Summary
Chronic myeloid leukaemia (CML) arises following transformation of a haemopoietic stem cell (HSC) by protein-tyrosine kinase BCR-ABL1. Direct inhibition of BCR-ABL1 kinase has revolutionized disease management, but fails to eradicate leukaemic stem cells (LSC), which maintain CML. LSC are independent of BCR-ABL1 for survival, providing a rationale to identify and target kinase-independent pathways. Here we show using proteomics, transcriptomics and network analyses, that in human LSC aberrantly expressed proteins, in both imatinib-responder and non-responder patients are modulated in concert with p53 and c-Myc regulation. Perturbation of both p53 and c-Myc, not BCR-ABL1 itself, leads to synergistic kill, differentiation and near elimination of transplantable human LSC in mice, whilst sparing normal HSC. This unbiased systems approach targeting connected nodes exemplifies a novel precision medicine strategy providing evidence that LSC can be eradicated.
Introduction
BCR-ABL1 is a chimeric oncogene arising from t(9;22)(q34;q11) chromosomal translocation. The resultant protein-tyrosine kinase (PTK) drives signalling events1 and transforms haemopoietic stem cells (HSC). BCR-ABL1 activity in HSC causes chronic myeloid leukaemia (CML) which if untreated, is fatal.
TK inhibitors (TKI), such as imatinib mesylate (IM), are standard CML treatment and have improved survival, illustrating justification for single-target therapies2. However, these drugs do not kill leukaemic stem cells (LSC) that maintain the disease3, resulting in ever-increasing costs to sustain remissions. TKI discontinuation in the best 10-20% of TKI-responders gave relapse rates of 50-60%, reinforcing the need to understand and target CML LSC4 with curative therapies. Recent studies suggest that LSC survival is BCR-ABL1-kinase independent5 and BCR-ABL1 has functionality beyond PTK activity explaining shortcomings of TKIs6.
We have applied systems biology approaches to patient material to identify key protein networks that perpetuate CML phenotype, aiming to elucidate potentially curative therapy. Using unbiased transcriptomic and proteomic analyses, transcription factors (TFs), p53 and c-Myc, are identified as having defining roles in CML LSC survival. We demonstrate an integral relationship between p53 and c-Myc in the maintenance of CML and importantly, the potential therapeutic advantage they provide as drug targets over BCR-ABL1 for eradication of CML LSC.
Results
p53 and c-Myc mediate the CML network
To interrogate perturbations in BCR-ABL1 signalling of potential therapeutic value, isobaric tag mass spectrometry (MS) was used to compare treatment-naïve CML and normal CD34+ cells. 58 proteins were consistently deregulated in three CML samples (Online Methods; Supplementary Table 1). Dijkstra’s algorithm7 and MetaCore™ knowledge base (https://portal.genego.com/) were used to identify p53 and c-Myc as central hubs (Supplementary Table 2) in a CML network of 30 proteins (Fig. 1a) predominantly downstream of the TFs, with significant enrichment for p53/c-Myc targets (Fisher exact test, p=0.001). Whilst the majority of proteins downstream of p53 were down-regulated, those downstream of c-Myc included proteins up or down-regulated in CML, in keeping with c-Myc as an activator and repressor of gene transcription8. The deregulated network suggests an altered dependency on p53 and c-Myc in CML CD34+ cells.
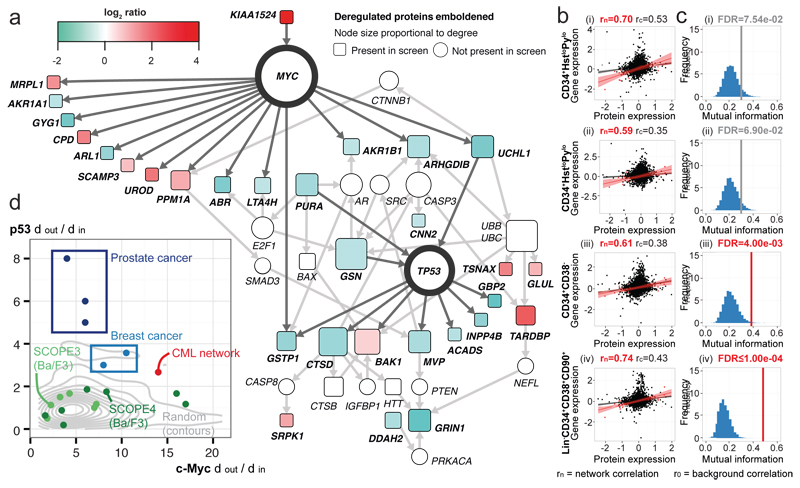
Figure 1.
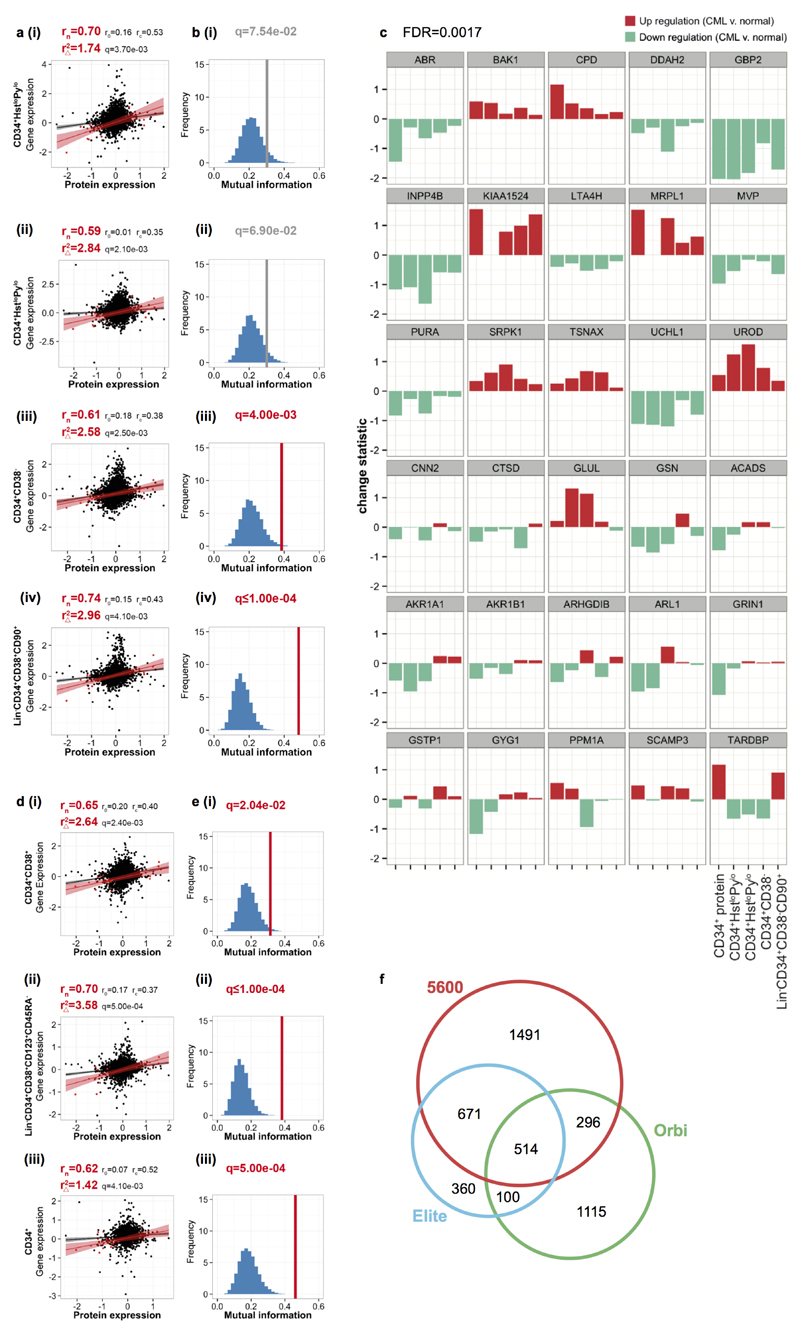
p53 and c-Myc network in CML regulation. (a) Network analysis reveals c-Myc and p53 central in a putative CML network. (b) Correlation between proteomic/transcriptomic deregulation in primitive (i-ii) CD34+HstloPylo (G0) (iii) CD34+CD38− (iv) Lin−CD34+CD38−CD90+ CML cells (●=all protein/genes; ●=network). (c) Gene/protein MI for the CML network (red FDR<0.05; grey FDR<0.10); FDR calculated using 10,000 re-samplings (blue histogram). (d) The out:in degree ratio for p53 and c-Myc in haematological PTK-regulated cell lines; other primary cancers and random protein networks.
This dataset represents the first relative quantitative comparison of CML to normal CD34+ cells using MS. Importantly CML initiating cells reside within the CD34+CD38−Lin− subpopulation and may differ to bulk CD34+ cells. To substantiate the CML proteome observations and investigate regulation in LSCs, we examined relevant, primary CML transcriptomic data. Network protein levels correlated well with respective gene levels, in both LSC (four independent datasets Fig. 1b; Extended Data Fig. 1a-c) and CD34+ progenitors (Extended Data Fig. 1d-e). Correlations were stronger for the 30 network candidates compared to all 58 deregulated proteins; seven datasets showed significant gain in r2 for network candidates (Extended Data Fig. 1a,d). The mutual information (MI) of proteomic/transcriptomic data for network proteins was significantly greater than random (Fig. 1c; Extended Data Fig. 1b,e). This consistent mRNA/protein correspondence, in both progenitors and LSC, confirmed the network was transcriptionally regulated, compatible with c-Myc and p53 function.
p53 and c-Myc play significant roles in oncogenesis and appear in many cancer networks. To distinguish true regulatory effectors, we assessed the bias towards outgoing vs. incoming signalling (degreeout/degreein or dout/din) for p53 and c-Myc. We generated networks from deregulated proteins in (i) primary MS datasets9–11; (ii) cell lines transduced with oncogenic PTKs driving haematological malignancies12; and from (iii) 50 randomly generated protein sets. Our network falls outside the expected random distribution and no other dataset exhibits greater downstream bias for p53 and c-Myc (Fig. 1d). These data support a novel network in, and unique to, CML centred on p53 and c-Myc.
Validation of network candidates
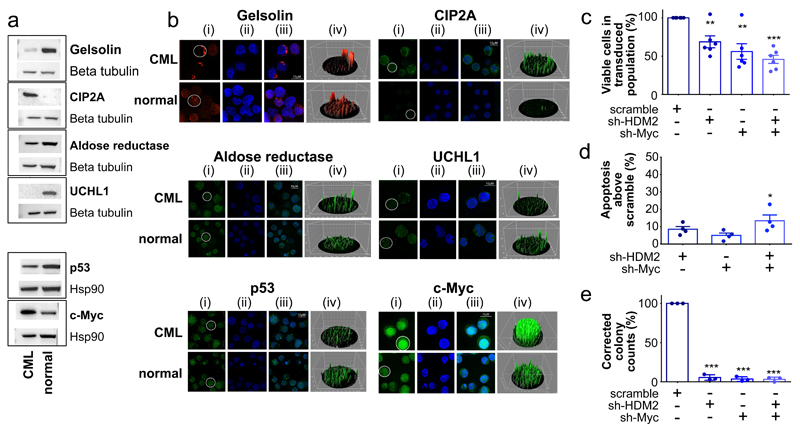
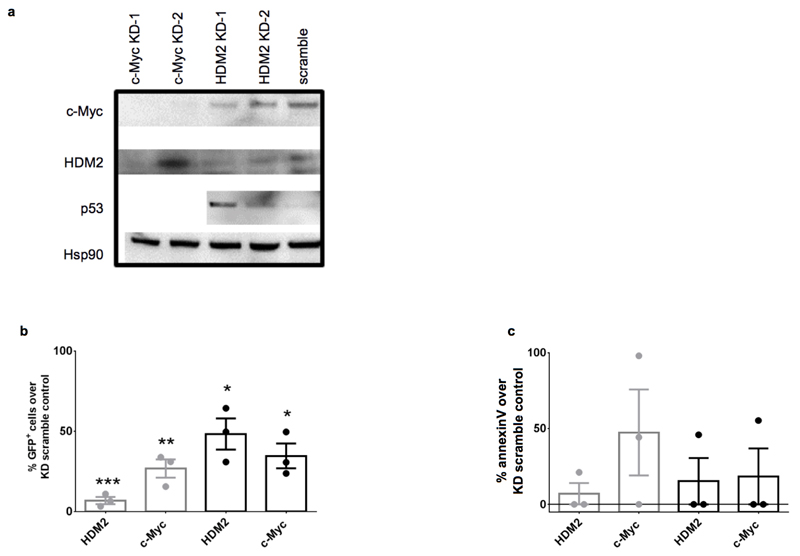
The CML network revealed well-characterised p53/c-Myc targets and proteins not previously associated with CML pathogenesis (Supplementary Table 3). To validate proteomic predictions (Fig. 1a), Gelsolin, CIP2A, UCHL1, aldose reductase, p53 and c-Myc were assessed using western blotting and immunofluorescence (IF) (Fig. 2a-b). Protein expression of gelsolin, CIP2A, UCHL1 and aldose reductase were consistent with CML network predictions (Fig. 2a). IF was confirmatory, highlighting the dramatic difference in CIP2A expression between normal and CML cells and intracellular localisation of gelsolin and aldose reductase (Fig. 2b). CML cells also expressed increased c-Myc and decreased p53 (Fig. 2a-b), correlating well with appropriate modulation of downstream targets. We therefore hypothesised that simultaneous p53 activation and c-Myc inhibition would kill LSC. To assess dual hub requirement for CML survival, lentiviral shRNA constructs (Extended Data Fig. 2a) were employed. Knockdown (kd) of HDM2 (E3 ligase/negative regulator for p53), c-Myc, or both, in CML CD34+ cells reduced viability and enhanced apoptosis; the combined effects were synergistic. In colony-forming cell (CFC) assays, effects were more dramatic with single or combined kd, strengthening the hypothesis that p53 and c-Myc are critical for CML cells’ survival (Fig. 2c-e; Extended Data Fig. 2b-c). We then investigated synergistic interactions between p53 and c-Myc testing clinically-tractable inhibitors.
Figure 2.
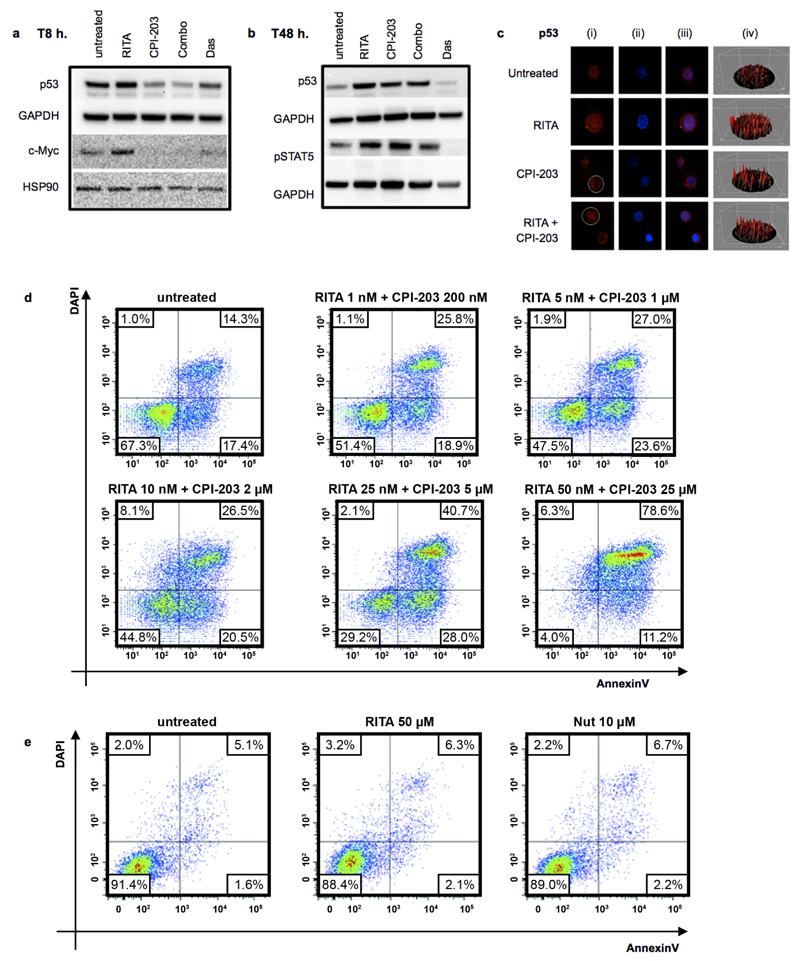
Validation of proteomic network. (a) Network proteins, p53 and c-Myc western blots using CML and normal CD34+ cells. For gel source data, see Supplementary Figure 1. (b) Network proteins validated by IF in CML and normal CD34+ cells labelled (i) green or red (ii) nucleus (blue) using DAPI (iii) overlays of images (iv) 3D fluorescent signal. (c) CML CD34+ cells post HDM2, c-Myc, or scramble kd (n=6 PS). (d) Apoptosis post kd (n=4 PS). (e) CFC from kd (n=3 PS). Values normalised to scrambled control, mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
RITA and CPI-203 synergize CML CD34+ kill
To target identified hubs, we selected RITA, which binds p53 and blocks its degradation, and CPI-203, a bromodomain and extra terminal protein (BET) inhibitor hindering transcription by disrupting chromatin-dependent signal transduction13,14. As anticipated15, c-Myc was down-regulated 8 hours (h.) post CPI-203 treatment (Extended Data Fig. 3a). CPI-203 also reduced p53 at 8h. RITA subtly increased p53 by 8h., further enhanced by 24-48h. Dasatinib (Das) at 150nM (≥achievable in patients to fully inhibit Bcr-Abl16) gradually reduced p53 levels and inhibited phosphorylation of STAT5 as previously observed.17,18 RITA with CPI-203 for 8h. reduced both p53 and c-Myc, suggesting a dominant effect of CPI-203 at this early time point, but by 48h. markedly increased p53 (Extended Data Fig. 3a-c).
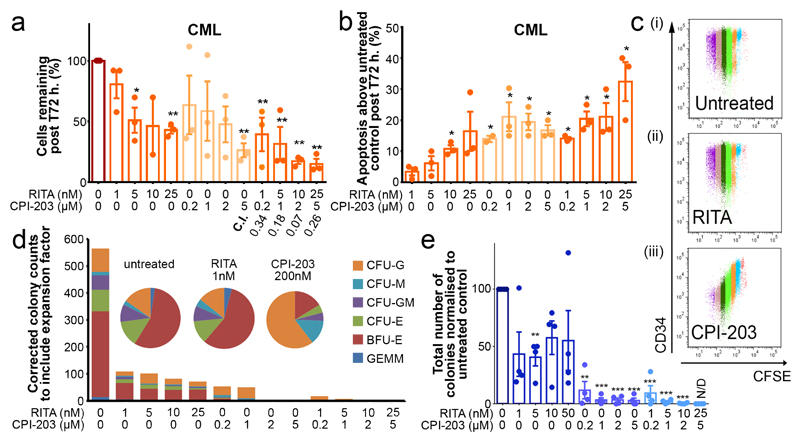
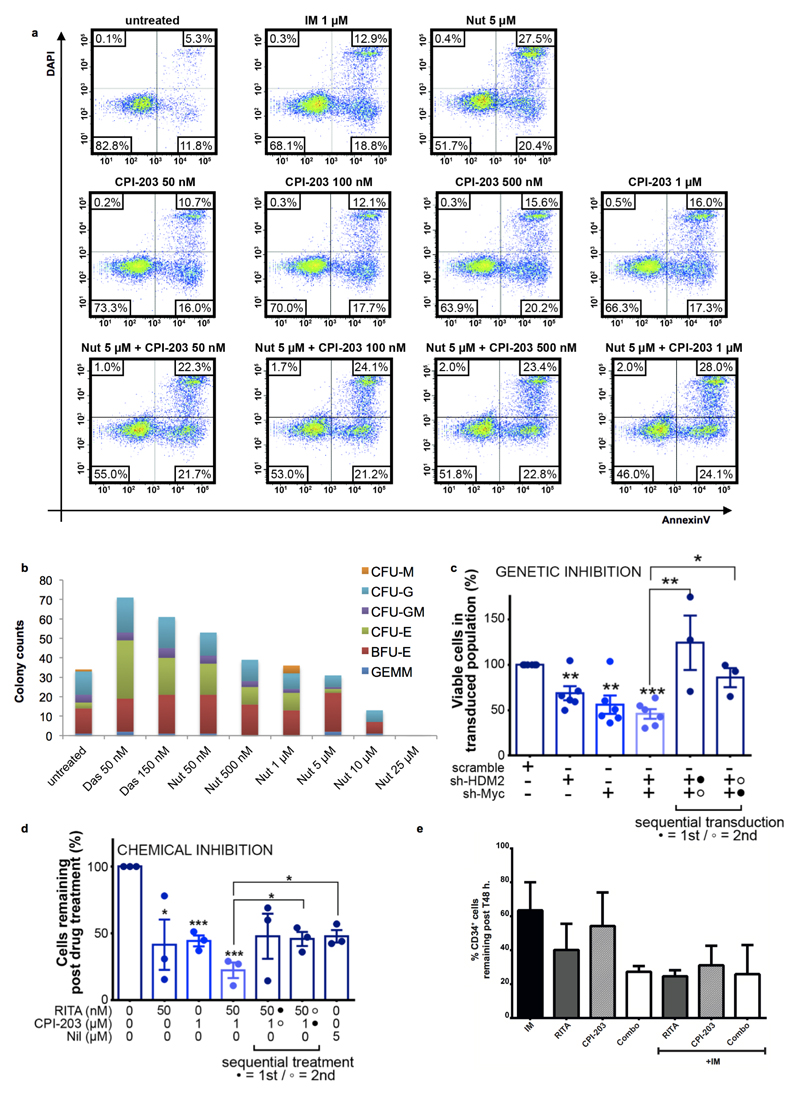
RITA or CPI-203 treatment of CML CD34+ cells for 72h. reduced viability in a concentration-dependent manner and induced significant apoptosis; combining drugs resulted in further significant reductions in these parameters (Fig. 3a-b; Extended Data Fig. 3d). CFSE (cell division tracker)/CD34-labelling showed that as CML cells divided in the presence of CPI-203, there was clear and rapid loss of CD34 expression not seen with RITA (Fig. 3c), suggesting that c-Myc (a predominant target of CPI-20315) inhibition induces differentiation of CML CD34+ cells. Differentiation was further suggested by skewing of the morphology, size and number of CFC (Fig. 3d). RITA decreased CFC but did not affect colony types (Fig. 3d-e). By inducing apoptosis and differentiation, the drugs may synergise and enhance elimination of CML. By measuring drug-dose response19, combination therapy was potently synergistic with combination indices (C.I.) from 0.07-0.34 (Fig. 3a). Nutlin-3a (Nut), another HDM2 inhibitor, produced similar results. The effects of RITA and Nut were p53 dependent as K562 cells lacking p53 were non-responsive (Extended Data Fig. 3e, 4a-b). Since CPI-203±RITA reduced p53 at the early time point, sequential inhibition of HDM2 and c-Myc was tested using chemical and genetic approaches. Neither inhibition of HDM2 prior to c-Myc nor vice versa improved cell kill compared to simultaneous kd or drug inhibition, or compared to Nilotinib (Nil) (Extended Data Fig. 4c-d).
Figure 3.
Modulation of p53 and c-Myc demonstrates CML sensitivity. (a) Drug titrations: cell viability with combination indices (C.I.) (n=3 PS), (b) apoptosis (n=3 PS). (c) CFSE/CD34 labelled cells. Cell divisions are multi-coloured. Representative of n=3 PS. (d) CFC from treated cells. Representative of n=3 PS. (e) Averaged CFC (n=3 PS). One experiment represented in (d), mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
CML patients receive a TKI, irrespective of response. We therefore assessed RITA and CPI-203 effects in IM-pre-treated CML CD34+ cells. IM neither ameliorated nor enhanced the efficacy of RITA and/or CPI-203 (Extended Data Fig. 4e).
RITA and CPI-203 eliminate LSC
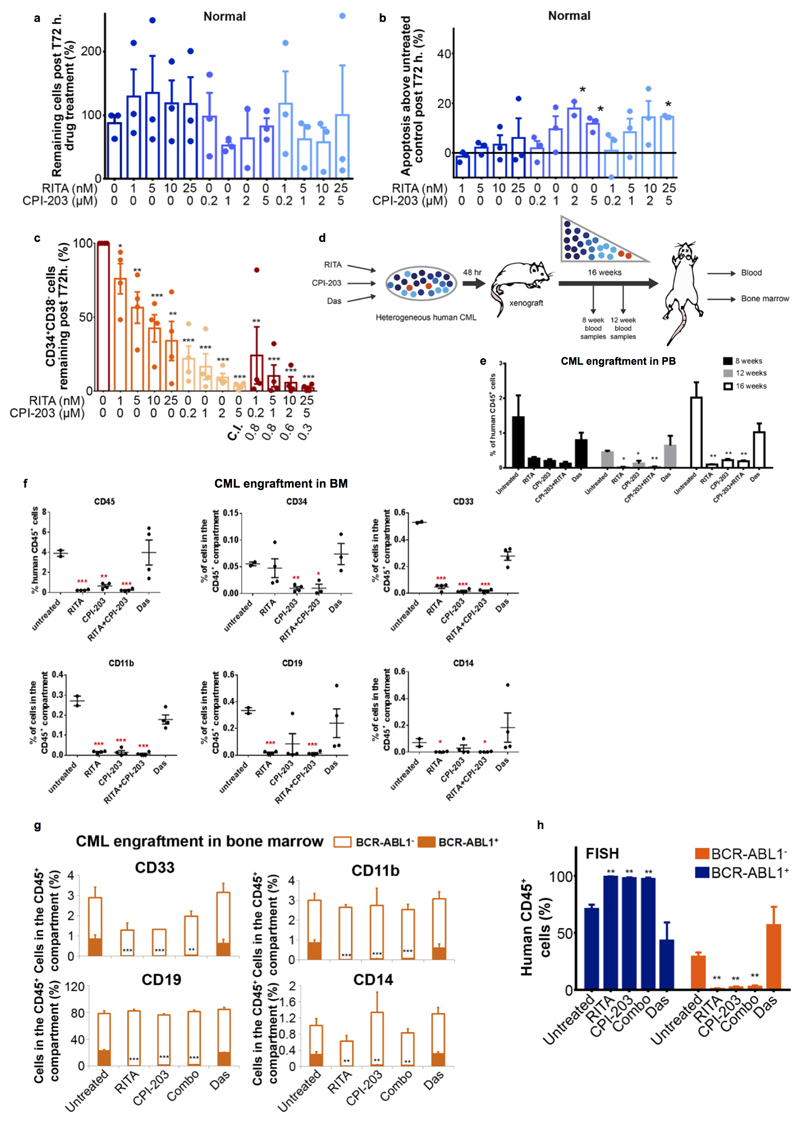
The CML network suggested an altered dependency on p53/c-Myc signalling. We therefore hypothesised that normal cells may be less susceptible to the drug combination. Treatment of normal CD34+ cells with single agents or combination had no significant effects on cell counts. However, increased apoptosis was observed at higher CPI-203 concentrations (2/5μM) and with the highest combination (RITA 25nM, CPI-203 5μM; (Extended Data Fig. 5a-b)). In CML cells apoptosis was observed with all four CPI-203 and combination concentrations (Fig. 3b), confirming a therapeutic window.
To confirm in silico predictions (Fig. 1b-c, Extended Data Fig. 1c) that altered dependency on p53 and c-Myc extended to primitive LSC, we exposed CML LSC to the drug combination. LSC were defined as either CFSEmax or CD34+CD38−. As shown previously16, in comparison to untreated control, over 5 days the CFSEmax population persisted in response to Das and Nil, but was significantly reduced by CPI-203 alone and by combination treatment (Fig. 3c, 4a-b). Over 72h. RITA with CPI-203 was also effective in synergistically eliminating residual CD34+CD38- cells (C.I.=0.3-0.8, Extended Data Fig. 5c).
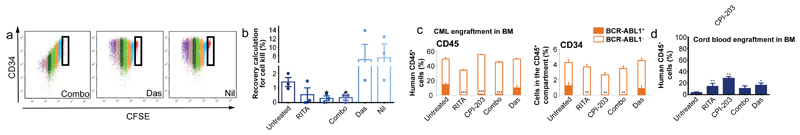
Figure 4.
p53/c-Myc abrogation in normal/primitive CML cells. (a) CFSE/CD34 labelled CML cells. (b) Recovery of CFSEmax CML cells after 5 days treatment (n=3 PS). (c) BM analyses of human CML, replicated twice (2 PS), with a minimum of n=6 mice/arm. (d) BM analyses of human Cord Blood replicated once n=5 mice/arm; mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
HSC and LSC are most stringently defined by their engraftment capacity at 16 weeks. We exposed CML CD34+ cells to RITA, CPI-203, the combination or Das for 48h. prior to transplantation into sub-lethally irradiated NSG mice (Extended Data Fig. 5d). Human CD45+ cells were detectable in peripheral blood (PB) at 8, 12 and 16 weeks and in bone marrow (BM) at 16 weeks post-transplantation. Das had no significant effect on NSG repopulating CML LSC representing the most primitive long-term engrafting cells. In contrast RITA, CPI-203 and the combination, reduced engraftment as indicated by the decreased CD45, CD34, CD33, CD11b, CD19 and CD14+ cells (Extended Data Fig. 5e-f). Using a CML sample known to engraft both BCR-ABL1+ and BCR-ABL1- cells by D-FISH, there was a marked decrease in the long-term-engrafting potential of RITA, CPI-203 or combination treated leukaemic cells, with no significant effect on non-leukaemic populations (Fig. 4c-d, Extended Data Fig. 5g-h). Experiments using cord blood CD34+ cells confirmed the selectivity of RITA and CPI-203 for BCR-ABL1+ v. BCR-ABL1- stem cells (Fig. 4d).
Mechanism of LSC kill and clinical scope
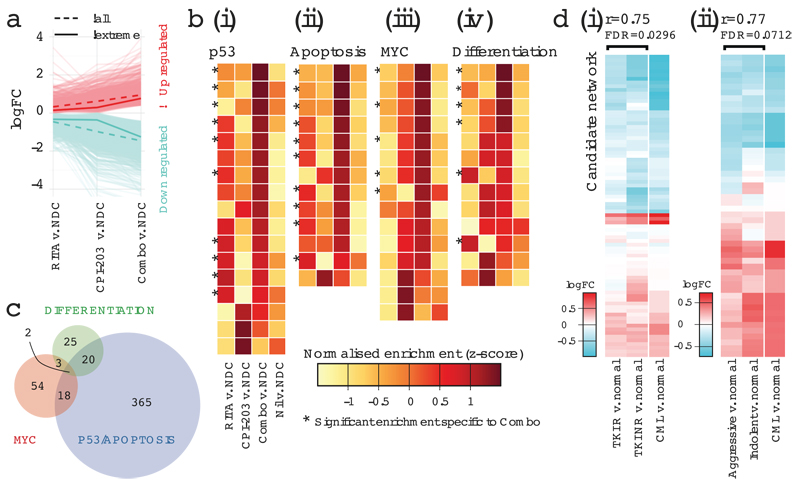
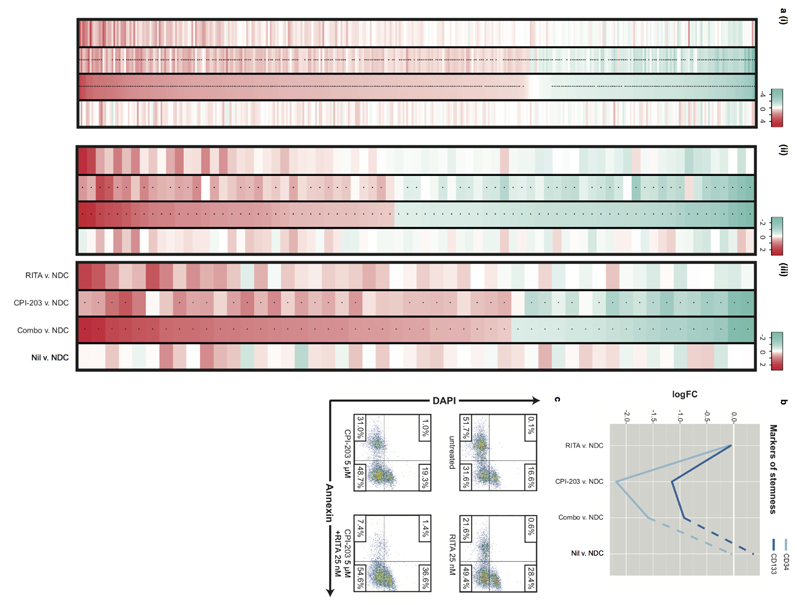
To understand the mechanism(s) underlying reduction of CML stem and progenitor cells in response to RITA and CPI-203, RNA-seq was performed. 2,134 of the 12,248 genes sequenced were identified as synergistically modulated by combination treatment; 166 demonstrated extreme synergy (Fig. 5a). Moreover, 81% of the genes differentially expressed in response to the combination were deregulated in the same direction with RITA or CPI-203 (χ²(1)=891.93, p<0.01). While transcriptional responses to RITA or CPI-203 were enriched for p53/apoptosis or c-Myc/differentiation (not found with Nil) respectively, the combination induced enhanced or additional enrichment of these molecular signatures and pathways (Fig. 5b; Extended Data Fig. 6a-c; Supplementary Tables 5-7). Furthermore, stem/progenitor markers CD34 and CD133 were dramatically down-regulated by CPI-203 and the combination, but not by RITA or Nil (Extended Data Fig. 7b). The enrichment in p53/apoptosis, c-Myc and differentiation pathways paralleled the in vitro phenotypic effects observed. Limited overlap in gene membership of the signatures identified in silico demonstrates that distinct molecular components contribute to single and combined drug responses (Fig. 5c).
Figure 5.
Mechanism and clinical relevance of treatment. (a) Molecular synergy for 100nM RITA, 1µM CPI-203 and RITA+CPI-203 (Combo) 24h. treatment; mean (μ) expression of ‘all’ and ‘extreme’ synergistic genes summarized as indicated. (b) Enrichment of (i) p53 (ii) apoptosis (iii) MYC (iv) differentiation MSigDB signatures. (c) Gene membership of three functional signatures. (d) Comparison of transcriptional profiles of (i) R/NR-CML (TKI-responder/TKI-non-responder) vs. normal and (ii) Aggressive/Indolent CML vs. normal for our candidate network (Fig. 1a).
CML stem cell persistence is an issue for all CML patients, however many also exhibit or acquire TKI-resistance or demonstrate a more aggressive clinical phenotype20. These represent patients in whom novel agents targeting p53 and c-Myc would first be tested. To investigate whether the deregulated p53/c-Myc network is present in both TKI-responder (TKI-R) and TKI-non-responder (TKI-NR) patients21 and in more advanced forms of CML20 we considered data from CD34+ CML cells derived from suitable patient cohorts. Transcriptional expression of the network components was highly correlated across all CML v. normal comparisons, irrespective of TKI response or clinical phenotype (Fig. 5d). In keeping with these in silico data, CD34+ cells from a TKI-NR patient showed high levels of apoptosis after treatment with RITA and/or CPI-203 (Extended Data Fig. 7c), suggesting that these drugs should be of therapeutic value for such patients.
RG7112/7388 and CPI-203/0610 therapy
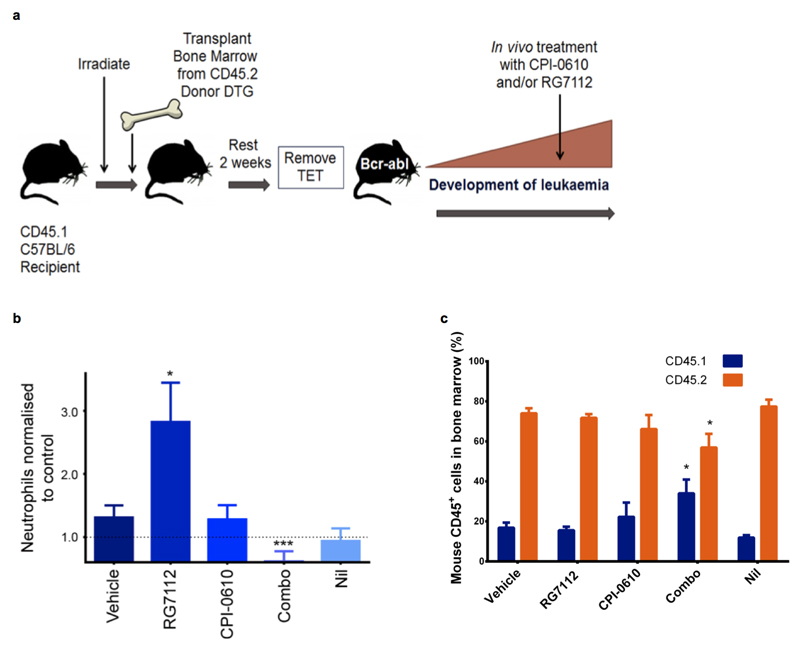
To progress the drug combination towards the clinic we employed complementary pre-clinical mouse models and introduced RG7112/7388, HDM2 inhibitors22 and CPI-0610, a BET inhibitor23; drugs already advanced in clinical trials in humans. In the SCL-tTA-BCR-ABL double transgenic (DTG) leukaemia mouse model BCR-ABL1, driven from the stem cell promoter (SCL), is inducible in HSC by tetracycline withdrawal (tTA), resulting in a transplantable CML-like disease with increased myeloid counts and splenomegaly16,17,24.
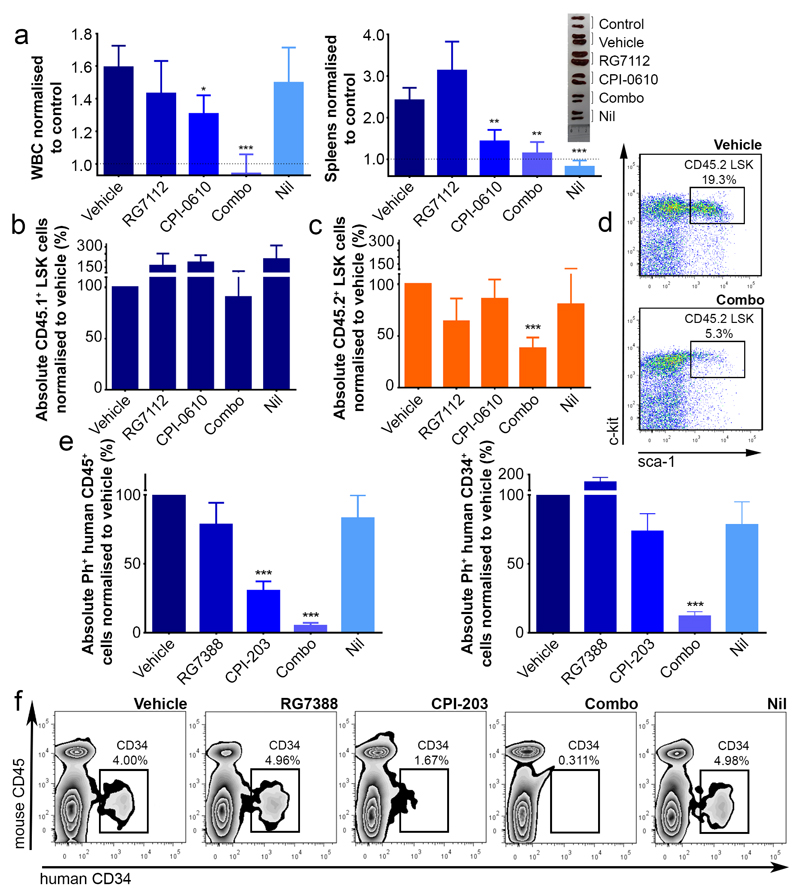
Following irradiation C57BL/6 CD45.1+ mice were used as recipients and CD45.2+ mice as DTG BM donors (Extended Data Fig. 8a). Following transplantation (to synchronize leukaemia development and assess transplantable LSC) CML was induced. In mice and/or rats, RG7112 at 50-200mg/kg and CPI-0610 at 15-60mg/kg have demonstrated on-target effects in tumours22,23 Excellent tolerability was achieved with modest doses of RG7112 (50mg/kg once daily), CPI-0610 (15mg/kg twice daily, both for 4 weeks), selected to demonstrate synergy. White blood cell and neutrophil counts returned to non-leukaemic control levels with the drug combination, but not with single treatments (Fig. 6a; Extended Data Fig. 8b). Whilst CPI-0610, Nil and the combination significantly reduced spleen size (Fig. 6b), only the combination significantly reduced donor leukaemic CD45.2+ cells in BM (protected by the niche), whilst simultaneously allowing recovery of host normal CD45.1+ cells; CD45.1:CD45.2 ratio modulated from 20:80 (untreated) to 40:60 (combination) (Extended Fig. 8c). At the stem cell level donor leukaemic Lin−Sca-1+c-Kit+ (CD45.2+ LSK) cells were reduced by >60% by the combination, whilst host LSK were unaffected. None of the single arms reduced LSK (Fig. 6b-d) supporting synergistic effects demonstrated in vitro.
Figure 6.
Targeting p53 and c-Myc in CML elicits synergistic kill in BCR-ABL1+ LSC. (a) WBC, spleen weights – normalised to control (·····) (experiments replicated twice, minimum n=7 mice/arm; vehicle=no drug control). (b-d) BM stained for CD45.1/2 and further gated on Lin−Sca-1+c-Kit+ (LSK). Drug treatments (experiments replicated twice, minimum n=5 mice/arm). (e-f) NSG mice in vivo treatment: BM stained for human Ph+ CD45+ (left) and further gated on CD34+ cells (right). (g) representative CD34+ dotplots (experiments replicated twice (2 PS), minimum n=9 mice/arm); mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
To validate these therapeutic in vivo effects extended to human CML, two cohorts of sub-lethally irradiated NSG mice were transplanted with independent CML CD34+ samples and treated with RG7388 and CPI-203 (75-100mg/kg and 6-7.5mg/kg, respectively) for 3-4 weeks. Of the single agents only CPI-203 showed a consistent effect. The drug combination, however, eliminated 95% of Ph+ CD45+ and 88% of CD45+CD34+ subsets (Fig. 6e-f). These results were significant as compared to vehicle (p<0.001) or Nil (p=0.0016 [CD45+]; p=0.0047 [CD45+CD34+]) and as compared to RG7388 (p=0.0017 [CD45+]; p=0.0004 [CD45+CD34+]) or CPI-203 (p=0.0046 [CD45+]; p=0.0008 [CD45+CD34+]), respectively (Fig. 6e-f), again suggesting a high degree of synergy.
Discussion
This work demonstrates the potential of unbiased, systems approaches to uncover new therapeutic options by analysing primitive stem cell subsets from primary material. We identified that p53 and c-Myc work together with BCR-ABL1 to shape leukaemia stem cell phenotype and show that modulation of both p53 and c-Myc is critical to drive synergistic enhancement of apoptosis and differentiation seen in vitro, in vivo, at the stem cell level and the molecular level by RNA-seq.
p53 and c-Myc have individually been identified as proteins in CML pathobiology17,25–30 and cancer31–33, but have not previously been considered for dual targeting. In recent CML studies, enhanced LSC kill converged on p53 as the mediator of apoptosis17,25,34. CML LSC are also susceptible to enhancement or depletion of c-Myc. Following deletion of the E3 ligase Fbxw7, c-Myc increases with p53, resulting in cell cycle entry and p53-dependent apoptosis29,30. However Fbxw7 may not represent a viable drug target based on its role in haemopoiesis, tissue stem cells and importantly as a tumour suppressor. Currently, there is interest in drugging the spliceosome machinery, particularly for Myc-driven cancers, however a therapeutic window remains to be established and exploited with well tolerated agents35,36. BET inhibition is a rapidly expanding market with multiple agents in phase 1-2 development. These agents are well tolerated and demonstrate efficacy in haematological malignancies. Resistance to BET inhibitors evolves through epigenetic mechanisms37, however our combination approach will be less susceptible to resistance.
Over the last decade CML has been transformed from a fatal cancer to a manageable disease with life-long therapy. Despite recognition that LSC prevent cure, the paradigm established by TKI means that novel drugs must be safe, supported by a clear therapeutic window and easy to administer. As a result few pre-clinical studies have reached the clinic and trials fail due to toxicity and poor recruitment24,38. CML is often regarded as an incomplex cancer, driven solely by BCR-ABL1, yet we do not understand why targeting BCR-ABL1 does not eradicate LSC, nor cure CML. Our work shows that BCR-ABL1 re-programs potent oncoproteins and tumour suppressors, to establish a signalling network that underlies the propagation of CML. Critically we identified that simultaneous perturbation of p53 and c-Myc, mechanistically driving modulation of p53, apoptosis, c-Myc and differentiation pathways, improved selective kill of LSC, as compared to TKI. Nil was ineffective against these pathways, potentially explaining why TKI are not sufficient to cure CML. The fact that the aberrant network was similarly regulated in TKI-R, TKI-NR, aggressive and indolent CML, coupled with the availability of well tolerated, oral agents in an advanced stage of development, now offers an entirely novel approach for the treatment of CML, with therapeutic potential to address CML LSC persistence and improve outcome for CML patients.
Online Methods
Patient samples
Patient samples (PS) were leukapheresis products taken at time of diagnosis with chronic phase CML, with written informed consent in accordance with the Declaration of Helsinki and approval of the Greater Glasgow and Clyde National Health Service Trust Institutional Review Board. CD34+ cells were enriched using CliniMACS (Miltenyi Biotec), with stem cell subsets purified by FACS. CML CD34+ samples were cultured in serum-free medium (SFM) supplemented with growth factors as described previously16. Normal CD34+ were CD34-enriched leukapheresis products or cord blood maintained as described for CML CD34+ samples. All PS and relevant clinical data are summarized in Supplementary Table 4. All in vitro work was performed with a minimum of 3 PS (3 biological replicates) unless otherwise indicated. Unless otherwise indicated each PS was analysed as an individual sample replicated once in an experiment.
Cell lines
The HeLa cell line was obtained from DSMZ (German Collection of Microorganisms and Cell Cultures)(originally deposited by ATCC). HeLa cells were sub-cultured in RPMI 1640 (10% FCS+ 2mM L-glutamine, 100units/ml Penicillin and 100mcg/ml Streptomycin (Gibco- Life Technologies)) (Passage 3). The K562 cell line (DSMZ) was sub-cultured in IMDM (10% FCS+ 2mM L-glutamine, 100units/ml Penicillin and 100µg/ml Streptomycin (Passage <6) the cell lines were not authenticated between passage 2-6. Cell lines were mycoplasma negative in DAPI, microbiological culture, RNA hybridization, and PCR assays.
CML cytoplasmic preparations for MS
CML and normal CD34+ cells were thawed and cultured overnight as described24. Cytoplasmic preparations were prepared using the Active Motif Nuclear Extraction Kit (Active Motif).
Materials
RITA (CAS 213261-59-7) No:10006426, Nutlin-3a (CAS 675576-98-4) No:18585 (Cayman Chemical) and Nil (CAS 641571-10-0) (Selleck.chem.com) were stored as per manufacturer’s instructions. CPI-203 and CPI-0610 were obtained from Constellation Pharmaceuticals and kept as a solid powder at room temperature (RT). Das (Selleck.chem.com) was kept as a stock solution (10mg/ml) in DMSO (Sigma-Aldrich) and prepared and stored in aliquots at −20°C. IM (LC Laboratories) was stored at 100mM in distilled water at 4°C. RG7112 and RG7388 were supplied by Roche. A list of all antibodies used are listed in Supplementary Table 8
Proteomics
Methods employed have been described12. 20µg of protein was isobarically tagged (iTRAQ™ reagent, ABSciex). Peptides were identified by RP-LC-MS/MS on three different instruments: ABSciex Q-STAR® Elite, Thermo LTQ Orbitrap Velos, ABSciex TripleTOF 5600. For the 5600 and Elite, dried peptide fractions were re-suspended in 15µl 3% (v/v) acetonitrile, 0.1% (v/v) formic acid and 20mM citric acid. For each analysis 5µl peptide sample was loaded onto a nanoACQUITY UPLC Symmetry C18 Trap (5µm, 180µm×20mm) and separation of the peptides was performed using nanoACQUITY UPLC BEH C18 Column (1.7µm, 75µm×250mm). For the Orbitrap, 10% of the peptide sample was loaded onto Acclaim PepMap µ-Precolumns, analytical separation of the peptides was performed using Acclaim PepMap RSLC C18 Columns. Data were acquired using the information dependent acquisition (IDA) protocol.
Elite and 5600 data were processed by a ‘Thorough’ search against the UniProtKB/SwissProt human database containing 532146 sequence entries using ProteinPilot™ Software 4.1, revision number 460, Paragon™ Algorithm 4.0.0.0. Orbitrap data were analysed using Proteome Discoverer 1.3. The data were searched using the MASCOT node of Proteome Discoverer with the UniProtKB database (release 2011_11). The proteins observed in the three datasets demonstrated that using multiple instruments enhanced coverage (Extended Data Fig. 1).
Analysis and integration of MS proteomic data
MS datasets were filtered for peptides observed in all channels (one normal sample was removed from all experiments due to poor labelling). Deregulated proteins were identified using a threshold of mean ±2SD on CML v. normal log2 ratios. This candidate list was refined to include only those proteins corroborated by (i) log ratio changes of ±0.5 in murine Ba/F3 ± BCR-ABL1 MS data12 (ii) a complementary CD34+ cell proteomics dataset39 and/or (iii) all instruments within the current experimental dataset. A parallel, manual inspection retained candidates if (i) log ratios were ≈±1.3; or (ii) log ratios were lower and neither alternative instrument reported differential expression in the opposite direction; this manual selection step was blinded. Together these filtering steps reduced the candidate list to 58 proteins (see Supplementary Table 1).
Formation of candidate network
The MetaCore™ implementation (13/06/12) of Dijkstra’s shortest path algorithm7, a general purpose algorithm that identifies the shortest paths between “seed” nodes of interest in a graph, was used to build a network around the 58 deregulated proteins (the graph used was the fully manually annotated MetaCore™ KB). Paths between seeds were limited to length=2 and all shortest paths of the minimum length were retained in the resulting network. Topology statistics (Supplementary Table 2) were calculated using the igraph package in R.
Transcriptomic data analysis
Three bulk CD34+ (1-3) and four primitive LSC (4-7) CML chronic phase v. normal datasets are discussed: (1) E-MTAB-2581: Affymetrix Human Gene 1.0 ST Array transcriptional data from newly diagnosed CML chronic phase and normal progenitor cells (CD34+CD38+) from G-CSF mobilized peripheral blood (PB). mRNA was extracted using the RNeasy Mini Kit (Qiagen) and DNase I treated on columns using the ribonuclease (RNase)-Free DNase Set (Qiagen). Affymetrix GeneChIP analysis was performed using 50ng of RNA to manufacturer’s instructions; (2) GSE4792740; (3) GSE555041; (4) E-MTAB-2581 (as described in (1) but for CD34+CD38− cells); (5) GSE4792740; (6) GSE2473942; (7) E-MTAB-250843.
CEL files were obtained directly from collaborators or public repositories. All transcriptional data were RMA normalised with the exception of GSE5550 for which only the VSN44 normalised data were available. Log2 scale expression values were analysed using limma45 (p value correction by Benjamini-Hochberg46). In calculating the correlations and MI statistics (Fig. 1b-c, Extended Data Fig. 1), multiple probesets corresponding to single genes were median averaged.
Calculating correlations across multiple datasets
Ensembl’s web services were used to map between (i) human and murine datasets via orthologs and (ii) human transcriptomic and proteomic datasets via HGNC symbols. The Bioconductor package biomaRt (v2.18.0) was used in R (v3.0.1). Pearson’s product moment correlation coefficients (r) were calculated across datasets for the 58 candidates and the 30 networked candidates (rc and rn respectively in Extended Data Fig. 1). Resulting r2 values quantify the proportion of variability captured by the linear relationship and r2Δ is defined as r2n/r2c, that is, the ratio of r2 for the 30 networked candidates to the r2 for the 58 candidates. FDRs for the r2Δ observed were generated by considering 10,000 random samplings of each dataset and counting the number of random samples meeting or exceeding the r2Δ and r2n statistics.
Calculation of MI
We calculated the MI values for 10,000 random subsets (of size 30) to generate a distribution of MI values that we would expect by chance (expression values binned between -3 and 3, bin width=0.1; entropy package in R); these are the values summarized by the distributions plots in Fig. 1c and Extended Data Fig. 1. The FDR values represent the proportion of random subsets that generate an MI greater than or equal to the MI for the network.
False discovery rate calculation assessing bulk proteomic/primitive transcriptional consistency
Of the 30 proteins in Extended Data Fig. 1, 21 showed consistency of deregulation in at least three of the primitive transcriptional datasets and the bulk proteomic dataset. To assess how likely it would be to observe such consistency by chance, the data were randomly permuted 10,000 times and permutations exhibiting similar deregulation consistency (i.e., deregulation correspondence across four datasets) recorded.
Topological analysis of p53/c-Myc in other MS datasets
Comparison of leukaemogenic PTK proteomic effects and three primary proteomics datasets, describing two types of breast cancer11 (ductal carcinoma in situ and invasive carcinoma), three types of prostate cancer10 (non-aggressive, aggressive and metastatic) and cervical9 cancer, were obtained directly from the authors. In each dataset, deregulated proteins were identified using z-scores ±2 when considering cancer v. normal log2 ratios and subjected to the same MetaCore™ network building process as described above. In the case of the cervical cancer dataset no network could be found and the data were removed from the analysis. In addition, 50 sets of 58 random proteins were generated from the list of all proteins observed across the three MS datasets and subjected to the same network building process. The ratio dout/din was calculated to quantify the bias of out-going to in-coming connections to/from p53 and c-Myc (Fig. 1d).
Generation of responder/non-responder and aggressive/indolent CML transcriptional profiles
The PB “validation” set TKI-R/TKI-NR samples in GEO dataset GSE14671 were integrated with a material-matched CML v. normal dataset (CD34+CD38+ cell data from E-MTAB-2581) using COMBAT47 (Bioconductor package inSilicoDb v1.10.1). All probeset-to-probeset mappings between the Affymetrix HG U133+2 and Affymetrix HuGe 1.0 ST chips (obtained via Bioconductor’s biomaRt package) were retained and used by COMBAT. The TKI-R and TKI-NR samples were compared to the integrated normal data (using limma) to generate logFC representing differential expression (see left two lanes in each panel of Fig. 5e). The pattern of differential expression in the TKI-R/TKI-NR v. normal comparisons was then compared to that of CML v. normal, as calculated separately by limma in the material-matched datasets (E-MTAB-2581) (see right lane in each panel of Fig. 5e). Transcriptional profiles for probe pairs corresponding to the 30 members of the candidate network were identified using HGNC symbols (data corresponding to TARDBP were removed due to the large number (66) of corresponding probe pairs). A FDR was calculated using 10,000 resamplings of the merged dataset to describe the likelihood of observing a correlation as high or greater by chance. The transcriptional data for the aggressive and indolent samples in ArrayExpress dataset E-MIMR-17 were processed as above, but integrated with the CML and normal CD34+ dataset GSE5550 using all probeset-to-probeset mappings between the Affymetrix U133a and Affymetrix HG Focus chips (again obtained using biomaRt).
Enrichment of MSigDB signatures
For the candidate network analysis (the results of which are shown in Supplementary Table 3), MSigDB signatures (C2: curated genesets) were accessed via the c2BroadSets object in GSVAdata v 1.0.0. using R. Enrichment scores were calculated using the hypergeometric distribution as implemented by dhyper(). Signatures corresponding to CML, c-Myc and p53 related biology were extracted using appropriate regular expressions on the signature name. In the RNA-seq analysis (Fig. 5b), the MSigDB signatures (C2: curated genesets) were identified as significantly differentially expressed in the TMM/VOOM normalised RNA-seq data using GSVA48 and limma49. Significant pathways were identified using FDR=0.05 threshold on corrected p values46. The p53, apoptosis, c-Myc and differentiation signatures were extracted using appropriate regular expressions on the signature name.
Enrichment of PANTHER pathways
The top 1500 differentially expressed genes (as ranked by increasing p value, calculated by limma49) were identified comparing treated/untreated samples. These genes and their logFCs for each arm were uploaded to PANTHER (http://www.pantherdb.org/) and subjected to a Mann-Whitney U Test50 to identify enrichment of PANTHER pathways. Fig. 5c shows enrichment results (without Bonferroni correction) including the hypothesised direction of pathway deregulation.
Cell counting and apoptosis assays
CML CD34+ cells were seeded at 1-2×106 cells/ml before drug treatment and counted by trypan blue (Sigma-Aldrich) exclusion. Apoptosis was quantified by staining with AnnexinV–APC and 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI). In specified experiments, CML and human cord CD34+ cells were labelled with CD34-APC and CD38-FITC or PerCP-Cy5.5 and sorted using the FACSAria (BD). The selected CD34+CD38− cells were analysed 72h. post drug treatments. To measure the dose-effect relationship of each drug and its combination and to determine synergy, combination indices (CI) were calculated using the Calcusyn software package (BioSoft). Except where documented, all results are expressed as a mean±minus standard error of mean (s.e.m.).
Colony forming cell assay
CD34+ cells were treated for 72h. at the indicated concentrations of RITA, CPI-203 and Das. Drug-treated cells (2,000 cells per plate) were washed and seeded in Methocult H4435 (STEMCELL Technologies). CML cells were transduced with lentivector constructs and sorted then washed and seeded into methylcellulose. Colonies were assessed 10-14 days after plating.
Western blotting
CD34+ cells were lysed in RIPA buffer with inhibitors and westerns performed by standard protocols.
Immunofluorescence microscopy
CML CD34+ cells were left untreated or treated with the indicated drugs for 24h. Cells were harvested and spotted onto slides coated with poly-l-lysine and fixed with 3.7% (w/v) formaldehyde and permeabilized using a 0.25% (w/v) Triton-PBS solution for 15min. Cells were blocked with 5% (w/v) BSA-PBS and stained with primary and secondary antibodies. Cells were concurrently stained with DAPI. Cells were imaged using a Zeiss Imager M1 AX10 fluorescence microscope (Carl Zeiss) and subjected to deconvolution (AxioVision software; Carl Zeiss) for image manipulation. Fluorescent signal was measured in 3D by Image Processing and Analysis in Java (Image J) program.
Tracking cell divisions
CFSE (Molecular Probes) /CD34 staining was performed and cell divisions identified as before3.
Lentivirus transduction
The pCMV-VSV-G and pCMV-HIV1 were provided by Dr John Rossi (Beckman Institute of City of Hope, Duarte, CA). The pLKO-GFP came from Professor Kamil Kranc (University of Edinburgh). The following optimized pLKO vectors were purchased from Open Biosystems and sub cloned into the pLKO-GFP vector:
-
1)
TRCN0000003380: MDM2 shRNA bacterial stock NM_002392.x-1495s1c151
-
2)
TRCN0000355728: MDM2 shRNA bacterial stock NM_002392.3-1496s21c1
-
3)
TRCN0000174055: c-Myc shRNA bacterial stock NM_002467.2-1377s1c252
-
4)
TRCN0000039642: c-Myc shRNA bacterial stock NM_002467.2-1377s1c153
Transduction of HeLa cell lines was performed at a MOI 1-10 with 70-95% of the cells expressing GFP after 48h. For transduction CD34+ cells were cultured in medium supplemented with growth factors (IL-3 25ng/mL, IL-6 10ng/mL, Flt-3L 100ng/mL, SCF 50ng/mL, TPO 100ng/mL) for 48h., followed by two exposures to concentrated virus-containing supernatants (MOI=5) via spinoculation. Cells were harvested 48h. post second transduction and analysed or sorted for GFP positivity.
Transduced viable cells (assessed as AnnexinV-/DAPI− percentages multiplied by the absolute cell count) are presented as a percentage of CML CD34+ cells transduced with scramble control.
Immunodeficient mouse engraftment
For the ex vivo drug studies CML (2×106 cells per mouse) or cord blood (2×105) CD34+ cells were cultured with the indicated drugs (RITA 70nM, CPI-203 1µM and Das 150nM). After 48h. cells were transplanted via tail vein into female 8-10-week-old sub-lethally irradiated (2.5Gy) NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ NSG mice (The Jackson Laboratory). Human cells were assessed by anti-human CD45 antibody analysed by flow cytometry. Specific cell subsets were detected using antibodies to human CD34, CD33, CD11b, CD14 and CD19 (See Mouse Antibody Table).
For the in vivo drug treated NSG experiments, CML (2×106 cells per mouse) CD34+ cells were transplanted via tail vein into female 8-10-week-old sub-lethally irradiated (2.5Gy) NSG mice (The Jackson Laboratory). After 4 weeks, mice were treated with RG7388 (75-100mg/kg, oral gavage once daily), CPI-203 (6-7.5mg/kg, intraperitoneal twice daily) or Nil (50mg/kg, oral gavage once daily) for 3-4 weeks. Two CML samples were assessed separately, each performed with 4-5 mice per drug arm per experiment. Results presented represent data from both experiments (each experiment normalised to vehicle). To quantify the frequency of BCR-ABL1+ cells within the engrafted human CD45+ cells, dual-fusion D-FISH was performed as previously described3.
Transgenic Mouse Model
Inducible (tetracycline (TET)-based) DTG (SCLtTAxBCR-ABL) donor mice in a C57BL/6 (CD45.2) background were a kind gift from Daniel G. Tenen. B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) recipients (a mixture of female and males between 8-10 weeks old) were purchased from Charles River Laboratories.
Bone marrow transplantation and analysis of disease
BM cells of DTG mice (1×106) were injected into the tail veins of 10-week-old irradiated (2 doses of 4.25Gy, 3 h. apart) recipients. TET was continued for 2 weeks after radiation. Tail veins bleeds were performed weekly post TET removal and Gr1/Mac1 percentages (flow cytometry), WBC, neutrophils and Hb (Hemovet) were monitored.
DTG In vivo drug treatment
Drugs were administered to DTG mice 5 weeks after transplantation, over a 4-week period. Nil 75mg/kg once daily, CPI-0610 15mg/kg twice daily and RG7112 50mg/kg once daily, all by oral gavage. For no drug control mice were administered the vehicles at the same concentrations and volumes as used for the combination arm.
Flow analysis
PB, BM and spleen cells were stained using appropriate antibodies and analysed using a FACSCanto or FACSAria machine (BD Biosciences).
Husbandry
All experiments were performed in accordance with the local ethical review panel, the UK Home Office Animals Scientific Procedures Act, 1986 and UKCCCR and NCRI guidelines.
Animals were kept in regulated facilities, monitored daily, and all experiments were carried out in compliance with UK Home Office guidelines. Mice were genotyped by Transnetyx (Cordova, TN).
RNA sequencing: sample preparation
CML CD34+ cells were seeded in 48-well plates at 1-2×106 cells/ml, before drug treatment (RITA 50nM, CPI-203 1µM, Nil 5µM) for 24h. After treatment, RNA was extracted using RNeasy Plus Mini Kit (Qiagen).
RNA sequencing: library generation
RNA-seq libraries were generated using TruSeq Stranded Total RNA (Part no. 15031048 Rev. E October 2013) kits. Ribosomal depletion was performed on 1ug of RNA using Ribo-Zero Gold prior to a heat fragmentation step aimed at producing libraries with an insert size between 120-200bp. cDNA was synthesized from the enriched and fragmented RNA using SuperScript II Reverse Transcriptase (Invitrogen) and random primers. The cDNA was converted into double stranded DNA in the presence of dUTP to prevent subsequent amplification of the second strand. Following 3’ adenylation and adaptor ligation libraries were subjected to 15 cycles of PCR to produce RNA-seq libraries. Prior to sequencing, RNA-seq libraries were qualified and quantified via Caliper’s LabChip GX (Part no. 122000) instrument using the DNA High Sensitivity Reagent kit (Product no. CLS760672). Quantification of libraries for clustering was performed using the KAPA Library Quantification Kits for Illumina sequencing platforms (Kit code KK4824) in combination with Life Technologies QuantStudio 7 real-time PCR instrument. Libraries were finally pooled in equimolar ratios and sequenced on Illumina’s NextSeq500 platform using 75bp paired-end high output runs. Data (fastq) are available using accession code PRJEB9942 at the EMBL Nucleotide Sequence Database (ENA, http://www.ebi.ac.uk/ena).
RNA sequencing: alignment and analysis
Sequencing reads were aligned to the genome (GRCh38/release 80 primary assembly as obtained via ftp.ensembl.org) using Subread (v1.4.6-p3)54. RNA-SeQC was used to confirm adequate mapping quality and gene-level counts were calculated using Subread’s featureCounts algorithm55. Count data for each arm were normalised independently by TMM56 (as implemented in the Bioconductor package edgeR) and VOOM57 (as implemented in the Bioconductor package limma). Genes with <3cpm in three samples were removed from further analysis. Differential expression was identified using limma49 (using Benjamini-Hochberg46 correction).
Definition of synergy
Genes were described as loosely synergistic if (1) RITA, CPI-203 and combined treatment all induced deregulation in the same direction; (2) the deregulation in response to the combined treatment was significant (q<0.05); (3) the deregulation induced by the combined treatment was greater than both RITA and CPI-203 in isolation. A more extreme definition had an additional criteria of the log ratio of the observed and additive effect being >0.6 (corresponding to 150% increase).
Statistical analysis
For in vitro experiments a minimum of 3 PS were chosen as a sample size to ensure adequate power. For all animal studies, each experiment was replicated twice in the laboratory with a minimum number of 5 mice per arm, unless indicated. NSG mice were excluded from analyses if they died of radiation poisoning (within 10 days of being irradiated, out of a 16 week procedure). For DTG mice, mice were excluded from analysis if leukemic cells (CD45.2) failed to engraft host mice (CD45.1 +) and therefore would not develop leukaemia. This was determined 1 week prior to drug treatment. Patient samples were only excluded if clinical data identified patient sample as entering blast crisis. Pre-established criteria also included if sample data point deviated 2 standard deviations from the mean, it was to be excluded, but this was not applied to the data in main or extended data. Group allocation to mice was done as mice were either purchased and subsequently numbered or weaned, to remove any investigator bias. For both NSG and DTG mice studies, all mice were randomly assigned treatment groups, ensuring all animals were of equal health and leukemic status, within normal variability. Mice were assessed at pre-determined time points: NSG mice were assessed at 8 and 16 weeks and DTG mice were assessed 4 weeks after drug treatment, so there was minimized bias as to mice assessing outcome. All mice were cared for equally in an unbiased fashion by animal technicians and investigator. No blinding was done.
Unless indicated, data are presented as the mean ±s.e.m. and p values were calculated by two-tailed Student’s t test using GraphPad Prism software. Significant statistical differences (*p<0.05, **p<0.01, ***p<0.001) are indicated.
Code availability
All computer code was implemented in R and is available from the authors upon request.
Extended Data
Extended Data Figure 1. BCR-ABL1 drives a proteomic signature mediated by p53 and c-Myc.
a-b, These figures are equivalent to Fig. 1b-c with additional information regarding the correlations calculated from the complete list of 58 candidate proteins (rc) in addition to the correlations for the candidate network (rn) and the background (r0). Also shown is the gain in r2 obtained for the candidate network as compared to the r2 obtained for the candidate list as a whole (r2Δ). FDR calculated from 10,000 resamplings. c, Expression changes of the network components (shown as bar plots) in the context of quiescent and primitive CML cells; data shown in each panel (L-R) are (1) CD34+ protein log2 ratios; (2) CD34+HstloPylo transcript logFC (E-MTAB-2508); (3) CD34+HstloPylo transcript logFC (GSE24739); (4) CD34+CD38− logFC (E-MTAB-2581); and (5) Lin−CD34+CD38−CD90+ logFC (GSE47927). Down-/up-regulation are indicated by turquoise/red respectively. Where multiple probesets were found for individual genes, the probeset corresponding to the maximal log ratio was selected. d-e, Correlation of the candidate network in progenitor (CD34+) CML cells: (i) CD34+CD38+ progenitor (ii) common myeloid progenitor Lin−CD34+CD38+CD123+CD45RA− (iii) CD34+ cells. As in a-b, correlations for the background (r0), candidate list (58 proteins, rc) and candidate network (Fig. 1a, rn) are shown. Also shown is the gain in r2 obtained for the candidate network as compared to the r2 obtained for the candidate list as a whole (r2Δ). FDR calculated from 10,000 resamplings; MI statistics corresponding to FDRs<0.05 are colored red (otherwise results are grey). f, A Venn diagram showing the overlap in protein identification of the three MS instruments: ABSciex Q-STAR® Elite (Elite), Thermo LTQ Orbitrap Velos (Orbi) and ABSciex TripleTOF 5600 (5600).
Extended Data Figure 2. Validation of network candidates.
a, HDM2 and c-Myc knockdown using shRNA constructs. Western blots of c-Myc, HDM2, p53 and Hsp90 in HeLa cells transduced with lentiviral constructs specific for either c-Myc (2 constructs), HDM2 (2 constructs) or scrambled control (1 construct). b-d, CML CD34+ cells were transduced with either lentiviral (GFP) shRNA constructs to HDM2 (constructs 1, 2), c-Myc (constructs 1, 2) or scramble control (1 construct). b, Transduced viable GFP+ cells (assessed as AnnexinV-/DAPI-/GFP+ percentages multiplied by the absolute cell count) are presented as a percentage of CML CD34+ cells transduced with scramble control (n=3 PS). c, Early apoptosis levels (assessed as AnnexinV+/DAPI-/GFP+) post transduction of CML CD34+ cells (n=3 PS) as described in b. Statistical significance was calculated by a two-tailed student t test and error bars represent the SEM.
Extended Data Figure 3. RITA and CPI-203 synergize to eliminate CML CD34+ cells.
a-b, Western blots of CML CD34+ cells untreated or treated with 50nM RITA; 1µM CPI-203; the combination of 50nM RITA and 1µM CPI-203 or 150nM Das for a, 8h. and b, 48h. c, p53 (red, nucleus in blue) 24h. post treatment in CML CD34+ cells. d, RITA, CPI-203 and combination drug treatment eliminates CD34+ CML cells through mechanisms likely dependent on apoptosis; after 72 h. of drug treatment apoptosis levels were assessed (AnnexinV/DAPI) using flow cytometry techniques. e, RITA or Nutlin3 (Nut) cannot induce death of K562 cells that lack p53. K562 cells were treated with either 50 µM RITA or 10 µM Nutlin3 (Nut) and after 72 h. of drug treatment, apoptosis levels were assessed (AnnexinV/DAPI) using flow cytometry techniques.
Extended Data Figure 4. RITA and CPI-203 synergize to eliminate CML CD34+ cells continued.
a, CD34+ CML cells were treated with Nutlin3 and CPI-203 for 72 h. with apoptosis levels assessed (AnnexinV/DAPI) using flow cytometry techniques. b, Treatment of CD34+ CML cells with Nutlin3 results in the elimination of early and late progenitor cells as assessed by functional colony forming capacity of drug-treated CML cells. c, Sequential drug treatments (n=3 PS; drug one for 24h. then both for 48h.). d, Sequential knockdown treatments (n=3 PS; knockdown one for 24h. then both for 48h.), mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001). e, CML CD34+ primary samples were pre-treated or not with IM (1 µM) for 8 h. followed by RITA (50 nM), CPI-203 (1 µM) or the combined treatment (RITA+CPI-203) for 72 h. (right 3 columns). Cell counts were obtained using trypan blue exclusion.
Extended Data Figure 5. RITA and CPI-203 selectively eliminate LSC.
a, Viable cell counts (n=3 PS); b, apoptosis in normal CD34+ cells (n=3 PS) in response to RITA and/or CPI-203. c, Gated CML CD34+CD38− cells 72h. Post-treatment (n=4 PS). d, Ex-vivo protocol for CML/cord blood CD34+ cells in NSG mice (n=5 mice/arm). e-f, Targeting p53 and c-Myc in CML eliminates NSG repopulating leukaemic stem cells. CML CD34+ cells were treated with RITA (70 nM) and/or CPI-203 (1 µM) or Das (150 nM) for 48 h. and recovered cells were injected intravenously into 8-12-week old, sub-lethally irradiated (2.5 Gy) NSG mice (2-4 mice per arm). e, Percentage of human CD45+ cell levels in peripheral blood (PB) at 8, 12 and 16 weeks. f, Percentages of human CD45, CD34, CD33, CD11b, CD19 and CD14 positive cells in the bone marrow at 16 weeks. g, CML BM analyses CD33, CD11b, CD19 and CD14 from a CML sample determined to engraft both BCR-ABL positive and negative cells. h, D-FISH analyses of (g) BM human engraftment studies performed twice (2 PS) with a minimum of n=6 mice/ arm; mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
Extended Data Figure 6. Mechanism of LSC elimination and clinical scope.
a, Enrichment of (i) p53; (ii), apoptosis; (iii), MYC; (iv), differentiation MSigDB signatures in the four treatment arms (columns named as per b). Figures a (i)-(iv) here are equivalent to Fig. 5b (i)-(iv), but with named MSigDB signatures. b, Enrichment of PANTHER pathways in the four treatment arms. Pathway enrichment calculated from (i) the top 1500 genes, as ranked by increasing p value; (ii), only those genes exhibiting an absolute FC of >0.5 in each arm; (iii), only those genes exhibiting a p value of >0.05 in each arm. c, Assessing molecular synergy of the combined RITA+CPI-203 treatment, as compared to the individual RITA and CPI-203 arms of the RNA-seq experiments in the three in silico functional signatures: (i) p53/apoptosis; (ii) MYC and (iii) differentiation. Mean expression is shown as a solid line.
Extended Data Figure 7. Mechanism of LSC elimination and clinical scope continued.
a, Gene expression patterns (logFC) shown for the members of the three broad signatures identified in silico: (i) p53/apoptosis; (ii) MYC and (iii) differentiation (*=q<0.05). Corresponding Expression data provided in Supplementary Tables 5-7. b, Differential expression of CD34 and CD133 (markers of stemness) in the four arms of the RNA-seq experiment. c, Apoptosis levels assessed (AnnexinV+/DAPI) using flow cytometry on a TKI-NR CD34+ sample after 72 h. treatment with RITA and CPI-203 as indicated.
Extended Data Figure 8. RG7112 and CPI-0610 as a combination decrease BCR-ABL+ cells.
a, DTG mice in vivo treatment: b, neutrophils – normalised to control (·····). c, Bone marrow (BM) cells stained for CD45.1/2. Drug treatment arms (minimum of n=7 mice) mean ± s.e.m. (P values: two-tailed student t test; *p<0.05, **p<0.01, ***p<0.001).
Supplemental Information
8 supplementary tables named Holyoake_Supplementary_Table1-8
1 supplementary figure (uncut western blots) Holyoake_Supplementary_Fig. 1
Acknowledgements
We thank all CML patients and UK haematology departments who contributed samples; Dr Alan Hair for sample processing; Jen Cassels for cell sorting; Dr Alison Michie and Dr Vignir Helgason for assisting with the in vivo research and providing cord blood samples; Dr Christine Wells, Dr Peter Bailey and Mr John Cole for discussions regarding the RNA-seq analysis. We acknowledge the Cancer Research UK (CR-UK) Glasgow Centre (C596/A18076) and the BSU facilities at the CR-UK Beatson Institute (C596/A17196) for providing animal care and housing facilities. We acknowledge Constellation Pharmaceuticals for providing CPI-203, CPI-0610 and part funding M.D., Roche for providing RG7112, RG7388 and part funding M.E.D. and the SPIRIT Trials Management Group for access to CML samples. This study was supported by the Glasgow and Manchester Experimental Cancer Medicine Centres (ECMC), which are funded by CR-UK and the Chief Scientist’s Office (Scotland). We acknowledge the funders who have contributed to this work: MRC stratified medicine infrastructure award (A.D.W.), CR-UK C11074/A11008 (F.P., L.E.M.H., T.L.H., A.D.W.); LLR08071 (S.A.A., E.C.); LLR11017 (M.C.); SCD/04 (M.C.); LLR13035 (S.A.A., K.D., A.D.W., and A.P.); LLR14005 (M.T.S., D.V.); KKL690 (L.E.P.); KKL698 (P.B.); LLR08004 (A.D.W., A.P. and A.J.W.); MRC CiC (M.E.D.); The Howat Foundation (FACS support); Friends of Paul O’Gorman (K.D. and FACS support); ELF 67954 (S.A.A.); BSH start up fund (S.A.A.); MR/K014854/1 (K.D.).
Footnotes
Authorship Contributions
ADW and TLH supervised the entire study and research. SAA, LEMH, ADW and TLH designed the research, analysed and interpreted data, and wrote the manuscript. SAA conceived the hypothesis, supervised in vivo research, prepared samples for proteomic and RNA-seq, performed all in vitro work including westerns, IF, cloning and kd studies, clonogenic studies, flow cytometry and all mouse in vivo studies including tissue processing-FISH preparation and slide interpretation, engraftment determination and analysis of primitive stem cell subsets. LEMH designed and performed all in silico work including global omics handling, integration and analysis (MS, RNA-seq and microarray data); network analyses; correlation/MI calculations; functional enrichment analyses and permutation experiments for calculation of p-values. EC performed proteomic work. AJKW performed proteomic work and generated relative proteomic quantification. MED performed virus preparation, prepared drug for in vivo work and assisted with in vivo studies. KD provided maintenance and care for all mouse colonies and assisted with in vivo work. PB, LEP provided assistance for in vivo studies and D-FISH preparation. FP, MTS provided assistance with in vivo studies. DV provided analysed datasets and analysed/interpretated RNA-seq data. SMG supervised and interpreted RNA-seq. CN performed RNA-seq experiments. KK/MC provided analysed datasets. AP supervised proteomic studies. All authors reviewed/edited the manuscript.
Data Deposition Statement – Accession codes for all data sets have been included in the online methods. For those that are not yet published detailed methods have been included in online methods.
Competing Interest Declaration–The work presented in Fig. 6 was in part supported by funding from Constellation Pharmaceuticals and Roche.
References
- 1.O’Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012;12:513–526. doi: 10.1038/nrc3317. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 3.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 4.Mahon FX, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. The lancet oncology. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 5.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neviani P, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013;123:4144–4157. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra EW. A note on two problems in connexion with graphs. Numerische Mathematik. 1995;1:269–271. [Google Scholar]
- 8.Van Dang C, McMahon SB. Emerging Concepts in the Analysis of Transcriptional Targets of the MYC Oncoprotein: Are the Targets Targetable? Genes & cancer. 2010;1:560–567. doi: 10.1177/1947601910379011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boichenko AP, et al. A panel of regulated proteins in serum from patients with cervical intraepithelial neoplasia and cervical cancer. Journal of proteome research. 2014;13:4995–5007. doi: 10.1021/pr500601w. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Molecular & cellular proteomics : MCP. 2014;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton CW, et al. Quantitative proteomic profiling of matched normal and tumor breast tissues. Journal of proteome research. 2010;9:3891–3902. doi: 10.1021/pr100113a. [DOI] [PubMed] [Google Scholar]
- 12.Pierce A, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Molecular & cellular proteomics : MCP. 2008;7:853–863. doi: 10.1074/mcp.M700251-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Issaeva N, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 14.Wong C, et al. The bromodomain and extra-terminal inhibitor CPI203 enhances the antiproliferative effects of rapamycin on human neuroendocrine tumors. Cell death & disease. 2014;5:e1450. doi: 10.1038/cddis.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton A, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah NP, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer cell. 2008;14:485–493. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 20.Yong AS, Szydlo RM, Goldman JM, Apperley JF, Melo JV. Molecular profiling of CD34+ cells identifies low expression of CD7, along with high expression of proteinase 3 or elastase, as predictors of longer survival in patients with CML. Blood. 2006;107:205–212. doi: 10.1182/blood-2005-05-2155. [DOI] [PubMed] [Google Scholar]
- 21.McWeeney SK, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010;115:315–325. doi: 10.1182/blood-2009-03-210732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tovar C, et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 23.Albrecht BK, et al. Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials. J Med Chem. 2016;59:1330–1339. doi: 10.1021/acs.jmedchem.5b01882. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanza F, Bi S. Role of p53 in leukemogenesis of chronic myeloid leukemia. Stem Cells. 1995;13:445–452. doi: 10.1002/stem.5530130416. [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Casares MT, et al. MYC antagonizes the differentiation induced by imatinib in chronic myeloid leukemia cells through downregulation of p27(KIP1) Oncogene. 2012 doi: 10.1038/onc.2012.246. [DOI] [PubMed] [Google Scholar]
- 28.Winter GE, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nature chemical biology. 2012;8:905–912. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeishi S, et al. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer cell. 2013;23:347–361. doi: 10.1016/j.ccr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Reavie L, et al. Regulation of c-Myc Ubiquitination Controls Chronic Myelogenous Leukemia Initiation and Progression. Cancer cell. 2013;23:362–375. doi: 10.1016/j.ccr.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 33.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Li H, Ho N, Li D, Li S. Scd1 plays a tumor-suppressive role in survival of leukemia stem cells and the development of chronic myeloid leukemia. Molecular and cellular biology. 2012;32:1776–1787. doi: 10.1128/MCB.05672-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu TY, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 37.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prost S, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARgamma agonists. Nature. 2015;525:380–383. doi: 10.1038/nature15248. [DOI] [PubMed] [Google Scholar]
- 39.Pizzatti L, Sa LA, de Souza JM, Bisch PM, Abdelhay E. Altered protein profile in chronic myeloid leukemia chronic phase identified by a comparative proteomic study. Biochim Biophys Acta. 2006;1764:929–942. doi: 10.1016/j.bbapap.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Cramer-Morales K, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–1304. doi: 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Blanco E, et al. Molecular signature of CD34(+) hematopoietic stem and progenitor cells of patients with CML in chronic phase. Leukemia. 2007;21:494–504. doi: 10.1038/sj.leu.2404549. [DOI] [PubMed] [Google Scholar]
- 42.Affer M, et al. Gene Expression Differences between Enriched Normal and Chronic Myelogenous Leukemia Quiescent Stem/Progenitor Cells and Correlations with Biological Abnormalities. Journal of oncology. 2011;2011:798592. doi: 10.1155/2011/798592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham SM, Vass JK, Holyoake TL, Graham GJ. Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells. 2007;25:3111–3120. doi: 10.1634/stemcells.2007-0250. [DOI] [PubMed] [Google Scholar]
- 44.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 45.Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. C V, Gentleman R, Dudoit S, Irizarry R, Huber W, editors. Springer; 2005. pp. 397–420. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 47.Johnson WE, Li C, A R. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 48.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. Bmc Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 50.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 51.Xu XL, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137:1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfer A, et al. MYC regulation of a "poor-prognosis" metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheth A, et al. Inhibition of human mitochondrial peptide deformylase causes apoptosis in c-myc-overexpressing hematopoietic cancers. Cell death & disease. 2014;5:e1152. doi: 10.1038/cddis.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.