Abstract
Cells frequently experience DNA damage that requires repair by homologous recombination (HR). Proteins involved in HR are carefully coordinated to ensure proper and efficient repair without interfering with normal cellular processes. In Saccharomyces cerevisiae, Rad55 functions in the early steps of HR and is regulated in response to DNA damage through phosphorylation by the Mec1 and Rad53 kinases of the DNA damage response. To further identify regulatory processes that target HR, we performed a high-throughput genetic interaction screen with RAD55 phosphorylation site mutants. Genes involved in the mRNA quality control process, nonsense-mediated decay (NMD), were found to genetically interact with rad55 phospho-site mutants. Further characterization revealed that RAD55 transcript and protein levels are regulated by NMD. Regulation of HR by NMD extends to multiple targets beyond RAD55, including RAD51, RAD54 and RAD57. Finally, we demonstrate that loss of NMD results in an increase in recombination rates and resistance to the DNA damaging agent methyl methanesulfonate, suggesting this pathway negatively regulates HR under normal growth conditions.
INTRODUCTION
Homologous recombination (HR) is a DNA repair and DNA damage tolerance pathway that functions to restart stalled/collapsed replication forks, ensures proper chromosome segregation during meiosis and restores chromosomal double-strand breaks (DSBs), interstrand crosslinks, and single-strand gaps (1–3). Defects in HR genes cause an increased sensitivity to DNA damaging agents, chromosomal instability and predisposition to cancer development. Additionally, failure to properly regulate HR can lead to aberrant or ectopic recombination and result in genomic rearrangements and loss of heterozygosity (4,5). The complexity of regulating HR is compounded by the fact that several alternative pathways including non-homologous end joining, microhomology mediated end joining and single-strand annealing (SSA) are capable of repairing a DSB. Moreover, translesion DNA synthesis and fork regression compete with HR during single-strand gap repair. In effect, execution of HR is regulated at multiple steps to ensure proper timing and coordination with other DNA repair pathways and cellular processes (6,7).
The signature reactions of HR, homology search and DNA strand exchange, are carried out by a highly conserved structure composed of ssDNA coated by Rad51 protein bound as a helical nucleoprotein filament (8). Assembly of Rad51 into a functional filament marks the critical point at which an ssDNA substrate is competent to perform HR. As such, multiple proteins involved in Rad51 filament formation are tightly regulated by post-translational modifications (9–14). Among those involved in Rad51 filament formation, Rad55 and Rad57, are regulated in response to DNA damage by phosphorylation (15–20).
Rad51 filament assembly and stabilization depends on a number of accessory proteins known as mediators. Understanding the function and regulation of mediator proteins has become particularly relevant to human health as an increasing number of these proteins have been identified as genuine tumor suppressors (e.g. BRCA2, RAD51C, RAD51D) (6). The use of budding yeast as a paradigm for mediator function has been instrumental in defining the role of human mediator proteins. In Saccharomyces cerevisiae, Rad51 filament assembly/stability depends on Rad52, Rad55-Rad57 and the SHU complex (Shu1, Shu2, Csm2, Psy3) (8,21–27). While much work has focused on the mechanisms of how these proteins function, little is known about their regulation and how regulation of mediators may ultimately influence DNA repair in the cell.
Rad55 and Rad57 are Rad51 paralogs that have a similar RecA core domain to Rad51 but lack the ability to form a filament on DNA or perform homology search and DNA strand invasion (1). Rad55 and Rad57 form a heterodimeric complex that promotes the Rad51 filament by overcoming the kinetic barrier to Rad51 binding imposed by the ssDNA binding protein RPA (21,23,28–30). Moreover, the Rad55-Rad57 paralog complex stabilizes Rad51 filaments against disruption by Srs2 activity (24). The role of Rad55-Rad57 appears to be particularly critical for recombination occurring at stalled/collapsed replication forks (30).
As a mediator of Rad51 filament formation, the Rad55-Rad57 heterodimer is ideally positioned as a regulatory target. To date, eight phosphorylation sites have been identified on Rad55 and a single site on Rad57 (19–20,31) (This work). Six of the sites on Rad55 have been shown to be specifically phosphorylated in response to DNA damage by DNA damage checkpoint kinases including Mec1 and Rad53, the S. cerevisiae orthologs of human ATR and CHK2, respectively (16,20) (Supplementary Figure S1A). In budding yeast, Mec1 and Rad53 play a central role in regulating the DNA damage response (DDR) signaling pathway. Mutation of serines 2, 8, 14 in Rad55 to non-phosphorylatable alanine residues results in increased sensitivity to DNA damage (19). In addition, such rad55-S2A,8,14A mutants exhibit a delay in the recovery of stalled replication forks resulting from treatment with the alkylating agent methyl-methane sulfonate (MMS) (19). These findings suggest that phosphorylation regulates the mediator function of Rad55-Rad57 and is critical to ensure proper DNA repair by HR.
To investigate regulatory processes that target Rad55 and influence Rad51 filament formation, we utilized rad55-phosphorylation site mutants in a quantitative, high-throughput genetic interaction screen known as an epistatic miniarray profile (E-MAP) (32). In this screen, we systematically assessed the phenotypes of double-mutants generated by crossing rad55- phosphorylation site point mutants to an array of gene deletion and DAmP (decreased abundance by mRNA perturbation) alleles, which represent every major cellular process (33). Here, we report the entire data set of the screen and focus on a novel interaction between HR and nonsense-mediated decay (NMD) that was identified in our screen.
NMD is a conserved RNA quality control pathway, which was discovered as a mechanism for degrading mRNAs that terminate prematurely during translation (34). However, emerging evidence implicates NMD in the broader regulation of mRNAs that do not contain a premature termination codon (35–42). The core reaction of NMD requires three conserved UPF (Up-frameshift) proteins (Upf1/Nam7, Upf2/Nmd2 and Upf3) (43). Nam7 is a conserved adenosine triphosphate-dependent, superfamily-1 RNA helicase that serves as the primary effector of NMD, whereas Nmd2 and Upf3 regulate Nam7 activity (34). Since NMD has been implicated as a mechanism for post-transcriptional regulation (35), we followed up on results from our screen implicating this pathway in regulating Rad55. To our surprise, we found that NMD plays a direct role in regulating HR. In NMD-deficient cells, we find that both RAD55 mRNA levels and Rad55 protein pools are significantly increased. Broader analysis of mRNA levels across a number of key HR genes shows that the effect of NMD targets the HR pathway beyond RAD55. Importantly, the activity of NMD toward HR transcripts establishes a previously unknown regulatory mechanism of HR in which NMD regulates the levels of several key HR factors through targeted mRNA decay.
MATERIALS AND METHODS
Yeast strains
All S. cerevisiae strains were in the W303 background with the exception of the strains utilizing SGA technology for the E-MAP screen (S288C background). The relevant genotypes are listed in Supplementary Tables S1 and 3, respectively. Epitope tagged Rad55 strains were generated by integration of a Myc9- TRP1K.lactis epitope cassette as previously described (44) to generate a C-terminal Myc9-epitope fusion with Rad55. Myc9-tagged Rad55 strains displayed the same sensitivity to MMS as untagged strains (Supplementary Figure S5) suggesting the Myc9-tagged Rad55 protein is fully functional in vivo. Mutagenic adaptamer polymerase chain reaction (PCR)-based allele replacement (45) was used to generate rad55-phospho-minus and potential phospho-mimic point mutants. Both WDHY2945 (rad55-S2,8,14,378A) and WHDY2569 (rad55-S2,8,14,19,20A) mutants were generated by mutating S378 and S19 and 20, respectively, to alanine in WDHY2016 (rad55-S2,8,14A). All point mutants were confirmed by genomic sequencing of the entire open reading frame of RAD55. Gene replacements were generated using a PCR-amplified natMX4 (noursiothricin resistance) disruption cassette as previously described (46) and confirmed by PCR.
Cultures and methyl methanesulfonate treatment
For all experiments, cultures were grown at 30°C in liquid rich media (YPD), unless otherwise specified. Overnight liquid cultures were diluted to OD600 = 0.1 and grown to mid-log phase (OD600 = 0.4–0.6) prior to experimental treatment. Acute MMS treatment was carried out by direct addition of MMS to liquid YPD at a final concentration of 0.075% and incubation at 30°C for a specified amount of time. To avoid variability between experiments, a single MMS stock was used in all cases. Cycloheximide chase experiments were performed by resuspending mid-log cultures to OD600 = 0.5 and adding cycloheximide to a final concentration of 200 μg/ml. At each time point, 4 ml of each culture was mixed with cold sodium azide (final conc. = 30 mM) and centrifuged at 4°C to pellet cells. Pellets were frozen in liquid nitrogen and stored at −80°C.
Epistatic miniarray profile techniques
The query strains used in the screen are listed in Supplementary Table S1 and all share the common background genotype MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ can1Δ::MATaPr-HIS3 lyp1Δ::MATαPr-LEU2. Integration cassettes for rad55 point mutant query strains were generated by adaptamer PCR (45). Briefly, genomic DNA containing the desired rad55 point mutations was PCR amplified using a reverse adaptamer primer that annealed 25-bp downstream from the Rad55 stop codon. In a parallel PCR, a natMX fragment was amplified using a forward primer containing complementary adaptamer sequence and a reverse primer containing sequence homologous to the region 26–66-bp downstream from the RAD55 STOP codon. A second round of adaptamer-mediated PCR was performed to fuse the two fragments together. The resulting DNA cassette was transformed into the query strain and proper integration was confirmed by PCR and DNA sequencing. We verified that the insertion of a natMX marker did not have an effect on RAD55 by comparing the DNA damage sensitivity of strains containing natMX strains to MMS to isogenic strains lacking the natMX insert (Supplementary Figure S2). The E-MAP deletion library is listed in Supplementary Table S2. The deletion library strain background is MATa his3Δ1 leu2Δ ura3Δ0 met15Δ0 LYS2+ CAN1+ LYP1+ YYY::kanMX.
Synthetic growth array, pinning, growth techniques and colony measurements were performed using previously established methods (32) with a modification to the double-mutant selection step where each array was pinned onto three double mutant selective media plates containing either 0, 0.001 or 0.01% MMS. Colony areas were measured using the HT colony grid analyzer JAVA program (47). Preprocessing, normalization and S-score computation was performed using the E-MAP toolbox for MATLAB (http://sourceforge.net/projects/emap-toolbox/) as previously described (32). Hierarchical clustering of MAP data was performed using the average linkage method in the Cluster 3.0 application (48). Images from hierarchical clustering were generated in Java Treeview (http://jtreeview.sourceforge.net/).
Genotoxin sensitivity
Plate assays were performed by making a 1:5 dilution series of late log-phase cultures and plating 4 μl onto YPD agar plates containing the indicated amount of MMS. For galactose induction experiments, cells were plated on complete supplement mixture plates lacking uracil and either 2% glucose or 2% galactose. Plates were incubated at 30°C and imaged daily for up to 7 days.
Recombination assay
Spontaneous recombination rates between direct repeats were measured using the method of the median as previously described (24,49). At least nine individual colonies from strains RHJY51 (wild-type) and RHJY52 (nam7Δ) were inoculated into 4 ml of YPAD and grown at 30°C for 2–3 days. Each culture was diluted and plated onto YPAD and CSM-Ade-Ura and counted after 3 days of growth. Recombination rates and 95% confidence intervals were calculated using the method of the median as previously described (49,50). Each experiment was repeated at least twice.
Immunoblotting
Ten OD600 units of cells from mid-log cultures were harvested and total protein extraction was performed by using the trichloroacetic acid precipitation method previously described (20). All protein samples were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, transferred to PVDF membranes and immunoblotted with either murine monoclonal anti-Myc antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or murine anti-3-phosphoglycerate kinase (3PGK) antibodies (Abcam, Cambridge, MA, USA).
RT-qPCR
A total of 10 OD600 units of Mid-log phase YPD cultures were harvested and total RNA was purified using the hot acid phenol method exactly as described in (51). Genomic DNA was eliminated by treating 500 ng of RNA sample with 1 Unit of RQ1 DNase (Promega) for 30 min at 37°C. The digest reaction was stopped by addition of 1 μl of RQ1 DNase stop solution (Promega) and subsequent incubation at 65°C for 10 min. Reverse transcription and real time qPCR was performed in a simultaneous reaction using the iTaq universal SYBR-green one-step kit (BioRad). A total of 10 ng of RNA template was used in a reaction volume of 20 μl. All RT-qPCR reactions were carried out in 96-well plate format on a Lightcycler 480 real time qPCR machine (Roche). Amplification was carried out by a reverse transcription step at 50°C for 10 min, a 95°C denaturing step for 1 min and 45 cycles of denaturing at 95°C for 15 s, annealing at 60°C for 15 s and extension at 72° for 15 s followed by a melting curve analysis. Primer sets used for qPCR are listed in Supplementary Table S4.
Mass spectrometry
Protein preparation, digestion, mass spectrometry protocols and analysis were performed as previously described (19,52,53).
RESULTS
E-MAP identifies genetic interactions with rad55 phosphorylation site mutants
To identify components that regulate Rad55 in response to DNA damage, we utilized an E-MAP approach (47) to quantitatively measure the global genetic interaction pattern of a complete open reading frame deletion of RAD55 (rad55Δ) and six rad55-phosphorylation site query mutants containing unique combinations of serine to alanine/aspartic acid point mutations (Supplementary Table S1). The particular rad55 phosphorylation sites were selected because their phosphorylation is induced after DNA damage. Different combination of sites were chosen based on their distinct effect on protein function but not protein stability (Serines 2, 8 and 14) (19) and on protein stability (Serines 19, 20 and 378) (Supplementary Figure S1) (54). Two phosphorylation sites that were not dependent on the induction of DNA damage, Rad55-S287 (31) and Rad55-S404 (this work) were not entered in this screen. The phenotype of site mutants in Rad55-S287 has not been tested (31). Mutations in RAD55-S404 do not show a discernible growth or survival phenotype in the absence or presence of DNA damage (data not shown). The set of rad55 phosphorylation site mutants (Supplementary Table S1) was crossed against a mutant array library containing ∼1200 single kanMX ORF replacement and DAmP alleles, which represent all major biological processes (33,55). We verified that the wild-type query mutant construct did not affect RAD55 gene function (Supplementary Figure S2). In order to measure DNA damage-induced genetic interactions that are absent under standard growth conditions, the double mutant arrays were plated on media containing varying MMS concentrations (0, 0.001 and 0.01% in YPD plates), similar to that previously described in Bandyopadhyay et al. (56). MMS induces stalling of replication forks and recombination is required for normal growth and survival under conditions of chronic exposure to MMS. The complete data set of measured genetic interaction scores for the E-MAP is made available in Supplementary Table S2.
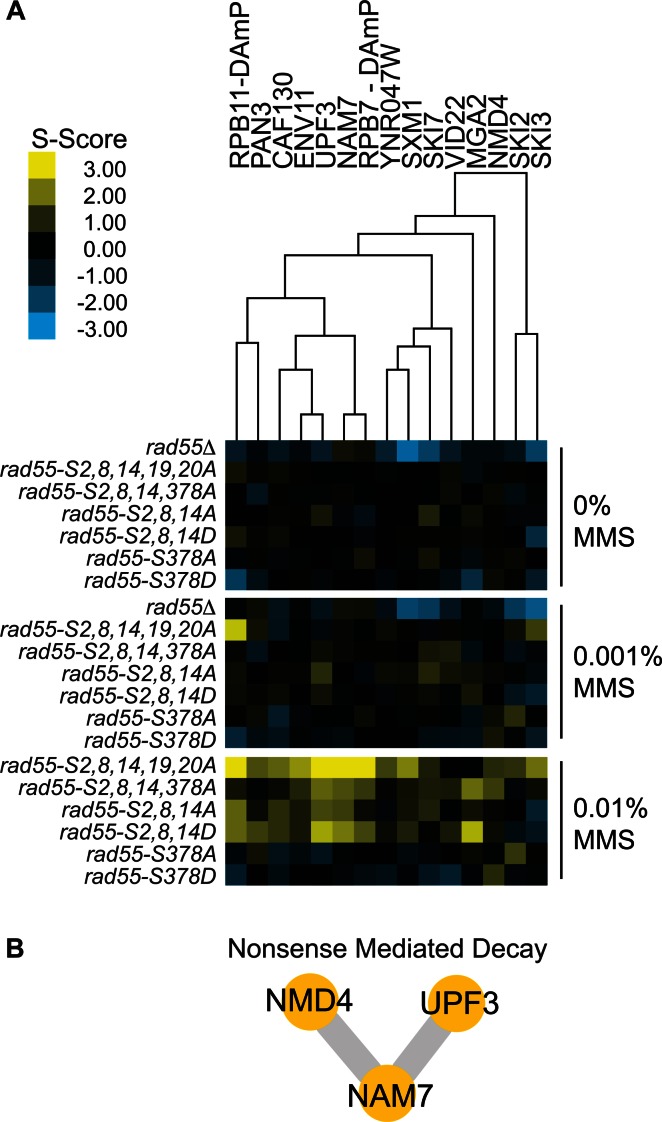
To identify processes uniquely involved in the regulation of Rad55 in response to DNA damage, we focused on genetic interactions meeting the following criteria: (i) the interaction is MMS-induced, (ii) the interaction with rad55 phosphorylation site mutants is consistent with suppression or epistasis (positive S-score), (iii) the positive interaction is unique to rad55-phosphorylation site mutants and absent with rad55Δ. Hierarchical clustering of the E-MAP dataset identified a subset of interactions meeting the above criteria (Figure 1A). This subset was almost exclusively identified at the highest dose of MMS (0.01%), and the same double mutants mostly contained neutral S-scores at an intermediate MMS dose (0.001%) and from cells grown on YPD alone. The severe sensitivity of rad55Δ mutants to MMS precluded our ability to quantify genetic interactions with this mutant on plates containing 0.01% MMS. However, because the sensitivity of rad55Δ to MMS is much greater than that of the phosphorylation site mutants used here, MMS-induced interactions only detected at the highest dose with the rad55- phosphorylation site mutants were often readily detected with rad55Δ at the intermediate MMS dose (0.001%). The subset of interactions shown in Figure 1 are unique in that only the rad55 phospho-site mutants show MMS-induced, positive genetic interactions while S-scores for the rad55Δ query strain were either neutral or negative at the MMS doses tested. Here, we focus on one functional pathway identified in this screen, NMD (Figure 1B). Supplementary Figure S3 shows three additional pathways (Ski complex, RNA PolII, Tbf1–Vid22 complex) identified with multiple hits using the above criteria.
Figure 1.
MMS-induced positive genetic interactions with RAD55 phosphorylation site mutants uncovers a novel link between Rad55 and nonsense-mediated decay (NMD). (A) Heat map displaying a subset of MMS-induced positive genetic interactions identified by E-MAP analysis and subsequently grouped by hierarchical clustering. (B) A network of previously curated physical protein interactions was generated using GeneMANIA for positively interacting array NMD genes identified in (A).
Some array mutants specifically had both a negative interaction with rad55Δ and a MMS-induced positive interaction with rad55-phosphorylation site mutants. The negative interactions were observed even in the absence of MMS. Array strain mutants falling into this category likely suppress rad55-phosphorylation site mutant phenotypes by a mechanism that depends on the presence of Rad55 protein while the absence of Rad55 in the cell (rad55Δ) results in synthetic sickness. Factors that regulate Rad55 protein levels would be expected to fall in to this category. The genes of this distinct class of interactions likely impact HR, but also function in other pathways required for cellular fitness in the absence of RAD55. We did not analyze this class of interactors further.
Nonsense-mediated decay pathway genetically interacts with RAD55
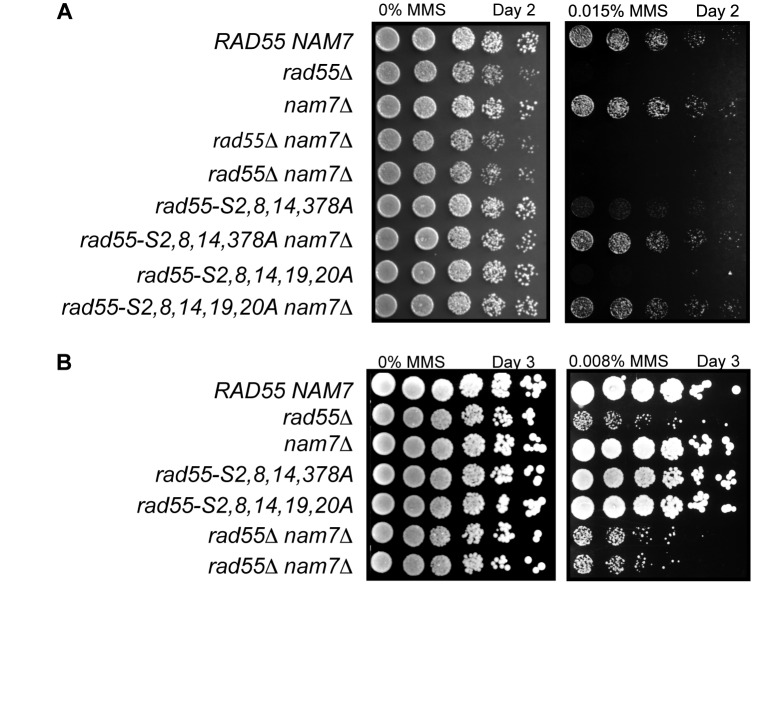
Among the strongest genetic interactors were mutants of the NMD pathway, including nam7, upf3 and nmd4. To validate the results from the E-MAP analysis, NAM7 was deleted in an independent strain background (W303), and the genetic interaction with rad55 mutants was recapitulated independently. In agreement with our E-MAP analysis, sensitivities of rad55-phosphorylation site mutants to MMS were suppressed by nam7Δ as measured by plate assays that record defects in growth and/or survival under genotoxic stress (Figure 2). Because rad55Δ strains are highly sensitive to DNA damage, assessment of the genetic interaction between rad55Δ and nam7Δ required substantially lower MMS concentrations (0.008% MMS) than the phospho-site mutants (0.015% MMS). At this lower MMS concentration the sensitivity of rad55-phosphorylation site mutants cannot be distinguished from wild-type. However, the effect of 0.008% MMS on rad55Δ strains is readily observed (Figure 2B), and sensitivity of rad55Δ nam7Δ was no different from that of rad55Δ (Figure 2B). These results independently validate our findings from the E-MAP screen that disruption of the NMD pathway specifically suppresses phospho-site mutants but not the deletion mutant of RAD55.
Figure 2.
Validation of E-MAP results showing suppression of rad55 phosphorylation site mutants by nam7Δ using an independent strain background. (A) Plate assay measuring the sensitivity of W303 strains with the indicated genotypes to MMS. Serial dilutions (1:5) of cultures were spotted onto YPD containing 0.015% MMS and grown at 30°C for 2 days prior to imaging. (B) Plate assay as in (A). In order to assess the genetic interaction between rad55Δ and nam7Δ, a concentration of 0.008% MMS was used and plates were imaged after 3 days of growth.
The core machinery of the NMD pathway consists of the Nam7–Upf3–Nmd2 complex (43). While Nam7 possesses the primary enzymatic activity required for NMD, Upf3 and Nmd2 act as accessory factors that are essential to Nam7 function and regulation (34). Nmd2 bridges the core NMD complex through physical interactions between both Nam7 and Upf3 and these interactions are required to stimulate Nam7 helicase and adenosine triphosphatase activities (57). We therefore hypothesized that disruption of any one of these components would result in similar genetic interactions with RAD55. Through hierarchical clustering, we found a high degree of similarity in the E-MAP profiles of nam7 and upf3 (Figure 1A). Though NMD2 was not included in the E-MAP dataset, we generated an nmd2Δ strain and tested its genetic interaction with rad55 mutants. As predicted, nmd2Δ displayed a genetic interaction pattern with rad55 mutants that mirrors those of nam7Δ and upf3Δ (Supplementary Figure S4). These results strongly establish a novel functional link between RAD55 and NMD.
The genetic interaction between nonsense-mediated decay and RAD55 requires Rad51-dependent homologous recombination
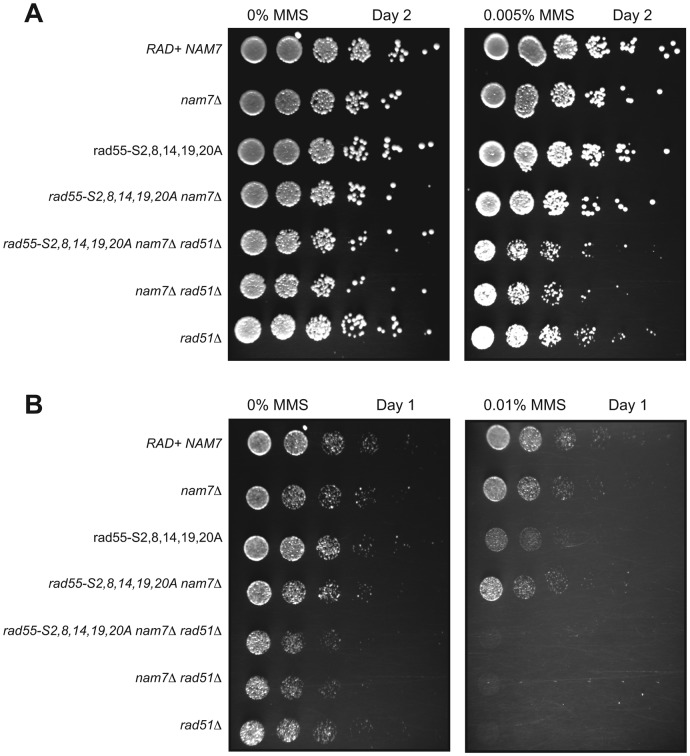
Rad55 functions by promoting the formation of Rad51 nucleoprotein filaments during the early stages of HR. The suppression of rad55 phosphorylation site-mutant sensitivity to MMS by deletion of NMD genes could be attributed to an increased ability to perform HR in the absence of NMD. Alternatively, it is possible that a parallel, Rad51-independent repair pathway that is ordinarily downregulated by NMD could become active in an NMD mutant and rescue the sensitivity to MMS caused by a rad55 mutation. To distinguish between these two models, we tested whether the suppression of rad55-phosphorylation site mutants by nam7Δ was dependent on RAD51. As with previous experiments, the differences in sensitivities between rad55-phosphorylation site mutants and rad51Δ mutants required that we use a range of MMS concentrations to observe all of the relative genetic effects. While clear suppression of rad55-S2,8,14,19,20A by nam7Δ was observed, a rad51Δ nam7Δ rad55-S2,8,14,19,20A triple mutant was no more sensitive to MMS than a rad51Δ nam7Δ double mutant or rad51Δ alone (Figure 3). These results demonstrate that rad51Δ is epistatic to the suppression of rad55-phosphorylation site mutants by nam7Δ. We conclude that NMD is directly affecting HR and the effect is not due to the derepression of an alternative DNA repair/tolerance pathway that bypasses the need for recombinational DNA repair.
Figure 3.
Suppression of rad55-phosphorylation site mutants by nam7Δ is RAD51-dependent. Plate assays were performed with W303 strains containing the indicated mutations. (A) YPD plates containing 0% MMS and 0.005% MMS were incubated at 30°C for 2 days. (B) YPD Plates containing 0 and 0.01% MMS were imaged after 1 day of growth at 30°C.
Rad55 steady state protein levels increase in NMD-deficient cells
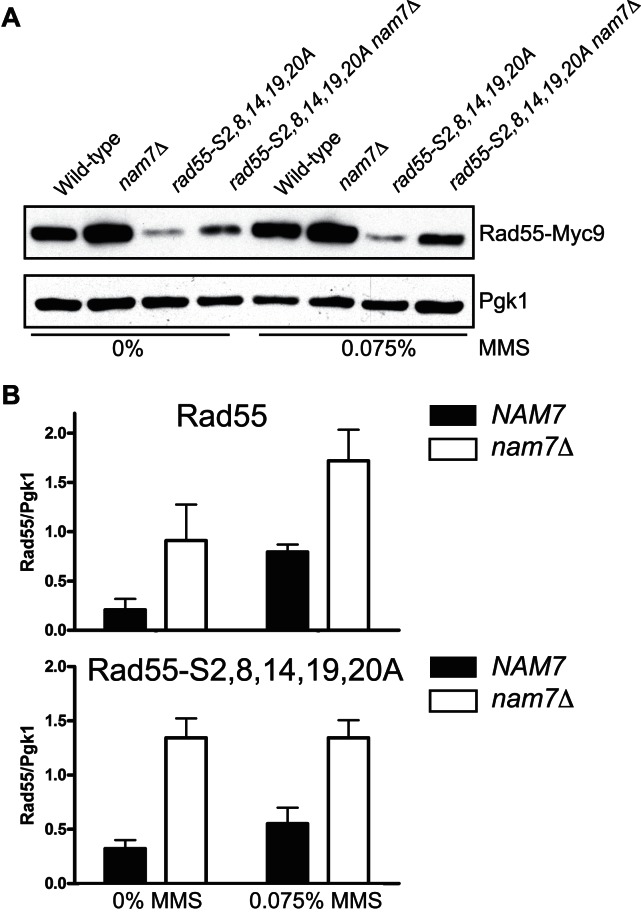
We wanted to test whether the absence of NMD affected Rad55 protein levels. Cellular Rad55 levels were assessed by immunoblot (Figure 4A). Because antibodies against native Rad55 are not of sufficient sensitivity for quantitative immunoblotting, we generated strains that expressed Rad55 with a C-terminal Myc9 epitope-tag from the native RAD55 locus. The addition of the Myc9 epitope tag did not interfere with Rad55 function as these strains were no more sensitive to MMS than wild-type (Supplementary Figure S5). The Rad55-S2,8,14,19,20A phosphorylation mutant is destabilized and results in greatly reduced protein levels compared to wild-type Rad55 (Figure 4A, Supplementary Figure S6). In asynchronous, mid-log growth cultures the steady state levels of Rad55 and Rad55-S2,8,14,19,20A were increased in nam7Δ strains 2–5-fold compared to strains with functional NMD both before and after 1 h treatment with 0.075% MMS (Figure 4A, Supplementary Figure S6). Quantitative measurements of these results are shown in Figure 4B. The observed increase in Rad55 protein levels is not explained by a difference in Rad55 protein turnover rate in wild-type compared to nam7Δ cells (Supplementary Figure S7). We conclude that NMD regulates the steady state levels of Rad55.
Figure 4.
Rad55 protein levels increase in NMD-deficient cells. (A) Representative immunoblots from mid-log cultures of WDHY2972 (RAD55-Myc9), WHDY3982 (RAD55-Myc9 nam7Δ), WDHY3449 (rad55-S2,8,14,19,20A-Myc9) and WDHY3980 (rad55-S2,8,14,19,20A nam7Δ). Cultures were either left untreated or treated with 0.075% MMS for 1 h. Antibodies against the Myc epitope were used to determine Rad55 levels. Membranes were reprobed with antibodies against Pgk1, which served as a loading control. (B) Plots of quantified immunoblot signals by densitometry. Rad55 signals were normalized to Pgk1 level from the same sample. The average normalized Rad55 levels of at least three experiments were plotted with error bars representing standard error of the mean. The Rad55 (upper panel) and Rad55-S2,8,14,19,20A (lower panel) samples were quantified from different exposures (see Supplementary Figure S6) and plotted in separate graphs.
We hypothesized that the suppression of MMS sensitivity in a nam7Δ rad55-S2,8,14,19,20A double mutant might be due to the accompanying increase in Rad55 protein level. As a test of this hypothesis, we reasoned that Rad55-S2,8,14,19,20A overexpressed might mimic the effect of losing NMD. Because Rad55 and Rad57 occurs as a 1:1 heterodimer complex, we overexpressed Rad55 (or Rad55-S2,8,14,19,20A) and Rad57 from a vector with the galactose-inducible bidirectional GAL1–10 promoter that was shown previously to maintain a 1:1 stoichiometry of the complex (54). Low levels of expression from this plasmid occur in the presence of glucose, which resulted in a slight increase in MMS resistance in rad55-S2,8,14,19,20A mutant strains containing the wild-type Rad55-Rad57 expression vector. However, when cells were plated on galactose, greater overexpression of Rad55-S2,8,14,19,20A-Rad57 partially suppressed the MMS sensitivity of a rad55-S2,8,14,19,20A mutant strain, though not to the same extent as overexpression of wild-type Rad55-Rad57 (Supplementary Figure S8). This result supports a mechanism in which increased Rad55 protein levels in NMD-deficient cells contributes to the observed suppression of rad55-S2,8,14,19,20A sensitivity to MMS. Importantly, overexpression of only Rad55-S2,8,14,19,20A-Rad57 did not elicit an equivalent degree of suppression of MMS sensitivity as an NMD mutant. This suggests that NMD regulates additional proteins that are required for MMS resistance.
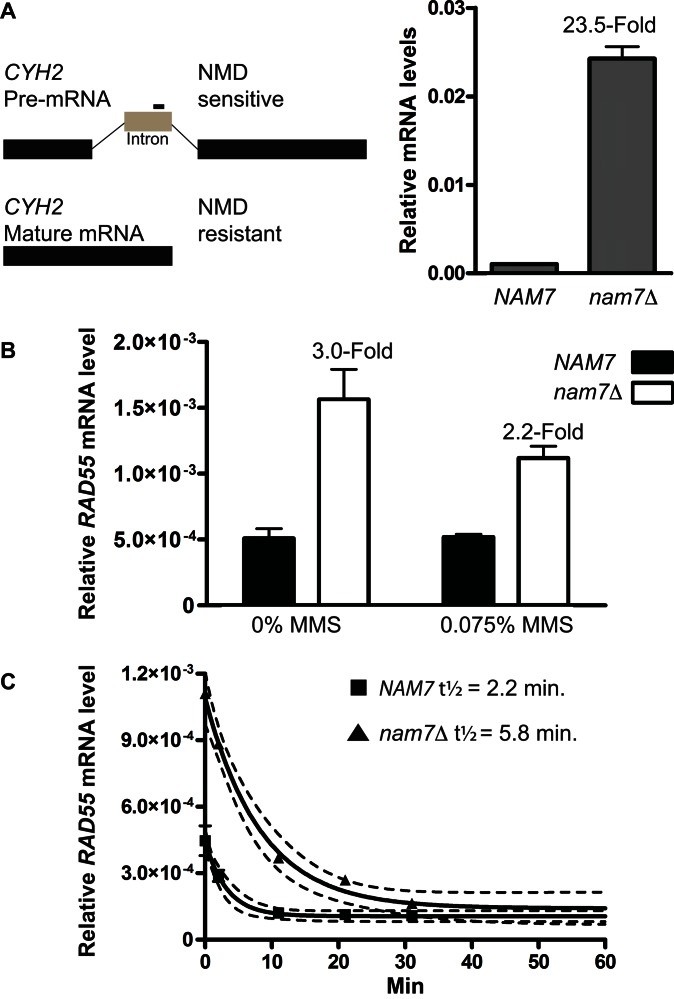
Nonsense-mediated decay regulates normal RAD55 mRNA levels
Because NMD acts as an mRNA quality control pathway, we tested whether the observed increase in Rad55 protein levels in NMD-deficient cells was due to an effect on RAD55 mRNA levels. RNA samples were prepared from mid-log phase cultures before and after MMS treatment, and mRNA levels were quantitatively measured using RT-qPCR. In NMD-mutant cells, the increase in abundance of CYH2 pre-mRNA can be used as a positive control for NMD-deficiency (51,58). CYH2 mRNA is inefficiently spliced, and the resulting pre-mRNA is exported to the cytoplasm where it is typically degraded in an NMD-dependent manner. Using primers that target the CYH2 pre-mRNA intron for amplification, we were able to measure the expected significant increase in CYH2 pre-mRNA abundance in nam7Δ cells compared to NMD-proficient cells by RT-qPCR (Figure 5A). The highly-abundant, non-coding scR1 RNA, which is unaffected by NMD, was used as a reference for normalization in all RT-qPCR experiments (51). Under the same culture conditions, we measured the effect of a nam7Δ mutation on RAD55 mRNA levels. Loss of NMD resulted in a statistically significant increase in the cellular level of RAD55 mRNA in untreated and MMS-treated samples (Figure 5B). We conclude that NMD regulates the steady state levels of RAD55 mRNA.
Figure 5.
Rad55 mRNA levels are regulated by NMD. (A) qPCR using primers specific to the intron region of CYH2 were used as a readout for NMD activity in NAM7 (WDHY2217) and nam7Δ (WDHY3865) strains. CYH2 levels were calculated relative to ScR1 transcript levels which are unaffected by NMD and serve as a baseline control for normalization. Each data point is the average of three independent experiments. Error bars represent standard error of the mean. Statistical significance was calculated by a two-tailed t-test and defined by a P-value < 0.05. (B) RAD55 mRNA levels were measured by qPCR using the same strains as in (A). Cells were grown to mid-log and treated with 0.075% MMS for one hour where indicated. Error bars represent standard error of the mean. Statistical significance was calculated by a two-tailed t-test and defined by a P-value < 0.05. (C) rpb1–1 shutoff experiment measuring RAD55 mRNA decay. Plotted are the average levels of RAD55 mRNA relative to ScRI as measured by qPCR in NAM7 (WDHY4057) and nam7Δ (WDHY4103) strains. Each data point represents the average of three independent samples collected at the indicated time after shifting cultures to the non-permissive temperature of 39°C. Non-linear regression using a first-order decay equation (solid lines, dotted lines represent the 95% confidence interval) was used to calculate the half-life of RAD55 mRNA in wild-type and nam7Δ strains.
Steady state mRNA levels are reflective of two independent processes, mRNA synthesis and degradation. Therefore, measuring steady state mRNA levels cannot distinguish whether RAD55 mRNA is directly targeted for degradation by NMD or whether the change in level is an indirect effect due to regulation of a secondary target that typically represses or activates transcription of RAD55. It is possible to identify direct NMD targets by measuring mRNA half-life. The half-life of an mRNA that is directly degraded by NMD will typically increase in an NMD-mutant cell whereas the half-life of an indirect target typically remains stable (35). To determine whether RAD55 mRNA is directly targeted for degradation by NMD, we measured the decay of RAD55 mRNA over time. To monitor mRNA decay we blocked new mRNA transcription using an rpb1–1 temperature sensitive allele (51). Mid-log cultures were shifted to the non-permissive temperature to inhibit RNA Pol II transcription, and mRNA levels were monitored over time. The turnover of RAD55 mRNA was rapid (t1/2 = 2.2 min., 95% CI = 1.5–4.2 min.) in cells with functional NMD. In nam7Δ cells, the rate of RAD55 mRNA turnover was reduced nearly 3-fold (t1/2 = 5.8 min., 95% CI = 4.2–9.1 min.) (Figure 5C). These results demonstrate that RAD55 transcripts are direct targets of NMD turnover.
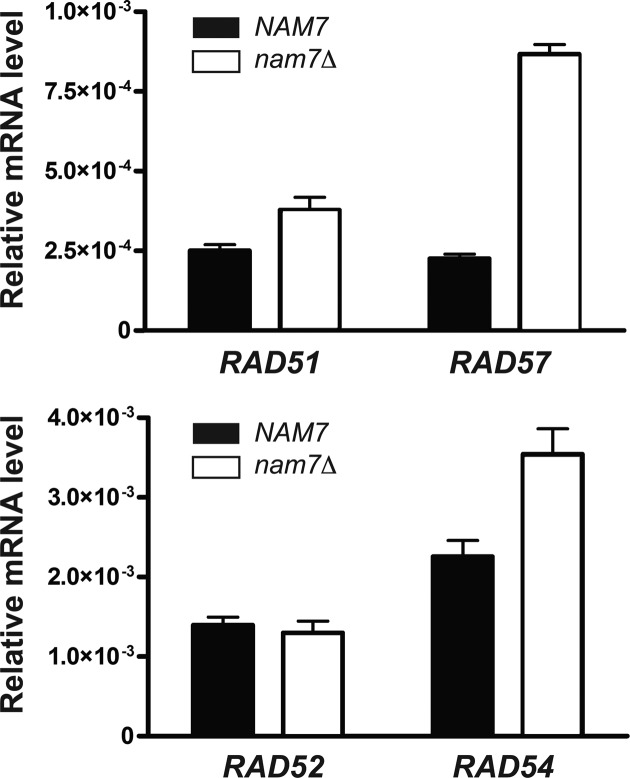
Nonsense-mediated decay regulates transcripts of homologous recombination genes
Our data (Supplementary Figure S8) shows that overexpression of Rad55-Rad57 alone does not result in the same degree of MMS resistance as an NMD mutant. This result suggests that additional genes required for increased MMS resistance are under NMD regulation. To understand more broadly how NMD may regulate genes involved in HR, steady state levels of mRNAs coding for proteins central to HR were measured by qPCR. The only known role of Rad55 is to promote Rad51 filament formation. Therefore, we wanted to test other genes exclusively involved in this step. Rad55 functions as a heterodimer with Rad57, and loss of nam7Δ resulted in a 3.8-fold increase of RAD57 mRNA levels, which was notably similar to the increase observed for RAD55 mRNA levels (Figure 6). Additionally, loss of NMD results in a statistically significant increase in Rad51 mRNA levels (Figure 6). Next, we tested several genes that function during Rad51 filament formation but also have additional roles in HR or DNA repair. Rad54 has early and late roles during HR. We consistently observed higher RAD54 mRNA levels in cells lacking NMD (Figure 6). In S. cerevisiae, mutants of RAD52 are among the most sensitive to ionizing radiation among genes involved in HR due to its multiple roles in HR and additional function in SSA (1). Interestingly, we observed no change in RAD52 mRNA levels in a nam7Δ mutant, suggesting it is excluded from regulation by NMD. Overall these data suggest that NMD acts to limit the levels of normal mRNA transcripts of genes involved in HR beyond just RAD55 and is particularly important for Rad51 filament regulation.
Figure 6.
NMD regulates the mRNA levels of multiple genes involved in homologous recombination (HR). qPCR was used to measure mRNA levels of RAD51, RAD57, RAD52 and RAD54 in WDHY2217 (NAM7) and WDHY3865 (nam7Δ). Relative mRNA levels were calculated by normalizing to ScRI RNA levels. Each plot represents the average of at least three individual experiments, error bars represent standard error of the mean. Statistical significance was calculated by a two-tailed t-test and defined by a P-value < 0.05.
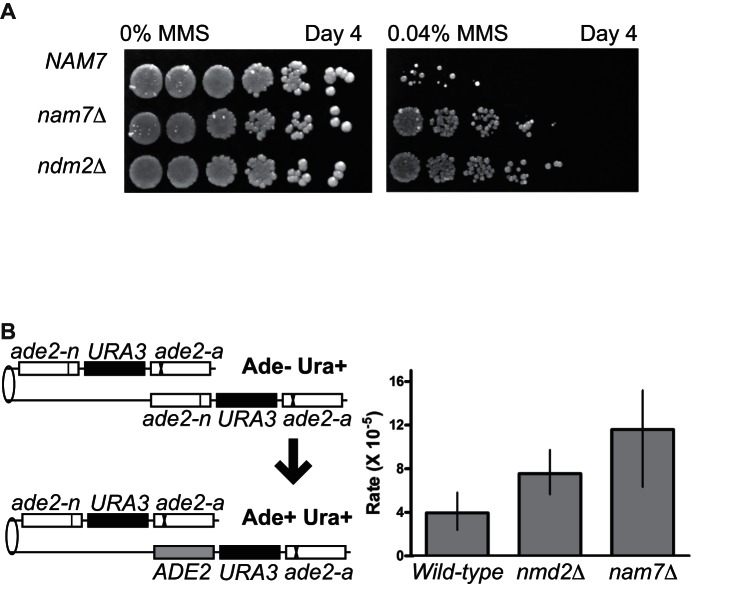
In vivo consequences of NMD on DNA repair
The loss of NMD results in increased resistance to MMS in the sensitized genetic background of a rad55-S2,8,14,19,20A mutant (Figure 2). We also wished to test whether loss of NMD conferred increased MMS resistance in an otherwise wild-type strain. Indeed, when MMS doses high enough to impact viability of a wild-type strain were used, cells with nam7Δ and nmd2Δ mutations showed substantially increased resistance to MMS treatment (Figure 7A). This result demonstrates that NMD has functional consequence on DNA repair in a wild-type strain, likely by regulating HR transcript levels.
Figure 7.
In vivo consequences of NMD on DNA damage resistance and HR. (A) Plate assay measuring the sensitivity of wild-type (WDHY2217), nam7Δ (WDHY3865) and nmd2Δ (WDHY4012) strains to MMS. Serial dilutions (1:5) of cultures were spotted onto YPD containing 0.04% MMS and grown at 30°C for 4 days prior to imaging. (B) Plot showing Ade+ Ura+ recombination rates of the indicated strains (wild-type: WDHY2371; nmd2Δ: WDHY4650; nam7Δ:WDHY4660). Rates were defined as the number of events/cell/division. Error bars represent the 95% confidence interval, non-overlapping of error bars between two samples was deemed statistically significant.
The increased resistance of NMD mutants to MMS is dependent on RAD51 (Figure 3), and we therefore hypothesized that NMD limits the amount of HR occurring in a cell. To measure the impact of NMD on rates of HR, we used a well characterized direct repeat recombination assay (59). This assay consists of two ade2 heteroalleles oriented in the same direction and separated by URA3 (Figure 7B). Spontaneous unequal sister chromatin gene conversion events between the ade2 alleles result in Ade+ Ura+ cells (59). These events are typically thought to be associated with repair of ssDNA gaps that are generated by replication forks during S-phase (30), similar to the majority of DNA damage induced by MMS treatment (60). We observed an increase in Ade+ Ura+ recombinants in NMD mutants. The recombination rate in strains lacking NAM7 increased 3-fold compared to wild-type, and 2-fold in nmd2Δ mutants cells (Figure 7B). The hyper-recombination effect in NMD-deficient cells is unlikely the consequence of increased spontaneous DNA damage. First, if NMD-deficient cells were accumulating increased spontaneous DNA damage, they would be expected to be hyper-sensitive to external genotoxic stress. To the contrary, we find that NMD-deficient cells are hyper-resistant against genotoxic stress induced by MMS (Figure 7A). Second, increased spontaneous DNA damage load in nmd mutants would predict negative genetic interactions with HR mutants, which were not observed (Figures 1–3). In sum, the increase in recombination rates in NMD-deficient cells supports a model in which NMD negatively regulates HR.
DISCUSSION
To obtain a greater understanding of mechanisms that regulate Rad55-Rad57 during HR, we combined our previous knowledge of Rad55 regulation with E-MAP technology. This approach uncovered an unexpected interaction between HR and the NMD mRNA quality control pathway. NMD acts to regulate cellular mRNA levels for genes encoding proteins that are essential for HR.
E-MAP identifies regulators of Rad55
We generated an E-MAP with the specific intention of obtaining an overview of regulatory pathways that interact with RAD55. The use of rad55 mutants that ablate specific damage-induced phosphorylation sites led to the identification of a set of positive genetic interactions that were unique to phospho-deficient rad55 mutants and absent in rad55Δ. A number of published genetic interaction datasets have included RAD55 and other HR genes, though all have exclusively utilized complete gene replacement mutants where the entire ORF of RAD55 was replaced with a selectable marker (56,61–63). While this approach has been highly valuable for studying genetic systems and the function of individual genes, the resulting interaction networks lack the phenotypic resolution to ascribe an observed genetic interaction to a particular domain or residue of Rad55 (64). Additionally, complete ORF deletions may mask certain genetic interactions, including those caused by changes in protein abundance or specific post-translational modifications. E-MAPs of domain or function-specific point mutants have previously been successfully used to study individual residue functions and post-translational modifications (33,65–67). Additionally, conditional E-MAPs have been used to identify interactions induced by various cellular perturbations (56,68). Here, we have combined the logic of both approaches to successfully identify genetic interactions that are unique to hypomorphic mutations.
Positive S-scores values represent alleviating genetic interactions, either in the form of epistasis or suppression (69). Here, we report one subset of MMS-induced positive interactions limited to rad55-phosphorylation site mutants. We envision these particular interactions to fit one of two possible scenarios, depending on whether the positive interaction is epistatic or suppressive in nature. First, an epistatic relationship may suggest that phosphorylated Rad55 acts in a unique role separate from unmodified Rad55, and mutation of other genes within this separate pathway masks the effect of Rad55 phosphorylation site mutants. Such a scenario could be theoretically possible if the phosphorylated pool of Rad55 specifically localizes to a unique class of DNA lesions induced by MMS. Second, suppression that is specific to rad55-phosphorylation site mutants but does not extend to rad55Δ mutants, may indicate an effect on Rad55 protein levels in the cell. Our data are consistent with this latter model as we observed a significant increase in Rad55 protein steady state levels in cells deficient in NMD.
NMD regulation of RAD55 mRNA and protein levels
NMD is best understood as a quality control pathway to degrade mRNAs containing premature termination codons (34). However, loss of NMD results in upregulation of 1–10% of all transcripts and accumulating evidence across multiple organisms suggests that NMD plays a broader role in the post-transcriptional regulation of normal mRNAs (35–42). We identified a novel positive genetic interaction specifically between rad55 phosphorylation site mutants and genes involved in NMD. Further testing independently confirmed the results obtained from the E-MAP screen showing that deletion of genes involved in NMD suppress the MMS-sensitive phenotype of rad55 phosphorylation site mutants. Previous studies using E-MAP and synthetic growth array technology failed to identify an interaction between NMD and RAD55 due to the fact that these screens exclusively utilized ORF deletion mutants equivalent to the rad55Δ mutant used in this study.
Disruption of the NMD pathway led to suppression of the MMS sensitivity of a rad55-S2,8,14,19,20A mutant and an increase in Rad55 mRNA and protein levels. The mRNA levels of key factors involved in Rad51 filament formation, including Rad51, Rad57 and Rad54 were also increased. The increase in Rad55 protein level provides a partial mechanism by which NMD mutants suppress the MMS sensitivity of rad55-S2,8,14,19,20A. Previous work has shown that phosphorylation of Rad55 is required for efficient DNA repair by HR after DNA damage (19). This enhancement of Rad55 function may be partially attributed to a stabilizing effect that phosphorylation imparts on the protein, and mutation of the five N-terminal phosphorylation sites of Rad55 results in lower protein levels (Figure 4A, Supplementary Figure S6). As a critical test of our model, we overexpressed Rad55-S2,8,14,19,20A from a galactose-inducible plasmid in a rad55-S2,8,14,19,20A mutant background and observed partial suppression of the MMS sensitivity. This result, in principle, demonstrates that limited Rad55-S2,8,14,19,20A pools is partially responsible for MMS sensitivity associated with the rad55-S2,8,14,19,20A mutant allele.
Loss of NMD results in a greater suppression of rad55-S2,8,14,19,20A suggesting that there are additional effects relevant to the phenotype that are not recapitulated by simply overexpressing the Rad55–Rad57 complex. The observed suppression of rad55-S2,8,14,19,20A MMS sensitivity was dependent on Rad51. Therefore, models in which deleting NAM7 upregulates other parallel DNA repair pathways, such as non-homologous end-joining or Rad51-independent SSA, are not sufficient to account for the Rad51-dependent suppression of rad55-phosphomutants in cells lacking NMD. Our data demonstrates that NMD regulates several important HR proteins involved in Rad51 filament formation (including Rad51 itself). It is possible that increased levels of one of these other factors is required to overcome the MMS sensitivity of phosphorylation-defective Rad55. However, it is also possible that the entire stoichiometry of the Rad51 filament is critical and that no single factor involved in Rad51 filament formation is responsible for the observed suppression in its entirety. The same effect of NMD on MMS sensitivity was also observed in strains that were wild-type for RAD55. These strains were similarly more resistant to MMS and also had increased steady state Rad55 mRNA and protein levels in the absence of NMD. Collectively, our data suggest that NMD limits mRNA and protein levels of key components of HR and that this is sufficient to control HR in a physiologically relevant manner.
Mechanism of NMD-dependent degradation of RAD55 mRNA
NMD appears to play a major role beyond targeting aberrant transcripts for degradation and may function as a general post-transcriptional regulatory pathway (35–42,70). While our studies directly implicate RAD55 mRNA as a target for NMD regulation, the mechanism by which NMD recognizes normal RAD55 transcripts will require further analysis. Though a number of possible targeting mechanisms exist (reviewed in 34), RAD55 mRNA contains a conspicuously large 3′-UTR. Transcripts in which termination codons are at an abnormally distant position upstream from the poly A-tail are targeted for degradation by NMD (71–74). In S. cerevisiae, 3′-UTR lengths are generally short with a median length of 120 nucleotides (75). However, transcripts from a number of genes with naturally long 3′-UTRs have been shown to be enriched for degradation by NMD (74,76). Based on tiling array experiments from Xu et al. (77), RAD55 has a 3′-UTR length of 987 nt, well above the typical 3′-UTR length in S. cerevisiae, and within the range of reported 3′-UTR lengths of transcripts targeted for NMD (76). We suggest this natural feature of RAD55 results in transcripts that are targeted for NMD, though experimental validation of this model is still required.
NMD as a regulator of DNA repair
We observe that loss of NMD has an impact on mRNA levels for several genes of the RAD52 epistasis group. Among those tested, RAD55 and RAD57 show the largest change in mRNA levels (∼3–5-fold) as a result of deactivating NMD, while both RAD51 and RAD54 also showed increases in mRNA levels. Additionally, we tested whether RAD55 mRNA is a direct target for NMD degradation by measuring its half-life using the RNA polymerase II temperature-sensitive allele rpb1–1. Consistent with many NMD targets, RAD55 mRNA is found in low abundance and has a very short half-life (35). The 2.6-fold increase in RAD55 mRNA half-life in NMD-deficient cells is similar to the effect observed on other targets of NMD degradation (35,36). While we observe NMD-dependent effects on the steady state of RAD57, RAD51 and RAD54 mRNA levels, we did not distinguish between a direct and indirect effect of NMD.
Our results implicate NMD as a general mechanism to regulate the transcripts of genes involved in HR and DNA repair. In agreement with our results, RAD57 was independently identified in a global screen as a target of NMD regulation but no additional information was reported (35). The same study included a number of genes that function in double strand break repair by non-homologous end-joining (NEJ1, POL4, DNL4,YKU80), DDR signaling (RED1, MEC3) and HR (MPH1, SHU2, RAD1, RAD57, SLX1) (35). A recent study in mammalian cells also showed that NMD ablation led to increased sensitivity to doxorubicin, a DNA intercalator used as a chemotherapeutic agent (78) in a process involving apoptosis. Both studies (78) (our work) reveal additional complexity in the cellular response to DNA damage involving NMD as a way to sculpt the transcriptome in response the genotoxic stress.
Other examples of post-transcriptional regulation of mRNA in response to DNA damage have recently been described. A recent study by Graber et al. (79) showed that treatment of cells with the UV mimetic, 4-nitroquinoline 1-oxide (4NQO) resulted in global, systematic changes in poly(A) site usage and mRNA isoform abundance in S. cerevisiae. Interestingly, 4NQO treatment resulted in an elongation of transcript isoforms for 1720 genes including RAD55, accounting for the majority of measured changes. One possibility is that DNA damage triggers a regulatory effect of NMD processes that lead to a shift toward elongated mRNA isoforms observed by Graber et al. Conversely, DNA damage-dependent regulation of polyadenylation site choice could shift a pool of NMD sensitive mRNAs to more stable isoforms that evade degradation by NMD.
The process of Rad51 filament formation is thought to be highly regulated as it marks a key commitment point and likely rate-limiting step to repairing a DNA lesion by HR. A number of steps involved in Rad51 filament formation, including the function of Rad55–Rad57, are known to be regulated by post-translational processes (6,20). This report extends the regulation of Rad51 filament formation to involve post-transcriptional regulation by NMD. Proteins that have a direct role in promoting Rad51 filaments are typically low in abundance, and this may be a mechanism to prevent aberrant recombination or interference with other nuclear processes in a way that could be detrimental to a cell. Indeed, our results show that NMD acts to limit HR in wild-type cells, as inferred from an increase in recombination rates and MMS resistance in cells lacking NMD (Figure 7B).
Supplementary Material
Acknowledgments
The authors thank Shuyi Wang for technical assistance, Michael Shales, Rinti Mukherjee, Damon Meyer and Aurèle Piazza for helpful discussion and comments on the manuscript, and Domenico Libri for the rpb1–1 strain.
Footnotes
Present address: Ryan Janke, Department of Molecular & Cell Biology, California Institute for Quantitative Biosciences, QB3, University of California, Berkeley, Berkeley, CA 94720-3220, USA.
FUNDING
US National Institutes of Health [CA92276 to W.D.H., GM084448, GM084279, GM081879, GM098101 to N.J.K., P41 GM103533, R01 MH067880 to J.R.Y.]; Training grant [T32 ES007059 to R.J., in part]. Funding for open access charge: NIH [CA92276].
Conflict of interest statement. None declared.
REFERENCES
- 1.Heyer W.D. Biochemistry of eukaryotic homologous recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X., Heyer W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 381–441. [Google Scholar]
- 4.Kasparek T.R., Humphrey T.C. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin. Cell Dev. Biol. 2011;22:886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 5.George C.M., Alani E. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit. Rev. Biochem. Mol. Biol. 2012;47:297–313. doi: 10.3109/10409238.2012.675644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyer W.-D., Ehmsen K.T., Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Liu J., Ehmsen K.T., Heyer W.D., Morrical S.W. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit. Rev. Biochem. Mol. Biol. 2011;46:240–270. doi: 10.3109/10409238.2011.576007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 10.Huertas P., Cortes-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 14.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 15.Sacher M., Pfander B., Hoege C., Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nature cell biology. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 16.Bashkirov V.I., King J.S., Bashkirova E.V., Schmuckli-Maurer J., Heyer W.-D. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen C.S., Hansen L.T., Dziegielewski J., Syljuasen R.G., Lundin C., Bartek J., Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 19.Herzberg K., Bashkirov V.I., Rolfsmeier M., Haghnazari E., McDonald W.H., Anderson S., Bashkirova E.V., Yates J.R., Heyer W.D. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell. Biol. 2006;26:8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janke R., Herzberg K., Rolfsmeier M., Mar J., Bashkirov V.I., Haghnazari E., Cantin G., Yates J.R., Heyer W.D. A truncated DNA-damage-signaling response is activated after DSB formation in the G1 phase of Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:2302–2313. doi: 10.1093/nar/gkp1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 22.Hays S.L., Firmenich A.A., Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson R., Symington L. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., Heyer W.D. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.New J.H., Sugiyama T., Zaitseva E., Kowalczykowski S.C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 26.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 27.Shinohara A., Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 28.Fortin S., Symington L.S. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugawara N., Wang X., Haber J.E. In Vivo Roles of Rad52, Rad54, and Rad55 Proteins in Rad51-Mediated Recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 30.Mozlin A.M., Fung C.W., Symington L.S. Role of the Sacharomyces cerevisiae Rad51 paralogs is sister chromatid recombination. Genetics. 2008;178:113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins S.R., Roguev A., Krogan N.J. Quantitative genetic interaction mapping using the E-MAP approach. In: Jonathan W, Christine G, Gerald RF, editors. Guideto Yeast Genetics: Functional Genomics, Proteomics, and Other System Analysis. Vol. 470. Amsterdam: Academic Press; 2010. pp. 205–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braberg H., Jin H., Moehle E.A., Chan Y.A., Wang S., Shales M., Benschop J.J., Morris J.H., Qiu C., Hu F., et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mole. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan Q., Zheng W., Tang S., Liu X., Zinkel R.A., Tsui K.-W., Yandell B.S., Culbertson M.R. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 37.Johansson M.J.O., He F., Spatrick P., Li C., Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 39.Rehwinkel J., Letunic I., Raes J., Bork P., Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isken O., Maquat L.E. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decourty L., Doyen A., Malabat C., Frachon E., Rispal D., Séraphin B., Feuerbach F., Jacquier A., Saveanu C. Long open reading frame transcripts escape nonsense-mediated mRNA decay in yeast. Cell Rep. 2014;6:593–598. doi: 10.1016/j.celrep.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 43.He F., Brown A.H., Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 45.Erdeniz N., Mortensen U.H., Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 47.Collins S.R., Schuldiner M., Krogan N.J., Weissman J.S. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lea D.E., Coulson C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 50.Spell R., Jinks-Robertson S. In: Genetic Recombination. Waldman A, editor. Vol. 262. Totowa: Humana Press; 2004. pp. 3–12. [Google Scholar]
- 51.Kebaara B.W., Baker K.E., Patefield K.D., Atkin A.L. Analysis of nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Curr. Protoc. Cell Biol. 2012 doi: 10.1002/0471143030.cb2703s54. Chapter 27, Unit 27.23. [DOI] [PubMed] [Google Scholar]
- 52.MacCoss M.J., McDonald W.H., Saraf A., Sadygov R., Clark J.M., Tasto J.J., Gould K.L., Wolters D., Washburn M., Weiss A., et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald W.H., Ohi R., Miyamoto D.T., Mitchison T.J., Yates Iii J.R. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 2002;219:245–251. [Google Scholar]
- 54.Bashkirov V.I., Herzberg K., Haghnazari E., Vlasenko A.S., Heyer W., Campbell J.L., a.P.M. DNA damage-induced phosphorylation of Rad55 protein as a sentinel for DNA damage checkpoint activation in S. cerevisiae. Methods Enzymol. 2006;409:166–182. doi: 10.1016/S0076-6879(05)09010-5. [DOI] [PubMed] [Google Scholar]
- 55.Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 56.Bandyopadhyay S., Mehta M., Kuo D., Sung M.K., Chuang R., Jaehnig E.J., Bodenmiller B., Licon K., Copeland W., Shales M., et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chamieh H., Ballut L., Bonneau F., Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 58.He F., Peltz S.W., Donahue J.L., Rosbash M., Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang K.N., Symington L.S. Mutation of the gene encoding protein kinase C 1 stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:6039–6045. doi: 10.1128/mcb.14.9.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Groth P., Auslander S., Majumder M.M., Schultz N., Johansson F., Petermann E., Helleday T. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J. Mol. Biol. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 63.Leung G.P., Aristizabal M.J., Krogan N.J., Kobor M.S. Conditional genetic interactions of RTT107, SLX4, and HRQ1 reveal dynamic networks upon DNA damage in Saccharomyces cerevisiae. G3 (Bethesda). 2014;4:1059–1069. doi: 10.1534/g3.114.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan C.J., Cimermancic P., Szpiech Z.A., Sali A., Hernandez R.D., Krogan N.J. High-resolution network biology: connecting sequence with function. Nat. Rev. Genet. 2013;14:865–879. doi: 10.1038/nrg3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuchs S.M., Kizer K.O., Braberg H., Krogan N.J., Strahl B.D. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 Di- and trimethylation at lysine 36. J. Biol. Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charles G.M., Chen C., Shih S.C., Collins S.R., Beltrao P., Zhang X., Sharma T., Tan S., Burlingame A.L., Krogan N.J., et al. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proc. Natl. Acad. Sci. 2011;108:10620–10625. doi: 10.1073/pnas.1019735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrison A.J., Kim J.-A., Person M.D., Highland J., Xiao J., Wehr T.S., Hensley S., Bao Y., Shen J., Collins S.R., et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 68.Guenole A., Srivas R., Vreeken K., Wang Z.Z., Wang S., Krogan N.J., Ideker T., van Attikum H. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol. Cell. 2013;49:346–358. doi: 10.1016/j.molcel.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beltrao P., Cagney G., Krogan N.J. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He F., Jacobson A. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 2015;49:339–366. doi: 10.1146/annurev-genet-112414-054639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 72.Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaborske J.M., Zeitler B., Culbertson M.R. Multiple transcripts from a 3′-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae. PLoS One. 2013;8:e80981. doi: 10.1371/journal.pone.0080981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kebaara B.W., Atkin A.L. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Popp M.W., Maquat L.E. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat. Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graber J.H., Nazeer F.I., Yeh P.-c., Kuehner J.N., Borikar S., Hoskinson D., Moore C.L. DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Res. 2013;23:1690–1703. doi: 10.1101/gr.144964.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.