Abstract
Nuclear-cytoplasmic transport through nuclear pore complexes is mediated by nuclear transport receptors. Previous reports have suggested that aberrant nuclear-cytoplasmic transport due to mutations or overexpression of nuclear pore complexes and nuclear transport receptors is closely linked to diseases. Nup214, a component of nuclear pore complexes, has been found as chimeric fusion proteins in leukemia. Among various Nup214 fusion proteins, SET-Nup214 and DEK-Nup214 have been shown to be engaged in tumorigenesis, but their oncogenic mechanisms remain unclear. In this study, we examined the functions of the Nup214 fusion proteins by focusing on their effects on nuclear-cytoplasmic transport. We found that SET-Nup214 and DEK-Nup214 interact with exportin-1 (XPO1)/CRM1 and nuclear RNA export factor 1 (NXF1)/TAP, which mediate leucine-rich nuclear export signal (NES)-dependent protein export and mRNA export, respectively. SET-Nup214 and DEK-Nup214 decreased the XPO1-mediated nuclear export of NES proteins such as cyclin B and proteins involved in the NF-κB signaling pathway by tethering XPO1 onto nuclear dots where Nup214 fusion proteins are localized. We also demonstrated that SET-Nup214 and DEK-Nup214 expression inhibited NF-κB-mediated transcription by abnormal tethering of the complex containing p65 and its inhibitor, IκB, in the nucleus. These results suggest that SET-Nup214 and DEK-Nup214 perturb the regulation of gene expression through alteration of the nuclear-cytoplasmic transport system.
INTRODUCTION
Biological macromolecules are transported between the nucleus and the cytoplasm in response to extracellular signals. Transport of molecules with a molecular mass greater than 40 kDa does not occur by simple diffusion but is generally facilitated by nuclear transport receptors (NTRs) through nuclear pore complexes (NPCs) embedded in the nuclear envelope (1–3). Controlled nuclear-cytoplasmic transport plays important roles in maintaining cellular integrity in eukaryotic cells. It is reported that aberrant subcellular localization of some proteins is associated with various cancer cases (4). p53 has nuclear localization signals (NLSs) and nuclear export signals (NESs), and the accumulation of p53 in the cytoplasm has been reported to be a prognostic indicator in cancer (5). The nuclear factor κB (NF-κB) transcription factor is observed mainly in the cytoplasm in normal cells, whereas in many cancer cells, it is localized largely in the nucleus (6).
In addition to aberrant subcellular localization of proteins in cancer, mutations of genes encoding NTRs and NPC components are found in various types of cancer (7). Mutations of exportin-5 (XPO5) and exportin-1 (XPO1)/CRM1, members of export receptors (8–10), are found in solid cancer and leukemia, respectively. Four nucleoporins—Nup98, Nup214/CAN, Nup358/RanBP2, and Tpr—have been reported to form chimeric proteins by chromosomal translocations mainly in leukemia (11–15). Nup214, located at the cytoplasmic filament of the NPC, interacts with NTRs to control macromolecular transport. set-nup214 and dek-nup214 have been identified in acute undifferentiated leukemia and acute myeloid leukemia (AML), respectively (16, 17) and have recently been found in several T-cell acute lymphoid leukemia (T-ALL) and AML patients (18, 19). SET-Nup214 has been found to bind to the hoxa locus and activate its expression (20). Ectopic expression of SET-Nup214 causes expansion of hematopoietic progenitors (21, 22) and blocks cell differentiation (23). Expression of DEK-Nup214 leads to the acceleration of protein synthesis (24), cell proliferation (25), and the development of leukemia in mice (26). However, the detailed functions of these fusion proteins in leukemogenesis remain unclear.
Nup214 interacts with various NTRs, such as importin-β (IPOB) (27, 28), exportin-T (XPOT) (29), XPO1 (30), nuclear RNA export factor 1 (NXF1)/TAP (31–34), NXF2, and NXF3 (35). The export of both mRNAs and proteins is severely reduced when the level of Nup214 expression is decreased (36–38), and ectopic overexpression of truncated Nup214 causes the accumulation of NES proteins in the nucleus in mammals (39). On the other hand, in Drosophila spp., nup214 deletion enhances the export of green fluorescent protein (GFP) fused with the NES (40). These results indicate that appropriate expression of Nup214 is critical for regulated export of macromolecules and that the generation of fusion genes containing Nup214 by chromosomal translocation may affect the nuclear-cytoplasmic transport system (41).
In previous studies, we and another group reported that both SET-Nup214 and DEK-Nup214 interact with XPO1. This interaction causes a change in XPO1 localization and consequently impairs correct localization of an artificial model protein containing a NES (28, 42, 43). Here we comprehensively analyzed the effects of SET-Nup214 and DEK-Nup214 expression on the functions of NTRs and their cargos. We found that SET-Nup214 and DEK-Nup214 interact with NXF1 and XPO1 and that they abrogate the XPO1 function to dampen nuclear export of endogenous NES proteins, such as cyclin B and proteins involved in the NF-κB pathway. In addition, we demonstrated that SET-Nup214 and DEK-Nup214 expression inhibited NF-κB-mediated transcription due to abnormal retention of complexes containing p65 and its inhibitor, IκB, in the nucleus. These results suggest the possibility that perturbation of proper nuclear-cytoplasmic shuttling of macromolecules by the expression of the Nup214 fusion proteins leads to various hematologic disorders.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells and HEK293T cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin. For transfection assays, GeneJuice (Merck KGaA, Germany) (immunofluorescence [IF] assay, luciferase assay, fluorescence recovery after photobleaching [FRAP] assay) or linear polyethylenimine (molecular weight, 25,000) (Polysciences, Inc.) (immunoprecipitation [IP] assay, chromatin immunoprecipitation [ChIP] assay) was used.
Plasmids.
For the construction of NTR expression vectors, cDNAs were prepared from total RNA derived from HeLa and HEK293T cells by reverse transcription with ReverTra Ace (Toyobo Co., Ltd., Japan) and oligo(dT)20. PCR amplification was performed using KOD FX (Toyobo Co., Ltd.). PCR fragments were inserted into pCHA (44). pCAGGS-HA-XPO1 was made by inserting a hemagglutinin (HA)-tagged XPO1 fragment, obtained by PCR using pXHC1 as a template, into pCAGGS. pCAGGS-SET-Nup214-3Flag and pCAGGS-DEK-Nup214-3Flag were made by inserting an amplified PCR fragment (the C-terminal fragment of Nup214 fused with three Flag tags), using pCAGGS-SET-Nup214 as a template, into pCAGGS-SET-Nup214 or pCAGGS-DEK-Nup214. For pCAGGS-3Flag-Nup214(1057-2090) and pCAGGS-3Flag-SET-Nup214(1637), Nup214(1057-2090) and SET-Nup214(1637) were digested from pCAGGS-Nup214(1057-2090) and pCAGGS-SET-Nup214(1637), respectively, and inserted into pCAGGS-3Flag. pmKate2-C-SET-Nup214 and pmKate2-C-DEK-Nup214 were constructed by excising pmKate2-C (Evrogen) and ligating it with SET-Nup214 and DEK-Nup214 excised from pCAGGS-SET-Nup214 and pCAGGS-DEK-Nup214. pEGFP-C1-NXF1 was constructed by the ligation of a fragment excised from pEGFP-C1 (Clontech Laboratories, Inc.) with an NXF1 fragment generated by PCR using pCHA-NXF1 as a template. To construct pNF-κB40-firefly luciferase, the interferon-stimulated response element (ISRE) of pISRE-TA-luc (Clontech Laboratories, Inc.) was removed, and a fragment of the NF-κB binding element from pNF-κB-SEAP (Clontech Laboratories, Inc.) was inserted. To construct pTA-Renilla luciferase, the ISRE of the pISRE-TA-luc vector was removed, and the firefly luciferase region was replaced with a fragment of the Renilla luciferase region, which was obtained from pRL-SV40 (Promega). The sequences of all fragments obtained by PCR were confirmed by sequencing analysis.
IP assay and Western blot analysis.
IP assays and Western blot analyses were performed as described previously (45). To detect chemiluminescence in Western blot analyses, a Chemi-Lumi One L (Nacalai Tesque, Inc., Japan) or ImmunoStar LD (Wako Pure Chemical Industries, Ltd., Japan) kit was used, and signals were observed using the LAS-4000 Mini system (GE Healthcare UK Ltd.) and were processed by Adobe Photoshop Elements (Adobe Systems).
IF assay, oligo(dT)-mediated in situ hybridization, and PLA.
IF assays were performed as described previously (45). For DNA staining, TO-PRO-3 Iodide (dilution, 1:5,000; Life Technologies) was used. For IF assays of spleen sections, sample sections with 2 μm thick were examined. Paraffin-embedded sections were deparaffinized and were rehydrated in xylene and ethanol. Samples were autoclaved for 5 min in 10 mM citrate buffer solution. After a wash with phosphate-buffered saline (PBS), samples were subjected to IF assays. In situ hybridization using oligo(dT) was performed according to a protocol described previously (46). A proximity ligation assay (PLA) was performed using a Duolink In Situ PLA kit (Sigma-Aldrich Co. LLC) according to the manufacturer's instructions after incubation with primary antibodies. After PLA, samples were incubated with Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 633-conjugated anti-rabbit IgG antibodies for 30 min to enable the observation of IκBα and p65, respectively. Samples were observed by use of an LSM5 Exciter confocal microscope with a Plan-Apochromat 63× objective lens (Carl Zeiss Microscopy GmbH, Germany). Images were processed by ZEN software (Carl Zeiss Microimaging GmbH, Germany). Statistical analyses were performed with Student's t test.
RNA extraction and reverse transcription (RT)-qPCR.
Total RNA was extracted using a MagExtractor-RNA kit (Toyobo Co., Ltd.). The experimental procedure was that described in the manufacturer's protocol. Total RNA (0.5 μg) was reverse transcribed by ReverTra Ace (Toyobo Co., Ltd.) and oligo(dT)20 for 60 min at 42°C. For quantitative PCR (qPCR), Fast SYBR green master mix (Roche Diagnostics GmbH) was mixed with reverse-transcribed samples and primers, and PCR was carried out by a Thermal Cycler Dice real-time system (TaKaRa Bio Inc., Japan). The primer sequences used were as follows: for A20, 5′-AAG CTG TGA AGA TAC GGG AGA-3′ and 5′-CGATGAGGGCTTTGTGGATGAT-3′; for IκBα/NFKBIA, 5′-CTCCGAGACTTTCGAGGAAATAC-3′ and 5′-GCCATTGTAGTTGGTAGCCTTCA-3′; and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-AGCCAAAAGGGTCATCATCTC-3′ and 5′-GGACTGTGGTCATGAGTCCTTC-3′. Statistical analyses were performed with Student's t test.
ChIP assay.
ChIP assays were carried out according to the manual provided by Merck, except for the sonication buffer used for Fig. 7D. The sonication buffer consisted of 640 mM KCl, 30 mM NaCl, 1% Triton X-100, 10 mM EDTA, 20 mM Tris-HCl (pH 7.9), and 20% glycerol. qPCR analysis was performed as described for RT-qPCR above. The primer sequences used were as follows: for A20, 5′-CAGCCCGACCCAGAGAGTCAC-3′ and 5′-CGGGCTCCAAGCTCGCTT-3′, and for IκBα/NFKBIA, 5′-ATTCAAATCGATCGTGGGAAAC-3′ and 5′-GGGAATTTCCAAGCCAGTCA-3′.
FIG 7.
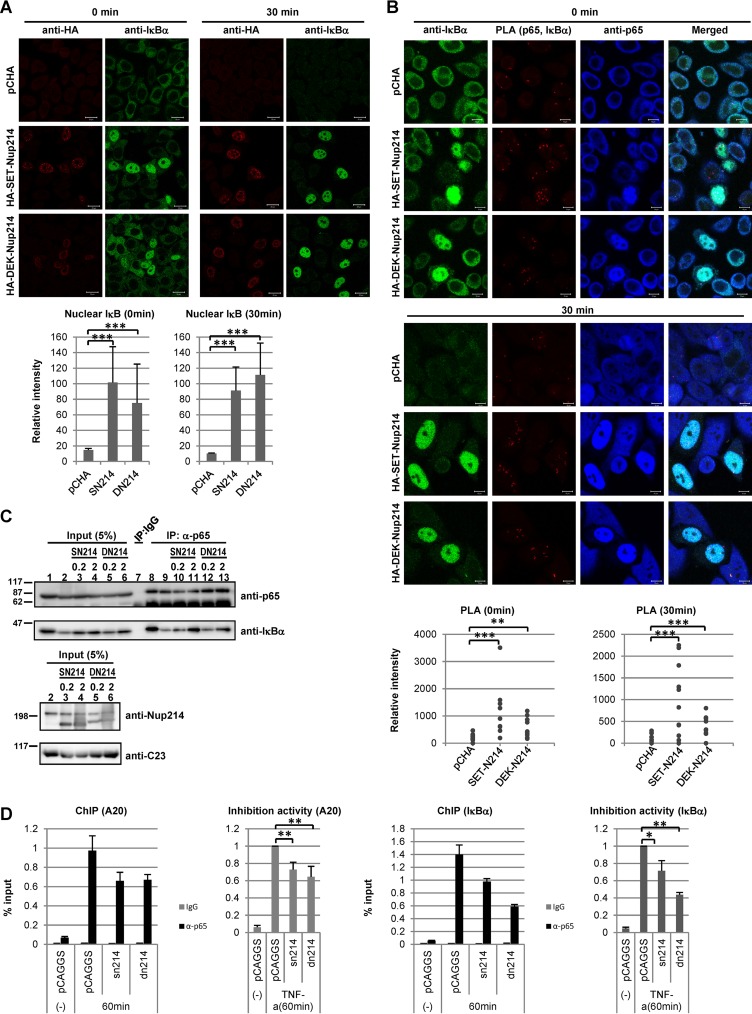
Interaction of p65 with IκBα or chromatin in the presence of stimuli. (A) HeLa cells were transfected with 1 μg of pCHA, HA-SET-Nup214, or HA-DEK-Nup214. At 2 days after transfection, cells were treated with TNF-α (10 ng/ml) for 30 min, and IF assays were performed using rabbit polyclonal anti-HA and anti-IκBα (L35A5) antibodies. Bars, 20 μm. The fluorescence intensities of nuclear IκBα in control cells, SET-Nup214-expressing cells, and DEK-Nup214-expressing cells were determined quantitatively using ImageJ software. ***, P < 0.001. (B) The protocol was the same as for panel A. After incubation with anti-p65 (ab7970) and anti-IκBα (L35A5), PLAs were performed. “Merged” panels are composite images of cells stained with Alexa Fluor 488, Detection Reagents Red (for PLA), and Alexa Fluor 633. Bars, 10 μm. The sum of the fluorescence intensities of PLA dots in each cell was quantitated using ImageJ software. **, P < 0.005; ***, P < 0.001. (C) HEK293T cells were transfected with 1 μg pCAGGS, pCAGGS-SET-Nup214 (SN214), or pCAGGS-DEK-Nup214 (DN214) (0.2 or 2 μg). At 2 days after transfection, cells were treated with TNF-α (20 ng/ml) for 30 min and were collected, and IP assays were conducted using anti-p65 (ab7970) and rabbit polyclonal IgG antibodies. Proteins in input lysates and immunoprecipitated samples were separated by 12.5% or 5% SDS-PAGE, and Western blot analyses were performed using anti-p65, anti-IκBα, anti-Nup214, and anti-C23 antibodies. Molecular weights (in thousands) of prestained markers are indicated on the left. (D) HEK293T cells were transfected with 2 μg pCAGGS, pCAGGS-SET-Nup214, or pCAGGS-DEK-Nup214. At 2 days after transfection, cells were either left untreated or treated with TNF-α (20 ng/ml) for 60 min and were then subjected to ChIP assays as for Fig. 6C. Left graphs show representative examples of ChIP-qPCR results, and right graphs show fold inhibition relative to the level of DNA immunoprecipitated from pCAGGS-transfected lysates by the anti-p65 antibody in the presence of TNF-α. Data in graphs on the right are means ± standard deviations for three independent experiments. *, P < 0.05; **P < 0.01.
RESULTS
SET-Nup214 and DEK-Nup214 interact with nuclear transport receptors.
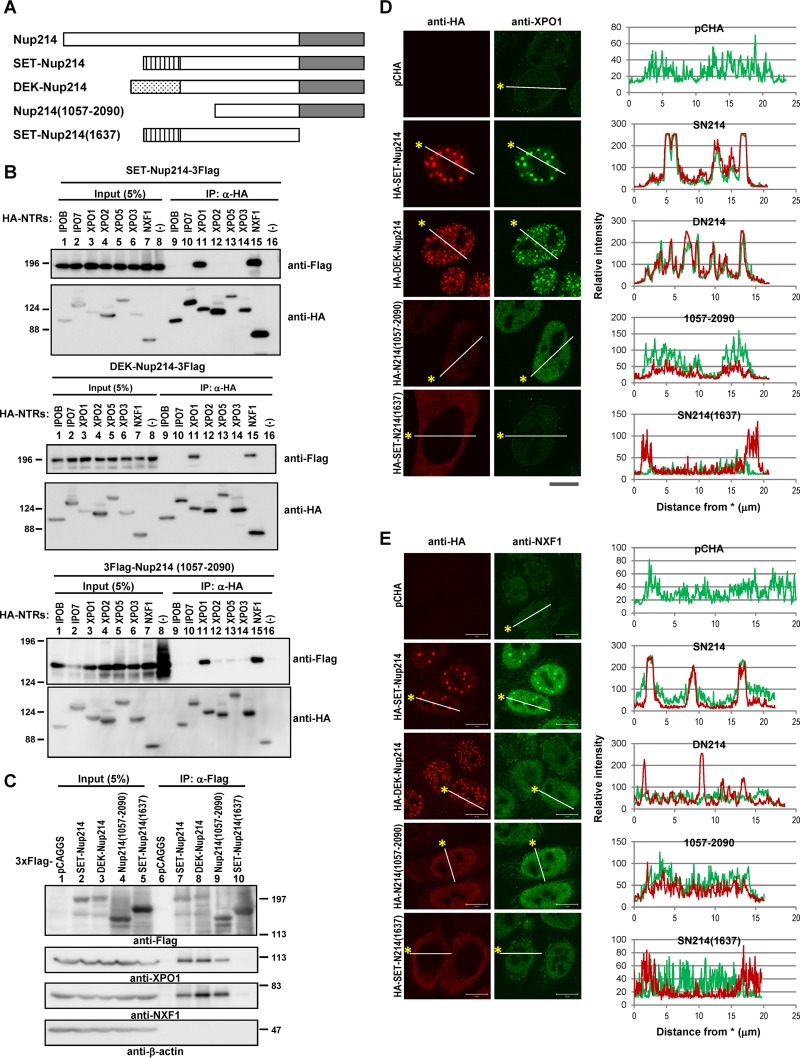
Nucleoporins can be categorized into three groups: transmembrane nucleoporins, scaffold nucleoporins, and nucleoporins containing phenylalanine-glycine repeats (FG-Nups). Nup214 is one of the FG-Nups and interacts with several NTRs through its FG repeat region. Because both SET-Nup214 and DEK-Nup214 contain the intact FG repeat region of Nup214 (Fig. 1A), it is possible that these fusion proteins interact with NTRs. Indeed, SET-Nup214 and DEK-Nup214 have been shown to interact with exportin-1 (XPO1) (42, 43). To determine whether SET-Nup214 and DEK-Nup214 bind to several NTRs other than XPO1, we first performed immunoprecipitation (IP) assays. We assessed the following seven well-known NTRs (with their functions given in parentheses): importin-β1 (IPOB) (import of NLS-containing proteins), importin-7 (IPO7) (import of proteins such as histone and mitogen-activated protein kinase [MAPK]), XPO1 (export of NES-containing proteins, snRNAs, and snoRNAs), exportin-2 (XPO2)/CSE1L (export of importin-α), exportin-5 (XPO5) (export of small RNAs), exportin-3 (XPO3)/exportin-t (export of tRNAs), and NXF1 (export of mRNAs). Among these NTRs, XPO1 and NXF1 bind to SET-Nup214 and DEK-Nup214 efficiently (Fig. 1B). Because SET-Nup214 and DEK-Nup214 have a shared Nup214 portion (amino acids 813 to 2090), which is composed of coiled-coil and FG repeat domains, we addressed whether the binding of SET-Nup214 and DEK-Nup214 with NTRs was mediated by the Nup214 portion. The C-terminally truncated Nup214(1057-2090) construct bound to XPO1 and NXF1 efficiently (Fig. 1B), demonstrating that the interaction between SET-Nup214 or DEK-Nup214 and XPO1 or NXF1 depends on the Nup214 portion. Reciprocal IP experiments confirmed the interaction between SET-Nup214, DEK-Nup214, or Nup214(1057-2090) and endogenous XPO1 or NXF1 (Fig. 1C).
FIG 1.
Interaction between SET-Nup214 and DEK-Nup214 proteins and NTRs. (A) Schematic representation of the SET-Nup214, DEK-Nup214, Nup214(1057-2090), and SET-Nup214(1637) constructs used in our study and of full-length Nup214. Striped rectangle, SET portion; stippled rectangle, DEK portion; shaded rectangle, FG repeat region. (B) HEK293T cells cultured in 6-well plates were transfected with 1 μg of pCHA-nuclear transport receptors (NTRs) or pCAGGS and 1 μg of pCAGGS-SET-Nup214-3Flag, DEK-Nup214-3Flag, or 3Flag-Nup214(1057-2090). At 2 days after transfection, cells were collected and were subjected to IP assays with 300 ng of anti-HA high affinity antibody (clone 3F10; Roche Diagnostics GmbH), and immunocomplexes were recovered with nProtein A Sepharose Fast Flow (GE Healthcare UK Ltd.). After IP assays, proteins in input lysates and immunoprecipitated samples were separated by 6% SDS-PAGE, and Western blot analyses were performed using anti-Flag M2 (2 μg/ml; Sigma-Aldrich Co. LLC) and anti-HA (clone 3F10; dilution, 1:1,000) antibodies. Molecular weights (in thousands) of prestained markers (Nacalai Tesque, Inc., Japan) are indicated on the left. (C) HEK293T cells cultured in 10-cm dishes were transfected with 5 μg of pCAGGS, SET-Nup214-3Flag, DEK-Nup214-3Flag, 3Flag-Nup214(1057-2090), or 3Flag-SET-Nup214(1637). At 2 days after transfection, cells were collected, and cell lysates were subjected to immunoprecipitation with anti-Flag M2–agarose affinity gel (Sigma-Aldrich Co. LLC). Proteins in input lysates and immunoprecipitated samples were separated by 6.5% SDS-PAGE, and Western blot analyses were performed using anti-Flag, anti-XPO1 (H-300; dilution, 1:1,000; Santa Cruz Biotechnology, Inc.), anti-NXF1 (53H8; dilution, 1:500; Santa Cruz Biotechnology, Inc.), and anti-β-actin (AC-15; dilution, 1:5,000; Sigma-Aldrich Co. LLC) antibodies. Molecular weights (in thousands) of prestained markers are indicated on the right. (D and E) HeLa cells cultured in 35-mm dishes were transfected with pCHA, HA-SET-Nup214, HA-DEK-Nup214, HA-Nup214(1057-2090), or HA-SET-Nup214(1637). At 2 days after transfection, cells were subjected to IF assays. The primary antibodies used were anti-HA (clone 3F10; dilution, 1:100) (D), anti-XPO1 (1:20) (D), rabbit polyclonal anti-HA (1:500) (E), and anti-NXF1 (1:20) (E). Bars, 10 μm. Graphs on the right represent the relative intensities of HA-tagged protein (red) and XPO1 (D) or NXF1 (E) (green).
SET-Nup214 and DEK-Nup214 affect the subcellular localization of nuclear transport receptors.
As we and another group reported previously (42, 43), SET-Nup214 and DEK-Nup214 are localized mainly in the nucleus as granular dots and influence the subcellular localization of XPO1. Because NXF1 was also coimmunoprecipitated with SET-Nup214 and DEK-Nup214, we next examined the subcellular localizations of both NTRs by indirect immunofluorescence (IF) assays. Endogenous XPO1 was located in the nucleus and the nuclear envelope, whereas it was localized markedly as granular dots upon expression of SET-Nup214 or DEK-Nup214 (Fig. 1D). XPO1 localization at the nuclear envelope in cells expressing Nup214(1057-2090) was reduced, and XPO1 was localized mainly in the nucleus, where Nup214(1057-2090) accumulated. Endogenous NXF1 was observed in the nuclei of control cells. On the other hand, in cells expressing SET-Nup214, NXF1 was found in nuclear dots where SET-Nup214 accumulated (Fig. 1E), although the accumulation was less than that of XPO1. In addition, in cells expressing DEK-Nup214, we could not find clear accumulation of NXF1 at the sites where DEK-Nup214 was accumulated. It was reported previously that the FG repeat region of Nup214 plays crucial roles in binding with NTRs (31, 32, 39, 42). Therefore, we next examined the importance of the FG repeat region of the fusion proteins for the changes in the localization of XPO1 and NFX1 by using the deletion mutant of SET-Nup214 termed SET-Nup214(1637), which lacks the FG repeat region of SET-Nup214. This mutant neither interacted with endogenous XPO1 and NXF1 nor changed their localization (Fig. 1C, lane 10, and Fig. 1D and E). These results indicate that expression of SET-Nup214 and DEK-Nup214 affects the localization patterns of both XPO1 and NXF1 by their physical interaction through the FG repeat region of Nup214.
NES proteins enhance dot formation by SET-Nup214 and DEK-Nup214.
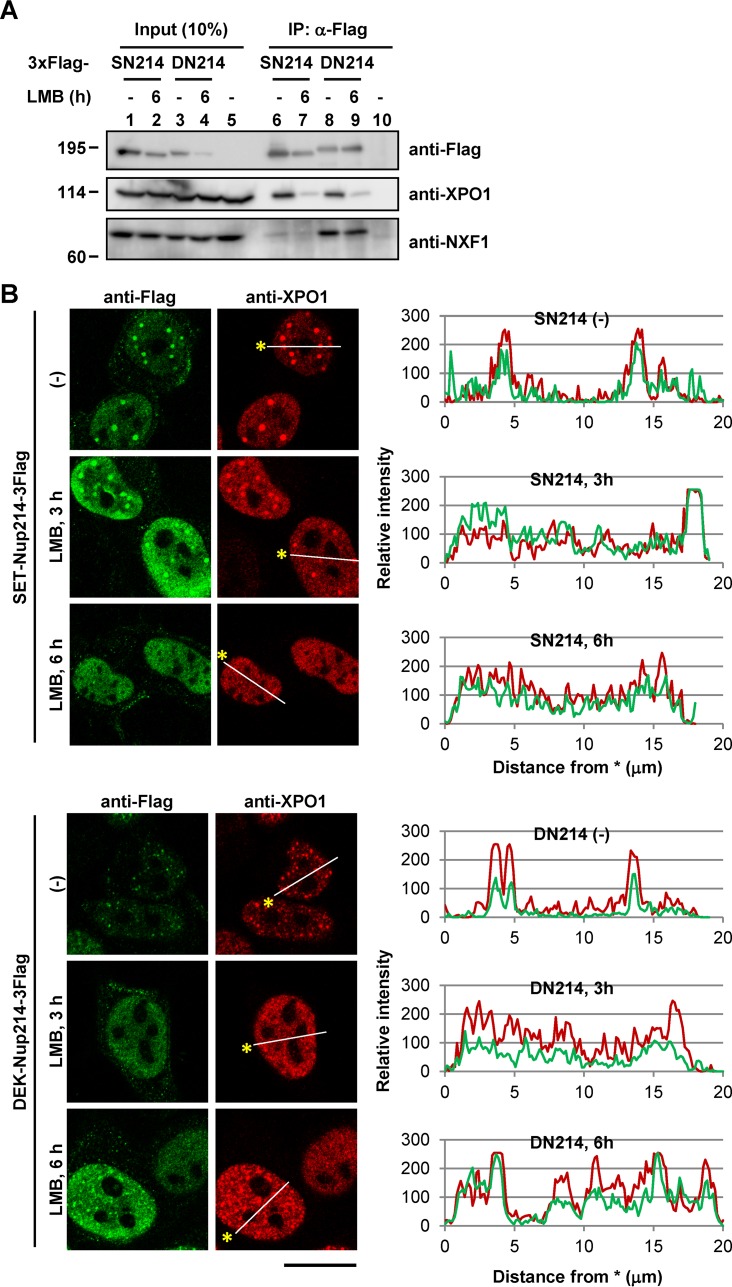
SET-Nup214 and DEK-Nup214 induced changes in the localization of XPO1 and NXF1 through their physical interactions (Fig. 1). NES proteins facilitate the interaction between XPO1 and Nup214 (38, 47, 48). To address the importance of NES proteins in the interaction between SET-Nup214 or DEK-Nup214 and NTRs, IP experiments were performed using cell lysates that were either left untreated or treated with leptomycin B (LMB), an inhibitor of binding between the NES and XPO1 (49). The amount of XPO1 coimmunoprecipitated with SET-Nup214 or DEK-Nup214 was decreased in LMB-treated cells (Fig. 2A). In addition, granular dots generated by SET-Nup214 or DEK-Nup214 decreased in size or disappeared after LMB addition (Fig. 2B). These results suggest that the interaction of XPO1 with SET-Nup214 or DEK-Nup214 is under the control of its binding with NES proteins and that the interaction between XPO1 and its cargos is indispensable for granular dot formation by SET-NUP214 and DEK-Nup214.
FIG 2.
Dependency of NES proteins on complex and dot formation by either SET-Nup214 or DEK-Nup214 with XPO1. (A) HeLa cells cultured in 35-mm dishes were transiently transfected with 1 μg of either pCAGGS-SET-Nup214-3Flag (SN214), DEK-Nup214-3Flag (DN214), or pCAGGS. At 2 days after transfection, cells were incubated in 5 ng/ml LMB (L-6100; LC Laboratories) for 6 h. After incubation, cells were collected and were subjected to IP assays using Flag M2 beads (lanes 6 to 10). Proteins in the input lysate and immunoprecipitated samples were separated by 6.5% SDS-PAGE, and Western blot analyses were performed using anti-Flag and anti-XPO1 antibodies as primary antibodies. Molecular weights (in thousands) of prestained markers are indicated on the left. (B) The protocol was the same as that for panel A. After cells were collected, IF analyses were performed. Anti-Flag M2 (1:1,000) and anti-XPO1 were used as primary antibodies. Bar, 20 μm. Graphs on the right represent the relative intensities of Flag-tagged protein (green) and XPO1 (red).
SET-Nup214 and DEK-Nup214 affect the subcellular localization of endogenous proteins harboring NES.
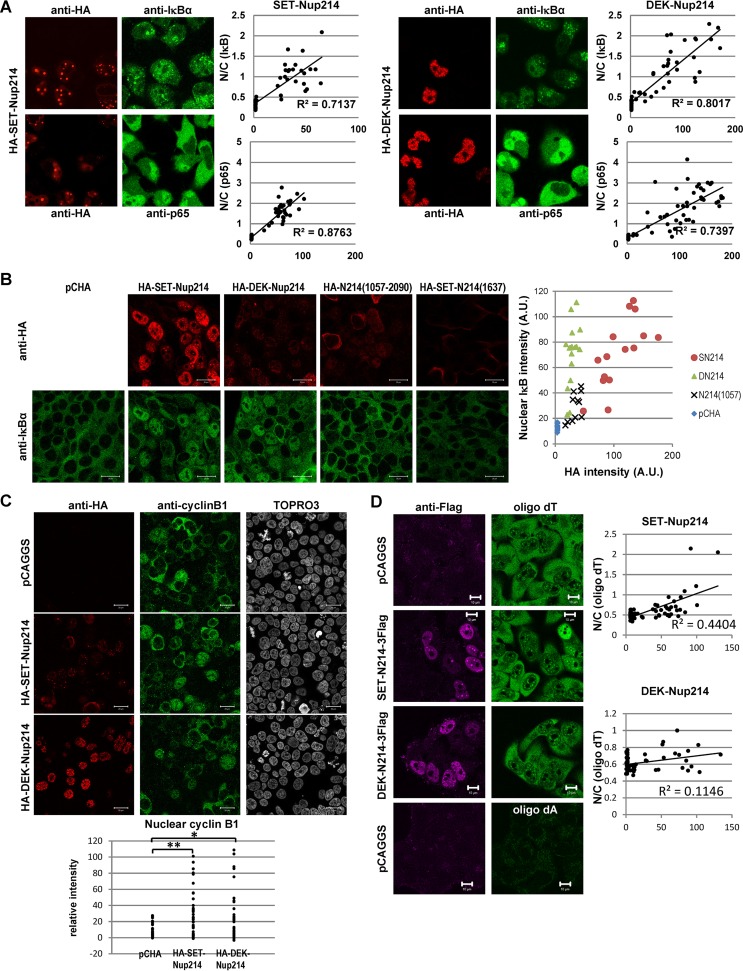
Since XPO1 accumulated in nuclear dots in cells expressing SET-Nup214 or DEK-Nup214 (Fig. 1D), it was reasonable to hypothesize that the intracellular availability of XPO1 may be decreased. We have found previously that enhanced green fluorescent protein (EGFP) fused to the NES of cyclic AMP (cAMP)-dependent protein kinase inhibitor (PKI) accumulated in the nuclei of cells expressing SET-Nup214 (43). However, it is not known whether the subcellular localization of endogenous NES proteins is actually affected by the expression of both SET-Nup214 and DEK-Nup214. Hence, we performed IF assays to observe the endogenous XPO1 cargo proteins IκBα and cyclin B1. IκBα (Fig. 3A) and cyclin B1 (Fig. 3C) were localized mainly in the cytoplasm in cells that did not express SET-Nup214 or DEK-Nup214. In contrast, they uniformly accumulated in the nucleus upon expression of SET-Nup214 or DEK-Nup214. Because the NF-κB transcription factor p65/RelA binds to IκBα in unstimulated cells, we examined the localization of p65 in cells expressing SET-Nup214 or DEK-Nup214. Interestingly, we found that the cytoplasmic localization of p65 was also disturbed by SET-Nup214 and DEK-Nup214. Quantitative analyses revealed that the ratio of the fluorescence intensity of IκBα or p65 in the nucleus to that in the cytoplasm increased significantly as the intensity of SET-Nup214 or DEK-Nup214 increased (Fig. 3A). When the C-terminal region of Nup214 [Nup214(1057-2090)] was expressed, nuclear accumulation of IκBα was also observed, although it was mild compared to that in cells expressing SET-Nup214 or DEK-Nup214 (Fig. 3B). This indicated that the C-terminal region of Nup214 can function as a dominant negative mutant of endogenous Nup214, as reported previously (39). From these observations, we conclude that SET-Nup214 and DEK-Nup214 change the subcellular localizations of endogenous proteins harboring NES by inhibiting the function of endogenous Nup214.
FIG 3.
Localization of endogenous proteins harboring the NES and mRNA. (A) (Images) HeLa cells were transiently transfected with pCHA-SET-Nup214 or HA-DEK-Nup214 and were subjected to IF assays using anti-HA (clone 3F10), anti-IκBα (C-21; dilution, 1:100; Santa Cruz Biotechnology, Inc.), or anti-p65 (PC137; dilution, 1:100; Calbiochem) antibodies. (Graphs) Fluorescence intensity was determined quantitatively using ImageJ software. Nuclear (N) and cytoplasmic (C) areas were selected manually. The mean intensity of HA-tagged protein in the nucleus minus the mean background intensity is shown along the x axis. The N/C ratio, shown along the y axis, is calculated as (mean intensity of IκBα or p65 in the nucleus − mean background intensity)/(mean intensity of IκBα or p65 in the cytoplasm − mean background intensity). (B) 293T cells were transfected with pCHA, HA-SET-Nup214, HA-DEK-Nup214, HA-Nup214(1057-2090), or HA-SET-Nup214(1637). Two days later, cells were subjected to an immunofluorescence assay using anti-HA (clone 3F10) and anti-IκBα (L35A5; dilution, 1:20; Cell Signaling Technology [CST], Inc.) antibodies. Bars, 20 μm. The fluorescence intensities of nuclear IκBα and HA-tagged protein in each sample were determined quantitatively using ImageJ software and were plotted. A.U., arbitrary units. (C) HeLa cells were transiently transfected with pCHA-SET-Nup214 or DEK-Nup214 and were subjected to IF assays using anti-HA (clone 3F10), anti-cyclin B1 (antibody 4138; dilution, 1:20; CST, Inc.), and TO-PRO-3. Bars, 20 μm. The fluorescence intensity of nuclear cyclin B1 in each cell was determined quantitatively using ImageJ software and was plotted. *, P < 0.005; **, P < 0.0005. (D) HeLa cells were transiently transfected with pCAGGS, SET-Nup214-3Flag, or DEK-Nup214-3Flag and were subjected to IF assays using anti-Flag M2 and to in situ hybridization assays with 10 ng/μl biotinylated oligo(dT)45 or oligo(dA)45 as a probe. Bars, 10 μm. The dot plots show fluorescence intensity, quantified using ImageJ software, as described for panel A.
SET-Nup214 has a small effect on poly(A) mRNA localization.
In addition to XPO1, the expression of SET-Nup214 affected the localization of NXF1, an NTR for mRNA (Fig. 1E). We assessed the subcellular localization pattern of mRNA by fluorescence in situ hybridization assays using oligo(dT) as a probe (Fig. 3D). In control cells, the oligo(dT) signal was observed both in the nucleus and in the cytoplasm, and the cytoplasmic intensity was higher than the nuclear intensity. In some cells expressing SET-Nup214, the ratio of the intensity of the signal of oligo(dT) in the nucleus to that in the cytoplasm is higher than the ratio in control cells. Quantitative analyses showed that cells highly expressing SET-Nup214 were prone to mRNA accumulation in the nucleus. However, the accumulation of mRNAs was less clear than that of proteins harboring NES, suggesting that the effect of SET-Nup214 on NXF1 function is lower than that on XPO1 function. In addition, we found little difference in the oligo(dT) staining pattern between control cells and cells expressing DEK-Nup214.
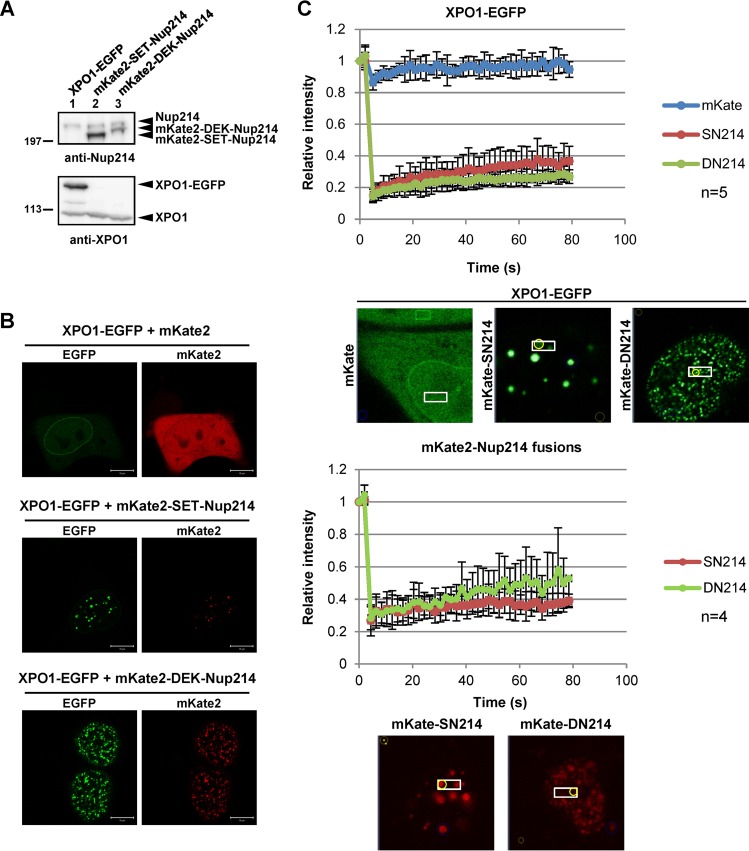
SET-Nup214 and DEK-Nup214 reduce the mobility of XPO1.
Fluorescence recovery after photobleaching (FRAP) analyses have shown that XPO1 is highly mobile in the cell (50). Since XPO1 is localized as granular dots in cells expressing SET-Nup214 or DEK-Nup214, it was possible that the XPO1 mobility was decreased. To test this, we performed FRAP assays for fluorescent protein-fused XPO1 (51), SET-Nup214, and DEK-Nup214. The expression of fluorescent proteins was confirmed by Western blot analyses and immunofluorescence microscopy (Fig. 4A and B). In the experiment for which results are shown Fig. 4C, the areas boxed in the images were bleached with a 488-nm laser line, and the intensity of the bleached area was monitored every 2 s. In control cells, the XPO1-EGFP fluorescence intensity in the bleached area was restored rapidly after bleaching. On the other hand, XPO1-EGFP accumulated in dots, and the recovery rate of fluorescence in the dots (circled in yellow in Fig. 4C) was significantly reduced by the coexpression of mKate2-SET-Nup214 or mKate2-DEK-Nup214. The intensities of mKate2-SET-Nup214 and mKate2-DEK-Nup214 were not efficiently recovered after photobleaching, and only a small fraction was recovered in 80 s (Fig. 4C). These results indicate that SET-Nup214 and DEK-Nup214 in the dots are not exchangeable efficiently but rather form stable complexes/aggregates to which XPO1 is attracted.
FIG 4.
Decreased mobility of XPO1. (A) HEK293T cells cultured in 35-mm dishes were transiently transfected with 1 μg pHCF1 (XPO1-EGFP expression vector), pmKate2C-SET-Nup214, or pmKate2C-DEK-Nup214. Samples were separated by 5% SDS-PAGE and were subjected to Western blot analyses using anti-Nup214 (dilution, 1:1,000) and anti-XPO1 antibodies. Molecular weights (in thousands) of prestained markers are indicated on the left. (B and C) HeLa cells were transfected with 1 μg pHCF1 and 1 μg either pmKate2C, pmKate2C-SET-Nup214, or pmKate2C-DEK-Nup214. (B) Typical localization patterns of XPO1-EGFP, mKate2, mKate2-SET-Nup214, and mKate2-DEK-Nup214 are shown. Bars, 10 μm. (C) Transfected cells were subjected to FRAP assays as described previously (82).
SET-Nup214 and DEK-Nup214 affect NF-κB transcriptional activity.
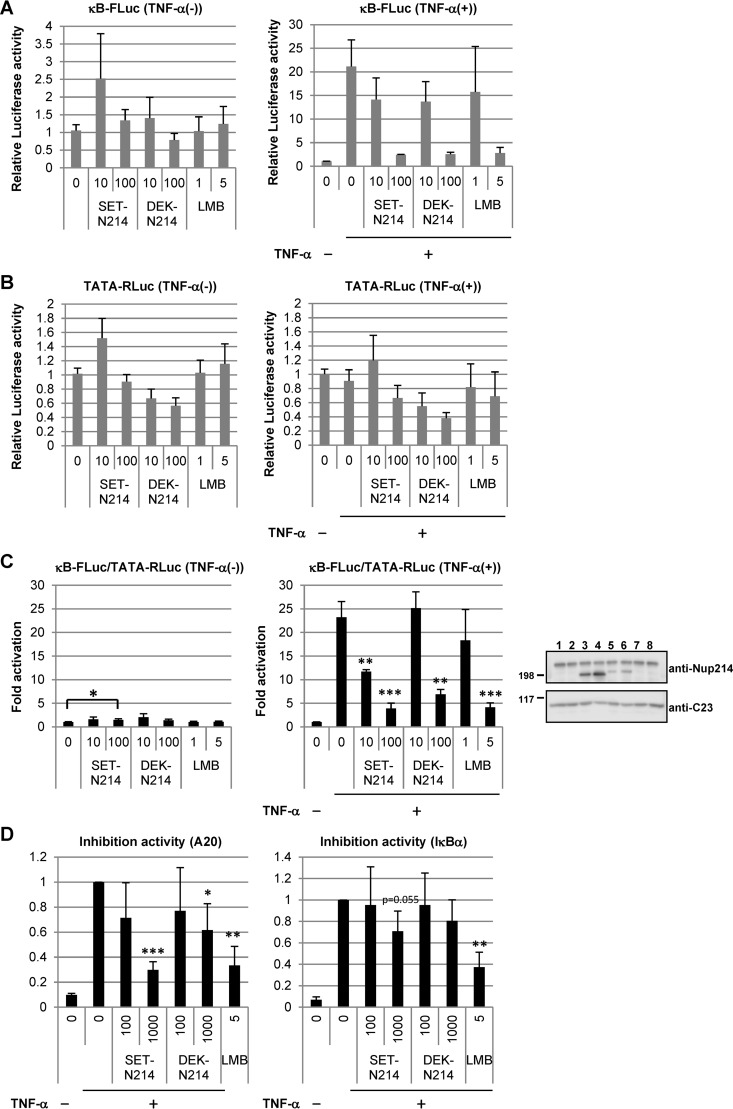
We presumed that the promotion of oncogenesis by the expression of SET-Nup214 and DEK-Nup214 was due, at least in part, to the deregulation of gene expression caused by aberrant localization of proteins and/or RNAs. The subcellular localizations of p65 and IκBα were changed upon expression of SET-Nup214 or DEK-Nup214 (Fig. 3A). It has been reported that Nup98 fusion proteins stimulate NFAT (nuclear factor of activated T cells)- and NF-κB-mediated transcription activities by impairing the function of XPO1 (52). Thus, we examined the effects of Nup214 fusion proteins on the NF-κB signaling pathway. The transcriptional activity of NF-κB was assessed first by reporter assays using firefly luciferase under the control of NF-κB (κB-FLuc). In the absence of tumor necrosis factor alpha (TNF-α), the fusion proteins did not affect luciferase activity (Fig. 5A to C, left graphs), although they induced nuclear accumulation of p65 (Fig. 3A). TNF-α treatment dramatically increased the reporter activity of κB-FLuc. This increase in transcriptional activity was markedly inhibited by the expression of SET-Nup214 or DEK-Nup214 (Fig. 5A to C, right graphs). As reported elsewhere (53–55), LMB treatment showed an inhibitory effect on the luciferase activity of NF-κB-mediated transcription in the presence of TNF-α (Fig. 5A to C, right graphs), while no significant effect was observed in the absence of TNF-α (Fig. 5A to C, left graphs). The effects of SET-Nup214 and DEK-Nup214 expression on the pTA-Renilla luciferase reporter under the control of the minimal promoter (TATA-RLuc) were less clear than those on κB-FLuc activity, suggesting that the effect of the expression of SET-Nup214 or DEK-Nup214 on NF-κB-mediated transcription was specific. We next evaluated the effects of SET-Nup214 and DEK-Nup214 on the transcription of endogenous NF-κB target genes A20 and IκBα by RT-qPCR. In agreement with the results of the reporter assays described above, set-nup214 and dek-nup214 diminished the amounts of A20 and IκBα mRNAs in a dose-dependent manner (Fig. 5D). Collectively, these results demonstrate that SET-Nup214 and DEK-Nup214 impair NF-κB transcriptional activity and that this impairment occurs when the NF-κB signaling pathway is activated.
FIG 5.
Effects of SET-Nup214 and DEK-Nup214 on NF-κB transcriptional activity. (A to C) HEK293T cells (3 × 104) cultured in 24-well plates were transfected with pNF-κB40-firefly luciferase (10 ng) and either pCAGGS-SET-Nup214 (SET-N214) or DEK-Nup214 (DEK-N214) (10 or 100 ng). pTA-Renilla luciferase (100 ng) was cotransfected for the normalization of transfection efficiency. At 2 days after transfection, cells were incubated with 1 ng/ml or 5 ng/ml LMB for 30 min. Then recombinant human TNF-α (catalog no. 300-01A; PeproTech) was added at a final concentration of 20 ng/ml; the mixture was incubated for 3 to 4 h; and cell lysates were subjected to luciferase assays using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luminescence was measured by a Centro XS3 LB 960 luminometer (Berthold Japan K.K.). Relative firefly luciferase activity (A), relative Renilla luciferase activity (B), and normalized luciferase activity (C) were expressed as fold activation relative to the conditions in the leftmost lane (assigned a value of 1). Data are means ± standard deviations for three independent experiments. (C) The P value was calculated relative to the first (left graph) or second (right graph) lane. *, P < 0.05; **, P < 0.005; ***, P < 0.001. Western blot analyses were performed using lysates prepared for luciferase assays in the presence of TNF-α. Anti-Nup214 and anti-C23 (D6; dilution, 1:1,000; Santa Cruz Biotechnology, Inc.) antibodies were used as primary antibodies. Molecular weights (in thousands) of prestained markers are indicated on the left. (D) HEK293T cells (3 × 105) cultured in 6-well plates were transfected with pCAGGS-SET-Nup214 (SET-N214) or DEK-Nup214 (DEK-N214) (100 or 1,000 ng). At 2 days after transfection, cells were incubated with or without 5 ng/ml LMB for 30 min, and TNF-α was added at a final concentration of 20 ng/ml. After TNF-α incubation for 3 to 4 h, cells were collected, and isolated RNAs were subjected to RT-qPCR in order to quantify A20 and IκBα mRNAs. These mRNA expression levels were normalized to the level of β-actin mRNA and are shown as fold inhibition relative to expression in the second lane, taken as 1. Data are means ± standard deviations for three independent experiments. The P value was calculated relative to the value in the second lane. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
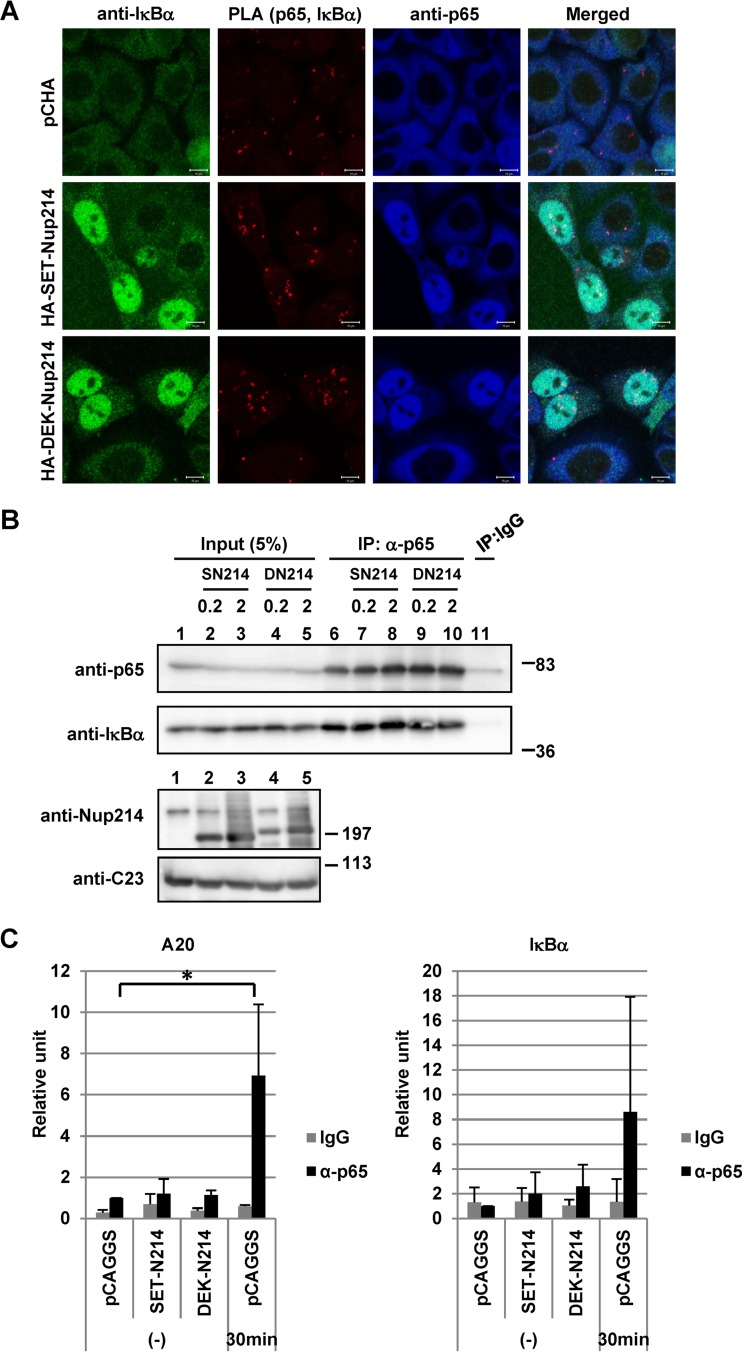
SET-Nup214 and DEK-Nup214 induce nuclear accumulation of the p65-IκBα complex in the absence of a stimulus.
In unstimulated cells, the majority of NF-κB transcription factors, such as p65 and p50, interact with IκB. Since p65 and p50 have NLSs, and IκBα has an NLS and an NES, the NF-κB–IκBα complex shuttles between the nucleus and the cytoplasm in an XPO1-dependent manner and is observed mainly in the cytoplasm (6, 53–58). In cells expressing SET-Nup214 and DEK-Nup214, NF-κB remained transcriptionally inactive (Fig. 5C, left graph), although p65 is located in the nucleus (Fig. 3A). Since IκBα is also localized in the nucleus, we hypothesized that the interaction between p65 and IκBα was maintained in the nucleus and that thus, NF-κB was kept inactive. To test this, we performed in situ proximity ligation assays (PLAs) and IP assays (Fig. 6A and B). In control cells, p65 and IκBα were observed in the cytoplasm, and cytoplasmic PLA signals were detected, indicating the proximity of p65 and IκBα in the cytoplasm. When cells were transfected with set-nup214 or dek-nup214, both p65 and IκBα were found in the nucleus (Fig. 3A), and nuclear PLA signals were observed in these cells (Fig. 6A). By IP assays, p65 was found to interact with IκBα, and this interaction was not affected by the absence or presence of SET-Nup214 or DEK-Nup214 (Fig. 6B). These results suggest that SET-Nup214 and DEK-Nup214 induced nuclear accumulation of the p65-IκBα complex but that the NF-κB signaling pathway was kept inactive, since the fusion proteins did not affect the interaction between p65 and IκBα. The binding of IκBα to p65 causes a release of p65 from DNA (59). Therefore, it was presumed that nuclear p65 bound by IκBα in the presence of SET-Nup214 or DEK-Nup214 could not bind to the target gene promoter. To confirm this notion, we performed chromatin immunoprecipitation assays. The levels of p65 that bound to A20 and IκBα promoter regions were increased by TNF-α treatment. However, the binding of p65 to these promoters was not enhanced by SET-Nup214 or DEK-Nup214 (Fig. 6C), although p65 was localized in the nucleus (Fig. 3A). These results support our notion that p65 was kept inactive in cells expressing SET-Nup214 and DEK-Nup214.
FIG 6.
Interaction of p65 with IκBα or chromatin in the nucleus. (A) HeLa cells cultured in 6-cm dishes were transfected with 2 μg pCHA, HA-SET-Nup214, or HA-DEK-Nup214. At 2 days after transfection, cells were collected and were subjected to IF assays and PLAs. Anti-p65 (ab7970; dilution, 1:100; Abcam) and anti-IκBα (L35A5; dilution, 1:30; Cell Signaling Technology, Inc.) antibodies were used as primary antibodies. “Merged” panels show composite images of cells stained with Alexa Fluor 488, Detection Reagents Red (for PLA), and Alexa Fluor 633. Bars, 10 μm. (B) HEK293T cells were transfected with pCAGGS, pCAGGS-SET-Nup214 (SN214), or pCAGGS-DEK-Nup214 (DN214) (0.2 or 2 μg) and were incubated for 2 days. Cells were collected, and IP assays were conducted using anti-p65 (ab7970) and rabbit polyclonal IgG antibodies (PP64B) (Merck KGaA, Germany). Proteins in input lysates and immunoprecipitated samples were separated by 10% or 5% SDS-PAGE, and Western blot analyses were performed using anti-p65, anti-IκBα, anti-Nup214, and anti-C23 antibodies. Molecular weights (in thousands) of prestained markers are indicated on the right. (C) HEK293T cells were transfected with 5 μg pCAGGS, pCAGGS-SET-Nup214 (SET-N214), or pCAGGS-DEK-Nup214 (DEK-N214). At 2 days after transfection, cells were either left untreated (−) or treated with TNF-α (20 ng/ml) for 30 min; they were then subjected to ChIP assays using 2 μg anti-IgG or anti-p65 (ab7970) antibodies to measure the binding of p65 to A20 or IκBα promoter regions. The levels of immunoprecipitated DNA were then normalized to the input DNA level. Results are shown as fold activation relative to the level of DNA immunoprecipitated from pCAGGS-transfected lysates by the anti-p65 antibody in the absence of TNF-α. Data are means ± standard deviations for three independent experiments. *, P < 0.05.
The p65-IκBα complex is maintained in the presence of a stimulus in cells expressing SET-Nup214 or DEK-Nup214.
Reporter assays and RT-qPCR showed that SET-Nup214 and DEK-Nup214 downregulate NF-κB transcriptional activity in the presence of TNF-α (Fig. 5C, right graph). IκBα is phosphorylated in the cytoplasm upon stimulation, followed by degradation by the ubiquitin proteasome system (60). NF-κB transcription factors are then freed from IκB, are localized in the nucleus, and execute target gene transcription. We predicted that the NF-κB–IκBα complex would be maintained by the expression of SET-Nup214 and DEK-Nup214 even after TNF-α addition. To test this, the localization patterns of IκBα and p65 were monitored, and PLAs and coimmunoprecipitation assays were performed. After the addition of TNF-α, IκBα levels in the cytoplasm of both control cells and SET-Nup214- or DEK-Nup214-expressing cells were markedly reduced (Fig. 7A), indicating the degradation of cytoplasmic IκBα. In contrast, IκBα was visible in the nuclei of SET-Nup214- and DEK-Nup214-expressing cells after TNF-α treatment (Fig. 7A). The restoration of the expression level of IκBα by SET-Nup214 or DEK-Nup214 was also confirmed by Western blotting (Fig. 7C, lanes 1 to 6). In cells expressing nuclear p65 and IκBα, PLA signals were detected both 0 and 30 min after TNF-α treatment, demonstrating that the interaction between p65 and IκBα was maintained in the nucleus after stimulation (Fig. 7B). The formation of the p65-IκBα complex in stimulated cells expressing SET-Nup214 or DEK-Nup214 was confirmed by an IP assay (Fig. 7C, lanes 7 to 13). Finally, to examine whether the recruitment of p65 to its target genes was affected by SET-Nup214 or DEK-Nup214, a ChIP assay was performed. It was demonstrated that the binding of p65 to the A20 and IκBα promoter regions was impaired in the presence of SET-Nup214 or DEK-Nup214 (Fig. 7D). These results indicate that SET-Nup214 and DEK-Nup214 reduce IκBα degradation by retaining it in the nucleus and maintain the p65-IκBα complex.
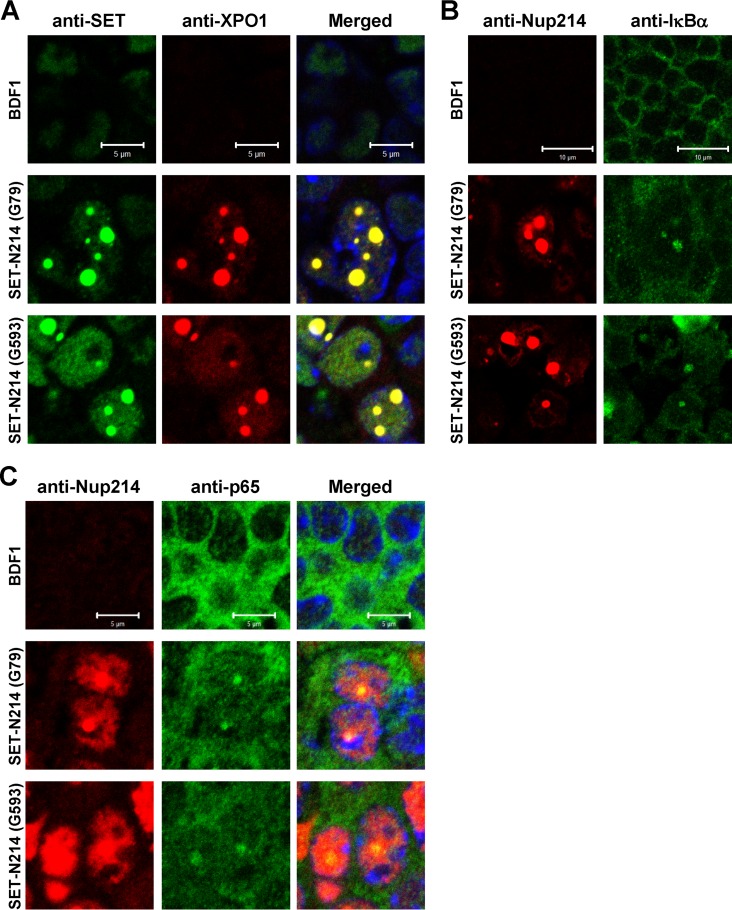
Subcellular localization of XPO1 and its cargos in set-nup214 transgenic mice.
Previously, we generated a transgenic mouse expressing set-nup214 (22). Although this mouse did not develop leukemia, it showed severe anemia and a halt in hematopoietic differentiation, both of which are frequently associated with leukemia. In order to understand the biological relevance of the results obtained from in vitro cell culture studies, we assessed the localization patterns of XPO1 and its cargos using spleen sections from set-nup214 transgenic mice. We observed that SET-Nup214 and XPO1 are colocalized in the nucleus as granular dots (Fig. 8A). In addition, it was found that IκBα (Fig. 8B) and p65 (Fig. 8C) are also localized in the dots. These results demonstrate that the localization pattern of XPO1 is affected, and its function could be impaired by SET-Nup214 in vivo as well.
FIG 8.
Subcellular localization of XPO1, IκBα, and p65 in the spleens of set-nup214 transgenic mice. Spleen sections of wild-type BDF1 and set-nup214 transgenic mice (lines G79 and G593) were subjected to IF assays. The primary antibodies used were anti-SET/TAF-Iβ (KM1721; dilution, 1:20) (A), anti-XPO1 (dilution, 1:100) (A), anti-Nup214 (dilution, 1:100) (B and C), anti-IκBα (L35A5; dilution, 1:20) (B), and anti-p65 (F6; dilution, 1:20) (C). “Merged” panels are composite images of cells stained with Alexa Fluor 488, Alexa Fluor 568, and TO-PRO-3. Bars, 5 μm (A and C) and 10 μm (B).
DISCUSSION
Interaction of SET-Nup214 and DEK-Nup214 with XPO1 and NFX1.
In this study, we examined the functions of SET-Nup214 and DEK-Nup214 in terms of their effects on the nuclear-cytoplasmic transport of proteins and RNAs. We found that among several NTRs, SET-Nup214 and DEK-Nup214 interact preferentially not only with XPO1 but also with NXF1 (Fig. 1B). These interactions were dependent on the FG repeat regions of SET-Nup214 and DEK-Nup214 (Fig. 1C). It was shown that individual FG-Nups have different preferences for various NTRs (61, 62). Hence, it is supposed that the affinities of the FG repeat regions of SET-Nup214 and DEK-Nup214 for NTRs also differ, and the affinity difference generates a binding preference. Nup214 interacts with XPO1 rather than Xpo-t, NXF1, or XPO2/CAS (29, 63). Ectopic expression of truncated Nup214 containing the FG repeat region has an inhibitory effect on the functions of a subset of NTRs, including XPO1 (39). Our results are consistent with previous findings and imply that the SET and DEK portions of SET-Nup214 or DEK-Nup214 do not affect the structure and function of the Nup214 portion for association with NTRs.
Effects of SET-Nup214 and DEK-Nup214 on the functions of XPO1 and NXF1.
We have demonstrated that SET-Nup214 and DEK-Nup214 associate with both XPO1 and NXF1. However, the effect of Nup214 fusion proteins on XPO1 function was different from that on NXF1 function. We showed that SET-Nup214 and DEK-Nup214 induce lower mobility of XPO1 and cause the accumulation of XPO1 cargos in the nucleus (Fig. 3A and C and 4C). In contrast, mRNA, which is an NXF1 cargo, did not accumulate in the nucleus of cells expressing SET-Nup214 and DEK-Nup214 (Fig. 3D). XPO1 was mainly incorporated into the dots where SET-Nup214 and DEK-Nup214 are located, whereas diffused NXF1 was observed in cells expressing SET-Nup214 or DEK-Nup214 (Fig. 1D and E). This differential localization of XPO1 and NXF1 in cells expressing SET-Nup214 or DEK-Nup214 could explain the different effects of SET-Nup214 and DEK-Nup214 on the functions of XPO1 and NXF1. It is assumed that SET-Nup214-XPO1 and DEK-Nup214-XPO1 complexes could be more stable than SET-Nup214-NXF1 and DEK-Nup214-NXF1 complexes.
SET-Nup214 and DEK-Nup214 form stable complexes with XPO1.
A question is raised as to how SET-Nup214 and DEK-Nup214 can form a stable complex with XPO1 to induce the accumulation of NES proteins in the nucleus. The interaction between Nup214 and XPO1 is stabilized when both RanGTP and NES proteins are incorporated (38, 47, 48). In agreement with this, we found that inhibition of the interaction between XPO1 and NES proteins by LMB leads to the disappearance of dots formed in the presence of SET-Nup214 and DEK-Nup214 (Fig. 2). From these results, we speculate that the nuclear dots are formed by the quaternary stable complex containing SET-Nup214 or DEK-Nup214, NES proteins, XPO1, and RanGTP. Furthermore, it is possible that other proteins play roles in the formation of the dots. The formation of RanGTP-XPO1-NES protein complexes is enhanced by Nup98 (64) and RanBP3 (65–67). Nup214 functions as a scaffold for the recruitment of several nucleoporins, such as Nup88, Nup358, Nup62, and Nup98 (38, 40, 68, 69). These proteins may facilitate stable complex formation induced by SET-Nup214 and DEK-Nup214. In spleen cells, these dots are much larger than those in cultured cells. It is likely that high expression of proteins constituting these dots in mouse spleen enlarges the SET-Nup214 nuclear dots.
Deregulation of transcription by SET-Nup214 or DEK-Nup214 and oncogenesis.
We found that IκBα is localized in the nucleus regardless of the presence or absence of TNF-α (Fig. 7A) in cells expressing SET-Nup214 or DEK-Nup214. It is presumed that nuclear accumulation of IκBα enables it to escape from IKKβ (IκB kinase β)-mediated phosphorylation after TNF-α addition, and thus, NF-κB transcriptional activity is repressed by the interaction with IκBα in the nucleus. In general, NF-κB induces the transcription of various genes related to inflammation, cell proliferation, invasion, etc. NF-κB inactivation is known to counteract oncogenesis or tumorigenesis, and NF-κB is an efficient therapeutic target for cancer (70, 71). In contrast, NF-κB has also been reported to have antioncogenic activities, such as induction of cellular senescence. p65−/− mouse embryonic fibroblasts (MEFs) bypass senescence (72), and knockdown of p65 induces chemoresistance in mouse lymphoma (73). SET-Nup214 and DEK-Nup214 impair the NF-κB pathway (Fig. 5). Along these lines, one can speculate that SET-Nup214 and DEK-Nup214 may promote the bypass of senescence via suppression of the NF-κB signaling pathway. In addition, several studies have documented the importance of the NF-κB pathway for hematopoiesis. Conditional knockout of the IKKβ gene upregulates interleukin 1 (IL-1) production and stimulates the proliferation of neutrophil progenitor, leading to neutrophilia and splenomegaly (74) (75). In addition, it has been reported that conditional knockout of p65 or IKKβ induces cell cycling of hematopoietic stem cells and increases their number (76, 77). These observations suggest that inhibition of the NF-κB pathway is one potential cause of the SET-Nup214-induced block of the differentiation of hematopoietic progenitor cells observed in SET-Nup214 transgenic mice. Furthermore, it has been demonstrated previously that the differentiation of U937 was inhibited by SET-Nup214 expression (23). Similarly, the differentiation of U937 was inhibited by the presence of an IKKβ inhibitor (78, 79). These results also suggest that the inhibition of the NF-κB pathway by SET-Nup214 is, at least in part, a potential cause of the U937 differentiation block induced by SET-Nup214.
In conclusion, SET-Nup214 and DEK-Nup214 interact with NTRs, and the interaction of either SET-Nup214 or DEK-Nup214 with XPO1 leads to a malfunction of transcriptional regulation by NF-κB. Until now, many proteins have been identified as XPO1 cargos (80, 81). In addition to IκBα and cyclin B1, these various cargos might accumulate in the nucleus when either SET-Nup214 or DEK-Nup214 is present. In fact, β-catenin has been reported to accumulate in the nuclei of set-nup214 transgenic mice (21). Because appropriate nuclear-cytoplasmic transport is required for cellular integrity, these disturbances of the localization of various proteins may lead synergistically to oncogenesis by SET-Nup214 or DEK-Nup214. To know which NES proteins are responsible for SET-NUP214- or DEK-Nup214-mediated oncogenesis, comprehensive postgenomic analyses are required.
ACKNOWLEDGMENTS
We thank Minoru Yoshida (RIKEN) for providing the pHCF1 vector. We are grateful to Catherine Ann Moroski-Erkul for critical reading of the manuscript.
This work was supported by JSPS KAKENHI grants 24790309 (to S.S.) and 25291001 (to K.N.).
REFERENCES
- 1.Walde S, Kehlenbach RH. 2010. The part and the whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol 20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Strambio-De-Castillia C, Niepel M, Rout MP. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11:490–501. doi: 10.1038/nrm29281. [DOI] [PubMed] [Google Scholar]
- 3.Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau TR, Way JC, Silver PA. 2004. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer 4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 5.O'Brate A, Giannakakou P. 2003. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist Updat 6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Oeckinghaus A, Ghosh S. 2009. The NF-κB family of transcription factors and its regulation. Cold Spring Harbor Perspect Biol 1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capelson M, Hetzer MW. 2009. The role of nuclear pores in gene regulation, development and disease. EMBO Rep 10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, Yamamoto H, Schwartz S Jr, Esteller M. 2010. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A, Hernandez L, Yague J, Nicolas P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjose S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpi JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigo R, Bayes M, Heath S, Gut M, et al. 2011. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A, Haferlach T, Kern W, Schnittger S. 2014. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 28:108–117. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Powers MA. 2009. Nuclear pore proteins and cancer. Semin Cell Dev Biol 20:620–630. doi: 10.1016/j.semcdb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler A, Hurt E. 2010. Gene regulation by nucleoporins and links to cancer. Mol Cell 38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 13.Funasaka T, Wong RW. 2011. The role of nuclear pore complex in tumor microenvironment and metastasis. Cancer Metastasis Rev 30:239–251. doi: 10.1007/s10555-011-9287-y. [DOI] [PubMed] [Google Scholar]
- 14.Simon DN, Rout MP. 2014. Cancer and the nuclear pore complex. Adv Exp Med Biol 773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- 15.Zhou MH, Yang QM. 2014. NUP214 fusion genes in acute leukemia. Oncol Lett 8:959–962. doi: 10.3892/ol.2014.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol 12:1687–1697. doi: 10.1128/MCB.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. 1992. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol 12:3346–3355. doi: 10.1128/MCB.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben Abdelali R, Roggy A, Leguay T, Cieslak A, Renneville A, Touzart A, Banos A, Randriamalala E, Caillot D, Lioure B, Devidas A, Mossafa H, Preudhomme C, Ifrah N, Dombret H, Macintyre E, Asnafi V. 2014. SET-NUP214 is a recurrent γδ lineage-specific fusion transcript associated with corticosteroid/chemotherapy resistance in adult T-ALL. Blood 123:1860–1863. doi: 10.1182/blood-2013-08-521518. [DOI] [PubMed] [Google Scholar]
- 19.Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, Adachi S, Auvrignon A, Beverloo HB, Cayuela JM, Chilton L, Fornerod M, de Haas V, Harrison CJ, Inaba H, Kaspers GJ, Liang DC, Locatelli F, Masetti R, Perot C, Raimondi SC, Reinhardt K, Tomizawa D, von Neuhoff N, Zecca M, Zwaan CM, van den Heuvel-Eibrink MM, Hasle H. 2014. t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica 99:865–872. doi: 10.3324/haematol.2013.098517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, Stubbs A, Cools J, Nagata K, Fornerod M, Buijs-Gladdines J, Horstmann M, van Wering ER, Soulier J, Pieters R, Meijerink JP. 2008. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 111:4668–4680. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozbek U, Kandilci A, van Baal S, Bonten J, Boyd K, Franken P, Fodde R, Grosveld GC. 2007. SET-CAN, the product of the t(9;9) in acute undifferentiated leukemia, causes expansion of early hematopoietic progenitors and hyperproliferation of stomach mucosa in transgenic mice. Am J Pathol 171:654–666. doi: 10.2353/ajpath.2007.060934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito S, Nouno K, Shimizu R, Yamamoto M, Nagata K. 2008. Impairment of erythroid and megakaryocytic differentiation by a leukemia-associated and t(9;9)-derived fusion gene product, SET/TAF-Iβ-CAN/Nup214. J Cell Physiol 214:322–333. doi: 10.1002/jcp.21199. [DOI] [PubMed] [Google Scholar]
- 23.Kandilci A, Mientjes E, Grosveld G. 2004. Effects of SET and SET-CAN on the differentiation of the human promonocytic cell line U937. Leukemia 18:337–340. doi: 10.1038/sj.leu.2403227. [DOI] [PubMed] [Google Scholar]
- 24.Ageberg M, Drott K, Olofsson T, Gullberg U, Lindmark A. 2008. Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis. Genes Chromosomes Cancer 47:276–287. doi: 10.1002/gcc.20531. [DOI] [PubMed] [Google Scholar]
- 25.Sanden C, Ageberg M, Petersson J, Lennartsson A, Gullberg U. 2013. Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC Cancer 13:440. doi: 10.1186/1471-2407-13-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oancea C, Ruster B, Henschler R, Puccetti E, Ruthardt M. 2010. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia 24:1910–1919. doi: 10.1038/leu.2010.180. [DOI] [PubMed] [Google Scholar]
- 27.Moroianu J, Hijikata M, Blobel G, Radu A. 1995. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci U S A 92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boer J, Bonten-Surtel J, Grosveld G. 1998. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol 18:1236–1247. doi: 10.1128/MCB.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuersten S, Arts GJ, Walther TC, Englmeier L, Mattaj IW. 2002. Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol Cell Biol 22:5708–5720. doi: 10.1128/MCB.22.16.5708-5720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J 18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA (New York, NY) 6:136–158. doi: 10.1017/S1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levesque L, Guzik B, Guan T, Coyle J, Black BE, Rekosh D, Hammarskjold ML, Paschal BM. 2001. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J Biol Chem 276:44953–44962. doi: 10.1074/jbc.M106558200. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol 22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol 20:8996–9008. doi: 10.1128/MCB.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Deursen J, Boer J, Kasper L, Grosveld G. 1996. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J 15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 37.Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. 2006. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60S preribosomal nuclear export. J Biol Chem 281:19378–19386. doi: 10.1074/jbc.M512585200. [DOI] [PubMed] [Google Scholar]
- 38.Hutten S, Kehlenbach RH. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol 26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roloff S, Spillner C, Kehlenbach RH. 2013. Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1. J Biol Chem 288:3952–3963. doi: 10.1074/jbc.M112.433243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. 2006. The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFκB activation in Drosophila. J Cell Sci 119:4409–4419. doi: 10.1242/jcs.03201. [DOI] [PubMed] [Google Scholar]
- 41.Takeda A, Yaseen NR. 2014. Nucleoporins and nucleocytoplasmic transport in hematologic malignancies. Semin Cancer Biol 27:3–10. doi: 10.1016/j.semcancer.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. 1996. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene 13:1801–1808. [PubMed] [Google Scholar]
- 43.Saito S, Miyaji-Yamaguchi M, Nagata K. 2004. Aberrant intracellular localization of SET-CAN fusion protein, associated with a leukemia, disorganizes nuclear export. Int J Cancer 111:501–507. doi: 10.1002/ijc.20296. [DOI] [PubMed] [Google Scholar]
- 44.Nagata K, Saito S, Okuwaki M, Kawase H, Furuya A, Kusano A, Hanai N, Okuda A, Kikuchi A. 1998. Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res 240:274–281. doi: 10.1006/excr.1997.3930. [DOI] [PubMed] [Google Scholar]
- 45.Numajiri Haruki A, Naito T, Nishie T, Saito S, Nagata K. 2011. Interferon-inducible antiviral protein MxA enhances cell death triggered by endoplasmic reticulum stress. J Interferon Cytokine Res 31:847–856. doi: 10.1089/jir.2010.0132. [DOI] [PubMed] [Google Scholar]
- 46.Herold A, Klymenko T, Izaurralde E. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA (New York, NY) 7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 47.Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol 19:6276–6285. doi: 10.1128/MCB.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol 145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 50.Daelemans D, Costes SV, Lockett S, Pavlakis GN. 2005. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol 25:728–739. doi: 10.1128/MCB.25.2.728-739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. 1997. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem 272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 52.Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR. 2010. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J Biol Chem 285:16248–16257. doi: 10.1074/jbc.M109.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson C, Van Antwerp D, Hope TJ. 1999. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J 18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez MS, Thompson J, Hay RT, Dargemont C. 1999. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J Biol Chem 274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 55.Huang TT, Kudo N, Yoshida M, Miyamoto S. 2000. A nuclear export signal in the N-terminal regulatory domain of IκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc Natl Acad Sci U S A 97:1014–1019. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tam WF, Lee LH, Davis L, Sen R. 2000. Cytoplasmic sequestration of Rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol 20:2269–2284. doi: 10.1128/MCB.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malek S, Chen Y, Huxford T, Ghosh G. 2001. IκBβ, but not IκBα, functions as a classical cytoplasmic inhibitor of NF-κB dimers by masking both NF-κB nuclear localization sequences in resting cells. J Biol Chem 276:45225–45235. doi: 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- 58.Hayden MS, Ghosh S. 2004. Signaling to NF-κB. Genes Dev 18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 59.Baeuerle PA, Baltimore D. 1988. IκB: a specific inhibitor of the NF-κB transcription factor. Science (New York, NY) 242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 60.Kanarek N, Ben-Neriah Y. 2012. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol Rev 246:77–94. doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 61.Allen NP, Huang L, Burlingame A, Rexach M. 2001. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem 276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 62.Terry LJ, Wente SR. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang Y, Bogerd HP, Cullen BR. 2000. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J Virol 74:5863–5871. doi: 10.1128/JVI.74.13.5863-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oka M, Asally M, Yasuda Y, Ogawa Y, Tachibana T, Yoneda Y. 2010. The mobile FG nucleoporin Nup98 is a cofactor for Crm1-dependent protein export. Mol Biol Cell 21:1885–1896. doi: 10.1091/mbc.E09-12-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U. 2001. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2:926–932. doi: 10.1093/embo-reports/kve200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG. 2001. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koyama M, Shirai N, Matsuura Y. 2014. Structural insights into how Yrb2p accelerates the assembly of the Xpo1p nuclear export complex. Cell Rep 9:983–995. doi: 10.1016/j.celrep.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 68.Bernad R, van der Velde H, Fornerod M, Pickersgill H. 2004. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol 24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwartz M, Travesa A, Martell SW, Forbes DJ. 2015. Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin. Nucleus (Austin, Tex) 6:40–54. doi: 10.1080/19491034.2015.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prasad S, Ravindran J, Aggarwal BB. 2010. NF-κB and cancer: how intimate is this relationship. Mol Cell Biochem 336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perkins ND. 2007. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol 8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 72.Wang J, Jacob NK, Ladner KJ, Beg A, Perko JD, Tanner SM, Liyanarachchi S, Fishel R, Guttridge DC. 2009. RelA/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep 10:1272–1278. doi: 10.1038/embor.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, Kogan SC, Lowe SW. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu LC, Enzler T, Seita J, Timmer AM, Lee CY, Lai TY, Yu GY, Lai LC, Temkin V, Sinzig U, Aung T, Nizet V, Weissman IL, Karin M. 2011. IL-1β-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKβ. Nat Immunol 12:144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mankan AK, Canli O, Schwitalla S, Ziegler P, Tschopp J, Korn T, Greten FR. 2011. TNF-α-dependent loss of IKKβ-deficient myeloid progenitors triggers a cytokine loop culminating in granulocytosis. Proc Natl Acad Sci U S A 108:6567–6572. doi: 10.1073/pnas.1018331108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein SJ, Baldwin AS. 2013. Deletion of the NF-κB subunit p65/RelA in the hematopoietic compartment leads to defects in hematopoietic stem cell function. Blood 121:5015–5024. doi: 10.1182/blood-2013-02-486142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Li L, Baldwin AS Jr, Friedman AD, Paz-Priel I. 2015. Loss of IKKβ but not NF-κB p65 skews differentiation towards myeloid over erythroid commitment and increases myeloid progenitor self-renewal and functional long-term hematopoietic stem cells. PLoS One 10:e0130441. doi: 10.1371/journal.pone.0130441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dai Y, Rahmani M, Grant S. 2003. An intact NF-κB pathway is required for histone deacetylase inhibitor-induced G1 arrest and maturation in U937 human myeloid leukemia cells. Cell Cycle (Georgetown, Tex) 2:467–472. [PubMed] [Google Scholar]
- 79.Song MG, Ryoo IG, Choi HY, Choi BH, Kim ST, Heo TH, Lee JY, Park PH, Kwak MK. 2015. NRF2 signaling negatively regulates phorbol-12-myristate-13-acetate (PMA)-induced differentiation of human monocytic U937 cells into pro-inflammatory macrophages. PLoS One 10:e0134235. doi: 10.1371/journal.pone.0134235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu SC, Huang HC, Horton P, Juan HF. 2013. ValidNESs: a database of validated leucine-rich nuclear export signals. Nucleic Acids Res 41:D338–D343. doi: 10.1093/nar/gks936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. 2013. Identification of CRM1-dependent nuclear export cargos using quantitative mass spectrometry. Mol Cell Proteomics 12:664–678. doi: 10.1074/mcp.M112.024877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okuwaki M, Sumi A, Hisaoka M, Saotome-Nakamura A, Akashi S, Nishimura Y, Nagata K. 2012. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res 40:4861–4878. doi: 10.1093/nar/gks162. [DOI] [PMC free article] [PubMed] [Google Scholar]