Abstract
The nitric oxide (NO) metabolites nitrite (NO2−) and nitrate (NO3−) can be quantified as an endpoint of endothelial function. We developed a LC-MS/MS method of measuring nitrite and nitrate isotopologues, which has a lower limit of quantification (LLOQ) of 1 nM. This method allows for isotopic labeling to differentiate newly formed nitrite and nitrate from nanomolar to micromolar background levels of nitrite and nitrate in biological matrices. This method utilizes 2,3-diaminonaphthalene (DAN) derivatization, which reacts with nitrite under acidic conditions to produce 2,3-naphthotriazole (NAT). NAT was chromatographically separated on a Shimadzu LC System with an Agilent Extend-C18 5 μm 2.1 × 150 mm column and detected using a multiple reaction monitoring (MRM) method on an ABSciex 3200 QTRAP mass spectrometer operated in positive mode. Mass spectrometry allows for the quantification of 14N-NAT (m/z 170.1) and 15N-NAT (m/z 171.1). Both nitrite and nitrate demonstrated a linear detector response (1 nM – 10 μM, 1 nM – 100 nM, respectively), and were unaffected by common interferences (Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), phenol red, and NADPH). This method requires minimal sample preparation, making it ideal for most biological applications. We applied this method to develop a cell culture model to study the development of nitrate tolerance in human endothelial cells (EA.hy926).
Keywords: Nitrite, Nitric oxide, Nitrate tolerance, LC-MS/MS, Glyceryl trinitrate, cell culture
1. Introduction
Nitric oxide (NO) is a signaling molecule and free radical that plays a diverse role in cardiovascular, immune, and nervous systems [1][2]. NO is produced from L-arginine by NO synthase (NOS) in mammals [1], yet it can also be produced via the reduction of nitrite (NO2−) and nitrate (NO3−) by xanthine oxidase (XO) [3; 4; 5; 6; 7]. Nitrite and nitrate are present in the environment and diet. NO is released by cells in the picomolar to nanomolar range [8], and has an estimated in vivo half-life of 3-4 seconds in human blood [9]. It is therefore very difficult to accurately measure, so its nitrite and nitrate metabolites are often measured as surrogates [10]. Measurement of nitrite and nitrate is the most suitable and practical method to assess NO synthesis in vivo [11]. Given that nitrite and nitrate can be reduced to NO by XO, nitrite and nitrate can be viewed as bioavailable storage pools for NO [12].
Nitrate tolerance
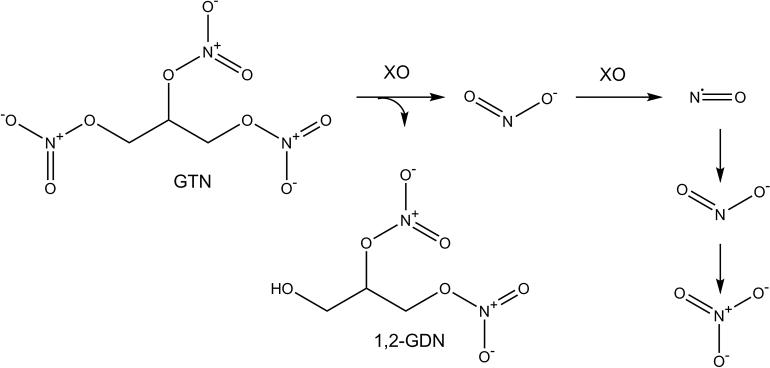
Glyceryl trinitrate (GTN), more commonly known as nitroglycerin, is used for the treatment of angina pectoris, coronary artery disease, acute myocardial infarction, hypertension, and congestive heart failure. GTN is a prodrug that must be bioactivated in endothelial cells to produce NO (Figure 1), which activates soluble guanylyl cyclase (sGC) to release the second messenger cyclic guanosine monophosphate (cGMP), stimulating the relaxation of smooth muscle cells [13; 14]. Continuous treatment with GTN results in nitrate tolerance after only a few weeks, which limits the long-term use and benefits of treatment [15]. Currently, nitrate tolerance is managed by implementing nitrate-free intervals, but this strategy could become obsolete when co-therapies that effectively prevent nitrate tolerance become available. The cause of nitrate tolerance and the mechanism of GTN bioactivation remain relatively unknown. In the present study we measure nitrite and nitrate as endpoints of GTN bioactivation in endothelial cells. Our intent is to determine whether cell culture and the measurement of nitrite and nitrate isotopologues can be used as an appropriate model to determine the mechanism of GTN bioactivation and the cause of nitrate tolerance.
Figure 1.
Glyceryl Trinitrate (GTN) is denitrified by xanthine oxidase (XO) in endothelial cells to produce 1,2-glyceryl dinitrate (1,2-GDN) and NO2−, which is reduced into NO and subsequently non-enzymatically oxidized to NO2− and NO3−.
Quantification of nitrite and nitrate
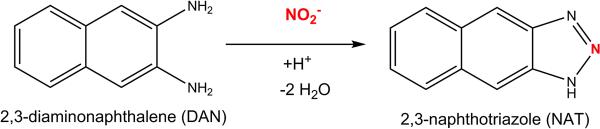
There are over 200 analytical methods for the measurement of nitrite and nitrate, yet a small portion of these can be applied to human biological fluids [11]. Current methods that are used for nitrite and nitrate analysis include fluorescence, chemiluminescence, high performance liquid chromatography (HPLC), capillary electrophoresis, colorimetric and ultraviolet (UV) spectrophotometric methods, and gas chromatography-mass spectrometry (GC-MS). The current methods have been extensively reviewed [8; 11; 16; 17; 18]. Mass spectrometry is the only analytical technique that is able to measure isotopologues of nitrite and nitrate, and is an indispensable analytical tool for reliable quantitative analysis of nitrite and nitrate [8]. In the present study, we modify a method of measuring nitrite with LC-MS/MS [19; 20], and expand upon it to include quantification of nitrate. We utilized the well-established 2,3-diaminonaphthalene (DAN) derivatization, which reacts with nitrite under acidic conditions to produce the highly fluorescent 2,3-naphthotriazole (NAT)[21](Figure 2). Measuring NAT with LC-MS/MS, instead of by fluorescence, allows for the use of isotope labeling which reduces the impact of high background levels of nitrite and nitrate in biological matrices.
Figure 2.
NO2− reacts with 2,3-diaminonaphthalene (DAN) under acidic conditions to produce 2,3-naphthotriazole (NAT).
This LC-MS/MS method is a significant contribution to the field because it is a sensitive and reproducible method for quantifying nitrite and nitrate isotopologues as metabolic products of GTN. This method is used to investigate the mechanism of nitrate tolerance in human endothelial cells, which is made possible with the use of 15N3-GTN.
2. Materials and Methods
2.1 Chemicals
2,3-diaminonaphthalene (DAN) was procured from TCI America (Portland, OR). 15N3-glyceryl trinitate (GTN) in acetonitrile (15N, 98%+), sodium 15N-nitrite (15N, 98%+), and sodium 15N-nitrate (15N, 98%+) were from Cambridge Isotope Laboratories (Tewksbery, MA). GTN, NADPH, sodium hydroxide, and nitrate reductase were from Sigma Aldrich (St. Louis, MO). Dulbecco's Modified Eagle Medium (DMEM), Hank's Buffered Sodium Salt (HBSS), trypsin-EDTA, fetal bovine serum (FBS), penicillin, and streptomycin were procured from Invitrogen (Carlsbad, CA). Ammonium bicarbonate, LCMS-grade water and methanol were purchased from J.T Baker (Center Valley, PA).
2.2 Endothelial cell culture and treatments with glyceryl trinitrate (GTN)
EA.hy926 human hybrid endothelial cells (American Type Culture Collection; Manassas, VA) were maintained in culture in T75 flasks with DMEM medium containing 4.5 g/L glucose and 84 mg/L L-arginine, and supplemented with 10% FBS and 1% penicillin-streptomycin (PS) in a humidified incubator at 37°C with 5% CO2. Endothelial cells were chosen because they have been previously shown to produce NO from nitrite, which caused changes in markers of intracellular nitrosation reactions [22]. Endothelial cells express xanthine oxidase, mitochondrial aldehyde dehydrogenase, and nitric oxide synthase, among others.
EA.hy926 cells were plated at a density of 5.0 × 105 cells per well in 6-well plates with 2 mL medium following a trypsinization protocol. Cells were returned to the incubator for 24 hours before treatment. Nitrate tolerance was induced in cells using a modified method from Hinz and colleagues [23]. In short, cells were given an initial 20 μM GTN challenge in phenol red-free DMEM with 1% FBS, and incubated for 5 hours. Medium was aspirated, washed once with 2 mL HBSS, then treated for 60 minutes at four concentration levels of 15N-GTN (1 μM – 20 μM) in 1 mL phenol red-free DMEM with 1% FBS. Results were compared to control cells, which were treated with 5 μL of acetonitrile as a vehicle control. The entire final volume (1 mL) was transferred to Eppendorf tubes and immediately frozen at −80°C until analysis.
2.3 Enzymatic reduction of nitrate into nitrite
Nitrate was enzymatically converted to nitrite by incubating with nitrate reductase [24; 25]. The enzymatic method of reducing nitrate to nitrite is preferred to cadmium reduction because it is non-toxic, and offers high specificity, simplicity, rapidity, and complete conversion of nitrate into nitrite [24; 25]. One hundred μL of the cell medium, or 15N-nitrate standard, was incubated with 10 μL of 1 U/mL nitrate reductase and 10 μL of 120 μM NADPH and incubated at room temperature for 60 minutes. There was complete conversion of nitrate into nitrite, as evidenced by conversion of known amounts of 15N standards of sodium nitrite and sodium nitrate. After incubation, samples were immediately derivatized for nitrite analysis.
2.4 2,3-Diaminonaphthalene (DAN) derivatization
DAN reacts with nitrite under acidic conditions to form NAT [21], which is detectable with mass spectrometry. For mass spectrometric analysis of NAT, the protocol developed by Misko et al. [26] was adapted. No filtration is necessary because it was determined that our culture media is not a source of interference for the measurement of nitrite and nitrate. A 100 μL sample aliquot (culture medium, nitrite standards, or nitrate-derived standards) was incubated at 24°C for 60 minutes with 10 μL of 316 μM DAN in 50% EtOH with 0.62 M HCl. The reaction was quenched with 5 μL of 2.8 M NaOH. In alkaline conditions, NAT is stable for at least 24 hours [25]. The samples were centrifuged at 16,000 x g for 1 minute, then transferred to glass LCMS vials (Microsolv, Eatontown, NJ) and analyzed for 14N-NAT and 15N-NAT.
2.5 Detection of 2,3-naphthotriazole (NAT) with LC-MS/MS
Mass spectrometric detection of 14N-NAT and 15N-NAT was performed on a Shimadzu Prominence LC system coupled to an AB Sciex 3200 QTRAP operated in positive ion mode. An Agilent Extend-C18 5 μm 2.1 × 150 mm column was maintained at 40°C with a flow rate of 0.2 mL/min (0-13 minutes) and 0.4 mL/min (13-25 minutes). Mobile phase A was 5 mM aqueous NH4HCO3, mobile phase B was 100% methanol, and the needle wash solvent was 50% methanol. The total run time was 25 minutes, column eluent was directed to MS between 8-12 minutes only, and injections were 15 μL. The solvent gradient profile started at 5% B for 4 minutes, changed linearly to 50% B over 1 minute, was held at 50% B for 8 minutes, changed linearly to 95% over 3 minutes, was held at 95% B for 5 minutes, changed linearly to 5% B over 3 minutes, and remained at 5% B to equilibrate the column. Retention time was monitored, and no significant shifts were observed throughout sample sets. A multiple reaction monitoring (MRM) method monitored the following transitions over 100 ms for the analyte indicated: m/z 171.1 → 115.1 for 15N-NAT, m/z 159.1 → 159.1for DAN, and m/z 170.1 → 115.1 for 14N-NAT. The collision voltages were 33 V for all except DAN, which was 5 V. Declustering, entrance, and collision cell exit potentials were 51, 11.5 and 4 V, respectively. The IonSpray voltage was 5500 V and the ion source temperature was 550 °C.
14N-NAT and 15N-NAT concentrations were quantified by integrating the peaks in Analyst 1.5.1 to obtain peak area. Peak area was converted to concentration (nM) by using individual seven-point concentration curves of the analytes. Nitrate concentration was obtained after reduction with nitrate reductase.
2.6 Method validation and quality control
Seven-point concentration curves (1 nM – 10 μM) for sodium 15N-nitrite and sodium 15N-nitrate were made in combination and individually in milli-Q water, 200 mM HEPES, and DMEM with 1% FBS. These were used to calculate LLOQ, linear range, percent recovery in DMEM, and percent conversion of nitrate reduction to nitrite. Background levels of nitrate and nitrite were measured in water, HEPES, and DMEM. Three concentrations (10 nM, 100 nM, and 1000 nM) of nitrite and nitrate isotopologues were analyzed ten times in water and DMEM to determine precision and accuracy. For every day of analysis of cell culture or enzyme samples, a seven-point concentration curve of nitrite in water was run to assess instrument performance. If a drop in sensitivity was observed, the instrument was cleaned and calibrated before analysis of samples. All samples were randomized to prevent batch effect, yet nitrite and nitrate samples were paired and run in sequence.
2.7 Statistical analysis
Results were analyzed using a one-way ANOVA, followed by Tukey post hoc analysis with a P-value of < 0.05 indicating significance. All statistical analyses were performed using GraphPad Prism 4.
3. Results
3.1 Quantification of nitrite and nitrate
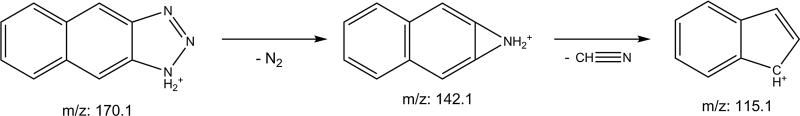
The derivatization product of NO2− is 2,3-naphthotriazole (NAT), which is detected by LC-MS/MS. 14N-NAT has a parent mass of 169.1 Da, which has an [M+H]+ mass of 170.1 Da. The major product ions of 14N-NAT are C10H8N+ (m/z 142.1) and C9H7+ (m/z 115.1). The proposed fragmentation pattern of 14N-NAT is in Figure 3. The transition of m/z 170.1→115.1 was chosen for quantitation because of the high abundance and specificity. 15N-NAT has a m/z of 171.1 and a quantitation transition of m/z 171.1→115.1.
Figure 3.
Proposed fragmentation pattern of 2,3-naphthotriazole (NAT). The molecular cation of 14N-NAT (m/z 170.1) is fragmented into the qualifier ion C10H8N+ (m/z 142.1), and undergoes additional fragmentation into C9H7+ (m/z 115.1) which is used for quantification.
14N-NAT and 15N-NAT co-elute at 9.60 minutes. There is a significant peak at 10.04 minutes that was identified as a degradation product of 14N-NAT and 15N-NAT. 14N-NAT and 15N-NAT degrade into their N-oxide metabolites (m/z 186 and 187)(Supplemental Figure 1), which give rise to protonated 14N-NAT and 15N-NAT species upon in-source loss of •OH (m/z 169 and 170). This “degradation peak” is larger in cell culture media samples, and increases over time if the samples remain at room temperature for several days. This demonstrates the necessity of measuring all derivatized samples within 24 hours of preparation.
3.2 Method Validation
The LC-MS/MS method of measuring nitrite and nitrate was analyzed for sensitivity, reproducibility, accuracy, precision, interferences, percent recovery, and carryover. Standard curves of nitrate and nitrite were used to determine LLOQ and the linear range. The LLOQ was defined as the lowest standard on the calibration curve at which the analyte peak was at least 5 times the response relative to blank response, with a precision of ≤ 20% RSD and an accuracy of 80-120%. 15N-Nitrite and 15N-nitrate had a LLOQ of 1 nM in both water and in DMEM (n=10). This value is within the same range as previous studies that have measured 15N-nitrite with LC-MS/MS, which reported LLOQ of 4 nM [19] and 5 nM [20]. We postulate that our quantitation limit is lower because different instrumentation and column, as well as our choice in solvent system. NAT is more stable under alkaline conditions, so using a solvent system with 0.1% formic acid [19; 20] may lead to degradation of NAT and a reduction in sensitivity.
15N-Nitrite had a linear range from 1 nM to 10 μM (correlation coefficient > 0.99, P<0.01), whereas 15N-nitrate was linear from 1 nM – 100 nM (correlation coefficient > 0.99, P<0.05) (n=5). Though nitrate is detectable at very low concentrations, it demonstrated a low precision, high level of variability, and low recovery at concentrations greater than 100 nM (correlation coefficient = 0.72, P>0.05). This is most likely due to the saturation of nitrate reductase. For these reasons, this method is only suitable to measure nitrate at nanomolar levels without further method development. However, increasing the amount of enzyme was not desirable due to potential interferences with mass spectrometry.
Precision and accuracy were analyzed by spiking known amounts (10 nM, 100 nM, 1000 nM) of 15NO2−, NO2−, 15NO3−, and NO3− into water and DMEM (phenol red-free, with 1% FBS and PS), n=10. Individual calibration curves (seven point, 1 nM – 10 μM) were used for quantitation. Accuracy is determined as the percentage deviation of calculated concentration from the nominal concentrations, and was calculated with the equation: . Precision is determined as the relative standard deviation (RSD) of the 10 measurements, and is calculated as . Accuracy and precision is presented in Table 1. Satisfactory accuracies were obtained for 14N-nitrite and 15N-nitrite in both DMEM and water, with biases less than 7.5% and 5.6%, respectively. Accuracy for 14N-nitrate and 15N-nitrate was satisfactory up to 100 nM (less than 7.3% and 4.7%, respectively) but was not acceptable for 1000 nM (up to 20.4% and 17.9%, respectively). There was good precision for 14N-nitrite (4.76 to 10.9%) and 15N-nitrite (2.41 to 9.17%) in water and DMEM. Precision for 14N-nitrate (6.32 to 9.43%) and 15N-nitrate (5.16 to 8.51%) was high at 100 nM and lower, but was unacceptably low for the 1000 nM concentration (up to 19.2 and 15.6%, respectively). The results demonstrate that using 15N labeled analytes improves accuracy and precision compared to the 14N analytes. Nitrate has very low recovery above 100 nM, and should not be quantified at high nanomolar to micromolar concentrations without further method validation. This method is precise and accurate for the measurement of 15N-nitrite and 15N-nitrate in both water and DMEM.
Table 1.
Three concentrations (10 nM, 100 nM, 1000 nM) of 15NO2-, NO2-, 15NO3-, and NO3- were spiked into water (n=10) and DMEM (n=10), and the concentration was calculated by comparing to a seven-point concentration curve. Accuracy is determined as the percentage deviation of calculated concentration from the nominal concentrations. Precision is determined as the relative standard deviation (RSD) of the 10 measurements. Blank values for 14N-nitrite and 14N-nitrate in DMEM and water are provided.
| Analyte | Isotope | Matrix | Concentration (nM) | Measured Concentration (nM) | Accuracy (%) | Precision (RSD, %) |
|---|---|---|---|---|---|---|
| Nitrite | 14N | Water | Blank | 91 | N/A | 8.71 |
| 10 | 9.22 | −7.80 | 9.43 | |||
| 100 | 96.3 | −3.70 | 5.38 | |||
| 1000 | 926 | −7.40 | 6.38 | |||
| DMEM | Blank | 339 | N/A | 10.3 | ||
| 10 | 9.25 | −7.50 | 10.9 | |||
| 100 | 101 | 1.00 | 4.76 | |||
| 1000 | 1060 | 6.00 | 9.06 | |||
| 15N | Water | 10 | 9.49 | −5.10 | 9.17 | |
| 100 | 94.7 | −5.30 | 2.41 | |||
| 1000 | 964 | −3.60 | 5.39 | |||
| DMEM | 10 | 10.1 | 1.00 | 4.42 | ||
| 100 | 96 | −4.00 | 3.07 | |||
| 1000 | 944 | −5.60 | 6.02 | |||
| Nitrate | 14N | Water | Blank | 141 | N/A | 12.3 |
| 10 | 9.43 | −5.70 | 6.32 | |||
| 100 | 92.7 | −7.30 | 10.4 | |||
| 1000 | 841 | −15.9 | 17.6 | |||
| DMEM | Blank | 467 | N/A | 13.2 | ||
| 10 | 9.31 | −6.90 | 8.59 | |||
| 100 | 94.7 | −5.30 | 9.43 | |||
| 1000 | 796 | −20.4 | 19.2 | |||
| 15N | Water | 10 | 9.61 | −3.90 | 7.26 | |
| 100 | 95.3 | −4.70 | 8.51 | |||
| 1000 | 884 | −11.6 | 15.6 | |||
| DMEM | 10 | 10.1 | 1.00 | 5.16 | ||
| 100 | 98.9 | −1.10 | 6.48 | |||
| 1000 | 821 | −17.9 | 12.4 |
Neither nitrite nor nitrate was found to interfere with each other. Percent recovery of nitrite and nitrate in the presence of the other analyte was 97% ± 4 and 98% ± 5, respectively. This demonstrates that we can measure nitrate and nitrite in a single sample without compromising the quantitation. Phenol-red free DMEM culture media with 1% FBS and PS was found to not be a source of interference and caused minimal matrix effects. There was high recovery of spiked 15N-nitrite and 15N-nitrate within their linear range, n=10. The percent recovery of nitrite and nitrate in DMEM for all concentration points was 91% ± 3 and 99% ±11, respectively. This demonstrates that we can analyze nitrite and nitrate in cell culture medium with minimal sample preparation.
Both NADPH and nitrate reductase were added individually to 15N-nitrite standards (5 points, 1 nM- 1 μM) to determine whether they would interfere with mass spectrometric detection of nitrite. NADPH and nitrate reductase did not affect either derivatization of nitrite, or mass spectrometric quantification of NAT. Samples incubated with 10 μL of 120 μM NADPH had a percent recovery of 100% ± 5, and samples with 10 μL of 1 U/mL nitrate reductase had a percent recovery of 99% ± 3. This indicates that at the concentrations tested, the presence of NADPH and nitrate reductase will not interfere with the recovery of nitrite from nitrate. In both water and DMEM, there was greater than 99% conversion of nitrate into nitrite within the linear range.
We quantified the concentration of nitrite in milli-Q water, 200 mM HEPES, phenol red-free DMEM with 1% FBS, and phenol red-free DMEM with 1% FBS that was used to culture 5 × 106 EA.hy926 cells in 2 mL medium for 24 hours at 37°C. The results are reported in Table 2. Background levels of nitrate exceeded our linear range of measurement, so these values are not reported. These high levels of nitrite and nitrate in both water and media demonstrate the necessity of using isotope labeling in cell culture experiments.
Table 2.
Nitrite was measured in 1) reverse osmosis water filtered with Millipore water filter, 2) 200 mM HEPES made from Milli-Q water, 3) phenol red-free Dulbecco's modified Eagle's medium (DMEM) with 1% FBS, and 4) phenol red-free DMEM with 1% FBS that was used to culture 5 × 106 EA.hy926 cells in 2 mL medium for 24 hours at 37°C. Nitrate was not measured because all values were above the linear range.
| Nitrite (nM) | |
|---|---|
| Millipore water | 91 ± 13 |
| 200 mM HEPES2 | 103 ± 15 |
| DMEM – blank3 | 339 ± 28 |
| DMEM - EA.hy9264 | 461 ± 61 |
Value are mean ± standard deviation., n=6
3.1 GTN dose-response: nitrate tolerant and control
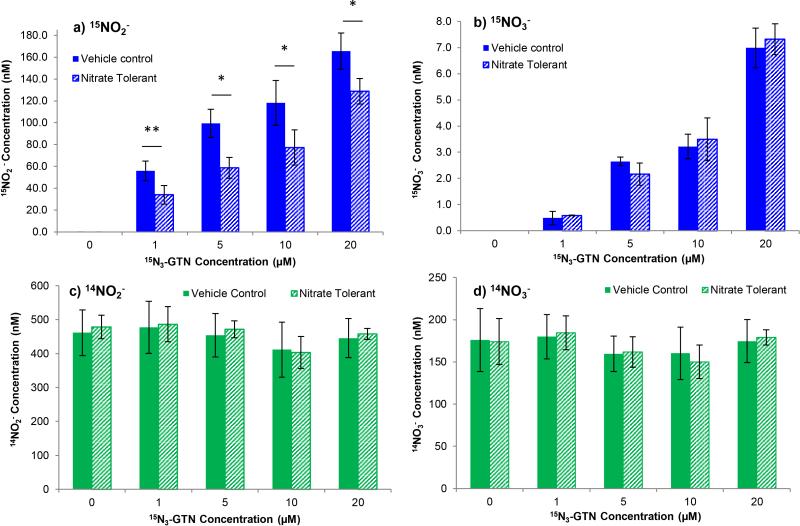
Nitrate tolerance was induced in EA.hy926 cells with a 5-hour pre-treatment with GTN. The endothelial cells bioactivated GTN to produce nitrite, whereas nitrate was not a significant product. Representative chromatograms are in Figure 4. A linear dose-response for 15N-nitrite and 15N-nitrate production was measured. GTN pre-treatment caused decreased production of 15N-nitrite compared to the vehicle control, at all 4 concentrations tested (1, 5, 10, 20 μM) (P= 0.006, 0.014, 0.047, 0.029) (Figure 5a). However, 15N-nitrate production was not altered by pre-treatment with GTN (P>0.05 for all concentration points) (Figure 5b). The concentration of nitrate was near the LLOQ, indicating that nitrate is not a significant metabolic product of GTN. There was no difference in 14N-nitrite and 14N-nitrate concentration between nitrate tolerant cells and control cells, demonstrating that there was no competition between GTN and 15N3-GTN that would invalidate results (Figure 5c and 5d). 14N-nitrite levels were similar for all concentration points, ranging from 411 to 487 nM, whereas 14N-nitrate was 150 to 184 nM. The lower levels of 14N-nitrate can be explained by the observation that these values were above our detection limit for nitrate, and thus suffer from incomplete conversion. This demonstrates that EA.hy926 cells can become nitrate tolerant, and may be a model for measuring the development of nitrate tolerance. This is a significant finding because it suggests that nitrate tolerance develops at the level of GTN bioactivation. The results indicate that the enzymes that are responsible for GTN metabolism may be inactivated or damaged by continuous GTN treatment.
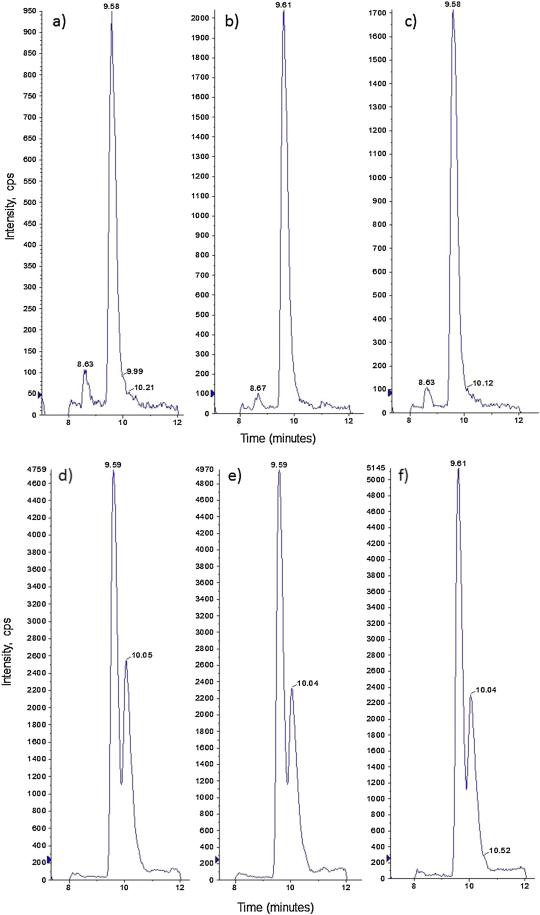
Figure 4.
Representative extracted ion-chromatograms (XIC) of 2,3-naphthotriazole (NAT). 15N-NAT (m/z 171.1→115.1) XIC is presented in a) 15NO2− standard in DMEM, b) 20 μM 15N3-GTN (vehicle control), and c) 20 μM 15N3-GTN (pre-treatment with GTN). These data demonstrate that pre-treatment with GTN resulted in reduced metabolism of 15N3-GTN into 15NO2−. 14N-NAT (m/z 170.1→115.1) is presented in d) 14N-nitrite standard in DMEM, e) 20 μM 15N3-GTN (Vehicle Control), and f) 20 μM 15N3-GTN (pre-treatment with GTN). These data demonstrate that pre-treatment with GTN did not cause a change in 14NO2− concentration.
Figure 5.
15NO2− and 15NO3− production from 15N3-GTN treatment in EA.hy926 endothelial cells. Cells were pre-treated with 20 μM glyceryl trinitrate (GTN), washed, and then treated with 15N3-GTN. Acetonitrile (5 μL) was used as a vehicle control. a) 15NO2− levels decreased in response to GTN pre-treatment, n=3 (P<0.05 at all concentration points). A dose-response curve with 15N3-GTN demonstrated linear production of 15NO2− in nanomolar levels. b) GTN pre-treatment had no significant effect on 15NO3− levels (P>0.5 at all concentration points). A dose-response curve with 15N3-GTN demonstrated linear production of 15NO3− in low nanomolar levels, indicating very low conversion of nitrite into nitrate. The reduction of nitrite to nitrate may occur nonenzymatically. c) 14NO2− concentration and d) 14NO3− concentration was not significantly affected (P>0.05 at all concentration points). This demonstrates that the pre-treatment was completely washed away and did not compete with 15N-GTN. The high level of 14NO2− and 14NO3− is a result of background levels of nitrite and nitrate, potentially from NO synthesis from L-arginine.
These results are supported by the MTT cell viability assay, which determined that 24-hour treatment with a wide range of GTN treatments (0.1 μM – 50 μM) does not cause significant levels of cell death (Supplemental Information Figure 2). In addition, the ATP Glo Assay to measure cell proliferation showed no significant differences between controls and multiple concentrations of GTN (1, 5, 10, 20 μM) (Supplemental Information Figure 3). Both assays demonstrated minor, yet insignificant, increases in cell viability/proliferation with GTN treatment. Therefore, the reduction in nitrite and nitrate production in nitrate tolerant cells is not caused by a reduced amount of viable cells.
4. Discussion
The results demonstrate that our method of measuring nitrite and nitrate is highly sensitive, can differentiate between isotopologues of nitrite and nitrate, is accurate and precise, is free of common interferences, and requires minimal sample preparation. HPLC methods that use fluorescence detection have detection limits in low nanomolar levels, however they are not able to distinguish between newly formed and pre-existing (i.e., background) nitrite and nitrate [25] [27],[28]. The ability of these methods to measure minute changes in nitrite and nitrate is questionable, particularly in light of the fact that we measured high nanomolar background levels of nitrite in cell culture media. Small changes in NO, and its metabolites, can have large physiological effects. We demonstrated that LC-MS/MS can measure changes in 15N-nitrite and 15N-nitrate concentrations in low nanomolar concentrations, which would undoubtedly be obscured without isotope labeling. Measuring NAT with LC-MS/MS, instead of by fluorescence, has the additional benefit of increased stability, because fluorescence quenching by biological components is not a concern. Though the merit of measuring nitrite and nitrate as surrogates for NO synthesis has been recently questioned [29], it remains the most suitable and practical method to assess NO production from GTN.
Despite the numerous benefits of measuring nitrite and nitrate with LC-MS/MS, there is one limitation that should be considered. This method requires for separate analysis of nitrite and nitrate, which increases sample preparation and analysis time. For many applications, this may not be a concern. We have demonstrated that nitrite is a better marker of GTN metabolism than nitrate, thus it may suffice to measure nitrite only. Human and mammalian in vivo studies indicate that circulating nitrite, rather than nitrate, reflect endothelial-dependent NO synthesis [30]. We acknowledge that methods that allow for simultaneous quantification of nitrite and nitrate may be preferable for human biological fluids. The GCMS method developed by Tsikas and coworkers, which uses pentafluorobenzyl derivatization, allows for the measurement of isotopes and has the benefit of being able to measure nitrite and nitrate simultaneously [31]. However, this GCMS method requires extensive sample preparation. The sensitivity of our LC-MS/MS method, as well as the ability to use isotope labeling, is necessary for mechanistic cell cultures studies.
One source of concern with this method could be that 15N-NAT and 14N-NAT only differ by a single mass unit. In theory, this could lead to issues with quantitation and interference with natural isotope abundance. We determined that 15N-NAT is not detectable in cell culture samples with no treatment, indicating that the natural abundance of 15N (0.36%) will not affect quantification. In addition, natural abundance of 13C could not be mistaken for 15N labeling because of the fragmentation pattern of NAT. There is a significant mass fragment peak at m/z 116.0, which results from the natural abundance of 13C1 labeled NAT (9.7%). Since we measure the m/z 171.1→115.1 transition for 15N-NAT, 13C labeling is excluded from detection.
The cell culture results demonstrate that endothelial cells are capable of metabolizing GTN into nitrite. Pre-treatment with GTN resulted in nitrate tolerance, as evidenced by reduced 15N-nitrite production. However, only low nanomolar levels of 15N-nitrate were measured, suggesting that it is not a significant metabolic product of GTN. These results indicate that this may be a good model to determine the mechanism of GTN bioactivation, as well as the cause of nitrate tolerance. Measuring nitrite as an endpoint of GTN metabolism is a valuable tool to determine the cause of nitrate tolerance. We hypothesize that inactivation of GTN-metabolizing enzymes, such as xanthine oxidase (XO), is the cause of nitrate tolerance.
5. Conclusion
LC-MS/MS is a sensitive and reproducible method of quantifying isotopologues of nitrite and nitrate, which will bring new insight into the phenomena of nitrate tolerance. We demonstrated that nitrate tolerance is associated with a significant decrease in nitrite production in human endothelial cells, which indicates inactivation of GTN-metabolizing enzymes.
Supplementary Material
Acknowledgments
This research was supported in part by the National Institute of Health Grant T32ES007060, the College of Pharmacy, and the Linus Pauling Institute at Oregon State University. We would like to acknowledge Jeffrey Morré for his help with mass spectrometry, Dr. Cristobal Miranda for his assistance with cell culture techniques, and Eunice Lee, Erik McEntire, and Eleonso Cristobal for their technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moncada S, Higgs EA. Endogenous nitric oxide: physiology, pathology and clinical relevance. Eur J Clin Invest. 1991;21:361–74. doi: 10.1111/j.1365-2362.1991.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 2.Culotta E, Koshland DE. NO news is good news. Science. 1992;258:1862–5. doi: 10.1126/science.1361684. [DOI] [PubMed] [Google Scholar]
- 3.Doel JJ, Godber BL, Goult TA, Eisenthal R, Harrison R. Reduction of organic nitrites to nitric oxide catalyzed by xanthine oxidase: possible role in metabolism of nitrovasodilators. Biochem Biophys Res Commun. 2000;270:880–5. doi: 10.1006/bbrc.2000.2534. [DOI] [PubMed] [Google Scholar]
- 4.Godber BL, Doel JJ, Sapkota GP, Blake DR, Stevens CR, Eisenthal R, Harrison R. Reduction of nitrite to nitric oxide catalyzed by xanthine oxidoreductase. J Biol Chem. 2000;275:7757–63. doi: 10.1074/jbc.275.11.7757. [DOI] [PubMed] [Google Scholar]
- 5.Millar TM, Stevens CR, Benjamin N, Eisenthal R, Harrison R, Blake DR. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998;427:225–8. doi: 10.1016/s0014-5793(98)00430-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Naughton D, Winyard PG, Benjamin N, Blake DR, Symons MC. Generation of nitric oxide by a nitrite reductase activity of xanthine oxidase: a potential pathway for nitric oxide formation in the absence of nitric oxide synthase activity. Biochem Biophys Res Commun. 1998;249:767–72. doi: 10.1006/bbrc.1998.9226. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Samouilov A, Liu X, Zweier JL. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrate reduction: evaluation of its role in nitrite and nitric oxide generation in anoxic tissues. Biochemistry. 2003;42:1150–9. doi: 10.1021/bi026385a. [DOI] [PubMed] [Google Scholar]
- 8.Ellis G, Adatia I, Yazdanpanah M, Makela SK. Nitrite and nitrate analyses: a clinical biochemistry perspective. Clin Biochem. 1998;31:195–220. doi: 10.1016/s0009-9120(98)00015-0. [DOI] [PubMed] [Google Scholar]
- 9.Czapski G, Goldstein S. The role of the reactions of .NO with superoxide and oxygen in biological systems: a kinetic approach. Free Radic Biol Med. 1995;19:785–94. doi: 10.1016/0891-5849(95)00081-8. [DOI] [PubMed] [Google Scholar]
- 10.Marzinzig M, Nussler AK, Stadler J, Marzinzig E, Barthlen W, Nussler NC, Beger HG, Morris SM, Brückner UB. Improved methods to measure end products of nitric oxide in biological fluids: nitrite, nitrate, and S-nitrosothiols. Nitric Oxide. 1997;1:177–89. doi: 10.1006/niox.1997.0116. [DOI] [PubMed] [Google Scholar]
- 11.Tsikas D. Methods of quantitative analysis of the nitric oxide metabolites nitrite and nitrate in human biological fluids. Free Radic Res. 2005;39:797–815. doi: 10.1080/10715760500053651. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg JO, Weitzberg E. NO generation from nitrite and its role in vascular control. Arterioscler Thromb Vasc Biol. 2005;25:915–22. doi: 10.1161/01.ATV.0000161048.72004.c2. [DOI] [PubMed] [Google Scholar]
- 13.Münzel T, Daiber A, Mülsch A. Explaining the phenomenon of nitrate tolerance. Circ Res. 2005;97:618–28. doi: 10.1161/01.RES.0000184694.03262.6d. [DOI] [PubMed] [Google Scholar]
- 14.Harrison R. Structure and function of xanthine oxidoreductase: where are we now? Free Radic Biol Med. 2002;33:774–97. doi: 10.1016/s0891-5849(02)00956-5. [DOI] [PubMed] [Google Scholar]
- 15.Abou-Mohamed G, Johnson JA, Jin L, El-Remessy AB, Do K, Kaesemeyer WH, Caldwell RB, Caldwell RW. Roles of superoxide, peroxynitrite, and protein kinase C in the development of tolerance to nitroglycerin. J Pharmacol Exp Ther. 2004;308:289–99. doi: 10.1124/jpet.103.056119. [DOI] [PubMed] [Google Scholar]
- 16.Jobgen WS, Jobgen SC, Li H, Meininger CJ, Wu G. Analysis of nitrite and nitrate in biological samples using high-performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:71–82. doi: 10.1016/j.jchromb.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Di Matteo V, Esposito E. Methods for the determination of nitrite by high-performance liquid chromatography with electrochemical detection. J Chromatogr A. 1997;789:213–9. doi: 10.1016/s0021-9673(97)00851-0. [DOI] [PubMed] [Google Scholar]
- 18.Tsikas D, Böhmer A, Mitschke A, Araujo P. Accurate measurement of nitrate, nitrite, and S- nitrosothiols in biological samples by mass spectrometry. Free Radic Biol Med. 2013;65:301–4. doi: 10.1016/j.freeradbiomed.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Shin S, Fung HL. Evaluation of an LC-MS/MS assay for 15N-nitrite for cellular studies of L-arginine action. J Pharm Biomed Anal. 2011;56:1127–31. doi: 10.1016/j.jpba.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batugo MR, Nardi CD, Johnson TR, Biondi S, Impagnatiello F. Determination of exogenous nitric oxide using stable labeled nitrogen coupled with liquid-chromatography-mass spectrometry. AAPS J. 2009 Abstract #1291. [Google Scholar]
- 21.Wiersma JH. 2,3-Diaminonaphthalene as a Spectrophotometric and Fluorometric Reagent for the Determination of Nitrite Ion. Anal. Lett. 1970:123–132. [Google Scholar]
- 22.May JM, Qu ZC, Li X. Nitrite generates an oxidant stress and increases nitric oxide in EA.hy926 endothelial cells. Free Radic Res. 2004;38:581–9. doi: 10.1080/10715760410001688366. [DOI] [PubMed] [Google Scholar]
- 23.Hinz B, Schröder H. Vitamin C attenuates nitrate tolerance independently of its antioxidant effect. FEBS Lett. 1998;428:97–9. doi: 10.1016/s0014-5793(98)00506-7. [DOI] [PubMed] [Google Scholar]
- 24.Wu GY, Brosnan JT. Macrophages can convert citrulline into arginine. Biochem J. 1992;281(Pt 1):45–8. doi: 10.1042/bj2810045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Meininger CJ, Wu G. Rapid determination of nitrite by reversed-phase high-performance liquid chromatography with fluorescence detection. J Chromatogr B Biomed Sci Appl. 2000;746:199–207. doi: 10.1016/s0378-4347(00)00328-5. [DOI] [PubMed] [Google Scholar]
- 26.Misko TP, Schilling RJ, Salvemini D, Moore WM, Currie MG. A fluorometric assay for the measurement of nitrite in biological samples. Anal Biochem. 1993;214:11–6. doi: 10.1006/abio.1993.1449. [DOI] [PubMed] [Google Scholar]
- 27.Gharavi N, El-Kadi AO. Measurement of nitric oxide in murine Hepatoma Hepa1c1c7 cells by reversed phase HPLC with fluorescence detection. J Pharm Pharm Sci. 2003;6:302–7. [PubMed] [Google Scholar]
- 28.Woitzik J, Abromeit N, Schaefer F. Measurement of nitric oxide metabolites in brain microdialysates by a sensitive fluorometric high-performance liquid chromatography assay. Anal Biochem. 2001;289:10–7. doi: 10.1006/abio.2000.4893. [DOI] [PubMed] [Google Scholar]
- 29.Tsikas D. Circulating and excretory nitrite and nitrate: their value as measures of nitric oxide synthesis, bioavailability and activity is inherently limited. Nitric Oxide. 2015;45:1–3. doi: 10.1016/j.niox.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Kleinbongard P, Dejam A, Lauer T, Jax T, Kerber S, Gharini P, Balzer J, Zotz RB, Scharf RE, Willers R, Schechter AN, Feelisch M, Kelm M. Plasma nitrite concentrations reflect the degree of endothelial dysfunction in humans. Free Radic Biol Med. 2006;40:295–302. doi: 10.1016/j.freeradbiomed.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 31.Tsikas D. Simultaneous derivatization and quantification of the nitric oxide metabolites nitrite and nitrate in biological fluids by gas chromatography/mass spectrometry. Anal Chem. 2000;72:4064–72. doi: 10.1021/ac9913255. [DOI] [PubMed] [Google Scholar]
- 32.Dikalov S, Fink B, Skatchkov M, Sommer O, Bassenge E. Formation of Reactive Oxygen Species in Various Vascular Cells During Glyceryltrinitrate Metabolism. J Cardiovasc Pharmacol Ther. 1998;3:51–62. doi: 10.1177/107424849800300107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.