ABSTRACT
The molecular pathways that govern how germ line fate is acquired is an area of intense investigation that has major implications for the development of assisted reproductive technologies, infertility interventions, and treatment of germ cell cancers. Transcriptional repression has emerged as a primary mechanism to ensure suppression of somatic growth programs in primordial germ cells. In this commentary, we address how xnd-1 illuminates our understanding of transcriptional repression and how it is coordinated with the germ cell differentiation program. We recently identified xnd-1 as a novel, early determinant of germ cell fates in Caenorhabditis elegans. Our study revealed that XND-1 is maternally deposited into early embryos where it is selectively enriched in the germ lineage and then exclusively found on chromatin in the germ lineage throughout development and into adulthood when it dissociates from chromosomes in late pachytene. This localization is consistent with a range of interesting germ cell defects that suggest xnd-1 is a pivotal determinant of germ cell characteristics. Loss of xnd-1 results in a unique “one PGC (primordial germ cell)” phenotype due to G2 cell cycle arrest of the germline precursor blastomere, P4, which predisposes the animal and its progeny for reduced fecundity. The sterility in xnd-1 mutants is correlated with an increase in the transcriptional activation-associated histone modification, dimethylation of histone H3 lysine 4 (H3K4me2), and aberrant expression of somatic transgenes but overlapping roles with nos-2 and nos-1 suggest that transcriptional repression is achieved by multiple redundant mechanisms.
KEYWORDS: epigenetics, germ line, Nanos, primordial germ cells, XND-1
Introduction
Germ cell development is a highly regulated, multi-step process that ensures the faithful transfer of genetic information to the next generation. Germ cells are derived from primordial germ cells which are set apart from somatic lineages during early embryogenesis by inheriting specialized cytoplasm called “germ plasm” containing maternally encoded mRNA and proteins. One of the hallmarks of developing PGCs is global transcriptional repression1-3 This repression is thought to be essential to prevent the activation of somatic genes within the germ line.
In C. elegans, the germline lineage undergoes 4 asymmetric cell divisions to give rise to germline blastomere, P4, that then divides symmetrically to give rise to the PGCs, Z2 and Z34 (Fig. 1). The PGCs are arrested in the G2 stage of the cell cycle for the remainder of embryogenesis.5 The early P lineage (P1-P4) is kept transcriptionally quiescent by direct repression of RNA polymerase II C-terminal domain (CTD) phosphorylation by the maternally derived protein PIE-16. Global transcriptional quiescence in Z2 and Z3 is maintained by epigenetic reprogramming of the PGCs, leading to loss of the active histone mark H3K4me2 and acquisition of a condensed chromatin state.7 During this stage, zygotic expression initiates for a handful of germline genes that play key roles in germ cell differentiation, including transcripts for P-granules components (pgl-1) and the nanos ortholog, nos-1.8,9
Figure 1.
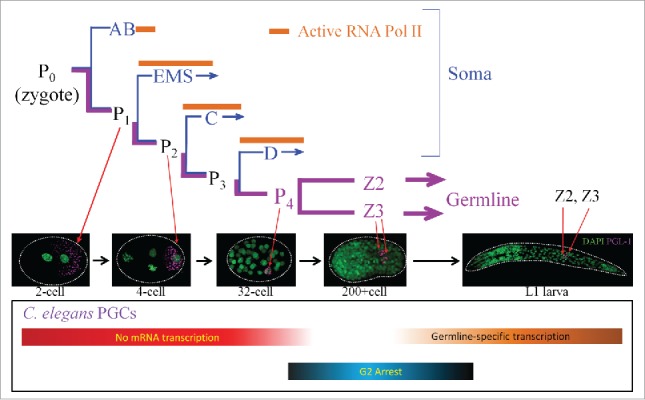
Embyonic divisions leading to the development of the germline. P lineage is depicted in magenta color, somatic lineages in blue. Embryos of indicated cell stages immunostained with anti-PGL-1 to mark germ lineage and the DNA dye DAPI to show embryonic nuclei were used to illustrate the developmental stages of the P cell. Bottom bar shows the approximate timing of key events during PGC development.
Zygotic gene activation and resumption of the cell cycle occurs post-hatching in response to feeding. However, the X chromosome remains transcriptionally silenced in oocytes, a repression that is required for normal germ cell development.10,11
XND-1 is among the earliest proteins to be expressed in the PGCs
During early embryogenesis, germ cell determinants are segregated away from the somatic lineage. Both active localization to the P lineage and degradation in the somatic cells lead to PGC enrichment of key determinants including P-granules, NOS-2, and the transcriptional repressor PIE-1.12 Characterization of XND-1 provides additional insight into strategies used to promote PGC enrichment. In situ hybridizations identify xnd-1 RNAs in all early embryonic blastomeres (NEXTDB; http://nematode.lab.nig.ac.jp). XND-1 proteins, by contrast, are found first in the cytoplasm of P4 and then quickly enriched in the nucleus upon division into Z2 and Z3 (Fig. 2). These observations suggest that xnd-1 maternal RNAs are actively repressed in all blastomeres prior to the 28-cell stage and only in somatic blastomeres thereafter. As shown in Figure 3, loss-of-function mutations of several RNA-binding proteins in the early embryo are responsible for this somatic repression as their removal causes XND-1 to misexpress in somatic blastomeres. Examination of the xnd-1 3'UTR identified putative binding sites for several of these proteins, suggesting they may directly repress xnd-1 expression. Whether somatic misexpression of XND-1 has a phenotypic consequence has not yet been ascertained. Yet, it is of note that misexpression of germline-specific P granule components in the intestine in synMuv B mutants leads to high temperature arrest, a phenotype that is suppressed by global regulators of germline chromatin including the MES-2/3/6 complex.13 The suppression of xnd-1 meiotic phenotypes by mes-2 and mes-314 raises the possibility of a similar consequence for XND-1 misexpression.
Figure 2.
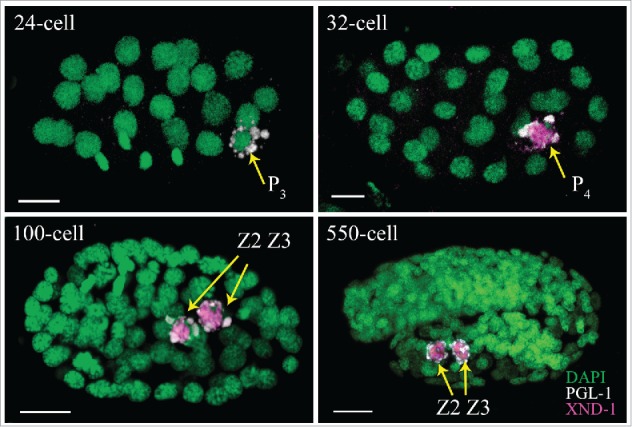
Expression of XND-1 during embryogenesis is limited to the P lineage. Embryos are oriented anterior to the left. XND-1 is marked in magenta, PGL-1 in white and DAPI in green. Scale bar: 5 µm. XND-1 is undetectable in the germline blastomere of<24-cell stage. At ∼32-cell stage, expression can be seen in the cytoplasm and nucleus of P4 and continues in the P4 daughters, Z2 and Z3, where it localizes to the nucleus and is associated with chromosomes.
Figure 3.
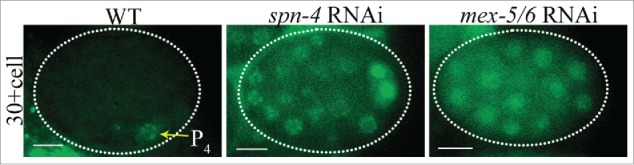
XND-1 is misexpressed upon depletion of RNA-binding proteins in early embryos. XND-1::GFP, shown in green, is expressed in P4 in wild-type embryos (left). XND-1 misexpression in somatic blastomeres in embryos treated with either spn-4 (middle) or mex-5/6 RNAi (right).
Zygotic expression of XND-1 is first detected around the 300-cell stage, making XND-1 one of the earliest expressed gene in the PGCs.15 This observation shows that zygotic transcription in the PGC starts earlier than had previously been anticipated and provides a tool for interrogating the molecular mechanisms that allow a subset of genes to overcome global repressive mechanisms in the PGC.
Nanos family members in C. elegans
Members of the nanos gene family members are conserved determinants of germline development.16 C. elegans has 4 Nanos homologs, nos-1, nos-2, nos-3 and T02G5.11. nos-2 is maternally deposited and is essential for incorporation of PGCs into the somatic gonad during embryogenesis and functions redundantly with nos-1 to promote PGC proliferation and survival during larval development.8 Nanos family members encode RNA binding proteins that associate with the CCR4/NOT deadenylation complexes to regulate target genes.17-19 Although a physical interaction between worm Nanos proteins and CCR4/NOT has not been established, genetic interactions between these genes suggest likely functional conservation. In Drosophila, the Nanos/CCR4/NOT complex is essential for repression of cyclin B and control of PGC cells proliferation.17 In the adult mouse germline, mei-P26, a modulator of micro-RNA dependent silencing, was recently identified as a target gene, linking CCR4/NOT function to transcriptional silencing.20 The functional targets of C. elegans NOS-1 and NOS-2 that are required for germ cell differentiation remain elusive and the partial sterile phenotypes seen in double mutants intimate the existence of additional germ cell determinants.
Progress in identifying downstream targets of nos-2 has been hampered by the lack of mutant alleles since the gene resides in the intron of the essential meiotic gene him-14. We generated a nos-2 null allele by rescuing the deletion allele with a fosmid containing the him-14 locus in which the nos-2 coding region was replaced with yellow fluorescent protein (YFP).15 This strain provided the first functional characterization of a nos-2 null allele in C. elegans. It also marks early PGCs with YFP providing a tool for in vivo imaging and PGC isolation studies. Recently, a CRISPR allele of nos-2 has also been created.21 Both nos-2 mutations confer only a minor sterility on their own, in contrast to reports of nos-2(RNAi).8 Since nos-2(RNAi) should also knock down expression of the T02G5.11, it remains possible that this predicted pseudogene retains some ancestral function.
Cell cycle regulation of PGCs
PGCs are arrested in G2 throughout the majority of embryogenesis, allowing them to rapidly initiate proliferation upon sensory input of food post-hatching. In xnd-1 mutant embryos, approximately half of post-gastrulation embryos contain a “single” PGC. Live imaging revealed that this “one PGC” phenotype results from a defect in P4 cell division, rather than the loss or death of Z2 or Z3.15 Unlike P4, the “single” PGCs appeared to have a 4N DNA content, indicating that P4 had progressed partially in its cell cycle prior to arrest.
Further examination of cell cycle-associated markers in one and 2 PGC animals revealed unexpected features of cell cycle arrest compared to other G2 stage cells. Specifically, one PGC arrest is independent of accumulation of cyclin B homolog, CYB-1, as its level appears the same in wild type and xnd-1 PGCs. The one PGC cells also accumulate CDK-1 containing inhibitory phosphorylation, pTyr15 (pCDK-1), a mark associated with G2 cell cycle arrest. In contrast, wild type or xnd-1 embryos with 2 PGCs never showed pCDK-1 accumulation, despite a 4N DNA content.15 Further comparison of the one and 2 PGC states may therefore reveal fundamental insights into distinct sub-stages of the G2 cell cycle. The accumulation of pCDK-1 in arrested single PGCs may reflect altered levels of CDK-1 regulatory components WEE-1 kinase or CDC25 phosphatase. The recent report of WEE-1 kinase GFP reporters22 should facilitate such analysis.
After hatching and in response to feeding, PGCs activate zygotic transcription and reenter the cell cycle. These events are associated with a spike in transcription-coupled DNA damage that needs to be repaired for proper division to ensue.23 In light of these studies, it is intriguing to speculate that xnd-1 or nos-1/2 may alter DNA damage repair in the L1 larvae leading to PGC loss and/or reduced proliferation. In support of this hypothesis, xnd-1 mutants are hypersensitive to gamma irradiation and the brood size defects of xnd-1 are partially suppressed by mutations in the checkpoint kinase, atm-1 (McClendon and Yanowitz, submitted).
Control of germ line size
Seminal studies by Kimble and White showed that ablation of either Z2 or Z3 led to the formation of 2 gonads, but both were smaller than their normally endowed counterparts.24 Consistent with this observation, one PGC xnd-1 mutant animals produced significantly smaller broods compared to their siblings with 2 PGCs. However, almost all xnd-1 germ lines are smaller than wild type yet only half of xnd-1 mutant animals show the one PGC defect. Therefore, we infer that xnd-1 function is required continuously for germ cell development. Indeed, our exploration of zygotic versus maternal contributions of xnd-1 showed that zygotic loss of xnd-1 (M+Z-) resulted in significantly smaller germline sizes.15
Dissection of maternal vs. zygotic functions indicated that zygotic xnd-1 can completely rescue the germ line size and brood size defects of one PGC animals, but cannot itself suppress the one PGC phenotype.15 While this result may seem to contradict the notion that one PGC predestines a worm for reduced fecundity, it may be understood in light of XND-1s role in regulating accumulation of histone post-translational modifications. We posit that zygotic expression of XND-1 protein may be sufficient to completely reset an altered chromatin state in one PGC animals. Additionally, we may explain the increase in germ line size if zygotic XND-1 proteins allow the singe PGCs to rapidly divide, acquire the Z2/Z3 state, and then proceed normally to produce a replete germ line. The unique cell cycle state of the one PGC animals may indeed leave the cells primed to divide if the appropriate signal(s) were present.
One of the most striking results of our analyses of one PGC animals is that they were more likely sire offspring that developed with one PGC.15 These results may be explained by a role for xnd-1 in establishing chromatin state(s) of germ cells. The observations that histone H2A lysine 5 acetylation is increased in the adult germ line14 and that H3K4me2 (see below) is affected in one PGC embryos15 suggest a mechanism by progressive accumulation of these marks could lead to cell cycle perturbation and altered cell fates.
Transcriptional repression in germ cell precursors
Suppression of new transcription in PGCs is important for their specification and to maintain germ cell fate. The labeling of somatic nuclei and not the germline nuclei with the mRNA precursor [H3]-uridine in Drosophila early embryos provided the first clue that germline specification might require transcriptional repression.25 Later, studies in C. elegans also concluded the same by using in situ hybridization to detect zygotic mRNAs in somatic, but not germline, blastomeres.26,27 Transcriptional quiescence in PGCs is achieved by direct inhibition of RNA polymerase II transcriptional initiation and elongation functions in early embryos by PIE-1 and Pgc in C. elegans and Drosophila, respectively.3 In C. elegans, loss of pie-1 function leads to mispatterning of embryos and loss of germline lineage.28,29 In Drosophila, loss of Pgc leads to degeneration of germ cells (pole cells) during mid-embryogenesis.30,31
Transcriptional quiescence in late C. elegans embryos is achieved by controlling chromatin state, specifically inhibition of histone H3K4me2, a mark of active chromatin, and retention of histone H3 lysine 9 methylation (H3K9me), a repressive chromatin mark.3 Evidence that these marks are important for fecundity came from studies of mutations in spr-5 and met-2 that encode the homologs of the H3K4me2 demethylase LSD1/KDM1 and H3K9me2 methyltransferase SETDB1, respectively. spr-5 and met-2 mutants gradually accumulate H3K4me2 and lose H3K9me over many generations, leading to misexpression of spermatogenesis-enriched genes, reduced brood sizes, and sterility.7,32,33 These studies highlight the importance of establishing an epigenetic “ground state” for germ cell development and illustrate the dramatic consequences of such an imbalance on fecundity.
In addition to global transcriptional silencing, specific repression of the X chromosome is critical for germ cell development. The X chromosome is repressed by the activity of 2 classes of chromatin proteins: the Polycomb group (PcG) chromatin repressors E(Z) MES-2/MES-3 and MES-610; and the autosome-enriched proteins MES-4 and MRG-1.11,34 The former impose H3K27 methylation on the X chromosome, leading to its silencing. The latter proteins methylate H3K36 on autosomes to prevent MES-2/3/6 binding. Loss of either set of genes leads to transcription activation of X chromosome in the PGCs and eventual loss of germ cells in developing larvae.10,11,34
In our analysis, we discovered that a significant fraction of xnd-1 embryos showed dramatic increase in the accumulation of H3K4me2 in the Z2/Z3.15 The claim of increased accumulation of H3K4me2 in PGCs of xnd-1 mutant embryos is further strengthened by the misexpression of at least 2 neuronal markers in the germline of xnd-1 adults.15 This analysis suggests that sterility in xnd-1 animals might result from their inability to maintain germline silencing.
Functional redundancy in germ line differentiation pathways
Since normal specification and differentiation of the germ line is critical for the maintenance of species, evolution has employed multiple, redundant mechanisms to ensure proper repression of transcription, so as to make germ cells with the potential to transmit correct genetic information. A small subset of xnd-1 mutant animals develops without a germ line, a phenotype that is exacerbated by further loss of nos-2 and nos-1.15 Detailed examination of these triple mutant embryos identified a fraction without PGCs, suggesting overlapping roles in germ cell specification. In all but 2% of the remaining animals, PGCs are present but cannot elaborate a functional germ line. Those remaining, rare, fertile, triple-mutant animals imply the existence of additional determinants and underscore the robustness of differentiation pathways for ensuring fertility.
Identifying the downstream targets of NOS-1, NOS-2 and XND-1 is the critical next step to understand their functional relationship in germ cell differentiation. One possibility is the XND-1 and the NOS proteins share target genes with XND-1 regulating their transcription in the nucleus and NOS-1/2 regulating their translational efficiency in the cytoplasm. Alternatively, XND-1 and NOS-1/2 may regulate independent sets of genes both of which are required for germ line development. XND-1 protein sequence provides no hints as its function, although its localization to autosomes in the developing and adult germ cells hints at a role as a chromatin regulator. The association of Nanos proteins with the CCR4/NOT complex that recently was shown to regulate heterochromatic loci in Saccromyces pombe35 raises the tantalizing possibility that Nanos family members may accordingly facilitate heterochromatinization of the germ line.
Our prior characterization of the meiotic phenotypes of xnd-1 mutations hints at potential mechanisms for xnd-1 function. In xnd-1 mutants, meiotic double-strand breaks (DSBs) levels are reduced resulting in nearly half of all nuclei lacking a DSB on the X chromosome. The COs that result from these DSBs are also shifted from the chromosome arms into the more active gene clusters. In addition, DSBs appear earlier and more synchronously than in wild type.14,36 These disparate data may be explained if xnd-1 alters chromatin state in the gene clusters, preventing the DSB machinery from accessing the open chromatin in gene promoters enriched in the clusters. In the absence of xnd-1, these regions would rapidly and efficiently recruit the DSB machinery, acting as a sink for these complexes and limiting breaks elsewhere in the genome. The X chromosome, which receives fewer DSBs,36 would be more sensitive to such a change. XND-1 may directly repress these loci or may work through the imposition of a chromatin state since the mutant shows altered accumulation of modified histones.14
Perspectives
Although analysis of germ line deficient mutations has long drawn our attention to transcriptional regulatory mechanism that repress somatic fates in germ cells (and vice versa), studies in mice and flies highlight the importance of additional pathways. Of particular importance in mice, and humans, is the repression of transposable elements whose aberrant activation can lead to uncontrolled DNA damage or insertional mutagenesis. In the N2, Bristol strain, the paucity of transposons makes these pathways potentially less critical to fecundity; but in the Bergerac strain and other wild strains, with high copy numbers of transposons,37 mechanisms that dampen transposition likely play a more central role. Key players in this process are components of the PIWI-piRNA pathway that are thought to maintain an epigenetic memory of gene expression38-41 and HRDE-1 that directs the trimethylation of H3K9.42 Understanding the interplay between the factors that transmit epigenetic information from one generation to the next and the factors, such as xnd-1 and nos-2, that regulate early events in germ cell proliferation and differentiation are likely to provide important insights into conserved processes controlling fertility.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Nicolas Macaisne for critical reading of the manuscript.
Funding
This work was supported by the Pennsylvania Department of Health—Formula Funds. Magee-Womens Research Institute, and a National Institutes of Health grant to J.L.Y. (GM1040070).
References
- [1].Pirrotta V. Silence in the germ. Cell 2002; 110:661-4; PMID:12297039; http://dx.doi.org/ 10.1016/S0092-8674(02)00967-4 [DOI] [PubMed] [Google Scholar]
- [2].Seydoux G, Schedl T. The germline in C. elegans: origins, proliferation, and silencing. Int Rev Cytol 2001; 203:139-85; PMID:11131515; http://dx.doi.org/ 10.1016/S0074-7696(01)03006-6 [DOI] [PubMed] [Google Scholar]
- [3].Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development 2008; 135:3817-27; PMID:18997110; http://dx.doi.org/ 10.1242/dev.022434 [DOI] [PubMed] [Google Scholar]
- [4].Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/ 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- [5].Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol 2006; 16:773-9; PMID:16631584; http://dx.doi.org/ 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- [6].Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 1997; 124:2191-201; PMID:9187145 [DOI] [PubMed] [Google Scholar]
- [7].Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell 2003; 5:747-57; PMID:14602075; http://dx.doi.org/ 10.1016/S1534-5807(03)00327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 1999; 126:4861-71; PMID:10518502 [DOI] [PubMed] [Google Scholar]
- [9].Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 2004; 167:645-61; PMID:15238518; http://dx.doi.org/ 10.1534/genetics.103.023093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol 2004; 14:1639-43; PMID:15380065; http://dx.doi.org/ 10.1016/j.cub.2004.08.062 [DOI] [PubMed] [Google Scholar]
- [11].Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 2006; 133:3907-17; PMID:16968818; http://dx.doi.org/ 10.1242/dev.02584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang JT, Seydoux G. Germ Cell Specification. In: Schedl T, ed. New York, NY: Springer New York, 2013:17-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Petrella LN, Wang W, Spike Ca, Rechtsteiner A, Reinke V, Strome S. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 2011; 138:1069-79; PMID:21343362; http://dx.doi.org/ 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 2010; 467:839-43; PMID:20944745; http://dx.doi.org/ 10.1038/nature09429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mainpal R, Nance J, Yanowitz JL. A germ cell determinant reveals parallel pathways for germ line development in Caenorhabditis elegans. Development 2015; 142:3571-82; PMID:26395476; http://dx.doi.org/ 10.1242/dev.125732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science 2003; 301:1239-41; http://dx.doi.org/ 10.1126/science.1085222 [DOI] [PubMed] [Google Scholar]
- [17].Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 2007; 134:1519-27; PMID:17360772; http://dx.doi.org/ 10.1242/dev.002212 [DOI] [PubMed] [Google Scholar]
- [18].Suzuki A, Igarashi K, Aisaki K-I, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci U S A 2010; 107:3594-9; PMID:20133598; http://dx.doi.org/ 10.1073/pnas.0908664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PloS One 2012; 7:e33558-e; PMID:22448252; http://dx.doi.org/ 10.1371/journal.pone.0033558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep 2013; 1:411-24; PMID:24286029; http://dx.doi.org/ 10.1016/j.stemcr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, Seydoux G. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 2014; 198:1347-56; PMID:25249454; http://dx.doi.org/ 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Allen AK, Nesmith JE, Golden A. An RNAi-based suppressor screen identifies interactors of the Myt1 ortholog of Caenorhabditis elegans. G3 2014; 4:2329-43; PMID:25298536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Butuci M, Williams AB, Wong MM, Kramer B, Michael WM. Zygotic Genome Activation Triggers Chromosome Damage and Checkpoint Signaling in C. elegans Primordial Germ Cells. Dev Cell 2015; 34:85-95; PMID:26073019; http://dx.doi.org/ 10.1016/j.devcel.2015.04.019 [DOI] [PubMed] [Google Scholar]
- [24].Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol 1981; 81:208-19; PMID:7202837; http://dx.doi.org/ 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- [25].Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol 1976; 49:425-37; PMID:817947; http://dx.doi.org/ 10.1016/0012-1606(76)90185-8 [DOI] [PubMed] [Google Scholar]
- [26].Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 1996; 382:713-6; PMID:8751441; http://dx.doi.org/ 10.1038/382713a0 [DOI] [PubMed] [Google Scholar]
- [27].Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 1994; 120:2823-34; PMID:7607073 [DOI] [PubMed] [Google Scholar]
- [28].Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell 1992; 70:163-76; PMID:1623520; http://dx.doi.org/ 10.1016/0092-8674(92)90542-K [DOI] [PubMed] [Google Scholar]
- [29].Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature 1996; 382:710-2; PMID:8751440; http://dx.doi.org/ 10.1038/382710a0 [DOI] [PubMed] [Google Scholar]
- [30].Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 2008; 451:730-3; PMID:18200011; http://dx.doi.org/ 10.1038/nature06498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol 2004; 14:159-65; PMID:14738740; http://dx.doi.org/ 10.1016/j.cub.2003.12.036 [DOI] [PubMed] [Google Scholar]
- [32].Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 2009; 137:308-20; PMID:19379696; http://dx.doi.org/ 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Strome S, Lehmann R. Germ versus soma decisions: lessons from flies and worms. Science 2007; 316:392-3; PMID:17446385; http://dx.doi.org/ 10.1126/science.1140846 [DOI] [PubMed] [Google Scholar]
- [34].Takasaki T, Liu Z, Habara Y, Nishiwaki K, Nakayama J, Inoue K, Sakamoto H, Strome S. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development 2007; 134:757-67; PMID:17215300; http://dx.doi.org/ 10.1242/dev.02771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cotobal C, Rodriguez-Lopez M, Duncan C, Hasan A, Yamashita A, Yamamoto M, Bähler J, Mata J. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenetics Chromatin 2015; 8:28; PMID:26279681; http://dx.doi.org/ 10.1186/s13072-015-0018-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gao J, Kim HM, Elia AE, Elledge SJ, Colaiacovo MP. NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during C. elegans Meiosis. PLoS Genet 2015; 11:e1005029; PMID:25768301; http://dx.doi.org/ 10.1371/journal.pgen.1005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Moerman DG, Waterston RH. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics 1984; 108:859-77; PMID:6096205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012; 150:65-77; PMID:22738726; http://dx.doi.org/ 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al.. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012; 150:88-99; PMID:22738725; http://dx.doi.org/ 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 2012; 337:574-8; PMID:22700655; http://dx.doi.org/ 10.1126/science.1220952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].de Albuquerque BF, Placentino M, Ketting RF. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev Cell 2015; 34:448-56; PMID:26279485; http://dx.doi.org/ 10.1016/j.devcel.2015.07.010 [DOI] [PubMed] [Google Scholar]
- [42].Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012; 489:447-51; PMID:22810588; http://dx.doi.org/ 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]


