Abstract
While folding or performing functions, a protein can sample a rich set of conformational space. However, experimentally capturing all of the important motions with sufficient detail to allow a mechanistic description of their dynamics is nontrivial since such conformational events often occur over a wide range of time and length scales. Therefore, many methods have been employed to assess protein conformational dynamics and, depending on the nature of the conformational transition in question, some may be more advantageous than others. Herein, we describe our recent efforts, and also those of others, wherever appropriate, to use infrared- and fluorescence-based techniques to interrogate protein folding and functional dynamics. Specifically, we focus on discussing how to use extrinsic spectroscopic probes to enhance the structural resolution of these techniques and how to exploit various cross-linking strategies to acquire dynamic and mechanistic information that was previously difficult to attain.
INTROUDUCTION
The biological function of a protein is primarily defined by its structure, which in turn is encoded in its amino acid sequence. Thus, tremendous effort has been devoted to deciphering this sequence-structure relationship or, in other words, how proteins fold.1-4 In addition, rather than a simple series of static structural building blocks, folded proteins are dynamic objects; they undergo a varying degree of spontaneous conformational fluctuations and, when other molecules or interactions are present, their structures can be induced to change. Such conformational flexibilities or dynamics are often essential for a protein to carry out its function. Hence, another active area of research in biochemistry and biophysics is to understand the structure-dynamics-function relationship of proteins.5-8 While we now know a great deal about how proteins fold as well as the role of dynamics in function, many fundamental questions still remain. This is because the free energy landscape and hence the conformational dynamics, for any given protein, depends on a large number of degrees of freedom, making it difficult to experimentally capture and dissect the many underlying factors at play. In this regard, we have employed a divide-and-conquer approach to study the protein folding problem as well as protein functional dynamics, using infrared (IR) and fluorescence spectroscopic techniques.
The folded state of a protein consists of well-packed sidechains and secondary structures. Thus, the ultimate goal of the experimental study of protein folding dynamics is to create a molecular ‘movie’ to depict, in a frame-by-frame manner and with all necessary mechanistic details, how and when those structural elements are formed to produce the folded conformation. Although achieving this goal has proven to be difficult, significant progress has been made over the past decade toward elucidating some of the fundamental factors governing protein folding dynamics and mechanisms and, in some cases, creating low-resolution folding movies (i.e., by following one or few experimental observables). This includes, but is not limited to, measurement of the folding transition-path time,9,10 assessment of the role of specific and nonspecific interactions as well as native and nonnative contacts in controlling protein folding dynamics,11,12 detection of folding intermediates,13,14 uncovering parallel folding pathways,15-18 and identification of the structural origin of slow conformational diffusion in protein folding dynamics.12,19 In the context of those developments and advancements, herein we describe our contributions to the field. Specifically, we focus on discussing our recent efforts in developing new strategies to extract more specific structural and/or mechanistic information from conventional protein folding experiments, which include 1) using a multi-probe approach to enhance the structural resolution of folding kinetic measurements; 2) using a varying-initial-potential temperature-jump (VIPT-jump) method to distinguish between different folding scenarios; and 3) using various cross-linkers to facilitate determination of the attempt frequency of barrier crossing, to measure the magnitude of internal friction, and to engineer a transition state (TS) analog. In addition, we provide a brief summary of our recent studies in interrogating the effect of naturally occurring osmolytes (or cosolvents) on protein hydration and conformational dynamics using linear and nonlinear IR spectroscopic methods as well as an example wherein we use a single-molecule fluorescence technique to assess protein functional dynamics.
METHODS
The three key methods used to obtain the experimental results discussed in this article are (A) nanosecond transient IR spectroscopy, (B) two-dimensional IR (2D IR) spectroscopy, and (C) fluorescence correlation spectroscopy (FCS). Specifically, all the results related to protein folding dynamics were acquired with method A and the results related to the cosolvent effect and the M2 proton channel were acquired with either method B or C.
Nanosecond transient IR setup
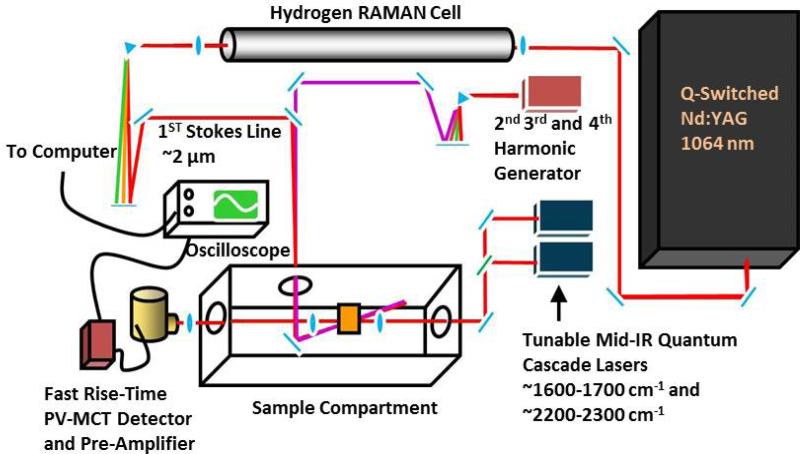
As shown (Figure 1),20 the transient IR absorption apparatus consists of a nanosecond pump source and a continuous wave (CW) quantum cascade (QC) IR laser (Daylight Solutions, CA) to serve as the probe source. For the laser-induced temperature-jump (T-jump) experiments, the wavelength of the pump pulse (~3 ns) was tuned to ~1.9 μm via Raman shifting the fundamental output of an Nd-YAG laser (Infinity, Coherent Radiation, CA) using H2. For the other transient IR measurements discussed, the third harmonic of the Minilite II Nd-YAG laser (Continuum Electro-Optic, Inc., CA) was used as the pump pulse. The pump-induced change in the probe signal was measured by a 50 MHz liquid nitrogen cooled HgCdTe (MCT) detector (Kolmar Technologies, MA) and signal digitization was accomplished by a digital oscilloscope (Tektronix, OR). The sample holder was constructed with two CaF2 windows and a 50 μm spacer. For most experiments, the sample concentration was in the range of 1-4 mM.
Figure 1.
Schematic diagram of the nanosecond transient IR setup. Adapted from Ref. 20 with permission.
2D IR setup
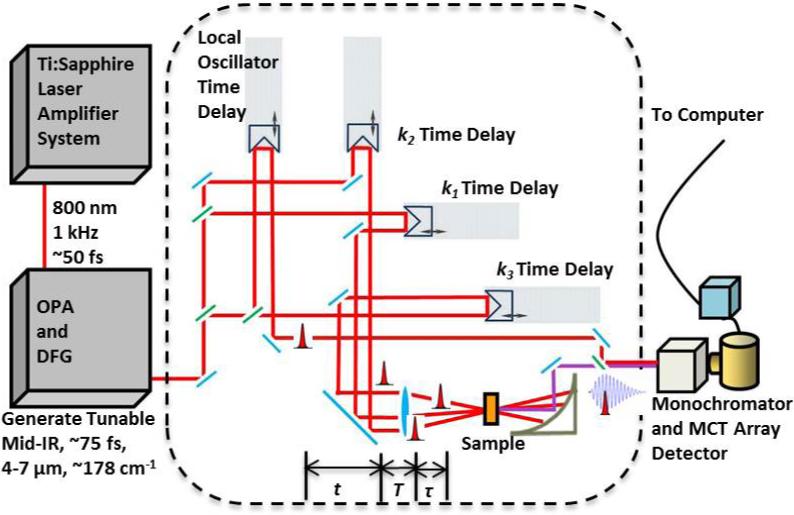
As shown (Figure 2), the 2D IR photon echo signal (−k1 + k2 + k3) is produced by focusing three ultrafast (<100 fs) IR pulses (k1, k2, and k3) onto the sample in a box-car geometry.21 Detection of this emitted signal was achieved via heterodyning its field with that of the local oscillator (LO) pulse. Specifically, this mixed signal at each set of delay times (i.e., τ and T) was dispersed by a monochromator and the spectral interferometry signals were detected by a 64-element liquid nitrogen cooled MCT array detector (InfraRed Associates, FL). Subsequent data analysis includes the Fourier transformation of the emitted signal acquired at a specific delay time T along two time axes, namely τ and t, which spreads the photon echo signal into two frequencies, ωτ and ωt,. The sample concentration was typically between 10-50 mM.
Figure 2.
Schematic diagram of the 2D-IR setup.
FCS setup
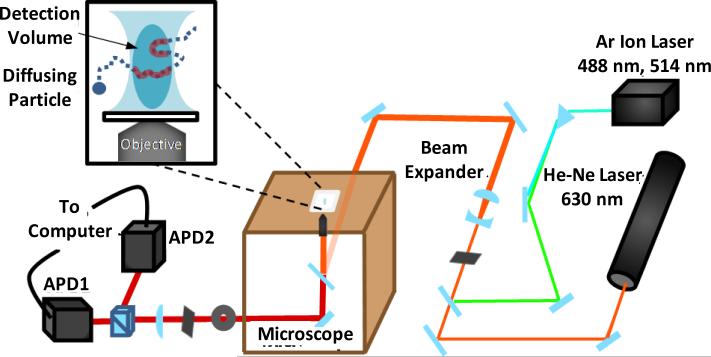
A schematic diagram of our FCS setup is shown in Figure 3. The excitation light was derived from either an argon ion laser (Spectra-Physics, CA) or a helium-neon laser (JDS Uniphase Corporation, CA), depending on the fluorophore used. Light focusing was achieved by a confocal microscope (Nikon Eclipse TE 300) equipped with an oil immersion objective (100× or 60×) and the confocal volume was defined by a 50 μm pinhole. A Hanbury-Brown and Twiss setup, which involves a 50/50 nonpolarizing beamsplitter (Newport, CA) and two avalanche photodiodes (APDs) (Perkin Elmer, NJ), was used to increase the time resolution. Cross-correlation was accomplished by a Flex 03-LQ-01 autocorrelation card (Correlator.com, NJ). The sample concentration was typically in the range of 1 - 10 nM.
Figure 3.
Schematic diagram of the FCS Setup.
RESULTS AND DISSCUSSION
Protein folding energy landscape theory22,23 provides a conceptual framework for us to understand why and how an unfolded protein can spontaneously find its folded state, via a conformational diffusion search on a multi-dimensional free energy surface. However, there is currently no experimental technique that is capable of capturing the entire trajectory of this process with sufficient temporal and structural resolution. In fact, most experimental studies of protein folding kinetics performed so far have relied on one observable, such as the fluorescence of a tryptophan residue, which limits the description of the folding dynamics in question to only one coordinate or dimension. Thus, over the past ten years many studies have been devoted to expand our capability to capture protein conformational motions occurring at different time and length scales, different sites, and different structural levels, and to extract specific dynamic and mechanistic information. Below, we first describe our efforts in this regard and then highlight a few examples whereby IR or fluorescence spectroscopy was used to delineate hydration or conformational dynamics that are important for the function of the protein system in question.
Strategies to enhance structural resolution
Both vibrational and fluorescence spectroscopies have been widely used to monitor the kinetics of protein folding.24,25 However, in most cases, they are used to follow the time evolution of a conformational ensemble or population, rather than to probe the development of a specific structure or structural element. This is because the most frequently used naturally occurring or intrinsic spectroscopic probes, such as Trp in fluorescence studies26,27 and the amide I band in linear IR measurements,28 only offer certain and limited structural information. Thus, a common strategy to enhance the structural resolution of these types of spectroscopic techniques is to exploit various extrinsic probes,29,30 especially those that can form an interacting pair for example, via electronic or vibrational coupling.
For instance, in a proof-of-principle study,31 Culik et al. demonstrated the feasibility of monitoring the folding kinetics of an individual secondary structural element in a protein using IR spectroscopy. While proteins display a large number of vibrational transitions, arising from both sidechains and backbone units,32 only a few have been used as structural probes in protein folding studies, among which, the amide I vibrational mode (consisting of mostly backbone C=O stretching vibrations) is the most utilized. Although the amide I band of a protein contains rich structural information, distinguishing contributions from individual secondary structural components is not always easy and sometimes even impossible, due to spectral overlap.33 Culik et al. showed that by selectively isotopic-labeling the backbone carbonyls of a discrete structural element in a protein, such as an α-helix, which separates its amide I band from those arising from other secondary structures, it is possible to independently follow its structural evolution during a folding or unfolding process, hence increasing the structural resolution of the study.31 Similarly, by following the transient signal arising from an isotopically-labeled amide unit located in a solvent protected region on the second helix of the three-helix bundle villin headpiece subdomain (HP36), Dyer and coworkers34 were able to monitor the dynamics of solvent penetration into this region, before global protein unfolding. With a similar approach, they also detected at least partial helical formation in the early folding intermediate state of the B domain of staphylococcal protein.35 More recently, Chen and coworkers36 demonstrated that such an isotopic-labeling approach can render site-specific interrogation of the structure and orientation of individual residues in a membrane-bound peptide using sum frequency generation vibrational spectroscopy. Furthermore, others have shown that it is also viable to achieve a higher structural sensitivity in IR studies of protein structure, folding, and aggregation by using vibrational coupling between two vibrational transitions, introduced via site-specific isotopic labeling.37-48
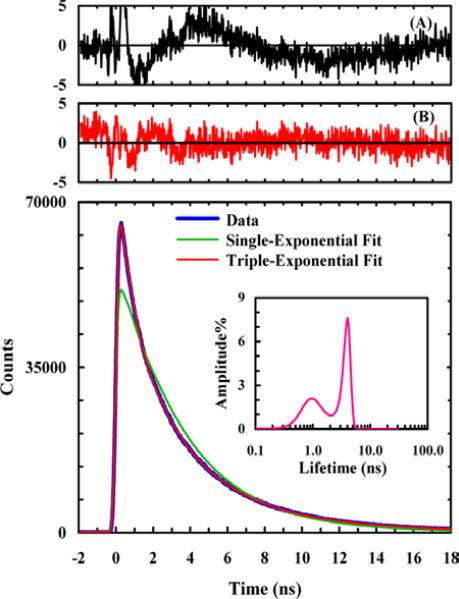
Electronic coupling leading to either energy or electron transfer between two molecules has long been exploited to provide structural information in biological studies, including protein folding. Recently, we expanded the utility of this method, specifically in protein folding studies, by introducing new, unnatural amino acid-based fluorescence resonance energy transfer (FRET) and fluorophore-quencher pairs. These include FRET pairs composed of either p-cyano-phenylalanine (PheCN) and Trp49 or PheCN and 7-aza-tryptophan (7AW),50 and a fluorophore-quencher pair composed of PheCN and selenomethionine (SeMet).51 One advantage of using such amino acid-based spectroscopic probes is that they can be easily incorporated into proteins and also minimize any potential perturbation to the native structure. The Förster distances (R0) of these FRET pairs are between 16-18 Å, making them useful to probe the formation of both tertiary and secondary structures. For example, Serrano et al.52 employed both the PheCN-Trp and the PheCN-7AW FRET pairs to explore the conformational heterogeneity in the folded state of the villin headpiece subdomain (HP35) using time-resolved fluorescence measurements. As the intrinsic fluorescence decay of PheCN follows first-order kinetics, any deviation from this single-exponential behavior indicates the existence of different protein conformations that have distinct donor-acceptor distances. As shown (Figure 4), the PheCN fluorescence decay of a HP35 mutant (i.e., HP35-AP) where the PheCN-7AW pair was placed on the same helix (i.e., helix-3) can be best described by a triple-exponential function, indicating that this helix can sample different conformations, even in the folded potential well of HP35. While the study of Serrano et al. was carried out under equilibrium conditions, it nevertheless demonstrated the feasibility of using such amino acid-based FRET pairs to directly monitor the formation of individual α-helices in protein folding dynamics. In addition, Rogers et al.50 extended the utility of these FRET pairs by showing that the PheCN-Trp and PheCN-7AW pairs could be used simultaneously as a parallel FRET system to obtain more structural information from a single fluorescence measurement.
Figure 4.
PheCN fluorescence decay kinetics of HP35-AP in 20 mM phosphate buffer (pH 7), which can be adequately described by a triple-exponential function as judged by the residuals of a double-exponential fit (A) and a triple-exponential fit (B) as well as the lifetime distribution (inset). Reprinted with permission from Ref. 52. Copyright 2012, the American Chemical Society.
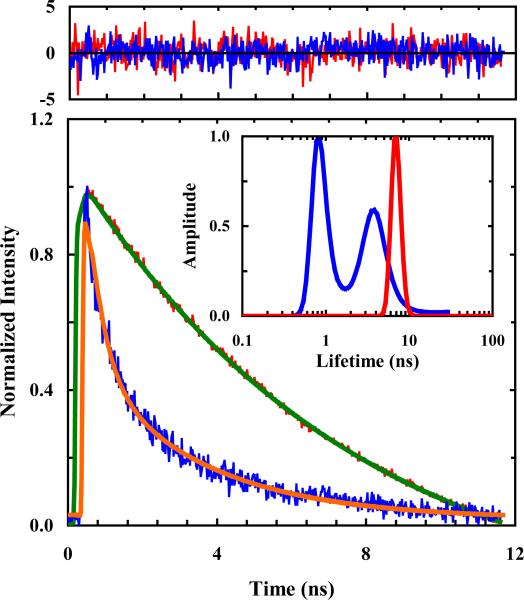
Like FRET, fluorescence quenching via electron transfer provides another convenient means to measure distances or distance changes in proteins. However, as electron transfer requires overlap of electronic orbitals, fluorophore-quencher pairs constructed based on this mechanism are uniquely useful in characterizing short distances.53 As indicated (Figure 5), Mintzer et al.51 recently demonstrated that SeMet is an efficient quencher of PheCN fluorescence and the underlying quenching rate constant (kQ) was determined to be kQ = k0exp(−β(r – a0)), where k0 = 42.6 ns−1, β = 1.6 Å−1, and a0 = 7.0 Å. Using this fluorophore-quencher pair, they further explored the conformational distribution of a series of short polyproline (Pro) peptides consisting of 1-4 Pro residues and found, unexpectedly, that these peptides do not sample all-trans configurations. Subsequently, Raleigh and coworkers54 showed that this fluorophore-quencher pair can be used to probe local helix structure formation in both peptides and proteins.
Figure 5.
Fluorescence decay kinetics of Gly-PheCN-Gly (red) and SeMet-PheCN (blue), showing the quenching effect of SeMet. The Gly-PheCN-Gly data can be fit by a single-exponential function (green) with a time constant of 7.5 ns, whereas those of SeMet-PheCN can be fit by a double-exponential function (orange) with time constants (amplitudes) of 2.0 ns (47%) and 0.2 ns (53%). Residuals of these fits (top panel) and results obtained from a maximum entropy analysis (inset) also support these assessments. Reproduced from Ref. 51 with permission from the PCCP Owner Societies.
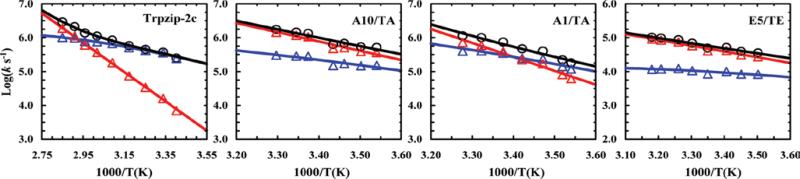
Another novel application of unnatural amino acids is to interrogate the dynamics of backbone-backbone hydrogen bond (BB-HB) formation.55 While the nativeness of a specific sidechain at the folding TS can be conveniently assessed by ϕ-value analysis56 via sidechain mutation, there has been much less choice to do the same for BB-HB. Following the work of Kiefhaber and coworkers,57,58 recently we have shown that replacing a native oxoamide (O) with a thioamide (T), which is a weaker hydrogen bond acceptor in comparison, constitutes a useful approach to alter the strength of a specific BB-HB, thus allowing determination of its role in the folding dynamics via ϕ-value analysis. Specifically, we studied the thermal stability and folding kinetics of three mutants of a β-hairpin, Trpzip-2c (sequence: NH2-AWAWENGKWAWA-CONH2), with an O-to-T mutation at Ala1, Ala10, and Glu5. Specifically, these mutations weaken three BB-HBs located at the turn (i.e., Ala1), middle (i.e., Ala10), and terminal (i.e., Glu5) regions of the β-hairpin. As shown (Figure 6), we found that only the O-to-T mutation at Glu5 position leads to a significant change in the folding time of the β-hairpin, in comparison to that of the wild type sequence (i.e., increased from 3.2 μs to 100.6 μs at 25 °C). This indicates that the BB-HB thus perturbed is formed in the TS. Since this BB-HB is located immediately next to the turn region, this finding is consistent with the notion that the turn formation is the rate-limiting step in β-hairpin folding.59-62 Because BB-HB is a key structural element of proteins, we believe that this O-to-T mutational approach will expand our ability to study the role of specific hydrogen bonding interactions in protein folding and conformational dynamics.
Figure 6.
Arrhenius plot of the folding (blue), unfolding (red), and relaxation (black) rate constants of Trpzip-2c and its O-to-T mutants. Reprinted with permission from Ref. 55. Copyright 2012, the American Chemical Society.
Strategies to increase information content
Kinetic measurements can provide accurate information about protein folding rates. However, in many cases important dynamic and mechanistic insights cannot be directly revealed by conventional kinetic experiments. Below, we describe several strategies that are useful in uncovering some hidden details of the folding free energy landscape of interest.
VIPT-jump method
Different folding mechanisms can be characterized according to the number, position, and height of the free energy barriers located between the folded and unfolded states along a given conformational coordinate. However, information obtained from traditional kinetics measurements of protein folding and/or unfolding is not always easy or sufficient to distinguish between different folding scenarios, for example two-state and downhill folding processes. In an effort to alleviate this limitation, Lin et al.63 devised a strategy, named as the VIPT-jump method, for extracting additional mechanistic information from a conventional T-jump experiment. This method involves measurements of T-jump kinetics with varying initial temperatures (Ti) but a fixed final temperature (Tf). The working principle is based on the notion that as distinct folding free energy surfaces show a different temperature dependence, the folding mechanisms can be distinguished by its dependence on Ti in the VIPT-jump kinetics. For example, a two-state folding mechanism will only give rise to a change in the amplitude of the kinetic signal when Ti is varied but the relaxation rate will remain constant, whereas a barrierless folding process,64 as shown (Figure 7), will produce VIPT-jump kinetics with different relaxation rates. While this method is developed in the context of T-jump measurements, it can be extended to other techniques where a different physical parameter, such as pressure, pH, or denaturant concentration, is used to manipulate the folding-unfolding equilibrium.
Figure 7.
For the free energy surfaces shown in the inset, a Langevin dynamics simulation indicates that the T-jump induced relaxation kinetics for a given final temperature also depend on the initial temperature. Reprinted with permission from Ref. 63. Copyright 2013, the American Chemical Society.
Novel application of cross-linking
Cross-linking is commonly used to stabilize the folded state of proteins and peptides. Recently, we have shown that this strategy can also be exploited to provide insights into protein folding dynamics. Below we highlight three such applications where we utilized (1) an m-xylene cross-linker to help assess the magnitude of internal friction, (2) a photo-responsive azobenzene cross-linker to modulate the attempt frequency, and (3) a disulfide cross-linker to create a TS analog.
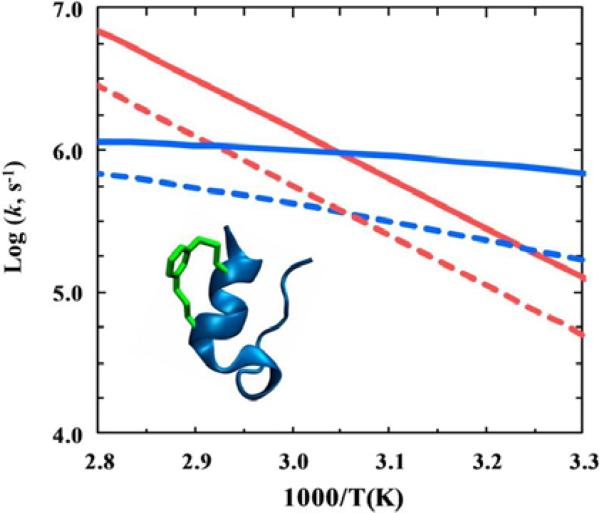
Protein folding is subject to both external (i.e., from solvent) and internal (i.e., from backbone and sidechains) frictional forces.65-73 While the frictional effect arising from the solvent is relatively easy to quantify, that arising from a protein's backbone and sidechains is more difficult to characterize. In a recent study, Markiewicz et al.74,75 showed that it is possible to estimate the magnitude of internal friction arising from a short structural segment by selectively increasing the local mass density of a protein via addition of a cross-linker. Specifically, they demonstrated the feasibility of this approach by characterizing the stability and folding/unfolding kinetics of a cross-linked variant of the mini-protein Trp-cage,76 4-8-CL-Trp-cage, where an m-xylene cross-linker77 is anchored to the protein's α-helix between positions 4 and 8 (Figure 8). They found that the thermal melting temperature (Tm) of 4-8-CL-Trp-cage (54.1 °C) was almost identical to that of the wild type (55.0 °C), which suggests that in this case the inclusion of the m-xylene cross-linker did not change the stability of the fold in any significant manner. Interestingly, their kinetic results showed that at 35 °C, there was a significant decrease in both the folding rate (by a factor of 3.8) and unfolding rate (by a factor of 2.5) of 4-8-CL-Trp-cage, in comparison to that of the wild type (Figure 8). Taken together, these findings suggest that the m-xylene cross-linker does not change the folding free energy barrier, but instead increases the frictional drag on the folding/unfolding. Adopting a theoretical model developed by Thirumalai, Straub, and coworkers,78 they further calculated that the m-xylene cross-linker increases in the roughness79 of the folding energy landscape by around 0.4-1.0 kBT.
Figure 8.
Arrhenius plot of the folding (blue) and the unfolding (red) rate constants of Trp-cage 10b (solid lines) and 4-8-CL-Trp-cage peptide (dashed lines). The inset shows the cartoon of the latter which contains an m-xylene cross-linker. Reproduced with permission from Ref. 75. Copyright 2013, the American Chemical Society.
In addition to internal friction, the attempt frequency of barrier crossing is an important aspect of protein folding dynamics. According to Kramers’ theory,80 the rate constant (k) of a barrier-crossing process, as shown below,
| (1) |
where R is the gas constant and T is the absolute temperature, depends not only on the barrier height (ΔG≠) but also on the curvatures of the reactant () and TS () potential wells and the friction coefficient (γ). Specifically, the pre-exponential factor, k0, constitutes the so called attempt frequency in TS theory. Interestingly, the k0 value for protein folding is typically several orders of magnitude smaller than that of chemical reactions involving small molecules,81 indicative of smaller ωR and/or ωB values for folding. This notion inspired us to design an approach to tune the attempt frequency of protein folding by rigidifying the TS via a cross-linker, as this would lead to an increase of ωB and hence k0, provided that the cross-linker does not change ΔG≠. Abaskharon et al.82 tested this hypothesis using a photoactivatable azobenzene cross-linker and the 10b variant of the mini-protein Trp-cage. As the single α-helix in Trp-cage is formed in the TS,83-86 the azobenzene cross-linker, when appropriately attached to this α-helix (i.e., between residues 1 and 8), can initiate α-helix formation and also impose a geometric constraint on the TS upon photoswitching the azobenzene moiety from the trans to cis conformation. Abaskharon et al. found that the photo-induced folding kinetics of this cross-linked variant of Trp-cage 10b (referred to as 10b-azob) follow a double-exponential function with time constants of 90 ns and 1.1 μs. While the longer time component corresponds to the folding time of the wildtype Trp-cage 10b protein,31 further control experiments on a truncated version of 10b-azob, which consisted of only the α-helical segment, showed that those two exponentials arose from two parallel folding pathways, with the fast component corresponding to a more rigid TS. By further assuming that both pathways have identical or comparable free energy barriers, they determined the value of k0 to be 6.9 × 106 s−1 for the unconstrained Trp-cage.
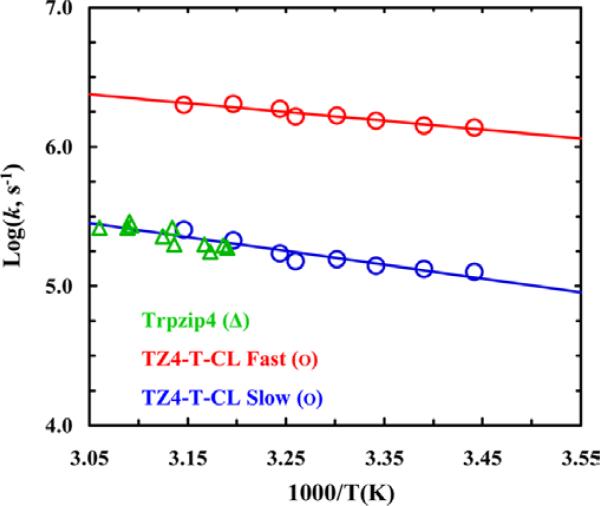
The rate of protein folding is also affected by the dynamics underlying the transition from the TS to the folded state and, while the structure of the folding TS can be characterized by the ϕ-value method,55 these dynamics are more difficult, if not impossible, to investigate. In a proof-of-principle study,87 we showed that it is possible, at least for simple protein systems, to engineer TS analogs for assessing these type of dynamics. Using a tryptophan zipper,88 Trpzip4, as a model, we aimed to stabilize a specific native contact formed in the TS using a suitable covalent bond. Since previous studies have shown that the β-turn of this hairpin is formed in the folding TS,59,60,89-98 we proposed to generate a thermodynamically stable analog of this TS by using a disulfide cross-linker to stabilize the β-turn. Specifically, a disulfide bond was introduced via two cysteine residues placed at the native Aps6 and Thr11 positions, whose amide groups are involved in the formation of the first BB-HB outside the β-turn. Our T-jump IR kinetic measurements indicated that this cross-linked variant of Trpzip4, named as TZ4-T-CL, exhibits double-exponential relaxation kinetics. As shown (Figure 9), the rate constant of the slow component is similar to the relaxation rate constant of Trpzip459 (at the same temperature), whereas that of the fast component is approximately an order of magnitude larger. Based on results obtained with another Trpzip4 variant where the disulfide cross-linker was introduced at the termini of the β-hairpin, as well as molecular dynamics simulations, we concluded that the fast kinetic component arises from a population ensemble that contains a native or native-like β-turn structure and that its ultrafast folding rate, (500 ns)−1, is a manifestation of a barrierless process. We also found, based on an empirical analysis, that the downhill folding time (τ) of a two-stranded β-sheet consisting of nH hydrogen bonds exhibits a power law dependence on nH, namely, , with τ0= 20 ns and α = 2.3.
Figure 9.
Arrhenius plot of the fast and slow relaxation rate constant of TZ4-T-CL. The green circles represent the relaxation rate constants of the wild-type Trpzip4. Reprinted with permission from Ref. 87. Copyright 2012, the American Chemical Society.
Cosolvent effect on protein dynamics
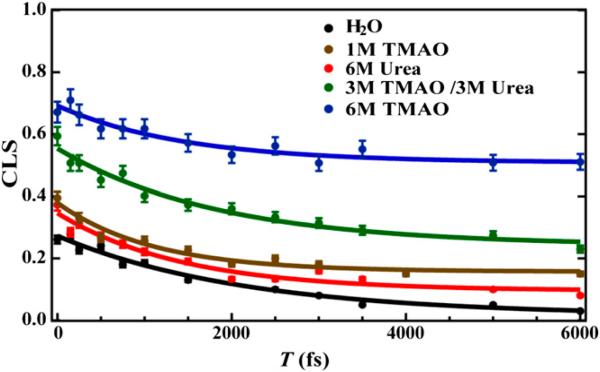
A wide range of small molecules are used by nature as osmolytes to cope with dehydration stress. Interestingly, some of them, such as urea, are protein denaturants, and some of them, such as trimethylamine N-oxide (TMAO), are protein protectants. Therefore, many studies have been devoted to elucidating how such protein denaturing and protecting ability is achieved.99-104 Capitalizing on the sensitivity of an IR probe, i.e., PheCN,105 to hydration, we investigated the effect of urea and TMAO on the strength and dynamics of hydrogen bonds (HBs) formed between water and protein sidechains using a PheCN-containing peptide and IR spectroscopic techniques. Our linear IR results106 indicate that both osmolytes act to weaken the strength of such HBs. This is an interesting finding as the behavior of urea is inconsistent with its protein-denaturing ability, which suggests that the ability of urea to denature proteins does not arise from a simple indirect mechanism or by altering the hydrogen bonding structure of water. On the other hand, the IR result obtained with TMAO is not only in line with its protein-protecting role, but also indicates that it can affect protein conformational dynamics as the C≡N stretching vibrational band of PheCN in a peptide environment becomes significantly broader when TMAO is present. To further investigate the nature of this spectral broadening, we carried out 2D IR measurements on the C≡N stretching vibration of PheCN under different solvent conditions and in different peptide environments.107 By measuring the spectral diffusion dynamics of an IR mode, 2D IR spectroscopy is able to assess the underlying dynamics of the events that lead to frequency fluctuations.108,109 As shown (Figure 10), the spectral diffusion dynamics of the C≡N probe in a short peptide, as measured by the time dependence of the inverse of the center line slope (CLS) of its 2D IR spectrum, show a strong dependence on solvent. It is clear that TMAO significantly increases the offset of the inverse CLS plot, indicating that it can effectively slow down or even suppress some conformational motions that lead to local environmental changes of the C≡N probe. In other words, this result suggests that TMAO can increase the protein rigidity and hence stability by acting as a nanocrowder, a notion that is consistent with several simulation studies.110,111 Interestingly, we find that another commonly used cosolvent, trifluoroethanol (TFE), which prompts α-helix formation, can also behave as a nanocrowder at certain percentages.112 As demonstrated in the case of TMAO, we believe that probing the spectral diffusion dynamics of an appropriate site-specific IR probe using 2D IR spectroscopy is a useful strategy to help elucidate the mechanism of action of the cosolvent in question. Currently, we are utilizing this strategy to interrogate the specific interactions between a cosolvent, such as urea and other Hofmeister ions, and a backbone or sidechain unit of proteins.
Figure 10.
CLS−1 versus T plots of a tripeptide (i.e., Gly-PheCN-Gly) measured under different solvent conditions, as indicated. The solid lines are fits of the data to the equation: CLS−1(T) = A·e(−t / τ) + B. Reprinted with permission from Ref. 107. Copyright 2014, Proceedings of the National Academy Sciences.
Dynamics of the M2 proton channel
Because of its importance in the viral life cycle, the M2 proton channel of the influenza A virus has been extensively studied and also pursued as a drug target.113 As water is required to facilitate proton diffusion, elucidating the water dynamics and distribution within the channel under different pH conditions is thus important for understanding the proton conduction mechanism of the M2 channel. Using a truncated version of the full length M2 protein (i.e., M2TM), which has been shown by DeGrado and coworkers114 to be functional in model membranes, Hochstrasser and coworkers115 showed that, through examination of the spectral diffusion dynamics of the amide I′ bands arising from isotopically labeled, pore-lining amide carbonyls (i.e., Gly34) via 2D IR spectroscopy, those water molecules located in the drug-binding site above the charged His37 tetrad, behave ice-like when the channel is closed (i.e., at pH 8) and bulk-like when the channel is open (i.e., at pH 6.2). In addition, they showed that the binding of channel-blocking drugs, such as rimantadine and 7,7-spiran amine, can affect the dynamics of the water in this pocket.116 Interestingly, drug binding leads to an increase in the mobility of the water, suggesting an entropic effect that favors the drug-protein interaction. In the context of these previous studies, an interesting future direction would be to examine the pH-dependent hydration status and dynamics near the C-terminal region of the M2TM channel, using either a backbone or sidechain IR probe, such as 5-cyanotryptophan (TrpCN),117 or, alternatively, using the fluorescence of Trp41 as a water probe.118,119
The Trp41 gate plays an important role in defining the unidirectional proton conduction function of the M2 channel.120 To characterize its conformational dynamics, we employed the technique of FCS and a Trp-oxamine dye photoinduced electron transfer (PET) pair.121 When sufficiently close, the fluorescence of the oxamine dye molecule, specifically Atto 655, is quenched by the Trp. Thus, the dynamics underlying the changes in the dye fluorescence intensity, due to changes in the separation distance of the pair, can be probed by FCS. Specifically, we introduced an Atto 655 maleimide dye at position 40 of the M2TM sequence via a Lys to Cys mutation.122 Then, channel samples were prepared in model membranes by mixing labeled and unlabeled peptides in a ratio such that a maximum of one dye-labeled peptide was present in each membrane-bound tetramer. The FCS curves of this proton channel obtained at different pH values (5.0 - 7.5) consist of four exponentials and one diffusion term. Through various control experiments, we were able to identify the PET component, which has a time constant of 673 ± 80 μs. Interestingly, the proton conducting rate of the M2 channel at low pH is estimated to be (~1000 protons/s),123-125 which is comparable to the conformational fluctuation rate of the Trp41 gate as measured by FCS-PET. Thus, this finding suggests that the Trp41 tetrad may play a key role in regulating the proton conductance of the M2 channel, besides its known role in preventing proton back flux.
SUMMARY
Proteins are polymers with many unusual and fascinating properties. In particular, their ability to spontaneously fold into a well-defined three dimensional structure in aqueous solution has inspired many researchers to study the so called protein folding problem, i.e., how this ability is achieved. Moreover, their ability to undergo a wide variety of conformational changes, either spontaneously or in response to an external stimulus, such as a change in an environmental condition or a molecular binding event, is equally amazing. As such, proteins have become the new playgrounds for many physical chemists, especially those who use spectroscopic methods to study molecular structures and dynamics. In this article, we discussed several examples, based mainly on our recent studies, of using linear and nonlinear IR spectroscopic methods, as well as fluorescence correlation spectroscopy, to interrogate protein conformational dynamics, in relation to either folding or function. One key point is that incorporating one or multiple extrinsic IR or fluorescent probes, via isotopic labeling or unnatural amino acid mutation, into the protein system of interest is a useful approach to improve the structural resolution and achieve site specificity in these type of measurements. Another is that the novel use of various cross-linkers can open up new avenues to study some fundamental questions in protein folding and conformational transitions.
ACKNOWLEDGMENT
We thank the National Institutes of Health (GM-065978 and P41-GM104605) for funding. M.R.H. is an NSF Graduate Research Fellow (DGE-1321851). We also wish to thank the current and former members of the Gai group as well as our collaborators for their contributions.
BIOGRAPHIES
 Bei Ding is a postdoctoral fellow in the Chemistry Department at the University of Pennsylvania. She received her B.S. degree in Chemistry from Peking University and her Ph.D. degree in Chemistry along with her MSE degree in Electrical Engineering (Optics and Photonics) from the University of Michigan, Ann Arbor. Currently she is interested in using site-specific vibrational spectroscopy to study peptide structure and dynamics.
Bei Ding is a postdoctoral fellow in the Chemistry Department at the University of Pennsylvania. She received her B.S. degree in Chemistry from Peking University and her Ph.D. degree in Chemistry along with her MSE degree in Electrical Engineering (Optics and Photonics) from the University of Michigan, Ann Arbor. Currently she is interested in using site-specific vibrational spectroscopy to study peptide structure and dynamics.
 Mary Rose Hilaire is a Ph.D. candidate in the graduate group of Chemistry at the University of Pennsylvania and a NSF Graduate Research Fellow. She graduated with a B.S. in Chemistry from Temple University in 2012. Her research interests include using non-natural amino acids to probe protein folding and protein-ligand binding events using fluorescence and infrared spectroscopies.
Mary Rose Hilaire is a Ph.D. candidate in the graduate group of Chemistry at the University of Pennsylvania and a NSF Graduate Research Fellow. She graduated with a B.S. in Chemistry from Temple University in 2012. Her research interests include using non-natural amino acids to probe protein folding and protein-ligand binding events using fluorescence and infrared spectroscopies.
 Feng Gai is the Edmund J. and Louise W. Kahn Endowed Term Professor of Chemistry and also directs The Ultrafast Optical Processes Laboratory at the University of Pennsylvania. His current research interests focus on protein folding dynamics and mechanisms, the protein structure-dynamics-function relationship, as well as the development of new spectroscopic probes and methods. http://gaigroup.chem.upenn.edu/
Feng Gai is the Edmund J. and Louise W. Kahn Endowed Term Professor of Chemistry and also directs The Ultrafast Optical Processes Laboratory at the University of Pennsylvania. His current research interests focus on protein folding dynamics and mechanisms, the protein structure-dynamics-function relationship, as well as the development of new spectroscopic probes and methods. http://gaigroup.chem.upenn.edu/
REFERENCES
- 1.Levinthal C. Are There Pathways for Protein Folding? J. Chim. Phys. Phys.-Chim. Biol. 1968;65:44–45. [Google Scholar]
- 2.Anfinsen CB. Principles that Govern the Folding of Protein Chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of Protein Folding: The Energy Landscape Perspective. Annu. Rev. Phys. Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Dill KA, Ozkan SB, Shell MS, Weikl TR. The Protein Folding Problem. Annu. Rev. Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warshel A. Electrostatic Basis of Structure-Function Correlation in Proteins. Acc. Chem. Res. 1981;14:284–290. [Google Scholar]
- 6.Henzler-Wildman K, Kern D. Dynamic Personalities of Proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 7.Hanoian P, Liu CT, Hammes-Schiffer S, Benkovic S. Perspectives on Electrostatics and Conformational Motions in Enzyme Catalysis. Acc. Chem. Res. 2015;48:482–489. doi: 10.1021/ar500390e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitford PC, Onuchic JN. What Protein Folding Teaches Us About Biological Function and Molecular Machines. Curr. Opin. Struct. Biol. 2015;30:57–62. doi: 10.1016/j.sbi.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Chung HS, McHale K, Louis JM, Eaton WA. Single-Molecule Fluorescence Experiments Determine Protein Folding Transition Path Times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HS, Eaton WA. Single-Molecule Fluorescence Probes Dynamics of Barrier Crossing. Nature. 2013;502:685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Best RB, Hummer G, Eaton WA. Native Contacts Determine Protein Folding Mechanisms in Atomistic Simulations. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H, Piana-Agostinetti S, Shaw D, Eaton W. Structural Origin of Slow Diffusion in Protein Folding. Science. 2015;349:1504–1510. doi: 10.1126/science.aab1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roder H, Elove GA, Englander SW. Structure Characterization of Folding Intermediates in Cytochrome c by H-Exchange Labeling and Proton NMR. Nature. 1988;335:700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadokura H, Beckwith J. Detecting Folding Intermediates of a Protein as It Passes through the Bacterial Translocation Channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright CF, Lindorff-Larsen K, Randles LG, Clarke J. Parallel Protein-Unfolding Pathways Revealed and Mapped. Nat. Struct. Biol. 2003;10:658–662. doi: 10.1038/nsb947. [DOI] [PubMed] [Google Scholar]
- 16.Jagannathan B, Elms PJ, Bustamante C, Marqusee S. Direct Observation of a Force-Induced Switch in the Anisotropic Mechanical Unfolding Pathway of a Protein. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17820–17825. doi: 10.1073/pnas.1201800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksel T, Barrick D. Direct Observation of Parallel Folding Pathways Revealed Using a Symmetric Repeat Protein System. Biophys. J. 2014;107:220–232. doi: 10.1016/j.bpj.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guinn EJ, Jagannathan B, Marqusee S. Single-Molecule Chemo-Mechanical Unfolding Reveals Multiple Transition State Barriers in a Small Single-Domain Protein. Nat. Commun. 2015;6:6861. doi: 10.1038/ncomms7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Dee DR, Liu X, Brigley AM, Sosova I, Woodside MT. Protein Misfolding Occurs by Slow Diffusion across Multiple Barriers in a Rough Energy Landscape. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8308–8313. doi: 10.1073/pnas.1419197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano AL. Ph.D. Dissertation. University of Pennsylvania; Philadelphia, PA: 2013. Time-Resolved Infrared and Fluorescence Spectroscopic Studies of Protein Dyanmics and Structure. [Google Scholar]
- 21.Asplund MC, Zanni MT, Hochstrasser RM. Two-Dimensional Infrared Spectroscopy of Peptides by Phase-Controlled Femtosecond Vibrational Photon Echoes. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8219–8224. doi: 10.1073/pnas.140227997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, Pathways, and the Energy Landscape of Protein Folding: A Synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 23.Dill KA, Chan HS. From Levinthal to Pathways to Funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 24.Eftink MR, Shastry MC. R. Fluorescence Spectroscopy. Methods Enzymol. 1997;278:258–286. doi: 10.1016/s0076-6879(97)78014-5. [DOI] [PubMed] [Google Scholar]
- 25.Callender RH, Dyer RB, Gilmanshin R, Woodruff WH. Fast Events in Protein Folding: The Time Evolution of Primary Processes. Annu. Rev. Phys. Chem. 1998;49:173–202. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 26.Royer CA. Probing Protein Folding and Conformational Transitions with Fluorescence. Chem. Rev. 2006;106:1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- 27.Zhong D. Hydration Dynamics and Coupled Water-Protein Fluctuations Probes by Intrinsic Tryptophan. Adv. Chem. Phys. 2009;143:83–149. [Google Scholar]
- 28.Serrano AL, Waegele MM, Gai F. Spectroscopic Studies of Protein Folding: Linear and Nonlinear Methods. Protein Sci. 2012;21:157–170. doi: 10.1002/pro.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Pazos IM, Zhang W, Culik RM, Gai F. Site-Specific Infrared Probes of Proteins. Annu. Rev. Phys. Chem. 2015;66:357–377. doi: 10.1146/annurev-physchem-040214-121802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waegele MM, Culik RM, Gai F. Site-Specific Spectroscopic Reporters of the Local Electric Field, Hydration, Structure, and Dynamics of Biomolecules. J. Phys. Chem. Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Culik RM, Serrano AL, Bunagan MR, Gai F. Achieving Secondary Structural Resolution in Kinetic Measurements of Protein Folding: A Case Study of the Folding Mechanism of Trp-Cage. Angew. Chemie - Int. Ed. 2011;50:10884–10887. doi: 10.1002/anie.201104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barth A, Zscherp C. What Vibrations Tell about Proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang W, Cui RZ, Silva D-A, Huang X. Simulating the T-Jump-Triggered Unfolding Dynamics of trpzip2 Peptide and Its Time-Resolved IR and Two-Dimensional IR Signals Using the Markov State Model Approach. J. Phys. Chem. B. 2011;115:5415–5424. doi: 10.1021/jp109592b. [DOI] [PubMed] [Google Scholar]
- 34.Brewer SH, Song B, Raleigh DP, Dyer RB. Residue Specific Resolution of Protein Folding Dynamics Using Isotope-Edited Infrared Temperature Jump Spectroscopy. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 35.Davis CM, Cooper AK, Dyer RB. Fast Helix Formation in the B Domain of Protein A Revealed by Site-Specific Infrared Probes. Biochemistry. 2015;54:1758–1766. doi: 10.1021/acs.biochem.5b00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding B, Panahi A, Ho JJ, Laaser JE, Brooks CL, Zanni MT, Chen Z. Probing Site-Specific Structural Information of Peptides at Model Membrane Interface in Situ. J. Am. Chem. Soc. 2015;137:10190–10198. doi: 10.1021/jacs.5b04024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woutersen S, Hamm P. Isotope-Edited Two-Dimensional Vibrational Spectroscopy of Trialanine in Aqueous Solution. J. Chem. Phys. 2001;114:2727–2737. [Google Scholar]
- 38.Fang C, Wang J, Charnley AK, Barber-Armstrong W, Smith AB, Decatur SM, Hochstrasser RM. Two-Dimensional Infrared Measurements of the Coupling between Amide Modes of an α-Helix. Chem. Phys. Lett. 2003;382:586–592. [Google Scholar]
- 39.Huang R, Kubelka J, Barber-Armstrong W, Silva RAGD, Decatur SM, Keiderling TA. Nature of Vibrational Coupling in Helical Peptides: An Isotopic Labeling Study. J. Am. Chem. Soc. 2004;126:2346–2354. doi: 10.1021/ja037998t. [DOI] [PubMed] [Google Scholar]
- 40.Decatur SM. Elucidation of Residue-Level Structure and Dynamics of Polypeptides via Isotope-Edited Infrared Spectroscopy. Acc. Chem. Res. 2006;39:169–175. doi: 10.1021/ar050135f. [DOI] [PubMed] [Google Scholar]
- 41.Fang C, Senes A, Cristian L, DeGrado WF, Hochstrasser RM. Amide Vibrations Are Delocalized across the Hydrophobic Interface of a Transmembrane Helix Dimer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16740–16745. doi: 10.1073/pnas.0608243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauser K, Ridderbusch O, Roy A, Hellerbach A, Huang R, Keiderling TA. Comparison of Isotopic Substitution Methods for Equilibrium and T-Jump Infrared Studies of β-Hairpin Peptide Conformation. J. Phys. Chem. B. 2010;114:11628–11637. doi: 10.1021/jp1028245. [DOI] [PubMed] [Google Scholar]
- 43.Remorino A, Korendovych IV, Wu Y, DeGrado WF, Hochstrasser RM. Residue-Specific Vibrational Echoes Yield 3D Structures of a Transmembrane Helix Dimer. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woys AM, Almeida AM, Wang L, Chiu C-C, McGovern M, de Pablo JJ, Skinner JL, Gellman SH, Zanni MT. Parallel β-Sheet Vibrational Couplings Revealed by 2D IR Spectroscopy of an Isotopically Labeled Macrocycle: Quantitative Benchmark for the Interpretation of Amyloid and Protein Infrared Spectra. J. Am. Chem. Soc. 2012;134:19118–19128. doi: 10.1021/ja3074962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones KC, Peng CS, Tokmakoff A. Folding of a Heterogeneous β-Hairpin Peptide from Temperature-Jump 2D IR Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai JK, Kubelka GS, Kubelka J. Sequence, Structure, and Cooperativity in Folding of Elementary Protein Structure Motifs. Proc. Natl. Acad. Sci. U. S. A. 2015;113:9890–9895. doi: 10.1073/pnas.1506309112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul C, Wang J, Wimley WC, Hochstrasser RM, Axelsen PH. Vibrational Coupling, Isotopic Editing, and β-sheet Structure in a Membrane-Bound Polypeptide. J. Am. Chem. Soc. 2004;126:5843–5850. doi: 10.1021/ja038869f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brauner JW, Dugan C, Mendelsohn R. 13C Isotope Labeling of Hydrophobic Peptides. Origin of the Anomalous Intensity Distribution in the Infrared Amide I Spectral Region of β-Sheet Structures. J. Am. Chem. Soc. 2000;122:677–683. [Google Scholar]
- 49.Tucker MJ, Oyola R, Gai F. Conformational Distribution of a 14-Residue Peptide in Solution: A Fluorescence Resonance Energy Transfer Study. J. Phys. Chem. B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 50.Rogers JMG, Lippert LG, Gai F. Non-Natural Amino Acid Fluorophores for One- and Two-Step Fluorescence Resonance Energy Transfer Applications. Anal. Biochem. 2010;399:182–189. doi: 10.1016/j.ab.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mintzer MR, Troxler T, Gai F. p-Cyanophenylalanine and Selenomethionine Constitute a Useful Fluorophore-Quencher Pair for Short Distance Measurements: Application to Polyproline Peptides. Phys. Chem. Chem. Phys. 2015;17:7881–7887. doi: 10.1039/c5cp00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano AL, Bilsel O, Gai F. Native State Conformational Heterogeneity of HP35 Revealed by Time-Resolved FRET. J. Phys. Chem. B. 2012;116:10631–10638. doi: 10.1021/jp211296e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakowicz JR. Principles of Fluorescence Spectroscopy. third edit. Springer; New York: 2006. [Google Scholar]
- 54.Peran I, Watson MD, Bilsel O, Raleigh DP. Selenomethionine, p-Cyanophenylalanine Pairs Provide a Convenient, Sensitive, Non-Perturbing Fluorescent Probe of Local Helical Structure. Chem. Commun. 2016;52:2055–2058. doi: 10.1039/c5cc08232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Culik RM, Jo H, DeGrado WF, Gai F. Using Thioamides to Site-Specifically Interrogate the Dynamics of Hydrogen Bond Formation in β-Sheet Folding. J. Am. Chem. Soc. 2012;134:8026–8029. doi: 10.1021/ja301681v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matouschek A, Kellis JT, Serrano L, Fersht AR. Mapping the Transition State and Pathway of Protein Folding by Protein Engineering. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 57.Reiner A, Wildemann D, Fischer G, Kiefhaber T. Effect of Thioxopeptide Bonds on α-Helix Structure and Stability. J. Am. Chem. Soc. 2008;130:8079–8084. doi: 10.1021/ja8015044. [DOI] [PubMed] [Google Scholar]
- 58.Bachmann A, Wildemann D, Praetorius F, Fischer G, Kiefhaber T. Mapping Backbone and Side-Chain Interactions in the Transition State of a Coupled Protein Folding and Binding Reaction. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3952–3957. doi: 10.1073/pnas.1012668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du D, Zhu Y, Huang C-Y, Gai F. Understanding the Key Factors That Control the Rate of β-Hairpin Folding. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15915–15920. doi: 10.1073/pnas.0405904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du D, Tucker MJ, Gai F. Understanding the Mechanism of β-Hairpin Folding via Ф-Value Analysis. Biochemistry. 2006;45:2668–2678. doi: 10.1021/bi052039s. [DOI] [PubMed] [Google Scholar]
- 61.Best RB, Mittal J. Microscopic Events in β-Hairpin Folding from Alternative Unfolded Ensembles. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11087–11092. doi: 10.1073/pnas.1016685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lane TJ, Bowman GR, Beauchamp K, Voelz VA, Pande VS. Markov State Model Reveals Folding and Functional Dynamics in Ultra-Long MD Trajectories. J. Am. Chem. Soc. 2011;133:18413–18419. doi: 10.1021/ja207470h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin CW, Culik RM, Gai F. Using VIP T-Jump to Distinguish between Different Folding 23 Mechanisms: Application to BBL and a Trpzip. J. Am. Chem. Soc. 2013;135:7668–7673. doi: 10.1021/ja401473m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadqi M, Fushman D, Muñoz V. Atom-by-Atom Analysis of Global Downhill Protein Folding. Nature. 2006;442:317–321. doi: 10.1038/nature04859. [DOI] [PubMed] [Google Scholar]
- 65.Plaxco KW, Baker D. Limited Internal Friction in the Rate-Limiting Step of a Two-State Protein Folding Reaction. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13591–13596. doi: 10.1073/pnas.95.23.13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagner C, Kiefhaber T. Intermediates Can Accelerate Protein Folding. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6716–6721. doi: 10.1073/pnas.96.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiu L, Hagen SJ. A Limiting Speed for Protein Folding at Low Solvent Viscosity. J. Am. Chem. Soc. 2004;126:3398–3399. doi: 10.1021/ja049966r. [DOI] [PubMed] [Google Scholar]
- 68.Frauenfelder H, Fenimore PW, Chen G, McMahon BH. Protein Folding Is Slaved to Solvent Motions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15469–15472. doi: 10.1073/pnas.0607168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chahine J, Oliveira RJ, Leite VBP, Wang J. Configuration-Dependent Diffusion Can Shift the Kinetic Transition State and Barrier Height of Protein Folding. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14646–14651. doi: 10.1073/pnas.0606506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherman E, Haran G. Fluorescence Correlation Spectroscopy of Fast Chain Dynamics Within Denatured Protein L. Chemphyschem. 2011;12:696–703. doi: 10.1002/cphc.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borgia A, Wensley BG, Soranno A, Nettels D, Borgia MB, Hoffmann A, Pfeil SH, Lipman EA, Clarke J, Schuler B. Localizing Internal Friction along the Reaction Coordinate of Protein Folding by Combining Ensemble and Single-Molecule Fluorescence Spectroscopy. Nat. Commun. 2012;3:1195. doi: 10.1038/ncomms2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulz JCF, Schmidt L, Best RB, Dzubiella J, Netz RR. Peptide Chain Dynamics in Light and Heavy Water: Zooming in on Internal Friction. J. Am. Chem. Soc. 2012;134:6273–6279. doi: 10.1021/ja211494h. [DOI] [PubMed] [Google Scholar]
- 73.Soranno A, Buchli B, Nettels D, Cheng RR, Muller-Spath S, Pfeil SH, Hoffmann A, Lipman EA, Makarov DE, Schuler B. Quantifying Internal Friction in Unfolded and Intrinsically Disordered Proteins with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Markiewicz BN, Jo H, Culik RM, DeGrado WF, Gai F. Assessment of Local Friction in Protein Folding Dynamics Using a Helix Cross-Linker. J. Phys. Chem. B. 2013;117:14688–14696. doi: 10.1021/jp409334h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Markiewicz BN, Culik RM, Gai F. Tightening up the Structure, Lighting up the Pathway: Application of Molecular Constraints and Light to Manipulate Protein Folding, Self-Assembly and Function. Sci. China Chem. 2014;57:1615–1624. doi: 10.1007/s11426-014-5225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-Residue Protein. Nat. Struct. Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 77.Jo H, Meinhardt N, Wu Y, Kulkarni S, Hu X, Low KE, Davies PL, DeGrado WF, Greenbaum DC. Development of α-Helical Calpain Probes by Mimicking a Natural Protein- Protein Interaction. J. Am. Chem. Soc. 2012;134:17704–17713. doi: 10.1021/ja307599z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sagnella DE, Straub JE, Thirumalai D. Time Scales and Pathways for Kinetic Energy Relaxation in Solvated Proteins: Application to Carbonmonoxy Myoglobin. J. Chem. Phys. 2000;113:7702–7711. [Google Scholar]
- 79.Zwanzig R. Diffusion in a Rough Potential. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hänggi P, Talkner P, Borkovec M. Reaction-Rate Theory: Fifty Years after Kramers. Rev. Mod. Phys. 1990;62:251–341. [Google Scholar]
- 81.Grote RF, Hynes JT. The Stable States Picture of Chemical Reactions. II. Rate Constants for Condensed and Gas Phase Reaction Models. J. Chem. Phys. 1980;73:2715–2732. [Google Scholar]
- 82.Abaskharon RM, Culik RM, Woolley GA, Gai F. Tuning the Attempt Frequency of Protein Folding Dynamics via Transition-State Rigidification: Application to Trp-Cage. J. Phys. Chem. Lett. 2015;6:521–526. doi: 10.1021/jz502654q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Juraszek J, Bolhuis PG. Rate Constant and Reaction Coordinate of Trp-Cage Folding in Explicit Water. Biophys. J. 2008;95:4246–4257. doi: 10.1529/biophysj.108.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paschek D, Hempel S, García AE. Computing the Stability Diagram of the Trp-Cage Miniprotein. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17754–17759. doi: 10.1073/pnas.0804775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Best RB, Mittal J. Balance between α and β Structures in Ab Initio Protein Folding. J. Phys. Chem. B. 2010;114:8790–8798. doi: 10.1021/jp102575b. [DOI] [PubMed] [Google Scholar]
- 86.Shao Q, Shi J, Zhu W. Enhanced Sampling Molecular Dynamics Simulation Captures Experimentally Suggested Intermediate and Unfolded States in the Folding Pathway of Trp-Cage Miniprotein. J. Chem. Phys. 2012;137:125103. doi: 10.1063/1.4754656. [DOI] [PubMed] [Google Scholar]
- 87.Markiewicz BN, Yang L, Culik RM, Gao YQ, Gai F. How Quickly Can a β-Hairpin Fold from Its Transition State? J. Phys. Chem. B. 2014;118:3317–3325. doi: 10.1021/jp500774q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith HO, Adams MD, Craig J. Tryptophan Zippers: Stable, Monomeric β-Hairpins. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muñoz V, Thompson PA, Hofrichter J, Eaton WA. Folding Dynamics and Mechanism of β-Hairpin Formation. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 90.Chen RP-Y, Huang JJ-T, Chen H-L, Jan H, Velusamy M, Lee C-T, Fann W, Larsen RW, Chan SI. Measuring the Refolding of β-Sheets with Different Turn Sequences on a Nanosecond Time Scale. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7305–7310. doi: 10.1073/pnas.0304922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dyer RB, Maness SJ, Peterson ES, Franzen S, Fesinmeyer RM, Andersen NH. The Mechanism of β-Hairpin Formation. Biochemistry. 2004;43:11560–11566. doi: 10.1021/bi049177m. [DOI] [PubMed] [Google Scholar]
- 92.Yang WY, Pitera JW, Swope WC, Gruebele M. Heterogeneous Folding of the Trpzip Hairpin: Full Atom Simulation and Experiment. J. Mol. Biol. 2004;336:241–251. doi: 10.1016/j.jmb.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 93.Pitera JW, Haque I, Swope WC. Absence of Reptation in the High-Temperature Folding of the Trpzip2 β-Hairpin Peptide. J. Chem. Phys. 2006;124:18–21. doi: 10.1063/1.2190226. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J, Qin M, Wang W. Folding Mechanism of β-Hairpins Studied by Replica Exchange Molecular Simulations. Proteins Struct. Funct. Genet. 2006;62:672–685. doi: 10.1002/prot.20813. [DOI] [PubMed] [Google Scholar]
- 95.Narayanan R, Pelakh L, Hagen SJ. Solvent Friction Changes the Folding Pathway of the Tryptophan Zipper TZ2. J. Mol. Biol. 2009;390:538–546. doi: 10.1016/j.jmb.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 96.Smith AW, Lessing J, Ganim Z, Peng CS, Tokmakoff A, Roy S, Jansen TLC, Knoester J. Melting of a β-Hairpin Peptide Using Isotope-Edited 2D IR Spectroscopy and Simulations. J. Phys. Chem. B. 2010;114:10913–10924. doi: 10.1021/jp104017h. [DOI] [PubMed] [Google Scholar]
- 97.Huang JJ-T, Larsen RW, Chan SI. The Interplay of Turn Formation and Hydrophobic Interactions on the Early Kinetic Events in Protein Folding. Chem. Commun. 2012;48:487. doi: 10.1039/c1cc13278d. [DOI] [PubMed] [Google Scholar]
- 98.Deeg AA, Rampp MS, Popp A, Pilles BM, Schrader TE, Moroder L, Hauser K, Zinth W. Isomerization- and Temperature-Jump-Induced Dynamics of a Photoswitchable β-Hairpin. Chem. - A Eur. J. 2014;20:694–703. doi: 10.1002/chem.201303189. [DOI] [PubMed] [Google Scholar]
- 99.Canchi DR, García AE. Cosolvent Effects on Protein Stability. Annu. Rev. Phys. Chem. 2013;64:273–293. doi: 10.1146/annurev-physchem-040412-110156. [DOI] [PubMed] [Google Scholar]
- 100.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT. Quantifying Why Urea is a Protein Denaturant, Whereas Glycine Betaine is a Protein Stabilizer. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16932–16937. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharp KA, Madan B, Manas E, Vanderkooi JM. Water Structure Changes Induced by Hydrophobic and Polar Solutes Revealed by Simulations and Infrared Spectroscopy. J. Chem. Phys. 2001;114:1791–1796. [Google Scholar]
- 102.Auton M, Holthauzen LMF, Bolen DW. Anatomy of Energetic Changes Accompanying Urea-Induced Protein Denaturation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rezus YLA, Bakker HJ. Effect of Urea on the Structural Dynamics of Water. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18417–18420. doi: 10.1073/pnas.0606538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei H, Fan Y, Gao YQ. Effects of Urea, Tetramethyl Urea, and Trimethylamine N-oxide on Aqueous Solution Structure and Solvation of Protein Backbones: A Molecular Dynamics Simulation Study. J. Phys. Chem. B. 2010:557–568. doi: 10.1021/jp9084926. [DOI] [PubMed] [Google Scholar]
- 105.Getahun Z, Huang CY, Wang T, De León B, DeGrado WF, Gai F. Using Nitrile-Derivatized Amino Acids as Infrared Probes of Local Environment. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 106.Pazos IM, Gai F. Solute's Perspective on How Trimethylamine Oxide, Urea, and Guanidine Hydrochloride Affect Water's Hydrogen Bonding Ability. J. Phys. Chem. B. 2012;116:12473–12478. doi: 10.1021/jp307414s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma J, Pazos IM, Gai F. Microscopic Insights into the Protein-Stabilizing Effect of Trimethylamine N-Oxide (TMAO). Proc. Natl. Acad. Sci. U. S. A. 2014;111:8476–8481. doi: 10.1073/pnas.1403224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts ST, Loparo JJ, Tokmakoff A. Characterization of Spectral Diffusion from Two-Dimensional Line Shapes. J. Chem. Phys. 2006;125:084502. doi: 10.1063/1.2232271. [DOI] [PubMed] [Google Scholar]
- 109.Kwak K, Park S, Finkelstein IJ, Fayer MD. Frequency-Frequency Correlation Functions and Apodization in Two-Dimensional Infrared Vibrational Echo Spectroscopy: A New Approach. J. Chem. Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 110.Cho SS, Reddy G, Straub JE, Thirumalai D. Entropic Stabilization of Proteins by TMAO. J. Phys. Chem. B. 2011;115:13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Levine ZA, Larini L, LaPointe NE, Feinstein SC, Shea J-E. Regulation and Aggregation of Intrinsically Disordered Peptides. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2758–2763. doi: 10.1073/pnas.1418155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Culik RM, Abaskharon RM, Pazos IM, Gai F. Experimental Validation of the Role of Trifluoroethanol as a Nanocrowder. J. Phys. Chem. B. 2014;118:11455–11461. doi: 10.1021/jp508056w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang J, Qiu JX, Soto C, Degrado WF. Structural and Dynamic Mechanisms for the Function and Inhibition of the M2 Proton Channel from Influenza A Virus. Curr. Opin. Struct. Biol. 2011;21:68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Identification of the Functional Core of the Influenza A Virus A/M2 Proton-Selective Ion Channel. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghosh A, Qiu J, Degrado WF, Hochstrasser RM. Tidal Surge in the M2 Proton Channel Sensed by 2D IR Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghosh A, Wang J, Moroz YS, Korendovych IV, Zanni M, Degrado WF, Gai F, Hochstrasser RM. 2D IR Spectroscopy Reveals the Role of Water in the Binding of Channel-Blocking Drugs to the Influenza M2 Channel. J. Chem. Phys. 2014;140:235105. doi: 10.1063/1.4881188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang W, Markiewicz BN, Doerksen RS, Smith AB, III, Gai F. C≡N Stretching Vibration of 5-Cyanotryptophan as an Infrared Probe of Protein Local Environment: What Determines Its Frequency? Phys. Chem. Chem. Phys. 2016;18:7027–7034. doi: 10.1039/c5cp04413h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jia M, Yang J, Qin Y, Wang D, Pan H, Wang L, Xu J, Zhong D. Determination of Protein Surface Hydration by Systematic Charge Mutations. J. Phys. Chem. Lett. 2015;6:5100–5105. doi: 10.1021/acs.jpclett.5b02530. [DOI] [PubMed] [Google Scholar]
- 119.Zhang L, Yang Y, Kao YT, Wang L, Zhong D. Protein Hydration Dynamics and Molecular Mechanism of Coupled Water-Protein Fluctuations. J. Am. Chem. Soc. 2009;131:10677–10691. doi: 10.1021/ja902918p. [DOI] [PubMed] [Google Scholar]
- 120.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The Gate of the Influenza Virus M2 Proton Channel Is Formed by a Single Tryptophan Residue. J. Biol. Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 121.Marm N, Knemeyer J, Sauer M, Wolfrum J, Marme N. Inter- and Intramolecular Fluorescence Quenching of Organic Dyes by Tryptophan. Bioconjug. Chem. 2003:1133–1139. doi: 10.1021/bc0341324. [DOI] [PubMed] [Google Scholar]
- 122.Rogers JMG, Polishchuk AL, Guo L, Wang J, Degrado WF, Gai F. Photoinduced Electron Transfer and Fluorophore Motion as a Probe of the Conformational Dynamics of Membrane Proteins: Application to the Influenza a M2 Proton Channel. Langmuir. 2011;27:3815–3821. doi: 10.1021/la200480d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mould JA, Li H, Dudlak CS, Lear JD, Pekosz A, Lamb RA, Pinto LH. Mechanism for Proton Conduction of the M 2 Ion Channel of Influenza A Virus. J. Biol. Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 124.Lin T-II, Schroeder C. Definitive Assignment of Proton Selectivity and Attoampere Unitary Current to the M2 Ion Channel Protein of Influenza A Virus. J. Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moffat JC, Vijayvergiya V, Gao PF, Cross TA, Woodbury DJ, Busath DD. Proton Transport through Influenza A Virus M2 Protein Reconstituted in Vesicles. Biophys. J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]