Abstract
Spectral Localization by Imaging (SLIM) based magnetic resonance spectroscopy (MRS) provides a new framework that overcomes major limitations of current MRS techniques, which allow only rectangular voxel shapes that do not conform the shapes of brain structures or lesions. However, the restrictive assumption of compartmental homogeneity in SLIM can lead to localization errors, thus its applications have been very limited to date. SLIM-based localization is subject to errors due to inhomogeneous B0 and B1 fields, particularly in organs with complex compartmental geometry including the human brain. The limitations of SLIM were overcome through the development and implementation of B0-Adjusted and Sensitivity-Encoded SLIM (BASE-SLIM) that includes corrections for inhomogeneities of both B0 and B1 fields throughout the volume of interest. In this study, we demonstrate significantly improved localization accuracy in compartments with arbitrary shapes and reliable quantification of metabolite concentrations in gray and white matter of the human brain using the BASE-SLIM technique.
Keywords: magnetic resonance spectroscopy, spectral localization by imaging, compartmental inhomogeneity, human brain, gray matter, white matter
Graphical Abstract
Introduction
In vivo magnetic resonance spectroscopy (MRS) has been increasingly utilized to quantify steady-state metabolite concentrations and to investigate metabolic activity of the living brain in health and disease. Clinical MRS is a valuable tool for diagnosis, assessing disease progression and treatment efficacy, and identifying underlying disease mechanisms in various neurological and neurodegenerative disorders. However, clinical MRS findings have often been inconclusive or inconsistent, partly due to the difficulty of correcting for significant intra-voxel variation of the tissue composition of gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) within the volume of interest (VOI), which complicates the assessment of metabolite concentrations. The limitations of current MRS techniques stem from not only the presence of different tissue types but also the partial volume effect in single voxel spectroscopy (SVS) and both partial volume and point spread function effects in magnetic resonance spectroscopic imaging (MRSI) due to the brain structures or lesions smaller than the MRS voxel size.
Spectral localization by imaging (SLIM) provides a basis for acquiring MR spectra from homogeneous tissue compartments with arbitrary shapes without partial volume effects by taking advantage of information from high-resolution MRI (Hu et al., 1988; Liang and Lauterbur, 1993). Whereas conventional MRSI reconstructs voxel-based spectral information using the Fourier transformation, SLIM reconstructs compartment-based spectral information by solving a non-Fourier, over-determined system, utilizing information about tissue compartments derived from the anatomical structures visible in MRI. Thus, SLIM promises to overcome limitations of MRSI, namely the extreme under-sampling of the underlying complex, convoluted anatomy of the brain. Thus far, the applications of SLIM have been rather limited, often using non-proton MRS with compartments of relatively simple anatomical shapes such as the human leg and heart (Hu et al., 1988; von Kienlin et al., 2001). In contrast, 1H MRS of the brain using SLIM presents highly stringent requirements for water and lipid suppression as well as challenges posed by much more complex compartmental shapes such as GM and WM.
The major limitation of SLIM stems from the assumption of spatial uniformity of each compartment in the theory, which is violated in practice by inhomogeneities of the static magnetic field (B0) and radiofrequency magnetic field (B1) in a given volume of interest. This issue has been partially addressed by various extensions of SLIM, including GSLIM (Liang and Lauterbur, 1991), SLOOP (Vonkienlin and Mejia, 1991), SPLASH (An et al., 2011), BSLIM (Khalidov et al., 2007), NL-CSI (Bashir and Yablonskiy, 2006), and SPREAD (Dong and Peterson, 2009). Among proposed extensions of SLIM, only a few have demonstrated its application to the human brain in a limited scope, with no demonstration of a compartmental separation between GM and WM to date.
In this study, we propose a B0-adjusted and sensitivity-encoded SLIM (BASE-SLIM) technique that addresses inhomogeneities of both B0 and B1 fields, which enables us to acquire 1H MRS signals from each tissue type, GM and WM, in the human brain with minimal inter-compartmental cross-contamination.
Theory
Theoretical background of SLIM and BASE-SLIM
In MR experiments, the signal can be generally expressed as
| [Eq. 1] |
where ρ̂(r, t) is the apparent MR spin density, kn, n = 1…N, is the serialized k-space vector, r is a spatial coordinate vector, and f(r) is the local precession frequency. The apparent MR spin density ρ̂(r, t) includes physical spin density ρ(r, t) with time dependent chemical shift information as well as B1 transmit and receive inhomogeneity information. The SLIM theory presumes that the MR spin density ρ(r, t) can be modeled as a collection of compartmental spin densities, ρm(t), m = 1…M, which are uniform throughout each compartment, without considering the presence of B0 or B1 inhomogeneities. The acquired MR signals, sn(t), can then be described by a transformation (Gnm) of the compartmental signals, cm(t) ≡ A ρm(t), where A is a constant composed of receiver gain and signal detection efficiency. Thus, the SLIM equation is described as
| [Eq. 2] |
where n is the phase encoding index, m the compartment index, and Gnm the geometry matrix with the size of N×M (N ≥ M). Gnm is defined as
| [Eq. 3] |
where ξm is a binary compartment function defined as ξm(r) = 1 when r ∈ Dm, otherwise ξm(r) = 0, and Dm indicates the mth compartment. When traditional Fourier-encoding is used, the values of Gnm correspond to a subset of the k-space values obtained by discrete Fourier transformation of the compartmental masks. Using the fast Fourier transform (FFT) to calculate Gnm, the computation speed is greatly enhanced compared with conventional numerical integration methods in Eq. 3. The final SLIM-reconstructed compartmental signals, cm(t), are obtained from the least-squares solutions of Eq. 2 as
| [Eq. 4] |
where * denotes the complex conjugate transform.
The limitations of SLIM regarding the effects of compartmental inhomogeneities have been previously investigated (Hu and Wu, 1993; Liang and Lauterbur, 1991). The SLIM theory presumes ρ̂(r, t) to be equal to ρ(r, t) and f(r) to be constant in Eq. 1. In practice, the B1 and B0 fields are the dominant source of compartmental inhomogeneity, which can be measured through high resolution MRI. BASE-SLIM extends the SLIM model by incorporating B0 and B1 field inhomogeneity information. Thus, Eq. 2 becomes
| [Eq. 5] |
where n′ = 1…N′ enumerates N steps of k-space encoding for each receiver coil where N′ = N×Nc and Nc is the number of coils. The BASE-SLIM matrix, Hn′m(t), is defined as
| [Eq. 6] |
where B1c(r), c = 1…Nc, are the complex-valued sensitivity maps of Nc coils, γΔB0(r) the angular frequency deviation associated with B0 field inhomogeniety, and t = 0…(Nt−1)Δt, where Nt is the number of time points and Δt the dwell time. When there is significant spatial variation of the B1 transmit field, the B1c(r) term can be modified to include the product of the transmit and receive B1 field information. The BASE-SLIM-reconstructed compartmental signals, cm(t), are obtained from the least-squares solutions of Eq. 6 as
| [Eq. 7] |
with Nt solutions compared with a single solution in SLIM. BASE-SLIM equations intrinsically apply to three-dimensional k-space encoded MRSI data. In the case of two-dimensional phase encoding, the three-dimensional binary compartment functions, ξm(r), are effectively averaged over the slice volume of interest, resulting in fractional values. With three-dimensional phase encoding, three-dimensional compartment functions will allow spectral localization via all three k-space dimensions.
Coil combination for BASE-SLIM
When using multiple receive coils, MR signals from each individual coil can be combined by normalizing the signals from each coil to achieve equal signal amplitudes among coils (Eq. 7). However, this approach does not take into account different noise contributions among coils, thereby yielding sub-optimal SNR of the combined BASE-SLIM spectra. To address this issue, BASE-SLIM reconstruction was performed on each coil separately, and then compartmental spectra from all receive coils were combined by incorporating the measured SNR of each spectrum based on noise and signal power in the frequency domain (Bydder et al., 2002; Wright and Wald, 1997).
The signal contribution of each coil to each BASE-SLIM compartment can be obtained by performing BASE-SLIM reconstruction for each coil, yielding Nc compartmental signals, cm,c(t), per compartment. Fourier transformation and zero-order phasing of cm,c(t) yield a set of compartmental spectra, Cm,c(f). Final compartmental spectra, Ĉm(f), are given by a weighted summation,
| [Eq. 8] |
where Wm,c are the complex coil weighting factors determined from SNR values for each coil and each compartment, and Bm are the compartmental normalization factors to remove weighting bias of the signal amplitudes Sm,c, as
| [Eq. 9] |
The SNR was estimated by taking the ratio of the amplitude of a major metabolite signal, e.g., creatine (Cr) or N-acetylaspartate (NAA), to the root-mean-square level of a neighboring spectral band containing only noise, e.g., in 10 – 12 ppm range.
Materials and Methods
Numerical Simulation
The effects of B0 and B1 inhomogeneities on SLIM based reconstruction were evaluated using synthesized k-space data with simulated B0 and B1 fields, and GM and WM maps of the human brain. A singlet with Lorenzian lineshapes centered at 3.0 ppm was assigned to GM and that at 2.0 ppm to WM, in order to examine compartmental contamination as well as line-shape distortions due to B0 and B1 inhomogeneities (Fig. 1). The parameters used to synthesize k-space data were: FOV = 20 x 20 cm and matrix size = 16 x 16. The B0 map consisted of a 10 μT/m gradient along the x-axis (left to right) with a value of zero at the center of the x-axis (Fig. 1). The B1 map consisted of the transverse magnetic field produced by a circular loop coil with 10 cm diameter positioned 10 cm below the center of the image (posterior to the brain). The synthetized B0 and B1 field maps are shown in Fig. 1. Compartments consisted of the two-dimensional segmented GM and WM tissue masks. The effects of B0 and B1 corrections were evaluated separately by performing spectral reconstructions with and without correcting for B0 and/or B1 inhomogeneities.
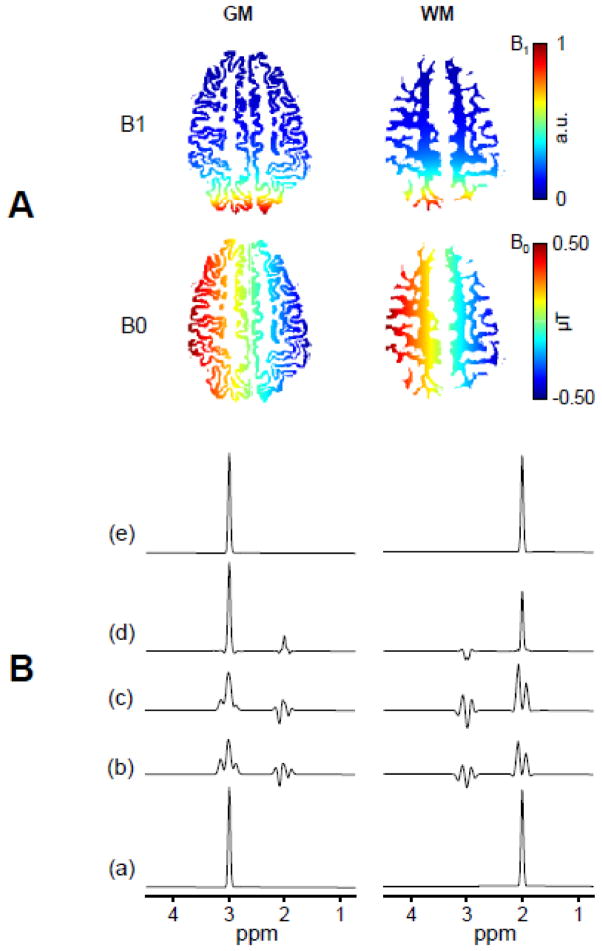
Figure 1. Simulations of SLIM and BASE-SLIM reconstruction in the presence of inhomogeneous B0 and B1 fields.
(A) Input B0 (bottom) and B1 (top) field maps of GM and WM used for simulations of SLIM and BASE-SLIM reconstruction. The B0 field consisted of a 1.0 μT offset and 10 μT/m gradient along the y-axis. The B1 field consisted of the transverse magnetic field generated by a 10 cm diameter surface coil located 10 cm left from the center of FOV. B0 and B1 maps are superimposed on GM (left) and WM (right) compartments.
(B) Input spectra (a) consisted of a singlet at 3 ppm for GM and at 2 ppm for WM. (b) Reconstructed spectra using SLIM without B0 and B1 corrections; (c) Reconstructed spectra with B0 correction only; and (d) those with B1 correction only. Reconstructed spectra using BASE-SLIM with both B0 and B1 corrections (e) were identical to the original input spectra (a), demonstrating effective corrections of B0 and B1 inhomogeneities. Reconstructed spectra without both B0 and B1 corrections resulted in various degrees of lineshape distortion and compartmental cross-contamination due to B0 and B1 inhomogeneities.
NMR methods
All experiments were performed on a 3 T MR system (Skyra, Siemens AG, Erlangen, Germany) with a 70 cm open bore horizontal magnet using a body transmit and a 16-channel head array receive coils. Shimming was performed with vendor-supplied field mapping routines in combination with manual first-order shim corrections. High-resolution three-dimensional anatomical MR images were acquired using the magnetization prepared rapid acquisition of gradient echo (MPRAGE) sequence (TR/TE = 2000/2.98 ms, TI = 900 ms, spatial resolution = 1 mm3). 1H MRSI was acquired using a custom-optimized semi-adiabatic localization by adiabatic selective refocusing (semi-LASER) sequence (TE/TR = 35/1600 ms, 16 × 16 phase encoding with elliptical k-space coverage (149 k-space points), FOV = 20 × 20 cm2, slab thickness = 2 cm, VOI = 80 × 100 mm2) (Scheenen et al., 2008) with a 1H MRSI slab positioned in the frontal to parietal regions superior to the corpus callosum. Water suppression was performed using a combination of variable power RF pulses with optimized relaxation delays (VAPOR) method (Tkac et al., 1999). 1H MRSI of unsuppressed water was acquired for quantification of metabolites. B0 mapping was performed using a gradient echo sequence (TE = 4.92/7.38 ms, TR =400 ms, FOV =192 x 192 mm2, spatial resolution = 3 x 3 x 3 mm3). B1 mapping was performed using a gradient echo sequence (TE/TR = 2.07/132 ms, spatial resolution = 3 x 3 x 3 mm3) with two acquisions: (1) body coil transmit and receive, and (2) body coil transmit and head coil receive. B1 inhomogeneity generated during transmission was considered negligible at 3 T when using a large body transmit coil. Thus, only B1 inhomogeneity generated during reception, i.e., coil sensitivity, was considered for B1 inhomogeneity corrections in BASE-SLIM. B1 (or coil sensitivity) maps were calculated by complex division of MR images from each receive coil element by the body coil MR images. The computed B1 maps were further processed with spatial smoothing and conditioning procedures adapted from the algorithm used in sensitivity encoding (SENSE) reconstruction (Pruessmann et al., 1999).
Phantom studies
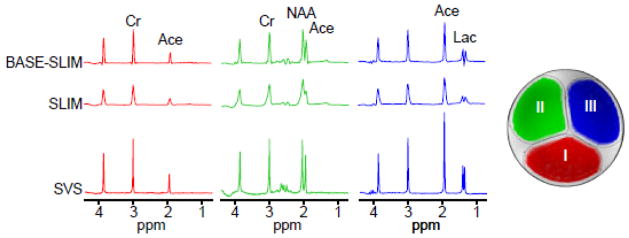
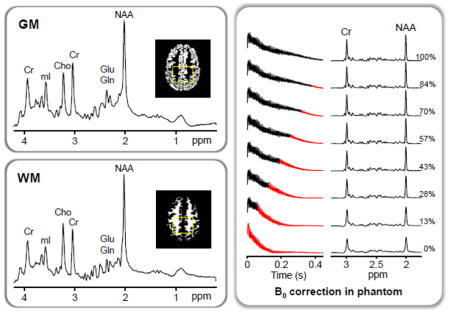
Phantoms with one or multiple compartments were constructed with outer dimensions comparable to the human head to evaluate the performance of BASE-SLIM reconstructions. A three-compartment cylindrical phantom was used 1) to compare SLIM and BASE-SLIM reconstructions under B0 and B1 inhomogeneities, and 2) to test the feasibility of BASE-SLIM reconstruction using only B1 inhomegeneity information. Each compartment of the three-compartment phantom (15 cm diameter) contained (I) 10 mM Cr and 5 mM sodium acetate, (II) 10 mM Cr, 10 mM sodium acetate and 10 mM NAA, and (III) 10 mM Cr, 20 mM sodium acetate and 10 mM lactate (Fig. 2 inset). To simulate B0 inhomogeneity present in the brain in vivo, first order shim coil currents were adjusted to create a linear magnetic field gradient of 4.0 μT/m along the x-axis. SLIM reconstruction was performed using only the acquired MRSI k-space data and compartment maps, while BASE-SLIM reconstruction was performed using the MRSI k-space data, compartment maps, and B0 and B1 maps. Each compartment was manually delineated based on the outlines of the inner container walls. Reconstructed SLIM and BASE-SLIM spectra in each compartment were compared with SVS spectra in each compartment (voxel size = 2.5 x 2.5 x 2.5 cm3).
Figure 2.
SLIM and BASE-SLIM reconstruction in a three-compartment phantom (see the inset transverse MRI). Spectra from both BASE-SLIM (top) and SLIM (middle) were reconstructed from whole compartments (approximately 80 ml per compartment) using full k-space data (16 x 16, circular k-space sampling), compared to SVS (bottom) from small VOIs (16 ml) located at the center of each compartment. Compartments contained various mixture of solutions with (I) 10 mM Cr and 5 mM sodium acetate, (II) 10 mM Cr, 10 mM sodium acetate and 10 mM NAA, and (III) 10 mM Cr, 20 mM sodium acetate and 10 mM lactate. Spectral linewidth and SNR of BASE-SLIM reconstructed spectra were improved by 58% and 46% compared with those of SLIM, respectively.
Signal-to-noise ratio (SNR) of phantom spectra were measured by the ratio of peak amplitude of Cr at 3 ppm to the root mean square (RMS) value of noise between 10.0 ppm and 12.00 ppm. Linewidths were measured at full width at half maximum (FWHM) of the Cr peak. Each spectrum was normalized by the peak amplitude of the corresponding unsuppressed water spectrum from each compartment in order to compare amplitudes across compartments. Reported SNR and linewidth values were averaged over all three compartments.
To demonstrate the feasibility of single-shot localization of compartmental spectra without any phase encoding, BASE-SLIM reconstruction was performed using only the center of k-space data, which were extracted from the full MRSI k-space data. The effects of B1 inhomogeneity on SLIM and BASE-SLIM reconstructed spectra were examined by comparing areas under the curve of the Cr signal at 3.0 ppm (with a bandwidth of 0.5 ppm) from each compartment and each coil. Signal intensity variations without B1 corrections in BASE-SLIM were calculated based on the B1 maps of each coil at each compartment.
A single-compartment spherical phantom (17 cm diameter) was used to characterize B0 correction in BASE-SLIM. The single-compartment phantom contained a solution of 10 mM NAA, 10 mM Cr and 10 mM glutathione (GSH). The B0 field inhomogeneity was adjusted to 5.0 μT/m along the y-axis by adjusting the first-order shim currents. B0 correction was evaluated by varying the number of time points processed in the time-dependent BASE-SLIM equation (Eq. 7) and measuring the improvement of lineshapes.
In vivo human studies
Eleven healthy subjects (6 female and 5 male; 29 ± 4 years old, mean ± SD) were studied according to the study protocol approved by the Institutional Review Board of the University of Kansas Medical Center. The details of the study were explained to all subjects and written informed consent was obtained prior to their participation in the study. All subjects were positioned supine on the MRI patient table with the head positioned inside a 16-channel head coil. A series of MR scans were performed including 1H MRSI, B0 and B1 maps and MPRAGE.
Image segmentation
Three-dimensional MPRAGE images were co-registered to the MRSI data set using in-house written software in MATLAB (Matlab R2009B, MathWorks, Inc., Natick, MA, USA). To improve the quality of automatic segmentation, 1) bias correction was applied to the MPRAGE images using an image domain normalization method (Sled et al., 1998) and 2) tissue probability maps from the international consortium for brain mapping (ICBM) 152 brain atlas, containing spatial prior information for GM, WM and CSF were co-registered to the bias-corrected MPRAGE images to improve segmentation performance, using the reference ICBM 152 T1-weighted 0.5 mm isotropic resolution image (Grabner et al., 2006). Brain tissue segmentation was performed on the bias-corrected MPRAGE images using SPM8 (Wellcome Department of Imaging Neuroscience, London, UK) (Ashburner and Friston, 2005; SPM8, 2009).
BASE-SLIM reconstruction
BASE-SLIM reconstruction was performed using the acquired MRSI k-space data, B0 and B1 maps, and GM and WM segmentation maps. The detailed BASE-SLIM reconstruction steps are: 1) co-register all MRI data to match the orientation of MRSI; 2) generate compartment masks incorporating slice selection profiles used in the MRSI; 3) calculate Hn′m(t) in Eq. 6 using B0 and B1 maps, GM and WM segmentation maps, and compartment masks; 4) calculate compartmental signals in Eq. 7 for each coil using the least squares matrix inversion; and 5) perform coil combination for optimal SNR in each compartmental spectrum based on Eq. 8. Raw k-space data of MRSI, B0 and B1 scans were obtained directly from the scanner without any post-processing except the automatic noise de-correlation performed by the scanner software for multi-channel data. Least squares matrix inversion in the fourth step above was performed using the singular value decomposition method for efficient calculation of the inverse for small size matrices. Singular values of the matrix at time t gradually diminish compared with t = 0 due to the k-space dispersion effect of B0 inhomogeneities. In order to mitigate noise amplification at the tail portion of the free induction decay (FID) and to minimize the data processing time, the B0 term was kept constant without further update after the time point at which the singular value reached below 20% of the initial value. In addition, the signal was attenuated in proportion to the time-varying singular values for each compartment to counteract a time-varying undesired amplification factor.
The computation time for BASE-SLIM reconstruction processing was minimized by calculating Hn′m(t) in Eq. 6 using the FFT as follows. High-resolution compartment mask images modified with B0 and B1 terms were resampled to have image sizes of a power of two (i.e. 256 × 256 from 192 × 256 original dimensions) using trilinear interpolation in the image domane. The FFTs of the mask images were calculated, and the central (12 × 12 or 16 × 16) sets of values corresponding to the k-space encoding vectors of MRSI were extracted. In order to reduce the demands on computer memory resources, only a portion of the total array, i.e., image volume with x×y×z× compartments was loaded and processed while looping through the time and coil indices. To further reduce the memory requirement and to improve the computation speed of time-consuming operations such as resampling and smoothing for B1 mapping, the MRI resolution was resampled by 1/2. Using the time- and memory-efficient implementation, BASE-SLIM reconstruction required less than 15 minutes on a personal computer (Intel Core i7, quad-core, 2.6 GHz) with the parameters of 128 × 128 × 8 MR image size, 8 compartments, 256 k-space steps, 128 time points and 16 coil channels.
Quantification of metabolites in GM and WM of the human brain: BASE-SLIM and MRSI
Metabolite concentrations were quantified using the Linear Combination of Model in vitro spectra (LCModel) analysis (Provencher, 2001) with the unsuppressed water signal as a concentration reference for both MRSI and BASE-SLIM reconstructions. Quantified metabolites included NAA, total creatine, total choline (tCho), myo-inositol (mI), and glutamate+glutamine (Glu+Gln). Identical BASE-SLIM reconstruction procedures were applied to both metabolite and unsuppressed water signals to ensure the same compartmental definition, and B0 and B1 inhomogeneity effects. MRSI data analysis was performed using voxels with tissue fraction greater than 75%. To compare MRSI and BASE-SLIM data from the same spatial regions, the GM and WM compartments in BASE-SLIM were selected to match the VOI boundaries of the selected MRSI data.
For the MRSI data, metabolite concentrations in GM and WM were obtained by extrapolation using linear regression analysis of metabolite concentrations obtained from LCModel from the included MRSI voxels as follows. First, the volume fractions of GM and WM in each voxel were calculated by performing a low-pass filtering operation on the high-resolution segmented MR images to account for the point spread function in the low resolution MRSI reconstruction as previously described (Choi et al., 2006; Weber-Fahr et al., 2002). Briefly, the point spread function correction steps were: 1) multiplication of the slice selection profiles of MRSI with the high resolution segmented MR images; 2) Fourier transformation of segmented images into k-space; 3) removal of higher-frequency k-space data to match the small MRSI k-space sampling points in MRSI; and 4) inverse Fourier transformation of k-space data back to the real space. Then, the final metabolite concentrations in GM and WM were obtained by extrapolating the linear regression of metabolite concentrations over the GM fraction, |GM|/(|GM| + |WM|), of the selected MRSI voxels, where |GM| and |WM| are absolute values of GM and WM volumes in each voxel, respectively.
Results
Simulations of SLIM and BASE-SLIM
The effects of B0 and B1 inhomogeneities on SLIM and BASE-SLIM reconstructions were demonstrated using numerical simulations (Fig. 1). B0 and B1 maps overlaid on the GM and WM compartments showed field variations in the range of −0.5 – 0.5 μT/m and 0 – 1.0 (a.u.), respectively (Fig. 1A). Compared with input spectra with a single peak for GM at 3 ppm and for WM at 2 ppm (Fig. 1B(a)), ordinary SLIM reconstruction without any correction for B0 and B1 inhomgeneities resulted in significant inter-compartmental signal contamination in both GM and WM spectra, with visible lineshape distortions and lower peak amplitudes at ~2 ppm in GM and at ~3 ppm in WM. The effects of individual B0 and B1 inhomogeneity correction are shown in Fig. 1B(c)–(d). When B0 inhomogeneity correction was not applied, severe inter-compartmental signal contamination and spectral distortions were present (Fig. 1B(c)) similarly to those in Fig. 1B(b) with slight improvement. When B1 inhomogeneities were not corrected, inter-compartmental signal contamination and spectral distortions were less severe than those in Figs. 1B(b) and (c). In contrast, when both B0 and B1 inhomogeneities were corrected using full BASE-SLIM reconstruction, GM and WM spectra from both compartments were identical to the input spectra with full intensity without any spectral distortions (Fig. 1B(e)).
Phantom experiments for SLIM and BASE-SLIM
Comparisons of spectral reconstructions using SLIM and BASE-SLIM were performed on a three-compartment phantom (Fig. 2). Both SLIM and BASE-SLIM spectra were reconstructed from the entire compartments, while the SVS spectra were measured from 2.5 x 2.5 x 2.5 cm3 voxels located at the center of each compartment. The SLIM reconstruction resulted in significant frequency shifts and lineshape distortions in spectra compared with those from SVS. The BASE-SLIM reconstruction resulted in the spectral patterns that closely matched those of SVS from all three compartments, indicating accurate correction of B0 and B1 inhomogeneities. BASE-SLIM spectra showed 46% improvement in average SNR and 58% (or 4.8 Hz) reduction in average linewidths compared with SLIM spectra.
BASE-SLIM reconstruction was also performed using only the center of k-space data, i.e., single-shot data, in order to evaluate its ability to separate compartmental spectra using only B1 field maps of the 16 receive coils (Fig. 3). The B1 maps of the center slice of the VOI are shown for all receive coils with the corresponding single-shot spectra overlaid on the maps. The three BASE-SLIM compartmental spectra reconstructed from the single-shot data showed substantially the expected spectral patterns of metabolites contained in each compartment, with some residual inter-compartmental spectral leakage (Fig. 3, bottom trace). The acetate signal at 1.9 ppm was present in all compartments and correctly corresponded to the concentration in each compartment. The lactate signal near 1.3 ppm was present mostly in compartment III, while the NAA signal at 2.0 ppm near the acetate signal appeared mainly in compartment II. Minor spectral leakages of lactate and NAA were observed across all compartments.
Figure 3. BASE-SLIM reconstruction for three compartments using only B1 (coil sensitivity) information.
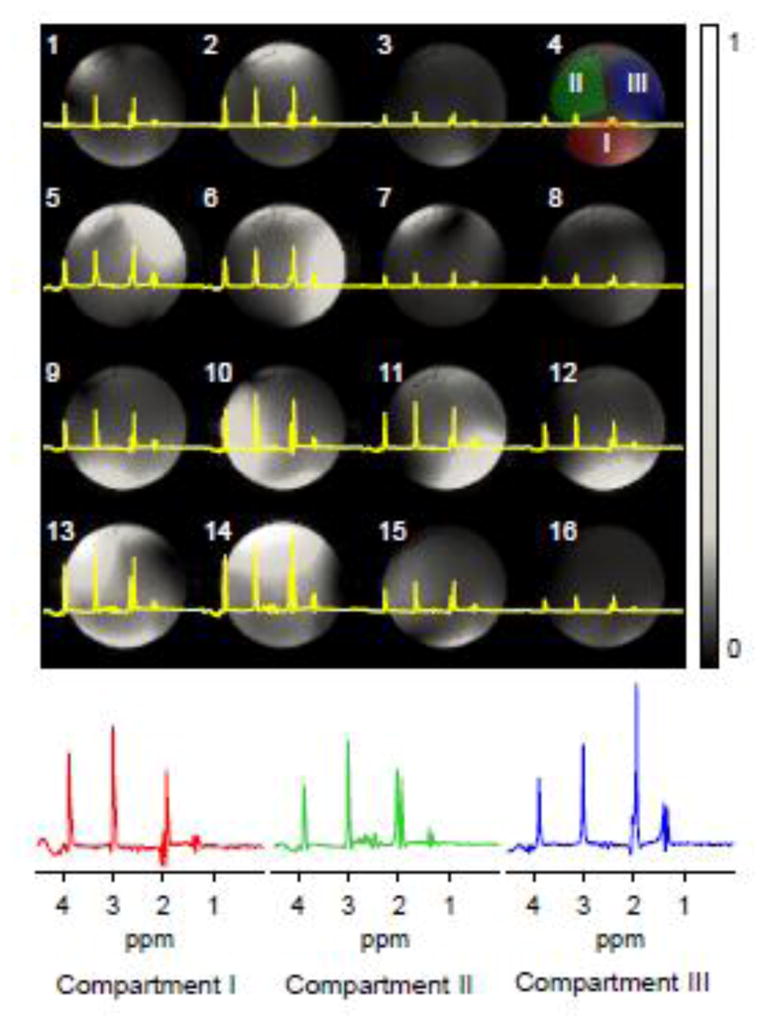
(A) Spectra acquired using 16 receive coils overlaid on respective B1 maps. Coil ID is indicated at the left top corner of each image. Central k-space data without phase encoding was extracted from the full k-space data used in Fig. 2. Compartment masks (I, II, and III) are shown on the B1 map of coil 4 (top right). Gray scale bar indicates normalized B1 values.
(B) BASE-SLIM reconstructed spectra from each compartment I, II, and III using only coil sensitivity information. Spectra are color-coded to match the color of each compartmental mask shown in (A).
The effect of B1 correction in BASE-SLIM reconstruction was evaluated in the presence of B1 inhomogeneities of multiple receive coils. The areas of the Cr peak at 3 ppm, normalized by the mean value over 16 coils, were plotted for BASE-SLIM and SLIM reconstructed spectra from all 16 coils (Fig. 4). The signal for each coil from SLIM reconstruction (without B1 correction) varied significantly among receive coils, while the signal from BASE-SLIM reconstruction (with B1 correction) was very consistent across all 16 receive coils and three compartments. Variations in the signal in SLIM reconstruction reflect coil sensitivity variations among receive coils for each compartment. The estimated signal bias, calculated by averaging the B1 field maps over the respective compartments (Fig. 4, dashed lines), closely matched those in SLIM reconstruction.
Figure 4.
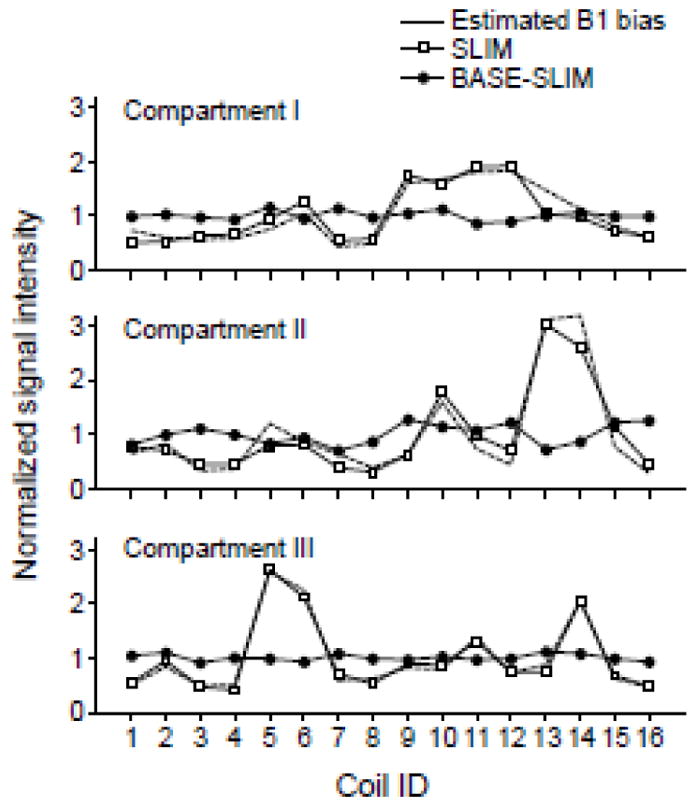
Comparison of signal amplitude variations among receive coils in reconstructed spectra using SLIM and BASE-SLIM. Signal amplitudes were measured from the area under the creatine peak at 3 ppm from each coil and each compartment. The area under the peak was normalized to a mean value of 1 for both SLIM and BASE-SLIM. Estimated B1 bias was calculated by averaging B1 values in each receive coil and each compartment. B1 bias values were normalized to mean value of 1 over all receive coils. Coil IDs are identical to those shown in Fig. 3.
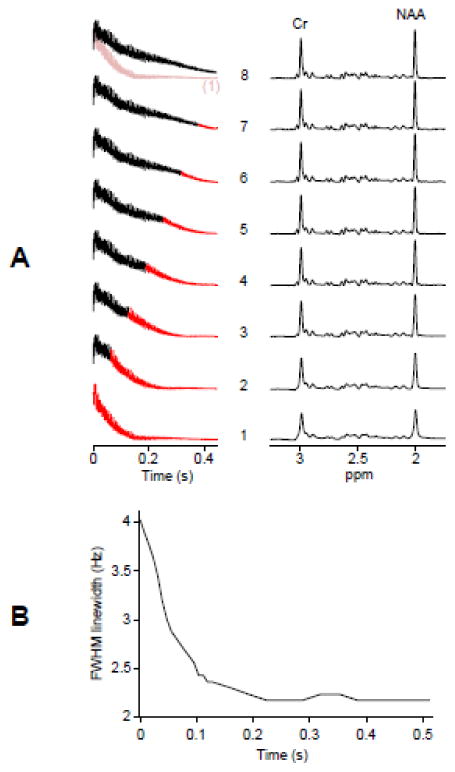
The effect of B0 correction in BASE-SLIM reconstruction was evaluated using a single-compartment spherical phantom (Fig. 5). The duration for B0 correction varied from zero to the full length of the FID with the B0 corrected portion indicated in black and the uncorrected portion in red (Fig. 5A left). Slower T2* decay is clearly visible in the B0 corrected FIDs compared with the B0-uncorrected FID (Fig. 5A, bottom trace). As the B0 corrected portion increased, the linewidth of corresponding spectra decreased and the peak amplitudes increased, reflecting slower T2* decay in FIDs (Fig. 5A right). The spectral linewidth of NAA showed an initial rapid decrease followed by a plateau beyond approximately 200 ms of processing (Fig. 5B).
Figure 5. The effect of B0 correction in BASE-SLIM reconstruction.
(A) Time domain signals (left) of a spherical single compartment phantom containing 10 mM Cr, 10 mM NAA, and 10 mM GSH with varying duration of B0 correction and corresponding BASE-SLIM reconstructed spectra (right). B0 inhomogeneity was generated by applying 5.0 μT/m gradient along the y-axis. The duration for B0 correction was varied from 0 (bottom trace, B0-uncorrected FID) to the full length (top trace, fully B0-corrected FID) of FID. The B0-uncorrected FID (top trace, pale red) is shown in comparison with the fully B0-corrected FID (top trace, black). Intermediary steps of B0 correction are shown between the top and bottom traces of FIDs. Time points with the reconstruction matrix held constant (no B0 phase update) are shown in red in each FID trace.
(B) Measured linewidth in FWHM of the NAA peak at 2 ppm from all the spectra in (A) with various durations of B0 correction. The FWHM values showed a rapid decrease followed by a plateau beyond approximately 200 ms of B0 correction.
In vivo experiments for SLIM and BASE-SLIM
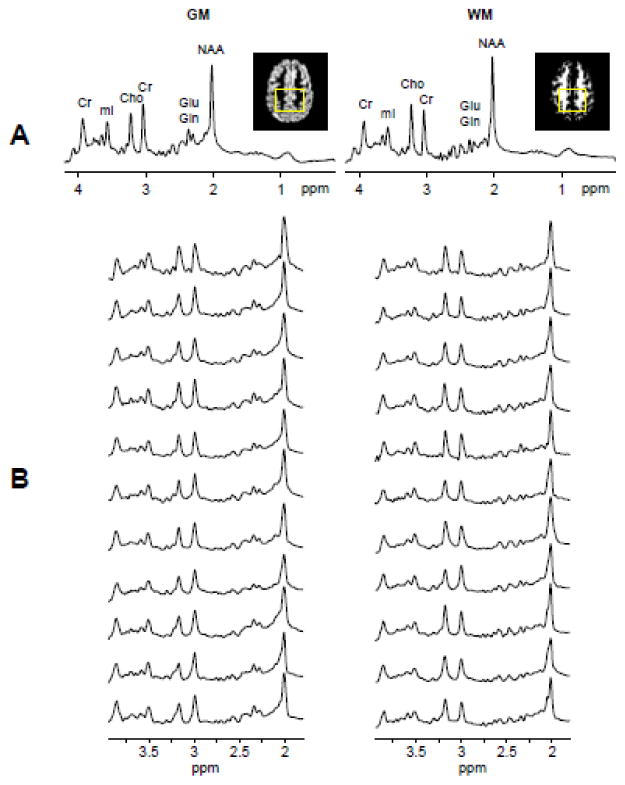
Successful BASE-SLIM reconstruction was achieved to obtain tissue-type specific spectra from GM and WM in the human brain (Fig. 6). Distinctive spectral patterns of GM and WM were observed in all spectra acquired from the fronto-parietal regions of eleven subjects: lower ratios of tCho (3.17 ppm) to Cr (3.03 ppm) signals and higher Glu+Gln signals at around 2.25 ppm in GM, compared with those in WM (Fig. 6B). The characteristic spectral patterns of GM and WM are clearly visible in averaged spectra of all subjects (Fig. 6A). When comparing the performance of BASE-SLIM and SLIM reconstructions, BASE-SLIM showed a 19% (or 1.01 Hz) narrower linewidth and 4% greater SNR of the Cr peak at 3.0 ppm compared with SLIM. Table 1 (top) shows metabolite concentrations in GM and WM quantified from BASE-SLIM reconstructed spectra using LCModel: NAA concentration did not differ between GM and WM (p > 0.05) while tCho concentration in GM was 23% (p < 0.0001) lower than those in WM. Cr, Glu+Gln, and mI concentrations in GM were 16% (p = 0.01), 144% (p = 0.0001), and 14% (p = 0.02) higher than those in WM, respectively. Table 1 (bottom) shows metabolite concentrations in GM and WM quantified from the MRSI regression analysis: NAA concentration did not differ between GM and WM (p = 0.2) while tCho concentration in GM was 15% (p = 0.006) lower than that in WM. Cr, Glu+Gln, and mI concentrations in GM were 39% (p < 0.0001), 169% (p < 0.0001), and 38% (p = 0.0005) higher compared with those in WM, respectively. Representative MRSI regression results of NAA, Cr, tCho, and Glu+Gln show examples of the concentration estimates from each subject with the MRSI regression results shown in regression lines and the BASE-SLIM results indicated in open squares (Fig. 7). Metabolite concentrations in GM and WM obtained from MRSI regression analyses and BASE-SLIM showed a good correspondence across all the metabolites (concordance correlation coefficient = 0.94) with the exception of Glu+Gln. Glu+Gln concentrations from the MRSI regression analysis tended to be higher than those from BASE-SLIM (Fig. 8).
Figure 6. GM and WM spectra of human subjects using BASE-SLIM.
(A) Averaged spectra of GM and WM from all eleven subjects show distinctive spectral patterns expected from GM and WM. Inset images show the representative GM and WM maps at the center of the MRSI slab with the rectangular compartmental boundary used for BASE-SLIM reconstruction. The averaged spectra were normalized using unsuppressed water signals in BASE-SLIM reconstructed spectra in each GM and WM compartment.
(B) Individual GM and WM spectra reconstructed using BASE-SLIM showed consistent spectral patterns of GM and WM in all subjects.
Table 1.
Metabolite concentrations in fronto-parietal GM and WM using BASE-SLIM (top) and MRSI regression analyses (bottom). All concentrations were determined using unsuppressed water signals as the concentration reference and are shown in mean and standard deviations in eleven subjects. Ratios between metabolite concentrations in GM and WM were calculated to characterize GM and WM concentration differences of each metabolite.
| BASE-SLIM
| |||||
|---|---|---|---|---|---|
| NAA | Cr | tCho | Glu+Gln | mI | |
| GM | 7.48 ± 0.56 | 5.30 ± 0.70 | 1.41 ± 0.16 | 10.13 ± 2.16 | 4.77 ± 0.39 |
| WM | 8.11 ± 0.83 | 4.57 ± 0.41 | 1.83 ± 0.18 | 4.16 ± 1.22 | 4.19 ± 0.42 |
|
| |||||
| GM/WM | 0.92 | 1.16 | 0.77 | 2.44 | 1.14 |
| MRSI
| |||||
|---|---|---|---|---|---|
| NAA | Cr | tCho | Glu+Gln | mI | |
| GM | 7.67 ± 0.97 | 5.99 ± 0.52 | 1.50 ± 0.15 | 13.69 ± 1.57 | 5.52 ± 0.58 |
| WM | 7.08 ± 0.72 | 4.32 ± 0.33 | 1.76 ± 0.17 | 5.10 ± 1.76 | 4.01 ± 0.57 |
|
| |||||
| GM/WM | 1.08 | 1.39 | 0.85 | 2.69 | 1.38 |
Figure 7.
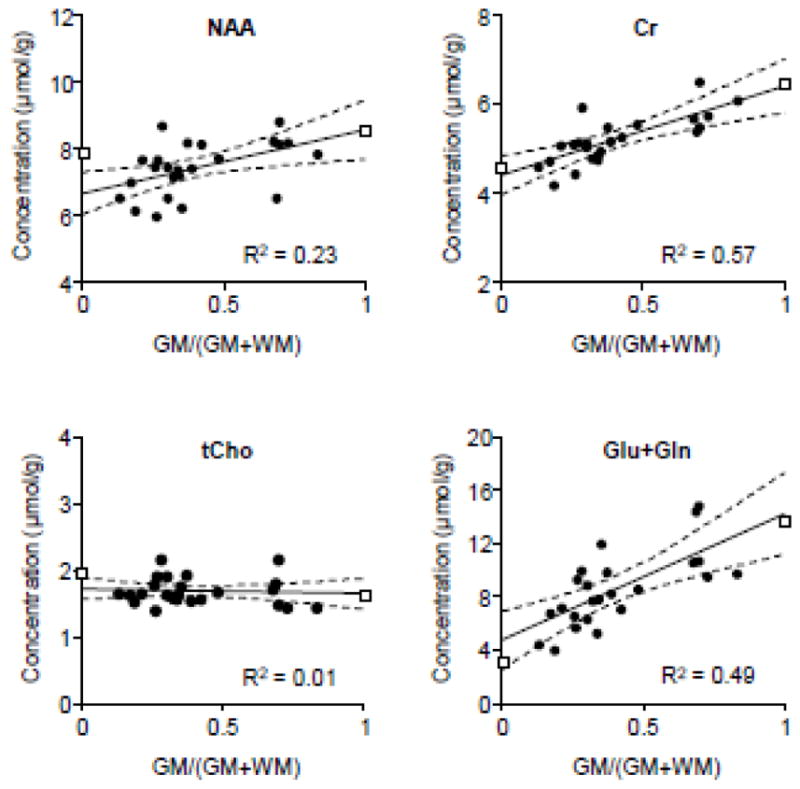
An example of estimated metabolite concentrations in GM and WM using BASE-SLIM and MRSI regression. Metabolite concentrations were determined using LCModel for each MRSI voxel. Tissue GM fractions in each MRSI voxel were calculated by down-sampling high resolution GM and WM segmented images. Linear regressions of metabolite concentrations are shown in solid black lines and 95% confidence intervals in dashed lines. The ends of each regression line represent extrapolated WM and GM values of MRSI regression. Metabolite concentrations in GM and WM using BASE-SLIM reconstruction are shown in open-squares at GM fractions of one and zero, respectively. BASE-SLIM used the external boundary matching the collection of voxels included in the MRSI regression.
Figure 8.
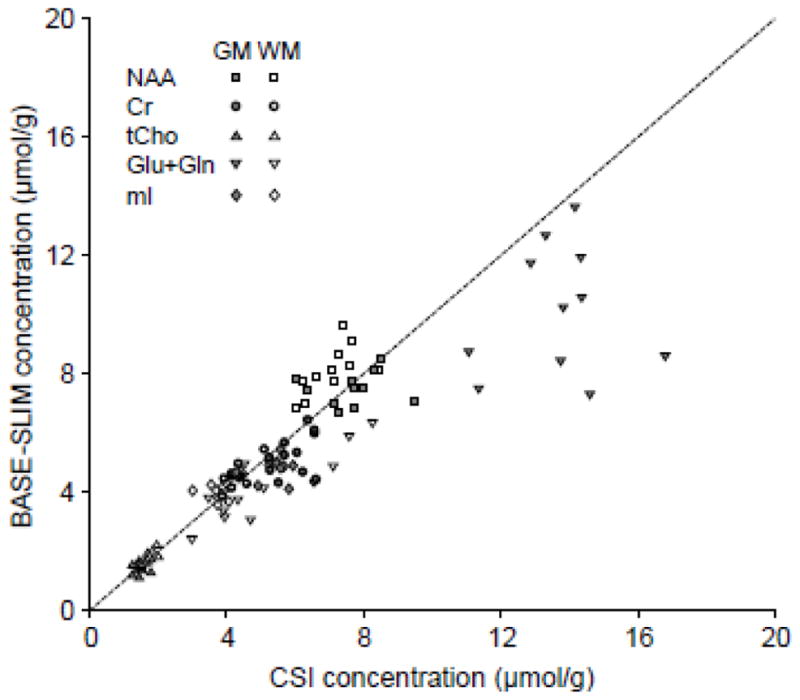
Comparisons of metabolite concentrations obtained using BASE-SLIM and MRSI regression analyses. Filled symbols indicate metabolite concentrations in GM and open symbols indicate those in WM. The dashed line indicates the line of identity. Most values lie close to the identity line, except Glu+Gln in GM. The concordance correlation coefficient was 0.94 for all metabolite concentration values from BASE-SLIM and MRSI regression, excluding Glu+Gln.
Discussion
SLIM had a long standing promise of localized MRS from any compartmental shapes, given that the metabolite concentration in each compartment is uniform. This study demonstrates that the promise of SLIM could be achievable for clinical applications using the proposed BASE-SLIM technique by overcoming major shortcomings of the basic SLIM theory. BASE-SLIM allowed successful reconstruction of compartmental 1H MR spectra of GM and WM with minimum inter-compartmental contamination, which is readily applicable for other organs and tissues as they are in general less complex in shape and tissue compositions compared with the human brain. The superior spectral quality was clearly demonstrated by improved lineshapes and line widths in BASE-SLIM reconstructed MR spectra, which was achieved by incorporating both B0 and B1 inhomogeneities in the physical model for the MR signals in SLIM. The results of numerical simulation and phantom experiments demonstrated significant influences of both B0 and B1 inhomogeneities on inter-compartmental signal leakage and spectral distortion in SLIM, demonstrating the necessity and importance of handling both B0 and B1 inhomogeneity issues to achieve the reliable spectral localization of given compartments with minimum cross-contamination. This requirement is especially stringent and imperative for the most intricate tissue boundaries such as GM and WM in the human brain.
The metabolite concentrations in GM and WM using BASE-SLIM closely matched those using regression analyses of MRSI voxels even though the two methods are distinctly different, which supports the validity of the BASE-SLIM reconstruction method. While major metabolite concentrations usually reported in clinical MRS studies such as NAA, Cr, tCho and mI showed an excellent match, the outcomes of Glu+Gln quantification were somewhat different between the two methods. The estimated concentrations of Glu+Gln tended to be higher using MRSI regression. This discrepancy could be due to large extrapolation errors in MRSI regression of Glu+Gln as imposed by a relatively narrow range of GM fraction values (0.39 ± 0.19, mean ± SD) and significant concentration differences between GM and WM.
The concentration ratios of GM to WM in the present study were in excellent agreement with the literature. All the ratios were within about one standard deviation of the average from the reported values in comparable regions of the brain. The mean concentration ratios of GM to WM from literature are: NAA: 1.07 ± 0.18; Cr: 1.35 ± 0.27; tCho: 0.83 ± 0.10; Glu+Gln: 2.03 ± 0.52; mI: 1.48 ± 0.16) (Ding et al., 2015; Maudsley et al., 2009; McLean et al., 2000; Schuff et al., 2001). However, it is in general rather difficult to make direct comparisons of absolute metabolite concentrations among studies due to technical differences in quantification methods including data analysis programs and correction of T1 and T2 relaxations, as well as differences in subjects’ age and sex, and selected brain regions.
Overall, BASE-SLIM provides important advantages over MRSI in determining metabolite concentrations in complex contoured compartments such as GM and WM in terms of reliability and flexibility. MRSI regression, currently the most commonly used approach, relies on quantification of relatively weaker signals from relatively small voxels followed by extrapolation to determine metabolite concentrations in GM and WM. This approach is very susceptible to errors in metabolite quantification due to the compounding of uncertainties. The quantification errors can be even more pronounced as the individual MRSI voxel size increases, because the range of GM fraction values will be accordingly reduced, resulting in larger uncertainties in the extrapolated values. Furthermore, this approach cannot be reliably applied to the metabolites with low concentrations because the high variability of the quantified metabolite concentrations due to low SNR results in high uncertainties of extrapolated GM and WM concentrations. In contrast, BASE-SLIM does not suffer from such drawbacks of MRSI regression because it does not require any linear extrapolation. Instead, it optimally combines all available k-space signals to produce compartmental spectra that can be directly quantified as GM and WM tissue signals or any given compartments. Thus, the major advantage of BASE-SLIM is more reliable quantification of metabolites in GM and WM with minimum compartmental cross-contamination when the underlying assumption of compartmental homogeneity is satisfied.
BASE-SLIM offers the capability and flexibility to choose a wide range of compartment sizes in order to obtain tissue-type specific MR spectra unlike SVS that requires small enough voxels to fit into only GM or WM tissue. Like SVS, the same is true for MRSI when the regression analysis is not applied to obtain metabolite concentrations in GM and WM. Furthermore, through B0 and B1 correction procedures, BASE-SLIM affords the reconstruction of MR spectra from large compartments without significant degradation of spectral quality, which cannot be otherwise achieved using other approaches such as SVS, MRSI and SLIM. The ability to select larger compartments provides the benefits of higher SNR compared with these other approaches, allowing the possibility of reducing the scan time and also applications of BASE-SLIM to metabolites with low concentrations including gamma-aminobutyric acid (GABA), GSH and vitamin C.
Determining metabolite concentrations in GM using SVS and MRSI is especially challenging because of the complex anatomical shapes of GM, e.g., cortical GM with thickness of 2–4 mm and complex three-dimensional folding patterns, and difficulties in fitting relatively large rectangular voxels (order of 10 x 10 x 10 mm3 for very high resolution MRSI with long scan time) in the cortical area with the inevitable partial volume effect in both SVS and MRSI. Accurate measurement of metabolite concentrations in GM and WM is critical in the case of metabolites with very large differences between GM and WM concentrations, e.g., Glu+Gln and GABA, because small variation of GM fraction values in SVS and MRSI voxels will significantly alter metabolite quantification in a given voxel. For example, when the GM fraction of each spectroscopy voxel changes from 0.4 to 0.3, estimated concentration changes for Glu+Gln and GABA are about −7% and −18%, respectively, based on about twofold concentration differences of Glu+Gln (current data) and about eight-fold differences of GABA between GM and WM (Choi et al., 2006). These apparent concentration differences due to the varying voxel composition could easily mask possible alterations in metabolite concentrations seen in various neurological and neuropsychiatric disorders.
Another advantage of BASE-SLIM is its capability to accelerate MRS data acquisition using only coil sensitivity information without any k-space encoding as demonstrated in this study (Fig. 3), and found similarly in previous results of SPLASH (An et al., 2011). In BASE-SLIM, coil sensitivity differences among receiver coils can function similarly to k-space encoding using gradients. Therefore, it is feasible to acquire accurate compartmental spectra without any k-space encoding with an increased number of receiver channels in clinical scanners, provided a sufficient SNR can be achieved through averaging and/or choosing large enough compartmental sizes.
When implementing B0 correction in BASE-SLIM, the accuracy of B0 maps is critical because inaccurate B0 values in the B0 maps lead to distorted FID and distorted spectral lineshapes. Due to the nature of the B0 term in the BASE-SLIM matrix, Hn′m(t) in Eq. 6, any phase errors from incorrect B0 values gradually accumulate over the duration of FID. For example, the accumulated phase errors at the end of the FID in a typical MRSI data acquisition are about 180° with the FID duration of 512 ms and the B0 error of 1 Hz, leading to signal cancellations. One of the most obvious sources of B0 map inaccuracy is the discrepancy between B0 inhomogeneity during MRS scans and B0 mapping, mostly caused by the subject movement. Thus, strategies to reduce these types of errors in the B0 maps are necessary for accurate BASE-SLIM reconstruction.
Another important aspect to consider in successful BASE-SLIM reconstruction is reducing possible noise amplification caused by B0 correction. In general, the B0 correction algorithm in BASE-SLIM and other reconstruction techniques such as BSLIM and NL-CSI introduces a time dependent gain factor that increases during the FID duration. This gain factor is related to the inverse of singular values of the in Eq. 7, such that the singular values decrease over time as the effects of dephasing accumulate in the BASE-SLIM matrix, Hn′m(t). The potential for the undue noise amplification becomes more problematic for the time points in FID where the signal intensity is comparable to or lower than the noise level, especially in the tail part of FID. The approach used to mitigate the noise amplification in the current BASE-SLIM implementation is using a shortened duration of B0 correction rather than the full duration of FID, while utilizing the BASE-SLIM matrix, Hn′m(t) stored from the cut-off time point to process the remainder of FID. The cut-off time point was determined based on the mean singular value of the matrix, , normalized to that at t = 0, which was set to 20% for all in vivo data. To reduce any possible lineshape distortion due to abrupt discontinuation of B0 correction, the time t of near the cut-off point was varied inverse-exponentially to approach the cut-off point asymptotically. In addition, the FID time course was scaled based on the SVD values in order to counteract excess signal scaling and thus maintain the physically based natural decay and even noise level throughout the FID.
In this study, unsuppressed water MRSI data from a separate acquisition were used as a reference to quantify metabolite concentrations. This approach to acquire a separate water MRSI can be time-consuming and impractical especially for high resolution MRSI. Recent technical developments provided strategies to shorten or even remove the time consuming separate water reference MRSI scans by using proton-density weighted MRI (Maudsley et al., 2009), by accelerated MRSI using parallel imaging techniques (Birch et al., 2015), by embedding water reference scans within the MRSI data acquisition (Maudsley et al., 2009), or by acquiring MRSI without water suppression (Dong, 2015). Among these approaches, the embedded water reference scans and MRSI without water suppression can provide a reference for metabolite concentrations as well as coil phase information necessary for optimal coil combination of MRSI data when multi-channel receive coils are used. These approaches for conventional MRSI is equally applicable to BASE-SLIM to reduce scan times.
One limitation of the current BASE-SLIM reconstruction is not including inhomogeneity information of the transmit B1 field (B1+) in the reconstruction model. However, it is expected that the inhomogeneity of B1+ at 3 T in small regions of interest, e.g., human heads, is relatively small when the body transmit coil is used. In cases when B1+ inhomogeneity is significant such as with higher magnetic fields (> 3T) or when using local transmit RF coils, it is necessary to include B1+ maps in BASE-SLIM to accurately reconstruct MR spectra from given compartments.
Another limitation of the current BASE-SLIM is not including possible regional concentration variations within compartments, i.e., endogenous inhomogeneities of neurochemical distributions. This type of inhomogeneity might result in an inter-compartmental spectral leakage and can lead to inaccurate reconstruction, although the magnitude of the leakage is expected to be smaller than that from concentration differences between tissue types. One approach to alleviate this limitation is to reduce compartment sizes in BASE-SLIM, which reduces variations of metabolite concentrations within each compartment, but this will also reduce the compartmental SNR. A strategy to improve BASE-SLIM includes the use of more complex mathematical modeling that incorporates unknown distributions of metabolite concentrations within each compartment, e.g., GSLIM (Liang and Lauterbur, 1991). However, incorporation of GSLIM would require much longer computation time due to its computational complexity being combined with the already computation intensive B0 correction algorithm in BASE-SLIM.
Conclusions
BASE-SLIM, a new spectral localization technique, enables us to obtain accurate and robust MR spectra from compartments with complex shapes such as GM and WM in the human brain, and thus tissue-type specific MR spectroscopy from arbitrary shaped VOIs can be achieved. The proposed BASE-SLIM overcomes limitations of conventional SLIM by incorporating B0 and B1 inhomogeneity information in the reconstruction model. BASE-SLIM also permits utilization of spatial encoding by multiple receive coils to acquire localized spectra in a single shot or in a significantly reduced scan time by removing or reducing the time-taking conventional k-space encoding using gradients. Thus, the clinical application of BASE-SLIM should allow reliable assessment of tissue-type or region specific concentrations of metabolites and biomarkers in health and disease.
Highlights.
Accurate spatial localization of arbitrary shaped VOI is possible using BASE-SLIM.
Robust acquisition of tissue-type specific MR spectra is achieved using BASE-SLIM.
Single-shot localization is achievable without k-space encoding using BASE-SLIM.
BASE-SLIM requires fewer spatial encoding steps than FFT based MRSI.
1H MR spectra of gray and white matter in the human brain are reliably measured.
Acknowledgments
This study was in part supported in part by a grant from the NIH (8R01 EB00315 to IYC). The Hoglund Brain Imaging Center is partly supported by grants from the NIH (1S10 RR029577) and the Hoglund Family Foundation.
Abbreviations
- B0
static magnetic field
- B1
radiofrequency magnetic field
- B1+
transmit radiofrequency magnetic field
- BASE-SLIM
B0-adjusted and sensitivity-encoded spectral localization by imaging
- BSLIM
spectral localization by imaging with explicit B0 field inhomogeneity compensation
- Cr
creatine
- CSF
cerebrospinal fluid
- FFT
fast Fourier transform
- FID
free induction decay
- FOV
field of view
- FWHM
full width at half maximum
- GABA
gamma-aminobutyric acid
- Gln
glutamine
- Glu
glutamate
- GM
gray matter
- GSH
glutathione
- GSLIM
generalized spectral localization by imaging
- ICBM
international consortium for brain mapping
- LCModel
linear combination of model in vitro spectra
- mI
myo-inositol
- MPRAGE
magnetization prepared rapid acquisition of gradient echo
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance spectroscopic imaging
- NAA
N-acetylaspartate
- NL-CSI
natural linewidth chemical shift imaging
- RMS
root mean square
- Semi-LASER
semi-adiabatic localization by adiabatic selective refocusing
- SENSE
sensitivity encoding
- SLIM
spectral localization by imaging
- SLOOP
spectral localization with optimal point spread function
- SNR
signal-to-noise ratio
- SPLASH
spectral localization achieved by sensitivity heterogeneity
- SPREAD
spectral resolution amelioration by deconvolution
- SVS
single voxel spectroscopy
- T1
longitudinal relaxation time
- T2
transverse relaxation time
- TI
inversion time
- tCho
total choline
- TE
echo time
- TR
repetition time
- VAPOR
a combination of variable power RF pulses with optimized relaxation delays
- VOI
volume of interest
- WM
white matter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An L, Warach S, Shen J. Spectral localization by imaging using multielement receiver coils. Magn Reson Med. 2011;66:1–10. doi: 10.1002/mrm.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bashir A, Yablonskiy DA. Natural linewidth chemical shift imaging (NL-CSI) Magn Reson Med. 2006;56:7–18. doi: 10.1002/mrm.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch R, Peet AC, Arvanitis TN, Wilson M. Sensitivity encoding for fast 1H MR spectroscopic imaging water reference acquisition. Magn Reson Med. 2015;73:2081–2086. doi: 10.1002/mrm.25355. [DOI] [PubMed] [Google Scholar]
- Bydder M, Larkman DJ, Hajnal JV. Combination of signals from array coils using image-based estimation of coil sensitivity profiles. Magn Reson Med. 2002;47:539–548. doi: 10.1002/mrm.10092. [DOI] [PubMed] [Google Scholar]
- Choi IY, Lee SP, Merkle H, Shen J. In vivo detection of gray and white matter differences in GABA concentration in the human brain. Neuroimage. 2006;33:85–93. doi: 10.1016/j.neuroimage.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Ding XQ, Maudsley AA, Sabati M, Sheriff S, Dellani PR, Lanfermann H. Reproducibility and reliability of short-TE whole-brain MR spectroscopic imaging of human brain at 3T. Magn Reson Med. 2015;73:921–928. doi: 10.1002/mrm.25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z. Proton MRS and MRSI of the brain without water suppression. Prog Nucl Magn Reson Spectrosc. 2015;86–87:65–79. doi: 10.1016/j.pnmrs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Dong Z, Peterson BS. Spectral resolution amelioration by deconvolution (SPREAD) in MR spectroscopic imaging. J Magn Reson Imaging. 2009;29:1395–1405. doi: 10.1002/jmri.21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med Image Comput Comput Assist Interv. 2006;9:58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- Hu X, Levin DN, Lauterbur PC, Spraggins T. SLIM: spectral localization by imaging. Magn Reson Med. 1988;8:314–322. doi: 10.1002/mrm.1910080308. [DOI] [PubMed] [Google Scholar]
- Hu X, Wu Z. SLIM revisited. IEEE Trans Med Imaging. 1993;12:583–587. doi: 10.1109/42.241887. [DOI] [PubMed] [Google Scholar]
- Khalidov I, Van De Ville D, Jacob M, Lazeyras F, Unser M. BSLIM: spectral localization by imaging with explicit B0 field inhomogeneity compensation. IEEE Trans Med Imaging. 2007;26:990–1000. doi: 10.1109/TMI.2007.897385. [DOI] [PubMed] [Google Scholar]
- Liang ZP, Lauterbur PC. A generalized series approach to MR spectroscopic imaging. IEEE Trans Med Imaging. 1991;10:132–137. doi: 10.1109/42.79470. [DOI] [PubMed] [Google Scholar]
- Liang ZP, Lauterbur PC. A Theoretical-Analysis of the Slim Technique. Journal of Magnetic Resonance Series B. 1993;102:54–60. [Google Scholar]
- Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MA, Woermann FG, Barker GJ, Duncan JS. Quantitative analysis of short echo time (1)H-MRSI of cerebral gray and white matter. Magn Reson Med. 2000;44:401–411. doi: 10.1002/1522-2594(200009)44:3<401::aid-mrm10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. Nmr in Biomedicine. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- Schuff N, Ezekiel F, Gamst AC, Amend DL, Capizzano AA, Maudsley AA, Weiner MW. Region and tissue differences of metabolites in normally aged brain using multislice 1H magnetic resonance spectroscopic imaging. Magn Reson Med. 2001;45:899–907. doi: 10.1002/mrm.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- SPM8. SPM8. Wellcome Department of Imaging Neuroscience; London, United Kingdom: 2009. [Google Scholar]
- Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41:649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- von Kienlin M, Beer M, Greiser A, Hahn D, Harre K, Kostler H, Landschutz W, Pabst T, Sandstede J, Neubauer S. Advances in human cardiac P-31-MR spectroscopy: SLOOP and clinical applications. Journal of Magnetic Resonance Imaging. 2001;13:521–527. doi: 10.1002/jmri.1074. [DOI] [PubMed] [Google Scholar]
- Vonkienlin M, Mejia R. Spectral Localization with Optimal Pointspread Function. Journal of Magnetic Resonance. 1991;94:268–287. [Google Scholar]
- Weber-Fahr W, Ende G, Braus DF, Bachert P, Soher BJ, Henn FA, Buchel C. A fully automated method for tissue segmentation and CSF-correction of proton MRSI metabolites corroborates abnormal hippocampal NAA in schizophrenia. Neuroimage. 2002;16:49–60. doi: 10.1006/nimg.2002.1057. [DOI] [PubMed] [Google Scholar]
- Wright SM, Wald LL. Theory and application of array coils in MR spectroscopy. Nmr in Biomedicine. 1997;10:394–410. doi: 10.1002/(sici)1099-1492(199712)10:8<394::aid-nbm494>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]