Abstract
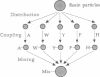
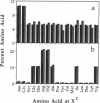
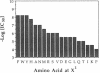
A fully automated peptide synthesizer has been constructed that is capable of the simultaneous synthesis of up to 36 individual peptides and the synthesis of equimolar peptide mixtures. The instrument consists of an array of reaction vessels, a series of solenoid valves to control liquid flow, and a Zymark robot to deliver solvents and reagents; all components are computer controlled and coordinated. Equimolar peptide mixtures are obtained by algorithms that automate the mixing and distribution of peptide-resin particles. This technology was used to synthesize a library of 361 peptides, generated by randomizing two critical binding residues of a 10-mer epitope known to bind an anti-human immuno-deficiency virus gp120 monoclonal antibody. Each critical residue was substituted with 19 amino acids consisting of all the natural amino acids except cysteine. The library was synthesized as 19 pools, each containing 19 peptides. Each pool was screened in a solution-phase competition ELISA assay. The 12 most inhibitory peptides in the library were isolated by a rapid affinity-selection method and were identified by mass spectrometry and amino acid analysis. The binding properties of these 12 selected peptides were verified by synthesis and assay of the individual peptides. The two critical residues investigated were found to contribute independently to antibody binding.
Keywords: chemical diversity, peptide library, multiple-peptide synthesis
Full text
PDF
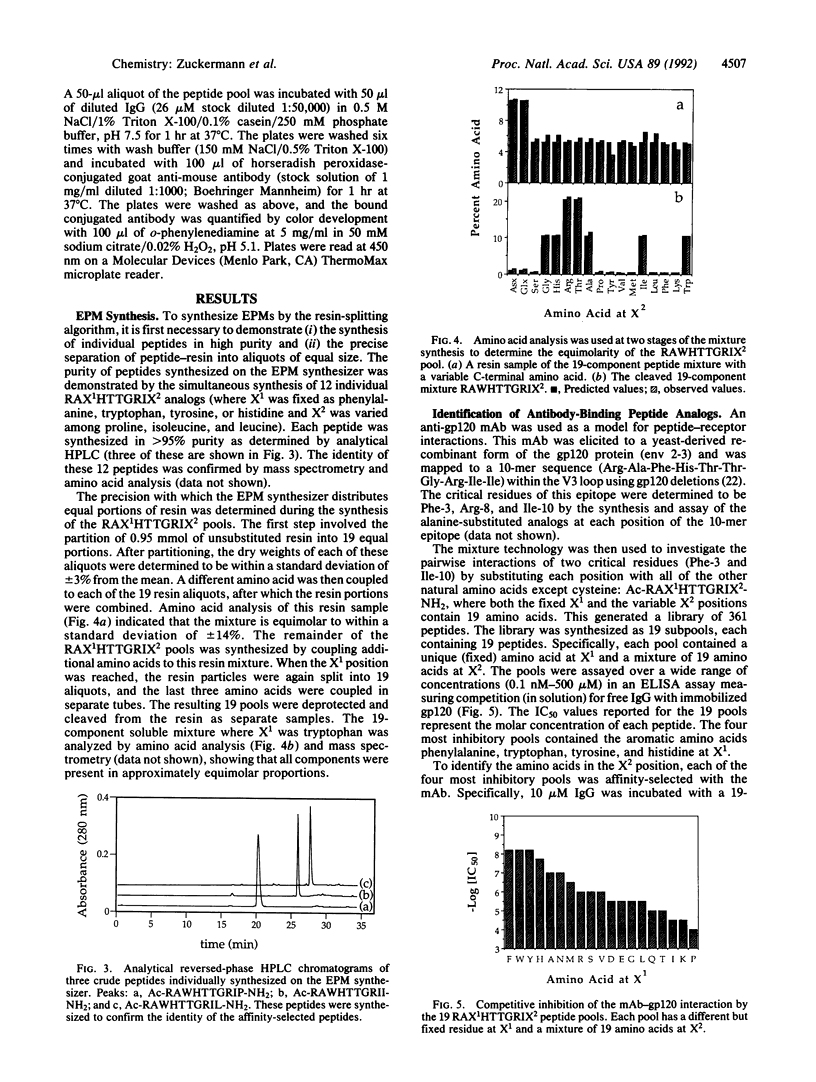
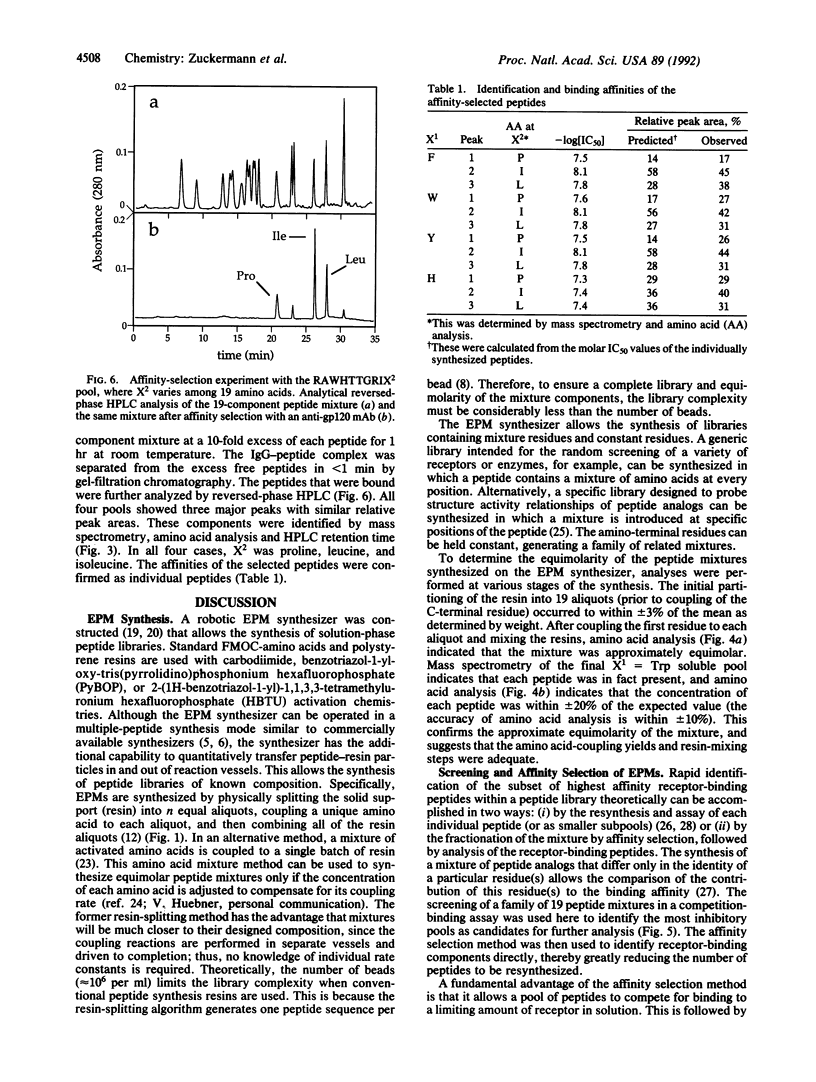

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Chapman B. S., Thayer R. M., Vincent K. A., Haigwood N. L. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991 Jul 25;19(14):3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwirla S. E., Peters E. A., Barrett R. W., Dower W. J. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J. J., Panganiban L. C., Devlin P. E. Random peptide libraries: a source of specific protein binding molecules. Science. 1990 Jul 27;249(4967):404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Fields G. B., Noble R. L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990 Mar;35(3):161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991 Feb 15;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Furka A., Sebestyén F., Asgedom M., Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991 Jun;37(6):487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Geysen H. M., Meloen R. H., Barteling S. J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geysen H. M., Rodda S. J., Mason T. J. A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol Immunol. 1986 Jul;23(7):709–715. doi: 10.1016/0161-5890(86)90081-7. [DOI] [PubMed] [Google Scholar]
- Haigwood N. L., Shuster J. R., Moore G. K., Lee H., Skiles P. V., Higgins K. W., Barr P. J., George-Nascimento C., Steimer K. S. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res Hum Retroviruses. 1990 Jul;6(7):855–869. doi: 10.1089/aid.1990.6.855. [DOI] [PubMed] [Google Scholar]
- Houghten R. A. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5131–5135. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghten R. A., Pinilla C., Blondelle S. E., Appel J. R., Dooley C. T., Cuervo J. H. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991 Nov 7;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- King D. S., Fields C. G., Fields G. B. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Pept Protein Res. 1990 Sep;36(3):255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Lam K. S., Salmon S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., Knapp R. J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991 Nov 7;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Petithory J. R., Masiarz F. R., Kirsch J. F., Santi D. V., Malcolm B. A. A rapid method for determination of endoproteinase substrate specificity: specificity of the 3C proteinase from hepatitis A virus. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11510–11514. doi: 10.1073/pnas.88.24.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. Searching for peptide ligands with an epitope library. Science. 1990 Jul 27;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Tjoeng F. S., Towery D. S., Bulock J. W., Whipple D. E., Fok K. F., Williams M. H., Zupec M. E., Adams S. P. Multiple peptide synthesis using a single support (MPS3). Int J Pept Protein Res. 1990 Feb;35(2):141–146. doi: 10.1111/j.1399-3011.1990.tb00249.x. [DOI] [PubMed] [Google Scholar]
- Wells J. A. Additivity of mutational effects in proteins. Biochemistry. 1990 Sep 18;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Wetzel R. Learning from the immune system: laboratory methods for creating and refining molecular diversity in polypeptides. Protein Eng. 1991 Apr;4(4):371–374. doi: 10.1093/protein/4.4.371. [DOI] [PubMed] [Google Scholar]