Abstract
DbpA is a DEAD-box RNA helicase implicated in Escherichia coli large ribosomal subunit assembly. Previous studies have shown that when the ATPase and helicase inactive DbpA construct, R331A, is expressed in E. coli cells, a large ribosomal subunit intermediate accumulates. The large subunit intermediate migrates as a 45S particle in a sucrose gradient. Here, using a number of structural and fluorescent assays, we investigate the ribosome profiles of cells lacking wild-type DbpA and overexpressing the R331A DbpA construct. Our data show that in addition to the 45S particle previously described, 27S and 35S particles are also present in the ribosome profiles of cells overexpressing R331A DbpA. The 27S, 35S, and 45S independently convert to the 50S subunit, suggesting that ribosome assembly in the presence of R331A and the absence of wild-type DbpA occurs via multiple pathways.
Keywords: DbpA, DEAD-box protein, ribosome maturation factor, RNA helicase, ribosome assembly
INTRODUCTION
In Escherichia coli (E. coli) cells, the ribosome assembly process involves the interaction of 55 proteins and three RNA molecules and consumes up to 40% of the cell's energy (Maguire 2009). In order to fulfill the cell's demand for new proteins and to avoid wasting valuable cellular energy, ribosome assembly needs to occur rapidly and accurately. To ensure this, cells use a number of different protein and RNA maturation factors (El Hage et al. 2001; Charollais et al. 2003, 2004; El Hage and Alix 2004; Jain 2008). One class of RNA maturation factors used by the cell during ribosome assembly is DEAD-box RNA helicases (Cordin et al. 2006; Linder 2006; Pyle 2008; Pan and Russell 2010; Shajani et al. 2011; Henn et al. 2012; Putnam and Jankowsky 2013). In vitro, these enzymes use the energy of ATP binding and hydrolysis to unwind short RNA double helices. In vivo, this unwinding activity is believed to facilitate RNA structural rearrangements during the ribosome assembly process (Shajani et al. 2011). E. coli contains five DEAD-box RNA helicases, with four of them (SrmB, DeaD, RhlE, and DbpA) involved in ribosome assembly (Iost and Dreyfus 2006; Jain 2008; Iost et al. 2013).
DbpA (DEAD-box-protein A), which is the focus of this study, is implicated in assembly of the large ribosomal subunit. DbpA binds tightly and specifically to helix 92 of the 23S ribosomal RNA (rRNA), which is located in the peptidyl transferase center of the ribosome (Nicol and Fuller-Pace 1995; Diges and Uhlenbeck 2001; Tsu et al. 2001). The C-terminal domain of DbpA is responsible for its tight and specific binding to helix 92, while the N-terminal RecA-like domains carry out the DbpA protein's ATPase and nonprocessive helicase activity.
It has been previously demonstrated that no changes in growth phenotype are observed when the dbpA gene is deleted from the E. coli chromosome (Baba et al. 2006; Iost and Dreyfus 2006; Elles and Uhlenbeck 2008; Peil et al. 2008). One plausible explanation for the dbpA deletion data is that because correct ribosome assembly is such a crucial process for cell survival, the cell performs this assembly through multiple pathways (Jain 2008; Shajani et al. 2011). In this way, when one of the maturation factors required for ribosome assembly is missing, the assembly can be completed by an alternative pathway and aided by a different maturation factor. In addition to E. coli, the dbpA gene is present in many other bacteria (López-Ramírez et al. 2011), suggesting that the DbpA protein could be essential for ribosome assembly under currently undetermined conditions, or when another ribosomal maturation factor is missing.
Interestingly, while no phenotype was observed when the dbpA gene was removed from the E. coli genome, overexpression of ATPase and helicase inactive DbpA, R331A, in the background of endogenous wild-type DbpA produced a cold-sensitive slow-growth phenotype (Elles and Uhlenbeck 2008). More importantly, the ribosome profiles of cells overexpressing R331A DbpA show the accumulation of a 45S particle, which is a precursor of the large ribosomal subunit. The accumulation of this particle is believed to be a consequence of the inability of R331A DbpA to facilitate ribosomal RNA structural rearrangement, while preventing wild-type DbpA and other maturation factors, which may perform a similar function to DbpA, access to the large subunit's ribosomal RNA (Elles and Uhlenbeck 2008; Elles et al. 2009).
The protein composition of the 45S particle has previously been investigated by mass spectrometry. The 45S particle lacks proteins that are added late during ribosome assembly, indicating that the 45S particle is a late-stage intermediate on the large subunit assembly pathway. Currently, it is not known whether the 45S particle is able to form the 50S large subunit over time in vivo, or whether the 45S particle is a dead-end intermediate. If the 45S particle is a dead-end intermediate, whether or not the particle is marked for degradation is also unknown. The experiments outlined here investigate the fate of the 45S intermediate particle in the large ribosomal subunit assembly pathway when helicase inactive R331A DbpA is present and the wild-type dbpA gene is removed from the E. coli chromosome.
RESULTS
Determination of RNA composition of separated ribosomal particles
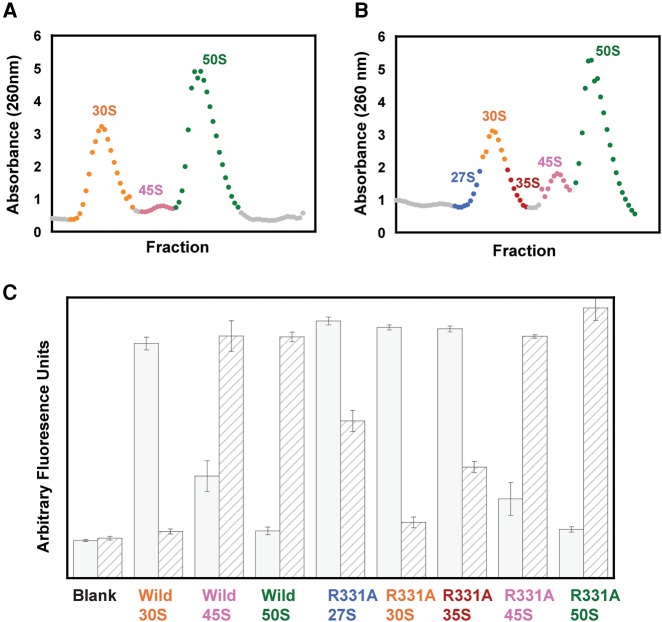
The ribosome profiles of the cells overexpressing wild-type and R331A DbpA were obtained via sucrose gradients as described in the Materials and Methods section below. Figure 1A and B show ribosomal RNA absorbance at 260 nm versus the sucrose gradient fractions for cells overexpressing the wild-type DbpA and R331A constructs, respectively. Both profiles have 30S, 45S, and 50S particles present; however, the 45S particle is considerably more abundant in the cells overexpressing R331A DbpA. This agrees with previous experimental work (Elles et al. 2009). Interestingly, in cells overexpressing R331A DbpA, the 30S peak is broader and contains left and right shoulders. To determine whether the 30S shoulders, which migrate as particles of about 27S and 35S on the sucrose gradient (Fig. 1B), are large ribosome subunit precursors, the RNA composition of the ribosome profiles of cells overexpressing wild-type or R331A DbpA was investigated via a binary deoxyribozyme (biDz) sensor assay. This sensitive fluorescence assay has been used successfully in the past to detect bacterial rRNA both in vitro and in vivo (Gerasimova and Kolpashchikov 2013; Gerasimova et al. 2015).
FIGURE 1.
Characterization of ribosomal RNA in E. coli cells overexpressing wild-type or R331A DbpA. (A) Ribosome profile of the cells overexpressing wild-type DbpA. (B) Ribosome profile of the cells overexpressing R331A DbpA. The ribosome profiles of the cells overexpressing wild-type or R331A DbpA contained only 1 mM MgCl2, which resulted in the ribosomes dissociating into separate subunits. (C) Fluorescent assay with binary deoxyribozyme (biDz) sensors. The fluorescence signals for biDz-16S and biDz-23S are shown as solid gray bars and striped bars, respectively. The amount of RNA used for each particle was normalized to match the fluorescent output of the dominant RNA species (16S rRNA in 27S, 30S, and 35S peak fractions, and 23S rRNA in 45S and 50S fractions) in the particle. Thus, the ratio of 23S:16S rRNA for each particle, which will not change with concentration, should be used to compare the particles. These data show that 23S RNA is present both in the right and the left shoulders of the 30S peaks on the cells overexpressing R331A DbpA; no 23S RNA was observed migrating with the middle fractions from 30S peak of the cells overexpressing R331A DbpA or the whole 30S peak from the cells overexpressing wild-type DbpA.
To perform rRNA content comparison between different fractions of the ribosomal profiles from cells overexpressing wild-type or R331A DbpA, two biDz sensors were used—one targeting E. coli 23S rRNA (biDz-23S) and another specifically recognizing 16S rRNA (biDz-16S) (Fig. 1C). As expected, no difference in the fluorescent response of both biDz-23S and biDz-16S was observed for the fractions corresponding to the 45S and 50S peaks in both types of cells: 50S fractions consisted entirely of 23S rRNA, and 45S fractions contained 23S rRNA and slight amounts of 16S rRNA. At the same time, the fractions of 30S peaks, and 27S and 35S shoulders of the ribosome profile from cells overexpressing wild-type or R331A DbpA demonstrated different rRNA composition. The fraction of 30S peak (whole 30S peak in the cells overexpressing wild-type DbpA and the center peak in the cells overexpressing R331A DbpA) had no 23S rRNA present. The 27S and 35S fractions from cells overexpressing R331A DbpA had a predominantly biDz-16S signal, like those in the presence of 30S fractions; however, these fractions also triggered significant fluorescent increase for biDz-23S, indicating that the 27S and 35S particles are precursors of the 50S ribosome subunit. This observation is in agreement with the data from previous primer extension experiments suggesting the presence of large subunit precursors that sediment in a sucrose gradient near the 30S particle in the cells overexpressing R331A DbpA (Elles et al. 2009).
In order to further ascertain that in the cells overexpressing R331A DbpA two distinct large subunit precursor particles migrate in a sucrose gradient near the 30S small subunit, RNA normalized to the same absorbance at 260 nm from individual, rather than concentrated, fractions across the entire 30S peak were tested with the biDz sensor specific to the 23S rRNA (Fig. 2). The biDz sensor experiments show an uneven distribution of 23S rRNA across the 30S peak, with the left and right shoulder containing a higher amount of 23S rRNA. This is consistent with the presence in the cells overexpressing R331A DbpA of two distinct large subunit precursors, which migrate in a sucrose gradient as particles of about 27S and 35S. On the other hand, in the cells overexpressing the wild-type DbpA protein, the biDz sensor assay shows an even distribution of the 23S rRNA on all the 30S RNA fractions, suggesting that in these cells 27S and 35S large subunit intermediate particles do not accumulate (Fig. 2).
FIGURE 2.
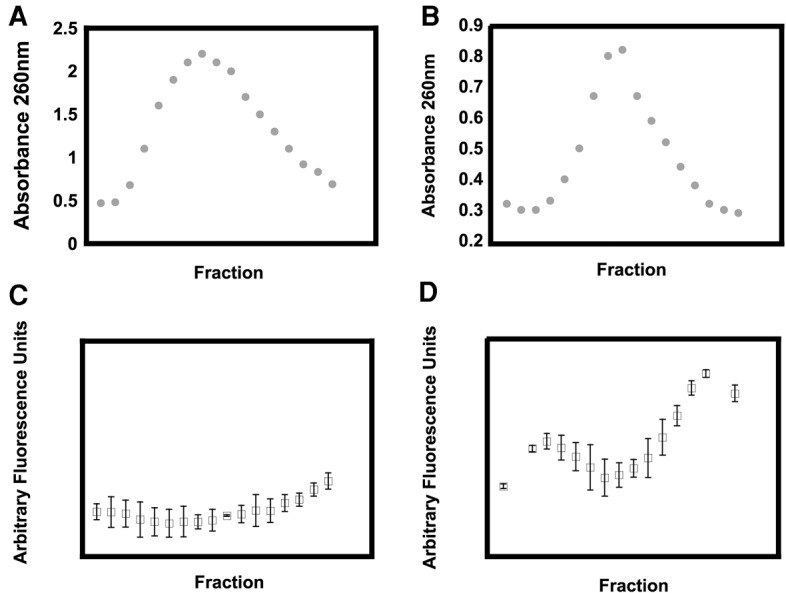
(A,B) The UV (absorbance 260 nm) profile of the small subunit peak from wild-type (A) and R331A (B) cells. (C,D) The 23S rRNA biDz fluorescence profile of the fractions from A and B.
Having established in the cells overexpressing R331A DbpA the presence of two new and distinct 50S precursors, we proceeded to further investigate their structural and physical properties. First, we investigated the new particles 5′ end 23S rRNA processing and compared it to the 5′ end 23S rRNA processing observed in large subunit ribosomal particles accumulated in the cells overexpressing R331A or wild-type DbpA. The 5′ end processing of 23S rRNA was investigated by 32P-labeled primer extension (Fig. 3). The maturation of the 5′ end 23S rRNA is poorly understood; however, it is believed that a particle's tertiary RNA structure and protein composition dictate the 23S rRNA maturation extent (Gutgsell and Jain 2010; Shajani et al. 2011). The primer extension reactions clearly show a difference in 23S RNA maturation between all four large subunit precursors—27S, 35S, 45S, and 50S—accumulated in the cells overexpressing R331A DbpA, suggesting that all of these particles have different RNA structure and/or protein compositions as implied from their differences in sedimentation coefficients. The primer extension experiments show that the 45S particle accumulated in the cells overexpressing R331A DbpA contains a larger fraction of mature 23S rRNA than the 27S particle. On the other hand, the 27S particle contains a larger fraction of mature 23S rRNA than the 35S particle. Thus, it stands to reason that during the ribosome assembly process the pool of 45S particles cannot feed into the pool of 27S or the 35S particles. By the same logic, the pool of 27S particle could not feed into the pool of 35S particles.
FIGURE 3.
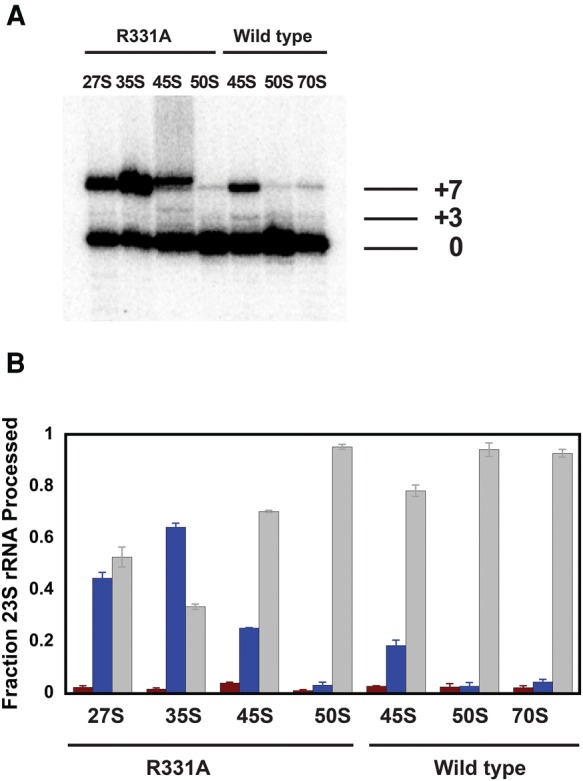
(A) A representative gel showing 5′ maturation of the rRNA for the various particles. The mature, +3, and +7 bands are marked. (B) The fraction of +7 (blue), +3 (red), and maturely (gray) processed 23S rRNA for each particle investigated. The fractions for the +3, +7 and mature population of each particle represent the means from two independent experiments and the errors are standard deviations from those means. The fractions for the +3, +7, and mature population were calculated from the ratio of radioactive counts on a specific band over the sum of counts on the bands representing +3, +7, and mature populations.
Interestingly, while both the 45S particles accumulated in the cells overexpressing R331A DbpA and wild-type DbpA migrate in the sucrose gradient with a similar sedimentation coefficient, they show different extents of maturation. The 45S particle accumulated in the cells overexpressing wild-type DbpA contains considerably more fully matured 23S rRNA than the 45S particle accumulated in the cells overexpressing R331A DbpA. Hence, these two particles are structurally different.
Subsequently we investigated the ability of different particles to stimulate the ATPase activity of the wild-type DbpA protein (Table 1). In order for a particle to stimulate the ATPase activity of DbpA, the DbpA C-terminal RNA-binding domain must have access to hairpin 92 of 23S rRNA and the DbpA catalytic core must have access to naked RNA structures. This means that variations in the DbpA protein's ATPase activity could be a consequence of three physical phenomena: (i) Ribosomal proteins could obstruct the DbpA C-terminal RNA binding domain access to hairpin 92, (ii) ribosomal proteins are loaded on the RNA regions that serve as substrates for the DbpA catalytic core, and (iii) the helix and hairpin 92 are not properly formed.
TABLE 1.
ATPase activity of DbpA stimulated by incubation with various large subunit precursor particles
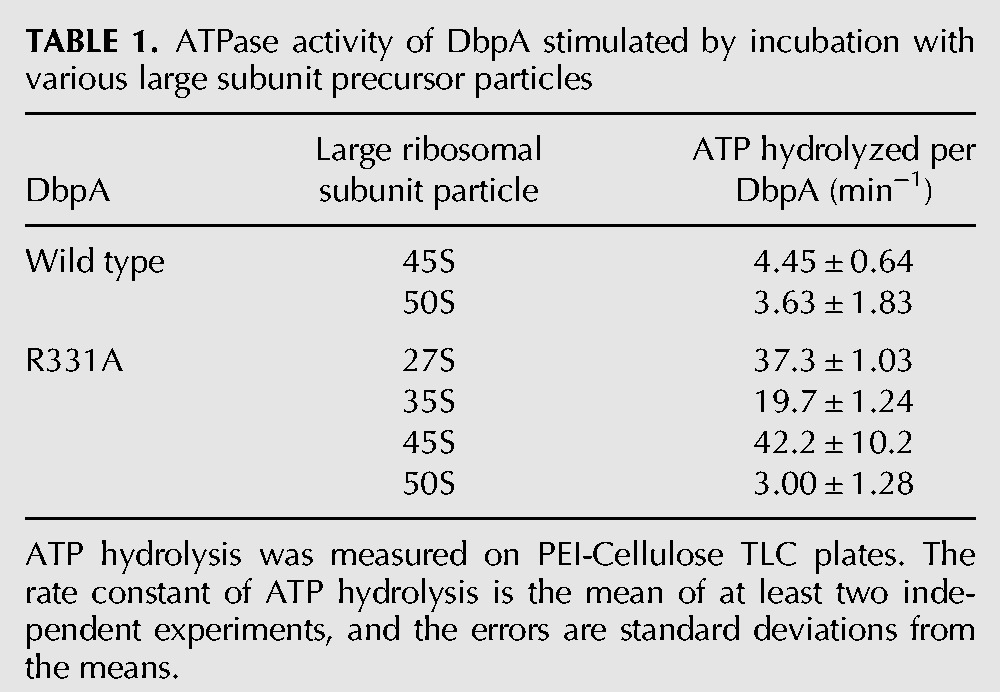
As shown previously, the 50S large ribosomal subunit present in the cells overexpressing R331A DbpA or the wild-type DbpA do not stimulate the purified wild-type DbpA protein's ATPase activity (Tsu and Uhlenbeck 1998; Elles et al. 2009). The reason these particles do not stimulate the ATPase activity of the purified wild-type DbpA protein is that helix and hairpin 92 are not solvent accessible in these particles (Ban et al. 2000; Yusupov et al. 2001). Hydroxyl radical footprinting data show that in the 45S particle accumulated in the cells overexpressing R331A DbpA, the helix and hairpin 92 are solvent accessible (Arai et al. 2015). Consequently, the experiments presented here and previous studies show that in the cells overexpressing R331A DbpA, the 45S particle supports the ATPase activity of the purified wild-type DbpA protein (Elles et al. 2009). On the other hand, we demonstrate here that the 45S particle accumulated in the cells expressing wild-type DbpA does not support the ATPase activity of the purified wild-type DbpA protein (Table 1). Thus, the stimulation of the ATPase activity data further suggests that the 45S particles accumulating in cells overexpressing wild-type and R331A DbpA have different RNA structures and/or protein compositions.
The 27S particles accumulating in the cells overexpressing R331A DbpA stimulate the ATPase activity of the wild-type DbpA protein twice as much as the 35S particles accumulating in those cells. Hence, the ATPase activation data agree with sucrose gradient migration, biDz fluorescent assay, and primer extension experiments that the 27S and 35S particles are two distinct intermediate particles with different RNA structures and/or protein compositions.
Kinetics of the intermediates during large subunit assembly
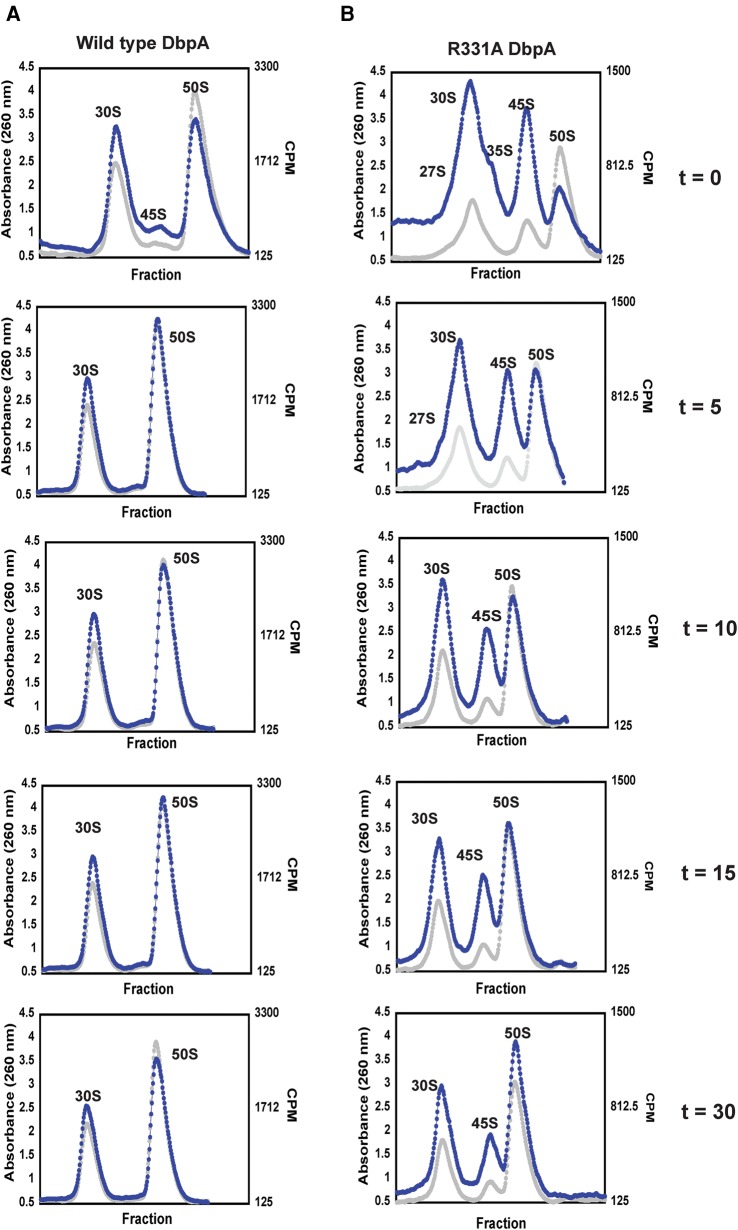
To investigate if the 27S, 35S, and 45S large subunit intermediate particles form the 50S large subunit over time, we radioactively labeled RNA in vivo for 10 min, stopped the transcription of new RNA by the addition of rifampicin, and followed the change in the radioactive counts of the separated ribosomal particles over time. This pulse labeling technique was used previously to investigate the conversion of a 40S particle, which accumulates in cells lacking DeaD, into the 50S subunit (Peil et al. 2008). The results of the pulse-labeling experiments are shown in Figure 4, where the RNA absorbance at 260 nm and the counts per minute (CPM) of radioactively labeled RNA are overlaid.
FIGURE 4.
Time course of ribosome assembly in cells overexpressing wild-type and R331A DbpA. (A) The time course of ribosome profiles of cells overexpressing wild-type DbpA. (B) Time course of ribosome profiles of cells overexpressing R331A DbpA. The absorbance readings are shown as gray circles and the CPM of 3H labeled RNA as blue circles. The ribosome profiles of both wild-type and R331A DbpA were performed at 0, 5, 10, 15, and 30 min after addition of rifampicin, which stops new RNA transcription. These data show that over time and with different observed microscopic rate constants (Table 3) the 27S, 35S, and 45S particles accumulating in the cells overexpressing R331A DbpA and 45S particle accumulating in the cells overexpressing wild-type DbpA form the 50S subunit.
In cells overexpressing the wild-type DbpA, a small amount of radioactively labeled 45S particle is observed, and this particle converts to the 50S large subunit with an observed microscopic rate constant of 0.17 ± 0.08 min−1 (Fig. 4; Table 3). The observed microscopic rate constant was calculated by fitting the data on Table 2 to a single exponential equation. More importantly, no radioactively labeled 27S and 35S particles were observed in those cells, consistent with the biDz assay (Figs. 1C, 2).
TABLE 3.
Kinetics of particle conversion
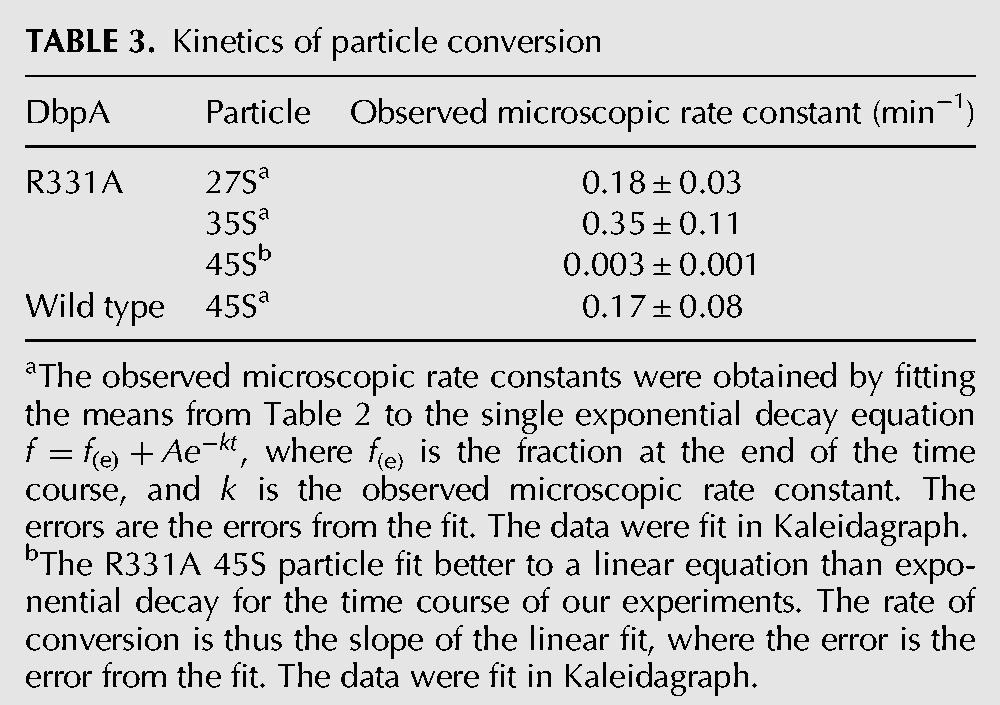
TABLE 2.
Relative abundance of 3H labeled ribosomal particles with time

The extent of radioactively labeled 50S particles during the labeling phase of our experiment (Fig. 4; Table 2, time, 0 min) is considerably smaller in the cells overexpressing R331A DbpA when compared with the cells overexpressing wild-type DbpA. Hence, the overall large subunit assembly pathway is slower in the cells overexpressing R331A DbpA compared to wild-type DbpA (Fig. 4; Table 2). Consequently, at any time fewer active 70S ribosomes are present and less protein is synthesized in the R331A DbpA overexpressing cells than in wild-type DbpA overexpressing cells. This explains the observed slow-growth phenotype of cells overexpressing R331A DbpA when compared to the cells overexpressing the wild-type DbpA (doubling time for cells overexpressing the R331A DbpA is 68 ± 11 min, doubling time for the cells overexpressing wild-type DbpA is 31 ± 12 min).
In the cells overexpressing R331A DbpA, radiolabeled 27S, 35S, and 45S particles are observed in addition to the 30S and 50S particles (Fig. 4B). The pool of radiolabeled 27S, 35S, and 45S particles decreases over time with the concomitant increase of the pool of radiolabeled 50S particles. This combined with the observation that the 27S, 35S, and 45S are not completely processed particles, while the 50S particle is (Fig. 3), strongly suggests that the 27S, 35S, and 45S particles are not turnover products of the 50S particle. The observed microscopic rate constants for the rearrangements of the 27S, 35S, and 45S particles are 0.18 ± 0.03 min−1, 0.35 ± 0.11 min−1, and 0.003 ± 0.001 min−1, respectively (Table 3). Consequently, the radiolabeled pool of 27S and 35S particles completely shifts to the 50S particle during the time course of our experiment, while only 20% of the 45S particle pool shifts to the 50S particle.
Since only 20% of the 45S intermediate accumulated in the cells overexpressing R331A DbpA rearranges to form the 50S subunit, it is possible that this 45S particle population consists of two populations, a dead-end population, which never rearranges to form the 50S subunit, and a population that rapidly forms the 50S subunit. However, the pulse labeling experiments show that the 45S particle radiolabeled counts consistently decrease during the entire time course of the experiments, which would be incompatible with the 45S particle consisting of a rapidly converting intermediate population and a dead-end intermediate population. If the 45S particle accumulated in the cells overexpressing R331A DbpA contained a mixture of a rapid converting and dead-end intermediates, then the radiolabel counts on the 45S intermediate would decrease initially fast, and after that initial decrease no change on the 45S particle radiolabel counts would occur.
The question remains, do all the intermediate particles present in the cells overexpressing R331A DbpA rearrange to form the 50S subunit via the same pathway; in other words, do these intermediates feed into each other's pools, or do they rearrange to form the 50S subunit via independent pathways? Because the 45S particle contains more mature 23S RNA than the 27S and 35S particles, the pool of the 45S particle cannot feed into the pool of the 27S and 35S particles. Moreover, the 27S particle contains a larger extent of mature 23S RNA than the 35S particle; hence, the pool of the 27S particle cannot feed into the pool of the 35S particle. Consequently, if there were only one pathway for the large subunit ribosome assembly in the cells overexpressing R331A DbpA, the order of particle conversion would be the 35S particle rearranging to the 27S particle, then the 27S particle rearranging to the 45S particle and subsequently the 45S particle forming the 50S subunit. The 45S particle rearranges to form the 50S subunit considerably slower than the disappearance of 27S and 35S labeled counts (Fig. 4; Table 3). If the above hypothetical pathway were correct, an increase in the pool of radioactive 45S labeled particles would be observed during the time course of our experiments. We do not observe this increase; hence, the 45S particle must be formed from a pathway that does not involve either the 27S particle or the 35S particle, implying that there are at least two different pathways of large subunit ribosome assembly in cells expressing R331A DbpA: one pathway involving the 45S particles, and another pathway involving the 27S and 35S particles.
Two alternative possibilities exist concerning the fate of the 27S and 35S particles on large subunit assembly. The first possibility is that the 35S particle rearranges to form the 27S particle, and subsequently the 27S particle forms the 50S subunit. Alternatively, the 35S and the 27S can rearrange to form the 50S particle via independent pathways. Since the 35S pool of labeled particles depletes faster than the 27S pool of labeled particles (Fig. 4; Table 3), if the 35S particle pool feeds into the 27S particle pool, then the 27S particle would accumulate over time. An accumulation of the 27S particle is not observed; hence, the 35S particle rearranges to form the 50S large subunit by a pathway that involves neither the 27S nor the 45S particles, suggesting that ribosome assembly in the cells expressing R331A DbpA occurs by at least three independent pathways.
Previous work has shown that the 45S particle isolated from cells overexpressing R331A DbpA is unable to form the 50S subunit when incubated with wild-type purified DbpA and ATP (Elles et al. 2009). Moreover, when the lysate made from cells overexpressing R331A DbpA was incubated with excess wild-type DbpA and ATP, the ribosome profiles of the treated lysate were identical to the ribosome profiles of R331A overexpressing cells untreated lysate (Elles et al. 2009). Hence, in vitro the 45S particle accumulated in cells overexpressing R331A appears to be a dead-end intermediate. Our experiments show that in vivo, within the time frame of our experiment ∼20% of the 45S population converts to the 50S large subunit (Fig. 4B; Table 2). Thus, in vivo and in vitro pathways of 45S particle conversion to the 50S large subunit are different. This is consistent with previous results suggesting that ribosome assembly in vitro and in vivo occurs via different pathways (Connolly and Culver 2009; Shajani et al. 2011).
DISCUSSION
The data presented here show that three large subunit intermediate particles accumulate in cells lacking wild-type DbpA and overexpressing R331A DbpA. The three intermediate particles migrate in a sucrose gradient with different sedimentation coefficients 27S, 35S, and 45S, they have different rRNA composition and degrees of 23S rRNA maturation, and they stimulate the ATPase activity of wild-type DbpA to different extents. Combined, these data suggest that the three intermediate particles accumulated in the cells overexpressing R331A DbpA are structurally different. The intermediates that migrate in a sucrose gradient as 27S and 35S particles readily rearrange to form the 50S particle, while the 45S particle slowly rearranges to form the 50S particle (Fig. 4; Table 3). The observation that the radiolabeled counts in the pool of the 45S particle shifts to the pool of the 50S particle, suggests that the 45S particle is not marked for degradation by the cell and is probably not considered toxic (Deutscher 2006). Similar behavior was observed for the 40S particle accumulated in the cells lacking the DEAD-box protein DeaD: This large ribosomal subunit intermediate also rearranges slowly to form the 50S particle and is not degraded by cellular machinery (Peil et al. 2008).
The presence of three classes of large subunit intermediate particles, 27S, 35S, and 45S, which independently and with different observed microscopic rates convert to the 50S subunit (Fig. 4; Table 3), indicates that in the cells overexpressing R331A DbpA and lacking endogenous DbpA large subunit assembly occurs by at least three different pathways. Multiple pathways of large subunit assembly were also suggested in a previous study investigating the role of DEAD-box proteins RhlE, SrmB, and DeaD on large subunit ribosome assembly (Jain 2008). In this study, the absence of RhlE worsened the slow growth phenotype of the cells lacking the DeaD protein; however, it improved the slow-growth phenotype of the cells lacking the SrmB protein, implying that the presence or lack of a DEAD-box protein in cells may influence the large subunit assembly pathway of choice.
A wealth of previous experimental data has shown that DbpA is a modular protein; the catalytic activity of DbpA is completely housed in its N-terminal domains, while its specific RNA binding activity is housed in its C-terminal domain (Karginov and Uhlenbeck 2004; Karginov et al. 2005). Indeed, changing arginine 331 to alanine in the DbpA protein's catalytic core, while abolishing this construct's ability to hydrolyze ATP and perform unwinding, has no effect on the R331A DbpA protein's affinity for RNA molecules containing hairpin 92 (Elles and Uhlenbeck 2008). Consequently, during in vivo large subunit assembly R331A and wild-type DbpA would be expected to tether to the same RNA intermediate structures and wild-type DbpA should sample the same assembly pathway intermediates as R331A. Therefore, wild-type DbpA, in vivo, may participate in at least three pathways of large subunit assembly.
The precise points on the large subunit assembly pathways at which R331A or wild-type DbpA acts remain unknown. The accumulation of 27S, 35S, and 45S particles in the cells overexpressing R331A DbpA combined with the observation that no phenotype is observed when the dbpA gene is removed for the E. coli genome suggests two possibilities. The first possibility is that R331A DbpA acts directly on the intermediate particles. However, because R331A DbpA cannot perform RNA unwinding it cannot facilitate the RNA isomerization steps required for the conversion of 27S, 35S, and 45S particles to the 50S particle. Once R331A leaves the intermediate particles, the RNA isomerization steps required for the formation of the 50S subunit could be facilitated by another maturation factor (Elles et al. 2009; Shajani et al. 2011). Due to the fact that ribosome assembly is a very coordinated process (Connolly and Culver 2009; Shajani et al. 2011), the delay in RNA structural rearrangement caused by R331A DbpA binding to the ribosome and its inability to perform unwinding, may promote the formation of alternative incorrect RNA structures or incorrect binding of ribosomal proteins, which could stabilize the intermediate particles. The more stable intermediates would require a longer time to rearrange and form the 50S subunit. Consequently, these intermediates accumulate in the cells overexpressing R331A DbpA. The other possibility is that R331A DbpA binds to the precursors of 27S, 35S, or 45S particles and the inability of R331A DbpA to perform its function promotes misassembled intermediate particle formation. Of course it is possible that DbpA binds to certain intermediate particles directly and binds to the precursor of the other intermediate particles.
A ribosomal particle could travel in a sucrose gradient as a particle with a large sedimentation coefficient if it contains a large number of ribosomal proteins, or if it has an incorrectly extended RNA structure (Nierhaus et al. 1973; Dohme and Nierhaus 1976; Charollais et al. 2003). Determination of the 45S particle's protein composition showed that this particle is a late stage large subunit ribosomal intermediate (Elles et al. 2009). The R331A DbpA could bind to the 23S rRNA before the 45S particle is formed; under this scenario R331A DbpA acts on the early stages of the large subunit ribosome assembly process involving the 45S particle. Alternatively, R331A DbpA could directly load onto the 45S particle; under this scenario R331A DbpA acts on the late stages of the large subunit ribosome assembly process involving the 45S particle. Consequently, it is not known when R331A DbpA acts on the large subunit assembly pathway involving the 45S particle. Based on their sedimentation coefficients, the 27S and 35S particles should be earlier large ribosomal subunit intermediate particles. A late large subunit ribosomal particle that contains a large number of ribosomal proteins and a considerable amount of properly formed RNA interactions would not travel as a particle with a small sedimentation coefficient. Therefore, in pathways that involve 27S and 35S particles, R331A DbpA seems to interact with the 23S rRNA in the earlier stages of large subunit assembly.
Future investigations of the RNA secondary and tertiary structure in the 27S, 35S, and 45S particles present in the cells overexpressing R331A DbpA, and the 45S particle accumulated in the cells overexpressing the wild-type DbpA, could elucidate the precise RNA isomerization steps that the DbpA protein facilitates and the effect of these structural isomerizations on the large subunit assembly process as a whole.
MATERIALS AND METHODS
Reagents
Reagents used for the preparation of cell growth media, including antibiotics, were obtained from Fisher Scientific. DNA oligomers used during the recombineering process and strands of biDz sensors were obtained from Integrated DNA Technologies. The recombineering plasmid pSIM9 was a gift from the Court Laboratory at the National Cancer Institute (Thomason et al. 2007).
Preparation of E. coli strains used in this study
To ensure no interference from the genomic expression of wild-type DbpA, the genomic copy of the dbpA gene was replaced with the kanamycin resistance gene (kanR) as previously explained (Thomason et al. 2007). The strain used for recombineering was E. coli BLR (DE3) obtained from Invitrogen. The E. coli BLR (DE3) ΔdbpA:kanR cells were used for all the experiments outlined in this work. These cells were transformed with the pET3a vectors bearing the coding sequence of either wild-type DbpA or R331A DbpA constructs (Elles and Uhlenbeck 2008).
Pulse-labeling experiments
Pulse chase experiments were carried out by culturing colonies from plates no more than ∼16 h old into 500 mL of a modified liquid LB media. This media contained 10 g/L sodium chloride, 10 g/L tryptone, and 1 g/L yeast extract, as well as 100 µg/mL carbenicillin and 34 µg/mL chloramphenicol. Cell cultures were grown to an OD600 of 0.2 at 37°C. After reaching the desired OD, 120 μCi of [5,6-3H]-Uridine was added to the cultures, and allowed an incubation of 10 min. After incubation, rifampicin was added to the culture to reach a final concentration of 500 µg/mL. The addition of rifampicin was considered to be time t = 0 min. At time points t = 0, 5, 10, 15, and 30 min, 80 mL of the culture was collected and poured over ice to rapidly cool the cells. Collection of data at time points longer than ∼30 min was complicated by cell death and degradation of the ribosome particles beginning at ∼45 min after addition of rifampicin. Collected fractions were centrifuged at 6500 max G for 10 min at 4°C.
The collected cell pellets were resuspended in 400 µL ribosomal lysis buffer (20 mM HEPES-KOH pH 7.5, 30 mM NH4Cl, 1 mM MgCl2, and 4 mM BME). The resuspended cells were incubated on ice with lysozyme for 20 min, then flash frozen in liquid nitrogen and thawed in room temperature water. The freeze/thaw cycle was repeated two more times, then 300 µL of a detergent buffer (0.5% Brij, 0.5% deoxycholate) with DNase I was added to the raw lysate. The lysate was incubated on ice for 30 min, then placed in a desktop centrifuge at 16,000 max G for 20 min at 4°C. The clarified lysate was transferred to new microcentrifuge tubes. The OD260 of the lysate was measured before preparing sucrose gradients.
Separation of ribosomal particles
The ribosomal particles were separated by ultracentrifugation in a sucrose gradient as previously described with a few modifications (Elles and Uhlenbeck 2008). Thirty milliliters of 20%–40% sucrose gradients (20 mM HEPES KOH pH 7.5, 150 mM NH4Cl, 1 mM MgCl2, and 4 mM BME) were prepared using a Biocomp Gradient Master. The clarified lysate was layered on top of each gradient, normalized to an amount of 45 OD260. Prepared gradients were placed in a Beckman LE-80 ultracentrifuge using a SW-32 rotor and centrifuged at 32k RPM at 4°C for 16 h. After centrifugation, gradients were fractionated using a Teledyne R1 Fraction Collector and a Brandel SYR-101 Syringe Pump with a 70% sucrose push from below.
Fractions were collected in Corning UV-transparent 96-well plates. Collected plates were measured for UV absorbance at 260 nm, including pathlength measurements to normalize differences in drop volumes. Finally, the collected fractions were suspended in OptiPhase Supermix liquid scintillation cocktail obtained from PerkinElmer, and tritium CPM was measured using a PerkinElmer MicroBeta liquid scintillation counter. To reduce the effect of instrument noise in the liquid scintillation measurements, the CPM data were passed through an adjacent averaging filter using a 10-point window in the data management software, Origin.
Fluorescent assay with 16S rRNA- and 23S rRNA-specific biDz sensors
In order to determine the presence of 16S and 23S rRNA in the ribosome profile fractions, two binary deoxyribozyme (biDz) sensors were used—biDz-16S targeting a fragment of E. coli 16S rRNA (nucleotides 193–240) and biDz-23S targeting a fragment of E. coli 23S rRNA (nucleotides 451–500) (Woese et al. 1980; Noller et al. 1981). The sequences of the sensor strands and the fluorogenic reporter have been previously published (Gerasimova and Kolpashchikov 2013; Gerasimova et al. 2015). For the assay, aliquots of the ribosome profile fractions were mixed with strands a and b of either of the sensors (15 nM each), and fluorogenic reporter substrate (200 nM) in a buffer containing 50 mM HEPES (pH 7.4), 50 mM MgCl2, 20 mM KCl, 120 mM NaCl, 0.03% Triton X-100 and 1% DMSO and heated at 95°C for 3 min. After centrifugation, the samples were incubated at 54°C for 1 h followed by measuring their fluorescence at 517 nm upon excitation at 485 nm in a 3-mm cuvette. The amount of the concentrated ribosome profile fractions in the sample was chosen to give the maximum fluorescent signal for the sensor targeting the predominant rRNA in the sample (16s rRNA in 27s, 30s, and 35s peak fractions and 23s rRNA in 45s and 50s peak fractions). As a consequence, the ratio of 23s:16s rRNA for each particle, which is unchanging with concentration, yields information about the rRNA composition in each segment of the gradient. When assaying individual ribosomal profile fractions, the same amounts of RNA based on A260 were used for the assay.
Primer extension assay
Investigation of the 5′ maturation of the rRNA for the intermediate particles was done using end P32-labeled primer extension. As previously described, a primer corresponding to residues 35–55 of the 23s rRNA was used (Elles et al. 2009). The particles were collected in a 20%–40% linear sucrose gradient, concentrated by ultracentrifugation, and the rRNA was purified by phenol/chloroform extraction. The extension reaction was run using Superscript II reverse transcriptase, purchased from Invitrogen, following the manufacturer's instructions. The reactions were then resolved on an 8 M urea 20% 19:1 polyacrylamide gel. The gels were dried, exposed to BioRad intensifying screens, and scanned in a BioRad Personal Molecular Imager. The images were quantified using BioRad's QuantityOne basic software.
ATPase assay
ATPase activity of the particles was investigated using PEI-Cellulose TLC plates as previously described, with the only adaptation being the use of 0.25 μCi γ-[32P]-ATP and a 750 mM LiCl 1 M Acetic Acid running buffer for the PEI-Cellulose TLC plates (Elles et al. 2009). Two microliter aliquots were removed from the reaction every 2 min and quenched with one microliter 500 mM EDTA over the course of a 20-min reaction, an additional time point at 30 min was taken. The rate of ATP hydrolysis was calculated using the slopes of a linear fit for the time course.
ACKNOWLEDGMENTS
E.K. thanks O. Uhlenbeck for many helpful discussions concerning this work. This work was supported by National Institutes of Health (5R21CA175625-02 to E.K.) and the University of Central Florida (in-house grant to E.K.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055137.115.
REFERENCES
- Arai T, Ishiguro K, Kimura S, Sakaguchi Y, Suzuki T, Suzuki T. 2015. Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc Natl Acad Sci 112: E4707–E4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 48: 1253–1265. [DOI] [PubMed] [Google Scholar]
- Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res 32: 2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K, Culver G. 2009. Deconstructing ribosome construction. Trends Biochem Sci 34: 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. 2006. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 34: 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diges CM, Uhlenbeck OC. 2001. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J 20: 5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohme F, Nierhaus KH. 1976. Total reconstitution and assembly of 50 S subunits from Escherichia coli ribosomes in vitro. J Mol Biol 107: 585–599. [DOI] [PubMed] [Google Scholar]
- El Hage A, Alix JH. 2004. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol Microbiol 51: 189–201. [DOI] [PubMed] [Google Scholar]
- El Hage A, Sbai M, Alix JH. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol Gen Genet 264: 796–808. [DOI] [PubMed] [Google Scholar]
- Elles LM, Uhlenbeck OC. 2008. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res 36: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elles LMS, Sykes MT, Williamson JR, Uhlenbeck OC. 2009. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res 37: 6503–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova YV, Kolpashchikov DM. 2013. Folding of 16S rRNA in a signal-producing structure for the detection of bacteria. Angew Chem Int Ed Engl 52: 10586–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova YV, Yakovchuk P, Dedkova LM, Hecht SM, Kolpashchikov DM. 2015. Expedited quantification of mutant ribosomal RNA by binary deoxyribozyme (BiDz) sensors. RNA 21: 1834–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgsell NS, Jain C. 2010. Coordinated regulation of 23S rRNA maturation in Escherichia coli. J Bacteriol 192: 1405–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Bradley MJ, De La Cruz EM. 2012. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys 41: 247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Dreyfus M. 2006. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res 34: 4189–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Bizebard T, Dreyfus M. 2013. Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. Biochim Biophys Acta 1829: 866–877. [DOI] [PubMed] [Google Scholar]
- Jain C. 2008. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 14: 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Uhlenbeck OC. 2004. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res 32: 3028–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. 2005. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem 280: 35499–35505. [DOI] [PubMed] [Google Scholar]
- Linder P. 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34: 4168–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ramírez V, Alcaraz LD, Moreno-Hagelsieb G, Olmedo-Álvarez G. 2011. Phylogenetic distribution and evolutionary history of bacterial DEAD-box proteins. J Mol Evol 72: 413–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire BA. 2009. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol Mol Biol Rev 73: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol SM, Fuller-Pace FV. 1995. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc Natl Acad Sci 92: 11681–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH, Bordasch K, Homann HE. 1973. Ribosomal proteins. 43. In vivo assembly of Escherichia coli ribosomal proteins. J Mol Biol 74: 587–597. [DOI] [PubMed] [Google Scholar]
- Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, et al. 1981. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res 9: 6167–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Russell R. 2010. Roles of DEAD-box proteins in RNA and RNP folding. RNA Biol 7: 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Virumäe K, Remme J. 2008. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J 275: 3772–3782. [DOI] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. 2013. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta 1829: 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37: 317–336. [DOI] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526. [DOI] [PubMed] [Google Scholar]
- Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 78: 1.16.1–1.16.24. [DOI] [PubMed] [Google Scholar]
- Tsu CA, Uhlenbeck OC. 1998. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 37: 16989–16996. [DOI] [PubMed] [Google Scholar]
- Tsu CA, Kossen K, Uhlenbeck OC. 2001. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 7: 702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, et al. 1980. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res 8: 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]