers of significant genes or methylation loci are shown. Enrichment p value was calculated using 2-sided Fisher's exact test.
| Data | Tissue | Fructose signature | DHA signature | Overlap | Background | Fold enrichment | p value |
|---|---|---|---|---|---|---|---|
| RNAseq | Hypothalamus | 734 | 910 | 374 | 17,435 | 9.76 | 7.6E − 305 |
| RNAseq | Hippocampus | 206 | 569 | 118 | 17,411 | 17.53 | 1.9E − 124 |
| RRBS | Hypothalamus | 1544 | 1665 | 381 | 6,686,093 | 984.54 | 0 |
| RRBS | Hippocampus | 1872 | 1957 | 557 | 8,742,773 | 1329.25 | 0 |
3.3. Functional Categorization of Fructose and DHA Transcriptomic Signatures
We found enrichment of both tissue-specific and shared biological pathways between brain regions and between treatment groups (select pathways shown in Fig. 2d; full details in Dataset S2). Fructose and DHA signatures shared similar over-represented pathways, although the direction of expression changes of the overlapping genes was predominantly opposite (exemplified in Fig. 2d insert; details in Dataset S1). The pathways unique to hypothalamus included those highly relevant to cardiometabolic, inflammatory, and neuronal functions. The pathway affected in the hippocampus was “neurological processes”. Those affected in both regions were related to extracellular matrix (ECM), adhesion, complement, and insulin-like growth factor (IGF) signaling.
3.4. Induction of DNA Methylomic Changes by Fructose and Normalization by DHA
To identify epigenomic changes which may play a role in the transcriptomic alternation associated with fructose and DHA, we examined the DNA methylome using RRBS that measures millions of potential DNA methylation sites at single base resolution. We found that fructose consumption induced a large-scale switch of the methylation patterns in both brain regions: 734 and 810 of differentially methylated loci (DMLs) showed hypomethylation and hypermethylation in hypothalamus, respectively; 972 and 900 DMLs showed hypomethylation and hypermethylation in hippocampus at a FDR < 5%, respectively (Fig. 2c). DHA largely reversed the fructose-induced methylation changes in these two brain regions (Fig. 2c; Table 1).
3.5. Relationship Between Differential Methylation and Differential Expression
To explore the potential relationship between DMLs and gene expression, we mapped the fructose DMLs in each brain region to adjacent genes within 10 kb distance. We found 50% of DMLs to be located in the intergenic regions, 34% in the gene body (including 5′-UTR, intron, coding sequence, and 3′-UTR), and 16% in the upstream or downstream regions of the genes, with a paucity of DMLs in the promoter and transcription start site (Fig. S1; Table S2). Assessing the methylome-transcriptome relationship revealed co-localization of DMLs and gene signatures: 74 out of the 734 (10.1%) hypothalamic signature genes and 18 out of the 206 (8.7%) hippocampal signature genes co-localized with DMLs within 50 kb distance (Table S3), suggesting cis-regulation of gene expression by the local DMLs. Notably, some of the genes with co-localizing DMLs are transcription factors (Aff3, Junb, and Zbtb16) or epigenetic factors (Parp9) (Dataset S3), which could in turn trans-regulate downstream target genes. Additionally, microRNAs adjacent (within 20 kb or 50 kb) to the DMLs had the potential to regulate 36%–45% and 17%–45% of hypothalamic and hippocampal signatures which were the predicted target genes of the corresponding microRNAs (Table S3; details of the microRNAs in Dataset S3). These results suggest that fructose-induced DNA methylation changes could contribute to transcriptional alteration through both cis and trans-regulatory mechanisms.
3.6. Perturbation of Gene Subnetworks by Fructose and Normalization by DHA
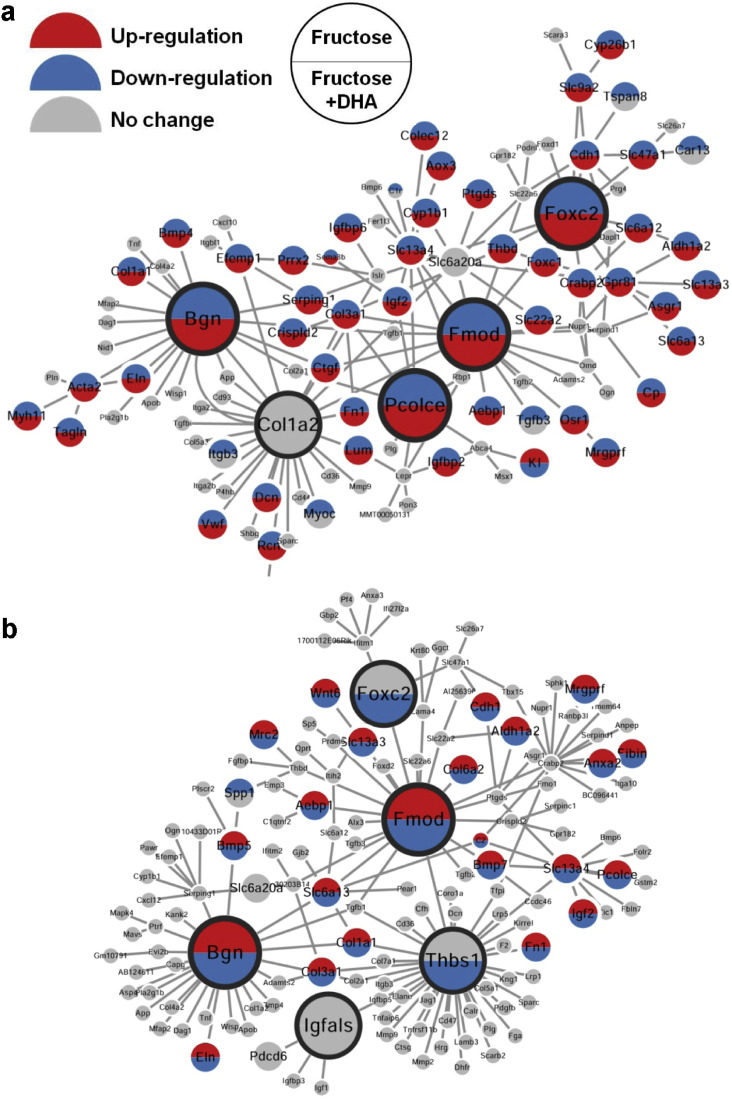
To explore the gene–gene relations among the fructose signature genes and to identify key perturbation points of fructose, we employed two types of networks - protein–protein interaction (PPI) networks (Keshava Prasad et al., 2009) that capture a majority of the well-known interactions between proteins, and data-driven Bayesian networks (BNs) (Zhu et al., 2004, Zhu et al., 2008) that elucidate gene–gene regulatory relationships (detailed in Materials and Methods). Using these networks and a network topology-based key driver analysis (KDA), we identified multiple candidate key driver (KD) genes whose network neighboring genes highly overlapped with the fructose signature genes (Dataset S4). As shown in Fig. 3, the top 5 KDs for each brain region form central hubs connecting many fructose signature genes in coherent gene subnetworks, which contained genes involved in diverse processes such as the ECM (e.g., Col6a2, Lama4, Thbs1, Fmod, Bgn), small molecule transporters (e.g., Slc6a13, Slc22a2, Slc22a6, Slc13a3), and forkhead transcription factors (e.g., Foxc2, Foxd2). Between the subnetworks of the two brain compartments, Fmod, Bgn, and Foxc2 were consistent KDs revealed from our data-driven approach (Dataset S4). Remarkably, these KDs and subnetworks were largely reversed by DHA supplementation (Fig. 3).
Fig. 3.
Gene subnetworks and top network key drivers (KDs) of fructose and DHA. (a) KDs and gene subnetwork in hypothalamus. (b) KDs and gene subnetwork in hippocampus. Larger nodes depict KDs; grey nodes are network genes in the neighborhood of KDs that are not affected by fructose or DHA; top and bottom halves of each node, if colored, denotes genes affected by fructose (top) and DHA (bottom), respectively; red and blue colors denotes increased and decreased expression, respectively. Direction of change for the fructose group is determined by comparison with the control group; direction of change for the DHA + fructose group is determined by comparison with the fructose group.
3.7. Validation of Bgn and Fmod as Key Regulators of MetDs and Behavioral Aberrations in KO Mouse Models
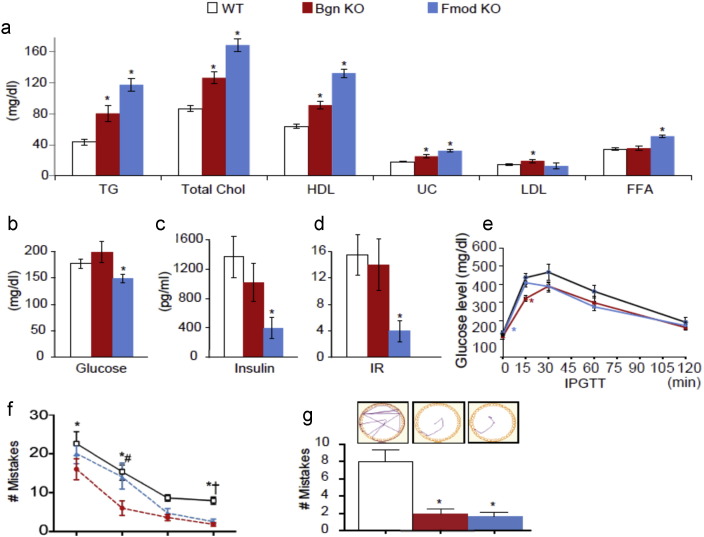
To evaluate our network KD prediction, we used Bgn and Fmod KO mice (Xu et al., 1998, Svensson et al., 1999) to test the effect of perturbing these KDs on phenotypes related to MetDs and cognitive function. Compared to wild type (WT) controls, we observed significantly elevated triglyceride, total cholesterol, HDL cholesterol, and un-esterified cholesterol in the plasma of both Bgn and Fmod KO mice, significant increases in LDL cholesterol in the Bgn KO, and elevated free fatty acids in the Fmod KO (Fig. 4a). We also found decreased levels of plasma glucose, insulin, as well as insulin resistance index in Fmod KO (Fig. 4b–d). The intraperitoneal glucose tolerance test of Bgn KO demonstrated significantly lower plasma glucose at 15 min after glucose injection and a similar trend in the Fmod KO (Fig. 4e).
Fig. 4.
Phenotypic validation of Bgn and Fmod using knockout (KO) mice. (a) Lipid traits including triglyceride (TG), total cholesterol, HDL cholesterol, un-esterified cholesterol (UC), LDL cholesterol and free fatty acids (FFA). (b) Glucose. (c) Insulin. (d) Insulin resistance (IR) measured by insulin sensitivity index. (e) Intraperitoneal glucose tolerance test (IPGTT). (f) Mistakes made during the Barnes Maze test in four days of the learning phase. (g) Track plots and mistakes made during Barnes Maze test for spatial memory. Two-sided Student's t-test was used to test statistical difference between knockout mice (Bgn KO or Fmod KO) and wild-type (WT) mice for analyses in (a)–(g). Two-way ANOVA with Holm-Sidak post hoc analysis for multiple comparisons was performed for the learning curves in panel (f) with *WT vs. Bgn KO; #Bgn KO vs. Fmod KO; and †WT vs. Fmod KO. Error bars in the plots are standard errors. ⁎p < 0.05. Sample size n = 8–16.
To assess the potential influence of Bgn and Fmod for spatial learning and memory, we tested the animals in the Barnes Maze. Because Fmod and Bgn KOs have been previously shown to have impaired tendon and joints which may affect latency time (Ameye et al., 2002, Chakravarti, 2002, Jepsen et al., 2002), we analyzed the Barnes Maze data by number of mistakes to eliminate the potential confound originated by differences in mobility. In addition, number of mistakes has been demonstrated as a more accurate measure of cognitive abilities in mice (O'leary and Brown, 2013). During the learning phase, there was a significant effect of animal type (F2,35 = 10.14; p = 0.0003 by 2-way ANOVA) and day (F3,105 = 40.44; p < 0.0001 by 2-way ANOVA) on the number of mistakes made in the Barnes Maze (Fig. 4f). Post hoc analysis revealed that both Bgn KO and Fmod KO made fewer mistakes compared with WT during the learning and the memory phases. Representative track plot analysis show a difference in strategy to find the escape hole (Fig. 4g, inset): the WT animals use a trial and error approach while the Bgn and Fmod KO animals follow a more direct path.
3.8. Relevance of Fructose Signatures and KDs to Human Diseases
To assess the relevance of our nutrigenomic signals from the rodent models to human pathophysiology, we compared the fructose signature genes as well as the KDs to human GWAS of MetDs and brain related diseases or traits. We found numerous overlapping genes between fructose signature genes and top GWAS hits in the GWAS catalog for a broad range of brain disorders and MetDs-related diseases or traits (Dataset S4). Importantly, KDs such as Fmod, Col1a2, Col18a1, and Dcn were GWAS candidate genes associating with obesity, cardiovascular disease, metabolic syndrome, and brain morphology and cognition, respectively. When using full sets of GWAS results we had accessed for plasma lipids, T2D, systolic and diastolic blood pressure traits, cognitive traits, and bipolar disorder, we found that human orthologs of the fructose signature genes derived from both hypothalamus and the hippocampus were significantly enriched for genetic polymorphisms showing stronger associations with these diseases or traits (Table 2).
Table 2.
Enrichment of human GWAS signals in fructose signatures and KDs by SNP set enrichment analysis (SSEA).
| Tissue | GWAS disease/trait | GWAS studya | p-Value (KS test)b | p-Value (Fisher's exact testb |
|---|---|---|---|---|
| Hypothalamus |
HDL cholesterol | GLGC | 4.35E − 05 | 1.25E − 12 |
| LDL cholesterol | GLGC | 1.05E − 03 | 1.90E − 05 | |
| Total cholesterol | GLGC | 8.51E − 06 | 3.65E − 14 | |
| Triglycerides | GLGC | 5.88E − 04 | 3.09E − 09 | |
| Diastolic blood pressure | ICBP | 2.40E − 05 | 7.55E − 05 | |
| Systolic blood pressure | ICBP | 3.06E − 03 | 1.16E − 02 | |
| Type 2 diabetes | DIAGRAM + | 2.07E − 02 | 3.39E − 02 | |
| Cognitive function | FHS | 1.78E − 02 | 3.18E − 03 | |
| Bipolar disorder |
WTCCC |
3.06E − 02 |
4.04E − 02 |
|
| Hippocampus | HDL cholesterol | GLGC | 6.48E − 05 | 2.02E − 07 |
| LDL cholesterol | GLGC | 2.98E − 05 | 2.16E − 04 | |
| Total cholesterol | GLGC | 2.37E − 05 | 4.19E − 13 | |
| Triglycerides | GLGC | 1.34E − 02 | 6.55E − 06 | |
| Diastolic blood pressure | ICBP | 4.52E − 01 | 4.59E − 03 | |
| Cognitive function | FHS | 9.43E − 03 | 4.28E − 02 | |
| Schizophrenia | CATIE | 2.26E − 02 | 3.02E − 01 | |
GLGC: Global Lipids Genetics Consortium; ICBP: The International Consortium for Blood Pressure; DIAGRAM +: The Diabetes Genetics Replication And Meta-analysis Consortium; FHS: Framingham Heart Study; WTCCC: Wellcome Trust Case Control Consortium; CATIE: Clinical Antipsychotic Trials for Intervention Effectiveness.
Bolded p values are those passed Bonferroni-corrected p < 0.05.
4. Discussion
In this systems nutrigenomics study, we show that fructose remodels fundamental aspects of gene regulation such as differential DNA methylation, differential gene expression, alternative splicing, and implications for microRNA alterations, all of which have the potential to impact pathogenesis. In addition, we found that fructose alters the organization of genes in brain region-specific networks interrelating cell metabolism, immune function, inflammation, and cell communication via key regulators such as the extracellular matrix genes Bgn and Fmod. Remarkably, we found that DHA supplementation was capable to reverse the genomic, epigenomic, network, and phenotypic perturbations induced by fructose via the same key drivers.
Our findings indicate that fructose and DHA induce transcriptomic reprogramming by engaging core transcription factors, splicing factors, and epigenetic modifications. For instance, fructose altered classic transcription factors such as Tcf7l2 (the most significant and robust signal for T2D from human GWAS) (Voight et al., 2010) and Zbtb16. Additionally, > 20% of the fructose and DHA signature genes only showed changes at isoform or alternative splicing levels but not overall expression, and alternative splicing factors such as Bicc1, Prpf31, and Rbpms were among the signature genes, supporting the importance of RNA splicing in the pathogenesis of human diseases highlighted recently (Xiong et al., 2015). The involvement of epigenetic mechanisms in fructose and DHA actions was substantiated by the significant alterations in the methylome of hypothalamus and hippocampus. cis-Regulation of transcriptome by DNA methylation was evidenced by co-localization of a subset of signature genes with local DMLs; trans-regulation mechanism was supported by the co-localization of DMLs with a number of transcriptional regulators (e.g., Aff3, Junb, Zbtb16, and Parp9) and microRNAs (e.g., rno-miR-421, rno-miR-143) whose predicted target genes were enriched among the signature genes. An increasing body of evidence indicates that neurological and psychiatric disorders have an epigenetic component (Jakovcevski and Akbarian, 2012, Tsankova et al., 2007). In particular, disruption in cell metabolism seems to be a driving force for epigenetic changes associated with cognitive function (Tyagi et al., 2015). The current results showing that DHA can counteract the broad transcriptomic and methylomic alterations induced by fructose imply that diet has the ability to regulate the susceptibility to disease by modulating the multifaceted transcriptional machinery.
Our studies also revealed broad impact of fructose and DHA on fundamental cell processes encompassing metabolism, inflammation, cell communication, and neuronal signaling, as well as critical differences between the two brain regions. For instance, the fructose and glucose transporter genes Slc2a5 and Slc2a4, and a broader category of pathways including insulin signaling, cardiomyopathy, transforming growth factor (TGF) beta signaling, mitogen-activated protein kinases (MAPK) signaling, platelet-derived growth factor (PDGF) signaling, toll like receptor (TLR) signaling, brain-derived neurotropic factor (BDNF) signaling, neural cell adhesion molecule (NCAM) signaling, and axon guidance were perturbed only in the hypothalamus. In fact, most of the hypothalamic pathways have been recently implicated in leptin resistance, hypothalamic deregulation and peripheral metabolism impairment (Milanski et al., 2012, Yan et al., 2014). The prominent response of the hypothalamus to diets is consistent with its role as the master metabolic sensor and regulator, and the current results suggest that these actions intermingle with inflammatory and cognitive signals. In contrast, the effects of fructose on the hippocampus were prominent in pathways engaged in cognitive function and neurological processes. For instance, hippocampal signature genes Pdc, Chrnb4, and Gch1 were previously implicated in schizophrenia (Sullivan et al., 2008), addictive behavior (Picciotto and Kenny, 2013), attention and vigilance (Yasuda et al., 2014), and early-onset Parkinson's disease (Cederfjall et al., 2013). The biological pathways affected in both brain regions were related to ECM, focal adhesion, complement cascade, smooth muscle contraction, and IGF signaling, which have been implicated in both metabolic and brain disorders (Soleman et al., 2013, Frischknecht and Gundelfinger, 2012, Chen et al., 2011, Phieler et al., 2013, Skaper, 2007).
A highlight of the study was the identification of the extracellular matrix genes Bgn and Fmod as key intermediates of the fructose effects on the gene network and phenotypic alterations, and these findings were corroborated in two knockout mouse models. Small leucine-rich proteoglycans Fmod and Bgn consistently emerged as KDs in both brain regions for both dietary components, and the expression levels of both genes were also significantly altered by fructose and DHA. Fmod and Bgn are important structural components of the ECM. The role of the ECM in the regulation of neurite outgrowth, and neural functionality and plasticity (Dityatev et al., 2010) as well as metabolic diseases (Soleman et al., 2013, Frischknecht and Gundelfinger, 2012, Chan et al., 2014) is becoming increasingly recognized. Our study provides unique nutrigenomic-based evidence that proteoglycans are likely main orchestrators of important metabolic and cognitive processes in the brain.
To assess the translatability of our rodent findings to human pathophysiology, we found significant intersections between the fructose signatures or the network KDs identified in our rodent models and human GWAS hits for neurological and neurodegenerative diseases, psychiatric diseases, and various metabolic diseases. As human GWAS implies a causal role of the candidate genes based on genetic evidence, the overlapping signatures or network KDs in response to fructose likely play causal roles in the development of MetDs or brain disorders. The same genes also likely mediate the beneficial actions of DHA.
The strength of our study lies in the use of powerful systems nutrigenomic approach to comprehensively characterize phenotypic and multi-tissue transcriptomic and methylomic profiles of fructose and DHA, followed by network modeling, in vivo validation, and human data integration. The highly integrative nature not only enables a comprehensive and in-depth exploration of the molecular mechanisms mediating the actions of nutrition on health and disease, but also pinpoints critical regulators of the biological pathways and provides compelling evidence for the potential causal role of the identified genes and pathways in human diseases.
We note the following limitations of the current study. First, the Bgn and Fmod KO animals used are not brain-specific. It is possible that the phenotypic changes in peripheral metabolism observed in these models are the results of perturbations of these genes outside of the brain. Future tissue-specific studies are warranted to further investigate the action sites of these ECM genes. Second, the behavioral changes observed in our systemic treatment studies of fructose and DHA as well as in the genetically modified animal models of Bgn and Fmod could be influenced by numerous variables that are either not investigated or difficult to tease out. Third, we only focused on two brain regions in the current study and future investigations of additional tissues and cell types are warranted.
The staggering increase in the prevalence of MetDs, its negative impact on brain function and pathogenesis of brain disorders, and the role played by fructose consumption as a dietary perturbation in these processes are becoming major concerns for public health and basic and clinical science. Our study provides molecular evidence supporting the ability of fructose to disrupt critical genes and fundamental physiological processes such as insulin, IGF, TLR, TGF beta, BDNF, MAPK, NCAM, and PDGF signaling pathways and neurological processes. The majority of these pathways have been implicated in both MetDs and brain disorders. In addition, genes affected by fructose significantly overlapped with human GWAS genes for brain disorders (e.g., Alzheimer's disease, attention deficient hyperactive disorder, depression, addiction, Parkinson's disease) and MetDs-related diseases (e.g., blood pressure, cardiovascular disease, lipids, obesity, metabolic syndrome, and T2D). Our results point to the capacity of fructose to reprogram molecular substrates underlying pathology. The remarkable capacity of DHA supplementation to restore genomic, epigenomic, pathway, network, and phenotypic traits vulnerable to the effects of fructose provides multi-level support for the potential of omega-3 fatty acids as a nutritional remedy for metabolic and brain dysregulation. This points to an effective means to guide personalized medicine by matching the disease-promoting molecular signatures with an agent capable of reversing the patterns.
In summary, we show that DHA reverses biological pathways and gene networks perturbed by fructose via transcriptional regulators (such as transcription factors, epigenetic regulators, splicing factors) and essential network regulators (such as Bgn and Fmod) that may control the balance between health and disease. Our study reveals critical information supporting an action of ECM in orchestrating brain function through gene network organizations. Our experimental validation highlights the predictive power of our systems genomics approaches in identifying vulnerable disease genes and mechanisms. In addition, our paradigm based on the integration between rodent studies and human GWAS serves as a precedent to apply nutrigenomics principles to predict disease susceptibility and to guide personalized medicine.
The following are the supplementary data related to this article.
Supplementary information including Supplementary Materials and Methods, Supplementary Figures, and Supplementary Tables.
Transcriptomic signatures of fructose and DHA in hypothalamus and hippocampus tissues, and comparisons between tissues and treatment.
Biological pathways over-represented in the fructose and DHA transcriptomic signatures.
Differential DNA methylation loci of fructose and DHA in hypothalamus and hippocampus tissues, and comparisons between tissues and treatment.
Key drivers of the fructose and DHA networks and intersections with human GWAS.
Role of Funding Sources
XY is supported by NIH grant R01DK104363, American Heart Association Scientist Development Grant 13SDG17290032, Leducq Foundation, Hellman Fellows Award, UCLA Faculty Research Grant, and UCLA CTSI Grant UL1TR000124. FGP is supported by NIH grants R01DK104363, R01NS050465 and The Letten Foundation. MFY was supported in part by the Intramural Program of the NIDCR, NIH. XX is supported by NIH grants R01HG006264 and U01HG007013, and NSF grant 1262134. GW was supported by NSFC (91019016), NBRPC (2012CB316503), and the China Scholarship Council. None of the funding sources had any role in the design and conduct of the study, the collection, analysis, or interpretation of the data, or the preparation, review, or approval of the manuscript.
Conflicts of Interests
The authors declare no conflict of interests.
Author Contributions
QM contributed to study design, project coordination, experiments, data analyses, writing and editing of the manuscript; XY and FGP designed and directed the study, and contributed to data interpretation, writing and editing of the manuscript; EN contributed to experiments, data analysis, and writing of the manuscript; ZY, YZ, RA, YZhuang, ET, AM, QZ, JL, MM, LO, WG, JZ, BZ, MP, TMK, XX, and MFY contributed to experiments and/or data analyses, and reviewed and edited the manuscript.
Acknowledgements
We thank Dr. Oldberg Ake from Experimental Medical Sciences, University of Lund, Lund, Sweden for kindly providing the Fmod knockout mouse model for this study.
Contributor Information
Fernando Gomez-Pinilla, Email: fgomezpi@mednet.ucla.edu.
Xia Yang, Email: xyang123@ucla.edu.
References
- Agrawal R., Gomez-Pinilla F. ‘Metabolic syndrome’ in the brain: deficiency in omega-3 fatty acid exacerbates dysfunctions in insulin receptor signalling and cognition. J. Physiol. 2012;590:2485–2499. doi: 10.1113/jphysiol.2012.230078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R., Noble E., Vergnes L., Ying Z., Reue K., Gomez-Pinilla F. Dietary fructose aggravates the pathobiology of traumatic brain injury by influencing energy homeostasis and plasticity. J. Cereb. Blood Flow Metab. 2015 doi: 10.1177/0271678X15606719. (Online before print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameye L., Aria D., Jepsen K., Oldberg A., Xu T., Young M.F. Abnormal collagen fibrils in tendons of biglycan/fibromodulin-deficient mice lead to gait impairment, ectopic ossification, and osteoarthritis. FASEB J. 2002;16:673–680. doi: 10.1096/fj.01-0848com. [DOI] [PubMed] [Google Scholar]
- Bomfim T.R., Forny-Germano L., Sathler L.B., Brito-Moreira J., Houzel J.C., Decker H., Silverman M.A., Kazi H., Melo H.M., Mcclean P.L., Holscher C., Arnold S.E., Talbot K., Klein W.L., Munoz D.P., Ferreira S.T., De Felice F.G. An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer's disease-associated Abeta oligomers. J. Clin. Invest. 2012;122:1339–1353. doi: 10.1172/JCI57256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer A.A., Stanhope K.L., Graham J.L., Cummings B.P., Ampah S.B., Saville B.R., Havel P.J. Fish oil supplementation ameliorates fructose-induced hypertriglyceridemia and insulin resistance in adult male rhesus macaques. J. Nutr. 2014;144:5–11. doi: 10.3945/jn.113.178061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederfjall E., Nilsson N., Sahin G., Chu Y., Nikitidou E., Bjorklund T., Kordower J.H., Kirik D. Continuous DOPA synthesis from a single AAV: dosing and efficacy in models of Parkinson's disease. Sci. Rep. 2013;3:2157. doi: 10.1038/srep02157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti S. Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj. J. 2002;19:287–293. doi: 10.1023/A:1025348417078. [DOI] [PubMed] [Google Scholar]
- Chan K.H., Huang Y.T., Meng Q., Wu C., Reiner A., Sobel E.M., Tinker L., Lusis A.J., Yang X., Liu S. Shared molecular pathways and gene networks for cardiovascular disease and type 2 diabetes mellitus in women across diverse ethnicities. Circ. Cardiovasc. Genet. 2014;7:911–919. doi: 10.1161/CIRCGENETICS.114.000676. [DOI] [PubMed] [Google Scholar]
- Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., Gewirtz A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., Macneil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., Leonardson A., Castellini L.W., Wang S., Champy M.F., Zhang B., Emilsson V., Doss S., Ghazalpour A., Horvath S., Drake T.A., Lusis A.J., Schadt E.E. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Y., Stern S.A., Garcia-Osta A., Saunier-Rebori B., Pollonini G., Bambah-Mukku D., Blitzer R.D., Alberini C.M. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caterina R. n-3 fatty acids in cardiovascular disease. N. Engl. J. Med. 2011;364:2439–2450. doi: 10.1056/NEJMra1008153. [DOI] [PubMed] [Google Scholar]
- De Graaf A.A., Freidig A.P., De Roos B., Jamshidi N., Heinemann M., Rullmann J.A., Hall K.D., Adiels M., Van Ommen B. Nutritional systems biology modeling: from molecular mechanisms to physiology. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A., Schachner M., Sonderegger P. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat. Rev. Neurosci. 2010;11:735–746. doi: 10.1038/nrn2898. [DOI] [PubMed] [Google Scholar]
- Farooqui A.A., Farooqui T., Panza F., Frisardi V. Metabolic syndrome as a risk factor for neurological disorders. Cell. Mol. Life Sci. 2012;69:741–762. doi: 10.1007/s00018-011-0840-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht R., Gundelfinger E.D. The brain's extracellular matrix and its role in synaptic plasticity. Adv. Exp. Med. Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- Guo W., Fiziev P., Yan W., Cokus S., Sun X., Zhang M.Q., Chen P.Y., Pellegrini M. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski M., Akbarian S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K.J., Wu F., Peragallo J.H., Paul J., Roberts L., Ezura Y., Oldberg A., Birk D.E., Chakravarti S. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem. 2002;277:35532–35540. doi: 10.1074/jbc.M205398200. [DOI] [PubMed] [Google Scholar]
- Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D.S., Sebastian A., Rani S., Ray S., Harrys Kishore C.J., Kanth S., Ahmed M., Kashyap M.K., Mohmood R., Ramachandra Y.L., Krishna V., Rahiman B.A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., Pandey A. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowette K., Roosen L., Tack J., Vanden Berghe P. Effects of high-fructose diets on central appetite signaling and cognitive function. Front. Nutr. 2015;2:5. doi: 10.3389/fnut.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig R.H., Schmidt L.A., Brindis C.D. Public health: the toxic truth about sugar. Nature. 2012;482:27–29. doi: 10.1038/482027a. [DOI] [PubMed] [Google Scholar]
- Lyssiotis C.A., Cantley L.C. Metabolic syndrome: F stands for fructose and fat. Nature. 2013;502:181–182. doi: 10.1038/502181a. [DOI] [PubMed] [Google Scholar]
- Makinen V.P., Civelek M., Meng Q., Zhang B., Zhu J., Levian C., Huan T., Segre A.V., Ghosh S., Vivar J., Nikpay M., Stewart A.F., Nelson C.P., Willenborg C., Erdmann J., Blakenberg S., O'donnell C.J., Marz W., Laaksonen R., Epstein S.E., Kathiresan S., Shah S.H., Hazen S.L., Reilly M.P., Lusis A.J., Samani N.J., Schunkert H., Quertermous T., Mcpherson R., Yang X., Assimes T.L. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanski M., Arruda A.P., Coope A., Ignacio-Souza L.M., Nunez C.E., Roman E.A., Romanatto T., Pascoal L.B., Caricilli A.M., Torsoni M.A., Prada P.O., Saad M.J., Velloso L.A. Inhibition of hypothalamic inflammation reverses diet-induced insulin resistance in the liver. Diabetes. 2012;61:1455–1462. doi: 10.2337/db11-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., Mcelwee J., Zhang B., Zhang C., Lamb J.R., Xie T., Suver C., Molony C., Melquist S., Johnson A.D., Fan G., Stone D.J., Schadt E.E., Casaccia P., Emilsson V., Zhu J. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014;10:743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer J.W. Metabolic syndrome and mental illness. Am. J. Manag. Care. 2007;13:S170–S177. [PubMed] [Google Scholar]
- O'leary T.P., Brown R.E. Optimization of apparatus design and behavioral measures for the assessment of visuo-spatial learning and memory of mice on the Barnes maze. Learn. Mem. 2013;20:85–96. doi: 10.1101/lm.028076.112. [DOI] [PubMed] [Google Scholar]
- Panagiotou G., Nielsen J. Nutritional systems biology: definitions and approaches. Annu. Rev. Nutr. 2009;29:329–339. doi: 10.1146/annurev-nutr-080508-141138. [DOI] [PubMed] [Google Scholar]
- Phieler J., Garcia-Martin R., Lambris J.D., Chavakis T. The role of the complement system in metabolic organs and metabolic diseases. Semin. Immunol. 2013;25:47–53. doi: 10.1016/j.smim.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto M.R., Kenny P.J. Molecular mechanisms underlying behaviors related to nicotine addiction. Cold Spring Harb. Perspect. Med. 2013;3:a012112. doi: 10.1101/cshperspect.a012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H., Fujita R., Qiang L., Cheng R., Lee J.H., Abeliovich A. Integrative genomics identifies APOE epsilon4 effectors in Alzheimer's disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- Skaper S.D. The brain as a target for inflammatory processes and neuroprotective strategies. Ann. N. Y. Acad. Sci. 2007;1122:23–34. doi: 10.1196/annals.1403.002. [DOI] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleman S., Filippov M.A., Dityatev A., Fawcett J.W. Targeting the neural extracellular matrix in neurological disorders. Neuroscience. 2013;253:194–213. doi: 10.1016/j.neuroscience.2013.08.050. [DOI] [PubMed] [Google Scholar]
- Steffen B.T., Steffen L.M., Zhou X., Ouyang P., Weir N.L., Tsai M.Y. n-3 Fatty acids attenuate the risk of diabetes associated with elevated serum nonesterified fatty acids: the multi-ethnic study of atherosclerosis. Diabetes Care. 2015;38:575–580. doi: 10.2337/dc14-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J., Korem T., Zeevi D., Zilberman-Schapira G., Thaiss C.A., Maza O., Israeli D., Zmora N., Gilad S., Weinberger A., Kuperman Y., Harmelin A., Kolodkin-Gal I., Shapiro H., Halpern Z., Segal E., Elinav E. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Lin D., Tzeng J.Y., Van Den Oord E., Perkins D., Stroup T.S., Wagner M., Lee S., Wright F.A., Zou F., Liu W., Downing A.M., Lieberman J., Close S.L. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol. Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson L., Aszodi A., Reinholt F.P., Fassler R., Heinegard D., Oldberg A. Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J. Biol. Chem. 1999;274:9636–9647. doi: 10.1074/jbc.274.14.9636. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N., Renthal W., Kumar A., Nestler E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tyagi E., Zhuang Y., Agrawal R., Ying Z., Gomez-Pinilla F. Interactive actions of Bdnf methylation and cell metabolism for building neural resilience under the influence of diet. Neurobiol. Dis. 2015;73:307–318. doi: 10.1016/j.nbd.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen J.K., Mursu J., Voutilainen S., Uusitupa M., Tuomainen T.P. Serum omega-3 polyunsaturated fatty acids and risk of incident type 2 diabetes in men: the Kuopio ischemic heart disease risk factor study. Diabetes Care. 2014;37:189–196. doi: 10.2337/dc13-1504. [DOI] [PubMed] [Google Scholar]
- Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., Mcculloch L.J., Ferreira T., Grallert H., Amin N., Wu G., Willer C.J., Raychaudhuri S., Mccarroll S.A., Langenberg C., Hofmann O.M., Dupuis J., Qi L., Segre A.V., Van Hoek M., Navarro P., Ardlie K., Balkau B., Benediktsson R., Bennett A.J., Blagieva R., Boerwinkle E., Bonnycastle L.L., Bengtsson Bostrom K., Bravenboer B., Bumpstead S., Burtt N.P., Charpentier G., Chines P.S., Cornelis M., Couper D.J., Crawford G., Doney A.S., Elliott K.S., Elliott A.L., Erdos M.R., Fox C.S., Franklin C.S., Ganser M., Gieger C., Grarup N., Green T., Griffin S., Groves C.J., Guiducci C., Hadjadj S., Hassanali N., Herder C., Isomaa B., Jackson A.U., Johnson P.R., Jorgensen T., Kao W.H., Klopp N., Kong A., Kraft P., Kuusisto J., Lauritzen T., Li M., Lieverse A., Lindgren C.M., Lyssenko V., Marre M., Meitinger T., Midthjell K., Morken M.A., Narisu N., Nilsson P., Owen K.R., Payne F., Perry J.R., Petersen A.K., Platou C., Proenca C., Prokopenko I., Rathmann W., Rayner N.W., Robertson N.R., Rocheleau G., Roden M., Sampson M.J., Saxena R., Shields B.M., Shrader P., Sigurdsson G., Sparso T., Strassburger K., Stringham H.M., Sun Q., Swift A.J., Thorand B. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., Morris Q., Barash Y., Krainer A.R., Jojic N., Scherer S.W., Blencowe B.J., Frey B.J. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Bianco P., Fisher L.W., Longenecker G., Smith E., Goldstein S., Bonadio J., Boskey A., Heegaard A.M., Sommer B., Satomura K., Dominguez P., Zhao C., Kulkarni A.B., Robey P.G., Young M.F. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat. Genet. 1998;20:78–82. doi: 10.1038/1746. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang H., Yin Y., Li J., Tang Y., Purkayastha S., Li L., Cai D. Obesity- and aging-induced excess of central transforming growth factor-beta potentiates diabetic development via an RNA stress response. Nat. Med. 2014;20:1001–1008. doi: 10.1038/nm.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Deignan J.L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R.R., Karlsson J., Castellani L.W., Mumick S., Wang K., Xie T., Coon M., Zhang C., Estrada-Smith D., Farber C.R., Wang S.S., Van Nas A., Ghazalpour A., Zhang B., Macneil D.J., Lamb J.R., Dipple K.M., Reitman M.L., Mehrabian M., Lum P.Y., Schadt E.E., Lusis A.J., Drake T.A. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang B., Molony C., Chudin E., Hao K., Zhu J., Gaedigk A., Suver C., Zhong H., Leeder J.S., Guengerich F.P., Strom S.C., Schuetz E., Rushmore T.H., Ulrich R.G., Slatter J.G., Schadt E.E., Kasarskis A., Lum P.Y. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20:1020–1036. doi: 10.1101/gr.103341.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda Y., Hashimoto R., Ohi K., Yamamori H., Fujimoto M., Umeda-Yano S., Fujino H., Fukunaga M., Horiguchi M., Takeda M., Ichinose H. A functional polymorphism of the GTP cyclohydrolase 1 gene predicts attention performance. Neurosci. Lett. 2014;566:46–49. doi: 10.1016/j.neulet.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., Mcelwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., Macdonald M.E., Lamb J.R., Bennett D.A., Molony C., Stone D.J., Gudnason V., Myers A.J., Schadt E.E., Neumann H., Zhu J., Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Barrere-Cain R.E., Yang X. Nutritional systems biology of type 2 diabetes. Genes Nutr. 2015;10:481. doi: 10.1007/s12263-015-0481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Lum P.Y., Lamb J., Guhathakurta D., Edwards S.W., Thieringer R., Berger J.P., Wu M.S., Thompson J., Sachs A.B., Schadt E.E. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet. Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang B., Smith E.N., Drees B., Brem R.B., Kruglyak L., Bumgarner R.E., Schadt E.E. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information including Supplementary Materials and Methods, Supplementary Figures, and Supplementary Tables.
Transcriptomic signatures of fructose and DHA in hypothalamus and hippocampus tissues, and comparisons between tissues and treatment.
Biological pathways over-represented in the fructose and DHA transcriptomic signatures.
Differential DNA methylation loci of fructose and DHA in hypothalamus and hippocampus tissues, and comparisons between tissues and treatment.
Key drivers of the fructose and DHA networks and intersections with human GWAS.