Abstract
The decision by embryonic ectoderm to give rise to epidermal versus neural derivatives is the result of signaling events during blastula and gastrula stages. However, there also is evidence in Xenopus that cleavage stage blastomeres contain maternally derived molecules that bias them toward a neural fate. We used a blastomere explant culture assay to test whether maternally deposited transcription factors bias 16-cell blastomere precursors of epidermal or neural ectoderm to express early zygotic neural genes in the absence of gastrulation interactions or exogenously supplied signaling factors. We found that Foxd4l1, Zic2, Gmnn and Sox11 each induced explants made from ventral, epidermis-producing blastomeres to express early neural genes, and that at least some of the Foxd4l1 and Zic2 activity is required at cleavage stages. Similarly, providing extra Foxd4l1 or Zic2 to explants made from dorsal, neural plate-producing blastomeres significantly increased expression of early neural genes, whereas knocking down either significantly reduced them. These results show that maternally delivered transcription factors bias cleavage stage blastomeres to a neural fate. We demonstrate that mouse and human homologues of Foxd4l1 have similar functional domains compared to the frog protein, as well as conserved transcriptional activities when expressed in Xenopus embryos and blastomere explants.
Keywords: Foxd5, Foxd4, Foxd4l1.1, Foxd4l1, Geminin, Sox11, Zic2, Sox2, Zic1, neural induction
Introduction
It has long been known that the vertebrate nervous system develops during gastrulation in response to dorsal mesoderm (the “Organizer”; Spemann and Mangold, 1923-reprinted in 2001) that secretes inhibitors of the BMP and Wnt pathways (Khokha et al., 2005; De Robertis, 2006; Levine and Brivanlou, 2007; Wills et al., 2010; Pinho et al., 2011; Itoh and Sokol, 2014; Pera et al., 2014). As a result, the newly induced neural ectoderm expresses numerous transcription factors that prevent it from becoming epidermis (reviewed in Sasai, 1998; Rogers et al., 2009; Moody et al., 2013; Lee et al., 2014). However, several observations suggest that earlier interactions also are required. For example, dorsal cells in the blastula express transcription factors and signaling factors that are required for both the Organizer and neural ectoderm to form in the gastrula (Kuroda et al., 2004; Reversade and De Robertis, 2005; Ishibashi et al., 2008; Suduo et al., 2012; Klein and Moody, 2015).
An important mechanism that influences cell fate prior to gastrulation is the region-specific inheritance of maternally synthesized mRNAs and proteins (Davidson, 1990; Sullivan et al., 1998; King et al., 2005; Heasman, 2006; White and Heasman, 2008; Abrams and Mullins, 2009; Cuyendall and Houston, 2010). There is abundant evidence in Xenopus that the ectodermal germ layer forms in part due to animal pole-enriched maternal transcripts that both promote ectoderm and oppose mesoderm germ layer formation (Sasai, 1998; Zhang et al., 2003; Zhang et al., 2004; Dupont et al., 2005; Zhang and Klymkowsky, 2007; Cha et al., 2012; Xu et al., 2012; Bates et al., 2013). Interestingly, several of the maternal transcripts enriched in the animal hemisphere are later expressed in the nascent neural ectoderm, and are required at that time for nervous system formation. It is not known whether these transcripts also function prior to gastrulation to influence cells to become part of the nervous system. Some embryological studies in amphibians support this notion, however. For example, deletions of animal blastomeres cause disruptions of the neural tube (Kageura and Yamana, 1984; Gallagher et al., 1991; Kageura, 1995), transplantation of animal blastomeres that normally give rise to parts of the nervous system maintain their neural fate in ectopic locations (Kageura and Yamana, 1986; Kageura, 1990; Gallagher et al., 1991), and in explant culture (Gallagher et al., 1991; Hainski and Moody, 1992; Pandur et al., 2002). In fact, transfer of total RNA from neural precursor blastomeres can cause epidermis precursor blastomeres to contribute to ectopic neural tubes (Hainski and Moody, 1992). Although several screens have identified animal hemisphere specific maternal transcripts (e.g., Cha et al., 2012; Bates et al., 2013; Grant et al., 2014), they have not identified the specific gene product(s) responsible for instructing cleavage stage blastomeres to autonomously express a neural fate.
This study examined whether epidermal precursor blastomeres can be made to express neural plate genes in the absence of germ layer interactions by introducing four candidate transcription factors. These factors were selected because each is known to promote a neural ectodermal fate as well as oppose an epidermal fate at gastrulation, and their maternal transcripts are found in animal blastomeres. Foxd4l1 (aka Foxd5) is required for the expression of eleven other neural transcription factors and directly induces three of them (geminin [gmnn], zic2, sox11) when ectopically expressed in an epidermal lineage (Yan et al., 2009). At gastrula stages, these four transcription factors cooperatively maintain the neural ectoderm in an immature, proliferative state, regulate the transition to neural plate stem cells, and delay neuronal differentiation (Sullivan et al., 2001; Yan et al., 2009; reviewed in Moody et al., 2013). Since each of them also is expressed as a maternal transcript (Kroll et al., 1998; Nakata et al., 1998; Koyano et al., 1999; Sullivan et al., 2001; Hyodo-Miura et al., 2002; Yanai et al., 2011; Peshkin et al., 2015; www.xenbase.org), we took advantage of the detailed Xenopus 16-cell fate map (Moody, 1987) to misexpress them in the blastomere precursors of either epidermis or neural plate, culture the blastomeres as explants in the absence of interactions from neighboring cells or exogenous growth factors (Fig. 1), and test for early (i.e., zygotic) neural gene expression. We found that each of these four transcripts ectopically induced early neural genes in ventral epidermal progenitor explants in the absence of increased mesoderm gene expression. Second, increased levels of either Foxd4l1 or Zic2 in dorsal neural plate progenitor explants significantly increased their expression of early neural and dorsal mesoderm genes above their normal low levels. Conversely, knocking down endogenous levels of either Foxd4l1 or Zic2 reduced early neural gene expression, indicating that they are required for the endogenous capacity of dorsal-animal blastomeres to express neural plate genes. Finally, we demonstrate a functional conservation of Foxd4/Foxd4-like genes across tetrapod species by targeting the expression of either the single mouse homologue (mFoxd4) or two human homologues (hFOXD4, hFOXD4L1) in frog embryos or in blastomere explants. Together, these experiments demonstrate that some animal-enriched maternally delivered transcription factors play important roles in biasing cleavage stage blastomeres towards a neural fate. They further show that the ability of Foxd4/Foxd4-like genes to promote early neural ectoderm is conserved across tetrapods, including human.
Figure 1.

Explants can be made from cleavage stage blastomeres of known fates. (A) When 16-cell Xenopus embryos cleave in a regular pattern, the dorsal midline blastomeres (D11) will faithfully give rise to neural plate (np) and the ventral midline blastomeres (V11) will give rise to epidermis (epi). (B) The same embryo has gone through one cell division, i.e., is at the 32-cell stage. (C) The four daughters (outlined in B) of the midline D11 blastomeres (labeled in A) have been manually dissected and transferred to explant culture.
Results
Are maternal neural transcription factor mRNAs differentially distributed in animal blastomeres?
Transcripts encoding four neural transcription factors (nTFs) (foxd4l1.1a [hereafter called foxd4l1], zic2, gmnn, sox11) are reported to be maternally deposited in the egg and detectable in the animal hemisphere, but not vegetal hemisphere, prior to the onset of zygotic transcription at the mid-blastula transition (MBT) (Hiraoka et al., 1997; Brewster et al., 1998; Kroll et al., 1998; Sullivan et al., 2001; Houston and Wylie, 2005; Fujimi et al., 2012; Yanai et al., 2011; Peshkin et al., 2015; www.xenbase.org). We first assessed whether they are differentially distributed in the animal hemisphere of the 16-cell embryo by in situ hybridization (ISH). For each of the four nTFs, there was no detectable difference in ISH staining in the dorsal-animal neural plate progenitors (D11) versus the ventral-animal epidermis progenitors (V11) (Fig. 2). To detect potential quantitative differences, we compared the levels of each transcript in dissected D11 and V11 blastomeres by quantitative real-time PCR. Consistent with the ISH data, and with published RNA-seq data of 8-cell blastomeres (De Domenico et al., 2015), the dorsal/ventral ratios were not significantly different from 1.0 (range = 0.85-1.02; p>0.05, t-test). These results indicate that the neural versus epidermal fates of these two blastomeres is not due to differential inheritance of maternal nTF mRNAs.
Figure 2.

There is no detectable dorsal-ventral difference in mRNA localization of four neural transcription factors at cleavage stages. In situ hybridization detects the indicated mRNAs at the same relative intensities in the dorsal (top) and ventral (bottom) 16- to 32-cell animal blastomeres, indicating a lack of localization. Not shown: 1) there is no signal in vegetal hemisphere blastomeres; 2) similar results were observed when animal cap fragments were processed for ISH to ensure probe penetrance in the yolk-ladened cells; and 3) sense probes did not shown signal in either animal or vegetal hemispheres.
Can maternal nTFs cause epidermal precursors to express early zygotic neural genes?
To determine whether increasing the level of nTF protein in ventral blastomeres can convert the fate of these cells from epidermis to neural, we injected nTF mRNAs that can be immediately translated (due to 5′ and 3′ UTR sequences included in the pCS2+ vector), and then explanted blastomere pairs to develop in the absence of signaling from other cells or exogenous growth factors (Fig. 1). When siblings reached nascent neural ectoderm stages (st 11.5-12.5), explants were fixed and processed for the expression of two genes expressed only after the MBT in the nascent neural ectoderm and later in the definitive neural plate (sox2, zic1; Kuo et al. 1998; Mizuseki et al., 1998), or epidermis (K81 epidermal-specific cytokeratin; Jonas et al., 1985). Uninjected V11 explants never expressed sox2, rarely expressed zic1 but expressed K81 in every case (Fig. 3A). Foxd4l1 and Zic2 induced both early neural genes at significantly increased frequencies, whereas Gmnn preferentially induced sox2, and Sox11 only induced zic1 above controls (Fig. 3A, B). Foxd4l1 and Zic2 also reduced the frequency of explants that expressed K81, whereas Gmnn and Sox11 did not (Fig. 3A). However, in nearly every case for all four nTFs, the expression domain of K81 in the explant did not overlap with the nβGal lineage tracer, indicating that the original epidermal fate was preserved only in patches of cells that did not highly express the injected nTF (Fig. 3C).
Figure 3.

Neural transcription factors ectopically induce early neural genes in ventral-animal (V11) blastomere explants. (A) The percentage of V11 explants scored positive for early neural (sox2, zic1) or epidermis (K81) mRNAs after ectopic expression of one of the four neural transcription factors (Foxd4l1, Zic2, Sox11, Gmnn). The number above each bar indicates the number of explants scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to uninjected, control V11 explants. (B) Examples of explants microinjected with each neural transcription factor and assayed for each early neural gene. For sox2, all Foxd4l1- and all Gmnn-expressing explants are positive. For other panels, arrows indicate the positively stained explants. Pink cells are labeled with the nβGal lineage tracer. (C) Examples of explants assayed for an epidermis gene (K81). In uninjected, control V11 explants, K81 is expressed in all surface cells of the explant, whereas in neural transcription factor-expressing explants (Zic2, Gmnn), K81 expression (arrows) is only in nβGal-negative cells. (D) The percentage of V11 explants scored positive for mesoderm genes (bra, chd) after ectopic expression of one of the four neural transcription factors. The number above each bar indicates the number of explants scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to uninjected, control V11 explants. (E) Examples of explants microinjected with each neural transcription factor and assayed for each mesoderm gene. Arrows indicate the positively stained explants.
These results demonstrate that increasing the protein levels of each of the four nTFs in isolated blastomeres causes ventral-animal blastomeres to change fate from epidermal to neural. To determine whether this is mediated via the ectopic induction of dorsal mesoderm genes, we also assayed explants for pan-mesoderm (bra) or dorsal-specific mesoderm (chd) genes. Uninjected, control V11 explants express bra at low frequency and do not express chd (Fig. 3D, E), as expected from the 16-cell fate map (Moody, 1987). Foxd4l1, Zic2 and Sox11 significantly reduced bra expression, and did not induce ectopic chd expression; Gmnn did not significantly alter either mesoderm gene (Fig. 3D, E). These results indicate that the ability of the nTFs to induce early neural genes in epidermal precursors is direct rather than the result of ectopic dorsal mesoderm formation. This conclusion is consistent with reports that each of the nTFs has been associated with either an anti-BMP or anti-Wnt activity (Kroll et al., 1998; Yan et al., 2009; Yan et al., 2010; Pourebrahim et al., 2011; Fujimi et al., 2012).
Are maternal nTFs required for neural precursor expression of early neural genes?
Previous work showed that explants of dorsal-animal blastomeres have an intrinsic ability to differentiate into dorsal axial structures (Gallagher et al., 1991). However, when those experiments were published the only molecular marker available was an antibody that recognized the differentiated notochord; there was no information regarding the expression of early zygotic neural genes. Therefore, we tested whether increased levels of Foxd4l1 or Zic2 increases the frequency of early neural gene expression in D11 explants. We focused on these two nTFs because they had similar effects on the ectodermal and mesodermal genes in the V11 explant assays. Uninjected, control D11 explants expressed sox2 and zic1 at moderate frequencies (Fig. 4A). Increasing Foxd4l1 or Zic2 levels significantly increased the frequency of sox2 expression, but only Foxd4l1 significantly increased zic1 (Fig. 4A). Both Foxd4l1 and Zic2 significantly reduced the frequency of K81 expression; in every case the K81-positive cells in the explant had minimal overlap with the nβGal lineage tracer (Fig. 4B), indicating that even in explants scored positive, the nTFs repressed K81 expression. Interestingly, both bra and chd were expressed at higher frequencies in Foxd4l1- and Zic2-injected explants (Fig. 4C, D), in contrast to their effects in V11 explants. This increase in mesodermal gene expression is not unexpected because: 1) the D11 blastomere also is a major precursor of the Organizer and the notochord (Moody, 1987; Bauer et al., 1994); 2) previous work showed D11 explants autonomously differentiate into notochord (Gallagher et al., 1991); and 3) dorsal-animal blastomeres have a pre-MBT competence to respond to mesoderm inductive signals (Sokol and Melton, 1991; Kinoshita et al., 1993; Ding et al., 1998). Nonetheless, the frequency of D11 explants expressing early neural genes was higher than those expressing mesoderm genes, indicating that at least some explants express early neural genes independent of mesoderm.
Figure 4.

Neural transcription factors increase the frequency of early neural gene expression in dorsal-animal (D11) blastomere explants. (A) The percentage of D11 explants scored positive for early neural (sox2, zic1) or epidermis (K81) gene expression after increasing Foxd4l1 or Zic2 levels by microinjecting additional mRNA, or after decreasing endogenous levels by microinjecting translation blocking MOs (FoxMOs, ZicMOs). The number above each bar indicates the number of explants scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to uninjected, control D11 explants. (B) Examples of explants assayed for sox2 or K81. In uninjected, control D11 explants, sox2 is autonomously expressed in ∼40% of explants. In either Foxd4l1- or Zic2-expressing D11 explants this frequency increases to nearly every explant. Conversely, in MO-injected D11 explants the frequency is reduced significantly below control. Arrows point out positively stained explants. In both Foxd4l1- and Zic2-expressing explants, K81 is expressed (arrows) only in nβGal-negative cells. (C) The percentage of D11 explants scored positive for mesoderm genes (bra, chd) after increasing Foxd4l1 or Zic2 levels by microinjecting additional mRNA. The number above each bar indicates the number of explants scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to uninjected, control D11 explants. (D) Examples of explants assayed for bra or chd. Arrows point out positively stained explants.
To determine whether the intrinsic expression of sox2 and/or zic1 in D11 explants requires endogenous Foxd4l1 or Zic2, blastomeres were injected with translation-blocking antisense morpholino oligonucleotides (MOs) prior to explant culture; the specificity and efficacy of the MOs used have previously been published (Sullivan et al., 2001; Houston and Wylie, 2005; Fujimi et al., 2012; Supplemental Fig. 1A-C). Knock-down of endogenous Foxd4l1 (Foxd4l1.1a, Foxd4l1.1b and Foxd4l1.2) using two MOs significantly reduced the frequency of sox2 and zic1 expression below control explants (Fig. 4A, B); this reduction could be reversed by co-injecting an MO-insensitive mRNA (Supplemental Fig. 1D). While knock-down of endogenous Zic2 also reduced the frequency of sox2- and zic1-expressing explants, these effects did not reach statistical significance (Fig. 4A, B).
Is the ability of Foxd4l1 or Zic2 to induce early neural gene expression in epidermal precursors limited to cleavage stages?
Although Foxd4l1 and Zic2 ectopically induced early neural gene expression in the V11 epidermal precursors, their effects may have occurred at cleavage (maternal) and/or blastula (zygotic) stages because these exogenously supplied mRNAs are translated soon after mRNA microinjection and are stable due to the 5′ and 3′UTRs supplied by the pCS2+ vector. To address the timing of their effects, we made constructs that are fused to the human glucocorticoid receptor (hGR). Although the nTF-hGR fusion transcript is immediately translated, the protein is sequestered in the cytoplasm by heat shock proteins and cannot access the nucleus until the ligand, in this case exogenously applied dexamethasone (Dex), is presented (Mattioni et al., 1994; Kolm and Sive, 1995). As shown in Table 1, Dex treatment alone at blastula stages (st 8) does not induce the expression of either sox2 or zic1 in V11 explants. Blastula activation of Foxd4l1-hGR only slightly induced expression of either early neural gene, in significant contrast to the wild-type, early-expressed construct. Blastula activation of Zic2 also was significantly less effective at inducing zic1 compared to the wild type, early-expressed construct, whereas its induction of sox2 was not different. These results indicate that cleavage stage activity of Foxd4l1 and Zic2 are required for at least some of their inductive activity of early, zygotic neural genes.
Table 1. Frequency of zygotic neural plate gene expression in V11 explants.
| sox2 | zic1 | |
|---|---|---|
| uninjected | 0% (65) | 6% (50) |
| uninjected + Dex st 8 | 0% (50) | 0% (33) |
| Foxd4l1-wild type | 90% (40) | 50.8% (61) |
| Foxd4l1-hGR + Dex st 8 | 12.5% (48)* | 9.4% (32)* |
| Foxd4l1-hGR, no Dex | 0% (27) | 0% (21) |
| Zic2-wild type | 26.3% (38) | 40.3% (72) |
| Zic2-hGR + Dex st 8 | 43.6% (55) | 6.3% (32)* |
| Zic2-hGR, No Dex | 0% (19) | 2.9% (34) |
Legend: The percentage of V11 explants expressing either sox2 or zic1 at early neural plate stages (11.5-12.5). The number of explants analyzed is shown in parentheses.
Asterisk indicates a significant difference from wild-type frequency (p<0.05, Chi-square statistic).
Do mammalian homologues have the same functions in Xenopus assays?
To determine whether frog and mammalian Foxd4/Foxd4-like proteins are similar, we compared the amino acid sequence of Xenopus and human homologues by CLUSTAL analysis (Supplemental Fig. 2). The two Xenopus tropicalis proteins (Foxd4l1.1, Foxd4l1.2), the three Xenopus laevis proteins (Foxd4l1.1a, Foxd4l1.1b, Foxd4l1.2) and the two human proteins (hFOXD4; hFOXD4L1) all contain an N-terminal “acidic blob” region (Fig. 5A). We previously showed in frog that this domain is required for transcriptional activation of target genes (Neilson et al., 2012; Klein et al., 2013). The five Xenopus proteins and hFOXD4L1 also contain an Eh-1 motif (Fig. 5B) that can bind to Groucho-related (Grg/TLE) co-repressor proteins (Yaklichkin et al., 2007). We previously showed in frog that this motif is partially responsible for transcriptional repression of target genes (Neilson et al., 2012; Klein et al., 2013). However, there is a single nucleotide deletion in the hFOXD4 gene that introduces a frame shift that completely eliminates the Eh-1 motif (Fig. 5B). To test whether these differences in Eh-1 motifs in the human proteins affect their ability to bind Groucho-related proteins, we assayed the binding of Xenopus Grg4 to frog and mammalian Foxd4/Foxd4-like proteins by co-immunoprecipitation (Fig. 5C). While Xenopus laevis Foxd4l1 and mFoxd4 strongly bind with Grg4, the human homologues only bind weakly. However, the higher avidity of hFOXD4L1 compared to that of hFOXD4 is consistent with the latter's loss of the Eh-1 motif.
Figure 5.
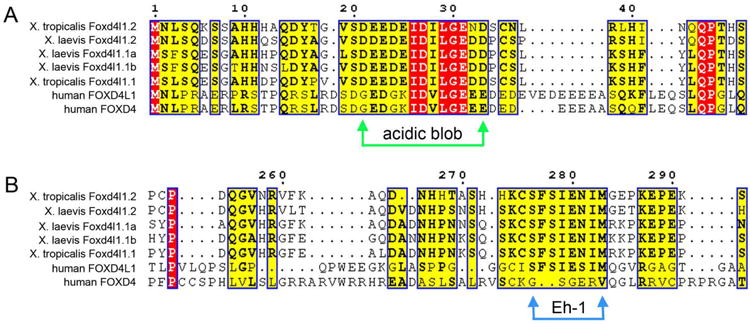
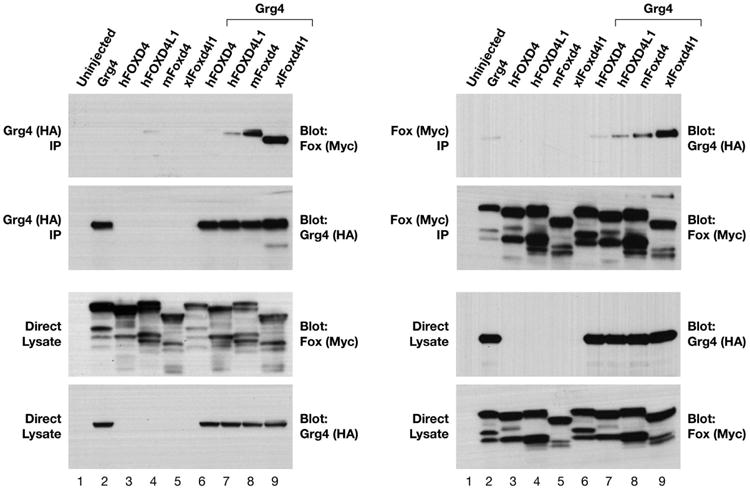
CLUSTAL analysis displayed in ESpript comparing frog and human Foxd4/Foxd4-like protein functional domains. (A) The three Xenopus laevis proteins (Foxd4l1.2, Foxd4l1.1a, Foxd4l1.1b), the two Xenopus tropicalis proteins (Foxd4l1.2, Foxd4l1.1) and the two human proteins (FOXD4L1, FOXD4) each contain an “acidic blob” domain in the N-terminal portion of the protein (between green arrows). Red blocks indicate identical amino acids (aa), yellow blocks indicate conserved aa, and bold aa within a yellow block indicate aa of the same type (e.g., acidic, hydrophobic, etc.). (B) The Eh-1 domain (between blue arrows), which binds the transcriptional repressor Groucho/Grg/TLE, is conserved in all of the proteins except human FOXD4. (C) Xenopus Grg4 binds to the Foxd4/Foxd4-like proteins from different species. Myc-tagged versions of Foxd4/Foxd4-like proteins from Xenopus, mouse and human were expressed in Xenopus oocytes either alone or along with HA-tagged wild-type Xenopus Grg4. Co-immunoprecipitation (IP) and Western blot (WB) analyses of oocyte lysates expressing HA- and Myc-tagged constructs are indicated (top left and right panels). All four constructs bind with Grg4 with the following avidities XlFoxd4l1>mFoxd4 >hFOXD4L1>hFOXD4. The control panels (2nd from top) show that the IPs contain similar levels of Grg4 (left) or Foxd4/Foxd4-like proteins (right), as do the direct lysates (bottom 2 panels on both left and right).
Since the frog and human Foxd4/Foxd4-like proteins contain similar functional domains but have different abilities to bind to frog Grg4, we tested whether the human proteins can functionally substitute for the Xenopus proteins in intact embryos. Our previous work showed that when Xenopus Foxd4l1 is ectopically expressed in the V11 lineage, gmnn, zic2 and sox11 are ectopically induced in non-neural ectoderm at high frequency (Yan et al., 2009). hFOXD4 and hFOXD4l1 each ectopically induced gmnn and zic2, but did not ectopically induce sox11 (Fig. 6A). Similarly, we previously reported that when additional Xenopus Foxd4l1 is expressed in the D11 lineage, the neural plate expression of gmnn and zic2 is up-regulated, whereas that of sox2, sox11 and zic1 is down-regulated (Yan et al., 2009). hFOXD4 and hFOXD4l1 also each up-regulated gmnn and zic2 (Fig. 6B), and down-regulated sox2, sox11 and zic1 (Fig. 6C). Thus, in whole embryo assays the human homologues have similar effects compared to the endogenous frog proteins; in another study we show the same for mFoxd4 (Sherman et al., 2016). However, in several of these assays the human proteins are not as effective as the Xenopus proteins (asterisks in Fig. 6), suggesting that they may not bind well to frog co-factors. Interestingly, the lack of an Eh-1 motif in hFOXD4 cannot be the reason for these differences since both human proteins show less robust effects on downstream gene expression.
Figure 6.
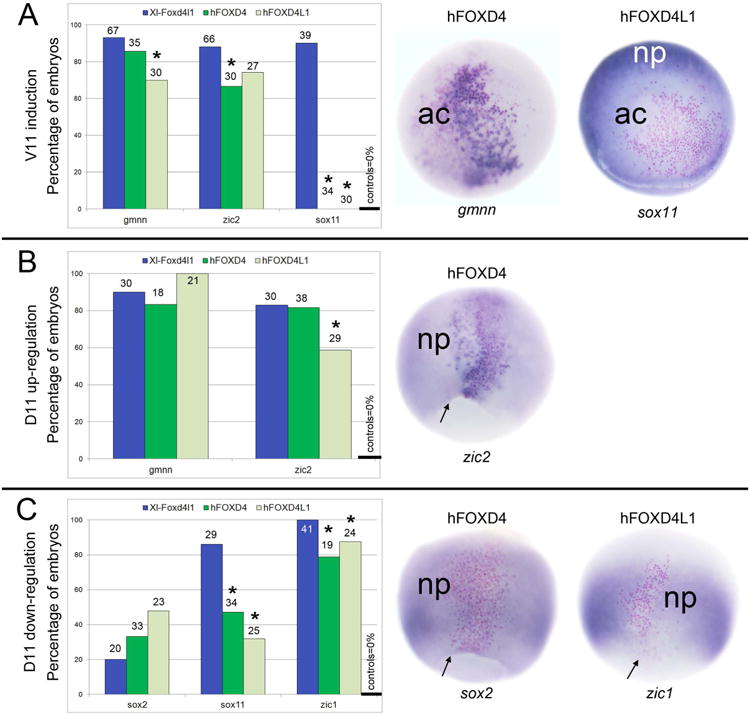
Similar to Xenopus Foxd4l1, human FOXD4 and FOXD4L1 can affect the expression of early neural genes when mis-expressed in Xenopus embryos. (A) Left: Ectopic expression of Xenopus Foxd4l1, human FOXD4 and human FOXD4L1 in the V11 lineage results in a high frequency of ectopic expression of gmnn and zic2. However, unlike Xenopus Foxd4l1, neither human protein ectopically induces sox11. In control embryos none of these genes are expressed in the V11 lineage (far right bar). The number above each bar indicates the number of embryos scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to ectopic expression of Xenopus Foxd4l1. Middle: an example of ectopic induction of gmnn (dark blue stain) in the animal cap ectoderm (ac). Right: an example of lack of induction of sox11 in the animal cap ectoderm (ac); endogenous sox11 expression can be seen in the neural plate (np). Red dots indicate the lineage-labeled cells expressing the microinjected mRNAs. Embryos are animal cap views with dorsal to the top. (B) Left: Expressing Xenopus Foxd4l1, human FOXD4 or human FOXD4L1 in the D11 lineage up-regulates gmnn and zic2 within that lineage at high frequencies. This change in gene expression is never seen in control embryos (far right bar). The number above each bar indicates the number of embryos scored. Asterisk (*) indicates a significant difference (p<0.05, Chi-square) compared to ectopic expression of Xenopus Foxd4l1. Middle: an example of up-regulation of zic2 (dark blue stain) within the lineage-labeled clone (red dots) compared to endogenous levels in the neural plate (np). Embryo is a dorsal view with animal to the top and blastopore lip marked by arrow. (C) Left: Expressing Xenopus Foxd4l1, human FOXD4 or human FOXD4L1 in the D11 lineage down-regulates sox2, sox11 and zic1 within that lineage; for sox11 and zic1 the frequencies are significantly less than Xenopus Foxd4l1 (asterisks; p<0.05, Chi-square). This change in gene expression is never seen in control embryos (far right bar). The number above each bar indicates the number of embryos scored. Middle: an example of reduced sox2 expression in the lineage-labeled clone (red dots) compared to endogenous expression in the adjacent neural plate (np). Right: an example of reduced zic1 expression in the lineage-labeled clone (red dots) compared to endogenous expression in the adjacent neural plate (np). Embryos are dorsal views with animal to the top and blastopore lips marked by arrows.
We also tested the activity of the single mouse homologue (mFoxd4) and the two human homologues in blastomere explant assays. In V11 explants, each of the three mammalian homologues increased zygotic sox2 expression over controls, but they were significantly less effective compared to the frog protein (Fig. 7A). Only mFoxd4 significantly increased zic1 expression in V11 explants, albeit less frequently than the frog protein. In D11 explants, mFoxd4 and hFOXD4 significantly increased sox2 expression, and none of the mammalian proteins significantly increased zic1 expression (Fig. 7B). In each case the frequencies were significantly smaller compared to the frog protein. These results indicate that while the mammalian proteins trend towards similar activities as the frog protein in blastomere explants, they are less effective, particularly for zic1.
Figure 7.

Mammalian Foxd4/Foxd4-like homologues can affect the expression of early neural genes in Xenopus blastomere explants. (A) The percentage of V11 explants that express early neural genes (sox2, zic1). The mammalian homologues cause ectopic expression at frequencies significantly greater than uninjected controls (*, p<0.05, Chi-square), but are not as effective as Xenopus protein (# indicates a significant difference [p<0.05, Chi-square] compared to Xenopus Foxd4l1-expressing explants). (B) The percentage of D11 explants that express early neural genes (sox2, zic1). The mFoxd4 and hFOXD4 increase sox2 expression at frequencies significantly greater than uninjected controls (*), but are not as effective as Xenopus protein (# indicates a significant difference [p<0.05, Chi-square] compared to Xenopus Foxd4l1-expressing explants).
Discussion
The differential distribution of maternal mRNAs and proteins can contribute to cell fate decisions in a variety of animals. In contrast, there is an abundance of evidence that the decision to give rise epidermal versus neural ectoderm is the result of signaling events (Wnt, BMP, FGF) that take place during blastula and gastrula stages (Khokha et al., 2005; De Robertis, 2006; Levine and Brivanlou, 2007; Wills et al., 2010; Pinho et al., 2011; Itoh and Sokol, 2014; Pera et al., 2014). Nonetheless, several studies indicate that in Xenopus the cleavage stage precursors of the neural ectoderm contain maternally derived molecules that allow them to autonomously present certain aspects of a neural fate prior to or independent of inductive signaling (e.g., Kageura, 1990; Gallagher et al., 1991; Hainski and Moody, 1992; Darras et al., 1997; Ding et al., 1998). Herein we provide further evidence for this autonomous proclivity towards a neural fate, which we call neural “bias”, by using a blastomere explant culture assay that assesses early zygotic neural gene expression in the absence of normal germ layer interactions and exogenous signaling factors. We show that Foxd4l1, Zic2, Gmnn and Sox11 can induce explants made from ventral, epidermis-producing blastomeres to express two early neural genes in the absence of mesoderm, and that at least some Foxd4l1 and Zic2 activity is required prior to blastula stages. Similarly, increased levels of either Foxd4l1 or Zic2 in explants made from dorsal, neural plate-producing blastomeres significantly increased the frequency at which early neural genes are expressed in dorsal explants, whereas, knock-down of either significantly reduces that frequency. These results indicate that Foxd4l1 or Zic2 are required for the endogenous capacity of dorsal-animal blastomeres to express early neural genes. This differential dorsal versus ventral competence to autonomously express early neural genes is similar to previous reports that Xenopus animal blastomeres are differentially competent to express dorsal versus ventral mesoderm (Sokol and Melton, 1991; Kinoshita et al., 1993; Ding et al., 1998).
In combination with several previous reports, these data suggest that neural cell fate is acquired in a series of steps (Fig. 8). At cleavage stages, dorsal-animal blastomeres contain molecules, including but not necessarily limited to nTFs, that bias them to a neural cell fate. Although we do not yet know the molecular basis for this bias, nTFs could repress BMP signaling, enhance maternal β-catenin signaling required for Organizer formation, or prime the chromatin for later transcriptional events. At blastula stages, maternal β-catenin becomes nuclear in the dorsal marginal cells that are the descendants of the dorsal-animal blastomeres (Schneider et al., 1996; Medina et al., 1997); in these cells β-catenin directly activates Siamois/Twin transcription factors (Carnac et al., 1996). These in turn directly activate the zygotic expression of some some nTFs (foxd4l1, gmnn, zic2) in these same cells (Klein and Moody, 2015). Finally, during gastrulation, signaling factors that also are directly activated by Siamois/Twin in the Organizer mesoderm diffuse towards the anterior pole to maintain and expand nTF expression in the nascent neural ectoderm (Laurent et al., 1997; Kessler, 1997; Kodjabachian and Lemaire, 2001; Bae et al., 2011; Reid et al., 2012). The nTFs then up-regulate early neural genes (e.g., sox2, zic1), which in turn act upstream of neural differentiation factors (e.g., irx1, ngnr1) and markers of committed neurons (e.g., tubb2) (Sullivan et al., 2001; Yan et al., 2009). This scheme poses that acquiring a committed neural cell fate relies upon sequential transcriptional and signaling events that over developmental time eventually produce differentiated neurons and glia. Our data indicate that the maternal nTFs promote an early neural state, but we do not know if they promote further neural differentiation because the zygotic neural markers we used do not define committed neurons (Wills et al., 2010). However, previous RT-PCR assays of Foxd4l1-expressing animal caps cultured to later stages do show expression of ngnr1 and tubb2 (Sullivan et al., 2001).
Figure 8.

A scheme for how neural cell fate may be acquired step-wise. At the 16-cell stage, dorsal-animal blastomeres (blue) contain molecules, such as nTFs, that bias them to a neural cell fate by an unknown mechanism (indicated by “?”). At blastula stages, maternal β-catenin becomes nuclear in the dorsal marginal cells that are the descendants of the dorsal-animal blastomeres (blue) to directly activate Siamois/Twin. These transcription factors in turn directly activate the zygotic expression of some some nTFs (foxd4l1, gmnn, zic2) in these same cells. During gastrulation, Siamois/Twin directly activate signaling factors in the Organizer mesoderm that diffuse towards the animal pole to promote nTF expression in the nascent neural ectoderm (blue). The nTFs then up-regulate early neural genes (e.g., sox2, zic1), which in turn act upstream of neural differentiation factors (e.g., irx1, ngnr1) and markers of committed neurons (e.g., tubb2b).
Neural fate “bias” is not due to differences in maternal transcript levels of nTFs
Since differential localization of maternal mRNA is a recognized mechanism by which adjacent cells in the embryo can attain different fates, we tested whether the nTF mRNAs were more abundant in D11 versus V11 blastomeres. The original papers describing the cloning of Foxd4l1, Zic2, Gmnn and Sox11 identified the presence of maternal transcripts in the animal hemisphere (Hiraoka et al., 1997; Brewster et al., 1998; Kroll et al., 1998; Sullivan et al., 2001;Houston and Wylie, 2005; Fujimi et al., 2012), which has recently been confirmed by microarray and RNA-seq assays (Yanai et al., 2011; Peshkin et al., 2015; www.xenbase.org). However, our results from both ISH and qPCR analyses do not show dorsal-ventral differences in their maternal nTF mRNAs; a recent RNA-seq study of dissected 8-cell blastomeres also did not detect significant dorsal versus ventral differences (De Demenico et al., 2015). Therefore, the intrinsic differences in fate between dorsal and ventral 16-cell blastomeres likely are due to differences in epigenetic chromatin states, endogenous chromatin remodeling factors, translational efficiencies, post-translational modifications, or metabolic activities that either affect responses to maternal Wnt signaling (Wills et al., 2010), pre-MBT transcription (e.g., Yang et al., 2002), or responses to later inductive signals such as inhibition of BMP, FGFs and Wnts (e.g., Sokol and Melton, 1991; Kinoshita et al., 1993; Ding et al., 1998).
It is most interesting that there are reports of cleavage stage post-transcriptional differences between 16-cell blastomeres. Protein synthesis studies identified unique protein bands/spots from dorsal versus ventral animal 16- and 32-cell blastomeres (Miyata et al., 1987; Klein and King, 1988). A recent proteomic analysis showed that Gmnn protein is significantly enriched in D11 versus V11 blastomeres (Lombard-Banek et al., 2016), and D11 and V11 blastomeres have metabolite differences that affect cell fate decisions (Onjiko et al., 2015). These results indicate that as technologies for detecting protein and small molecules at the single-cell level are improved, many more maternally derived molecules that bias blastomere fate decisions at cleavage stages are likely to be identified. It also will be important to use proteomic approaches to determine whether the differences we observed in the efficacies of the nTF mRNAs to ectopically induce early neural genes are due to differences in nTF activity or to differences in the translation of the exogenously supplied mRNAs.
Functional conservation of Foxd4/Foxd4-like proteins
Tetrapods, i.e., amphibians, reptiles, avians and mammals, are genomically highly related. Therefore, we were interested to discover whether the nTFs are functionally conserved. First, it is notable that the amphibians and mammals have different numbers of Foxd4/Foxd4-like genes. Xenopus laevis and Xenopus tropicalis each have two Foxd4l1 paralogues (Foxd4l1.1 and Foxd4l1.2); in Xenopus laevis Foxd4l1.1 is present as two homeologues (Foxd4l1.1a, Foxd4l1.1b) that are derived from the hybridization event that created this species (Matsudo et al., 2015). Mouse has a single gene (Foxd4) and human has two expressed paralogues (FOXD4, FOXD4L1) as well as five presumed pseudogenes (FOXD4L2-6) (Jackson et al., 2010). Comparison of the amino acid sequences shows that all eight expressed proteins contain an N-terminal acidic blob region that we previously showed is responsible for transcriptional activation of downstream neural target genes (Neilson et al., 2012; Klein et al., 2013). Seven of the proteins, i.e., all except FOXD4, contain an Eh-1 motif that binds to Grg-related proteins and is involved in transcriptional repression (Yaklichkin et al., 2007; Neilson et al., 2012; Klein et al., 2013). It was surprising that both human proteins only weakly bound to Xenopus Grg4, since one contains an Eh-1 and one does not. This cannot be due simply to expressing a mammalian protein in an amphibian cell because the mFoxd4 bound strongly to Xenopus Grg4.
We addressed whether the mammalian homologues had a similar, conserved functional activity compared to frog Foxd4l1 by expressing them in Xenopus embryos and blastomere explants. We show that the mammalian proteins have similar effects on gene expression, but their effects are less robust than the Xenopus protein. This observation is consistent with the mammalian proteins having less avidity for Grg4 binding. However, their reduced efficacy cannot be due entirely to Grg4 binding since FOXD4 does not contain an intact Eh-1 motif, and both FOXD4 and FOXD4L1 show weak Grg4 binding in co-immunoprecipitation assays. Based on our previous observation that an additional domain, i.e., a C-terminal alpha-helical structure, also contributes to the transcriptional repressive activity of Foxd4l1 (Klein et al., 2013), we suggest that this structure also is contributing to the species differences in phenotypes. Nonetheless, the similar abilities of frog and mammalian Foxd4/Foxd4-like proteins to affect neural gene expression indicate functional conservation across tetrapods. Considering that there is some evidence for asymmetry in mouse blastomeres and in blastocyst ectodermal competence (Zernika-Goetz, 2005; Zernika-Goetz, 2006; Li et al., 2013), it will therefore be exciting to test the function of Foxd4-like proteins, as well as other nTFs, in mammalian assay systems.
Methods
Cloning and sequence comparisons
Mouse Foxd4 (mFoxd4), human FOXD4 (hFOXD4), and human FOXD4L1 (hFOXD4L1) cDNAs were purchased (Dharmacon) and the ORFs subcloned by PCR into the Stu1/Xho1 sites of pCS2+ and myc-tagged pCS2+ (pCS2+-MT) vectors using standard techniques. The ORF of Xenopus laevis zic2 was excised from pCS2+Zic2 vector (Brewster et al., 1998) with EcoR1 and Not1 and subcloned into the pCS2+-hGR vector. The resulting plasmids were fully sequenced in both directions. Frog and mammalian Foxd4/Foxd4-like protein sequences were aligned using Clustal Omega and displayed using ESpript in order to compare the known functional domains of these proteins (Sievers et al., 2011; Robert and Gouet, 2014).
Obtaining embryos and microinjections
Fertilized Xenopus laevis eggs were obtained by gonadotropin-induced natural mating of adult frogs (Moody, 1999; Moody, 2000). The eggs were dejellied with 2% cysteine solution and selected at the 2-cell stage if the first cleavage furrow bisected the lightly pigmented region of the animal hemisphere to accurately identify the dorsal-ventral axis (Klein, 1987; Miyata et al., 1987). These selected embryos were cultured in 100% Steinberg's solution until the 16-cell stage, when each blastomere was microinjected with 1nl of either mRNAs or antisense morpholino oligonucleotides (MOs) according to standard methods (Moody, 1999; Moody, 2000). mRNAs encoding Xenopus foxd4l1.1a (100pg; Sullivan et al., 2001), foxd4l1.1a-hGR (100pg; Yan et al., 2009), n-geminin (gmnn, 100pg; Kroll et al., 1998), sox11 (100pg; provided by Tim Grammar and Elena Casey), zic2 (100pg; Brewster et al., 1998), zic2-hGR (100pg), mFoxd4 (75pg), hFOXD4 (100pg), hFOXD4L1 (100pg), and a nuclear-localized β-galactosidase (nβgal; 100pg) were synthesized in vitro (Ambion, mMessage, mMachine). Each transcription factor mRNA was mixed with nβgal mRNA and microinjected into either both dorsal-animal blastomeres (D11), which are the major precursors of the neural plate, or both ventral-animal blastomeres (V11), which are the major precursors of the epidermis (Moody, 1987; Moody and Kline, 1990) (Fig. 1). In some experiments, hFOXD4 or hFOXD4L1 mRNA, mixed with nβgal mRNA, was microinjected into a single D11 or V11 (Fig. 1) to test whether they can alter neural gene expression in the intact embryo, as previously shown for Xenopus foxD4l1 (Yan et al., 2009).
Morpholino antisense oligonucleotides (MOs)
Two translation-blocking MOs that bind to Xenopus laevis foxd4l1.1a, foxd4l1.1b and foxd4l1.2 mRNAs (FoxMOs) or two that bind to zic2 mRNAs (ZicMOs) were synthesized (Gene Tools, LLC), and co-injected in equimolar concentrations, as described above. The specificity and efficacy of the FoxMOs were published previously (Yan et al., 2009). As shown in Supplemental Figure 1, the FoxMOs bind to all three Xenopus laevis transcripts. One zic2 MO overlaps the sequences verified in previous studies to specifically and efficiently knock-down maternal and zygotic expression (Houston and Wylie, 2005; Fujima et al., 2012), whereas a second zic2 MO lies further upstream in the 5′UTR (Supplemental Figure 1). To demonstrate rescue of the MO phenotypes, mRNAs insensitive to MO binding were co-injected with the MOs, and D11 explants created as described below. For FoxMOs, the mFoxd4 mRNA was used for rescue; for ZicMOs, myc-tagged zic2 mRNA was used (Supplemental Figure 1).
Blastomere explants
Upon reaching the 32- to 64-cell stages, injected embryos were transferred to an agarose-coated Petri dish filled with 50% sterile Steinberg's solution. The midline descendants of the injected pair of blastomeres (either blastomere pairs of D11 or V11; Fig. 1) were dissected free of the embryo using fine forceps, as previously described (Grant et al., 2013). Each explant was transferred to 1× Modified Barth's Solution (MBS) in an agar-coated well of a 24-well culture plate and cultured at 14° C overnight until sibling embryos, incubated in adjacent wells, reached early gastrula (st 10.5) or early neural ectoderm (st 11.5-12.5) stages (Nieuwkoop and Faber, 1994). For those explants injected with hormone-inducible (h-GR) constructs, dexamethasone (Dex), a synthetic hormone that activates hGR-fusion proteins (Mattioni et al., 1994; Kolm and Sive, 1995), was added to the culture medium (final concentration = 4 μg/mL) at blastula (st 8-8.5) stages.
Fixation, histochemistry and in situ hybridization (ISH)
Explants and whole embryos were fixed in 4% paraformaldehyde in MEM buffer when sibling control embryos reached the appropriate stages. They were stained for nβGal histochemistry, processed for ISH and bleached according to standard procedures (Yan et al., 2009). Anti-sense RNA probes for maternal neural (foxd4l1, zic2, sox11, gmnn), early zygotic neural (sox2, zic1), mesoderm (bra, chd), and epidermis (K81) genes were synthesized in vitro (Ambion MEGAscript kits) as previously described (Sullivan et al., 2001; Yan et al., 2009). For ISH processing of whole embryos (Fig. 2), some were cut into animal and vegetal fragments to ensure that the probe could penetrate the yolky cells, and some were processed with sense probes to ensure that the ISH signal shown in Fig. 2 is not background. For ISH processing of explants, sibling control embryos were included in the sample vial throughout the process as positive controls for the probes. Each experiment was repeated in 2-5 independent trials with different sets of parents. Explants and embryo ISH samples were scored for positive gene expression independently by 2-3 of the authors, and the values reported are means of their independent scores. Explants were scored as positive if they were stained above the background level (unstained domain) of the whole embryos included in the same batch/vial. Differences in frequencies of positive gene expression were compared by Chi-squared statistics, with p<0.05 indicating a significant difference.
Quantitative real-time PCR
To measure endogenous levels of nTFs, uninjected V11 and D11 blastomere pairs were dissected from the embryo as above and collected in 500μL of Trizol. Five independent samples each containing 20 blastomere pairs were collected from 5 different sets of parents. Total RNA was extracted, residual genomic DNA was removed by DNAse treatment (Turbo DNA-free, Invitrogen), and random-hexamer primed cDNA was generated using ImPromp-II reverse transcriptase (Promega, Madison WI). qPCR reactions were assembled using a EpMotion 5070 liquid handling system (Eppendorf, Hauppauge, NY) that combines forward and reverse gene-specific primers (0.3 μM final concentration, Integrated DNA Technologies, Coralville, IA, as listed in Supplemental Table 1), with 7.5 μl of SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA) in a 14 μl reaction. qPCR analysis was performed using a CFX-384 Real-Time PCR Detection System. Raw CT values were normalized to the average of two reference genes (Odc1, Eef1a1) to establish expression levels (delta-CT); statistical differences in expression differences between samples (delta-delta-CT) were assessed by Student's t-test (p<0.05) for each independent analysis.
Co-immunoprecipitation
Oocytes were injected with mRNAs coding for human FOXD4, human FOXD4L1, murine Foxd4, Xenopus laevis Fox d4l1.1a and/or Xenopus laevis Grg4, and cultured at 18°C for 18 hours. For each immunoprecipitation reaction, 150 μl of lysate (15 oocyte equivalents) was mixed with 650 μl ice-cold TNSG lysis buffer and 1 μg of antibody (raised against HA or Myc; Applied Biological Materials) and incubated at 4°C for 2 h or overnight, after which 25 μl protein A/G sepharose beads (GE Healthcare) were added to the reaction and rotated in an orbital mixer 1 hr at 4°C. Beads were briefly pelleted at 4°C and rinsed 3 times with ice-cold TNSG lysis buffer. All residual buffer was removed with a flat pipette tip and beads were resuspended in 35 μl 2× TNSG sample buffer (TNSG Buffer: 137 mM NaCl, 10% glycerol, 1% NP40, 20 mM Tris (8.0); 4× sample buffer: 4 mL 10% SDS, 2 mL glycerol, 0.3086 g DTT, 0.00001 g Bromphenol Blue; 4× sample buffer was diluted to 2× in TNSG buffer). Samples were boiled at 100°C for 10 min prior to loading on Tris-glycine SDS-Polyacrylamide 10% gels. For expression checks, 15 μl (1.5 oocyte equivalents) lysate was prepared with 4× sample buffer and loaded on Tris-glycine SDS-Polyacrylamide 10% gels. Proteins were resolved by SDS/PAGE, transferred to Immobilon-P transfer membranes (Millipore) using standard methods, and blocked in Tris-buffered saline (25 mM Tris)+0.2% Tween-20 (TBST)+5% nonfat dry milk for at least 1 h to overnight at 4°C. Whenever possible, IP-Western blots were incubated with the following HRP-conjugated primary antibodies to reduce background: anti-HA-HRP-conjugated (Roche), and anti-Myc-HRP–conjugated (Millipore). Following antibody incubation, blots were rinsed with TBST, blotted with Pierce ECL Western Blotting Substrate (Thermo Scientific) and exposed to film.
Supplementary Material
Supplemental Figure 1: MO sequences and binding sites.
A. Xenopus laevis has three Foxd4-like genes: the two homeologues foxd4l1.1-a (NM_001088529; GI:147907360) and foxd4l1.1-b (NM_001088253; GI:147906672), and the paralogous Foxd4l1.2 (AJ242678; aka FXD12″)(www.xenbase.org). Black letters indicate their 5′ sequences with translational start sites highlighted in yellow. The MOs used herein are as follows: MO#1 (red) overlaps the coding region and translational start sites of foxd4l1.1-a and foxd4l1.1-b (one mis-match at 5′ end of foxd4l1.1-b indicated by blue highlight). There are five mis-matches within foxd4l1.2 (blue highlight). Therefore, we co-injected MO#2 (blue), which binds just upstream of the ATG of foxd4l1.2, but has several mismatches (blue highlights) with foxd4l1.1-a and foxd4l1.1-b. Also shown is the sequence of mouse Foxd4 (NM_008022.2 GI:261824060); note that neither MO is likely to bind due to the numerous mis-matches (blue highlights). Thus, the mouse Foxd4 mRNA was used to rescue the MO effects to demonstrate specificity (D, below).
B. Black letters indicate the sequence of zic2 mRNA (NM_001087724.1 GI:147902815) with translational start site highlighted in yellow. The MOs used herein are above the sequence: MO#1 (red) binds in the 5′UTR and MO#2 (blue) overlaps the coding region. MO#2 significantly overlaps with single MOs that in previous studies were shown to be specific and effective in knocking-down Xenopus Zic2 protein (green = Fujimi et al., 2012; orange = Houston and Wylie, 2005).
C. Sequence of the “rescue” zic2 mRNA (black letters) with the start of the zic2 coding region highlighted in yellow. MO#1 cannot bind because the endogenous 5′UTR has been replaced with pCS2+ vector sequence (not shown). MO#2 (blue) cannot block translation of RNA synthesized from the pCS2+-MT-zic2 construct because the start codon is separated by six copies of the myc-epitope tag (350 bases, 6th myc-epitope underlined) from the zic2 coding region (which starts at the yellow highlight). Morpholinos that bind more than 30 bases from the translational start codon do not block translation (www.gene-tools.com/choosing_the_optimal_target#TranslationalBlocking). Also, there are four mis-matched nucleotides (blue highlight) just upstream from the zic2 coding region.
D. Expression of MO-insensitive mRNAs (see A and C above) in D11 explants rescues the expression of sox2 in the presence of either FoxMOs or Zic2MOs. Asterisks (*) indicate a significant difference compared to MO-injections (p<0.05, Chi-square statistic). Numbers above the bars indicate the number of explants assayed.
Supplemental Figure 2: CLUSTAL analysis displayed in ESpript comparing the amino acid (aa) sequences of the three Xenopus laevis proteins (Foxd4l1.2, Foxd4l1.1a, Foxd4l1.1b), the two Xenopus tropicalis proteins (Foxd4l1.2, Foxd4l1.1) and the two human proteins (FOXD4L1, FOXD4). Red blocks indicate identical aa, yellow blocks indicate conserved aa, and bold aa within a yellow block indicate the same type of aa, e.g., acidic, hydrophobic, etc.
Acknowledgments
We thank our many colleagues in the Xenopus community for providing the expression and ISH plasmids used in this study. We thank Steven Klein for performing some of the microinjection experiments with the mammalian constructs, and for critical reading of the manuscript.
Funding sources: NSF grant MCB-1121711 (SAM)
GWU Luther Rice Undergraduate Research Fellowships (SG, MM, MH)
GWU School of Medicine and Health Sciences (GWIN Biomarker Discovery Facility; TEM)
NCI ZIA BC010006 20 (IOD)
References
- Abrams EW, Mullins MC. Early zebrafish development: it's in the maternal genes. Curr Opin Genet Dev. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Reid CD, Kessler DS. Siamois and Twin are redundant and essential in formation of the Spemann organizer. Dev Biol. 2011;352:367–381. doi: 10.1016/j.ydbio.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TJ, Vonica A, Heasman J, Brivanlou AH, Bell E. Coco regulates dorsoventral specification of germ layers via inhibition of TGFβ signalling. Development. 2013;140:4177–4181. doi: 10.1242/dev.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DV, Huang S, Moody SA. The cleavage stage origin of Spemann's Organizer: Analysis of the movements of blastomere clones before and during gastrulation in Xenopus. Development. 1994;120:1179–1189. doi: 10.1242/dev.120.5.1179. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38(suppl):D607–D612. doi: 10.1093/nar/gkp953. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Carnac G, Kodjabachian L, Gurdon JB, Lemaire P. The homeobox gene Siamois is a target of the Wnt dorsalization pathway and triggers organizer activity in the absence of mesoderm. Development. 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- Cha SW, McAdams M, Kormish J, Wylie C, Kofron M. Foxi2 is an animally localized maternal mRNA in Xenopus, and an activator of the zygotic ectoderm activator Foxi1e. PLoS One. 2012;7:e41782. doi: 10.1371/journal.pone.0041782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex. RNA Dev Dyn. 2010;239:1838–1848. doi: 10.1002/dvdy.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras S, Marikawa Y, Elinson RP, Lemaire P. Animal and vegetal pole cells of early Xenopus embryos respond differently to maternal dorsal determinants: implications for the patterning of the organizer. Development. 1997;124:4275–4286. doi: 10.1242/dev.124.21.4275. [DOI] [PubMed] [Google Scholar]
- Davidson EH. How embryos work: a comparative view of diverse modes of cell fate specification. Development. 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- De Domenico E, Owens ND, Grant IM, Gomes-Faria R, Gilchrist MJ. Molecular asymmetry in the 8-cell stage Xenopus tropicalis embryo described by single blastomere transcript sequencing. Dev Biol. 2015;408:252–68. doi: 10.1016/j.ydbio.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM. Spemann's organizer and self-regulation in amphibian embryos. Nature Reviews. 2006;7:296–302. doi: 10.1038/nrm1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Hausen P, Steinbeisser H. Pre-MBT patterning of early gene regulation in Xenopus: the role of the cortical rotation and mesoderm induction. Mech Dev. 1998;70:15–24. doi: 10.1016/s0925-4773(97)00163-9. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Fujimi TJ, Hatayama M, Aruga J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/β-catenin signaling pathway. Dev Biol. 2012;361:220–231. doi: 10.1016/j.ydbio.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Gallagher BC, Hainski AM, Moody SA. Autonomous differentiation of dorsal axial structures from an animal cap cleavage stage blastomere in Xenopus. Development. 1991;112:1103–1114. doi: 10.1242/dev.112.4.1103. [DOI] [PubMed] [Google Scholar]
- Grant PA, Herold M, Moody SA. Blastomere explants to test for cell fate commitment during embryonic development. J Vis Exp. 2013 Jan 26;(71) doi: 10.3791/4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Yan B, Johnson MA, Johnson DL, Moody SA. Novel animal pole-enriched maternal mRNAs are preferentially expressed in neural ectoderm. Dev Dyn. 2014;243:478–496. doi: 10.1002/dvdy.24082. [DOI] [PubMed] [Google Scholar]
- Hainski AM, Moody SA. Xenopus maternal RNAs from a dorsal animal blastomere induce a secondary axis in host embryos. Development. 1992;116:347–355. doi: 10.1242/dev.116.2.347. [DOI] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Komatsu N, Sakai Y, Ogawa M, Shiozawa M, Aiso S. XLS13A and XLS13B: SRY-related genes of Xenopus laevis. Gene. 1997;197:65–71. doi: 10.1016/s0378-1119(97)00242-4. [DOI] [PubMed] [Google Scholar]
- Houston DW, Wylie C. Maternal Xenopus Zic2 negatively regulates Nodal-related gene expression during antero-posteror patterning. Development. 2005;132:4845–4855. doi: 10.1242/dev.02066. [DOI] [PubMed] [Google Scholar]
- Hyodo-Miura J, Urushiyama S, Nagai S, Nishita M, Ueno N, Shibuya H. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells. 2002;7:487–496. doi: 10.1046/j.1365-2443.2002.00536.x. [DOI] [PubMed] [Google Scholar]
- Ishibashi H, Matsumura N, Hanafusa H, Matsumoto K, De Robertis EM, Kuroda H. Expression of Siamois and Twin in the blastula Chordin/Noggin signaling center is required for brain formation in Xenopus laevis embryos. Mech Dev. 2008;125:58–66. doi: 10.1016/j.mod.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Sokol SY. Early Development of Epidermis and Neural Tissue. In: Moody SA, editor. Principles of Developmental Genetics. 2nd. Elsevier; NY: 2014. pp. 189–201. [Google Scholar]
- Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas E, Sargent TD, Dawid IB. Epidermal keratin gene expressed in embryos of Xenopus laevis. Proc Natl Acad Sci U S A. 1985;82:5413–5417. doi: 10.1073/pnas.82.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageura H, Yamana K. Pattern regulation in defect embryos of Xenopus laevis. Devel Biol. 1984;101:410–415. doi: 10.1016/0012-1606(84)90155-6. [DOI] [PubMed] [Google Scholar]
- Kageura H, Yamana K. Pattern formation in 8-cell composite embryos of Xenopus laevis. J Embryol Exp Morphol. 1986;91:79–100. [PubMed] [Google Scholar]
- Kageura H. Spatial distribution of the capacity to initiate a secondary embryo in the 32-cell embryo of Xenopus laevis. Devel Biol. 1990;142:432–438. doi: 10.1016/0012-1606(90)90365-p. [DOI] [PubMed] [Google Scholar]
- Kageura H. Three regions of the 32-cell embryo of Xenopus laevis essential for formation of a complete tadpole. Devel Biol. 1995;170:376–386. doi: 10.1006/dbio.1995.1223. [DOI] [PubMed] [Google Scholar]
- Kessler DS. Siamois is required for formation of Spemann's organizer. PNAS USA. 1997;94:13017–13022. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Bessho T, Asashima M. Competence pre-pattern in the animal hemisphere of the 8-cell stage Xenopus embryo. Dev Biol. 1993;160:276–284. doi: 10.1006/dbio.1993.1305. [DOI] [PubMed] [Google Scholar]
- Klein SL. The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev Biol. 1987;120:299–304. doi: 10.1016/0012-1606(87)90127-8. [DOI] [PubMed] [Google Scholar]
- Klein SL, King ML. Correlations between cell fate and distribution of proteins that are synthesized before the midblastula transition in Xenopus. Roux's Arch Dev Biol. 1988;197:275–281. doi: 10.1007/BF00380021. [DOI] [PubMed] [Google Scholar]
- Klein SL, Moody SA. Early neural ectodermal genes are activated by Siamois and Twin during blastula stages. Genesis. 2015;53:308–320. doi: 10.1002/dvg.22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Neilson KM, Orban J, Yaklichkin S, Hoffbauer J, Mood K, Daar IO, Moody SA. Conserved structural domains in FoxD4L1, a neural forkhead box transcription factor, are required to repress or activate target genes. PLoS One. 2013;8:e61845. doi: 10.1371/journal.pone.0061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodjabachian L, Lemaire P. Siamois functions in the early blastula to induce Spemann's organizer. Mech Dev. 2001;108:71–79. doi: 10.1016/s0925-4773(01)00484-1. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Koyano S, Ito M, Takamatsu N, Takiguchi S, Shiba T. The Xenopus Sox3 gene is expressed in oocytes of early stages. Gene. 1997;188:101–107. doi: 10.1016/s0378-1119(96)00790-1. [DOI] [PubMed] [Google Scholar]
- Kuo JS, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Wessely O, De Robertis EM. Neural induction in Xenopus: requirement for ectodermal and endomesodermal signals via Chordin, Noggin, beta-Catenin, and Cerberus. PLoS Biol. 2004;2:0623–0634. doi: 10.1371/journal.pbio.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent MN, Blitz IL, Hashimoto C, Rothbacher U, Cho KW. The Xenopus homeobox gene twin mediates Wnt induction of goosecoid in establishment of Spemann's organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lee HS, Moody SA. Neural transcription factors: from embryos to neural stem cells. Mol Cells. 2014;37:705–712. doi: 10.14348/molcells.2014.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;308:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu C, Beichele S, Zhu Q, Song L, Lanner F, Jing N, Rossant J. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140:4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- Lombard-Banek C, Moody SA, Nemes P. Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew Chem Int Ed Engl. 2016 Jan 12; doi: 10.1002/anie.201510411. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M. A new nomenclature of Xenopus laevis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 2015;145:187–191. doi: 10.1159/000381292. [DOI] [PubMed] [Google Scholar]
- Mattioni T, Louvion JF, Picard D. Regulation of protein activities by fusion to steroid binding domains. Meth Cell Biol. 1994;43:335–352. doi: 10.1016/s0091-679x(08)60611-1. [DOI] [PubMed] [Google Scholar]
- Medina A, Wendler SR, Steinbesser H. Cortical rotation is required for the correct spatial expression of nr3, sia and gsc in Xenopus embryos. Int J Dev Biol. 1997;41:741–745. [PubMed] [Google Scholar]
- Miyata S, Kageura H, Kihara HK. Regional differences of proteins in isolated cells of early embryos of Xenopus laevis. Cell Differ. 1987;21:47–52. doi: 10.1016/0045-6039(87)90447-7. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Moody SA. Cell Lineage Analysis in Xenopus Embryos. In: Tuan RS, Lo CW, editors. Methods in Molecular Biology, Vol 135: Developmental Biology Protocols. Humana Press Inc.; Totowa, NJ: 1999. [DOI] [PubMed] [Google Scholar]
- Moody SA. Testing the Cell Fate Commitment of Single Blastomeres in Xenopus laevis. In: Richter J, editor. Advances in Molecular Biology. Oxford University Press; 2000. pp. 355–381. [Google Scholar]
- Moody SA, Klein SL, Karpinski BA, Maynard TM, LaMantia AS. On becoming neural: what the embryo can tell us about differentiating neural stem cells. Amer J Stem Cells. 2013;2:74–94. [PMC free article] [PubMed] [Google Scholar]
- Moody SA, Kline M. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat Embryol (Berl) 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Neilson KM, Klein SL, Mhaske P, Mood K, Daar IO, Moody SA. Specific domains of FoxD4/5 activate and repress neural transcription factor genes to control the progression of immature neural ectoderm to differentiating neural plate. Dev Biol. 2012;365:363–375. doi: 10.1016/j.ydbio.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North-Holland, Amsterdam: 1994. [Google Scholar]
- Onjiko RM, Moody SA, Nemes P. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A. 2015;112:6545–6550. doi: 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur PD, Sullivan SA, Moody SA. Multiple maternal influences on dorsal-ventral fate in Xenopus animal blastomeres. Devel Dynamics. 2002;225:581–587. doi: 10.1002/dvdy.10181. [DOI] [PubMed] [Google Scholar]
- Pera EM, Acosta H, Gouignard N, Climent M, Arregi I. Active signals, gradient formation and regional specificity in neural induction. Exp Cell Res. 2014;321:25–31. doi: 10.1016/j.yexcr.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Peshkin L, Wuhr M, Pearl E, Haas W, Freeman RM, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S, Simonsson PR, Trevers KE, Stower MJ, Sherlock WT, Khan M, Streit A, Sheng G, Stern CD. Distinct steps of neural induction revealed by Asterix, Obelix and TrkC, genes induced by different signals from the organizer. PLoS One. 2011;6:e19157. doi: 10.1371/journal.pone.0019157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, Langenberg T, Vleminckx K, Bellefroid E, Cassiman JJ, Tejpar S. Transcription factor Zic2 inhibits Wnt/β-catenin protein signaling. J Biol Chem. 2011;286:37732–37740. doi: 10.1074/jbc.M111.242826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CD, Zhang Y, Sheets MD, Kessler DS. Transcriptional integration of Wnt and Nodal pathways in establishment of the Spemann organizer. Dev Biol. 2012;368:231–241. doi: 10.1016/j.ydbio.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl Acids Res. 2014;42(W1):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Research C: Embryo Today. 2009;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Identifying the missing links: genes that connect neural induction and primary neurogenesis in vertebrate embryos. Neuron. 1998;21:455–458. doi: 10.1016/s0896-6273(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Schneider S, Steinbeisser H, Warga RM, Hausen P. Beta-catenin translocation into nuclei demarcates the dorsalizing center in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- Sherman JH, Karpinski B, Fralish M, Cappuzzo J, Dhindsa D, Thal A, Moody SA, LaMantia AS, Maynard TM. Mouse Foxd4 is essential for establishing neural cell fate and for neuronal differentiation. 2016 doi: 10.1002/dvg.23031. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molec Systems Biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S, Melton DA. Pre-existent pattern in Xenopus animal pole cells revealed by induction with activin. Nature. 1991;351:409–411. doi: 10.1038/351409a0. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int J Dev Biol. 2001;45:13–38. [PubMed] [Google Scholar]
- Suduo N, Yamamoto S, Ogino H, Taira M. Dynamic in vivo binding of transcription factors to cis-regulatory modules of cer and gsc in the stepwise formation of the Spemann-Mangold organizer. Development. 2012;139:1651–1661. doi: 10.1242/dev.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SA, Moore KB, Moody SA. Early events in frog blastomere fate determination. In: Moody SA, editor. Cell Lineage and Fate Determination. Academic Press; NY: 1998. pp. 297–321. [Google Scholar]
- Sullivan SA, Akers L, Moody SA. foxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transcriptional repression that is dependent on the C-terminal domain. Devel Biol. 2001;232:439–457. doi: 10.1006/dbio.2001.0191. [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zool. 2008;310B:73–84. doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- Wills AE, Choi VM, Bennett MJ, Khokha MK, Harland RM. BMP antagonists and FGF signaling contribute to different domains of the neural plate in Xenopus. Dev Biol. 2010;337:335–350. doi: 10.1016/j.ydbio.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Cheng F, Liang J, Wu W, Zhang J. Maternal xNorrin, a canonical Wnt signaling agonist and TGF-β antagonist, controls early neuroectoderm specification Xenopus. PLoS Biol. 2012;10:e1001286. doi: 10.1371/journal.pbio.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J Biol Chem. 2007;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. FoxD5 plays a critical upstream role in regulating neural fate and onset of differentiation. Dev Biol. 2009;329:80–95. doi: 10.1016/j.ydbio.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. Microarray identification of novel downstream targets of FoxD5, a critical component of the neural ectodermal transcriptional network. Dev Dyn. 2010;239:3467–3480. doi: 10.1002/dvdy.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the mid-blastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. Developmental cell biology: cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–28. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. The first cell-fate decisions in the mouse embryo: destiny is a matter of both chance and choice. Curr Opin Genet Dev. 2006;16:406–12. doi: 10.1016/j.gde.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Hernandez-Lagunas L, Simpson P, Stemple DL, Artinger KB, Klymkowsky MW. Repression of nodal expression by maternal B1-type SOXs regulates germ layer formation in Xenopus and zebrafish. Devel Biol. 2004;273:23–37. doi: 10.1016/j.ydbio.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Zhang C, Klymkowsky MW. The Sox axis, nodal signaling and germ layer specification. Differentiation. 2007;75:536–545. doi: 10.1111/j.1432-0436.2007.00190.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: MO sequences and binding sites.
A. Xenopus laevis has three Foxd4-like genes: the two homeologues foxd4l1.1-a (NM_001088529; GI:147907360) and foxd4l1.1-b (NM_001088253; GI:147906672), and the paralogous Foxd4l1.2 (AJ242678; aka FXD12″)(www.xenbase.org). Black letters indicate their 5′ sequences with translational start sites highlighted in yellow. The MOs used herein are as follows: MO#1 (red) overlaps the coding region and translational start sites of foxd4l1.1-a and foxd4l1.1-b (one mis-match at 5′ end of foxd4l1.1-b indicated by blue highlight). There are five mis-matches within foxd4l1.2 (blue highlight). Therefore, we co-injected MO#2 (blue), which binds just upstream of the ATG of foxd4l1.2, but has several mismatches (blue highlights) with foxd4l1.1-a and foxd4l1.1-b. Also shown is the sequence of mouse Foxd4 (NM_008022.2 GI:261824060); note that neither MO is likely to bind due to the numerous mis-matches (blue highlights). Thus, the mouse Foxd4 mRNA was used to rescue the MO effects to demonstrate specificity (D, below).
B. Black letters indicate the sequence of zic2 mRNA (NM_001087724.1 GI:147902815) with translational start site highlighted in yellow. The MOs used herein are above the sequence: MO#1 (red) binds in the 5′UTR and MO#2 (blue) overlaps the coding region. MO#2 significantly overlaps with single MOs that in previous studies were shown to be specific and effective in knocking-down Xenopus Zic2 protein (green = Fujimi et al., 2012; orange = Houston and Wylie, 2005).
C. Sequence of the “rescue” zic2 mRNA (black letters) with the start of the zic2 coding region highlighted in yellow. MO#1 cannot bind because the endogenous 5′UTR has been replaced with pCS2+ vector sequence (not shown). MO#2 (blue) cannot block translation of RNA synthesized from the pCS2+-MT-zic2 construct because the start codon is separated by six copies of the myc-epitope tag (350 bases, 6th myc-epitope underlined) from the zic2 coding region (which starts at the yellow highlight). Morpholinos that bind more than 30 bases from the translational start codon do not block translation (www.gene-tools.com/choosing_the_optimal_target#TranslationalBlocking). Also, there are four mis-matched nucleotides (blue highlight) just upstream from the zic2 coding region.
D. Expression of MO-insensitive mRNAs (see A and C above) in D11 explants rescues the expression of sox2 in the presence of either FoxMOs or Zic2MOs. Asterisks (*) indicate a significant difference compared to MO-injections (p<0.05, Chi-square statistic). Numbers above the bars indicate the number of explants assayed.
Supplemental Figure 2: CLUSTAL analysis displayed in ESpript comparing the amino acid (aa) sequences of the three Xenopus laevis proteins (Foxd4l1.2, Foxd4l1.1a, Foxd4l1.1b), the two Xenopus tropicalis proteins (Foxd4l1.2, Foxd4l1.1) and the two human proteins (FOXD4L1, FOXD4). Red blocks indicate identical aa, yellow blocks indicate conserved aa, and bold aa within a yellow block indicate the same type of aa, e.g., acidic, hydrophobic, etc.


