Abstract
Background
Advances in white matter tractography enhance neurosurgical planning and glioma resection, but is limited by biological variables such as edema, mass effect, and tract infiltration, or selection biases related to regions of interest (ROIs) or fractional anisotropy (FA) values.
Objective
To provide an automated tract identification paradigm that corrects for artifacts created by tumor edema and infiltration, as well as providing a consistent, accurate method of fiber tractography.
Methods
An automated tract identification paradigm was developed and evaluated for glioma surgery. A fiber bundle atlas was generated from six healthy participants. Fibers of a test set (including three healthy participants and ten patients with brain tumors) were clustered adaptively using this atlas. Reliability of identified tracts in both groups was assessed by comparison with two experts, using Cohen's kappa to quantify concurrence. We evaluated six major fiber bundles: cingulum bundle (CB), fornix (FR), uncinate fasciculus (UF), arcuate fasciculus (AF), inferior fronto-occipital fasciculus (IFOF), and inferior longitudinal fasciculus (ILF) – the latter three tracts mediating language function.
Results
The automated paradigm demonstrated a reliable and practical method to identify white mater tracts, despite mass effect, edema, and tract infiltration. When the tumor demonstrated significant mass effect or shift, the automated approach was useful to provide an initialization to guide the expert with identification of the specific tract of interest.
Conclusion
We report a reliable paradigm for automated identification of white matter pathways in patients with gliomas. This approach should enhance the neurosurgical objective of maximal safe resections.
Keywords: arcuate fasciculus, diffusion tensor imaging, fractional anisotropy, glioma, surgical planning, tractography
Surgical resection of gliomas continues to be a challenge due to their diffuse, infiltrative nature.1 Because of the survival benefits of maximal resection,2–5 an objective of neurosurgical oncology is to determine the optimal resection margin while preserving language, visual, and motor function. Consequently, the localization of eloquent cortical regions as well as white matter pathways in the tumor margin is essential to decrease patient morbidity.
Current surgical planning relies heavily on MRI to visualize anatomic structures.6,7 Diffusion tensor imaging (DTI)8 and fiber tractography9–11 are used to visualize the anatomic relationship between white matter fibers and the surgical target in order to guide the surgical approach and maximize the extent of resection while preserving function.1,6,12,13 In current clinical practice, pre-operative DTI-based tract identification is typically achieved by segmentation of the whole brain tractography with regions of interest (ROIs) selected by a surgeon or radiologist. The selection of these ROIs, however, becomes problematic when the fibers in the white matter are altered by edema, infiltration, mass effect, or shift. These challenges render the manual placement of ROIs time-consuming and introduce significant inter- and intra-expert variability. Moreover, the variability is compounded when isolating a tract across multiple time points, e.g. pre- and post-operatively.
Automated tract identification methods, based on fiber clustering, groups individual fibers into bundles depending on their shape and diffusion characteristics. Software-based automation has emerged as a promising alternative to the manual drawing of ROIs.14–19 Existing methods have been mostly applied to identifying healthy tracts, with only a few studies including patients with brain tumors.20,21 Furthermore, most methods utilize fiber shape and location and thus are inapplicable when the fibers are perturbed by mass effect or infiltration.
In this paper, we introduce an innovative, connectivity-based clustering method for the automated identification of white matter tracts, including those that are disrupted and/or displaced in the presence of mass effect and/or edema. The method addresses the subjectivity and variability of manual ROI placement. We demonstrate the applicability of this paradigm in pre-operative and treatment planning and compare the reliability of this technique with the current manual methodology. Besides the ability to identify eloquent tracts essential for surgical planning, the proposed technique can also be used in future studies to identify smaller tracts and evaluate them longitudinally, with the aim of studying treatment effects or performing customized surgery that protects nuanced function. In the following sections, we refer to each single pathway of tractography as fiber. Groups of fibers are called fiber bundles. We use the term tract to refer to a white matter (WM) structure of interest such as the arcuate or the corpus callosum, which can consist of a single (arcuate) or multiple (corpus callosum) fiber bundles.
METHODS
Participants
Institutional review board approval was obtained, with waiver of informed consent for retrospective review of medical records. The tract identification paradigm was assessed on two datasets comprised of healthy participants and tumor patients. The first dataset consisted of nine healthy participants (six males and three females, age 31.25 ± 4.2 years) imaged at three time points separated by two weeks. This dataset was used to confirm the accuracy and reproducibility of the algorithm in healthy controls, and served as the basis to create the atlas of fiber bundles. Six male participants were selected to generate the fiber bundle atlas, and the remaining three participants were used in testing. The second dataset consisted of 10 male patients (age 57.3 ± 18.3 years) with 8 patients being diagnosed with glioblastoma multiforme (GBM), one patient with a pleomorphic xanthoastrocytoma, and another with a meningioma. All patients developed tumors involving the left temporal or left temporal-parietal lobes, with one exception of a tumor in the left frontal lobe. Accordingly, given the location primarily in the eloquent language areas, we focused primarily on the following tracts: the arcuate fasciculus (AF), the inferior fronto-occipital fasciculus (IFOF), and the inferior longitudinal fasciculus (ILF).
MRI Acquisition
Data of healthy participants was acquired on a Siemens 3T VerioTM with a monopolar Stejskal-Tanner diffusion weighted spin-echo, echo-planar imaging sequence (TR/TE=14.8s/111ms, 2mm isotropic voxels, b = 1000 s/mm2 and 64 gradient directions). Data of the patients with gliomas were acquired using Siemens 3T TrioTim scanner, echo-planar imaging sequence (TR/TE=5s/86ms, 1.7×1.7×3 mm anisotropic voxels, b=1000 s/mm2, 30 gradient directions).
Automated Identification of Tracts
The paradigm starts with generation of a fiber bundle atlas that will emulate the expert, using white matter fibers of healthy individuals. The white matter tracts in any patient with a brain tumor are then extracted based on the definitions encoded in the atlas. In order to build an atlas, a connectivity-based representation of white matter fibers was created.18 To achieve this, the brain was first parcellated into 95 regions by mapping the Desikan atlas22 to each brain. For healthy participants, we used FreeSurfer23 for mapping the Desikan atlas to the diffusion space, whereas DRAMMS24 was used with patients due to its robustness to deformations induced by tumors. Whole brain fibers were generated using TrackVis25 (see Text, Supplemental Digital Content 1, which provides details of the methods used), and each voxel of a fiber was represented by a 95-dimensional vector encoding the connection probabilities to the 95 regions, generated using the probabilistic tractography tool probtrackx11,26. Finally, a fiber was represented by the average of these vectors for all of its voxels, termed the connectivity profile of the fiber. The connectivity-based representation of fibers is expected to be robust to minor changes in the parcellation of the cortex;18 therefore, possible challenges associated with registration of the atlas to the patient brains is not expected to effect the final tract extraction results critically.
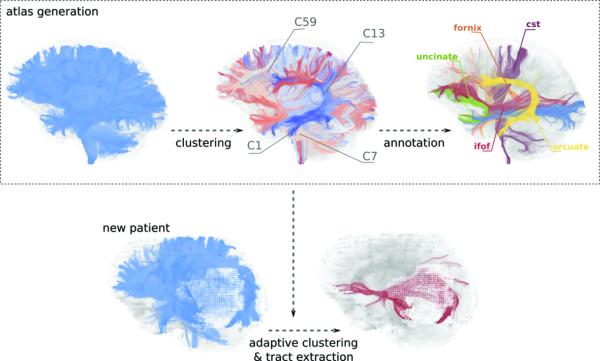
Six healthy participants were selected to generate a fiber bundle atlas. Whole brain fibers of all six participants were clustered based on their connectivity profiles. Fiber bundles in the atlas were annotated by an expert to identify the WM structures to which they belong. This atlas, which establishes automated correspondence between participants,27,28 was then used as a prior model for clustering remaining participants (patients and healthy controls) in a fully automated manner (Figure 1). A similar approach was established successfully for healthy participants.19 However, the distortion of WM fibers by edema and mass effect presented new challenges. Therefore, we developed a modified clustering algorithm to provide increased accuracy and stability (see Text, Supplemental Digital Content 1, which provides details of the methods used).
Figure 1.
Steps of automated tract identification. The WM fiber bundle atlas is generated by clustering fibers of several healthy participants. Atlas is then annotated according to the WM tracts that fiber bundles belong to. Fibers of a new patient are clustered adaptively by employing the atlas as a prior model. This procedure results in automated correspondence across fiber bundles of different patients that are adapted to the same atlas.
Evaluation of Automated Tract Identification
The automated tract identification results were evaluated via a comparison with those obtained from two experts by manual placement of ROIs. These experts identified six white matter tracts, selected for proximity to the lesion, favoring tracts that were pathologically affected and displaced. Both experts repeated the drawing of ROIs three times, separated by at least a day to estimate the intra-observer reproducibility. Cohen's kappa was used to quantify concurrence. Cohen's kappa takes values in the interval (0 – 1), where higher values indicate a better agreement. A kappa value of 0.41-0.60 is considered as moderate agreement, while 0.61-0.80 is considered substantial, and a score of 0.81-1.0 indicates near perfect agreement.29 Furthermore, we compared the selected tracts in terms of a scalar index derived from the diffusion tensor, namely fractional anisotropy (FA).
RESULTS
First, the reliability of the framework was assessed by quantitative empirical results based on comparisons of automatically identified white matter tracts to those identified with expert drawings. Then, we demonstrated the applicability of the proposed framework to surgical planning.
Reliability of tract identification
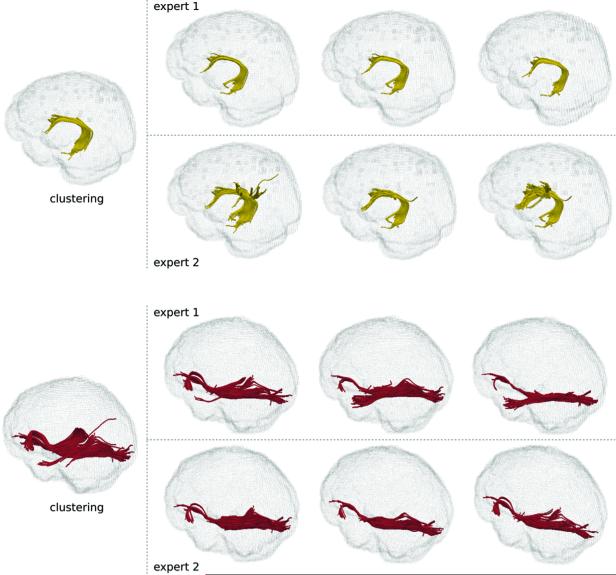
In order to validate the proposed methodology, we first evaluated the automatically identified white matter tracts in healthy participants. For example, we identified the IFOF and the AF, and then compared the results to those that were produced by experts using manual ROIs (Figure 2). A summary of quantified comparisons for healthy participants is found in Table 1. The variation in expert drawings, both for inter- and intra-user measures, was conspicuously high (Table 1), illustrating the difficulty for an expert to draw ROIs reproducibly, even in healthy participants. A detailed set of comparisons is provided as supplemental material (see Tables, Supplemental Digital Content 2-5, which provide additional experimental results).
Figure 2.
Comparison of the clustering results for the AF (top, yellow) and IFOF (bottom, red) to expert drawings for two healthy participants. Each expert repeated the drawing three times (columns) separated by at least a day. The high variation between expert drawings is evident. Overall agreement between the clustering results and the experts suggests a high reliability of the clustering paradigm.
Table 1.
Agreement between clustering and experts as quantified by Cohen's kappa for healthy controls and patients with brain tumors. Six tracts, namely the arcuate fasciculus (AF), the inferior fronto-occipital fasciculus (IFOF), the inferior longitudinal fasciculus (ILF), the cingulum bundle (CB), the fornix (FR), and the uncinate fasciculus (UF) were identified. Mean and standard deviation (in parentheses) are given. C: clustering, E1: expert 1, E2: expert 2, E: both experts (average of both). Intra-expert agreement quantifies the variation with the repeated drawings. The overall agreement between clustering and experts (C vs. E) is comparable to the agreement between experts (E1 vs. E2).
| Healthy Controls | Patients | ||||
|---|---|---|---|---|---|
| Bundle | C vs. E | E1 vs. E2 | E (Intra) | C vs. E | E1 vs. E2 |
| IFOF | 0.66 (0.05) | 0.61 (0.08) | 0.61 (0.14) | 0.49 (0.21) | 0.42 (0.22) |
| ILF | 0.62 (0.09) | 0.67 (0.04) | 0.69 (0.10) | 0.59 (0.17) | 0.55 (0.20) |
| AF | 0.64 (0.10) | 0.46 (0.16) | 0.78 (0.18) | 0.46 (0.21) | 0.49 (0.19) |
| UF | 0.60 (0.13) | 0.89 (0.03) | 0.91 (0.05) | N/A | |
| CB | 0.76 (0.05) | 0.74 (0.05) | 0.89 (0.04) | ||
| FR | 0.61 (0.07) | 0.59 (0.04) | 0.83 (0.12) | ||
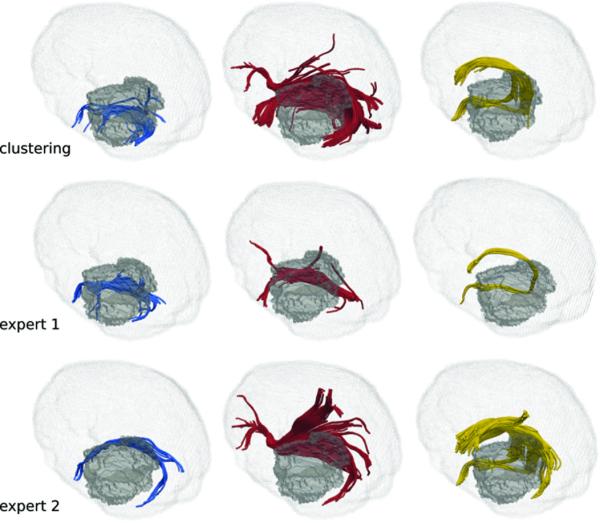
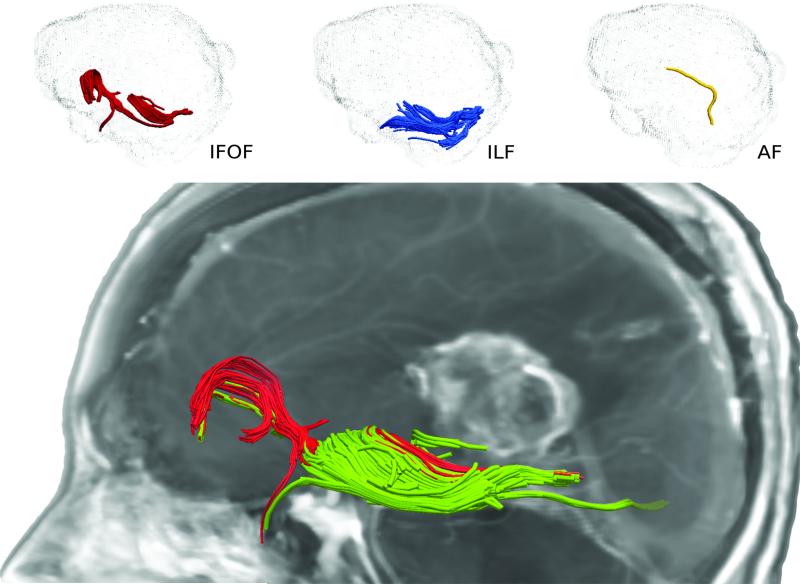
The difficulty in ROI identification is compounded in the presence of gliomas. This was demonstrated when experts placed ROIs to isolate the ILF, IFOF, and AF in patients with gliomas (Figure 3). Quantitative comparisons (Table 1) show that our method generated reliable results in patients with gliomas. Even with extreme deformation of the white matter, as in a patient with a prior surgical resection of a tumor, the white matter fibers within the internal capsule could be reliably visualized (Figure 4).
Figure 3.
Comparison of the clustering results for the ILF (first column), IFOF (second column), and the AF (third column) with the expert drawings for a patient with glioma. Edema volume is depicted by the gray shade. Due to high amount of deformation induced by mass effect, drawings of experts vary significantly as they need to find best inclusion and exclusion ROIs heuristically.
Figure 4.
Illustration of the WM fibers in the internal capsule in the atlas (first), a healthy participant (second), and a patient with a brain tumor and a prior surgical site (third). Surrounding edema around the tumor and resection volume is also shown. The part of the internal capsule that was reconstructed by the tractography was successfully identified by adaptive clustering in both healthy participant and the patient with the tumor despite the presence of large mass effect.
Automated tract identification in surgical planning
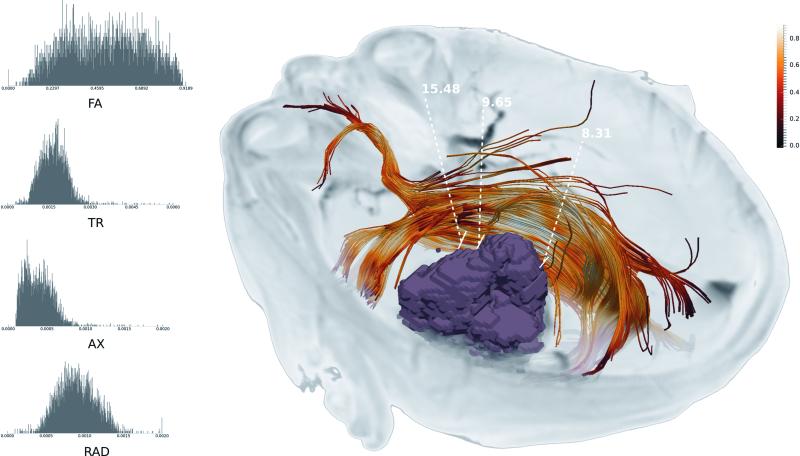
In surgical planning, the paradigm described herein can improve the visualization of white matter tracts in close proximity to the lesion/surgical target. Figure 5 demonstrates an automatically extracted IFOF, facilitating efficient visualization of this affected tract without any manual intervention. Additionally, our approach enables the selection of points along this tract, to further augment surgical planning. At each point, a DTI-based scalar index can be calculated, compared to a normative range, to estimate tract integrity. The surgeon can select individual points of interest along the fiber, and the distance to the tumor from each of these points is automatically provided (Figure 5), which can be used subsequently during the determination of the optimal resection margin.
Figure 5.
Tumor (purple mass) and surrounding WM tract (IFOF) are illustrated. The IFOF is overlaid with FA map to show how the tract is affected by the tumor. Distances between manually selected points on the tumor and the tract are shown. On the left side are distributions of several diffusion scalars along the tract.
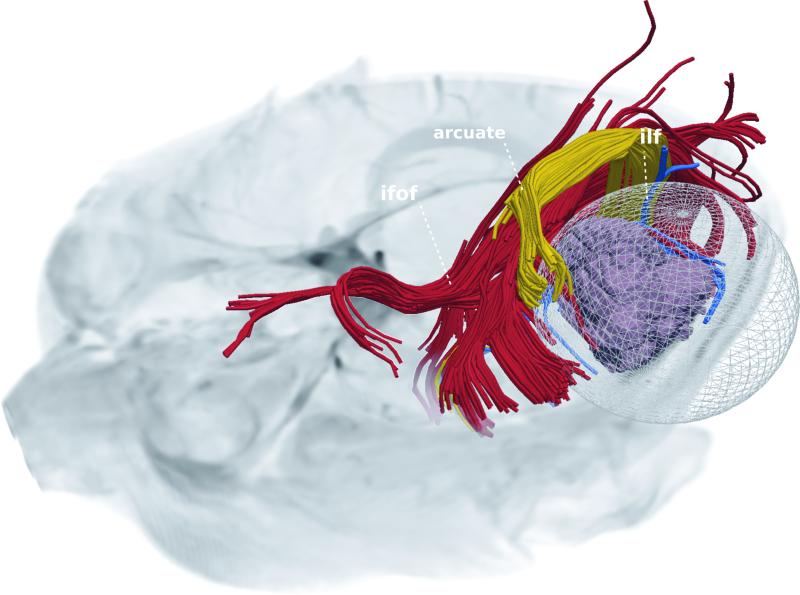
Figure 6 shows a proposed resection plan that was calculated purely based on distances between the tumor and selected WM tracts, for a patient with a glioma. This plan was likewise enhanced using automated tract identification. The tumor and three white matter tracts (AF, IFOF, ILF,) in proximity to the lesion were visualized. A proposed resection region was subsequently calculated, representing the tumor and the surrounding margin that could be resected with minimal or no damage to the surrounding tracts.
Figure 6.
Tumor (purple mass) and surrounding three WM tracts, namely IFOF, ILF, and AF are illustrated. The spherical volume depicts the maximal margin of resection that is estimated so that selected WM tracts are not affected. The estimated maximal margin depends on specific WM tracts selected.
Finally, the concept of an initialization for white matter tract identification was demonstrated in Figure 7. In the commonly encountered situation where white matter fibers are obscured due to excessive edema, mass effect, or infiltration, our paradigm provides an initial location of the tract. This location can be subsequently refined by an expert to provide a more detailed and accurate tract representation.
Figure 7.
A patient with a left temperoparietal periventricular GBM and resulting large mass effect. Clustering results for IFOF, ILF, and AF are shown on the top panel. Although tracts are only partially identified, these provide an initial estimate that was improved subsequently by experts (bottom panel, green fibers). Note that the partial fibers of tracts are captured successfully by the clustering algorithm.
DISCUSSION
Currently, white matter tract identification relies on the manual drawing of ROIs by experts. The manual placement of ROIs becomes especially challenging in the presence of human gliomas that are characterized by peritumoral vasogenic edema and mass effect. Anatomical landmarks, used by experts to identify tract location, can be obscured by these confounders. Moreover, tracts that are affected by tumor or edema can terminate prematurely during tractography.
In this work, we show that many of the limitations associated with contemporary tract identification techniques can be improved or resolved with the proposed paradigm. Our approach emulates an expert by defining an atlas of connectivity profiles of all the tracts in the brain. The connectivity-based fiber characterization is uniquely beneficial in automated identification of tracts, compared to traditional shape-based characterizations14–17 because fiber bundles can be fully or partially (in extreme cases) identified irrespective of changing fiber shape and location, or if the fiber is disrupted (Figure 3, Figure 4, and Figure 7), as long as they are reconstructed by the tractography algorithm that is used.
It should be noted that our approach is utilized once the tractography is performed, to identify any tract of interest from the whole brain tractography results. Hence, the proposed methodology should not be taken as resolving or eliminating, to any extent, intrinsic limitations of DTI and tractography such as the ones caused by edema and infiltration.30–32 Our results were produced using a tensor model and tractography method (see Text, Supplemental Digital Content 1, which provides details of the methods used) in order to demonstrate the potential of the proposed methodology. Thus, even better outcomes can be attained using more advanced and sophisticated image reconstruction schemes and tractography algorithms.33,34
Neurosurgeons rely on specialized navigation software during surgery for localization and identification of critical, though often indistinct, brain structures. The proposed methodology can augment such navigation capabilities by providing white matter information in a reproducible manner that is currently unavailable (Figure 5 and Figure 6). Furthermore, our approach goes beyond simple visualization of anatomical relationships. In Figure 5, we demonstrate how the proposed methodology could inform the surgeon on the state of the surrounding white matter tissue. In Figure 6, a safe “maximal resection” margin is estimated based on the selected white matter tracts of interest in proximity to a surgical target. Due to the speed and ease of automated identification of tracts, any number of tracts can now be visualized on the fly, without the need for a ROI-based plan. In current clinical practice, tracts required by the surgeon are mapped out in advance. By using the proposed tool, any tract can be interrogated at any time by any clinician. This is expected to provide immense flexibility for treatment planning, whether performed by radiologists, radiation oncologists, or surgeons.
The possible intrinsic limitations of DTI and tractography merit a few caveats. Although the proposed methodology can provide a reliable identification of any WM tract of interest, individual fibers may be incompletely reconstructed by the tractography algorithm used. Thus, critical decisions such as the maximal resection margin require constant evaluation by an expert (Figure 7). The final decision on the resection plan should be made by careful considerations of the limitations of tractography and potential contribution of complementary modalities such as fMRI, PET, MR spectroscopy, intraoperative ultrasound, electrophysiological monitoring, magnetoencephalography, or direct electrical stimulation of fiber tracts.
With standard methods of tract identification, the manual ROIs selected by experts can vary significantly, especially for complex tracts.35 We established that our methodology reliably generates reproducible white matter tracts by comparing automated results with those drawn manually by experts. The overall agreement of the results of automated clustering with the experts in healthy participants (Table 1 C vs. E) was comparable to the agreement between experts (Table 1 E1 vs. E2), underscoring the reliability of automated clustering as compared to manual identification by experts. Similar results were also observed in patients with brain tumors (Table 1). In expert generated tracts, the placement of ROIs was left to individual discretion, instead of defining a standard drawing protocol, to capture expert variability more realistically. Thus, the overall agreement between experts is expected to be lower than what is usually reported in studies where a standard drawing protocol is assumed for all the experts.29
In cases where fiber tractography cannot produce a complete set of fibers due to extensive edema or mass effect, our framework can be used to identify the approximate location of a given tract (Figure 7). This provides context for the expert when placing ROIs manually. This iterative and recursive improvement approach that interweaves the automated tract identification with expert refinement is beneficial even when the tracts can be identified successfully. This can be observed in Figure 2, Figure 3, and Figure 6 where part of ILF is clustered together with IFOF, or in Figure 7, where fibers from other bundles are clustered together with ILF, due to the way the tracts are defined in the atlas. Future improvement in the atlas will be made by increasing the number of participants used in atlas generation as well as incorporating annotations by several experts.
The WM tracts that are included in this study (AF, IFOF, ILF) were selected based on the expected difficulty of their identification in both healthy and tumor cases by experts, as well as their association with eloquent functions such as language.36,37 We selected the tracts that were affected by the tumors, as that demonstrated the capability and accuracy of the proposed methodology, in the presence of mass effect. Nevertheless, a future evaluation of the proposed technique can be performed on other clinically relevant tracts including the corticospinal tract or the optic radiation (OR). We could not include the OR in this study, as the 30 direction DTI data was fitted with a tensor model, which makes the identification of the OR difficult. However, the proposed technique can be applied to tracts produced by any diffusion reconstruction model (e.g. HARDI).38,39 Thus, complicated tracts like OR can be evaluated with a better dataset in a future study.
Beyond the preoperative surgical planning and intraoperative neurosurgical navigation, the ability to automate fiber tract extraction in a reproducible manner, avoiding confounding variables of mass effect, edema, and infiltration, has potential implications to quantitate white matter integrity and to link it with preservation of neurological function after resection of malignant brain tumors,40 or to monitor the recovery of function beyond the site of surgery as has been shown for visual pathways following resection of distant tumors.41 Future directions potentially include the use of automated tract identification to evaluate the disease trajectory,40,42–44 modulate radiation therapy42 or evaluate the response to therapy in patients with gliomas.45,46 Finally, the integrity of white matter tracts is increasingly being applied in the clinical neurosciences as a biomarker of injury in a range of diseases from traumatic brain injury47 to early Alzheimer's disease.48 The ability to use automated, probabilistic methods, has been shown to be more reliable than manual determinations by individual experts,49,50 as substantiated by the current report.
CONCLUSION
We have introduced a novel paradigm that can provide a standardized and reproducible automated tract extraction methodology that will enable DTI to advance as a clinical tool. This paradigm reduces many of the current technological limitations associated with the identification of WM tracts induced by peritumoral edema, glioma infiltration, and inter-observer variability. This approach is not only expected to enhance safety, but could also be used to evaluate the recovery of function after resection of gliomas in eloquent areas of the brain.
Automated identification of white matter tracts also facilitates longitudinal, quantitative characterization of white matter changes. Such sequential changes are currently difficult to assess due to a lack of reproducibility when identifying white matter tracts. Currently, the variability of fiber tracking is compounded due to the participation of multiple neuroradiologists and neurosurgeons in pre-operative planning. The automation of tract identification facilitates uniformity.
Ultimately, a combined approach of an automated tractography enhanced with surgical and radiological expertise may advance safety and efficacy of tumor resection. As the complexity of the brain inevitably requires expert judgment, the optimal results in a clinical setting would be achieved by using automated methodology, as described here, in conjunction with expert refinements. Such an approach represents a balanced scenario in which human and machine decisions go hand in hand to optimize outcomes of brain tumor surgery.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (R01-MH092862, PI: Ragini Verma and C-F Westin); University of Pennsylvania (CBICA-Radiology Internal Seed Grant, PI: Luke Macyszyn and Ragini Verma), and from the Chera Family Foundation (Brain Mapping Grant, PI: Steven Brem). Nickpreet Singh was supported by a Medical Student Summer Fellowship grant of the American Brain Tumor Association (ABTA).
ABBREVIATIONS
- AF
arcuate fasciculus
- CB
cingulum bundle
- DTI
diffusion tensor imaging
- FA
fractional anisotropy
- FR
fornix
- IFOF
inferior fronto-occipital fasciculus
- ILF
inferior longitudinal fasciculus
- OR
optic radiation
- ROI
region of interest
- UF
uncinate fasciculus
- WM
white matter
Footnotes
Disclosures: The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
REFERENCES
- 1.Abdullah KG, Lubelski D, Nucifora PGP, Brem S. Use of diffusion tensor imaging in glioma resection. Neurosurg Focus. 2013;34(4):E1. doi: 10.3171/2013.1.FOCUS12412. [DOI] [PubMed] [Google Scholar]
- 2.Brem SS, Bierman PJ, Brem H, et al. Central nervous system cancers. J Natl Compr Canc Netw. 2011;9(4):352–400. doi: 10.6004/jnccn.2011.0036. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62(4):753–764. doi: 10.1227/01.neu.0000318159.21731.cf. discussion 264-266. [DOI] [PubMed] [Google Scholar]
- 4.Eyüpoglu IY, Buchfelder M, Savaskan NE. Surgical resection of malignant gliomas-role in optimizing patient outcome. Nat Rev Neurol. 2013;9(3):141–151. doi: 10.1038/nrneurol.2012.279. [DOI] [PubMed] [Google Scholar]
- 5.Sanai N, Polley M-Y, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 6.Nimsky C, Ganslandt O, Hastreiter P, et al. Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery. 2005;56(1):130–137. doi: 10.1227/01.neu.0000144842.18771.30. [DOI] [PubMed] [Google Scholar]
- 7.Golby AJ, Kindlmann G, Norton I, Yarmarkovich A, Pieper S, Kikinis R. Interactive diffusion tensor tractography visualization for neurosurgical planning. Neurosurgery. 2011;68(2):496–505. doi: 10.1227/NEU.0b013e3182061ebb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basser PJ, Mattiello J, Lebihan D. MR Diffusion Tensor Spectroscopy and Imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 10.Mori S, van Zijl P. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15(7-8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 11.Behrens TEJ, Berg HJ, Jbabdi S, Rushworth MFS, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34(1):144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerner A, Mogensen MA, Kim PE, Shiroishi MS, Hwang DH, Law M. Clinical Applications of Diffusion Tensor Imaging. World Neurosurg. 2013 doi: 10.1016/j.wneu.2013.07.083. [DOI] [PubMed] [Google Scholar]
- 13.Nimsky C, Ganslandt O, Hastreiter P, et al. Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery. 2007;61(Supplement):178–186. doi: 10.1227/01.neu.0000279214.00139.3b. [DOI] [PubMed] [Google Scholar]
- 14.Wassermann D, Bloy L, Kanterakis E, Verma R, Deriche R. Unsupervised white matter fiber clustering and tract probability map generation: applications of a Gaussian process framework for white matter fibers. Neuroimage. 2010;51(1):228–241. doi: 10.1016/j.neuroimage.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddah M, Grimson WEL, Warfield SK, Wells WM. A unified framework for clustering and quantitative analysis of white matter fiber tracts. Med Image Anal. 2008;12(2):191–202. doi: 10.1016/j.media.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell LJ, Kubicki M, Shenton ME, Dreusicke MH, Grimson WEL, Westin CF. A method for clustering white matter fiber tracts. AJNR Am J Neuroradiol. 2006;27(5):1032–1036. [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G, Wang Q, Jia H, Shen D. Feature-based groupwise registration by hierarchical anatomical correspondence detection. Hum Brain Mapp. 2012;33(2):253–271. doi: 10.1002/hbm.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tunç B, Smith AR, Wasserman D, et al. Multinomial probabilistic fiber representation for connectivity driven clustering. In: Information Processing in Medical Imaging (IPMI). Lecture Notes in Computer Science. 2013:730–741. doi: 10.1007/978-3-642-38868-2_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tunç B, Parker WA, Ingalhalikar M, Verma R. Automated tract extraction via atlas based Adaptive Clustering. Neuroimage. 2014;102:596–607. doi: 10.1016/j.neuroimage.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Olivi A, Hertig SJ, van Zijl P, Mori S. Automated fiber tracking of human brain white matter using diffusion tensor imaging. Neuroimage. 2008;42(2):771–777. doi: 10.1016/j.neuroimage.2008.04.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Donnell LJ, Rigolo L, Norton I, Wells WM, Westin C-F, Golby AJ. fMRI-DTI modeling via landmark distance atlases for prediction and detection of fiber tracts. Neuroimage. 2012;60(1):456–470. doi: 10.1016/j.neuroimage.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(2) doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 24.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal. 2011;15(4):622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Benner T, Sorensen AG, Wedeen VJ. Diffusion Toolkit: A Software Package for Diffusion Imaging Data Processing and Tractography. Intl Soc Mag Reson Med. 2007:3720. [Google Scholar]
- 26.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell LJ, Westin C-F. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Trans Med Imaging. 2007;26(11):1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Grimson WEL, Westin C-F. Tractography segmentation using a hierarchical Dirichlet processes mixture model. Neuroimage. 2011;54(1):290–302. doi: 10.1016/j.neuroimage.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voineskos AN, O'Donnell LJ, Lobaugh NJ, et al. Quantitative examination of a novel clustering method using magnetic resonance diffusion tensor tractography. Neuroimage. 2009;45(2):370–376. doi: 10.1016/j.neuroimage.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White NS, McDonald C, McDonald CR, et al. Diffusion-weighted imaging in cancer: physical foundations and applications of restriction spectrum imaging. Cancer Res. 2014;74(17):4638–4652. doi: 10.1158/0008-5472.CAN-13-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Tie Y, Olubiyi O, et al. Reconstruction of the arcuate fasciculus for surgical planning in the setting of peritumoral edema using two-tensor unscented Kalman filter tractography. NeuroImage Clin. 2015;7:815–822. doi: 10.1016/j.nicl.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecoeur J, Caruyer E, Elliott M, Brem S, Macyszyn L, Verma R. Addressing the Challenge of Edema in Fiber Tracking. MICCAI 2014 DTI Tractography Chall. 2014 [Google Scholar]
- 33.Tournier J-D, Mori S, Leemans A. Diffusion tensor imaging and beyond. Magn Reson Med. 2011;65(6):1532–1556. doi: 10.1002/mrm.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farquharson S, Tournier J-D, Calamante F, et al. White matter fiber tractography: why we need to move beyond DTI. J Neurosurg. 2013;118(6):1367–1377. doi: 10.3171/2013.2.JNS121294. [DOI] [PubMed] [Google Scholar]
- 35.Bürgel U, Mädler B, Honey CR, Thron A, Gilsbach J, Coenen VA. Fiber tracking with distinct software tools results in a clear diversity in anatomical fiber tract portrayal. Cent Eur Neurosurg. 2009;70(1):27–35. doi: 10.1055/s-0028-1087212. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi Y, Kinoshita M, Nakada M, Hamada J. Correlation between language function and the left arcuate fasciculus detected by diffusion tensor imaging tractography after brain tumor surgery. J Neurosurg. 2012;117(5):839–843. doi: 10.3171/2012.8.JNS12348. [DOI] [PubMed] [Google Scholar]
- 37.Chang EF, Raygor KP, Berger MS. Contemporary model of language organization: an overview for neurosurgeons. J Neurosurg. 2015;122(2):250–261. doi: 10.3171/2014.10.JNS132647. [DOI] [PubMed] [Google Scholar]
- 38.Abhinav K, Yeh F-C, Mansouri A, Zadeh G, Fernandez-Miranda JC. High-definition fiber tractography for the evaluation of perilesional white matter tracts in high-grade glioma surgery. Neuro Oncol. 2015;17(9):1199–1209. doi: 10.1093/neuonc/nov113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Wang Y, Lu T, et al. Differences Between Generalized Q-Sampling Imaging and Diffusion Tensor Imaging in the Preoperative Visualization of the Nerve Fiber Tracts Within Peritumoral Edema in Brain. Neurosurgery. 2013;73(6):1044–1053. doi: 10.1227/NEU.0000000000000146. [DOI] [PubMed] [Google Scholar]
- 40.Brem S, Meyers CA, Palmer G, Booth-Jones M, Jain S, Ewend MG. Preservation of neurocognitive function and local control of 1 to 3 brain metastases treated with surgery and carmustine wafers. Cancer. 2013;119(21):3830–3838. doi: 10.1002/cncr.28307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul DA, Gaffin-Cahn E, Hintz EB, et al. White matter changes linked to visual recovery after nerve decompression. Sci Transl Med. 2014;6(266):266ra173. doi: 10.1126/scitranslmed.3010798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan AP, Asher IM, Davis D, Okunieff P, O'Dell WG. Evidence that MR diffusion tensor imaging (tractography) predicts the natural history of regional progression in patients irradiated conformally for primary brain tumors. Int J Radiat Oncol Biol Phys. 2008;71(5):1553–1562. doi: 10.1016/j.ijrobp.2008.04.017. doi:10.1016/j.ijrobp.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 43.Kallenberg K, Goldmann T, Menke J, et al. Glioma infiltration of the corpus callosum: early signs detected by DTI. J Neurooncol. 2013;112(2):217–222. doi: 10.1007/s11060-013-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson SM, Lam D, Babiak MC, et al. Transient aphasias after left hemisphere resective surgery. J Neurosurg. 2015;123(3):581–593. doi: 10.3171/2015.4.JNS141962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uh J, Merchant TE, Li Y, et al. Differences in brainstem fiber tract response to radiation: a longitudinal diffusion tensor imaging study. Int J Radiat Oncol Biol Phys. 2013;86(2):292–297. doi: 10.1016/j.ijrobp.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prabhu SP, Ng S, Vajapeyam S, et al. DTI assessment of the brainstem white matter tracts in pediatric BSG before and after therapy: a report from the Pediatric Brain Tumor Consortium. Childs Nerv Syst. 2011;27(1):11–18. doi: 10.1007/s00381-010-1323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Wang Z, Ding W, et al. Alterations in white matter microstructure as vulnerability factors and acquired signs of traffic accident-induced PTSD. PLoS One. 2013;8(12):e83473. doi: 10.1371/journal.pone.0083473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Xu Y, Zhu B, Kantarci K. The role of diffusion tensor imaging in detecting microstructural changes in prodromal Alzheimer's disease. CNS Neurosci Ther. 2014;20(1):3–9. doi: 10.1111/cns.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandelli ML, Berger MS, Bucci M, Berman JI, Amirbekian B, Henry RG. Quantifying accuracy and precision of diffusion MR tractography of the corticospinal tract in brain tumors. J Neurosurg. 2014;121(2):349–358. doi: 10.3171/2014.4.JNS131160. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y-J, Lo Y-C, Hsu Y-C, et al. Automatic whole brain tract-based analysis using predefined tracts in a diffusion spectrum imaging template and an accurate registration strategy. Hum Brain Mapp. 2015;36(9):3441–3458. doi: 10.1002/hbm.22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pieper S, Halle M, Kikinis R. 3D Slicer. IEEE International Symposium on Biomedical Imaging. 2004:632–635. [Google Scholar]
- 52.Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.