Abstract
Signal -transduction networks coordinate transcriptional programs activated by diverse extracellular stimuli, such as growth factors and cytokines. Cells receive multiple stimuli simultaneously, and mapping how activation of the integrated signaling network affects gene expression is a challenge. We stimulated colon adenocarcinoma cells with various combinations of the cytokine tumor necrosis factor (TNF) and the growth factors insulin and epidermal growth factor (EGF) to investigate signal integration and transcriptional crosstalk. We quantitatively linked the proteomic and transcriptomic data sets by implementing a structured computational approach called tensor partial least squares regression. This statistical model accurately predicted transcriptional signatures from signaling arising from single and combined stimuli and also predicted time-dependent contributions of signaling events. Specifically, the model predicted that an early-phase, Akt-associated signal downstream of insulin repressed a set of transcripts induced by TNF. Through bioinformatics and cell-based experiments, we identified the Akt-repressed signal as glycogen synthase kinase 3 (GSK3)–catalyzed phosphorylation of Ser37 on the long form of the transcription factor GATA6. Phosphorylation of GATA6 on Ser37 promoted its degradation, thereby preventing GATA6 from repressing transcripts that are induced by TNF and attenuated by insulin. Our analysis showed that predictive tensor modeling of proteomic and transcriptomic data sets can uncover pathway crosstalk that produces specific patterns of gene expression in cells receiving multiple stimuli.
INTRODUCTION
A goal of systems biology is to identify regulatory properties of biological networks by integrating experiments and kinetic models around a defined biological “parts list” (1). For signal transduction, such approaches have clarified the molecular pathways and regulatory events that can explain signaling thresholds (2–4), adaptation (5–8), and oscillations (9–15), as well as signal-induced cell -fate transitions (16–18). A complementary use of systems methods is for large-scale profiling and analysis of cellular signals in pursuit of integrative understanding or new molecular mechanisms (19–22). These applications favor data-driven models based on statistics or bioinformatics, which propose new links within the network that can be tested experimentally (23–28). Testing of a data-driven hypothesis about mechanism requires reductionist experimental approaches, even when the hypothesis itself emerges from network-level data (29, 30). Consequently, systems-level studies can yield findings that are immediately relevant to signal transduction biology (31).
Receptor-mediated signaling pathways are densely connected and exhibit nonadditive behaviors when two ligands activating different receptors are applied simultaneously (32–34). Downstream of receptor activation and signal transduction, complexity increases even further when considering the consequences of signaling on gene regulation (35–37). Fortunately, new signaling synergy or antagonism does not typically emerge with more than two inputs (38–41), suggesting that stimulus pairs are sufficient to assess potential crosstalk. For systems-level discovery, this type of in-depth cellular profiling must be complemented by data-driven modeling and extensive mechanistic follow-up.
Here, we examined crosstalk between a proinflammatory stimulus, tumor necrosis factor (TNF), and one of two growth factors, insulin or epidermal growth factor (EGF), with a focus on signaling -network activation and downstream regulation of the transcriptome. To examine the interface between signal -transduction and transcription networks at the systems level, we built upon a preexisting compendium of signaling events collected in the adenocarcinoma cell line HT-29, which were initially exposed to interferon- γ (IFNγ) and then stimulated with TNF, EGF, and insulin in various combinations (42, 43). Because TNF, EGF, and insulin trigger widespread and time-dependent changes in transcript abundance (44–47), we collected dynamic mRNA profiles after stimulation with single or paired stimuli and integrated these data with the previously measured intracellular signaling events (42, 43).
Because each signal and transcript was measured at every time point and stimulus condition, we could organize the signaling and transcriptomic data cubes as three-way tensors to build a model constrained by the structure of the data. Model analysis together with bioinformatics predicted a link between Akt–glycogen synthase kinase 3 (GSK3) signaling and the endodermal transcription factor GATA6. We found that GSK3 phosphorylation of Ser37 on the long form of GATA6 (GATA6L) accelerated its degradation in cells. The increased turnover reduced transcriptional repression by GATA6L of genes induced by TNF. Collectively, our results showed that GATA6L integrated growth factor–induced signaling activity and inflammatory transcriptional regulation.
RESULTS
Cytokine combinations elicit complex changes in signaling and transcript abundance
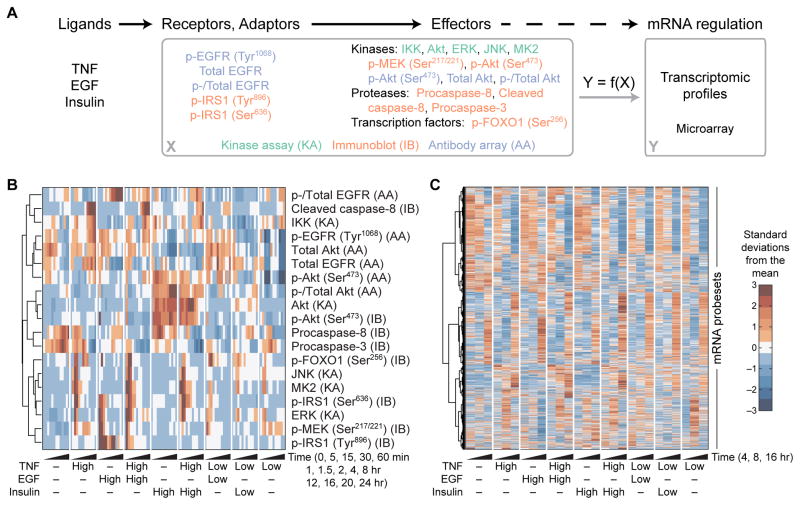
For the signaling pathway measurements, we used data from our own previous studies (42, 43), which enabled us to combine these data with the transcriptomic data generated here under the same experimental conditions. In the previous studies, HT-29 cells were exposed to IFNγ, which primes HT-29 cells to apoptose upon stimulation with TNF (48), and subsequently stimulated with saturating or subsaturating doses of TNF, EGF, or insulin alone or in combination. Lysates were profiled at 13 time points over 24 hours for 19 intracellular signaling events measured by kinase assay, immunoblot, or antibody array (Fig. 1, A and B) (49–51). These data provided quantitative, systematically collected information on phosphorylation-mediated regulatory events, changes in protein abundance, and cleavage-dependent protein activation.
Fig. 1. A compendium of ligand--induced signals and transcriptional responses.
(A) Overview of the experimental design. HT-29 cells were pretreated with IFNγ, stimulated with various combinations and concentrations of TNF, EGF, and insulin, and profiled for the indicated signaling receptors, adaptors, and effectors by kinase assay (KA), immunoblot (IB), or antibody array (AA) and for the associated transcriptomic signatures by microarray. The goal is to determine whether global ligand-induced mRNA regulatory states (Y) can be predicted from the upstream signaling network activation (X). (B) Hierarchical clustering of the signaling compendium for saturating (High) and subsaturating (Low) concentrations of TNF, EGF, and insulin (42, 43). Data are means of n = 3 to 6 independent biological replicates. (C) Hierarchical clustering of the dynamic transcriptomic responses resulting from the ligand combinations in (B). Data are means of n = 2 independent biological replicates.
To determine how the signaling events altered gene expression, we complemented the signaling compendium with a matched set of transcriptomic profiles (Fig. 1A). IFNγ-pretreated HT-29 cells were exposed to the same combinations of TNF-EGF-insulin and analyzed at a subset of the time points from the previous signaling studies (Fig. 1B). We collected transcriptomic profiles by microarray at intermediate-to-late times after cytokine simulation, thereby avoiding the bursts of immediate-early transcripts that are already well characterized (46, 47, 52). We selected the time points for transcriptomic analysis according to earlier modeling of the signaling compendium, which showed that signaling from 4 to 16 hours did not predict apoptosis accurately (42). We reasoned that the loss of predictive ability was because prolonged cytokine stimulation had transmitted the relevant information to the downstream transcriptional network.
The microarray data revealed extensive transcriptional alterations with time and stimulus condition (Fig. 1C). Among 14,541 probe sets identified as present in at least one sample, we identified significant changes in 10,319 with time, 4948 upon TNF stimulation, 75 upon EGF stimulation, and 15 upon insulin stimulation after correction for multiple hypothesis testing [four-way analysis of variance (ANOVA), false -discovery rate = 5%; file S1]. One unanticipated complication was that many transcript abundances changed with time in the mock stimulation condition lacking TNF, EGF, and insulin (Fig. 1C, leftmost column). We attributed these background transcriptional dynamics to the ongoing IFNγ exposure. Extensive background drifts in transcript abundance can confound interpretations from standard analyses of differentially expressed genes that focus on time-dependent changes (53, 54). Therefore, alternative methods were required to identify meaningful changes in transcriptional regulation and link them to the upstream signaling network.
Models of dynamic, multivariate data sets are properly structured as data tensors
Systematic biology experiments monitor the same signaling events or transcripts over multiple time points and across multiple stimulus conditions (19, 55). Each data point thus contains information about the various “modes” of its acquisition, for example, which stimulus was added (mode 1), the time after stimulus (mode 2), and the signal or transcript measured (mode 3). Additional modes are possible if multiple pharmacologic perturbations (56, 57) or cell types (58) are profiled systematically along with the modes listed above.
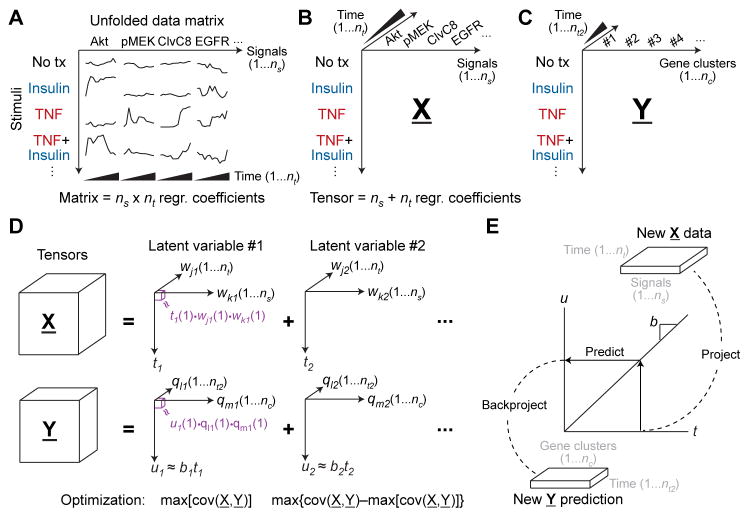
For systematic measurements, the acquisition modes create a data structure that is very powerful mathematically because it conveys how different data points are related to one another. This structure vanishes when, for example, a data cube is sliced along one of its modes and “unfolded” end-to-end as a series of matrices (Fig. 2A). When matrix-based algorithms are applied to unfolded data, each unfolded measurement variable is treated independently, and modes 2 and greater are lost. Using Fig. 2A as an example, Akt measurements at 2 and 4 hours after stimulation (same signal, two time points) are not handled any differently than Akt and epidermal growth factor receptor (EGFR) measurements at 2 hours after stimulation (two signals, same time point). The result of unfolding is a model that is less interpretable because of too many fitted regression coefficients (59).
Fig. 2. Structuring and modeling biological data sets as tensors.
(A) Structured data sets are conventionally unfolded with time to create a concatenated data matrix of ns signals and nt time points. Using the unfolded matrix, data-driven modeling approaches (20) treat each time point of each signal as a separate predictor variable, yielding ns × nt regression (regr) coefficients that must be inferred. (B) Recasting stimulus-signal-time data sets as a third-order tensor. The tensor structure (X) considers each time point as a predictor variable for all signals and each signal as a predictor variable for all time points, resulting in ns + nt regression coefficients and thus a more parsimonious model. (C) A dependent third-order transcriptomic tensor (Y) structured by stimulus, nc gene clusters, and nt2 time points. (D) Decomposing third-order data tensors as sums of latent variables composed of triple products. The decomposed tensor for each latent variable is reconstructed as the triple product (purple) of a scores vector (t or u) and two weight vectors (wj and wk or ql and qm). Latent variables are iteratively calculated to capture the maximum covariance between X and Y that remains from the preceding latent variable. X and Y are connected by a linear inner relationship between t and u with slope = b. (E) Prediction with tensor models involves projecting a new stimulus onto the latent variables of X, predicting the dependent scores vector u from the linear inner relationship (u = bt), and then backprojecting onto the latent variables of Y.
The alternative to unfolding is to retain data sets as cubes (three modes) or hypercubes (four or more modes) in the form of data “tensors” which are the higher-dimensional generalization of vectors (one mode) and matrices (two modes). For example, the TNF-EGF-insulin signaling compendium naturally organizes as a third-order tensor defined by stimulus, time point, and measured signaling event (Fig. 2B). The transcriptomic profiles likewise arrange as a third-order tensor according to stimulus, time point, and transcript or cluster of transcripts (Fig. 2C). Tensors reduce the parameterization of a data-driven model because free regression coefficients remain fixed across the other acquisition modes of each tensor (Fig. 2B) (59, 60). In this instance, the stimulus–time point–signaling tensor (the “regressor” tensor) is linked to the stimulus–time point–transcript tensor (the “regressand” tensor) by the regression coefficients.
Biological data tensors have been used successfully for unsupervised purposes, such as singular value decomposition (27), to analyze transcriptional kinetics during DNA replication origin firing (28) and to identify consistent copy number changes across different array-based comparative genomic hybridization platforms (61). Here, we sought a supervised method that could connect the signaling tensor to the transcriptomic tensor and predict gene expression patterns from signaling network dynamics. This application is ideal for the tensor generalization of partial least squares regression (PLSR), a matrix implementation that has been used widely to model signaling networks (20, 25, 58, 62–76).
Tensor PLSR [equivalently, “multilinear PLS” (59)] is an established method that creates a data-driven model by jointly factorizing an independent “predictor” tensor (X; here, the signaling tensor) and a dependent “predicted” tensor (Y; the transcriptomic tensor). X and Y are factorized as an element-by-element product of vectors, where the number of vector elements multiplied is equal to the number of dimensions in the data tensor (text S1). Thus, if X is a third-order tensor, then X(1,1,1) [the tensor element in X occupying the first position in mode 1 (stimulus), the first position in mode 2 (time point), and the first position in mode 3 (signal)] is factorized as: X(1,1,1) = t(1)• wj(1)•wk(1) (Fig. 2D, purple). In the factorization, t(1) is the first element of a “scores” vector (t) that relates to the stimulus conditions that are shared with the Y tensor. wj(1) and wk(1) are the first elements of two “weight” vectors (wj and wk) that relate to modes 2 and 3 of the tensor (here, time and signal). A similar calculation is performed for Y by factorizing it into its own scores (u) and weight (ql and qm) vectors. X and Y are linked by an “inner relationship” between their respective scores vectors: u = bt, where b is a linear regression coefficient determined by the model. The inner relationship implies that how a stimulus projects on t [through the signaling (wk) that occurs over time (wj)] is directly proportional to its projection on u and thus how that stimulus changes gene expression (qm) with time (ql).
The factorization of the two tensors is posed as a numerical optimization that seeks to capture as much of the inner relationship between X and Y as possible (details on the covariance maximization algorithm are described in text S1). The best first set of scores and weight vectors defines the first “latent variable” of the tensor PLSR model. Residual information (covariation) in X and Y not captured by the first latent variable is then subjected to a second factorization, which is optimized to capture as much covariance in the residual as possible (Fig. 2D). By repeating the algorithm, latent variables are iteratively calculated until there are no predictive inner relationships remaining between the X and Y data tensors (20, 62).
Predictions with a tensor PLSR model use wj and wk from each latent variable to project an X-like observation onto t (Fig. 2E). Then, the u = bt inner relationship is used to predict u, which is backprojected with ql and qm to yield a predicted set of values in the form of Y (time-dependent gene expression). The project-predict-backproject sequence is important for making independent predictions with new data and for cross-validation of the model to identify the optimum number of latent variables (20, 31, 62, 77).
Tensor PLSR modeling identifies predictive links between signaling and transcriptional dynamics
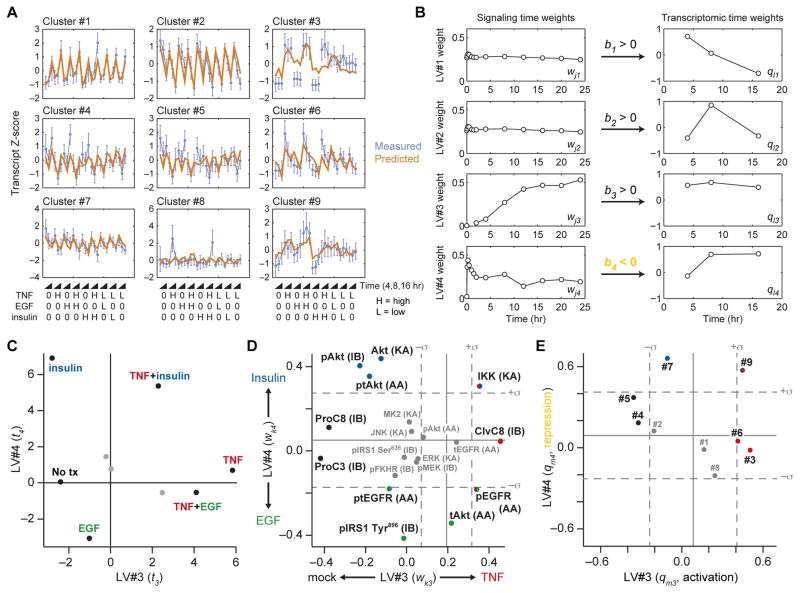
We first constructed a tensor PLSR model of three latent variables that predicted the 14,541 probe set fluorescence intensities of the transcriptomic data set. Although cross-validated predictions of the model were 99% accurate (fig. S1A), the model was strongly biased toward the differences in fluorescence intensities across probe sets. Consequently, changes in probe set intensities across treatment conditions were overlooked, and the resulting components of the model were uninterpretable (fig. S1, B to E). To focus on recurrent stimulus-dependent changes in transcript abundance shared by multiple genes, we condensed the transcriptomic data set by using the unbiased CLuster Identification via Connectivity Kernels (CLICK) algorithm (78). Among the transcripts profiled, CLICK identified nine separable clusters composed of dozens to hundreds of genes (file S2), the mean trajectories of which were organized as the Y data tensor (Fig. 2C). Using the entire signaling compendium as X, we constructed a tensor PLSR model of four latent variables that predicted gene -cluster dynamics to within 72% (Fig. 3A and fig. S2A). Although the model did not predict certain cytokine-induced changes for some gene clusters (Fig. 3A, see EGF and insulin stimuli of cluster #3), we considered the overall accuracy of predictions remarkable considering that the model involved ~10-fold fewer parameters than previous PLSR models of TNF-induced apoptosis (42, 62, 64).
Fig. 3. A tensor PLSR model linking ligand-induced signaling and changes in transcript abundance.
(A) Time-unfolded measurements of transcriptional clusters (blue) compared to cross-validated predictions of the tensor PLSR model (brown). Standardized z scores of measured transcriptional clusters are means ± SD of n = 897 (#1), 841 (#2), 119 (#3), 106 (#4), 66 (#5), 49 (#6), 42 (#7), 33 (#8), and 26 (#9) probe sets (file S2). High (H) indicates saturating concentration of ligand, 0 indicates absence of ligand, and low (L) indicates subsaturating concentration of ligand. (B) Latent variable time weights for the signaling and transcriptomic tensors. LV#4 has a negative inner relationship (orange), indicating that LV#4 signaling is anticorrelated with LV#4 transcription. (C to E) Projections of the indicated stimulus conditions (C), signals (D), and transcriptional clusters (E) onto LV#3 and LV#4. For (D) and (E), the null projections of reshuffled data tensors are means (solid gray) ± SD (dashed gray) of n = 500 randomizations (79). In (D), the type of assay used to measure the signaling protein is indicated in parentheses (see Fig. 1A for details). ClvC8, cleaved caspase 8; ProC3, procaspase 3; ProC8, procaspase 8; lowercase p prefix represents phosphorylated protein; lowercase t prefix represents total protein; lowercase pt prefix represents XXX.
Our principal motivation for building the tensor PLSR model was to use the model to reveal undiscovered mechanisms of how signaling alters gene expression. To identify which latent variables captured both signaling and gene -cluster dynamics, we analyzed the time weights (wj and ql) and inner regression coefficients for X and Y (Fig. 3B). The leading two latent variables (LV#1 and LV#2) harbored signaling time weights (wj1 and wj2) that were nearly constant from 0 to 24 hours, indicating that time-dependent changes in signaling did not determine the projection of X along LV#1 or LV#2. Accordingly, time weights for the gene clusters (ql1 and ql2) were time-variant and derived from the stimulus-independent transcriptional changes of clusters #1, #2, and #7 (Fig. 3, A and B), presumably resulting from IFNγ pretreatment. Because latent variables are calculated iteratively (Fig. 2D), LV#1 and LV#2 eliminated the TNF-, EGF-, and insulin-independent transcriptional changes, revealing paired signaling and gene -cluster dynamics in the third and fourth latent variables (LV#3 and LV#4). LV#3 harbored time weights of late-phase signaling (wk3) and sustained transcriptional activation (ql3). Conversely, LV#4 was weighted with early-phase signaling (wk4) and late-phase transcriptional regulation (ql4). The inner regression coefficient for LV#4 was negative (Fig. 3B, orange), implying a link between early-phase signaling and downstream transcriptional repression.
Focusing on LV#3 and LV#4, we evaluated the relationship between the treatment scores (t3 and t4; Fig. 3C). Relative to mock treatment, saturating TNF stimulation projected almost entirely along LV#3, suggesting that LV#3 represented a TNF “axis” In contrast to TNF, we found that EGF and insulin projected in opposite directions along LV#4, indicating that this latent variable distinguished between the two growth -factor stimuli. Combinatorial stimulations exhibited intermediate scores that approximately averaged the scores of the individual stimuli. For example, TNF+EGF projected positively along LV#3 (like TNF) and negatively along LV#4 (like EGF). The interpolated response observed here for gene regulation contrasts with previous work on apoptosis in which EGF and insulin each nonlinearly antagonized TNF-induced cell death (62).
To connect specific signals and gene clusters with the prevalent cytokine-induced dynamics, we evaluated the signaling and gene -cluster weights along LV#3 (TNF; late-phase signaling axis: wk3 and qm3) and LV#4 (EGF-insulin; early-phase signaling axis: wj4 and qm4) (Fig. 3, D and E). Multiple signals, such as Ser636-phosphorylated insulin receptor substrate-1 [pIRS1 (Ser636)], c-Jun N-terminal kinase (JNK) activity, and mitogen-activated protein kinase (MAPK)–activated protein kinase-2 (MK2) activity, were negligibly weighted (Fig. 3D), implying that these early-phase TNF-induced signals (Fig. 1B) were statistically uninformative for predicting Y (the transcriptional response). Gene cluster #1 was also unweighted along LV#3 and LV#4, because its dynamics were almost entirely captured by LV#1 and LV#2 (Fig. 3, A, B, and E, and fig. S2B). To filter the weight vectors further, we randomly shuffled the signaling, gene -cluster, and time information within each cytokine stimulation (mode 1 slice) of X and Y (79). With hundreds of shuffled tensor PLSR models, we estimated a null projection for the weight vectors of LV#3 and LV#4 (Fig. 3, D and E, gray line). We considered signals and gene clusters outside 1 SD (σ, gray dashed lines) of the null projection as weighted strongly enough to warrant further analysis (Fig. 3, D and E).
Among the strongest signaling weights, we found clear agreement with known mechanisms of signal transduction (Fig. 3D). For example, cleavage of apoptotic caspases [negative weighting for procaspase-8 and procaspase-3 and positive weighting of cleaved caspase-8] was strongly aligned along LV#3, which is consistent with late-phase caspase activation triggered by TNF (48, 80). Along LV#4, insulin stimulation coincided with positive weights for three complementary measures of Akt activation, a recognized effector pathway (81). Likewise, multiple measures of EGFR phosphorylation were weighted in a direction that corresponded to EGF stimulation. Also strongly associated with EGF signaling was phosphorylated insulin receptor substrate-1 [pIRS1 (Tyr896)], consistent with reports that this site may be directly phosphorylated by active EGFR (42, 82). Not all signaling events were associated with individual stimuli. For instance, the weights associated with inhibitor of nuclear factor-κB kinase (IKK) activation mapped not only to TNF but also to insulin stimulation, possibly because Akt signaling can activate IKK in certain contexts (83, 84). Similarly, phospho-EGFR (Tyr1068) projected with early-phase EGF and also late-phase TNF signaling. The latter is probably due to autocrine signaling by transforming growth factor-α, an EGFR ligand that is released after TNF stimulation (43, 85). Together, the weights of LV#3 and LV#4 provided a condensed map of the signaling compendium that was optimized for predicting the observed transcriptomic profiles.
The inner regression coefficient (b3) connecting X and Y along LV#3 was a positive value, indicating gene activation, whereas the inner regression coefficient b4 was negative, indicating that signaling along LV#4 resulted in gene repression (Fig. 3, B and E). Contrary to that of the signaling compendium, the projection of gene clusters along LV#3 and LV#4 was surprising (Fig. 3E). Amidst thousands of time-dependent transcriptional changes, few clusters were weighted toward specific stimuli. Clusters #3 and #6 were primarily weighted along LV#3, indicating an association with TNF stimulation. Accordingly, promoter analysis (86) of the transcripts in these two clusters revealed a strong overrepresentation of binding sites for nuclear factor κB (NF-κB) (fig. S2C). Cluster #7 mapped along LV#4 because of the mild suppression of transcripts observed with saturating insulin alone (Fig. 3, A and E). Only cluster #9 projected strongly along both latent variables, indicating that the transcripts in this cluster were induced by TNF and repressed by insulin (fig. S3). TNF antagonism of insulin function has been well documented in adipocytes (87, 88), but there are few reports of insulin antagonizing TNF (89). Given this predicted TNF and insulin “crosstalk cluster” we used the tensor PLSR model to investigate its mechanism of regulation by the upstream signaling network.
With respect to its latent variable projections, cluster #9 was cartographically most similar to IKK (Fig. 3, D and E). If these shared projections were indicative of mechanism, however, it would imply that early-phase IKK activity (downstream of TNF and insulin signaling) represses transcription of the crosstalk cluster, whereas late-phase IKK (downstream of TNF signaling) promotes it. Repress-then-activate kinetics are opposite of the prevailing view of IKK signaling (90). Accordingly, we found that TNF-induced responses of 85% of transcripts in cluster #9 were not significantly affected when a phosphorylation- and degradation-resistant mutant of IκBα was ectopically expressed in HT-29 cells (log-transformed Welch’s t test, false discovery rate = 15%; fig. S4). We therefore considered alternatives that were consistent with the tensor PLSR model.
One possible explanation was that the crosstalk cluster integrated two distinct signaling inputs. An activating input could arise from a TNF-specific signal that was either not measured or not projected strongly on LV#3 and LV#4. In parallel, the cluster could be transcriptionally inhibited by an insulin-specific signal, such as Akt (Fig. 3D), or a downstream effector pathway of Akt (91).
Promoter and signaling bioinformatics suggest a link between GSK3 and the crosstalk cluster through GATA6
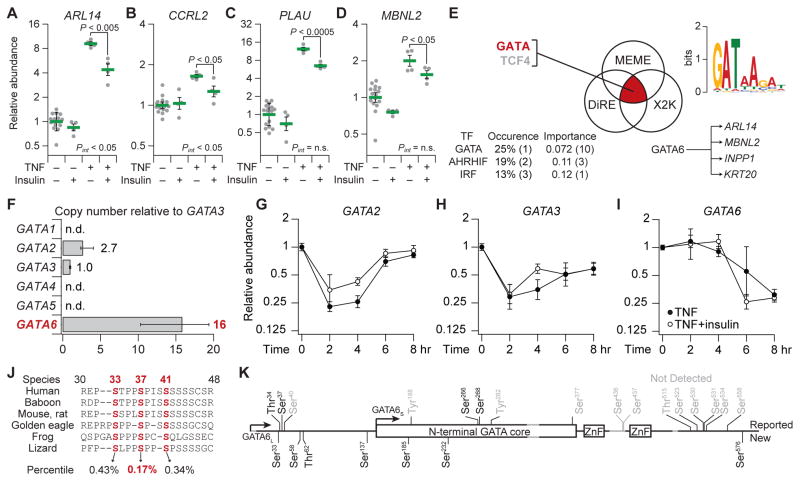
First, we determined the reliability and generality of TNF-insulin crosstalk among transcripts in cluster #9. We repeated the stimulation experiments with an independently obtained vial of HT-29 cells and assayed individual transcripts by quantitative reverse transcription polymerase chain reaction (qRT-PCR). This analysis confirmed the expression of more than 90% of the 22 cluster #9 transcripts (SPRR1B and PPARD from fig. S3 were false positives), and we detected an antagonistic interaction between TNF and insulin from 2 to 8 hours after stimulation (fig. S5). At individual time points for specific genes, we observed instances of antagonism represented by significant interaction P value (Pint < 0.05, two-way ANOVA; Fig. 4, A and B), an indication that TNF and insulin have nonadditive effects on the expression of those genes. We also observed nonlinear significant differences in gene expression for other transcripts even when the Pint was not significantly different (Fig. 4, C and D). The qRT-PCR data thus confirmed the microarray results and the tensor PLSR model, showing an early-phase suppression of TNF-induced cluster #9 genes by insulin (Fig. 3, B to E). Furthermore, these data indicated that the TNF-insulin crosstalk cannot be predicted by adding the effect of insulin to the TNF response.
Fig. 4. Multipronged bioinformatics of TNF-insulin crosstalk suggests posttranslational regulation from GSK3 to GATA6.
(A to D) qRT-PCR validation of selected cluster #9 transcripts upon pretreatment of HT-29 cells with IFNγ and stimulation with TNF with or without insulin for 2 hours (A and B) or 6 hours (C and D)). Data are geometric means ± log-transformed SEM of n = 4 or 16 biological replicates. Full cluster #9 data are shown in fig. S5. (E) Promoter bioinformatics (86, 92, 93) suggest GATA and TCF4 as candidate regulators of TNF-insulin crosstalk. (F) Relative copy number estimates (96, 97) for the six GATA isoforms in HT-29 cells. Data are medians ± range of n = 3 biological replicates. n.d., not detected. (G to I) Transcriptional dynamics of GATA isoforms in response to TNF and insulin. Data are geometric means ± log-transformed SEM of n = 4 or 8 biological replicates. (J) Scansite (98) identification of candidate GSK3 phosphorylation sites (red). Each site’s percentile rank is averaged across the indicated sequences. (K) Phospho-mass spectrometry identifies 11 phosphorylation sites on GATA6L. Previously unreported sites (New) are shown below the primary sequence, those consistent with reports in the literature (104) (Reported) are shown above, and reported sites not detected in this study are gray. Start methionines (arrows) for the long and short forms are indicated along with the conserved GATA core and zinc finger (ZnF) domains.
To identify candidate mediators of TNF-insulin crosstalk, we analyzed the expression-verified transcripts of cluster #9 with three orthogonal promoter -analysis algorithms (86, 92, 93). Only two transcription factors were suggested as candidate regulators by all three algorithms: T cell factor 4 (TCF4) and GATA (Fig. 4E). HT-29 cells harbor a truncating mutation in APC (adenomatous polyposis coli), a protein that inhibits the β-catenin pathway, and this truncation renders β-catenin and its transcriptional partner TCF4 constitutively active (94). Moreover, no changes in β-catenin localization were observed upon TNF simulation with or without insulin (fig. S6). Therefore, we focused on GATA, a family of six transcription factors that are important for development and differentiation (95). Using qRT-PCR (96, 97), we quantified the relative copy numbers of the GATA family and found that GATA6 was the most abundant isoform (Fig. 4F and fig. S7). Copies of GATA6 transcript also remained high during the early phase of TNF-only and TNF + insulin stimulation, whereas GATA2 and GATA3 were reduced two- to fourfold (Fig. 4, G to I). Notably, bioinformatic analysis (98) of the full-length GATA6 protein sequence uncovered a cluster of highly conserved serine residues that were candidate phosphorylation sites for GSK3 (Fig. 4J). Because GSK3 is a recognized substrate of Akt (81), these results suggested that GATA6 could be an insulin-dependent regulator of the crosstalk cluster.
Full-length GATA6 (GATA6L) is distinguished by a long N-terminal extension that is conserved across vertebrates but missing in other GATA family members (fig. S8). In addition, leaky ribosome scanning (99) onto an in-frame methionine (Met147) gives rise to a short form of GATA6 (GATA6S) that is comparable in size (~45 kD) to the other GATA isoforms. GATA6S lacks the candidate GSK3 phosphorylation sites that reside at the N terminus of GATA6L (Fig. 4J), raising the possibility of selective regulation of GATA6L by insulin.
Compared to GATA6S, GATA6L has been understudied because of early misannotations of nonhuman genomes and a lack of suitable reagents. Common plasmid repositories, including Addgene (100), the human ORFeome (101), and the Mammalian Gene Collection (102), have only the short form or lack the gene entirely. The unusually slow electrophoretic mobility of GATA6 has created additional confusion because commercial antibody vendors mistakenly label GATA6S as “GATA6” implying that the detected protein is the full-length form. As a result, the GATA6 literature is incredibly ambiguous, with many papers inadvertently focusing on GATA6S.
To determine whether the predicted GSK3 phosphorylation sites of GATA6L could be phosphorylated in HT-29 cells, we cloned the full-length gene with N-terminal FLAG and C-terminal AU1 tags into a doxycycline inducible lentivector (see Materials and Methods) (103). Stable HT-29 transductants were induced with doxycycline for 24 hours before lysis, and FLAG immunoprecipitates were subjected to phosphorylation analysis by mass spectrometry. We achieved 92% coverage of the GATA6L sequence and identified 11 phosphorylation sites, including 7 that had not been previously reported (Fig. 4K) (104). Among sites in the N-terminal extension specific to GATA6L, two (Ser33 and Ser37) were consistent with the bioinformatic predictions of GSK3 phosphorylation (Fig. 4J). Two N-terminal sites (Thr34 and Ser37) were also corroborated by a proteomics study of proline-directed phosphorylation in 293T cells (105). We concluded that Ser33, Thr34, and Ser37 were the leading candidates for GATA6L phosphorylation–mediated regulation by GSK3.
Perturbation of basophilic kinases differentially affects GATA6L and GATA6S
Studies of GATA6 phosphorylation have largely focused on its posttranslational regulation by MAPKs (106–108). However, GATA6 is reportedly phosphorylated on Ser436 (Ser290 in GATA6S) and stabilized upon prolonged mechanistic target of rapamycin complex 1 (mTORC1) inhibition with rapamycin and subsequent feedback activation of Akt2 in vascular smooth muscle cells (VSMCs) (109). Our mass spectrometry data on GATA6L did not include peptides containing Ser436; thus, we could not rule out a role for Akt as a GATA6 kinase activated by insulin stimulation.
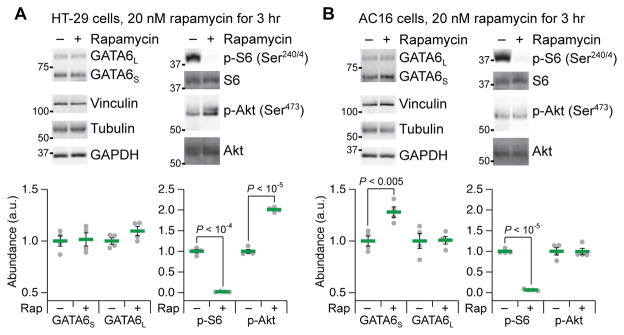
To determine whether Akt-catalyzed stabilization of GATA6 was relevant to our study, we treated HT-29 cells with the mTORC1 inhibitor rapamycin for 3 hours and monitored targets by quantitative immunoblotting (51). As expected, rapamycin eliminated phosphorylation of ribosomal protein S6 and increased phosphorylation of Akt by about twofold; however, the abundances of GATA6S and GATA6L were essentially unchanged (Fig. 5A). In the context of IFNγ pretreatment and TNF-insulin stimulation, we found that rapamycin treatment for 3 hours altered the distribution of GATA6 forms by decreasing the abundance of GATA6L relative to the abundance of GATA6S (fig. S9). The mechanism reported in VSMCs (109) might be restricted to mesodermal tissues, so we repeated the experiment in AC16 ventricular cardiomyocytes (110). In these cells, S6 phosphorylation disappeared without subsequent feedback activation of Akt, and yet, GATA6S abundance increased modestly after 3 hours as reported in VSMCs (Fig. 5B) (109). Critically, under the same conditions in AC16 cells, we did not observe any alterations in the abundance of GATA6L. These experiments illustrated that Akt feedback activation could be uncoupled from GATA6 stabilization, as could the posttranslational regulation of its long and short forms.
Fig. 5. GATA6L abundance is not altered by prolonged rapamycin treatment or feedback phosphorylation of Akt.
(A) HT-29 cells exhibit rapamycin-induced feedback phosphorylation of Akt, but do not stabilize GATA6. a.u., arbitrary units. (B) AC16 cells exhibit stabilization of GATA6S, but not GATA6L, without rapamycin-induced feedback phosphorylation of Akt. Vinculin,, tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as loading controls (51). Quantitative immunoblot data are means ± SEM of n = 4 biological replicates across two separate experiments.
GSK3-dependent phosphorylation of Ser37 accelerates GATA6L turnover
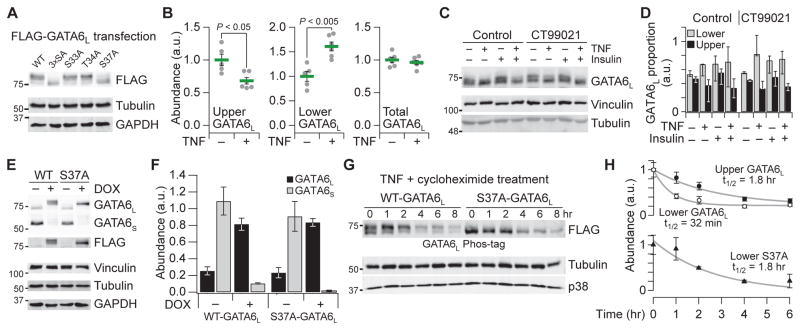
To assess the importance of the GATA6L phosphorylation sites, we transfected single alanine mutants of Ser33, Thr34, and Ser37 or the triple mutant (3×SA) into 293T cells, cells in which GATA6L phosphorylation has been detected previously (105). Compared to the wild-type allele, we noted a pronounced electrophoretic downshift in the major FLAG-immunoreactive band of the Ser37 and 3×SA mutants (Fig. 6A). The mobility shift was larger than that expected for a single phosphorylation site, suggesting that Ser37 phosphorylation was required for other phosphorylation events within GATA6L. Iterative phosphorylation-dependent phosphorylation is characteristic of many GSK3 substrates, such as glycogen synthase (GS) (111).
Fig. 6. Extensive phosphorylation of GATA6L is blocked by S37A mutation, reversed by TNF stimulation, and stabilized in HT-29 cells.
(A) Electrophoretic mobility of FLAG-tagged GATA6L is downshifted upon S37A mutation in lipofected 293T cells. (B) Phos-tag electrophoresis (112) reveals that TNF stimulation for 1 hour causes the dephosphorylation of GATA6L. (C and D) Phos-tag electrophoresis (C) and quantification (D) of the upper and lower forms of GATA6L in response to IFNγ sensitization for 24 hours, pretreatment with 1 μM CT99021 for 1 hour, and stimulation with TNF or insulin for one1 hour. Data are median proportion ± range of n = 3 biological replicates. (E and F) Doxycycline (DOX)–inducible addback in HT-29 cells replaces endogenous GATA6S with epitope-tagged GATA6L. Cells were treated with doxycycline (1 μg/ml) for 48 hours. (G and H) The less phosphorylated form of wild-type (WT) GATA6L is unstable. Cells were treated with TNF (100 ng/ml) + 50 μM cycloheximide for the indicated times, and half-lives were estimated by nonlinear least-squares curve fitting. Quantitative immunoblot data are means ± SEM of n = 3 (F and H) or 5 to 6 (B) biological replicates.
Careful inspection of endogenous GATA6L immunoreactivity in HT-29 extracts revealed a slower migrating species that was similar to the electrophoretic shifts observed in 293T cells transfected with the phosphorylation-deficient mutants. We isolated this species from the faster migrating GATA6L through Phos-tag electrophoresis (112) followed by Gaussian mixture modeling of the densitometric traces (see Materials and Methods). Acute TNF treatment reduced the upper form of GATA6L but with a concomitant increase in the lower form, such that total GATA6L abundance was not altered (Fig. 6B). Costimulation with insulin or pretreatment with the GSK3 inhibitor CT99021 (113) did not alter the GATA6L downshift, despite insulin increasing GSK3 phosphorylation and CT99021 decreasing GSK3 activity (Fig. 6, C and D, and fig. S10). Because GATA6 mRNA was not induced by TNF (Fig. 4I), these results indicated that GATA6L is dephosphorylated on some residues in response to TNF stimulation.
Our next goal was to evaluate the specific impact of Ser37 phosphorylation on GATA6L in HT-29 cells. One challenge was that the endogenous abundance of GATA6S was high compared to GATA6L (Fig. 5A), which could confound interpretations of ectopically expressed proteins. Therefore, we inducibly knocked down endogenous GATA6 with short hairpin RNA and added back epitope-tagged versions of wild-type or S37A GATA6L so that the abundance was comparable to total endogenous GATA6 (Fig. 6, E and F; see Materials and Methods). Doxycycline-induced addback in HT-29 cells recapitulated the electrophoretic mobilities of GATA6L that were observed in transfected 293T cells. The data suggested that the GATA6L modifications are not artifacts of overexpression, enabling use of the addback cells to examine the consequences of Ser37 phosphorylation.
Because GSK3 phosphorylation often accelerates substrate turnover (114), we combined the Phos- tag analysis with the HT-29 addback lines to estimate half-lives of wild-type and S37A GATA6L. We combined inhibition of protein synthesis with TNF stimulation to enrich for the lower migrating form of wild-type GATA6L in the addback cells. Under these conditions, we found that the half-life of more phosphorylated GATA6L was more than twice that of the wild-type GATA6L form that was less phosphorylated (Fig. 6, G and H). Surprisingly, the half-life of the S37A mutant was comparable to that of the more phosphorylated form of GATA6L, suggesting that phosphorylation of Ser37 without subsequent additional phosphorylation renders GATA6L unstable. Ser37 resides in the middle of a proline-glutamate-serine-threonine (PEST) degradation motif of GATA6L, and this motif scores more strongly as a PEST motif than those in other well-known unstable proteins (Fig. 4J and table S1) (115, 116). Ser37 phosphorylation might activate or expose the PEST sequence for rapid proteolytic degradation of GATA6L, whereas additional phosphorylation at other sites could inhibit substrate recognition (115).
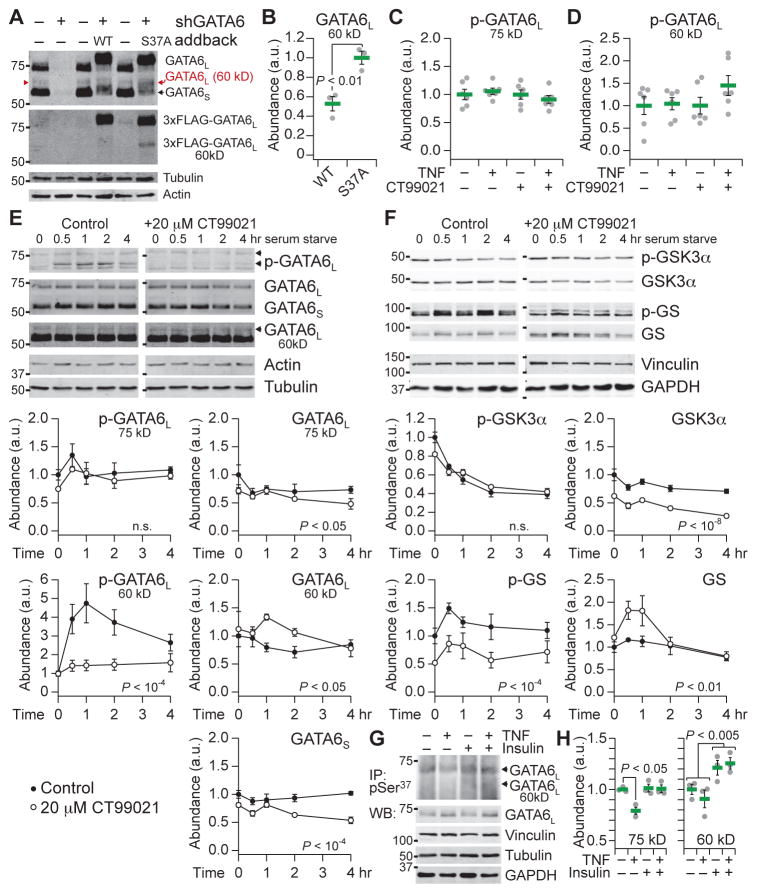
To monitor Ser37 phosphorylation specifically, we raised and affinity-purified a phospho-specific antibody against a monophosphorylated peptide fragment of the PEST sequence in GATA6L (see Materials and Methods; fig. S11). If Ser37 modification were a prerequisite for subsequent phosphorylation, then the antibody would capture this initial phosphorylation event of GATA6L, with the caveat that phosphorylation on Ser33 and Thr34 would ultimately disrupt the antibody epitope. To evaluate the phospho-Ser37 antibody, we reassessed the immunoreactivity of total GATA6. The predicted molecular weights of GATA6S and GATA6L are 45.4 and 60 kD, respectively. However, extensive posttranslational modifications (Fig. 4K) cause most GATA6S and GATA6L to run at an apparent molecular weight of ~54 kD and ~69 to 75 kD depending on electrophoresis conditions (Fig. 7A). Multiply phosphorylated GATA6S (~54 kD) can be misinterpreted as unmodified GATA6L (60 kD). Upon long exposure with a total GATA6 antibody, we revealed an additional immunoreactive band at ~60 kD that was eliminated by GATA6 knockdown and reconstituted with addback of wild-type GATA6L and the S37A mutant (Fig. 7A). In contrast to the 75-kD form (Fig. 6E), the ~60-kD form of wild-type GATA6L was significantly less abundant than the S37A mutant (Fig. 7B), consistent with decreased stability. We interpreted the ~60-kD band as the unmodified form of GATA6L.
Fig. 7. Phosphorylation and destabilization of endogenous GATA6L at 60 kD.
(A) Knockdown (shGATA6) and FLAG-tagged addback of GATA6L at ~60 kD (red). Samples were immunoblotted for total (modified and unmodified) GATA6 (upper), FLAG (lower), and the indicated loading controls. (B) Destabilization of the 60-kD form of WT GATA6L compared to the S37A-mutant addback cells. (C and D) Endogenous phospho-GATA6L (Ser37) immunoreactivity is not detectably affected by stimulation with TNF for 1 hour, inhibition with 20 μM CT99021 for 6 hours, or both. (E and F) Phosphorylation and destabilization of the 60-kD form of GATA6L upon serum starvation. Specificity was confirmed by preincubation of cells with 20 μM CT99021 for 1 hour before serum starvation. (G and H) Phospho-GATA6L (Ser37) immunoprecipitation and total GATA6 immunoblot of HT-29 cells pretreated with IFNγ and stimulated with TNF ± insulin for 1 hour. The gamma of the immunoprecipitation image is set to 1.5 to minimize background from the immunoprecipitating antibody heavy chain. Input (0.5% input%) of each immunoprecipitate was immunoblotted for total GATA6 and the indicated loading controls. See file S5 for details. Data are means ± SEM of n = 3 (B, E, F, and H) or 6 (C and D) biological replicates.
Endogenous Ser37 phosphorylation of the 75 and 60-kD GATA6L forms was not detectably altered in response to TNF treatment for 1 hour or CT99021 treatment for 6 hours (Fig. 7, C and D). However, the endogenous 60-kD phospho-GATA6L signal was barely above the detection limit and thus highly variable (coefficient of variation, ~40%), yielding only ~50% statistical power for detecting a 1.5-fold change. To evaluate phosphorylation of endogenous GATA6L on Ser37, we serum- starved the HT-29 cells to produce a stronger activation of GSK3 and confirmed specificity of the phosphorylation events with CT99021 pretreatment for 1 hour (Fig. 7, E and F). As expected, serum starvation reduced GSK3 phosphorylation (increasing GSK3 activity) and increased phosphorylation of GS (Fig. 7F). CT99021 reduced GS phosphorylation and total GSK3 abundance but also transiently increased total GS. Notably, within 1 hour of serum withdrawal, we observed a robust increase in the 60-kD form of phospho-GATA6L, which coincided with the timing of GSK3 dephosphorylation and was blocked by CT99021 in a dose-dependent manner (Fig. 7E and fig. S12). By contrast, phospho-Ser37 immunoreactivity of the 75-kD form of GATA6L was not altered by serum starvation or CT99021 treatment, suggesting that Ser37 was already stably phosphorylated in this form of GATA6L.
Total abundances of the different forms of GATA6L also showed dynamic changes. GATA6L at 75 kD and GATA6S decreased significantly with CT99021 treatment, whereas GATA6L at 60 kD increased compared to uninhibited control cells that were serum-starved (P < 0.05, two-way ANOVA). The time-dependent changes in 60-kD GATA6L abundance mirrored the changes in total GS and inversely correlated with changes in 60-kD GATA6L phosphorylation at Ser37 (Fig. 7, E and F). These experiments provide further evidence that Ser37 is a site phosphorylated by GSK3 and that phosphorylation at this site promotes turnover of GATA6L in the absence of phosphorylation at additional sites.
With greater confidence in the phospho-GATA6L (Ser37) antibody, we revisited the original biological context of IFNγ-pretreated HT-29 cells stimulated with TNF and insulin. To enable detection, we immunoprecipitated cell extracts with phospho-GATA6L (Ser37) antisera and immunoblotted for total GATA6 (Fig. 7G). An extended electrophoresis was required to separate the 60-kD form from the heavy chain of the immunoprecipitating antibody, causing a smear of immunoreactivity rather than a discrete band (see Materials and Methods). In response to TNF alone, we repeatedly observed a drop in 75-kD, but not 60-kD, GATA6L phosphorylation (Fig. 7H), corroborating the dephosphorylation previously noted by Phos-tag electrophoresis (Fig. 6B). Moreover, the reduction in 75-kD phospho-GATA6L (Ser37) was blocked by insulin costimulation, indicating a specific point of crosstalk between TNF and insulin. Insulin, by contrast, independently elevated the abundance of 60-kD GATA6L phosphorylation, suggesting that insulin delays the turnover of this form beyond its effect on Ser37 phosphorylation. We conclude that the phosphorylation of endogenous GATA6L at Ser37 is consistent with the antagonism and linear superposition in abundance of cluster #9 transcripts observed upon TNF + insulin stimulation (Fig. 4, A to D, and fig. S5).
GSK3-dependent phosphorylation of Ser37 alleviates GATA6L repression of transcripts in the crosstalk cluster
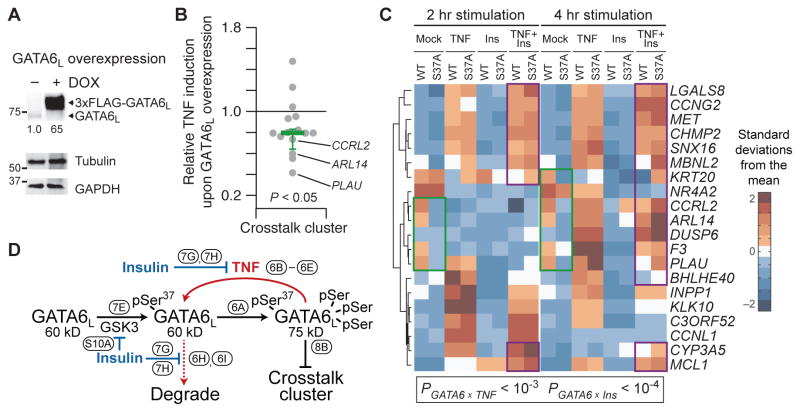
We investigated the role of GATA6L phosphorylation in the regulation of transcripts subjected to TNF-insulin crosstalk. We inducibly overexpressed wild-type GATA6L in HT-29 cells (Fig. 8A) before stimulation with TNF for 2 hours and transcriptomic profiling by microarray (see Materials and Methods). GATA6 can act as either a transcriptional activator or repressor (117), but we found that, in the GATA6L-overexpressing HT-29 cells, the effects of GATA6L were predominantly repressive. Without stimulation, GATA6L overexpression induced 51 transcripts and repressed 136 transcripts at a 5% false -discovery rate (P < 10−10, binomial test) (file S4). TNF stimulation increased the number of genes affected by GATA6L overexpression, but the bias toward repression persisted (317 induced transcripts versus 438 repressed transcripts; P < 10−5, binomial test) (file S4). Notably, the same repressive bias was observed for transcripts in the crosstalk cluster (Fig. 8B), and those with the strongest GATA6L-associated repression were among the clearest examples of TNF-insulin crosstalk (Fig. 4, A to C). Using chromatin immunoprecipitation (ChIP), we confirmed binding of GATA6L to consensus sites within the promoters of many TNF-insulin crosstalk genes (fig. S13), suggesting that repression is direct.
Fig. 8. S37A mutation of GATA6L mimics and competes with the repression of transcript abundance in the TNF-insulin crosstalk cluster.
(A) Doxycycline-inducible overexpression of WT GATA6L in HT-29 cells. Cells were treated with doxycycline (1 μg/ml) for 24 hours. (B) Ratio of TNF-induced transcript abundance for crosstalk cluster genes in the presence or absence of GATA6L overexpression. Data are mean ratios of n = 3 independent biological samples assessed by microarray profiling, with bias in the ratio assessed by two-sided binomial test. (C) S37A mutation of GATA6L mimics insulin stimulation (green) and antagonizes TNF-insulin crosstalk (purple). qRT-PCR data for cluster #9 genes in WT and S37A mutant (S37A) GATA6L addback cells pretreated with IFNγ and stimulated with TNF, insulin, or both for 2 or 4 hours. Data are row-standardized geometric means of n = 6 biological replicates across two separate experiments, with interactions between GATA6 status and TNF or insulin assessed by log-transformed five-way ANOVA with the following factors: GATA6, transcript, TNF, insulin, and time. (D) Three-state conceptual model for GATA6L regulation by TNF and insulin and its relation to the crosstalk cluster of transcripts. Ovals annotate the figure subpanels supporting the links depicted.
If GATA6L mediates the insulin-stimulated repression of the crosstalk cluster and is stabilized by inhibition of the GSK3 pathway, then phosphorylation of Ser37 would provide a mechanism for TNF-insulin crosstalk. Furthermore, the S37A mutant of GATA6L should mimic the effect of insulin on TNF-stimulated gene expression for transcripts in the crosstalk cluster and dampen the crosstalk observed when insulin is added to S37A mutant cells. We tested this prediction with the wild-type and S37A addback lines (Fig. 6, E and F), inducing GATA6L and then stimulating with TNF, insulin, or both. By qRT-PCR, we identified multiple instances in which S37A addback reduced transcript abundance similar to that observed in wild-type addback cells stimulated with insulin (Fig. 8C, green). We also found many examples in which TNF + insulin–induced transcript abundance was higher in S37A addback cells compared to wild-type addback cells (Fig. 8C, purple) and more similar to TNF-treated cells, suggesting reduced crosstalk. Analysis of the entire cluster #9 data set revealed significant interactions between GATA6 and TNF or insulin (interaction P < 10−3, five-way ANOVA), indicating that the Ser37 genotype alters the transcriptional response to both stimuli. Together, our data support a model whereby TNF promotes and insulin inhibits the formation and degradation of GATA6L monophosphorylated on Ser37 (Fig. 8D). This phosphoregulation is ultimately reflected by the abundance of transcripts in the crosstalk cluster.
Ser37 phosphorylation of different GATA6L forms is observed in diverse cell types
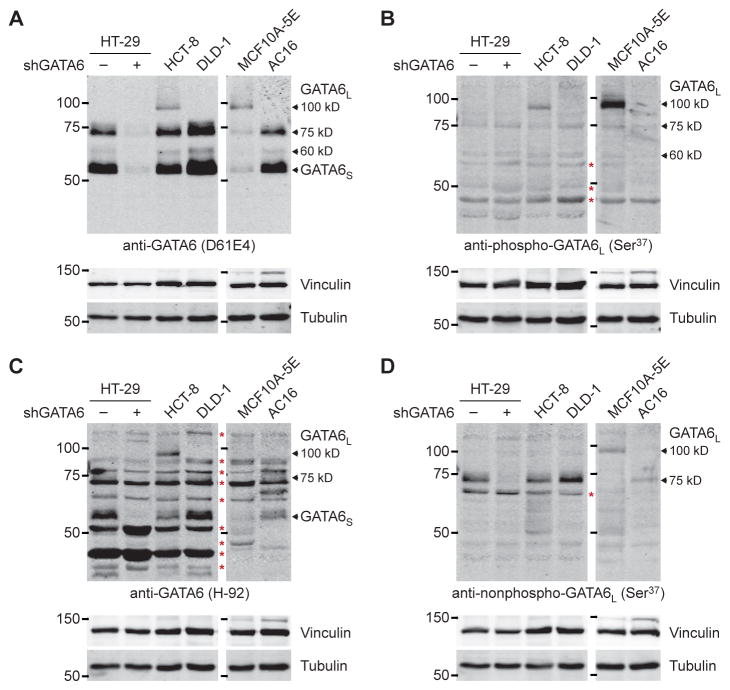
The model of GATA6L phosphorylation–mediated regulation (Fig. 8D) may be specific to HT-29 cells or could occur in other cell types. We immunoblotted various cell lines with the phospho-Ser37 antibody in comparison to affinity-purified antisera binding the nonphosphorylated peptide surrounding Ser37 (see Materials and Methods) and to other commercial GATA6 antibodies (100, 118). In HCT-8 and DLD-1 colorectal cancer lines, AC16 cardiomyocytes (110), and MCF10A-5E breast epithelial cells (119), we observed the 60 and 75-kD forms of GATA6L, which were phosphorylated to variable extents according to phospho-Ser37 immunoreactivity (Fig. 9, A and B). Multiple GATA6 antibodies also recognized another species at ~100 kD (Fig. 9, A to D), suggesting that an even more phosphorylated form of GATA6L may remain to be characterized. The aggregate number of reported phosphorylation sites on GATA6L now exceeds 20 (Fig. 4K).
Fig. 9. Diversity of GATA6L forms across different cell lineages.
(A to D) Arrows indicate the GATA6 forms confirmed earlier by knockdown or observed with multiple antibodies. Red asterisks indicate nonspecific bands. The MCF10A-5E and AC16 samples are on an immunoblot; HT-29, HCT-8, and DLD-1 are on another blot. The blots have been scaled to match. Data are representative of n ≥ 3 independent experiments.
DISCUSSION
Our study here introduces and implements tensor PLSR as an approach for structured biological data sets. Considering that signaling dynamics often occur in discrete temporal phases (49, 80, 120, 121), tensor PLSR provides an attractive means to deconstruct time -course data in a systematic manner. Although the mathematics has been established for decades (59), data types that can exploit the tensor framework are relatively new to cell signaling. For tensor generation, a multiplex technique that simply measures many genes or proteins is insufficient. The method must also be cost-effective, reproducible, and scalable for repeated use across multiple treatments, time points, and perturbations. The newest technologies rarely meet these criteria, prompting our use of long-established methods at a scale not typically considered.
We applied tensor PLSR with the goal of discovering molecular mechanisms that connect signaling to transcriptional regulation. Ideally, the mechanisms would involve proteins not originally included in the systematic data set. Systems-level studies rarely uncover these “hidden nodes” and validate them experimentally like we achieved here for GATA6L (25, 122). Testing model- or bioinformatics-derived predictions requires a skill set entirely different from the one needed to perform the analysis. Our findings argue for the benefits of dual training, where computationalists work at the bench and experimentalists use quantitative models, gaining an appreciation for the thematic similarities in each approach. For example, just as modeling assumptions should be subjected to falsification (31), we sought to challenge the prevailing biological assumptions about GATA6 and its different forms.
The deceptive electrophoretic mobilities of GATA6S and GATA6L have important implications for biological function. Although GATA6L is generally less abundant than GATA6S in most cell types, there is evidence that GATA6L is the more potent transcriptional regulator (99). GATA6 promotes the expression of the stem cell marker LGR5 in colorectal cancer (118, 123). Neither paper clarified whether the regulation occurs through GATA6L, GATA6S, or both. However, insulin-like growth factor inhibits GSK3 and promotes expansion of Lgr5+ stem cells in mice (81, 124). Our results indicate that one mechanism for this expansion is the stabilization of GATA6L.
The phosphorylation of Ser37 adds a GATA6L-specific mode of regulation to reports of posttranslational modifications that would presumably target both long and short forms (107–109). Although we were unable to reproduce the mechanism exactly (109), modification of Ser436 by Akt2 should coincide with the loss of Ser37 phosphorylation to stabilize GATA6L synergistically in contexts where both pathways operate. GATA6L phosphorylation–mediated regulation could prove important in endothelial cells, a TNF- and insulin-responsive cell type in which GSK3 and GATA6 interact as a complex (125). Our mass spectrometry study also uncovered other GATA6L-specific proline-directed modification sites (Thr62 and Ser137) that could function with the Ser266 site phosphorylated by extracellular signal–regulated kinase (ERK) (107, 108). Such complex layers of regulation should be expected of a transcription factor that is central to embryonic development and cell specification (95).
By coupling systematic experiments with statistical modeling approaches, such as tensor PLSR, one can identify relationships that would otherwise go unnoticed. Although it remains atypical to collect transcriptomic data as tensors, we expect widespread systematization of transcriptomics as expression -profiling costs drop. A model is just the first step, however, because the most surprising data-derived connections will require the identification of previously unrecognized mechanisms to explain them. These, in turn, require hypothesis-driven experiments with the best molecular -genetic and pharmacologic perturbations available. For understanding how gene expression is controlled by complex stimuli, the integration of molecular biology and systems biology has yet to be fully exploited.
MATERIALS AND METHODS
Cell culture
HT-29, 293T, DLD-1, and HCT-8 cells were obtained from the American Type Culture Collection (ATCC) and cultured according to their recommendations. The 5E clone of MCF10A cells was cultured as described previously (119). AC16 cells (110) were purchased from M. Davidson (Columbia University) and cultured in Dulbecco’s modified Eagle’s medium/F-12 medium (Life Technologies) with 12.5% tetracycline-free fetal bovine serum (Clontech) and penicillin-streptomycin (Gibco).
Cell stimulation
HT-29 cells were plated at 50,000 cells/cm2 for 24 hours, sensitized with human IFNγ (200 U/ml) for 24 hours (Roche), and then treated with TNF (100 ng/ml; PeproTech), insulin (500 ng/ml; Sigma), or both for the indicated times. HT-29 cells engineered to express GATA6L inducibly were treated with doxycycline (1 μg/ml) for 24 hours (overexpression) or 48 hours (addback) before cytokine stimulation.
Plasmids
Wild-type GATA6L was amplified by PCR from HT-29 RNA that had been reverse-transcribed with a GATA6-specific primer (CAAAAGCAGACACGAGTGGA). An N-terminal 3×FLAG tag and a C-terminal 3×AU1 tag were added by PCR before cloning into the Bam HI and Sal I sites of pBabe puro (126) or the Mfe I and Spe I sites of pEN_TTmiRc2 (103). The pEN_TT donor vector containing GATA6L was then recombined with the pSLIK Neo destination vector (103) by using LR Clonase (Invitrogen). The shGATA6 sequence (CCCAGACCACTTGCTATGAAA; #TRCN0000005390 from the RNAi Consortium) was cloned into tet-pLKO-puro (127) as described previously (97). S33A, T34A, S37A, and 3×SA point mutants were prepared by site-directed mutagenesis (QuikChange II XL, Agilent). RNA interference–resistant mutants of wild-type and S37A GATA6L were prepared by introducing four silent mutations into the sequence targeted by shGATA6, which were replaced with rare mammalian codons that would minimize ectopic expression. The phosphorylation- and degradation-resistant IκBα super-repressor plasmid has been previously described (128). All DNA constructs were verified by sequencing.
Production and purification of phospho-GATA6L (Ser37) antibody
The peptide sequence Ac-CREPSTPPpSPIS-amide was conjugated to keyhole limpet hemocyanin and used to immunize rabbits according to the manufacturer’s recommendations (Covance). Serum samples were tested by immunoblotting with positive and negative controls for phospho-GATA6L (Ser37). Serum pooled from the production and terminal bleeds was negatively selected on a CREPSTPPpSPIS peptide–conjugated N-hydroxysuccinimide (NHS)–Sepharose column. The bound immunoglobulin G (IgG) was eluted as the non–phospho-GATA6L custom antibody while the flow through was exposed to a second CREPSTPPpSPIS peptide–conjugated NHS Sepharose column. The bound IgG was eluted as the phospho-GATA6L (Ser37) antibody and used for detection by immunoblotting.
Lentiviral packaging and transduction
Lentiviruses were prepared in human embryonic kidney (HEK) 293T cells (ATCC) by calcium phosphate transfection of the lentivector together with psPAX2 and pMD.2G (Addgene). Lentiviral transduction of HT-29 cells was performed as described previously (96). Transduced cells were selected in growth medium containing 2 puromycin (2 μg/ml) or G418 (600 μg/ml) until control plates had cleared. For addback experiments, viral titers were reduced to ensure single-virion transductants that matched the endogenous protein abundance as closely as possible.
Microarray profiling
HT-29 cells were plated at 50,000 cells/cm2 for 24 hours and sensitized with IFNγ (200 U/ml; Roche) for 24 hours before stimulation with TNF (0, 5, or 100 ng/ml), EGF (0, 1, or 100 ng/ml), and insulin (0, 5, or 500 ng/ml) for 4, 8, or 16 hours. RNA isolation was performed with the RNeasy Mini Kit (Qiagen), and integrity of purified RNA was confirmed on a Bioanalyzer (Agilent). Preparation of labeled complementary RNA, hybridization to GeneChip Human Genome U133A Arrays (Affymetrix), microarray scanning, and microarray processing were performed as previously described (129).
For inducible GATA6L overexpression, stably transduced HT-29 cells were plated at 50,000 cells/cm2 for 24 hours, induced with doxycycline (1 μg/ml), and sensitized with IFNγ (200 U/ml Roche) for 24 hours before stimulation with TNF (100 ng/ml) for 2 hours. RNA was purified as described above and amplified with the Illumina TotalPrep-96 RNA Amplification Kit (Life Technologies) before hybridization to a HumanHT-12 v4 Expression BeadChip.
Hierarchical and CLICK clustering
One-way hierarchical clustering of the signaling and transcriptomic compendia was performed in MATLAB with the clustergram function using Euclidean distance and Ward’s linkage after row standardization. CLICK clustering was performed as described (78) with the default homogeneity parameter.
Tensor PLSR
Tensor PLSR was performed in MATLAB with version 2.02 of the NPLS Toolbox (130). The signaling compendium was structured by stimulus condition (mode 1), time point (mode 2), and measured signal (mode 3). The transcriptomic profiles were structured by stimulus condition (mode 1), time point (mode 2), and CLICK gene cluster (mode 3). Both data tensors were mean-centered along mode 1 and variance-scaled along modes 2 and 3 before calculation of latent variables (131). The scores and time weights of LV#4 of the signaling tensor were both multiplied by –1 to improve model interpretability. Randomized models were constructed in MATLAB with the shuffle matrix function applied within each stimulus condition before preprocessing and calculation of latent variables. The code for the tensor PLSR models is included in file S3.
Bioinformatic analyses of crosstalk cluster
The 20 transcripts from cluster #9 confirmed present by qRT-PCR were submitted to three promoter analysis algorithms. First, the proximal promoter of each transcript [defined as 2000 base pairs (bp) upstream and 500 bp downstream of the transcription start site] was collected from National Center for Biotechnology Information (NCBI) and used as an input set for MEME, which uses expectation maximization to define recurrent motifs in a set of sequences (132). The top five enriched motifs were searched against a database of 843 binding specificities (133) using TOMTOM (134) to identify known transcription factor recognition sequences. A GATA motif was also enriched when using 2000 bp of upstream sequence alone or 1500 bp of upstream sequence and 500 bp of downstream sequence.
Expression-verified cluster #9 transcripts were additionally analyzed with DiRE (86), which uses interspecies sequence conservation to define motifs that are searched against the TRANSFAC 10.2 database of roughly 400 transcription factor binding motifs. In DiRE, the occurrence metric reflects the overall frequency of a conserved binding motif in the input data set, whereas the importance metric reflects the specificity of the binding motif to the input data set compared to a background data set of 5000 randomly selected genes. The top 20 motifs based on occurrence were used as the DiRE predictions.
Last, X2K (92) was used to identify bioinformatic connections between cluster #9 transcripts and signaling pathways. X2K integrates the ChEA database (135) of transcription factor binding sites detected by ChIP, the JASPAR and TRANSFAC position weight matrices, as well as various protein-protein interaction and kinase-substrate databases to connect kinase signaling events to gene expression patterns. The top 20 transcription factors linked to signaling and cluster #9 transcripts in a 2011 analysis were used as the X2K predictions.
Quantitative RT-PCR
RNA from cultured cells was isolated with the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol. First-strand complementary DNA synthesis and qRT-PCR were performed as described (63). Parental HT-29 samples were normalized to the geometric mean of GAPDH, HINT1, PPIA, and PRDX6. GATA6L addback samples were normalized to the geometric mean of GAPDH, HINT1, PPIA, PRDX6, B2M, and GUSB. Primer sequences are available in table S2.
Mass spectrometry
HT-29 cells stably expressing doxycycline-inducible 3×FLAG-GATA6L were induced with 1 doxycycline (1 μg/ml) for 24 hours and lysed in NP-40 lysis buffer plus protease and phosphatase inhibitors (51). Protein extract (60 mg) in 6-ml volume was first cleared with 50 μl of mouse IgG–agarose beads (Sigma) for 1 hour at 4°C on a nutator. The cleared lysates were subjected to immunopurification using 80 μl of anti-FLAG M2 affinity gel (Sigma) for 3 to 4 hours followed by two washes with NP-40 lysis buffer, one wash with 500 mM NaCl, and one wash with tris-buffered saline. Immunoprecipitates were eluted with 3×FLAG peptide (500 ng/ml; Sigma) in 100 μl for 30 min at 4°C on a nutator. The eluate was concentrated using an Amicon ultra centrifugal filter (Millipore), and samples were prepared in dithiothreitol-containing Laemmli sample buffer and separated by SDS polyacrylamide gel electrophoresis on an 8% polyacrylamide gel followed by Coomassie brilliant blue staining. The stained bands were cut and subsequently reduced, alkylated, and digested with trypsin, chymotrypsin, or pepsin. Peptides from each enzymatic digestion were acrylamide-extracted and subjected to liquid chromatography–mass spectrometry on a Thermo Electron Orbitrap Velos ETD mass spectrometer system. The data were analyzed using the Sequest search algorithm against the International Protein Index human proteome database and the predicted GATA6 protein sequence. Full mass spectrometry details are available in texts S1 and S2.
Immunoblotting
Quantitative immunoblotting was performed as described previously in detail (51) with primary antibodies recognizing the following proteins or epitopes: phospho-GATA6L (Ser37, Covance, 1:1000 for crude antiserum and 1:500 after affinity purification), non–phospho-GATA6L (Ser37, Covance, 1:500), GATA6 (D61E4, Cell Signaling Technology #5851, 1:2000), GATA6 (H-92, Santa Cruz Biotechnology #9055, 1:600), phospho-GS (Ser641, Cell Signaling Technology #3891, 1:1000), GS (Cell Signaling Technology #3893, 1:1000), GSK3α (Cell Signaling Technology #9338, 1:1000), phospho-GSK3α (Ser21, Cell Signaling Technology #9316, 1:1000), phospho-Akt (Ser473, Cell Signaling Technology #4060, 1:1000), Akt (Cell Signaling Technology #9272, 1:1000), phospho-S6 (Ser240/244, Cell Signaling Technology #5364, 1:1000), S6 (54D2, Cell Signaling Technology #2317, 1:1000), FLAG (M2, Sigma #F1804, 1:10,000), β-actin (Ambion #4302, 1:5000), vinculin (Millipore #05-386, 1:10,000), GAPDH (Ambion #4300, 1:20,000), tubulin (Abcam #89984, 1:20,000), and p38 (C-20, Santa Cruz Biotechnology #535, 1:5000). Membrane blocking, antibody probing, and near-infrared fluorescence detection were performed as described (51), except for phospho-GATA6 (Ser37) immunoblotting, where blocking with 5% nonfat skim milk and use of tris-buffered saline buffers were required.
Phos-tag immunoblotting was performed on a 6% polyacrylamide gel containing 10 μM Phos-tag acrylamide AAL-107 (Wako Chemicals) and 0.1 μM MnCl2. Gels were run with WIDE-VIEW prestained protein markers under constant current (40 mA) for 170 min. Before electrophoretic transfer, gels were incubated with 1 mM EDTA in modified Towbin’s transfer buffer (51) for 15 min. Membrane blocking, antibody probing, and near-infrared fluorescence detection were then performed as described (51).
For Gaussian mixture modeling of GATA6L forms, raw 16-bit pixel intensities were integrated horizontally across each lane and then plotted along the vertical dimension. Using the fit function in MATLAB, the vertical trace was fit by nonlinear least-squares to the following function:
where f(x) is height of the vertical trace as a function of the vertical position x, b is a fixed background, w1 and w2 are the relative weights of the two bands, μ1 and μ2 are the mean vertical positions of the two bands, and σ2 is a shared variance for the two bands. Normalized versions of w1 and w2 were taken as the relative band densities for the two forms.
Phospho-GATA6L (Ser37) immunoprecipitation
HT-29 cells were plated at 75,000 cells/cm2, pretreated with human IFNγ for 24 hours (Roche), and then treated with TNF (100 ng/ml; PeproTech), insulin (500 ng/ml; Sigma), or both for 1 hour. Cells were lysed in NP-40 lysis buffer (51) supplemented with 10 mM sodium pyrophosphate and 30 mM sodium fluoride, and ~4 mg of cellular extract (adjusted according to total GATA6L abundance based on immunoblotting) was incubated with 10 μl of phospho-GATA6L (Ser37) antiserum overnight on a nutator at 4°C. The following day, 30 μl of Protein A/G Plus UltraLink resin (Thermo) was added to the immune complexes for 1 hour on a nutator at 4°C. Beads were washed twice with ice-cold supplemented NP-40 lysis buffer and twice with ice-cold phosphate-buffered saline (PBS) before elution in Laemmli sample buffer (136). Samples were electrophoresed on a 10% polyacrylamide gel for 3 to 5 hours (until the 50-kD marker reached the bottom of the gel) before proceeding with quantitative immunoblotting as described (51). Densitometry was performed by integrating the pixel intensity (without lane-specific background subtraction) of the 75-kD band and, for the 60-kD form, the intensity between the 75-kD band and the upper shoulder of the immunoprecipitating antibody heavy chain (file S5).
Chromatin immunoprecipitation
Five million wild-type and S37A GATA6L-addback HT-29 cells were seeded in 10-cm culture plates for 24 hours before inducing knockdown-addback with doxycycline (1 μg/ml) for 48 hours. Cells were fixed for 7 to 10 min by adding a 37% formaldehyde stock to the culture medium to a final concentration of 1%. Fixation was quenched with 1/20 volume of 2.5 M glycine for 7 to 10 min at room temperature. Cells were washed twice with cold PBS, scraped into 1 ml of PBS, and centrifuged at 400 relative centrifugal force (rcf) for 3 min. The cell pellets from four 10-cm plates were combined and lysed in ChIP lysis buffer (96) to a final volume of 1.5 ml. Lysates were incubated on ice for 10 min and then sonicated using a Branson digital sonifier for 5 min at 40% amplitude with 0.7 s “on” and 1.3 s “off” pulse cycles. After centrifugation at 14,000 rcf for 20 min, the supernatant was collected, and 20 μl of the soluble chromatin was retained as the input fraction. Soluble chromatin was diluted 10-fold in dilution buffer (96), precleared with 100 μl of mouse IgG–conjugated agarose beads (Sigma) for 4 hours at 4°C with constant agitation, and then incubated with 100 μl of anti-FLAG M2 affinity gel (Sigma) or mouse IgG–conjugated agarose beads overnight at 4°C with constant agitation. Agarose beads were collected and washed as previously described (96). DNA from the beads and the input fraction was eluted by reversing methylene cross-links with 500 μl of elution buffer (96) at 65°C for 5 hours. Samples were then treated with ribonuclease (100 μg/ml) for 30 min at 37°C and proteinase K (200 μg/ml) for 90 min at 50°C, followed by phenol-chloroform extraction. The aqueous fraction was ethanol-precipitated, washed once in 70% ethanol, air-dried, and dissolved in nuclease-free water. The samples were diluted 10-fold in nuclease-free water and quantified by PCR with primers designed for proximal promoter regions of selected crosstalk genes (123). Primer sequences are available in table S3.
Statistical analysis
Microarray data were analyzed by four-way ANOVA (factors: TNF, EGF, insulin, and time) in MATLAB with the anovan function at a false discovery rate of 5% (file S1). qRT-PCR data of cluster #9 genes were analyzed by four-way ANOVA (factors: transcript, TNF, insulin, and time) or, for GATA6L addback, by five-way ANOVA (factors: GATA6L genotype, transcript, TNF, insulin, and time) in MATLAB with the anovan function after log transformation. Half-lives of GATA6L forms were estimated by nonlinear least-squares curve fitting to the following function:
where g(t) is the relative band intensity as a function of time t, c is the scaling coefficient, b is a fixed background, and τ1/2 is the half-life. Differences in means were assessed by Welch’s t test, and differences in geometric means were assessed by Welch’s t test after log transformation. One- or two-sidedness was based on previous evidence or expectation for a directional change. Tests for enrichment were performed by binomial test. Differences between immunoblotting time courses were assessed by two-way ANOVA.
Supplementary Material
Fig. S1. Tensor PLSR modeling predicts overall transcript abundance but cannot link changes in transcript abundance to cytokine-induced signaling.
Fig. S2. Accuracy of tensor PLSR predictions.
Fig. S3. Induction of cluster #9 probe sets and repression by insulin.
Fig. S4. Disruption of NF-κB signaling does not widely affect the TNF-induced transcriptional response of cluster #9.
Fig. S5. Widespread TNF-insulin crosstalk among genes in transcriptional cluster #9.
Fig. S6. Immunolocalization of β-catenin is not altered by TNF stimulation or insulin costimulation in HT-29 cells.
Fig. S7. HT-29 cells lack GATA1, GATA4, and GATA5.
Fig. S8. Phylogeny of the human GATA family.
Fig. S9. Prolonged rapamycin treatment alters the proportion of GATA6S to GATA6L independently of TNF or insulin treatment.
Fig. S10. Insulin and CT99021 perturb GSK3 phosphorylation and activity.
Fig. S11. Phospho-GATA6L (Ser37) antiserum is specific for mobility-shifted wild-type GATA6L but not the S37A GATA6L mutant.
Fig. S12. GATA6L phosphorylation on Ser37 and GS phosphorylation on Ser641 are lost in a dose-dependent manner upon treatment with the GSK3 inhibitor, CT99021.
Fig. S13. GATA6L occupies GATA binding sites in the promoters of genes within the crosstalk cluster.
Table S1. Top-scoring PEST sequences in the indicated proteins according to PEST-FIND.
Table S2. qRT-PCR primer sequences.
Table S3. ChIP primer sequences.
File S1. Microarray probe sets differentially altered with time or by stimulation with TNF, EGF, or insulin.
File S2. Transcriptional clusters identified by CLICK.
File S3. Tensor PLSR model and associated files.
File S4. Transcripts differentially altered by wild-type GATA6L overexpression with or without TNF stimulation.
File S5. Raw files and densitometric analysis of phospho-GATA6L (Ser37) immunoprecipitation and total GATA6 immunoblots.
Text S1. Detailed description of tensor PLSR.
Text S2. Detailed description of GATA6L mass spectrometry.
Acknowledgments
We thank M. Luo of the BioMicro Center and J. Davis of the Center for Public Health Genomics for assistance with the microarray experiments, E. Jeffrey and N. Sherman of the W.M. Keck Biomedical Mass Spectrometry Laboratory for assistance with the mass spectrometry experiments, the laboratory of M. Adli for support with the ChIP experiments, M. Mayo for the plasmids, C. Borgman for copyediting, and I. Schulman, D. Brautigan, and T. Harris for comments on the manuscript.
Funding: This work was supported by the NIH (#1-DP2-OD006464), the Pew Charitable Trusts (#2008-000410-006), and The David and Lucile Packard Foundation (#2009-34710). The W.M. Keck Biomedical Mass Spectrometry Laboratory and The University of Virginia Biomedical Research Facility are funded by a grant from the University of Virginia’s School of Medicine.
Footnotes
Competing interests: K.A.J. is a paid consultant for Cell Signaling Technology and LI-COR Biosciences.
Data and materials availability: Microarray data are available through the NCBI Gene Expression Omnibus (GSE69639 and GSE72079).
Author contributions: Z.C. performed the qRT-PCR experiments and the promoter bioinformatics analyses, prepared the mass spectrometry samples, cloned and tested the GATA6 mutants, performed the Phos-tag and standard immunoblotting as well as immunoprecipitation, engineered the GATA6 addback lines, prepared the Illumina microarray experiments, and performed the ChIP-qPCR experiments. Z.Y. performed the GATA copy number analysis and assisted with the ChIP-qPCR experiments. Z.S. built the Gaussian mixture model of immunoblot data and analyzed the immunoblots relating to GATA6 half-life. S.L. assisted with cloning of GATA6L and testing shGATA6 hairpins. R.C.F. processed the Affymetrix microarray data and performed the CLICK clustering analysis. D.A.L. supervised the tensor PLSR model construction and supported the Affymetrix microarray study. K.A.J. prepared the Affymetrix microarray samples, built and analyzed the tensor PLSR model, performed the rapamycin, serum starvation, and immunofluorescence experiments, and was responsible for all statistical analysis. K.A.J. and Z.C. wrote the manuscript with input from R.C.F. and D.A.L.
REFERENCES AND NOTES
- 1.Ideker T, Galitski T, Hood L. A new approach to decoding life: Systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 3.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLOS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SY, Ferrell JE., Jr Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 6.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.English JG, Shellhammer JP, Malahe M, McCarter PC, Elston TC, Dohlman HG. MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast. Sci Signal. 2015;8:ra5. doi: 10.1126/scisignal.2005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldbeter A. Computational approaches to cellular rhythms. Nature. 2002;420:238–245. doi: 10.1038/nature01259. [DOI] [PubMed] [Google Scholar]
- 10.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH. Oscillations in NF-κB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 11.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, Sée V, White MRH. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 13.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerening JR, Kim SY, Ferrell JE., Jr Systems-level dissection of the cell-cycle oscillator: Bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 15.Meyer T, Stryer L. Molecular model for receptor-stimulated calcium spiking. Proc Natl Acad Sci USA. 1988;85:5051–5055. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novak B, Tyson JJ, Gyorffy B, Csikasz-Nagy A. Irreversible cell-cycle transitions are due to systems-level feedback. Nat Cell Biol. 2007;9:724–728. doi: 10.1038/ncb0707-724. [DOI] [PubMed] [Google Scholar]
- 17.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 18.Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLOS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7:803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- 20.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 21.Jaqaman K, Danuser G. Linking data to models: Data regression. Nat Rev Mol Cell Biol. 2006;7:813–819. doi: 10.1038/nrm2030. [DOI] [PubMed] [Google Scholar]
- 22.Vilela M, Danuser G. What’s wrong with correlative experiments? Nat Cell Biol. 2011;13:1011. doi: 10.1038/ncb2325. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding R, Pawson T. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326:1502–1509. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]
- 24.Bromberg KD, Ma’ayan A, Neves SR, Iyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen KJ, Garmaroudi FS, Zhang J, Lin J, Boroomand S, Zhang M, Luo Z, Yang D, Luo H, McManus BM, Janes KA. An ERK-p38 subnetwork coordinates host cell apoptosis and necrosis during coxsackievirus B3 infection. Cell Host Microbe. 2013;13:67–76. doi: 10.1016/j.chom.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alter O, Golub GH. Integrative analysis of genome-scale data by using pseudoinverse projection predicts novel correlation between DNA replication and RNA transcription. Proc Natl Acad Sci USA. 2004;101:16577–16582. doi: 10.1073/pnas.0406767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omberg L, Golub GH, Alter O. A tensor higher-order singular value decomposition for integrative analysis of DNA microarray data from different studies. Proc Natl Acad Sci USA. 2007;104:18371–18376. doi: 10.1073/pnas.0709146104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omberg L, Meyerson JR, Kobayashi K, Drury LS, Diffley JFX, Alter O. Global effects of DNA replication and DNA replication origin activity on eukaryotic gene expression. Mol Syst Biol. 2009;5:312. doi: 10.1038/msb.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorger PK. A reductionist’s systems biology: Opinion. Curr Opin Cell Biol. 2005;17:9–11. doi: 10.1016/j.ceb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Kirschner MW. The meaning of systems biology. Cell. 2005;121:503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Janes KA, Lauffenburger DA. Models of signalling networks - what cell biologists can gain from them and give to them. J Cell Sci. 2013;126:1913–1921. doi: 10.1242/jcs.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 35.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-beta signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee MS, Purvis JE, Brass LF, Diamond SL. Pairwise agonist scanning predicts cellular signaling responses to combinatorial stimuli. Nat Biotechnol. 2010;28:727–732. doi: 10.1038/nbt.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geva-Zatorsky N, Dekel E, Cohen AA, Danon T, Cohen L, Alon U. Protein dynamics in drug combinations: A linear superposition of individual-drug responses. Cell. 2010;140:643–651. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Hsueh RC, Natarajan M, Fraser I, Pond B, Liu J, Mumby S, Han H, Jiang LI, Simon MI, Taussig R, Sternweis PC. Deciphering signaling outcomes from a system of complex networks. Sci Signal. 2009;2:ra22. doi: 10.1126/scisignal.2000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janes KA. Paring down signaling complexity. Nat Biotechnol. 2010;28:681–682. doi: 10.1038/nbt0710-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 44.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 45.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 46.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 47.Rome S, Clément K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS, Riou JP, Laville M, Vidal H. Microarray profiling of human skeletal muscle reveals that insulin regulates approximately ~800 genes during a hyperinsulinemic clamp. J Biol Chem. 2003;278:18063–18068. doi: 10.1074/jbc.M300293200. [DOI] [PubMed] [Google Scholar]
- 48.Abreu-Martin MT, Vidrich A, Lynch DH, Targan SR. Divergent induction of apoptosis and IL-8 secretion in HT-29 cells in response to TNF-alpha and ligation of Fas antigen. J Immunol. 1995;155:4147–4154. [PubMed] [Google Scholar]
- 49.Janes KA, Albeck JG, Peng LX, Sorger PK, Lauffenburger DA, Yaffe MB. A high-throughput quantitative multiplex kinase assay for monitoring information flow in signaling networks: Application to sepsis-apoptosis. Mol Cell Proteomics. 2003;2:463–473. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc Natl Acad Sci USA. 2003;100:9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janes KA. An analysis of critical factors for quantitative immunoblotting. Sci Signal. 2015;8:rs2. doi: 10.1126/scisignal.2005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 53.Park T, Yi SG, Lee S, Lee SY, Yoo DH, Ahn JI, Lee YS. Statistical tests for identifying differentially expressed genes in time-course microarray experiments. Bioinformatics. 2003;19:694–703. doi: 10.1093/bioinformatics/btg068. [DOI] [PubMed] [Google Scholar]
- 54.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millard BL, Niepel M, Menden MP, Muhlich JL, Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nat Methods. 2011;8:487–493. doi: 10.1038/nmeth.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cosgrove BD, Alexopoulos LG, Hang T-c, Hendriks BS, Sorger PK, Griffith LG, Lauffenburger DA. Cytokine-associated drug toxicity in human hepatocytes is associated with signaling network dysregulation. Mol Biosyst. 2010;6:1195–1206. doi: 10.1039/b926287c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, Klamt S, Sorger PK. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, Macbeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bro R. Multiway calibration. Multilinear PLS. J Chemometr. 1996;10:47–61. [Google Scholar]
- 60.Kolda TG, Bader BW. Tensor decompositions and applications. SIAM Rev. 2009;51:455–500. [Google Scholar]
- 61.Sankaranarayanan P, Schomay TE, Aiello KA, Alter O. Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival. PLOS One. 2015;10:e0121396. doi: 10.1371/journal.pone.0121396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 63.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 64.Janes KA, Reinhardt HC, Yaffe MB. Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreeger PK, Mandhana R, Alford SK, Haigis KM, Lauffenburger DA. RAS mutations affect tumor necrosis factor–induced apoptosis in colon carcinoma cells via ERK-modulatory negative and positive feedback circuits along with non-ERK pathway effects. Cancer Res. 2009;69:8191–8199. doi: 10.1158/0008-5472.CAN-09-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-α-induced apoptosis and proliferation by MAPKs. Sci Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tentner AR, Lee MJ, Ostheimer GJ, Samson LD, Lauffenburger DA, Yaffe MB. Combined experimental and computational analysis of DNA damage signaling reveals context-dependent roles for Erk in apoptosis and G1/S arrest after genotoxic stress. Mol Syst Biol. 2012;8:568. doi: 10.1038/msb.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beyer EM, MacBeath G. Cross-talk between receptor tyrosine kinase and tumor necrosis factor-α signaling networks regulates apoptosis but not proliferation. Mol Cell Proteomics. 2012;11:M111.013292. doi: 10.1074/mcp.M111.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. Modeling HER2 effects on cell behavior from mass spectrometry phosphotyrosine data. PLOS Comput Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar D, Srikanth R, Ahlfors H, Lahesmaa R, Rao KVS. Capturing cell-fate decisions from the molecular signatures of a receptor-dependent signaling response. Mol Syst Biol. 2007;3:150. doi: 10.1038/msb4100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kemp ML, Wille L, Lewis CL, Nicholson LB, Lauffenburger DA. Quantitative network signal combinations downstream of TCR activation can predict IL-2 production response. J Immunol. 2007;178:4984–4992. doi: 10.4049/jimmunol.178.8.4984. [DOI] [PubMed] [Google Scholar]
- 72.Gordus A, Krall JA, Beyer EM, Kaushansky A, Wolf-Yadlin A, Sevecka M, Chang BH, Rush J, MacBeath G. Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol Syst Biol. 2009;5:235. doi: 10.1038/msb.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, Schoeberl B, Sorger PK. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal. 2013;6:ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kreeger PK. Using partial least squares regression to analyze cellular response data. Sci Signal. 2013;6:tr7. doi: 10.1126/scisignal.2003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jensen KJ, Janes KA. Modeling the latent dimensions of multivariate signaling datasets. Phys Biol. 2012;9:045004. doi: 10.1088/1478-3975/9/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11:544–561. doi: 10.1089/cmb.2004.11.544. [DOI] [PubMed] [Google Scholar]
- 77.Geladi P, Kowalski BR. Partial least-squares regression: A tutorial. Anal Chim Acta. 1986;185:1–17. [Google Scholar]
- 78.Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: A system for clustering and visualizing gene expression data. Bioinformatics. 2003;19:1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- 79.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: Evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cross DAE, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 82.Knowlden JM, Jones HE, Barrow D, Gee JMW, Nicholson RI, Hutcheson IR. Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signalling: Implication for gefitinib (‘Iressa’) response and resistance. Breast Cancer Res Treat. 2008;111:79–91. doi: 10.1007/s10549-007-9763-9. [DOI] [PubMed] [Google Scholar]
- 83.Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, Donner DB. NF-κB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 84.Gustin JA, Ozes ON, Akca H, Pincheira R, Mayo LD, Li Q, Guzman JR, Korgaonkar CK, Donner DB. Cell type-specific expression of the IκB kinases determines the significance of phosphatidylinositol 3-kinase/Akt signaling to NF-κB activation. J Biol Chem. 2004;279:1615–1620. doi: 10.1074/jbc.M306976200. [DOI] [PubMed] [Google Scholar]
- 85.Chen WN, Woodbury RL, Kathmann LE, Opresko LK, Zangar RC, Wiley HS, Thrall BD. Induced autocrine signaling through the epidermal growth factor receptor contributes to the response of mammary epithelial cells to tumor necrosis factor α. J Biol Chem. 2004;279:18488–18496. doi: 10.1074/jbc.M310874200. [DOI] [PubMed] [Google Scholar]
- 86.Gotea V, Ovcharenko I. DiRE: Identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 2008;36:W133–W139. doi: 10.1093/nar/gkn300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruan H, Miles PDG, Ladd CM, Ross K, Golub TR, Olefsky JM, Lodish HF. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-α. Diabetes. 2002;51:3176–3188. doi: 10.2337/diabetes.51.11.3176. [DOI] [PubMed] [Google Scholar]
- 88.Stephens JM, Pekala PH. Transcriptional repression of the GLUT4 and C/EBP genes in 3T3-L1 adipocytes by tumor necrosis factor-alpha. J Biol Chem. 1991;266:21839–21845. [PubMed] [Google Scholar]
- 89.Zheng JY, Zou JJ, Wang WZ, Feng XY, Shi YY, Zhao Y, Jin G, Liu ZM. Tumor necrosis factor-a increases angiopoietin-like protein 2 gene expression by activating Foxo1 in 3T3-L1 adipocytes. Mol Cell Endocrinol. 2011;339:120–129. doi: 10.1016/j.mce.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 90.Hayden MS, Ghosh S. Signaling to NF- B. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 91.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 92.Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 95.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Brugge JS, Janes KA. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc Natl Acad Sci USA. 2011;108:E803–E812. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bajikar SS, Fuchs C, Roller A, Theis FJ, Janes KA. Parameterizing cell-to-cell regulatory heterogeneities via stochastic transcriptional profiles. Proc Natl Acad Sci USA. 2014;111:E626–E635. doi: 10.1073/pnas.1311647111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ehrenberger T, Cantley LC, Yaffe MB. Computational prediction of protein-protein interactions. Methods Mol Biol. 2015;1278:57–75. doi: 10.1007/978-1-4939-2425-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takeda M, Obayashi K, Kobayashi A, Maeda M. A unique role of an amino terminal 16-residue region of long-type GATA-6. J Biochem. 2004;135:639–650. doi: 10.1093/jb/mvh077. [DOI] [PubMed] [Google Scholar]
- 100.Liang Q, De Windt LJ, Witt SA, Kimball TR, Markham BE, Molkentin JD. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 101.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, Johannessen CM, Silver SJ, Nguyen C, Murray RR, Hieronymus H, Balcha D, Fan C, Lin C, Ghamsari L, Vidal M, Hahn WC, Hill DE, Root DE. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA Mammalian Gene Collection Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, Fraser IDC. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 105.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ushijima H, Maeda M. cAMP-dependent proteolysis of GATA-6 is linked to JNK- signaling pathway. Biochem Biophys Res Commun. 2012;423:679–683. doi: 10.1016/j.bbrc.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 107.Kelly ML, Astsaturov A, Rhodes J, Chernoff J. A Pak1/Erk signaling module acts through Gata6 to regulate cardiovascular development in zebrafish. Dev Cell. 2014;29:350–359. doi: 10.1016/j.devcel.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Adachi Y, Shibai Y, Mitsushita J, Shang WH, Hirose K, Kamata T. Oncogenic Ras upregulates NADPH oxidase 1 gene expression through MEK-ERK-dependent phosphorylation of GATA-6. Oncogene. 2008;27:4921–4932. doi: 10.1038/onc.2008.133. [DOI] [PubMed] [Google Scholar]
- 109.Xie Y, Jin Y, Merenick BL, Ding M, Fetalvero KM, Wagner RJ, Mai A, Gleim S, Tucker DF, Birnbaum MJ, Ballif BA, Luciano AK, Sessa WC, Rzucidlo EM, Powell RJ, Hou L, Zhao H, Hwa J, Yu J, Martin KA. Phosphorylation of GATA-6 is required for vascular smooth muscle cell differentiation after mTORC1 inhibition. Sci Signal. 2015;8:ra44. doi: 10.1126/scisignal.2005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davidson MM, Nesti C, Palenzuela L, Walker WF, Hernandez E, Protas L, Hirano M, Isaac ND. Novel cell lines derived from adult human ventricular cardiomyocytes. J Mol Cell Cardiol. 2005;39:133–147. doi: 10.1016/j.yjmcc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 111.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 112.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 113.Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, Wagman AS, Hammond ME, Harrison SD. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 114.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of β-catenin by human Axin and its association with the APC tumor suppressor, #x003B2;-catenin and GSK3β. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 115.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 116.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 117.Wamaitha SE, del Valle I, Cho LTY, Wei Y, Fogarty NME, Blakeley P, Sherwood RI, Ji H, Niakan KK. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 2015;29:1239–1255. doi: 10.1101/gad.257071.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsuji S, Kawasaki Y, Furukawa S, Taniue K, Hayashi T, Okuno M, Hiyoshi M, Kitayama J, Akiyama T. The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nat Commun. 2014;5:3150. doi: 10.1038/ncomms4150. [DOI] [PubMed] [Google Scholar]
- 119.Janes KA, Wang CC, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7:311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sasagawa S, Ozaki Y-i, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 121.Roulston A, Reinhard C, Amiri P, Williams LT. Early activation of c-Jun N-terminal kinase and p38 kinase regulate cell survival in response to tumor necrosis factor α. J Biol Chem. 1998;273:10232–10239. doi: 10.1074/jbc.273.17.10232. [DOI] [PubMed] [Google Scholar]
- 122.Huang S-sC, Clarke DC, Gosline SJC, Labadorf A, Chouinard CR, Gordon W, Lauffenburger DA, Fraenkel E. Linking proteomic and transcriptional data through the interactome and epigenome reveals a map of oncogene-induced signaling. PLOS Comput Biol. 2013;9:e1002887. doi: 10.1371/journal.pcbi.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Whissell G, Montagni E, Martinelli P, Hernando-Momblona X, Sevillano M, Jung P, Cortina C, Calon A, Abuli A, Castells A, Castellvi-Bel S, Nacht AS, Sancho E, Stephan-Otto Attolini C, Vicent GP, Real FX, Batlle E. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- 124.Van Landeghem L, Santoro MA, Mah AT, Krebs AE, Dehmer JJ, McNaughton KK, Helmrath MA, Magness ST, Lund PK. IGF1 stimulates crypt expansion via differential activation of 2 intestinal stem cell populations. FASEB J. 2015;29:2828–2842. doi: 10.1096/fj.14-264010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsoyi K, Jang HJ, Nizamutdinova IT, Park K, Kim YM, Kim HJ, Seo HG, Lee JH, Chang KC. PTEN differentially regulates expressions of ICAM-1 and VCAM-1 through PI3K/Akt/GSK-3β/GATA-6 signaling pathways in TNF-α-activated human endothelial cells. Atherosclerosis. 2010;213:115–121. doi: 10.1016/j.atherosclerosis.2010.07.061. [DOI] [PubMed] [Google Scholar]
- 126.Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao Y-m, Lengauer C, Sellers WR, Benson JD. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 128.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, Hinkle G, Root DE, Roberts TM, Lander ES, Polyak K, Hahn WC. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 129.Kang BH, Jensen KJ, Hatch JA, Janes KA. Simultaneous profiling of 194 distinct receptor transcripts in human cells. Sci Signal. 2013;6:rs13. doi: 10.1126/scisignal.2003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Andersson CA, Bro R. The N-way Toolbox for MATLAB. Chemometr Intell Lab. 2000;52:1–4. [Google Scholar]
- 131.Bro R, Smilde AK. Centering and scaling in component analysis. J Chemometr. 2003;17:16–33. [Google Scholar]
- 132.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 133.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 134.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 137.Bro R. Parafac. PARAFAC. Tutorial and Applications. Chemometr Intell Lab. 1997;38:149–171. [Google Scholar]
- 138.Cattell RB. “Parallel proportional profiles” and other principles for determining the choice of factors by rotation. Psychometrika. 1944;9:267–283. [Google Scholar]
- 139.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 140.Hansen SK, Nerlov C, Zabel U, Verde P, Johnsen M, Baeuerle PA, Blasi F. A novel complex between the p65 subunit of NF-kappa B and c-Rel binds to a DNA element involved in the phorbol ester induction of the human urokinase gene. EMBO J. 1992;11:205–213. doi: 10.1002/j.1460-2075.1992.tb05043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Tensor PLSR modeling predicts overall transcript abundance but cannot link changes in transcript abundance to cytokine-induced signaling.
Fig. S2. Accuracy of tensor PLSR predictions.
Fig. S3. Induction of cluster #9 probe sets and repression by insulin.
Fig. S4. Disruption of NF-κB signaling does not widely affect the TNF-induced transcriptional response of cluster #9.
Fig. S5. Widespread TNF-insulin crosstalk among genes in transcriptional cluster #9.
Fig. S6. Immunolocalization of β-catenin is not altered by TNF stimulation or insulin costimulation in HT-29 cells.
Fig. S7. HT-29 cells lack GATA1, GATA4, and GATA5.
Fig. S8. Phylogeny of the human GATA family.
Fig. S9. Prolonged rapamycin treatment alters the proportion of GATA6S to GATA6L independently of TNF or insulin treatment.
Fig. S10. Insulin and CT99021 perturb GSK3 phosphorylation and activity.
Fig. S11. Phospho-GATA6L (Ser37) antiserum is specific for mobility-shifted wild-type GATA6L but not the S37A GATA6L mutant.
Fig. S12. GATA6L phosphorylation on Ser37 and GS phosphorylation on Ser641 are lost in a dose-dependent manner upon treatment with the GSK3 inhibitor, CT99021.
Fig. S13. GATA6L occupies GATA binding sites in the promoters of genes within the crosstalk cluster.
Table S1. Top-scoring PEST sequences in the indicated proteins according to PEST-FIND.
Table S2. qRT-PCR primer sequences.
Table S3. ChIP primer sequences.
File S1. Microarray probe sets differentially altered with time or by stimulation with TNF, EGF, or insulin.
File S2. Transcriptional clusters identified by CLICK.
File S3. Tensor PLSR model and associated files.
File S4. Transcripts differentially altered by wild-type GATA6L overexpression with or without TNF stimulation.
File S5. Raw files and densitometric analysis of phospho-GATA6L (Ser37) immunoprecipitation and total GATA6 immunoblots.
Text S1. Detailed description of tensor PLSR.
Text S2. Detailed description of GATA6L mass spectrometry.