Abstract
The life-cycle of Endogenous retroviruses (ERVs), also called long terminal repeat (LTR) retrotransposons, begins with transcription by RNA Pol II followed by reverse transcription and re-integration into the host genome. While most ERVs are relics of ancient integration events, “young” proviruses competent for retrotransposition- found in many mammals but not humans-represent an ongoing threat to host fitness. As a consequence, several restriction pathways have evolved to suppress their activity at both transcriptional and post-transcriptional stages of the viral life-cycle. Nevertheless, accumulating evidence has revealed that LTR sequences derived from distantly related ERVs have been exapted as regulatory sequences for many host genes in a wide range of cell types throughout mammalian evolution. Here, we focus on emerging themes from recent studies cataloguing the diversity of ERV LTRs acting as important transcriptional regulatory elements in mammals and explore the molecular features that likely account for LTR exaptation in developmental and tissue-specific gene regulation.
Introduction
Retrotransposons, which replicate via a transcription and reverse-transcription ‘copy-and-paste’ mechanism, account for greater than 40% of the human and mouse genomes (Venter et al., 2001; Waterston et al., 2002). These parasitic sequences can be classified into two major groups. Those lacking long-terminal repeats (LTRs), including long and short interspersed nuclear elements (LINEs and SINEs, respectively) and SINE-Variable number tandem repeat-Alu (SVA) elements, comprise ~30-35% of the genome, while those with LTRs, termed endogenous retroviruses (ERVs) or LTR retrotransposons comprise ~8% and 10% of the human and mouse genomes, respectively (Cordaux and Batzer, 2009; Friedli and Trono, 2015; Stocking and Kozak, 2008) (Figure 1A). ERVs are the descendants of exogenous retroviruses that integrated into the genome of germ cells. Most subsequently lost the ability to exit the host cell. Thus, those ERVs that may be defective for infection but are still competent for retrotransposition expand in their host genome by vertical transmission (Mager and Stoye, 2015; Magiorkinis et al., 2012). In addition to their 5’ and 3’ LTRs, which are identical in sequence following reverse transcription and integration, autonomous proviral elements typically harbour several ORFs that encode proteins essential for viral replication, including gag, which encodes a group-specific retroviral antigen and pol, which encodes the reverse transcriptase (Figure 1B). A third ORF encodes an envelope protein (env), although the vast majority of ERVs have truncated or mutated env sequences.
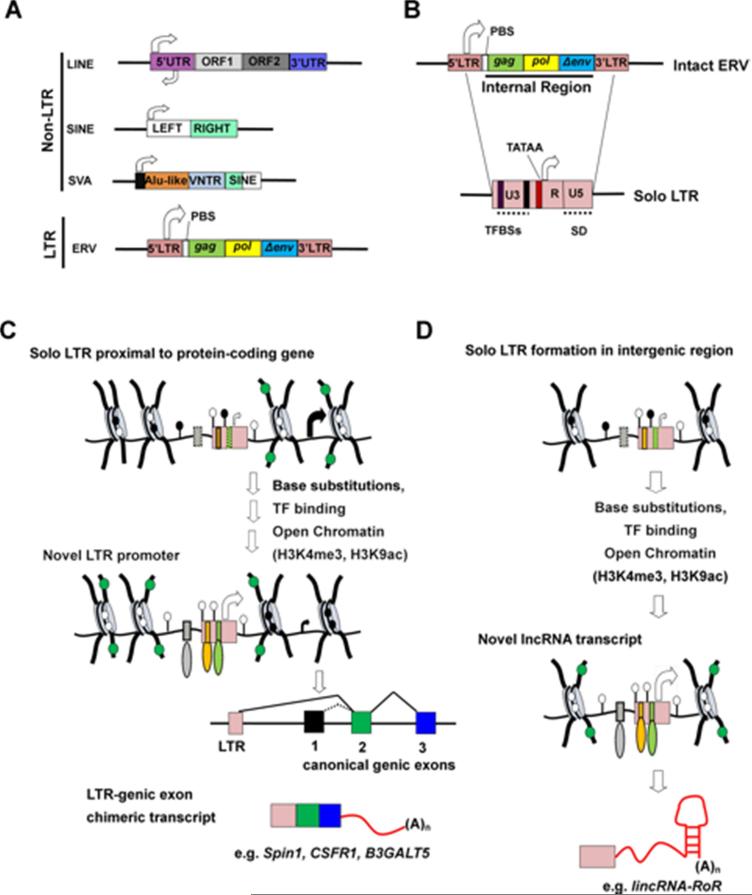
Figure 1. Structure of an intact ERV and solo LTR and the molecular mechanisms of LTR exaptation as protein-coding or lncRNA promoters.
(A) Schematic of non-LTR retrotransposons which include SINEs (i.e. Alus), LINEs (i.e. L1Hs), and SVAs (in humans) and LTR retrotransposons, which include many lineage/species-specific subfamilies. Most LINE elements are truncated at the 5’ end, thus lacking the 5’UTR promoter and TSS. (B) Full-lengthERVs have 5’ and 3’ LTRs, and an “internal” region that includes a primer-binding site (PBS) involved in priming reverse transcription, and retroviral ORFs gag, pol and a truncated or mutated env gene (Δenv). Recombination between 5’ and 3’ LTRs deletes the internal region, generating ‘solo’ LTRs (not to scale), which consist of unique 3’ (U3) and 5’ (U5) regions and a regulatory region (R) containing the TSS (white arrow). LTRs often harbour different combinations of TFBSs (green and orange rectangles) in addition to core Pol II promoter elements (such as TATA box shown in red) and may also contain a splice donor (SD) site (dashed line) within the U5 region. (C) LTR exaptation as a protein-coding gene promoter. In a developmental/tissue specific context, particularly in cell types undergoing epigenetic reprogramming (e.g. early embryo, placenta or germline) a hypomethylated solo LTR (pink rectangle) 5’ of a protein-coding gene (or in an intragenic region) may become exapted as a novel promoter (black circles represent DNA methylation). The process may involve base substitutions in a neighbouring near-consensus TFBS (grey rectangle with dash outline) and a near-consensus site within the LTR (green rectangle with dash outline), which then form a positive genetic interaction with another LTR-derived TFBS (orange rectangle) a mechanism termed ‘epistatic capture’ (Emera and Wagner, 2012a). This leads to synergy in the binding of several TFs (grey, orange and green ovals), deposition of “active” histone modifications, such as H3K4me3 and H3K9ac (green circles) and robust transcription initiation from the LTR-derived promoter. The canonical genic promoter may be DNA methylated as a consequence of such transcription. This process generates LTR-genic exon chimeric transcripts, where exon 1 is derived from the LTR and splicing occurs from the internal LTR SD (or from a cryptic SD site in the intervening genomic sequence downstream of the active LTR) to the first downstream exon with a splice-acceptor site, generally exon 2. Examples of such chimeric transcripts include Spin1 in mouse and CSFR1 and B3GALT5 in human. Arrow sizes indicate relative level of transcription from each promoter. (D) LTR exaptation as a promoter for a novel lncRNA. Through a process as in (C), a newly formed intergenic solo LTR without an SD site could initiate de novo lncRNA transcription, forming a novel lncRNA gene. An example of this is the lincRNA-RoR transcript.
While the general threat of insertional mutagenesis due to unmitigated ERV transcription and subsequent retrotransposition is minimized by epigenetic mechanisms, including DNA methylation, histone lysine methylation and small non-coding RNAs (Castro-Diaz et al., 2015; Wolf et al., 2015a), recent studies have revealed that ERVs have also played a prominent role in expanding the regulatory landscape of mammalian genomes (Cordaux and Batzer, 2009; Feschotte and Gilbert, 2012; Gifford et al., 2013; Jern and Coffin, 2008; Rebollo et al., 2012a). The ‘controlling element’ theory that TEs may participate in gene regulation was postulated over 60 years ago by Barbara McClintock (McClintock, 1950) and was later expanded upon by Britten and Davidson's gene battery hypothesis (Britten and Davidson, 1969). Genome-wide studies have indeed confirmed that species specific ERV LTRs exert regulatory effects on genes in many cell types during development to modulate the transcriptome (Cowley and Oakey, 2013; Gifford et al., 2013; Isbel and Whitelaw, 2012; Robbez-Masson and Rowe, 2015). However, the molecular mechanisms whereby these heterologous sequences are converted into regulatory elements for host genes remain obscure. Here, we highlight recent studies that have advanced our understanding of how LTR sequences are exapted into species-specific cis-regulatory elements. We begin by exploring why LTR retrotransposons are particularly suitable for co-option by the host and subsequently review recent experimental evidence supporting a model of reiterative exaptation of LTRs in mammals as tissue-specific promoters or enhancers for protein coding genes and long noncoding RNAs (lncRNAs). We conclude with a discussion of recent functional studies of the role of specific exapted LTRs in gene regulation and outstanding questions to be addressed in future studies.
Solo LTRs: autonomous regulatory modules
Several studies in mammals indicate that ERVs have been more frequently exapted as cis-regulatory elements relative to other transposable elements (Chuong et al., 2013; Jacques et al., 2013; Kannan et al., 2015; Kapusta et al., 2013; Kelley and Rinn, 2012; Sundaram et al., 2014; Xie et al., 2013). Consistent with these observations, ERVs have been reported to evolve more rapidly than other transposable elements, as evidenced by orthologous ERVs in humans and chimpanzees exhibiting signatures of directional selection since the human-chimp divergence ~5 million years ago (Gemmell et al., 2015). These observations are unlikely to be explained by the integration sites of ERVs, as retrotransposons of all types are most prevalent in intergenic regions and older LTR and LINE elements are underrepresented within 5 kb of gene promoters, perhaps due to their negative impact on expression of proximal genes and in turn host fitness (Medstrand et al., 2002). Rather, the frequent co-option of ERV sequences for gene regulation may be due to the relatively high probability of recombination between the 5’ and 3’ LTRs of intact proviruses, which deletes the internal region, leaving a single or ‘solo’ LTR at the original integration site (Belshaw et al., 2007) (Figure 1B). Recombination between 5’ and 3’ LTRs has generated an estimated 577,000 ‘solo’ LTRs in the human genome, representing the vast majority of annotated ERV sequences (Friedli and Trono, 2015). Notably, both full-length intact ERVs and solo LTRs are under-represented specifically in the sense orientation within introns, likely reflecting the generally deleterious effects of insertion of polyadenylation signals encoded by LTRs (Medstrand et al., 2002; Smit, 1999). As LTRs harbour the regulatory regions required for proviral transcription, generally including combinations of transcription factor binding sites (TFBSs), they have the intrinsic capacity to autonomously recruit cellular TFs and in turn to maximize transcription of proviral mRNA in specific cell types. Indeed, LTR-derived TFBSs are now known to have contributed up to ~20% of functional binding sites for many TFs in human and mouse (Sundaram et al., 2014), including p53, OCT4, SOX2 and NANOG (Bourque et al., 2008; Kunarso et al., 2010; Wang et al., 2007). In contrast with the regulatory properties of LTRs, the majority of LINE1 elements are truncated at the 5’end, which removes the regulatory region and canonical TSS (Figure 1A) of these RNA Pol II-driven elements, rendering them transcriptionally ‘dead on arrival’ (Cordaux and Batzer, 2009).
The presence of a conserved splice donor (SD) site within some classes of LTRs also likely contributes to the propensity for LTRs from specific families to be exapted as alternative promoters (Figure 1B). The consensus sequence of MaLR LTRs for example, including the MT subtypes (See Table 1 for classification of LTRs exapted as regulatory elements discussed here), harbor a conserved SD site that is utilized in many MT-initiated chimeric transcripts in oocytes (Peaston et al., 2004). Similarly, a primate-specific MaLR LTR, THE1B, which harbours an intact SD site, is aberrantly reactivated in Hodgkin's lymphoma and drives expression of CSF1R transcripts (Lamprecht et al., 2010). Alternatively, mutations within LTRs may generate novel SD sites, as is the case for the highly-expressed oocyte-specific Spin1 transcript, also driven by an MT LTR (Peaston et al., 2004). Furthermore, at specific loci, cryptic SD sites may be present in the flanking genomic sequence downstream of a transcriptionally active LTR. Regardless, the presence of an SD site within or immediately downstream of the LTR minimizes the length of the 5’ UTR. This decreases the likelihood that the transcript will contain a cryptic start codon upstream of the canonical start codon, thus preserving the native ORF in the resulting chimeric mRNA, and may stabilize the nascent RNA, as SD sites may compete with termination signals (Wu and Sharp, 2013).
Table 1.
Examples of LTRs exapted as regulatory elements in human and mouse and their classification.
| Species | ERV Class | ERV Family | Examples of LTR Subtypes |
|---|---|---|---|
| Mouse | I | ERV1 | LTR17 |
| I | ERV3 | MER77 | |
| II | ERVK | LTR10C, LTR10B, LTR13D5, BGLII | |
| III | MaLR | MT-A, MT-B, MT-C, ORR1A0 | |
| III | ERVL | MT2, MT2B, MT2C | |
| Humans | I | ERV1 | LTR7, MER39, MER41, LTR12C |
| I | ERV3 | MER21C | |
| I | ERV9 | LTR9 | |
| II | ERVK | LTR5 | |
| II | ERVK3 | LTR3B | |
| II | ERVK14 | LTR14B | |
| III | MaLR | THE1A, THE1B, THE1C, MER39 | |
| III | ERVL | MLT2A1, MLT2B3, LTR16A | |
Many intact ERVs are targeted for transcriptional silencing by the rapidly diversifying family of Krüppel-associated box zinc finger proteins (KRAB-ZFPs), which interact with the co-repressor KAP1 and the histone H3 lysine 9 (H3K9) methyltransferase SETDB1 (Liu et al., 2014; Matsui et al., 2010; Rowe et al., 2010; Turelli et al., 2014; Wolf et al., 2015b). Indeed, ChIP-seq analysis in mouse ESCs reveals that the solo LTRs of a subset of ERV families, including IAP solo LTRs, are marked by H3K9me3 (Karimi et al., 2011), indicating that for some ERVs, the LTR itself may be bound by specific KRAB-ZFPs. However, while the binding sites of only a few of the >300-400 KRAB-ZFPs in humans and mice have been studied, the majority characterized thus far recognize internal ERV sequences, including the primer binding site, 5’UTR, gag and 3’ polypurine tract regions (Rowe et al., 2010; Sadic et al., 2015; Wolf and Goff, 2009; Wolf et al., 2015b, Ecco et al., 2016). Since solo LTRs lack these internal sequences, they may escape the KRAB-ZFP/KAP1 silencing machinery directed at full-length elements, facilitating their exaptation as positive regulatory elements by the host.
Once all members of a particular ERV family are effectively silenced by the KRAB-ZFP/KAP1 repression system, the accumulation of inactivating mutations in replication competent proviruses, i.e. in functional viral protein-coding regions, would over time relieve the positive selective pressure for KRAB-ZFP recognition, allowing mutations to accumulate within the relevant KRAB-ZFP gene, ultimately modifying or ablating the DNA-binding specificity of the encoded protein regardless of whether it binds in the LTR or internal region. The remaining replication-incompetent full-length proviruses and solo-LTRs derived from these elements would no longer be recognized by a specific KRAB-ZFP, allowing for selection of LTRs as promoter or enhancer elements of nearby genes (Friedli and Trono, 2015). This does not exclude the possibility that there may be purifying selection of KRAB-ZFP binding sites within otherwise decaying ERV internal regions or LTRs, allowing for ERV exaptation for silencing of nearby genes (Ecco et al., 2016).
Consistent with the presence of TFBSs and their propensity to evade epigenetic silencing, many ERVs and LTRs exhibit tissue-specific expression patterns, especially during embryonic and germline development (Goke et al., 2015; Grow et al., 2015; Jacques et al., 2013; Okahara et al., 2004; Pavlicev et al., 2015; Peaston et al., 2004). Indeed, ERVs have likely been under selection to increase their odds of successful retrotransposition and vertical transmission and therefore exhibit high levels of transcription in the early embryo and reproductive tissues, including primordial germ cells (PGCs) and oocytes (Cohen et al., 2009; Peaston et al., 2004). Thus it is not surprising that the ERV families present in high copy number are also those competent for expression in the germline. Although H3K9me3 and/or DNA methylation play a role in silencing of ERVs in both undifferentiated and differentiated cell types, specific ERVs likely exploit global reprogramming of epigenetic states, such as during embryonic preimplantation development (Tomizawa et al., 2011; Ziller et al., 2013) or in the placenta (Chuong et al., 2013; Hon et al., 2013; Reiss et al., 2007; Xie et al., 2013) to promote their expression. During these developmental stages, the LTRs that have accumulated mutations that relieve selective pressure for KRAB-ZFP-based silencing in these cell types, or are otherwise not efficiently bound by KRAB-ZFPs due to low level of expression of the relevant KRAB-ZFP or their genomic context, would come under purifying selection for beneficial regulatory effects on neighbouring genes. Thus, the combination of autonomous RNA pol II promoter/enhancer activity conferred by intact TF binding sites and changes in the repertoire of ERVs bound by KRAB-ZFPs over evolutionary time are likely to provide a unique context for exaptation of solo LTRs for tissue-specific gene regulation.
From LTR to genic promoter
In support of this model, a substantial number of LTRs have been reported to function as tissue-specific primary or alternative promoters in a variety of mammalian cell types, including in the early mouse embryo, placenta, human and mouse pluripotent stem cells, mouse erythroid cells and growing mouse oocytes (Buzdin et al., 2006; Cohen et al., 2009; Faulkner et al., 2009; Fort et al., 2014; Karimi et al., 2011; Macfarlan et al., 2012; Mak et al., 2014; Peaston et al., 2004; Veselovska et al., 2015a; Wolf et al., 2015b). Notably, many of the LTRs that have apparently been exapted as genic promoters are not only lineage-specific but also show clear differences in transcriptional activity between cell types in the given species. For example, in mouse zygote and two-cell stage embryos, LTRs from the class III LTR retrotransposon MERVL drive expression of a cohort of stage-specific genes (Evsikov et al., 2004; Macfarlan et al., 2012; Maksakova et al., 2013; Peaston et al., 2004), whereas MaLR and ERVK family LTRs drive expression of many mouse oocyte-specific transcripts (Peaston et al., 2004; Veselovska et al., 2015b). Similarly, in human pluripotent stem cells, LTR7, derived from the primate-specific HERV-H, drives transcription of many pluripotency-associated lncRNAs (Durruthy-durruthy et al., 2016; Lu et al., 2014b; Wang et al., 2013). Furthermore, LTR3B, LTR14B, LTR12C, MLT2A1, THE1A and LTR5_Hs are expressed at discrete stages during the progression of human preimplantation embryo development from the zygote to the morula stage and serve as promoters for a class of previously unannotated transcripts that may serve important functions at these stages (Goke et al., 2015).
Early studies of the role of LTR elements as candidate genic promoters relied on single gene analyses using methods such as 5’RACE or PCR. Subsequently, higher-throughput approaches were developed, including those based on sequence mining of EST or RefSeq databases (Evsikov et al., 2004; van de Lagemaat et al., 2003; Lipatov et al., 2005; Medstrand et al., 2002; Peaston et al., 2004), or the combination of EST data with high-throughput sequencing by capped analysis of gene expression (CAGE) (Faulkner et al., 2009).
With the widespread use of next generation sequencing (NGS) technologies and complementary development of bioinformatics tools to exploit such datasets, novel transcripts, including those expressed at relatively low levels, can now be easily identified and enumerated. Indeed, LTR promoter usage in a given cell type can now be readily inferred genome-wide from RNA-seq data. Paired-end RNA-seq data in particular has been used to identify candidate chimeric transcripts (Karimi et al., 2011; Macfarlan et al., 2012). RNA-seq data has also been employed for de novo transcriptome assembly to identify LTR promoter usage in an unbiased manner in the developing oocyte and to identify novel chimeric transcripts initiating in RLTR10B in mouse testis (Isbel et al., 2015; Veselovska et al., 2015a). Recent technological advances have led to significant increases in library read-depth and standard read lengths, increasing the probability of mapping unique reads within such repetitive elements and in turn the identification of chimeric transcripts showing a broad range of expression levels.
In addition, as active LTR promoters exhibit the same chromatin modification patterns found at active genic promoters, including H3K4me3 and DNase I hypersensitivity, profiling of these features by NGS can also be exploited to identify candidate LTR promoters (Chuong et al., 2013; Jacques et al., 2013; Lynch et al., 2011, 2015; Veselovska et al., 2015b) (Figure 1C). For example, using ChIP-seq for H3K4me3 on cyclic AMP and progesterone-treated human decidualized stromal cells, Lynch et al. (2015) found that ~31% of active promoters mapped in those cells overlap with ancient mammalian TEs, including LTRs (Lynch et al., 2015). Similarly, analysis of DNAse I hypersensitivity data from a large panel of human embryonic, adult and cancer cell lines revealed that up to ~80% of LTRs are located in open chromatin regions in a cell type-specific manner (Jacques et al., 2013) and intersection with ENCODE H3K4me3 ChIP-seq data revealed that a subset of these LTRs are active promoters.
LTRs as tissue-specific genic promoters
Several LTRs derived from ancient proviruses that integrated near genes have likely been co-opted as regulatory elements, as indicated by strong purifying selection (Franchini et al., 2012; Lowe et al., 2007). The paucity of additional cases where LTR promoters/enhancers have been clearly shown to evolve under purifying selection may be due to weak selection or the fact that most instances of detectable LTR-derived regulatory elements are of recent origin (i.e. mouse or primate-specific), limiting the statistical power to observe signatures of purifying selection by sequence comparisons among different lineages. If LTR-driven transcription is beneficial in a specific cell type, persistence of its promoter activity will be under selective pressure and the expression pattern maintained in that lineage. For example, while the Dicer1 gene is driven from a CpG island promoter in most tissues where it is expressed, an oocyte-specific isoform in mice is driven by a rodent-specific intragenic MaLR solo LTR of the MT-C subtype (Flemr et al., 2013) (Table 1). Notably, deletion of the MT-C LTR alternative promoter abolishes Dicer1 expression in the oocyte and causes female sterility, providing strong evidence of the importance of this exapted LTR for host fitness. As this specific LTR is also present in the rat, the ancestral provirus must have integrated prior to the divergence of rats and mice, at least ~25 million years ago (Nei et al., 2001). In contrast with the highly active IAP and ETn/MusD families that are responsible for ~10% spontaneous mutations in laboratory mouse strains, there is no evidence for recent of de novo retrotransposition of MT-C elements (Maksakova et al., 2006; Rebollo et al., 2012b). Nevertheless, LTRs from MT-C as well as other MT subtypes are still clearly transcriptionally active specifically in oocytes, reflecting the innate tissue-specific expression profile of these nonautonomous MaLR elements (Evsikov et al., 2004; Peaston et al., 2004; Veselovska et al., 2015b). Similarly, the human metabolic gene B3GALT5 is expressed in many different tissues, but in the colon, a primate-specific MLT2B3 LTR promoter derived from the ERV-L family is utilized (Dunn et al., 2003).
A particularly dramatic example of the widespread exaptation of a specific LTR subtype in a specific tissue can be found in early mouse embryogenesis, where MT2 LTRs derived from mouse MERVL elements act as promoters for over 500 two-cell stage-specific gene transcripts (Macfarlan et al., 2012; Maksakova et al., 2013). Although the functions of most of these MT2 LTR chimeric transcripts remains to be determined, a subset may serve important roles in early mouse development, such as Tcstv1 and Tcstv3 which control telomere elongation and genome stability (Zhang et al., 2016), As with MT2 LTRs that serve as genic promoters, intact MERVL elements are also transcriptionally active at the zygote and two-cell stage, but are subsequently inactivated, at least in part as a consequence of a more repressive nuclear architecture instated during differentiation from totipotency to pluripotency, which is regulated by many chromatin modifiers (Hayashi et al., 2015; Hisada et al., 2012; Ishiuchi et al., 2015; Lu et al., 2014a; Macfarlan et al., 2012; Maksakova et al., 2013; Thompson et al., 2015). Thus, these LTR genic promoters likely retain the restricted tissue-specific expression pattern of the full-length ancestral provirus.
Further evidence for strong selective pressure for novel tissue-specific promoters of protein-coding genes can be inferred from the exaptation of different LTRs for orthologous genes in independent lineages. Emera and co-workers (2012) showed that MER39 and MER77 LTRs (Table 1) were independently exapted as novel promoters in primates and rodents, respectively, for the Prolactin gene, which is expressed in endometrial cells during pregnancy and essential for normal gestation (Emera et al., 2012). In addition, different LTRs have been independently exapted as promoters for the anti-apoptotic gene NAIP. In primates, testis-specific NAIP transcripts are driven by the MER21C LTR, while in rodents, Naip is expressed in many different tissues from ORR1E or MT-C LTRs (Romanish et al., 2007). Although these are isolated cases, many other instances of exaptation may have occurred earlier in mammalian evolution, with the regulatory elements in question no longer recognizable as LTRs.
In addition to promoting gene expression, the co-option of LTRs as promoters also provides the opportunity for TF-directed repression, as evidenced by a recent study which found that KLF3 enforces transcriptional repression of ORR1A0 LTR-driven transcripts in mouse fetal and adult erythroid cells (Mak et al., 2014). Whether suppression of such ORR1A0 LTR-driven chimeric transcripts serves only to prevent aberrant genic transcription emanating from the LTR remains to be determined. Chromatin modifiers may also direct the silencing of LTR-driven genes, similar to non-TE-derived genic promoters (Isbel et al., 2015; Karimi et al., 2011; Macfarlan et al., 2011; Wolf et al., 2015b). Thus, LTR promoters are apparently as versatile as typical genic promoters and may confer positive or negative regulatory functions on their cognate genes. Similarly, purifying selection for KRAB-ZFP-directed gene repression may reflect the persistence of ancient KRAB-ZFP binding sites in degenerate LTRs and/or non-repetitive regions near genes (Friedli and Trono, 2015).
Whether LTRs functioning as genic promoters generally exhibit substantial sequence differences relative to their ancestral sequence has not been systematically addressed. However, a recent study examining Prolactin expression in the placenta, which is driven by the MER39 LTR in various primate lineages but not in non-ape species (Emera and Wagner, 2012a), sheds some light on the role of “fine-tuning” of LTR promoters. While the ancestral MER39 LTR present in all primates and rodents possessed an intact ETS1 binding site at the time of integration, this LTR was a weak promoter in non-ape species and was replaced by the MER77 LTR as the major Prolactin promoter in mice (Emera and Wagner, 2012a). However, over millions of years of ape evolution, MER39 was gradually transformed into a strong promoter by selection for base substitution mutations that synergized with the ancestral ETS1 site in the LTR and consequently improved the strength of the promoter (Emera and Wagner, 2012a). Thus although the primordial LTR possessed a functional TFBS, it was likely inefficient to act as a promoter in the placenta and required a series of substitutions to refine its activity. This finding is consistent with previous work showing that species-specific expression of genes near TEs is positively correlated with the number of bound TFBSs in the TE, with a minimum of 2 bound TFBSs to detect the correlation (Xie et al., 2010). The mechanism termed ‘epistatic capture’ was proposed to describe the process by which a TE-derived TFBS comes under increased purifying selection as a consequence of epistatic interactions with nearby TFBSs refined by mutations over evolutionary time (Emera and Wagner, 2012b) (Figure 1C). Notably, this mechanism also accounts for the tissue specificity of LTR exaptation into promoters/enhancers, since the positive epistatic interactions between the TE-derived ancestral and newly derived TFBSs would be expected to occur only if they enhance recruitment of the TFs relevant to expression in that tissue. After the acquisition and selection for functional TFBSs within LTRs, the accumulation of additional mutations that are nonessential for their transcriptional activity will invariably lead to their progressive divergence from the ancestral sequence (Figure 1C). Indeed, LTRs co-opted as regulatory elements earlier in mammalian evolution may no longer be recognizable as repeat elements using conventional bioinformatics tools, raising the possibility that many more canonical gene promoters are actually derived from ancient LTRs.
While fewer cases have been identified, there is also evidence that recently integrated LTRs can function as cis-regulatory elements. Examples include mouse-specific LTR13D5 elements, which act as enhancers in the placenta, the primate-specific LTR9, which enhances β-globin gene expression and the primate MLT2B3, which drives B3GALT5 expression in the human colon (Chuong et al., 2013; Dunn et al., 2005; Pi et al., 2010). Thus, LTRs can also serve as ‘ready-made’ enhancers or promoters without substantial sequence modification, potentially contributing to rapid evolution of gene regulatory networks (Cohen et al., 2009).
LTRs in lncRNA expression
The role of TEs in lncRNA expression, function and evolution is just beginning to emerge (Kapusta and Feschotte, 2014). Recent genome-wide surveys have revealed that 75-80% of the ~10,000 annotated human lncRNAs contain TE sequences (Kannan et al., 2015; Kapusta et al., 2013; Kelley and Rinn, 2012). Furthermore, LTRs show considerable enrichment in lncRNA transcripts compared with non-LTR elements and other TEs in mouse and human (Kannan et al., 2015; Kapusta et al., 2013; Kelley and Rinn, 2012). While the majority of LTRs transcribed in lncRNAs serve as exons (Kannan et al., 2015), specific families have been co-opted as promoters. For example, many copies of the primate specific LTR7 derived from human HERVH are bound by pluripotency factors and function as essential regulatory elements in naïve pluripotent stem cells, likely by driving expression of specific lncRNAs (Durruthy-durruthy et al., 2016; Lu et al., 2014b; Ohnuki et al., 2014; Wang et al., 2013). In addition, genome-wide analysis suggests that many previously unannotated LTR-driven lncRNA transcripts are important for the maintenance of pluripotency in mouse and human (Fort et al., 2014). Deep sequencing of human preimplantation embryos has demonstrated the expression of stage-specific LTR-derived noncoding RNAs from a variety of ERV1, ERVK and ERVL family ERVs, however their functions remain to be determined (Goke et al., 2015; Grow et al., 2015) .
Due to the versatility of lncRNA biogenesis and function, there are likely to be fewer constraints upon the exaptation of LTRs as lncRNA promoters. In addition to the basic regulatory properties of LTRs relevant to promoters for protein-coding genes, novel lncRNA genes could arise de novo from solo LTRs in intergenic regions (Friedli and Trono, 2015; Kapusta and Feschotte, 2014), which would also not necessarily require an intact SD site (Figure 1D). Since conserved lncRNAs such as HOTAIR, lincRNA-RoR (a HERV-H derived lncRNA gene), lncRNA-p21 andTUNAR have been shown to regulate large cohorts of genes (Froberg et al., 2013; Huarte et al., 2010; Lin et al., 2014; Loewer et al., 2010; Rinn et al., 2007), a single LTR integration into a lncRNA gene has the potential to exert a broad regulatory effect on the transcriptome. Given the relatively rapid origins and turnover of lncRNA genes and their lack of high sequence conservation despite –in some cases- their clear functional conservation (Kapusta and Feschotte, 2014), it will be important to focus future investigations on the genome-wide contribution of LTRs to the genesis of novel lncRNA transcripts during mammalian evolution.
LTRs as tissue-specific enhancers
A number of recent studies have revealed that LTRs have also contributed substantially to the formation of enhancers during mammalian evolution (Emera and Wagner, 2012b; Friedli and Trono, 2015). In fact, the majority of LTRs contributing to placenta-specific gene expression in humans (Pavlicev et al., 2015) and species-specific expression in mouse placenta (Chuong et al., 2013) show signatures of enhancers rather than promoters. In light of the ability of enhancers to act over very long distances, the combinations of TFBSs present in LTRs and the general selection against ERV integrations near genic promoters (Medstrand et al., 2002), it is not surprising that LTRs have been co-opted as enhancers. Indeed, recent genome-wide surveys reveal that active LTR-derived enhancers exhibit the typical epigenomic signatures of active non-TE-derived enhancer elements, including enrichment of H3K4me1, H3K27ac, DNAse I hypersensitivity, DNA hypomethylation, depletion of repressive H3K9me3 and H3K27me3 and TF binding (Chuong et al., 2013; Fort et al., 2014; Jacques et al., 2013; Sundaram et al., 2014; Xie et al., 2013) (Figure 2). In addition to epigenomic signatures, candidate TE-derived enhancers can also be identified from comparative genomic analysis. Using the Marmoset genome, del Rosario et al. (2014) identified noncoding regions constrained in the anthropoid primate lineage that are unconstrained in other distantly related mammals and found 14,546 TE-derived regions covering ~4 Mb of genomic sequence that showed chromatin signatures of anthropoid lineage-specific enhancers, a subset of which were derived from LTRs (del Rosario et al., 2014).
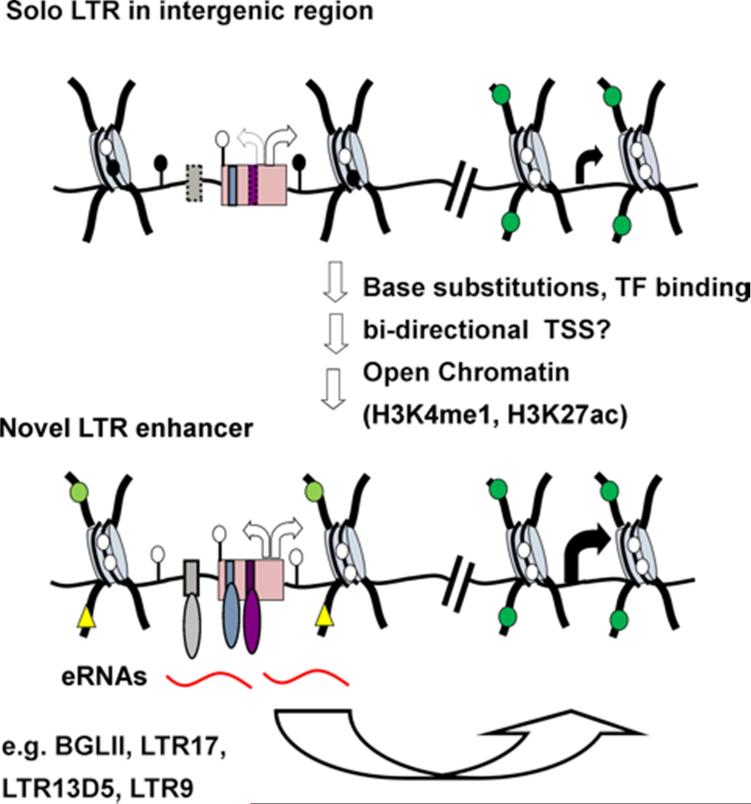
Figure 2. Molecular mechanisms of exaptation of LTRs as enhancers.
LTRs located both proximal or distal, (i.e. >10 kb) to a genic promoter may be exapted as enhancer elements. Such elements may be solo LTRs (shown) or intact ERVs. While these LTRs may have intrinsic enhancer activity, base substitutions that generate additional TF binding sites (potentially synergizing with pre-existing sites), may over time increase overall enhancer activity and/or refine tissue specificity. Note that in contrast with LTR-derived genic promoters, which are generally in the sense orientation, an LTR integrated in either orientation with respect to the relevant gene could be exapted as an enhancer. Robust enhancer activity also likely requires formation of an open chromatin structure and the generation of enhancer RNA transcripts (eRNAs) in the relevant cell type. The strong association of specific histone marks with enhancers, including H3K4me1 (light green circles) and H3K27ac (yellow triangles), the latter indicative of “active” enhancers, has been widely exploited to identify novel candidate enhancers, including within LTRs. Examples of LTR enhancers include BGLII and LTR17 and perhaps LTR13D5 and LTR9, but whether these latter two LTRs produce eRNAs has not been determined.
While candidate tissue-specific LTR-derived enhancer sequences have been reported by many groups based on the presence of specific TFBSs and/or epigenetic marks consistent with enhancer activity, only a few studies have performed confirmatory functional analyses of their activity. Using luciferase-based reporter assays, candidate LTR enhancers have been shown to increase expression from heterologous promoters in cell lines representing the tissue in which they are active, such as rat trophoblast stem cells or human 293T cells, respectively (Chuong et al., 2013; Xie et al., 2013). However, while these assays demonstrate potential for enhancer activity, they do not prove that the LTR-derived sequences function as enhancers in their native genomic context. To address this question, loss and/or gain-of-function analysis demonstrating putative enhancer function of LTRs have been employed in animal models. For example, deletion of the human LTR9 enhancer located ~100 kb upstream of the β-globin gene cluster abolishes β-globin gene expression in a transgenic mouse model (Pi et al., 2010). Similarly, transgenic mice were used to demonstrate bona fide enhancer activity of the novel primate-specific TE-derived ASC192 enhancer (del Rosario et al., 2014). More recently, CRISPR-mediated deletion was used to demonstrate the importance of the MER41 LTR as an enhancer of key innate immunity genes activated by interferons (Chuong et al., 2016). Further studies of candidate LTR-derived enhancers using genome-editing approaches will reveal the extent to which such elements influence target gene expression in vivo.
How are LTRs exapted as novel tissue-specific enhancers? In another example of LTR exaptation in different lineages, Franchini et al. (2011) showed that an ancient SINE and later a MaLR LTR were independently exapted as enhancers to control expression of the vertebrate Pomc gene in the pituitary and hypothalamus in independent lineages during mammalian evolution (Franchini et al., 2011). Interestingly, a recent functional analysis of these enhancers showed that while the ancient SINE-derived enhancer nPE2 is only required for ~20% of Pomc expression, the MaLR LTR-derived enhancer nPE1 is sufficient to drive ~80% of Pomc expression (Lam et al., 2015), suggesting that LTRs may be exapted as enhancers when large increases in gene expression are beneficial and thus selected. In addition to the mechanism of epistatic capture of novel TFBSs described above, which is relevant to both promoters and enhancers, the exaptation of LTRs as enhancers may depend on their capacity to produce bidirectional noncoding transcripts (Figure 2). Indeed enhancer function may require bidirectional transcription of distinct noncoding RNA species called enhancer RNAs (eRNAs) (Kim et al., 2015; Plank and Dean, 2014).
LTRs that serve as promoters for nuclear lncRNAs have been found in a variety of contexts (Faulkner et al., 2009; Herquel et al., 2013; Lu et al., 2014b), but whether they produce eRNAs had not been addressed. However, a recent comprehensive transcriptome analysis of pluripotent stem cells demonstrated that active BGLII and LTR17-derived enhancers do indeed express bidirectional eRNAs and many of these LTR-associated noncoding transcripts are important for the maintenance of ES cell pluripotency (Fort et al., 2014). While bi-directional transcription from LTRs has been reported, as in the case of the composite human LTR9/LTR16A that promotes tissue-specific expression of the DSCR4 and DSCR8 genes in opposing orientations (Dunn et al., 2006), most LTRs do not produce bi-directional transcripts. Therefore, these findings suggest that LTR exaptation as an enhancer may also depend on the acquisition of substitutions within or adjacent to the LTR that support eRNA transcription (Figure 2). Whether such mutations are distinct from those that support TF binding or acquisition of alternative features essential for enhancer function remains to be determined. Regardless, the number of enhancer-derived LTRs is likely to be high in certain cell types, with their activity restricted in other cell types by the establishment of repressive chromatin, such as the deposition of H3K9me3 by the KRABZFP/KAP1/SETDB1 system (Rowe et al., 2013).
Conclusions
In conclusion, recent transcriptomic and epigenomic studies have revealed that LTRs provide a plethora of novel gene regulatory elements, including tissue specific promoters and enhancers. Such LTRs are particularly prevalent in early embryonic development, germ cells and pluripotent stem cells, likely as a consequence of the relaxed epigenetic silencing in these cell types and regulatory regions optimized for expression in these tissues in the retroviral precursor (Fort et al., 2014; Xie et al., 2013). Future work will likely reveal whether these regions are generally further optimized for tissue-specific expression by the acquisition of TFBSs. In addition, although there are many candidate species-specific enhancers derived from LTRs, few studies have actually addressed their biological significance in vivo with rigorous functional analyses (de Souza et al., 2013). Using CRISPR technology, it is now feasible to inactivate or delete specific LTRs to determine their effects upon the host transcriptome (Yang et al., 2015), providing a powerful tool for systematic analyses of LTR-driven transcripts and candidate enhancers in different cell types and species. While ERVs show striking enrichment in lncRNAs (Kannan et al., 2015; Kapusta et al., 2013; Kelley and Rinn, 2012) and LTRs clearly drive expression of a subset of lncRNA transcripts that appear to play important developmental functions (Durruthy-durruthy et al., 2016; Fort et al., 2014), it remains to be determined what roles ERV-derived sequences generally play in lncRNA structure, function and evolution. Furthermore, caution must be exercised when interpreting the results of loss-of-function studies on lncRNAs due to the complex nature of their activities at the transcriptional and post-transcriptional levels (Bassett et al., 2014). Finally, while other retrotransposons have been exapted into both enhancers and insulators in humans (Jjingo et al., 2014; Wang et al., 2015) LTR-derived insulators have not been identified to date. Future investigations into these and related questions will further our understanding of the extent to which mammalian genomes have harnessed the latent regulatory potential of LTRs to control tissue-specific gene expression.
Acknowledgements
We wish to thank Dixie Mager and Cedric Feschotte for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflicts of interest.
Literature Cited
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-smith AC, Gingeras TR, Haerty W, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:1–14. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 2007;81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew J, Ruan Y, Wei C, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Buzdin A, Kovalskaya-alexandrova E, Gogvadze E, Sverdlov E. At Least 50 % of Human-Specific HERV-K ( HML-2 ) Long Terminal Repeats Serve In Vivo as Active Promoters for Host Nonrepetitive DNA Transcription †. J. Virol. 2006;80:10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz Na., Friedli M, Trono D. Drawing a fine line on endogenous retrovirus activity. Mob. Genet. Elements. 2015;5:1–6. doi: 10.1080/2159256X.2015.1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes : A critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer M. a. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, Oakey RJ. Transposable Elements Re-Wire and Fine-Tune the Transcriptome. PLoS Genet. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12841–12846. doi: 10.1073/pnas.2134464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 2006;366:335–342. doi: 10.1016/j.gene.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Durruthy-durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, Davila J, Mall M, Wong WH, Wysocka J, et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 2016;41:44–52. doi: 10.1038/ng.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev. Cell. 2016 doi: 10.1016/j.devcel.2016.02.024. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Wagner GP. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proc. Natl. Acad. Sci. U. S. A. 2012a;109:1–6. doi: 10.1073/pnas.1118566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Wagner GP. Transposable element recruitments in the mammalian placenta: impacts and mechanisms. Brief. Funct. Genomics. 2012b;11:267–276. doi: 10.1093/bfgp/els013. [DOI] [PubMed] [Google Scholar]
- Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D. Convergent Evolution of Endometrial Prolactin Expression in Primates , Mice , and Elephants Through the Independent Recruitment of Transposable Elements Research article. Mol. Biol. Evol. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004;105:240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, Noro Y, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Franchini LF, Lopez-Leal R, Nasif S, Beati P, Gelman DM, Low MJ, de Souza FJS, Rubinstein M. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15270–15275. doi: 10.1073/pnas.1104997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, de Souza FSJ, Low MJ, Rubinstein M. Positive selection of co-opted mobile genetic elements in a mammalian gene: If you can't beat them, join them. Mob. Genet. Elements. 2012;2:106–109. doi: 10.4161/mge.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Trono D. The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annu. Rev. Cell Dev. Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- Froberg JE, Yang L, Lee JT. Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 2013;425:3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell P, Hein J, Katzourakis A. Orthologous endogenous retroviruses exhibit directional selection since the chimp-human split. Retrovirology. 2015;12:52. doi: 10.1186/s12977-015-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, MacFarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J, Lu X, Chan Y, Ng H, Ly L, Sachs F, Szczerbinska I. Dynamic Transcription of Distinct Classes of Endogenous Retroviral Elements Marks Specific Populations of Early Human Embryonic Cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:222–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Maehara K, Harada A, Semba Y, Kudo K, Takahashi H, Oki S, Meno C, Ichiyanagi K, Akashi K, et al. Chd5 Regulates MuERV-L/MERVL Expression in Mouse Embryonic Stem Cells Via H3K27me3 Modification and Histone H3.1/H3.2. J. Cell. Biochem. 2015;117:780–792. doi: 10.1002/jcb.25368. [DOI] [PubMed] [Google Scholar]
- Herquel B, Ouararhni K, Martianov I, Gras S, Le, Ye T, Keime C, Lerouge T, Jost B, Cammas F, Losson R, et al. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat. Struct. Mol. Biol. 2013;20:339–346. doi: 10.1038/nsmb.2496. [DOI] [PubMed] [Google Scholar]
- Hisada K, Sanchez C, Endo T. a., Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. RYBP Represses Endogenous Retroviruses and Preimplantation- and Germ Line-Specific Genes in Mouse Embryonic Stem Cells. Mol. Cell. Biol. 2012;32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbel L, Whitelaw E. Endogenous retroviruses in mammals: An emerging picture of how ervs modify expression of adjacent genes. BioEssays. 2012;34:734–738. doi: 10.1002/bies.201200056. [DOI] [PubMed] [Google Scholar]
- Isbel L, Srivastava R, Oey H, Spurling A, Daxinger L, Puthalakath H, Whitelaw E. Trim33 Binds and Silences a Class of Young Endogenous Retroviruses in the Mouse Testis ; a Novel Component of the Arms Race between Retrotransposons and the Host Genome. PLoS Genet. 2015;11:e1005693. doi: 10.1371/journal.pgen.1005693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez- Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- Jacques P-E, Jeyakani J, Bourque G. The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements. PLoS Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Wang J, Mariño-ramírez L, Lunyak VV, Jordan IK. Mammalian-wide interspersed repeat ( MIR ) -derived enhancers and the regulation of human gene expression. Mob. DNA. 2014;5:14. doi: 10.1186/1759-8753-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Chernikova D, Rogodzin IB, Poliakov E, Managadze D, Koonin EV, Milanesi L. Transposable element insertions in long intergenic non-coding RNA genes. Front. Bioeng. Biotechnol. 2015;3:1–9. doi: 10.3389/fbioe.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires : mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova I. a, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015;7:a018622. doi: 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia N, Jeyakani J, Hwang C, Lu X, Chan Y, Ng H, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:6–8. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry J-R, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Lam DD, de Souza FSJ, Nasif S, Yamashita M, Lopez-Leal R, Otero-Corchon V, Meece K, Sampath H, Mercer AJ, Wardlaw SL, et al. Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold. PLoS Genet. 2015;11:e1004935. doi: 10.1371/journal.pgen.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Köchert K, Bouhlel MA, Richter J, Soler E, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010;16:571–579. doi: 10.1038/nm.2129. 1p following 579. [DOI] [PubMed] [Google Scholar]
- Lin N, Chang KY, Li Z, Gates K, Rana Z, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatov M, Lenkov K, Petrov DA, Bergman CM. Paucity of chimeric gene- transposable element transcripts in the Drosophila melanogaster genome. BMC Biol. 2005;18:24. doi: 10.1186/1741-7007-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh Y-H, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Haussler D. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS One. 2012;7:e43128. doi: 10.1371/journal.pone.0043128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014a;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay L, Jacques P-É, Göke J, Bourque G, Ng H-H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014b;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Nnamani MC, Brayer K, Plaza SL, Mazur EC. Ancient Transposable Elements Transformed the Uterine Regulatory Landscape and Transcriptome during the Evolution of Mammalian Pregnancy Article Ancient Transposable Elements Transformed the Uterine Regulatory Landscape and Transcriptome during the Evolut. Cell Rep. 2015;10:551–561. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015;3:1–20. doi: 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak KS, Burdach J, Norton LJ, Pearson RCM, Crossley M, Funnell APW. Repression of chimeric transcripts emanating from endogenous retrotransposons by a sequence-specific transcription factor. Genome Biol. 2014;15:R58. doi: 10.1186/gb-2014-15-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I. a, Romanish MT, Gagnier L, Dunn C. a, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, Lorincz MC. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova I. a, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U. S. A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL. Retroelement Distributions in the Human Genome: Variations Associated With Age and Proximity to Genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12426–12431. doi: 10.1073/pnas.1413299111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahara G, Matsubara S, Oda T, Sugimoto J, Jinno Y, Kanaya F. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics. 2004;84:982–990. doi: 10.1016/j.ygeno.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Pavlicev M, Hiratsuka K, Swaggart KA, Dunn C, Muglia L. Detecting Endogenous Retrovirus-Driven Tissue-Specific Gene Transcription. Genome Biol. Evol. 2015;7:1082–1097. doi: 10.1093/gbe/evv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Pi W, Zhu X, Wu M, Wang Y, Fulzele S, Eroglu A, Ling J. Long-range function of an intergenic retrotransposon. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12992–12997. doi: 10.1073/pnas.1004139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Dean A. Enhancer Function : Mechanistic and Genome-Wide Insights Come Together. Mol. Cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable Elements : An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annu. Rev. Genet. 2012a;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Zhang Y, Mager DL. Transposable elements : not as quiet as a mouse. Genome Biol. 2012b;13:159. doi: 10.1186/gb-2012-13-6-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D, Zhang Y, Mager DL. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 2007;35:4743–4754. doi: 10.1093/nar/gkm455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann S. a., Goodnough LH, Helms J. a., Farnham PJ, Segal E, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbez-Masson L, Rowe HM. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology. 2015;12:45. doi: 10.1186/s12977-015-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rosario RCH, Rayan NA, Prabhakar S. Noncoding origins of anthropoid traits and a new null model of transposon functionalization. Genome Res. 2014;24:1469–1484. doi: 10.1101/gr.168963.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadic D, Schmidt K, Groh S, Kondofersky I, Ellwart J, Fuchs C, Theis FJ, Schotta G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015;16:836–850. doi: 10.15252/embr.201439937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- de Souza S.J. De, Franchini F, Rubinstein M. Exaptation of Transposable Elements into Novel Cis -Regulatory Elements : Is the Evidence Always Strong ? Mol. Biol. Evol. 2013;30:1239–1251. doi: 10.1093/molbev/mst045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking C, Kozak C. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T, Mcclintock B, Britten RJ, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Dulberg V, Moon K, Foster LJ, Chen C, Karimi MM, Lorincz MC. hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells. PLoS Genet. 2015;11:e1004933. doi: 10.1371/journal.pgen.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014 doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015a;16:209. doi: 10.1186/s13059-015-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015b;16:209. doi: 10.1186/s13059-015-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2013;516:406–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- Wang J, Vicente-garcía C, Seruggia D, Moltó E, Fernandez-miñán A, Neto A. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4428–E4437. doi: 10.1073/pnas.1507253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within : Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob. DNA. 2015a;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer E-M, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015b;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Chen C, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Dan J, Wang H, Guo R, Mao J, Fu H, Wei X, Liu L. Tcstv1 and Tcstv3 elongate telomeres of mouse ES cells. Sci. Rep. 2016;6:19852. doi: 10.1038/srep19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT-Y, Kohlbacher O, De Jager PL, Rosen ED, Bennett D. a, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]