Abstract
Discriminating the gene target of a distal regulatory element from other nearby transcribed genes is a challenging problem with the potential to illuminate the causal underpinnings of complex diseases. We present TargetFinder, a computational method that reconstructs regulatory landscapes from genomic features along the genome. The resulting models accurately predict individual enhancer-promoter interactions across diverse cell lines with a false discovery rate up to fifteen times smaller than using the closest gene. By evaluating the genomic features driving this accuracy, we uncover interactions between structural proteins, transcription factors, epigenetic modifications, and transcription that together distinguish interacting from non-interacting enhancer-promoter pairs. Most of this signature is not proximal to the enhancers and promoters, but instead decorates the looping DNA. We conclude that complex but consistent combinations of marks on the one-dimensional genome encode the three-dimensional structure of fine-scale regulatory interactions.
Introduction
Genotyping, exome sequencing, and whole-genome sequencing have linked thousands of non-coding variants to traits in humans and other eukaryotes1–6. Non-coding variants are more likely to cause common disease than are non-synonymous coding variants7, and they can account for the vast majority of heritability8. Yet few non-coding mutations have been functionally characterized or mechanistically linked to human phenotypes7,9. Comparative10 and functional11–13 genomics, coupled with bioinformatics, are generating annotations of regulatory elements in many organisms and cell types14, as well as tools for exploring or predicting the impact of mutations in regulatory DNA15–18. However, this new information will only improve our understanding of disease and other phenotypes if we can accurately link functional non-coding elements to the genes, pathways, and cellular processes they regulate. This is a difficult problem because vertebrate promoters and their regulatory elements can be separated by thousands or millions of base pairs (bp)19. The closest promoter is usually not the true target in humans20, though this varies by species21, but remains a common heuristic for mapping target genes. Incorrectly mapping regulatory variants to genes prevents meaningful downstream studies.
Until recently, very few validated distal regulatory interactions were known. Hence, previous studies defined interactions indirectly via genomic proximity coupled with genetic associations (e.g., eQTLs22), gene expression14,23–25, or promoter chromatin state26,27. High-throughput methods for assaying chromatin interactions now exist, including paired-end tag sequencing (ChIA-PET)28 and extensions of the chromosome conformation capture (3C) assay29 (5C, Hi-C)30,31. When resolution is high enough to measure individual enhancer-promoter interactions32–35, Hi-C provides an opportunity to examine the genomic features that distinguish the true target of an enhancer from other nearby expressed genes. We hypothesized that modeling relationships between DNA sequences, structural proteins, transcription factors and epigenetic modifications that together predict looping chromatin might reveal novel protein functions and molecular mechanisms of distal gene regulation that are not immediately obvious from the Hi-C data itself.
We implemented an algorithm called TargetFinder that integrates hundreds of genomics datasets to identify the minimal subset necessary for accurately predicting individual enhancer-promoter interactions across the genome. We focused on enhancers due to their large impact on gene regulation36 and our ability to predict their locations genome-wide, though our approach works with other classes of regulatory elements. Our goal was to build a fine scale model capable of distinguishing individual enhancer-promoter pairs from amongst the many possible interactions within a topologically associating domain (TAD) or contact domain. Applying TargetFinder to six human ENCODE cell lines11 with high resolution Hi-C data32, we discovered that interacting enhancer-promoter pairs can be distinguished from noninteracting pairs within the same locus with extremely high accuracy. These analyses also showed that functional genomics data marking the window between the enhancer and promoter are more useful for identifying true interactions than are proximal marks at the enhancer and promoter. Exploration of this phenomenon revealed specific proteins and chemical modifications on the chromatin loop that bring an enhancer in contact with its target promoter and not with nearby active but non-targeted promoters. Thus, TargetFinder provides a framework for accurately assaying three-dimensional genomic interactions, as well as techniques for mining massive collections of experimental data to shed new light on the mechanisms of distal gene regulation.
Results
Distal enhancers physically interact with promoters of their target genes over long genomic distances while avoiding other nearby active and inactive promoters via precise chromatin looping. We hypothesized that transcription factors, histones, and architectural proteins might combine to distinguish these distal regulatory interactions from other regions of the genome. If so, it should be possible to computationally model known interactions from independent functional genomics data, and the most important genomic features in the model might shed light on the mechanisms of gene regulation in three dimensions.
Annotating the genomic features of regulatory interactions
We annotated enhancer-promoter interactions in six human ENCODE cell lines that have rich functional genomics data as well as high resolution interaction data produced by Rao et al32: K562 (mesoderm lineage cells from a leukemia patient), GM12878 (lymphoblastoid cells), HeLa-S3 (ectoderm lineage cells from a cervical cancer patient), HUVEC (umbilical vein endothelial cells), IMR90 (fetal lung fibroblasts), and NHEK (epidermal keratinocytes). We identified active promoters and enhancers in each cell line using segmentation-based annotations from ENCODE and Roadmap Epigenomics, as well as gene expression data from ENCODE (Supplementary Table S1). Enhancers are typically a few hundred base paris (bp) long, while promoters are mostly between 1–2 kilobases (Kb) (Supplementary Figures S1 to S3). Alternative enhancer and promoter definitions produce qualitatively similar results (Supplementary Text).
We annotated all enhancer-promoter pairs as interacting or non-interacting using high resolution genome-wide measurements of chromatin contacts in each line32, the majority of which were also detected by capture Hi-C35. Non-interacting pairs were sampled (20 per interacting pair) to have enhancer-promoter distances similar to interacting pairs, all of which were less than 2 megabases (Mb). To focus on distal regulatory enhancers, any pair separated by less than 10 Kb was dropped. We did not remove interactions crossing TAD boundaries, but most enhancer-promoter pairs occur within the same TAD (88% in GM12878, 77% in K56237). It is important to emphasize that by design all enhancers and promoters in our study, including those in non-interacting pairs, have marks of activation and open chromatin. The challenging question we address is whether interacting pairs have any distinguishing characteristics.
We generated features for all enhancer-promoter pairs in each line using functional genomics data such as measures of open chromatin, DNA methylation, gene expression, and chromatin immunoprecipitation followed by sequencing (ChIP-seq) for transcription factors (TFs), architectural proteins, and modified histones (Supplemental Table S2). We quantified signal at the promoter, at the enhancer, and in the genomic window between them. We also computed features for conserved synteny of the enhancer and promoter, as well as the similarity of TF and target gene annotations, which are associated with experimentally validated interactions25.
Finally, we created a “combined” dataset by pooling the enhancer-promoter pairs and features from 4 cell lines (K562, GM12878, HeLa-S3 and IMR90), which we used to discover features of looping chromatin that generalize across lines. Only features measured in all four lines were retained to avoid problems with missing data. NHEK and HUVEC had only ~ 20 datasets (versus > 50; Supplemental Table S2) and were hence excluded from the combined dataset.
No single feature distinguishes the true targets of active enhancers
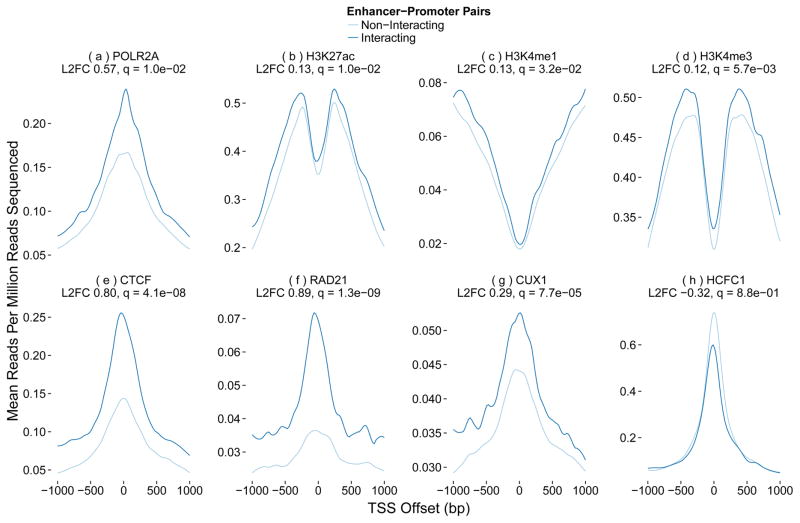
Signal profiles at enhancers and promoters show many expected differences between interacting and non-interacting pairs. These include higher Pol II signal at the transcription start site (TSS) of interacting promoters (Figure 1a) and enrichment of H3K27ac and H3K4me3 with depletion of H3K4me1 in regions flanking the TSS of interacting promoters (Figure 1b–d). Across cell types, CTCF and RAD21 are enriched near interacting promoters (Figure 1e–f). Structural proteins and their cofactors are also enriched nearby interacting enhancers (Figure 2).
Figure 1.
Predictive power of promoter proximal genomic features. Ratio of various ChIP-seq signals anchored at the transcription start sites (TSS) of interacting vs non-interacting promoters in K562, along with the log base 2 fold change (L2FC) and p-value corrected for multiple testing (q). All promoters have activating chromatin marks and show transcription. The top row shows expected patterns for promoter-associated marks at the TSS, such as a high ratio of H3K4me3 to H3K4me1. Some of these marks are enriched in interacting promoters, while others such as K4 methylation patterns are not. The second row shows TSS proximal patterns for several proteins associated with chromatin looping. CTCF and RAD21 are enriched at interacting promoters, while transcription factors CUX1 and HCFC1 are enriched and depleted, respectively.
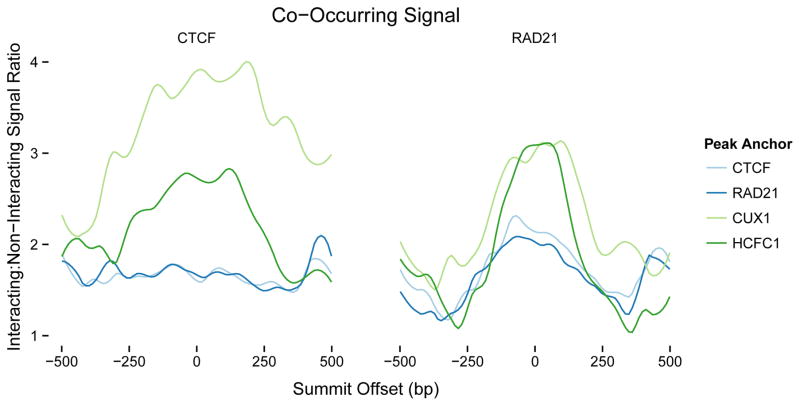
Figure 2.
Binding co-occurrence at enhancers enriches looping interactions. Ratio of CTCF and RAD21 ChIP-seq signals occurring within interacting enhancers vs non-interacting enhancers, anchored at peaks for CTCF, RAD21, and the transcription factors CUX1 and HCFC1 for the K562 cell line. CUX1 and HCFC1 are highly enriched at loop-associated enhancers when co-occurring with CTCF and RAD21. The context-dependence of protein binding is demonstrated by RAD21, which is not enriched at interacting promoters (Figure 1). Note that CTCF and RAD21 are already enriched at their respective peaks within interacting enhancers, but are further enriched when anchored at CUX1 or HCFC1. This visualizes how the co-occurrence of certain transcription factors increases the likelihood of looping interactions beyond CTCF or RAD21 peaks alone, helps interpret the predictive importance estimated by TargetFinder, and can identify novel looping factors.
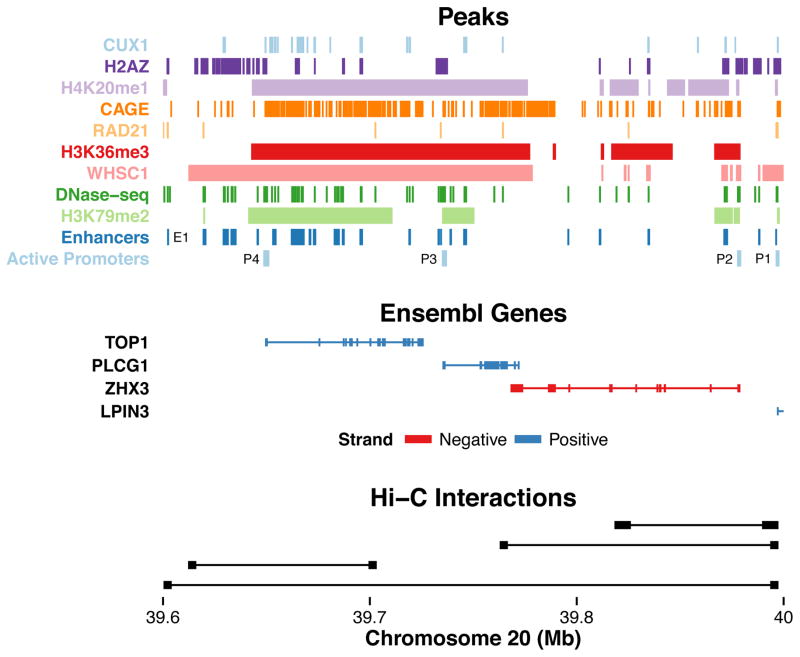
However, any given interaction has a complex combination of genomic features, some of which also occur at non-interacting pairs in the same locus. For example, LPIN3 has an enhancer that loops over approximately 400 Kb of intervening DNA containing the active promoters of TOP1, PLCG1, and ZHX3 in K562 (Figure 3). No single mark distinguishes LPIN3 from these alternate targets, though their gene bodies are covered by broad repressive marks (heterochromatin-associated H4K20me1) and by broad activating marks (elongation-associated H3K36me3). Notably, alternate promoters lack a RAD21, while ZHX3 and PLCG1 are lacking CUX1 which has been linked to both activation and repression. In GM12878, an intronic enhancer targeting CUTC loops over the promoter of ENTPD7, which has many activation marks but lacks RAD21 (Supplementary Figure S5). This complexity motivated us to model enhancer-promoter interactions as a function of diverse genomic signatures.
Figure 3.
Ensemble learning predicts enhancer-promoter pairs with high accuracy
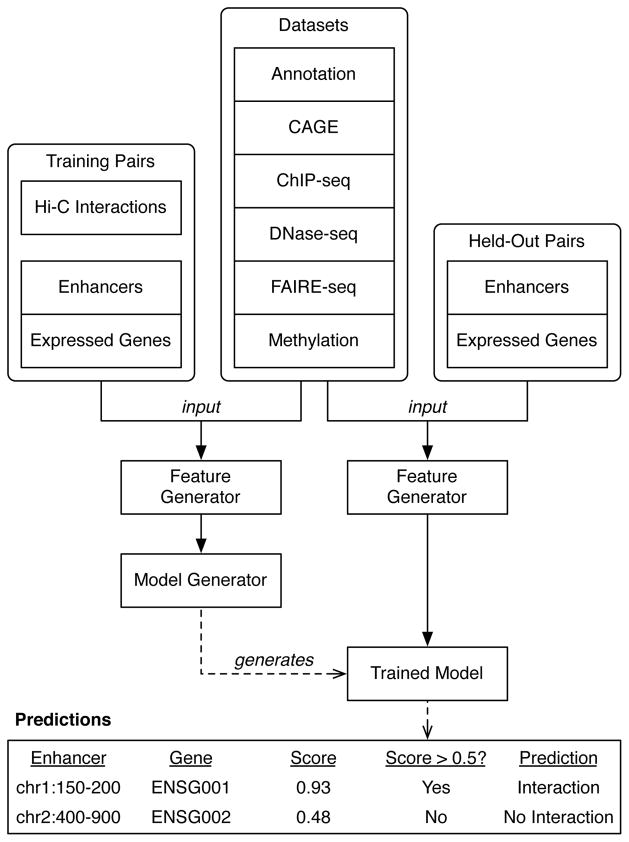
To quantitatively model the interaction status of enhancer-promoter pairs as a function of their genomic features, we built a machine learning pipeline called TargetFinder (Figure 4). The inputs are pairs of enhancers and promoters, annotated as interacting or not, and genomic features associated with each pair. The algorithm finds an optimal combination of features for distinguishing interacting from noninteracting pairs. Multiple machine learning techniques are implemented in the pipeline in a modular way so that performance can be optimized and conclusions can be tested for robustness to the prediction method. The outputs are a model for predicting if new enhancer-promoter pairs interact, assessments of model performance on held-out data, and estimates of each feature’s individual importance to the model as well as in combination with other features. The predictive contribution of different genomic regions and data types is explored by varying the feature set and quantifying predictive performance. By building models for many cell types, their shared and unique characteristics of looping chromatin can be discovered. The method is easily extended to other types of regulatory elements or interactions, such as promoter-promoter or enhancer-enhancer interactions.
Figure 4.
The TargetFinder pipeline. Features are generated from hundreds of diverse datasets for pairs of enhancers and promoters of expressed genes found to have significant Hi-C interactions (positives), as well as random pairs of enhancers and promoters without significant interactions (negatives). These labeled samples are used to train an ensemble classifier that predicts whether enhancer-promoter pairs from new or held-out samples interact, as well as estimate the importance of each feature for accurate prediction. Classifier predictions are probabilities, and a decision threshold (commonly 0.5 but may be adjusted) converts these to positive or negative prediction labels. This figure excludes selection of minimal predictor sets and evaluation of the accuracy of output predictions using held-out Hi-C interaction data.
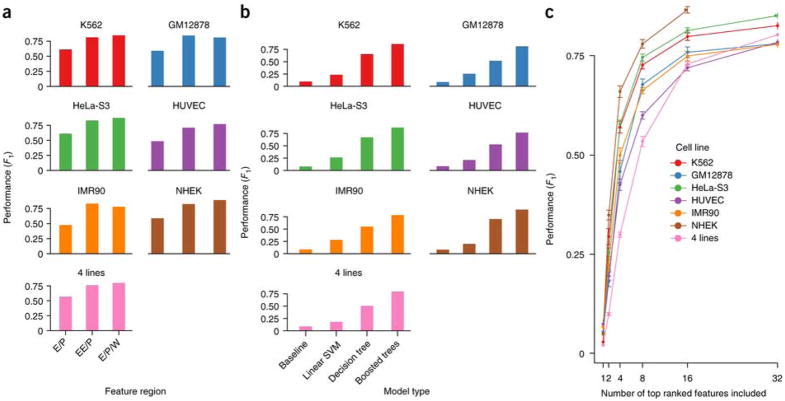
We hypothesized that ensemble learning algorithms would have the highest precision and recall on held-out data, because they are robust to over-fitting and account for non-linear feature interactions that could encode complex patterns of histone modifications and TF binding. Indeed, ensembles of boosted decision trees performed better than other methods and a random guessing null model on all cell lines and the combined data set (Figure 5a, Supplementary Table S3). Accuracy is high by all measures, especially given the noise in functional genomics data and the fact that some non-interacting pairs may be weakly interacting but below the significance cutoff (10% FDR32). TargetFinder with boosted trees achieves F1 between 77–90% (mean = 83%) and FDR between 8–15% (mean = 12%). By comparison, all commonly used bioinformatics methods have much higher FDR and lower recall. For example, using the closest active gene has an FDR of 53–77%20,38,39. The gain in predictive accuracy provided by ensemble learning is consistent across cell lines and in the combined data set (Supplementary Figure S6). This predictive accuracy demonstrates that there is rich information about chromatin looping in one-dimensional genomic datasets that are easier and less costly to collect than high-resolution Hi-C.
Figure 5.
TargetFinder performance by cell line, model type, and number of features. (a) Cross-validated performance of TargetFinder predictions for a baseline (random guessing null) model, a linear Support Vector Machine, a single decision tree, and a boosted ensemble of decision trees. Performance is given as a balance of precision and recall (F1), averaging 83% across cell lines and corresponding to a mean FDR of 12%. Ensemble methods utilize complex interactions between features to greatly increase the accuracy of predicted interactions. Performance is also high on a combined cell line comprised of K562, GM12878, HeLa-S3, and IMR90 datasets, with features restricted to datasets shared by all cell lines. (b) Performance of boosted trees using features for enhancers and promoters only (E/P), promoters and extended enhancers (EE/P), and enhancers/promoters plus the window between (E/P/W). (c) Recursive feature elimination (Methods) evaluates predictor subsets of size 1 up to the maximum per cell line and increasing by powers of 2 for computational efficiency. Near optimal performance was achieved using ~16 predictors for lineage-specific models as well as the combined model, while lower but acceptable performance required 8 predictors. The maximum feature subset size shown is 32 to enhance visibility of smaller feature subsets. NHEK lacks a measurement at subset size 32 since it has fewer than 32 total features. (Error bars = s.e.m.)
Variable importance highlights key datasets for predicting interactions
We next asked if the ability of TargetFinder to predict enhancer-promoter interactions depends on a particular subset of the features. By omitting different categories of features and evaluating performance with cross-validation, we learned that synteny and gene annotations contribute little to predictive accuracy. We therefore proceeded to evaluate models using only functional genomics features.
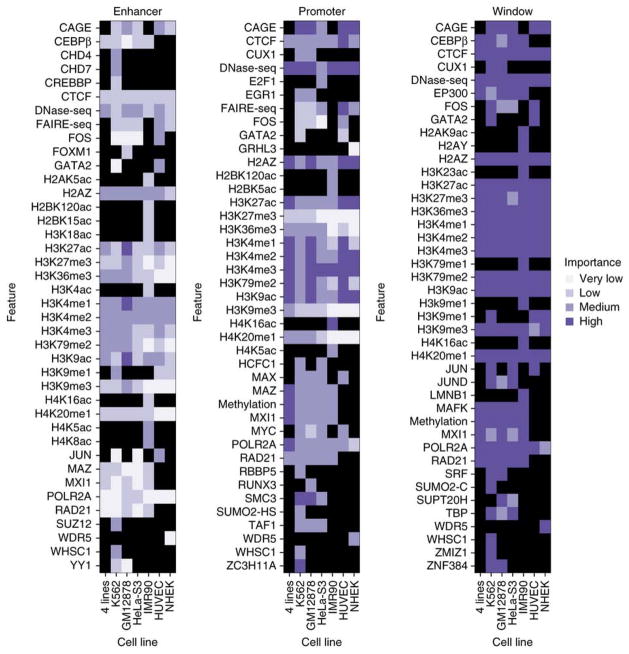
To derive mechanistic insights from the model, TargetFinder estimates feature importance for each genomics dataset within enhancer, promoter, and window regions (Methods). Decision trees inherently estimate predictive importance when deciding which features to split; importance is estimated per feature per tree, then averaged across all trees in the ensemble (Methods). This enabled us to deeply explore the genomic data associated with chromatin loops and revealed several interesting patterns.
The most predictive features that were robust across cell lines were DNA methylation, activation- and elongation-associated histone marks, binding of structural proteins, open chromatin, proteins related to repression (MXI1/MAZ/MAFK), and Cap Analysis of Gene Expression (CAGE) (Figure 6). Other trends emerged across many but not all cell lines, including the importance of the activator protein 1 (AP-1) complex40. Features differ in importance across cell lines for many reasons, including real functional differences (e.g., different co-factors), lack of expression (e.g., tissue-specific TFs), and differences in lab protocols and antibody qualities (Supplementary Figure S7). Interestingly, though there is some overlap with known looping factors such as CTCF and cohesin, features predictive of individual enhancer-promoter interactions are largely different than those used to identify TAD boundaries and large-scale chromatin organization37. This points to different molecular mechanisms operating across these scales.
Figure 6.
Predictive importance of genomic features by region. Predictive importance (Methods) across cell lines and regions. Importance is discretized by quartiles, and grid entries are colored black when a dataset is unavailable in a cell line. The highest average importance is assigned to features in the window region, followed by promoters. Promoter methylation and POLR2A are more important in the the combined “4 Lines” classifier (K562/GM12878/HeLa-S3/IMR90) than individual cell lines. Highly predictive features such as CAGE are available in most but not all cell lines needed for inclusion in the combined model. Certain TFs are available in multiple cell lines but are not universally predictive, such as FOS in the window region. Other TFs are only available in a single cell line but are highly predictive, such as WHSC1 and ZMIZ1 in the window region of K562 and RUNX3 in the window region of GM12878.
Proteins bound between enhancers and promoters predict if they interact
TargetFinder mines a diverse collection of hundreds of genomic features to build its models. To determine if such a large feature set is needed, we applied recursive feature elimination (Methods). Nearly optimal performance requires only ~16 features (Figure 5c), with performance varying by cell line due to differences in the number of enhancer-promoter pairs as well as the quality and quantity of functional genomics data (Supplementary Text).
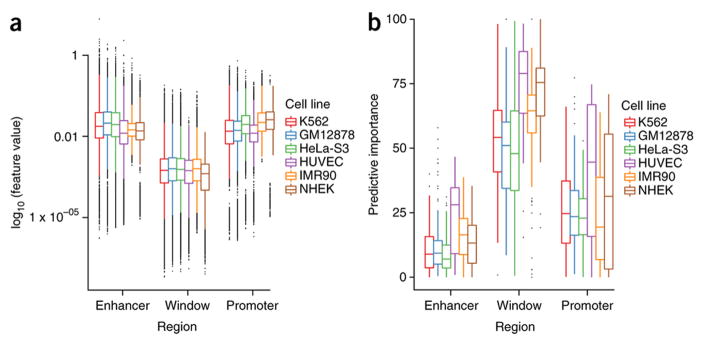
Many of the top features for each line and the combined model are from the genomic window between the enhancer and the promoter, rather than proximal signals at the regulatory elements (Figure 7a). This is true despite the fact that average signal (e.g., ChIP-seq peak density) is higher at enhancers and promoters for most features (Figure 7b). To further validate the importance of features marking the looping chromatin, we retrained TargetFinder with two alternative sets of features per cell line. The first included features for the enhancer and promoter only (EP), and the second included features for an extended enhancer (utilizing 3 Kb of flanking sequence) and a non-extended promoter (EEP) to test the hypothesis that only the enhancer-proximal part of the window is important for predicting looping. We found a large performance gap when using only the enhancer and promoter, without marks flanking the enhancer or in the window (Figure 5b). This indicates there is significant information relevant to looping interactions outside the enhancers and promoters themselves, which we observe consistently across cell lines (Figure S6). Performance was better for EPW than EEP, especially after accounting for the lower dimensionality of EEP (2 regions versus 3 per genomics dataset), which generally improves the performance of machine learning models. Using smaller windows around the enhancers for EEP resulted in lower performance, showing that the signal is not immediately next to the enhancer. Thus, signals relevant to looping are located throughout the genomic window between an enhancer and promoter, but especially within 3 Kb of the enhancer.
Figure 7.
Feature values and predictive importance for enhancer, promoter, and window regions. Despite having the lowest feature values, the predictive importance of the window dominates that of enhancer and promoter regions. (Error bars = 1.5 * interquartile range)
The surprising discovery that the interaction status of an enhancer-promoter pair can be predicted with high accuracy using protein binding and epigenetic marks on DNA between them, plus a few proximal marks, made sense when we examined the specific window features and combinations thereof that the model ranked most important. Some window features are directly involved in chromatin looping including CTCF, the cohesin complex (SMC3/RAD21), and zinc finger proteins such as ZNF384 and ZNF143. The latter interacts with CTCF to provide sequence specificity for chromatin interactions41 by binding lineage-specific TFs at interacting promoters (e.g., HCFC1 in HeLa-S342). Other window features impact the likelihood that additional promoters in the locus are the true targets of an enhancer. For example, RNA polymerase II (Pol II) at a promoter is not predictive by itself because it can indicate either active transcription or a gene that is poised for rapid activation. Such non-targets are distinguished by a lack of activators or co-activators43 as well as elongation-associated histone marks H3K36me3 and H3K79me2. When these features occur in the window between an enhancer and a promoter, they increase the likelihood that an intervening promoter may be the true target. On the other hand, the presence of heterochromatin, PRC2 silencing44, and various insulators in the window suggest that intervening genes are unavailable for binding and are therefore associated with non-interacting pairs in our analyses (Supplementary Figures S11 and S12). However, note that many interacting pairs have different architectures and are exceptions to this trend, including the distal enhancer of LPIN3 in Figure 3. This emphasizes that TargetFinder accurately predicts interactions by learning complex genomic signatures across loci.
Window features do not directly encode distance between the enhancer and promoter, though they may serve as a kind of proxy for active chromatin or domain boundaries. To offset this possibility, we matched the distance distributions for interacting and non-interacting pairs and normalized features by the length of the region. TargetFinder has high precision and recall largely independent of enhancer-promoter interaction distances in the range of 10 Kb to 2 Mb (Supplementary Figure S8). In fact, performance often increases with interaction distance, which is consistent with window features encoding information about contact domain boundaries. Indeed, domain boundaries are significantly enriched in non-interacting pairs compared to interacting pairs separated by similar distances (Supplementary Figure S4). Window-associated marks may also be proxies for relevant but unassayed histone modifications marking alternate targets45.
TargetFinder identifies complex interactions between DNA-binding proteins and epigenetic marks
The complex patterns of co-occurrence between DNA-binding proteins and known looping factors reveal mechanistic insights into the looping process itself. For example, we found that CUX1 and HCFC1 interact with CTCF and RAD21 within enhancers to increase the likelihood of looping interactions in K562 (Figure 2). Interestingly, CUX1 is also significantly enriched at interacting promoters (Figure 1g), while HCFC1 is not (Figure 1h). The importance of co-factors extends beyond this example. TargetFinder identified numerous cell type-specific TFs with high feature importance that increase the probability of an enhancer being involved in an interaction when they co-occur nearby the enhancer with CTCF and/or RAD21. This emerged only because we quantified features separately at enhancer, promoter, and window regions.
We also learned that proteins performing multiple functions are rarely predictive on their own. Instead, TargetFinder learns to utilize co-factors that determine their function. For example, histone acetyltransferase EP300 is rarely a top ranked feature, despite its strong association with active enhancers due to its ability to acetylate H3K2746. However, EP300 is correlated with highly predictive cofactors such as C/EBPβ that phosphorylates and modulates the activity of EP300, as well as translocates it to specific gene regions47. The high predictive importance of C/EBPβ may thus be due to its ability to determine the localized activity of EP300.
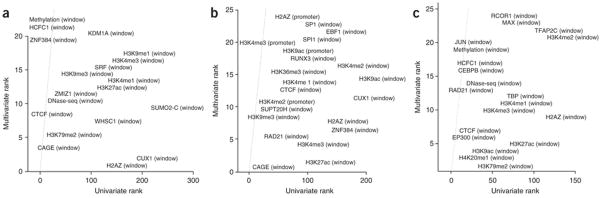
To further explore such context dependence, we plotted the predictive rank of an individual feature against its predictive rank when combined with other features (Figure 8). We observe many off-diagonal features that are not useful on their own (larger rank) but are extremely predictive (lower rank) in combination with additional features. In K562, for example, these include WHSC1, SUMO2, CUX1, and H2AZ. The latter two were assayed in other cell lines and show a similar pattern. Across cell lines, large rank changes commonly include activating histone marks such as H3K9ac and H2AZ that may help distinguish active from poised enhancers and promoters within window regions that cannot be discriminated by single activation marks. The elevated importance of H2AZ might also be explained by the link between H2A ubiquitination and polycomb silencing48. Chromatin modifiers such as methyl- and acetyltransferases also appear to disambiguate the state of enhancers and alternate promoter targets.
Figure 8.
Identification of complex interactions between DNA-binding proteins and epigenetic marks. Scatterplot of univariate feature significance (two-sample Kolmogorov-Smirnov test) versus multivariate feature importance (estimated via a boosted trees classifier) for three cell lines. In order to highlight datasets that are predictive in combination with other features (multivariate) but not predictive alone (univariate), only features with a multivariate rank less than 25 and univariate rank greater than 25 are shown. For example, the lower right corner of K562 shows H2AZ, WHSC1, CUX1, and SUMO2 are among the top 10 predictive features when the co-localization of other proteins is known. H2AZ has similar context-dependent importance in GM12878 and HeLa-S3. Many features predictive in one or more cell lines are not assayed uniformly and thus cannot be included in the combined model (ex: HCFC1, CUX1, SUMO2).
TargetFinder efficiently screens new datasets for relevance to chromatin looping
Motivated by results showing histone modifications can be predicted by TF binding45, we sought to determine if predictive TFs were proxies for important but unassayed post-translational modifications such as ubiquitination or sumoylation. We utilized genome-wide Small Ubiquitin-like Modifier (SUMO) ChIP-seq data for heat shocked and non-shocked K562 cells49 to evaluate the utility of sumoylation for predicting enhancer-promoter interactions. SUMO proteins are involved in protein stability and transcriptional regulation50, and CTCF post-translationally modified by SUMO proteins 1–3 organizes repressive chromatin domains51. When added to the TargetFinder K562 model, sumoylation in the window between an enhancer and promoter is a top predictor of interactions—nearly as important as CTCF. Thus, increased accuracy and insight into mechanisms of looping chromatin will be gained as additional genomic features are measured across many cell lines.
Discussion
This study demonstrates that complex genomic signatures strongly distinguish the true targets of active enhancers from other active but non-interacting promoters in the same loci. These signatures are primarily based on patterns of protein binding and epigenetic modifications on the looping chromatin. A unique feature of our approach is the combination of high resolution genome-wide Hi-C interaction data32 with the vast functional genomics datasets provided by the ENCODE and Roadmap Epigenomics projects partitioned by enhancer, promoter, and window regions. By integrating these diverse datasets and examining their relevance to enhancer-promoter interactions, we computed the most predictive datasets and highlighted the complex interplay between regulatory proteins and DNA in the three-dimensional genome.
Our ability to accurately predict interactions up to 2 Mb apart at high resolution, the identification of minimal sets of predictive features quantified by genomic region, as well as a focus on high resolution intra- rather than inter-TAD interactions, distinguishes TargetFinder from previous work. Machine learning has been shown to accurately identify TADs and other larger chromatin structures (e.g., A and B compartments) from two-dimensional genomic data37, but it has not yet been applied to such fine scale interactions within TADs.
How does TargetFinder distinguish targets from non-target promoters in the same locus?
Our careful examination of many enhancer-promoter pairs across cell lines suggests several broad rules influence TargetFinder’s score of an enhancer-promoter interaction: 1) do the enhancer and other nearby enhancers look particularly active? 2) does the target look like it is actively elongating? 3) is the target promoter cell type-specific? 4) do other promoters near the target have repressive marks or marks of paused polymerase? 5) is another pair interacting within the window? 6) does the interaction appear to cross a contact domain? and 7) are there marks of chromatin remodelers or architectural proteins in the window, or cohesion complex adjacent to the promoter and enhancer, that might facilitate looping interactions?
While some of the predictive accuracy of TargetFinder derives from genomic features that are limited to one or a few cell types, many of the top ranked features are similar across cell types and in the combined model. For example, members of the cohesin complex (SMC3/RAD21) and CTCF are highly predictive, as is CAGE when it is assayed. DNA methylation and Pol II have elevated importance in the combined cell line where the model was trained on fewer datasets that excluded some TFs and other features measured in only a subset of cell lines. Marks of heterochromatin and elongation are also consistently important. These robust, general features of looping chromatin promise to be useful assays for predicting regulatory interactions in new cell types, perhaps in combination with cell type specific regulators. They also suggest that as these predictive features are assayed in more cell types, we may be able to develop a generic TargetFinder model that could perform accurate in silico Hi-C on independent cell types that do not have genome-wide high-resolution chromatin interaction data. To do so will require rigorous normalization, because TargetFinder relies on numeric values of genomic data being comparable across datasets.
Many functional genomics experiments are unexpectedly informative about chromatin interactions
We identified numerous features whose role in distal enhancer-promoter interactions may be under-appreciated. These include the DNA binding proteins CUX1, ZNF384, SUPT20H, RUNX3, SPI1, SP1, EBF1, RCOR1, MAX, TFAP2C, HCFC1, C/EBPβ, JUND, TBP, SRF, ZMIZ1, and WHSC1 (Figure 8). Most of these are predictive only in combination with other features, some of which have roles in chromatin structure. For instance, several interact with the cohesin complex and ZNF143, which was recently shown to provide sequence specificity to cohesin-associated chromatin looping41. Predictive TFs often belong to activating or repressive complexes such as AP-1, AP-2γ, or PRC2, or are chromatin modifiers such as methyl- and acetyltransferases that help determine if enhancers or promoters are in an active or poised state. These general trends are consistent across cell types, but the particular TFs that provide a predictive boost are often specific to a small number of cell lines. In addition, we identified several more general predictors of looping chromatin. Sumoylation is a combinatorially predictive post-translational modification not assayed by ENCODE or Roadmap Epigenomics. The activating marks H2AZ/H3K9ac and elongation marks H3K36me3/H3K79me2 were also especially useful for chromatin loop prediction, more so than many of the well-known histone marks necessary for ChromHMM/Segway annotations of promoters and enhancers. CAGE is also a consistently top-ranked feature, providing information on the activation state of annotated enhancers and alternate targets in the window that is complementary to ChIP-seq assays.
Many of the top features utilized by TargetFinder are not predictive on their own. The association of these combinatorially predictive features with chromatin looping has been established to varying degrees, though our discovery that they provide specificity to interaction predictions is novel. Examples include SRF which regulates FOS52 and interacts with C/EBPβ53, TFAP2C (AP-2γ) which is a pioneer factor associated with estrogen receptor binding events and FOXA1 expression54, ZMIZ1 (hZimp10) which promotes expression and sumoylation of the androgen receptor55, and KDM1A which interacts with RCOR1 to demethylate H3K456. We identified several other proteins with poor univariate importance that nonetheless have known roles in chromatin looping and were highly ranked by TargetFinder. These include SP157,58, SPI1 (PU.1)59,60, HCFC1 which co-localizes with looping factor ZNF14342, and TBP whose TAF3 subunit is recruited by CTCF to distal promoters61 and which is linked with long range interactions62. Finally, WHSC1 (NSD2) is a histone methyltransferase of H3K36me3 and therefore is associated with predictive marks of elongation63. Thus, changes in univariate versus multivariate predictive rank recapitulate known protein interactions as well as identify under-appreciated or potentially novel biological interactions, often involving cell line-specific TFs.
DNA between interacting enhancers and promoters carries a distinct genomic signature
Our predictive accuracy and biological insights depended critically on the decision to include genomic data from the window between each enhancer and promoter in the analyses. We discovered these window features dominated those encoding chromatin states at the promoter and enhancer themselves. Because all enhancers and promoters we studied, including non-interacting pairs, had sufficient activation marks to be called by ChromHMM/Segway, our analysis revealed more subtle and complex genomic signatures that distinguish regulatory targets from poised, paused, or insulated promoters. The genomic signature of looping DNA has several components. First, interacting pairs are depleted for insulator and contact domain crossings in the window (Supplementary Figures S4 and S12), particularly for more distal interactions. Second, interacting pairs are depleted for cohesin complex bound to the window (Supplementary Figure S9), although it is prevalent near the enhancer and promoter. Third, DNA between interacting enhancers and promoters tends to lack activating TFs and epigenetic marks of elongation (Supplementary Figure S10) which could indicate the presence of an alternative promoter target, and indeed is depleted for active promoters (Supplementary Figure S14). On the other hand, windows do contain epigenetic marks associated with heterochromatin (Supplementary Figure S11), polycomb-associated proteins, and co-factors of CTCF associated with its insulator function. Looping interactions in the window are highly enriched (Supplementary Figure S13), strongly supporting existing evidence for TADs or contact domains and suggesting window features may be a proxy for domain membership.
These results are more relevant to looping models of interaction than alternatives such as facilitated tracking64. Polycomb complexes appear to play several roles in distinguishing nearby targets. For example, PRC2-targeted CpG islands are enriched for REST and CUX1 binding motifs, both transcriptional repressors65 with high predictive importance. In Drosophila, cohesin co-localizes with PRC1 at promoters and interacts to control gene silencing66. Given the conservation of PRC between flies and humans67, this has implications for the interaction of cohesin and PRC for mammalian gene silencing and thus discrimination of target promoters. Also, distal enhancers may sometimes serve to clear PRC from CpG islands68. Elongation has recently been shown to spatially segregate genes in the Hoxd locus present in separate TADs69, suggesting its role in inter-TAD gene clusters could contribute to its predictive importance. Finally, recent work shows that cohesin spatially clusters enhancers70 and is consistent with our observation that the presence of active marks at nearby enhancers often increase the likelihood of interaction. These are several of many possible explanations for the ability of window-based features to predict distal enhancer-promoter interactions with high precision and recall—explanations that may be refined by analysis of new functional genomics datasets.
Materials and Methods
All code and data is accessible via https://github.com/shwhalen/targetfinder.
Identification of regulatory elements
TSS-containing promoter regions and strong and weak enhancer regions were identified using combined ENCODE Segway71 and ChromHMM72 annotations for K562, GM12878, HeLa-S3, and HUVEC and Roadmap Epigenomics ChromHMM annotations for NHEK and IMR90. Enhancers closer than 10 Kb to the nearest promoter were discarded to focus the model on distal interactions. Promoters were retained if actively transcribed (mean FPKM > 0.373 with irreproducible discovery rate < 0.174) in each cell line using GENCODE75 version 19 annotations and RNA-seq data from the ENCODE portal (http://encodeproject.org/data/annotations). Promoter and enhancer counts per line are in Supplemental Table S1.
Chromatin interactions
Interacting enhancer-promoter pairs were annotated using high-resolution genome-wide Hi-C data (10% FDR, GEO accession GSE63525)32. These were assigned to one of 5 bins based on the distance between enhancer and promoter, such that each bin had the same number of interactions. Noninteracting enhancer-promoter pairs were assigned to their corresponding distance bin, then subsampled within each bin using 20 negatives per positive (Supplemental Table S1). Performance was similar without distance matching, losing approximately 1% F1 per 250,000 additional samples (a total loss of 6% F1 for K562).
Genomic features
Functional genomics data for each cell line were downloaded from ENCODE, Roadmap Epigenomics, or GEO; details and accessions are given in Supplemental Table S2. Peak calls for ENCODE data were obtained from GEO; raw reads for Roadmap Epigenomics and GEO datasets were obtained, quality trimmed using fastq-mcf, aligned to hg19 using bowtie276, and peak called using macs277 with default parameters. Peaks were intersected with promoter, enhancer, extended enhancer, and window regions. The strength of all peaks in a region, or counts of methylated bases in a region, were summed and divided by the length of the region in bp to generate features.
Software implementation
TargetFinder was implemented in Python using the scikit-learn machine learning library78, the pandas analytics library79, and bedtools80. We used DummyClassifier to measure baseline performance, LinearSVC for a linear Support Vector Machine81, DecisionTreeClassifier for a single decision tree82, and GradientBoostingClassifier for a decision tree ensemble83. The linear SVM was fit with parameter class weight = “balanced” as part of a Pipeline with a StandardScaler pre-processing step. The boosting classifier was fit with parameters n_estimators = 4000, learning_rate = 0.1, max_depth = 5, and max_features = “log2”. Models were fit with sample weights inversely proportional to class balance in order to prevent over-fitting the negative class. Identical parameters were used per cell line. Results were consistent with an alternative implementation in R (Supplemental Text).
All models were evaluated using 10-fold cross-validation where data is divided into 10 non-overlapping training and test sets. Performance was measured using multiple metrics, and the average over all test sets is reported. Feature importances were computed by scikit-learn using the method of Hastie et al.84 accessible via the feature_importances_ attribute of eligible models. The following pseudocode summarizes their implementation:
ensemble_importances = zeros(total_features)
for each tree in ensemble:
tree_importances = zeros(total_features)
for each node in tree:
if node is not a leaf:
tree_importances[node.feature_index] +=
node.sample_count * node.impurity -
node.left_child.sample_count * node.left_child.impurity -
node.right_child.sample_count * node.right_child.impurity
ensemble_importances += tree_importances / total_samples
ensemble_importances /= total_trees
where zeros(n) initializes an array of n zeros, total features is the total number of features in the dataset, node.feature index is the index of the feature used to split samples at a node, node.sample count is the number of samples present at a node before splitting, node.impurity is a measure of error (here, gini impurity), and node.left child and node.right child point to the children of a node. Overall, this method sums the weighted reduction in impurity when splitting on each feature across all trees in the ensemble, normalized by the number of samples per tree and total number of trees. Models were fit 10 times, each with a different random number seed, in order to better estimate the mean and variance of feature importances.
Recursive Feature Elimination (RFE)85 was used to estimate the optimal number of features via nested cross-validation86). Within each training set during “outer” cross-validation, feature importances are initially estimated using all features. The performance of the top n features is then estimated from “inner” cross-validation on the training set, with n increasing from 1 to the number of features by powers of 2. Finally, the best performing subset identified via inner cross-validation is evaluated against the outer test set to obtain an unbiased performance estimate.
Supplementary Material
Figure S1: Distribution of active enhancer lengths for each cell line.
Figure S2: Distribution of active promoter lengths for each cell line.
Figure S3: Distribution of genomic distance between interacting enhancers and promoters for each cell line.
Figure S4: Average number of contact domains (estimated by the arrowhead algorithm of Rao et al.32) crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs.
Figure S5: Significant peaks for the top 12 predictive datasets of an interacting promoter (P1) and enhancer (E1) in GM12878, separated by other active promoters and enhancers. Active enhancers are segments marked “E” by combined ChromHMM/Segway annotations, and active promoters are segments marked “TSS” and expressed in GM12878 with RPKM > 0.3. Ensembl genes are also displayed, with introns denoted as thin lines and exons as squares. Left and right fragments of the Hi-C assay are also shown to visually confirm E1 interacts with P1. This figure demonstrates the potential complexities of interpreting interactions. First, note that P1 is the target of multiple regulatory elements, one of which was not classified by ChromHMM/Segway as an active enhancer. This regulatory element lacks a RAD21 peak commonly associated with looping and also has a heterochromatin-associated H3K9me3 mark. However, RAD21 is not always present at looping interactions, and H3K9me3 is also associated with activation in cancer cells94. Second, note that E1 is an intronic enhancer, existing within the CUTC gene but interacting with P1 via long-range looping. Finally, note that non-target P2 lacks RAD21 but otherwise has many activation-associated marks. Thus, the looping interaction of an enhancer and promoter is defined by the complex interplay of protein binding and other features that often cannot be explained by the presence or absence of a single feature.
Figure S6: Held-out performance by cell line, model type, and region. Performance was measured separately for classifiers using features at enhancer and promoter loci (E/P), features at extended enhancer (+/− 3 Kb) and promoter loci (EE/P), and features using the full “window” between enhancer and promoter in addition to the enhancer and promoter loci themselves (E/P/W). This figure demonstrates the importance of capturing feature interactions, as well as binding activity outside the enhancer and promoter. Much of the relevant activity flanks the enhancer, though use of the complete window typically maximizes performance. Baseline performance was estimated by generating random predictions from the empirical distribution of class labels. Reported performance is a balance of precision and recall (F1) averaged over 10 cross-validation folds.
Figure S7: Consistency of CTCF ChIP-seq signals performed at 3 different institutions across 3 ENCODE cell lines. Signals are centered at transcription start sites and displayed as the ratio of signal at interacting vs non-interacting sites. This figure shows how variability between, and even within, cell lines contributes to the difficulty of using a model trained on one cell line to predict interactions in another.
Figure S8: Performance as a function of distance between enhancer and promoter. Samples are assigned to distance bins such that each bin has an equal number of interacting pairs. Performance is often higher for extremely distal interaction bins since TAD boundaries reduce the probability of interaction, and such boundaries are more likely to occur within the window between distal enhancer-promoter pairs. (Error bars = 95% c.i.)
Figure S9: Average number of RAD21 peak summits crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This suggests looping interactions within the window of a candidate enhancer-promoter pair act as a kind of insulator that decreases the likelihood of interaction.
Figure S10: Average number of H3K36me3 peaks crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for all but the first distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This suggests elongation within the window of a candidate enhancer-promoter pair decreases the likelihood of interaction.
Figure S11: Average number of H4K20me1 peaks crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This shows repression-associated heterochromatin is depleted in the window of interacting enhancer-promoter pairs.
Figure S12: Average number of CTCF peak summits crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This reinforces current evidence that interactions preferentially occur within TADs or contact domains demarcated by CTCF acting as an insulator to prevent spurious enhancer-promoter interactions.
Figure S13: Average number of looping interactions crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This supports the idea that interactions are enriched within TADs or contact domains and that window features may be a proxy for such domains.
Figure S14: Average number of active promoters crossedby interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. Active promoters within the window of a candidate interaction offer alternate targets for the enhancer and thus decrease the likelihood of interaction.
Figure S15: Cluster heatmap of the correlation between the top 16 predictive features for K562.
Figure S16: Cluster heatmap of the correlation between the top 16 predictive features for GM12878.
Figure S17: Cluster heatmap of the correlation between the top 16 predictive features for HeLa-S3.
Figure S18: Cluster heatmap of the correlation between the top 16 predictive features for HUVEC.
Figure S19: Cluster heatmap of the correlation between the top 16 predictive features for IMR90.
Figure S20: Cluster heatmap of the correlation between the top 16 predictive features for NHEK.
Table S1: Counts of promoters, enhancers, interacting pairs, and non-interacting pairs per cell line.
Table S2: Datasets and accession ids for each cell line, with blanks indicating unavailability.
Table S3: TargetFinder performance on held out data. Counts of true and false positives and negatives are given for each classifier and cell line, as well as several summary metrics including area under the ROC curve, Matthews correlation coefficient (φ), and a balance of precision and recall (F1). Performance was measured separately for classifiers using features at enhancer and promoter loci (E/P), features at extended enhancer (+/− 3 Kb) and promoter loci (EE/P), and features using the full window between enhancer and promoter in addition to the enhancer and promoter loci themselves (E/P/W). This table demonstrates the importance of binding activity outside the enhancer and promoter for accurate prediction of interactions, and that much of this activity occurs in a relatively small region around the enhancer – though the complete window typically maximizes performance. Baseline performance was estimated by generating random predictions from the empirical distribution of class labels.
Acknowledgments
This project was supported by the Bench to Bassinet Program of the NHLBI (U01HL098179, UM1HL098179), NIH/NHLBI (HL089707), the San Simeon Fund, and the Gladstone Institutes.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010;20:311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov NN, et al. Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol. 2006;60:69–85. doi: 10.1007/s11103-005-2564-9. [DOI] [PubMed] [Google Scholar]
- 4.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 5.Massouras A, et al. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 2012;8:e1003055. doi: 10.1371/journal.pgen.1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang R, et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 2014;15:R25. doi: 10.1186/gb-2014-15-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gusev A, et al. Regulatory variants explain much more heritability than coding variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 10.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lettice LA. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 20.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvon EZ, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Rendon A, Wernisch L. Transcription factor and chromatin features predict genes associated with eQTLs. Nucleic Acids Res. 2013;41:1450–1463. doi: 10.1093/nar/gks1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip KY, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rödelsperger C, et al. Integrative analysis of genomic, functional and protein interaction data predicts long-range enhancer-target gene interactions. Nucleic Acids Res. 2011;39:2492–2502. doi: 10.1093/nar/gkq1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilczynski B, Liu YH, Yeo ZX, Furlong EEM. Predicting spatial and temporal gene expression using an integrative model of transcription factor occupancy and chromatin state. PLoS Comput Biol. 2012;8:e1002798. doi: 10.1371/journal.pcbi.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 30.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Wit E, de Laat W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015 doi: 10.1101/gr.185272.114. gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 36.Maston GA, Evans SK, Green MR. Transcriptional Regulatory Elements in the Human Genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 37.Moore BL, Aitken S, Semple CA. Integrative modeling reveals the principles of multi-scale chromatin boundary formation in human nuclear organization. Genome Biol. 2015;16:110. doi: 10.1186/s13059-015-0661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6:85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 41.Bailey SD, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaud J, et al. HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. Genome Res. 2013;23:907–916. doi: 10.1101/gr.150078.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benveniste D, Sonntag HJ, Sanguinetti G, Sproul D. Transcription factor binding predicts histone modifications in human cell lines. Proc Natl Acad Sci U S A. 2014;111:13367–13372. doi: 10.1073/pnas.1412081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz C, et al. Recruitment of p300 by C/EBPbeta triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 2003;22:882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 49.Niskanen EA, et al. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hay RT. SUMO: A History of Modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 51.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujioka S, et al. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–7819. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanlon M, Sealy L. Ras regulates the association of serum response factor and CCAAT/enhancer-binding protein beta. J Biol Chem. 1999;274:14224–14228. doi: 10.1074/jbc.274.20.14224. [DOI] [PubMed] [Google Scholar]
- 54.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12:381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 55.Sharma M, et al. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003;22:6101–6114. doi: 10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S. Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc Natl Acad Sci U S A. 2014;111:8071–8076. doi: 10.1073/pnas.1404292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nolis IK, et al. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deshane J, et al. Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. J Biol Chem. 2010;285:16476–16486. doi: 10.1074/jbc.M109.058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Listman JA, et al. Conserved ETS domain arginines mediate DNA binding, nuclear localization, and a novel mode of bZIP interaction. J Biol Chem. 2005;280:41421–41428. doi: 10.1074/jbc.M509143200. [DOI] [PubMed] [Google Scholar]
- 60.van Riel B, Rosenbauer F. Epigenetic control of hematopoiesis: the PU. 1 chromatin connection. Biol Chem. 2014;395:1265–1274. doi: 10.1515/hsz-2014-0195. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertolino E, Singh H. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol Cell. 2002;10:397–407. doi: 10.1016/s1097-2765(02)00597-x. [DOI] [PubMed] [Google Scholar]
- 63.Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 64.Blackwood EM, Kadonaga JT. Going the Distance: A Current View of Enhancer Action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 65.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10:413–424. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dorsett D, Kassis JA. Checks and balances between cohesin and polycomb in gene silencing and transcription. Curr Biol. 2014;24:R535–9. doi: 10.1016/j.cub.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levine SS, et al. The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vernimmen D, et al. Polycomb eviction as a new distant enhancer function. Genes Dev. 2011;25:1583–1588. doi: 10.1101/gad.16985411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fabre PJ, et al. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc Natl Acad Sci U S A. 2015;112:201517972. doi: 10.1073/pnas.1517972112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ing-Simmons E, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25 doi: 10.1101/gr.184986.114. gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffman MM, et al. Unsupervised Pattern Discovery in Human Chromatin Structure Through Genomic Segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–1779. [Google Scholar]
- 75.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 79.McKinney W. Python for Data Analysis. O’Reilly; 2012. [Google Scholar]
- 80.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burges CJC. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min Knowl Discov. 1998;2:121–167. [Google Scholar]
- 82.Kingsford C, Salzberg SL. What are decision trees? Nat Biotechnol. 2008;26:1011–1013. doi: 10.1038/nbt0908-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friedman JH. Stochastic Gradient Boosting. Comput Stat Data Anal. 2002;38:367–378. [Google Scholar]
- 84.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer; 2009. [Google Scholar]
- 85.Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 86.Ambroise C, McLachlan GJ. Selection Bias in Gene Extraction on the Basis of Microarray Gene-Expression Data. Proc Natl Acad Sci U S A. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- 88.Law A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 89.Ridgeway G. Generalized boosted models: A guide to the gbm package. 2005. [Google Scholar]
- 90.Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 91.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan J, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 93.Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining; 1996. pp. 226–231. [Google Scholar]
- 94.Wiencke JK, Zheng S, Morrison Z, Yeh RF. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2008;27:2412–2421. doi: 10.1038/sj.onc.1210895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Distribution of active enhancer lengths for each cell line.
Figure S2: Distribution of active promoter lengths for each cell line.
Figure S3: Distribution of genomic distance between interacting enhancers and promoters for each cell line.
Figure S4: Average number of contact domains (estimated by the arrowhead algorithm of Rao et al.32) crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs.
Figure S5: Significant peaks for the top 12 predictive datasets of an interacting promoter (P1) and enhancer (E1) in GM12878, separated by other active promoters and enhancers. Active enhancers are segments marked “E” by combined ChromHMM/Segway annotations, and active promoters are segments marked “TSS” and expressed in GM12878 with RPKM > 0.3. Ensembl genes are also displayed, with introns denoted as thin lines and exons as squares. Left and right fragments of the Hi-C assay are also shown to visually confirm E1 interacts with P1. This figure demonstrates the potential complexities of interpreting interactions. First, note that P1 is the target of multiple regulatory elements, one of which was not classified by ChromHMM/Segway as an active enhancer. This regulatory element lacks a RAD21 peak commonly associated with looping and also has a heterochromatin-associated H3K9me3 mark. However, RAD21 is not always present at looping interactions, and H3K9me3 is also associated with activation in cancer cells94. Second, note that E1 is an intronic enhancer, existing within the CUTC gene but interacting with P1 via long-range looping. Finally, note that non-target P2 lacks RAD21 but otherwise has many activation-associated marks. Thus, the looping interaction of an enhancer and promoter is defined by the complex interplay of protein binding and other features that often cannot be explained by the presence or absence of a single feature.
Figure S6: Held-out performance by cell line, model type, and region. Performance was measured separately for classifiers using features at enhancer and promoter loci (E/P), features at extended enhancer (+/− 3 Kb) and promoter loci (EE/P), and features using the full “window” between enhancer and promoter in addition to the enhancer and promoter loci themselves (E/P/W). This figure demonstrates the importance of capturing feature interactions, as well as binding activity outside the enhancer and promoter. Much of the relevant activity flanks the enhancer, though use of the complete window typically maximizes performance. Baseline performance was estimated by generating random predictions from the empirical distribution of class labels. Reported performance is a balance of precision and recall (F1) averaged over 10 cross-validation folds.
Figure S7: Consistency of CTCF ChIP-seq signals performed at 3 different institutions across 3 ENCODE cell lines. Signals are centered at transcription start sites and displayed as the ratio of signal at interacting vs non-interacting sites. This figure shows how variability between, and even within, cell lines contributes to the difficulty of using a model trained on one cell line to predict interactions in another.
Figure S8: Performance as a function of distance between enhancer and promoter. Samples are assigned to distance bins such that each bin has an equal number of interacting pairs. Performance is often higher for extremely distal interaction bins since TAD boundaries reduce the probability of interaction, and such boundaries are more likely to occur within the window between distal enhancer-promoter pairs. (Error bars = 95% c.i.)
Figure S9: Average number of RAD21 peak summits crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This suggests looping interactions within the window of a candidate enhancer-promoter pair act as a kind of insulator that decreases the likelihood of interaction.
Figure S10: Average number of H3K36me3 peaks crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for all but the first distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This suggests elongation within the window of a candidate enhancer-promoter pair decreases the likelihood of interaction.
Figure S11: Average number of H4K20me1 peaks crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This shows repression-associated heterochromatin is depleted in the window of interacting enhancer-promoter pairs.
Figure S12: Average number of CTCF peak summits crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This reinforces current evidence that interactions preferentially occur within TADs or contact domains demarcated by CTCF acting as an insulator to prevent spurious enhancer-promoter interactions.
Figure S13: Average number of looping interactions crossed by interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. This supports the idea that interactions are enriched within TADs or contact domains and that window features may be a proxy for such domains.
Figure S14: Average number of active promoters crossedby interacting and non-interacting enhancer-promoter pairs, grouped by distance bin. The difference in mean number of crossings is significant for each distance bin (two-sample Wilcoxon test) and increases for more distal pairs. Active promoters within the window of a candidate interaction offer alternate targets for the enhancer and thus decrease the likelihood of interaction.
Figure S15: Cluster heatmap of the correlation between the top 16 predictive features for K562.
Figure S16: Cluster heatmap of the correlation between the top 16 predictive features for GM12878.
Figure S17: Cluster heatmap of the correlation between the top 16 predictive features for HeLa-S3.
Figure S18: Cluster heatmap of the correlation between the top 16 predictive features for HUVEC.
Figure S19: Cluster heatmap of the correlation between the top 16 predictive features for IMR90.
Figure S20: Cluster heatmap of the correlation between the top 16 predictive features for NHEK.
Table S1: Counts of promoters, enhancers, interacting pairs, and non-interacting pairs per cell line.
Table S2: Datasets and accession ids for each cell line, with blanks indicating unavailability.
Table S3: TargetFinder performance on held out data. Counts of true and false positives and negatives are given for each classifier and cell line, as well as several summary metrics including area under the ROC curve, Matthews correlation coefficient (φ), and a balance of precision and recall (F1). Performance was measured separately for classifiers using features at enhancer and promoter loci (E/P), features at extended enhancer (+/− 3 Kb) and promoter loci (EE/P), and features using the full window between enhancer and promoter in addition to the enhancer and promoter loci themselves (E/P/W). This table demonstrates the importance of binding activity outside the enhancer and promoter for accurate prediction of interactions, and that much of this activity occurs in a relatively small region around the enhancer – though the complete window typically maximizes performance. Baseline performance was estimated by generating random predictions from the empirical distribution of class labels.