Abstract
The craniofacial skeletal structures that comprise the human head develop from multiple tissues that converge to form the bones and cartilage of the face. Because of their complex development and morphogenesis, many human birth defects arise due to disruptions in these cellular populations. Thus, determining how these structures normally develop is vital if we are to gain a deeper understanding of craniofacial birth defects and devise treatment and prevention options. In this review, we will focus on how animal model systems have been used historically and in an ongoing context to enhance our understanding of human craniofacial development. We do this by first highlighting “animal to man” approaches: that is, how animal models are being utilized to understand fundamental mechanisms of craniofacial development. We discuss emerging technologies, including high throughput sequencing and genome editing, and new animal repository resources, and how their application can revolutionize the future of animal models in craniofacial research. Secondly, we highlight “man to animal” approaches, including the current use of animal models to test the function of candidate human disease variants. Specifically, we outline a common workflow deployed after discovery of a potentially disease causing variant based on a select set of recent examples in which human mutations are investigated in vivo using animal models. Collectively, these topics will provide a pipeline for the use of animal models in understanding human craniofacial development and disease for clinical geneticist and basic researchers alike.
Introduction
The development of the vertebrate head requires the integration of multiple structures the vertebral column, brain, sensory organs, jaws, and associated nerves, muscles, blood vessels and skeletal elements - into a functional whole. This is a complex process relying on precise control of several critical regulatory pathways – and it is perhaps unsurprising therefore that craniofacial abnormalities are one of the most common classes of human birth defects (World Health Organization, 2004). Human craniofacial disorders are often very complex and multifactorial, with origins during the first trimester of gestation so that it is difficult to determine their underlying development and progression. Thus, understanding how the face and skull develops relies on consistent animal models that are both genetically and morphologically relevant to human development. Table 1 summarizes the strengths and weaknesses of the major vertebrate model systems frog, zebrafish, chick and mouse - used to investigate the development of the head and associated craniofacial skeleton. There have been a number of excellent reviews on vertebrate head and face development and we refer the reader to these for an in depth understanding of the tissues and pathways involved (Bush and Jiang, 2012; Chai and Maxson, 2006; Cordero et al., 2011; Dixon et al., 2011; Santagati and Rijli, 2003). In this review, after a brief overview of head formation, we will instead focus on how model systems have informed our understanding of human craniofacial development as well as how recent advances are likely to accelerate the pace of discovery. First, we highlight “animal to man” approaches, specifically, how animal models have helped uncover fundamental mechanisms of craniofacial development. This section also discusses how emerging technologies, including high throughput sequencing, genome editing, and concerted mutagenesis efforts will revolutionize the future of animal models in craniofacial research. Secondly, we highlight recent “man to animal” approaches, namely the way animal models have been employed to test the function of candidate genes and associated cis-regulatory elements implicated in human craniofacial disorders. Collectively, these two complementary approaches provide a framework for how animal models can be employed from both the perspective of basic research and clinical genetics to gain a systems level understanding of human craniofacial development and its associated disorders.
Table 1.
Animal models routinely utilized to study craniofacial development
Table outlining various strengths and weaknesses of common animal models utilized in craniofacial research. For a thorough description of each model system see (Slack, 2013) and (Rossant and Tam, 2002).
| Model system | Strengths | Weaknesses |
|---|---|---|
| Mus musculus (mouse) |
|
|
| Gallus gallus (chicken) |
|
|
| Danio rerio (zebrafish) |
|
|
| Xenopus laevis/ tropicalis (frog) |
|
|
The formation of a specialized multipotent cell type – the neural crest (NC) – is central to the evolution and development of the vertebrate head (Bronner and LeDouarin, 2012; Green et al., 2015; Le Douarin and Dupin, 2012). These cells arise at the neural plate border, undergo an epithelial to mesenchymal transformation, and then migrate to the periphery where they form multiple structures. In the context of facial development, these cells migrate into the facial prominences and pharyngeal arches where they form the majority of the head mesenchyme. Once there, the neural crest cells (NCCs) interact with the surrounding mesoderm, endoderm and ectoderm to pattern the head and form the majority of the craniofacial skeleton. The cartilages and bones of this skeletal framework can be divided into the neurocranium (skull vault and skull base) and the viscerocranium, which forms the palate, ear, jaw and supporting structures. Fate mapping analyses have indicated that the viscerocranium is NCC in origin, while the neurocranium has a dual origin from the NCCs and mesoderm (Kague et al., 2012; Morriss-Kay, 2001; Noden and Trainor, 2005; Rossant and Tam, 2002). Thus, disruptions in NC development are often central to our understanding of the etiology of human craniofacial defects (Snider and Mishina, 2014; Zhang et al., 2014). Such defects can be either autonomous to the NC, such as formation, migration, cell proliferation, cell survival and differentiation or be caused by defects in the surrounding tissues, including the ectoderm, endoderm and cranial placodes, that provide critical signaling contexts to the NCCs. Therefore, most studies of human and animal craniofacial pathology must be interpreted and tested in the context of NC development and function.
I. “Animal to man”: using animal model systems to identify new genes driving craniofacial development
Animal models provide a vital platform for understanding key processes during development, delivering generally consistent genetic backgrounds, multiple replicates and extensive information concerning their embryology. More recently, advances in genomics and bioinformatics have accelerated the identification of genes that control craniofacial development as well as regulatory processes that go awry in disease. Historically, this approach began with the occasional spontaneous mutant that appeared in various animal breeds, then proceeded through larger forward genetic screens using radiation or chemicals to increase the rate of mutagenesis, and has now grown to encompass direct genome editing methods for making mutations for every gene in an organism. Here we will briefly describe the strengths and weakness of these previous approaches and describe current state of the art methodologies for gene function discovery that are accelerating the pace of scientific research in craniofacial development, including: 1) high throughput sequencing, which enables mutants to be readily pinpointed and; 2) genome editing to test gene function.
Historic approaches: Forward and reverse genetic methods to screen for craniofacial phenotypes
Forward genetic approaches represent a systematic and unbiased means to screen for abnormal phenotypes, and subsequently identify the responsible genetic lesion. This approach uses various ways to induce DNA damage, including: gamma irradiation, which tends to generate large chromosomal abnormalities such as deletions or translocations; viruses, transposons, or transgenes that cause insertional mutagenesis; and chemicals, particularly N-ethyl-N-nitrosourea (ENU), that result in single base pair changes. The benefit of such screens is their unbiased nature, in that they are able to identify new genes, new alleles of known genes, or new cis-regulatory sequences that influence a given phenotype. The downsides are that large-scale screens are labor intensive as well as costly in mouse due to per diems - and so tend to be focused on a particular time-point or developmental system. Moreover, although it was relatively simple to identify major chromosomal rearrangements, and to a lesser extent the sites of insertional mutagenesis, it often took years to identify the causative mutation induced by ENU. Now, with the advent of high throughput sequencing platforms and bioinformatics analyses based on available standard genomic sequence (discussed more below), identifying the potential mutation is far more rapid. Nevertheless, one further consideration of forward genetic screens that can be time-consuming is the importance of performing a non-complementation analysis with a second independent mutant allele of the gene identified to ensure the assignment is correct. Despite these caveats, as summarized in the next sections, forward genetic screens in mouse and zebrafish have been invaluable for the identification of genes involved in craniofacial development.
Mouse screens
The mouse has been used for decades as an important model for understanding gene function. However, the advent of mouse transgenic and embryonic stem (ES) cell technologies quickly provided new means to modify the mouse genome by the insertion of exogenous DNA. Originally, transgene insertion into a locus of interest for craniofacial development was not used as a screen but occurred by chance (Dennis et al., 2012; Meisler, 1992). Later, this mutagenic property was exploited in gene trap approaches in ES cells, which were then subsequently used to generate novel mouse strains and examine for defects in craniofacial development (Hildebrand and Soriano, 1999). These approaches have been successful, but tend to be biased for the integration site and have not been especially high-throughput at the level of novel mouse strain generation. In contrast, ENU mutagenesis is highly mutagenic in the mouse germ line and far less biased in its targeting of the genome (Caspary, 2010; Justice et al., 1999; Probst and Justice, 2010; Stottmann and Beier, 2014; Stottmann and Beier, 2010). Despite these advantages, efforts to harness ENU mutagenesis for major phenotypic screening efforts in this species only began in earnest in the 1990’s. A number of different experimental designs have been adopted to identify ENU-induced phenotypes including screens for dominant or recessive mutations (Caspary, 2010; Handschuh et al., 2014; Nolan et al., 2000; Probst and Justice, 2010; Stottmann and Beier, 2010). The experimental protocol has been amenable to both large and small-scale screens, but none have reached genome-wide saturation due to the considerable resources that would be required. Nevertheless, screens focusing on dysmorphic head and/or craniofacial phenotypes such as cranial neural tube closure, holoprosencephaly, micrognathia, agnathia, and orofacial clefting have identified many new genes and alleles that affect craniofacial development (Caruana et al., 2013; Feng et al., 2013; Nolan et al., 2000; Sandell et al., 2011). Perhaps one of the most important insights that has come from mouse ENU mutagenesis is the unexpected link between cilia formation and function in Hedgehog-dependent pathologies such as holoprosencephaly. Indeed, ENU mutagenesis has been instrumental in the identification of genes and the establishment of mouse models relevant to human ciliopathies. For example, two independent studies identified recessive mutations in Mks1, and showed that loss of this gene impacted the assembly of functional cilia. Moreover, these mouse mutants had phenotypes similar to those seen in human Meckel Syndrome, Type 1 (Cui et al., 2011; Norris and Grimes, 2012; Weatherbee et al., 2009), including craniofacial defects, and the mutation in the Mks1krc allele was also detected at an equivalent position in MKS1 for an affected human patient (Norris and Grimes, 2012). Similarly, the ENU-induced Cauli phenotype, which results in missing or reduced facial cartilage and bone among other defects, is caused by a mutation in Ift40 (Miller et al., 2013). This intraflagellar transport protein gene is also mutated in one type of human ciliopathy termed Short-Rib Thoracic Dysplasia (Norris and Grimes, 2012). Additional ENU mutations have provided new alleles that impact facial development by disrupting signaling through the Wnt, Hh, Tgfb, Fgf, and retinoic acid pathways and are valuable models to understand genetic and environmental influences on human craniofacial birth defects (Bjork et al., 2010; Feng et al., 2013; Handschuh et al., 2014; Sandell et al., 2011; Sandell et al., 2007). Of particular note, a dominant ENU screen identified the mouse line batface (Bfc), which presented with a shorter and broader face and was caused by a mutation in Ctnnb1, the gene encoding β-catenin (Nolan et al., 2000). Further analysis identified several individuals with mutations in human CTNNB1 that presented with both craniofacial and neurological defects similar to those seen in Bfc (Tucci et al., 2014). In sum, ENU based forward genetic approaches in mice have been fruitful in the identification of new genes and alleles driving craniofacial development and in providing new models of human craniofacial disorders.
Zebrafish screens
The zebrafish was employed for the first large scale ENU-based forward genetic screens that approached saturation in any vertebrate model system (Driever et al., 1996; Haffter et al., 1996). These screens were focused on overall embryonic morphology, with mutants affecting craniofacial morphology and the craniofacial skeleton being one class of interest. Several mutations were identified that produced craniofacial malformations including within edn1 (sucker), tfap2a (lockjaw/mont blanc) and tbx1 (van gogh) (Barrallo-Gimeno et al., 2004; Knight et al., 2003; Miller et al., 2000; Piotrowski et al., 2003; Piotrowski et al., 1996; Schilling et al., 1996). Additionally, mutations were identified which gave NC specific phenotypes, including within foxd3 (mother superior) (Montero-Balaguer et al., 2006; Neuhauss et al., 1996). Importantly, these screening approaches identified distinct alleles displaying similar phenotypes, leading to detailed gene regulatory networks that subsequently informed mouse and human clinical data. For example, a set of “ventral” craniofacial phenotypes was associated with alterations in genes of the Endothelin signaling pathway (Schilling et al., 1996). Notably, mutation of zebrafish plcb3 disrupted pharyngeal arch patterning downstream of edn1 (Walker et al., 2007) and helped identify mutations in EDN1 and PLCB4 in human auriculo-condylar syndrome (Gordon et al., 2013; Rieder et al., 2012). In addition to ENU, other mutation strategies that have been employed successfully include a viral insertion screen that identified mutations in many developmental processes including craniofacial formation (Amsterdam et al., 1999) and a transposase mediated protein trap screen (Trinh le et al., 2011). Subsequent analysis of mutations in the latter screen identified Rbms3, an RNA binding protein, as an important factor NCC survival (Jayasena and Bronner, 2012).
Note that many of these initial screens were focused on embryogenesis and so mutations that impacted the craniofacial complex specifically in adults would not have been identified. Additionally, genes important for craniofacial development may have been missed if their mutation caused early embryonic lethality. A further consideration is that a large maternal effect component can mask loss of the gene in the zygote, and more recent zebrafish screens have identified several maternal affect genes that impact formation of the developing head (Dosch et al., 2004; Wagner et al., 2004). In addition, because the zebrafish species has a partially duplicated genome, some genes that have a paralog with a redundant function would also be missed (Amores et al., 1998; Postlethwait et al., 1998). Nevertheless, the fact that many genes important for craniofacial development have been identified in such zebrafish screens demonstrates their success. Indeed several subsequent ENU-based approaches, including TILLING, maternal affect screens and adult screens, have extended their utility (Dosch et al., 2004; Moens et al., 2008; Wagner et al., 2004). For example, screens for mutations in adult skeletal development demonstrated that chordin functions in adult axial skeletal patterning and craniofacial development (Fisher and Halpern, 1999), barx1 for joint formation (Nichols et al., 2013), and gata3 for neurocranium development (Sheehan-Rooney et al., 2013). Additionally, adult mineralization mutants have been identified (Andreeva et al., 2011), and one which display a loss of cartilage structures due to increased apoptosis and reduced cell proliferation was subsequently mapped to wdr43, which is involved in nucleolar function suggesting that the mutant phenotype is a ribosomopathy (Zhao et al., 2014).
The last 20 years have seen considerable effort in forward genetic screens in the mouse and zebrafish that have resulted in significant advances in our understanding of craniofacial genetics and development. However, the advent of cheaper and faster methods to conduct reverse genetic analysis in these two species means that the number of novel target genes likely to be identified by forward genetics is ever diminishing. So, do major forward genetic approaches still have a bright future? In the short term, the answer is undoubtedly yes as many craniofacial mutants identified via ENU mutagenesis remain to be identified and characterized. We also suspect that the answer will also be a qualified yes in the long term as forward genetic screens can be used for focused analyses of specific genome regions or in association with other genes or markers to screen for modifiers. Finally, it should also be noted that the high-throughput mouse gene knockout efforts are designed to generate null alleles, whereas ENU mutagenesis has the capability of creating more subtle dominant negative, hypermorphic, or hypomorphic mutations and reveal the role of a gene in craniofacial development that would otherwise be missed using full knockout approaches (Handschuh et al., 2014; Nolan et al., 2000).
Additional species – a new horizon in genetic approaches to craniofacial development
In the past, the chick and Xenopus model systems have mainly been utilized as experimental models due to factors such as the size of the egg and the ease of embryo manipulation rather than for any advantages of their genetics. Indeed, the standard chick and Xenopus species have relatively long generation times compared with zebrafish and mouse, and also have drawbacks with genomics, such as allotetraploidy in X. laevis, that hamper their suitability for genetic screens. However, the recent focus on X. tropicalis as a model system has provided developmental biologists with an exciting new species that can be used for genetic analysis (Chung et al., 2014; Goda et al., 2006; Tomlinson et al., 2005; Yergeau et al., 2012). Specifically, X. tropicalis has a generation time comparable with zebrafish and mouse, possesses a diploid genome, and has a brood-size significantly larger than these other organisms. A small number of screens have already been performed in the Xenopus system using either ENU or transposon-based mutagenesis approaches and have begun to identify genes involved in development (Chung et al., 2014; Goda et al., 2006; Tomlinson et al., 2005; Yergeau et al., 2012). Therefore, Xenopus has significant potential to identify new genes and processes required for craniofacial development. Here the impact is likely to be on the earlier stages of craniofacial development, particularly the contribution of the NC and ectodermal placodes to formation and patterning of the head, as ossification and remodeling of the skull to an adult form only occurs later during metamorphosis.
In contrast to the revolution occurring in Xenopus genetics, genetic screens in avians remain problematic even though the genome sequences for species including the chick, duck, and quail are now available. Nevertheless, the study of mutant birds and the genetics and genomics of avians has not been entirely fruitless in our understanding of craniofacial development. First, in common with many domesticated animals or animal model systems, the occasional mutant will appear which will attract the interest of breeders and fanciers (Delany, 2004). Mutants with craniofacial association in chick are typified by the talpid strains, two of which are now known to be ciliopathies (Buxton et al., 2004; Chang et al., 2014), as well as other mutants that are associated with feather patterning (e.g. crested) (Wang et al., 2012), and in quail by hereditary multiple malformation (Tsudzuki et al., 1998). Second, the natural variation of beak morphology in Darwin’s finches, coupled with the relatively rapid evolution of these species, provides a means to make predictions from the available genomic data with respect to the genetic control of beak shape and size that can then be tested by experimental manipulation (Abzhanov et al., 2006; Abzhanov et al., 2004; Lamichhaney et al., 2015). The dog provides an additional example of the convergence of genomics with genetic variation in facial shape. Dogs have been selected to have diverse facial morphologies, including traits such as different shape of the skull dome, long narrow snouts or brachycephaly (Schoenebeck and Ostrander, 2014). These relate to craniofacial conditions in human including craniosynostosis, and genetic approaches such as genome-wide association studies (GWAS) have begun to identify candidate genes associated with these specific morphologies (Schoenebeck and Ostrander, 2014). In addition, some dog breeds are prone to orofacial clefting, such as the Nova Scotia Duck Tolling Retriever, and genetic mapping of the prospective causative mutation can potentially provide insight into genes underlying similar human conditions (Wolf et al., 2015). It would be remiss to omit certain cat breeds since these can also have novel phenotypic features, such as ear curling in the Scottish fold, which may be relevant to diseases such as osteochondrodysplasias that can impact the craniofacial skeleton (Hubler et al., 2004).
Coming full circle, the nexus of facial shape variation, genetics, genomics and disease associations in the dog is beginning to take hold in the mouse, with recent applications also to human. At the level of gross examination by a human, one mouse face may look identical to another, but there are distinct differences in the underlying facial shape of various Mus musculus strains and subspecies that can be revealed at a quantitative level by detailed geometric morphometric analysis (Maga et al., 2015). Mouse resources such as the Collaborative Cross provide well-defined reference populations of inbred strains in which subtle variation in facial shape revealed by morphometrics can be assessed in terms of the specific genetic background to reveal potential associations of a particular trait with one or more genes (Hochheiser et al., 2011; Maga et al., 2015). In this respect, morphometric analysis has been used to explore QTLs in the mouse underlying the size and shape of the skull (Maga et al., 2015) and the mandible (Dohmoto et al., 2002; Leamy et al., 2008; Leamy et al., 2000; Leamy et al., 2002), as well as to explore the basis of orofacial clefting (Young et al., 2007). Detailed morphological analysis may also be vital to detect and quantify slight but significant changes in facial shape that may be associated with multigenic gene traits or with alterations in specific cis-regulatory elements (Attanasio et al., 2013; Uslu et al., 2014). Thus, one likely development in the near term is that a combination of morphometrics and genetics/genomics will be needed to understand the more subtle aspects of facial morphology that influence susceptibility to craniofacial defects in specific human families and populations.
A candidate approach, using reverse genetics to decode craniofacial gene and cis-regulatory function
An important approach to consider in using animal models is reverse genetics or a candidate-based approach to perturb gene or cis-regulatory element function. First, a presumed candidate gene(s) or cis-regulatory enhancer element is identified from various approaches. For a gene, this may be through expression profiling (e.g. microarray or RNAseq) of relevant tissues [see (Bhattacherjee et al., 2007; Brunskill et al., 2014; Feng et al., 2009; Mukhopadhyay et al., 2010)], co-localization in a shared structure or pathway, or human disease mapping approaches (see Section II). For a cis-regulatory enhancer element, identification may be though a variety of means, including evolutionary conservation, distinct localization of chromatin marks in the genome, or reporter based functional assays. Then, gene function is disrupted in animal models by several possible knock-out or knockdown technologies. Targeted gene knockout technologies, in which the genomic locus of the gene of interest is specifically disrupted, have been standard in the mouse for some time, but have only recently become possible in the zebrafish and frog, in large part due to emerging genome-editing technologies (see below). Instead, it has been more common to manipulate the zebrafish, Xenopus, and chick model systems using various gene knockdown technologies. Such knockdown approaches include specially modified antisense oligonucleotides (i.e. morpholinos) to create ‘morphant’ embryos, siRNA application, and the injection or electroporation of dominant negative constructs into early stage embryos. In specific circumstances it has also been possible to target particular pathways with chemical inhibitor or activator treatment. These strategies have been successful in identifying important genes involved in NC development and craniofacial morphogenesis for both protein coding genes (mRNAs) and microRNAs (miRNAs). Given the depth and broad application of these approaches in animal model systems they are not covered in detail here. However, Section II of this review highlights a variety of these approaches in animal models as a result of mutations and variants identified in human patients.
The New Frontier - how application of emerging technologies and repositories will facilitate rapid animal model generation and characterization
The previous section highlighted a variety of approaches that have been utilized to investigate gene or cis-regulatory enhancer function in animal models of craniofacial development. However, new technologies and international efforts are now taking shape that are already transforming the speed and utility of unbiased forward genetic screens and reverse genetic approaches. Here, we briefly summarize some of these advances, including 1) sequencing and bioinformatic technologies, 2) genome-editing technologies, and 3) genome-wide knock-out and phenotyping efforts, with a focus on how their application will shape the future of animal models in craniofacial research.
The advent of traditional Sanger sequencing made the option of DNA sequencing commonplace in most laboratories during the 1980’s. However, new sequencing technologies, facilitating unparalleled depth, speed, cost-effectiveness, and accuracy, are quickly emerging. Such NextGen sequencing capabilities, alongside the computational and bioinformatic support needed to process the data, are found at most major research institutions or readily accessible through various commercial enterprises [for a recent review see (Reuter et al., 2015)]. These new instruments provide the ability to rapidly sequence an entire genome and align the resulting data to standard genomes for all the model organisms discussed in this review, as well as to the human genome. Although there are still some important gaps in the genomes of various species – the complex rRNA gene loci being a notable example relevant to craniofacial development – this power to assess the genetic code at such resolution is already changing the landscape of feasibility on both the human genetics and animal model front. First, these technologies are assisting in the identification of rare mutations as well as common variants contributing to craniofacial disorders (Khandelwal et al., 2013; Leslie et al., 2015b). To date, the majority of these changes have been identified using whole-exome sequencing, restricting the region of interest to protein coding sequences. Indeed, for a majority of the examples covered below in Section II of this review, whole-exome-sequencing was ultimately the means by which a mutation was identified. However, the study by Leslie EJ et al (Leslie et al., 2015b) highlights the use of targeted genome-sequencing, extending mutational analysis beyond just coding sequence into non-coding regions. Their studies were based on prioritized regions of the genome, but as this technology becomes commonplace, sequenced regions will undoubtedly encompass almost the entire genome. One clear consequence of this limitless sequencing capability will be a quickly expanding catalogue of human mutations, including common and rare variants in coding and non-coding sequence, and a corresponding increase in animal models needed to dissect the functional consequence of those mutations. As one example of a response to this need, a new sophisticated zebrafish based model was recently generated to assess the effect of identified non-coding variants on gene expression in vivo (Bhatia et al., 2015). This system is based upon a direct comparison of fluorescent reporter signals driven by wild-type and variant cis-acting elements, and was used to characterize regulatory sequence variants associated with human facial clefting and Pierre Robin Sequence. A second flourishing application of these new sequencing capabilities is the rapid identification of causative mutations responsible for phenotypes in forward genetic screens (Andrews et al., 2012; Henke et al., 2013a, b). As described above, what was once a laborious endeavor can now be streamlined, allowing a more high-throughput approach to identification of novel alleles driving craniofacial development. Collectively, advances in sequencing should add clarity to the genetics of craniofacial development both from human and animal model perspectives, and may ultimately increase the need for additional animal models.
Along with advances in DNA sequencing, advances in tools available for genome editing, particularly in model organisms, have been rapidly adopted (Sander and Joung, 2014). Most notable is the bacterial based type II clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system, which was preceded by both zinc finger nucleases (ZFNs) and Transcription activator-like effector nucleases (TALENs). Owing to their ease of construction as well as their ability to target a precise location of a genome the CRISPR/Cas system has become the genome-editing tool of choice. Reviewed extensively elsewhere, this system uses a short guide-RNA to target an enzymatic endonuclease to desired locations of the genome, and can be readily employed in vertebrate species [see (Sander and Joung, 2014)]. Upon targeted DNA cleavage by the endonuclease, conserved cellular repair mechanisms are deployed, often error prone, resulting in deletions or insertions at the target site, referred to as non-homologous end-joining (NHEJ). Alternatively, homologous-directed repair (HDR) can occur in the presence of suitable homologous sequence, resulting in incorporation of novel DNA fragments into the target site (Sander and Joung, 2014).
How will novel gene editing technologies shape the future of animal models in craniofacial research? First, they are revolutionizing reverse genetic approaches, particularly in the Xenopus and zebrafish, model systems that previously lacked simple and efficient gene targeting technologies (Hisano et al., 2014; Sander and Joung, 2014; Schmitt et al., 2014; Varshney et al., 2015). Instead, in the absence of an available mutant line, these non-mammalian systems often relied on anti-sense Morpholino technology to transiently “knockdown” gene function. Such Morpholinos are designed to inhibit translation or RNA splicing, and it is not always possible to assess their direct effect on specific gene expression with the tools at hand. Concerns have also been raised concerning off-target effects and non-specific toxicity of Morpholinos, most notably in zebrafish exemplified by a recent study examining vascular development in which up to 80% of gene mutations made via other approaches failed to recapitulate the morphant phenotype (Kok et al., 2015; Law and Sargent, 2014). However, the general applicability of the findings of Kok et al to other Morpholino based studies remains controversial (Blum et al., 2015; Rossi et al., 2015). In particular, Rossi et al. find that compensatory pathways are activated in genetically modified zebrafish, but not in morphants (Rossi et al., 2015). Under these circumstances, specific Morpholino-based gene knockdown may uncover critical developmental processes related to gene function that are masked by regulatory feedback mechanisms in mutants. Nevertheless, it is likely that a consensus on the relative merits and drawbacks of these reagents will only become apparent after further study. In the meantime, we note that there are well-established guidelines for the experimental design when using Morpholinos that will strengthen the conclusions in the absence of a targeted mutation (Blum et al., 2015; Eisen and Smith, 2008; Stainier et al., 2015). There are also specific considerations for the use of Morpholinos in the context of craniofacial development in the zebrafish system. First, a common off-target effect is edema of the heart, which via its proximity to the craniofacial complex can cause disruption to the posterior ceratobranchial cartilages as a secondary pathology rather than a direct effect. Second, several morphant craniofacial phenotypes can be sometimes be altered by adjusting p53 expression levels indicating either a non-specific p53 dependent apoptotic mechanism of action or potentially a specific genetic interaction with p53 in the development of the pathology (Jones et al., 2008; Melvin et al., 2013; Robu et al., 2007); both can make interpretation of the phenotypes difficult.
A second application of these novel gene targeting technologies will be precision mutagenesis, largely owing to HDR-based mechanisms, not only in mouse but also in other model organisms including zebrafish (Hisano et al., 2015). Although HDR is less prevalent than NHEJ using these novel endonuclease approaches, it still provides an effective means to generate specific modifications, down to single base pair resolution (Wijshake et al., 2014). Such advances will be important in deciphering allele specific modifications identified in human patients, contingent on some degree of conservation between species. For example, the effect of a single non-synonymous SNP identified in a human patient could be recapitulated in an animal model to assess functional consequences of hypomorphic or dominant alleles on craniofacial development.
Lastly, these technologies will also facilitate rapid genome modification at multiple loci simultaneously, given their exquisite targeting capabilities (Cong et al., 2013; Wang et al., 2013). This ability will obviate lengthy mating schemes required to combine multiple alleles. Such capabilities will become critical in rapidly constructing gene-networks driving craniofacial development, as more interacting genes are identified in humans with craniofacial disorders. Given the rapid adoption of these technologies, they should quickly revolutionize the genetic power of animal models in craniofacial research. However, as with any new technique, there are precautions that should be considered. In several species, CRISPR/Cas can cause insertions or deletions in spurious positions causing off target affects (Cho et al., 2014). Thus, careful confirmation of the desired mutation as well as breeding out potential non-specific mutations will be required. In this context, the rapid adoption and simplicity of the CRISPR/Cas system has made it the current system of choice for targeted mutagenesis and many refinements are being tested to make this system less prone to off target affects as well as adaptable to a variety of uses beyond simple gene targeting in early embryogenesis. For example, tissue specific use of genome editing technologies will be required to avoid the early embryonic lethality of many genes involved in craniofacial development and these approaches have already begun in the zebrafish field (Ablain et al., 2015).
Finally, along with advances in DNA sequencing and gene editing technologies, a concerted effort has been made to increase animal repositories harboring targeted “knock-outs”, or conditional alleles, of all human orthologs, most notably in mouse and zebrafish model systems (summarized in Table 2). In mice, this effort is largely represented by groups in North America and Europe that comprise the International Knockout Mouse Consortium (IKMC) (Bradley et al., 2012). Together, these groups have made a strategic initiative to generate targeted ES-cells of all known protein coding mouse genes. The current design of the targeting vector for homologous recombination in ES cells is versatile, allowing the production of LacZ reporter alleles, conditional alleles and knockout alleles (Skarnes et al., 2011; West et al., 2015). These properties of the targeting vector are very pertinent to research in craniofacial biology as they enable rapid determination of the gene’s expression pattern. Moreover, the conditional alleles allow genes to be removed in specific tissues of the head, such as the NC, ectoderm, or mesoderm and/or at specific times in the pre- or post-natal period. This latter consideration is critical if the null allele would result in lethality early in embryogenesis before it was possible to study head development. For any investigator, the ease of obtaining ES cells or mouse resources from these homologous recombination-based approaches for a gene of interest must now be weighed against the speed and simplicity of making a straight null allele using the CRISPR/Cas system.
Table 2.
Useful databases and repositories for animal model systems and craniofacial development
Summary of available animal model repositories and craniofacial databases. A compiled list of a variety of resources available to the craniofacial community, including animal model repositories as well as craniofacial focused datasets. The organization/resource is listed in the left column, a brief description of the resources’ main goals in the middle column, and a url address and reference for the given resource in the right column.
| Name | Content (as described by organization/repository) |
|---|---|
| International Knockout Mouse Consortium (IKMC) | “The members of the International Knockout Mouse Consortium (IKMC) are working together to mutate all protein-coding genes in the mouse using a combination of gene trapping and gene targeting in C57BL/6 mouse embryonic stem (ES) cells” |
| International Mouse Phenotyping Consortium (IMPC) | “The goal of the International Mouse Phenotyping Consortium (IMPC) is to discover functional insight for every gene by generating and systematically phenotyping 20,000 knockout mouse strains” |
| Knockout Mouse Project (KOMP) - member of IKMC | “The Knockout Mouse Project (KOMP) is a trans-NIH initiative that aims to generate a comprehensive and public resource comprised of mouse embryonic stem (ES) cells containing a null mutation in every gene in the mouse genome.” |
| European Conditional Mouse Mutagenesis Program (EUCOMM) - member of IKMC |
|
| North American Conditional Mouse Mutagenesis project (NorCOMM) - member of IKMC | “A large-scale research initiative focused on developing and distributing a library of mouse embryonic stem (ES) cell lines carrying single gene trapped or targeted mutations across the mouse genome.” |
| Mouse Genome Informatics (MGI)/Mouse Genome Database (MGD) | “MGI is the international database resource for the laboratory mouse, providing integrated genetic, genomic, and biological data to facilitate the study of human health and disease.” |
| Mutant Mouse Resource Research Centers (MMRRC) | “The MMRRC distributes and cryopreserves scientifically valuable, genetically engineered mouse strains and mouse ES cell lines with potential value for the genetics and biomedical research community. We are a national network of breeding and distribution facilities plus an information coordinating center serving together as NIH’s premier repository of spontaneous and induced mutant mouse and cell lines. The MMRRC is supported by the National Institutes of Health.” |
| The e-Mouse Atlas Project (EMAP) Electronic Mouse Atlas of Gene Expression (EMAGE) |
|
Another import facet of the IKMC is that they work in conjunction with the International Mouse Phenotyping Consortium (IMPC) to subject the homozygous and heterozygous mutants generated to a battery of phenotyping tests (http://www.mousephenotype.org) (Brown and Moore, 2012a, b). The site also provides a database that links the mouse models generated with potential human disease associations. In the context of the craniofacial complex, adult mice of both sexes are analyzed at the level of gross morphology as well as by X-ray analysis for defects in the size and shape of the skeleton, as well as the number, shape, and color of the teeth, among other phenotypes. Furthermore, with the recent establishment of NorCOMM2 and KOMP2, there is now also a pipeline to assess mice that die before weaning using various approaches that are relevant to craniofacial biology, including μCT analysis during embryogenesis (http://dmm.biologists.org/content/6/3/571.full). In sum, the databases maintained by Jax (http://www.informatics.jax.org) and the IMPC, among others, provide access to information on the availability of mice or ES cell resources available for studying a gene of interest, as well as any available expression data, including from LacZ insertion, or phenotyping information.
Similar efforts have gained traction in the zebrafish community as well, although not to the degree of the mouse repositories (Varshney and Burgess, 2014; Varshney et al., 2013a; Varshney et al., 2013b; Varshney et al., 2015). In addition, concerted efforts to compile craniofacial data, across broad scientific fields, are also underway, notably through the FaceBase Consortium (https://www.facebase.org) (Hochheiser et al., 2011) and Jackson Laboratories to maintain specific stocks of craniofacial mutations (https://www.jax.org/research-and-faculty/tools/mouse-resource-for-craniofacial-research). Such resources provide a comprehensive database of developmental, anatomical, and genetic information related to both human and animal models of craniofacial development (Brinkley et al., 2013a; Brinkley et al., 2013b; Eames et al., 2013).
In sum, the tools and resources emerging paint a bright future for craniofacial researchers and geneticists alike. Supplemented with more traditional methods, highlighted above, advances in sequencing, genome editing, and large-scale animal model repositories will provide the resources necessary to understand the genetics of craniofacial development and its disruption in associated congenital malformations. This should be evident in the years to come by a wave of newly identified genetic variants in humans with craniofacial disorders and a concomitant surge of sophisticated animal models, utilizing powerful cutting-edge genetics, to decipher the effects of those variants on craniofacial development.
II. Man to animal: using animal model systems to assign function to genetic elements associated with craniofacial disorders in humans
Detecting genomic alterations in human patients with craniofacial disorders
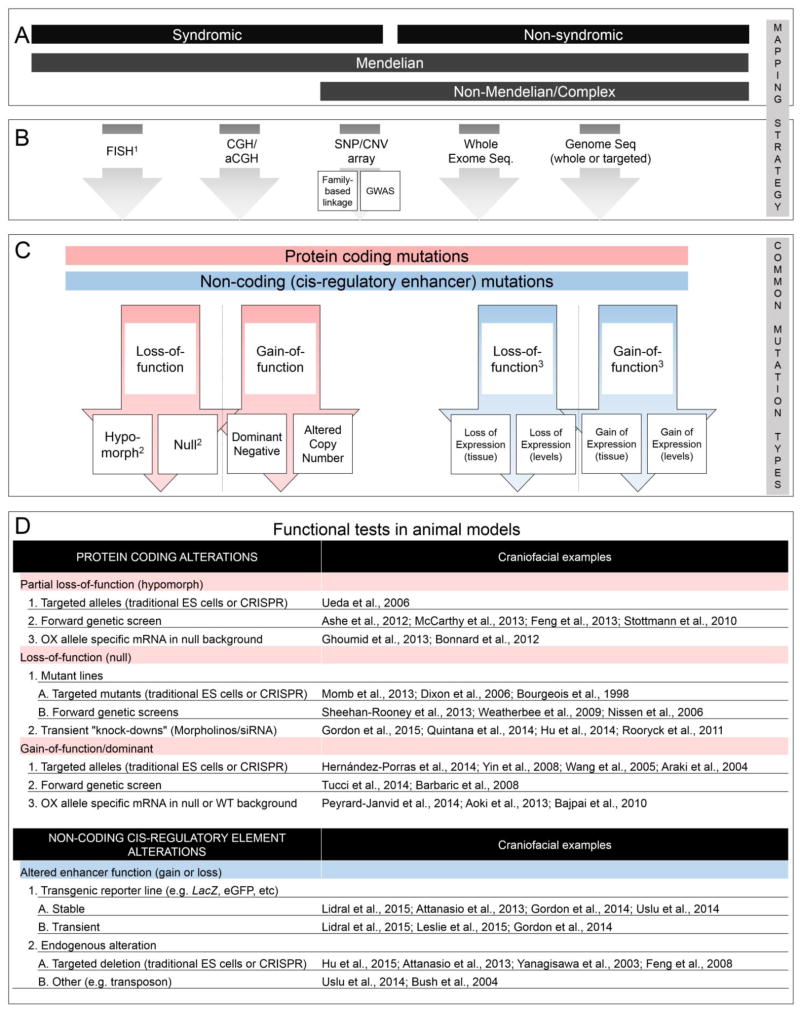
Human craniofacial disorders encompass a range of phenotypes that can have complex etiologies, ranging from simple Mendelian conditions to more genetically complex polygenic scenarios, involving gene-gene and gene-environment interactions. These disorders can often be classified as either syndromic, involving anomalies in multiple craniofacial structures (plus additional tissues), or non-syndromic, involving a craniofacial anomaly in isolation (such as cleft lip and/or palate, CL/P). In general, syndromic cases tend to follow simple Mendelian-based inheritance patterns, whereas non-syndromic cases (such as isolated cleft lip and/or palate) group into more complex gene/environment etiologies [reviewed in (Murthy and Bhaskar, 2009)] (Figure 1A).
Figure 1.
Systematic workflow of human craniofacial developmental disorder gene discovery and functional testing in animal models. (A) Summary of broad categories of human craniofacial developmental disorders. (B) Summary of common approaches used to detect genomic alterations in human patients. (C) Summary of potential (not all) mutation types identified in human patients. (D) Summary of functional tests, along with referenced examples, currently available and commonly utilized for testing gene function in animal model systems. 1FISH is often used when a known syndrome is suspected, whereas other mapping strategies are often unbiased genome-wide approaches. 2Null and hypomorphic mutations can be further classified as haploinsufficient or haplosufficient. 3Loss or gain of a repressive factor could have the opposite consequence. Note, although not a functional test, expression analysis is an important component of determining in what tissue and at what developmental time-point a gene or cis-regulatory element functions (not shown). Abbreviations: aCGH, array comparative genomic hybridization; CGH, comparative genomic hybridization; CNV, copy number variation; FISH, Fluorescence in-situ Hybridization; GWAS, genome wide association study; OX, over-express; Seq; Sequencing; SNP, single nucleotide polymorphism.
Given the phenotypic and underlying genetic heterogeneity of these disorders, a variety of mapping tools have been deployed to isolate the mutation(s) responsible for the developmental defect observed (Figure 1B). These have been thoroughly reviewed elsewhere in the context of craniofacial anomalies (Khandelwal et al., 2013), and each approach, although often suited for isolating a distinct class of genetic lesions, is routinely superseded by a more refined approach as costs decrease and technologies evolve. Briefly, common mapping methods have included chromosomal karyotyping, fluorescent in situ hybridization (FISH), comparative genomic hybridization (CGH), array CGH, as well as single nucleotide polymorphism (SNP) and copy number variation (CNV) arrays. The genome-wide resolution of SNP and CNV arrays have made possible more powerful approaches such as family-based linkage analyses as well as genome-wide association studies (GWAS), providing an opportunity to pinpoint loci associated with complex (non-Mendelian) craniofacial disorders (such as CL/P). In turn, these methods are being supplemented or replaced due to advances in DNA sequencing technology, notably exome sequencing and in some instances genome sequencing. Exome sequencing provides the means to scan for variation in all coding sequence of the genome and enables detection of alterations at the nucleotide level, including within flanking sequences associated with mRNA splicing. Targeted or whole genome sequencing extends the level of detection beyond coding regions to cis-regulatory elements and non-coding RNAs, which are undoubtedly a key component of several craniofacial disorders. Moreover, unlike previous mapping studies which needed large family groups to pinpoint the affected chromosomal interval, NextGen sequencing enables a much smaller number of related individuals to be interrogated to identify a potential candidate mutation. In a practical sense, a combination of the “classical” methods with these newer, sequence-based methods, are utilized and are quickly providing a catalogue of genomic alterations in both isolated and multiple congenital craniofacial disorders.
Using animal models to decipher the effects of identified mutations
The approaches listed above provide the means to identify potentially causative mutations responsible for the observed craniofacial phenotype in a human patient. However, once identified, functional tests are required to determine conclusively the consequences of the specific genetic alteration. Although in vitro based assays provide a convenient system to test biochemical and molecular based properties of the mutation, only animal models provide a powerful way to dissect the role of the mutation in a functional context during development. Although the power of using animal models to understand human developmental disorders conceptually is a straightforward approach, framing the appropriate question to ask and designing experiments to answer those questions can be a more difficult endeavor. For example, several human syndromes that have associated craniofacial defects are caused by large chromosomal abnormalities (e.g. Down syndrome and DiGeorge syndrome). Modeling these types of defects require complex chromosomal engineering and interpretation of the results in the animal model system are not always straightforward. It should also be stressed that animal models do not always present with the same phenotype as a human even when they contain the equivalent genetic lesions. For example, PVRL1 is mutated in Cleft lip/palate-Ectodermal Dysplasia Syndrome, which includes CL/P, but mice lacking this gene instead develop eye and tooth enamel defects (Barron et al., 2008; Inagaki et al., 2005). Furthermore, differences in craniofacial morphology between humans and model systems may also cause phenotypic differences. Thus, many gene mutations responsible for CL/P in humans may result in cleft palate only in mice due to differences in craniofacial morphology (Dixon et al., 2011). Data should also be interpreted keeping in mind that different inbred and outbred animal strains are not equivalent genetically, and a particular mutation may yield a completely different phenotype when placed onto a different strain background. With these caveats in mind, once a genetic lesion has been identified, animal models provide a powerful utility to systematically address, 1) what the molecular function of that gene/element is during normal craniofacial development and, 2) the molecular mechanisms responsible for the defects observed. Each model system has advantages and disadvantages (see Table 1) and the choice of which to use for a new problem is a balance of cost, available expertise, and suitability of the animal model for the question under consideration. For example, the mouse is the closest animal model to human and has more closely matched anatomical structures, such as the secondary palate and teeth. Therefore, this model is the gold standard for testing human cis-acting sequence function in driving tissue specific expression patterns. However, other models provide cheaper and more readily accessible embryos for analysis, and also provide a more rapid initial assessment of gene function. Often, a combination of disparate animal models, each utilized for its unique strength, will provide a clearer understanding of the developmental disorder at hand and potential treatment options. We also note that suitable animal model systems can also be used to study gene:environment interactions, for example between ethanol and genes involved in craniofacial anomalies in the zebrafish model system (McCarthy et al., 2013) and in chick embryos (Ahlgren et al., 2002), specifically reviewed elsewhere (McCarthy and Eberhart, 2014).
At the outset, it is valuable to obtain spatial and temporal information concerning the expression pattern associated with the gene under analysis as this can often provide insight into the potential role of the sequence in craniofacial development. Considerable knowledge regarding gene expression patterns can now be found in several public repositories and databases that have already imaged and/or catalogued expression patterns based upon timing and tissue (see Table 2, for example MGI/GXD and Facebase for mouse and ZFIN for zebrafish). Alternatively, if insufficient data exists for a gene of interest, detailed information can be obtained using standard approaches, such as in situ hybridization and/or immunohistochemistry to obtain a precise expression summary relevant to craniofacial development. In certain instances, it may also be possible to take advantage of LacZ knock-in reporter alleles for the gene of interest if mouse resources for such reagents are available through public repositories (e.g. EUCOMM, see Table 2). With respect to mutations in non-coding regions, several studies have now begun to identify potential cis-regulatory elements involved in gene expression during facial development by cataloging genome-wide chromatin modifications associated with enhancer elements in specific tissues or cell types (Hochheiser et al., 2011; Prescott et al., 2015). These datasets provide a valuable resource to determine if a particular non-coding mutation linked with human pathology might be associated with a possible enhancer. In several instances these potential enhancer elements have also been tested for their ability to direct tissue-specific expression to the developing mouse face providing an additional level of information concerning their possible regulatory importance [the VISTA Enhancer Browser (http://enhancer.lbl.gov/) (Visel et al., 2007), as well as within Facebase].
Finally, in developing an animal model system based on a human mutation it is critical to consider the mode of inheritance and nature of the pathology – dominant, recessive, partially penetrant, variable expressivity – as well as the nature of the allele - null, hypomorphic, neomorphic, dominant negative – as these considerations will affect allele design in an animal model (see Figure 1C). In the recent past, the discovery of a human mutation associated with a particular pathology and the generation of pertinent animal models to address the consequences of such mutations were frequently represented in separate publications. Classic examples of such analyses include the gene mutations associated with various types of craniosynostosis [reviewed in (Governale, 2015)]. Now, it is becoming more common for the identification of a possible causative mutation in affected individuals to be combined with evidence of a causative role based on studies in an animal model system in a single publication. We highlight a small set of such examples in the following section and provide additional examples in Table 3. Although far from exhaustive, we chose these examples because they cover a range of: 1) craniofacial disorders (syndromic and non-syndromic); 2) gene identification approaches; 3) allelic alterations (i.e. protein coding - null, hypomorphic, dominant-negative and non-coding); 4) animal model systems; and 5) experimental approaches. For the readers’ reference, additional studies not covered have also been summarized in Table 3. Collectively, these studies provide a framework and workflow for the initial functional assessment of genes or non-coding elements associated with human craniofacial disorders (Figure 1A–D).
Table 3.
Recent examples of “human to animal” craniofacial studies
A select list of recent (since 2010) studies in which, first a genetic lesion is detected in a human patient with either a syndromic or non-syndromic craniofacial condition and secondarily an animal model is utilized in the same study to further understand the consequences of that lesion on craniofacial development.
| Disorder | Mutation identification approach | Gene(s) | Model organism | Assays |
|---|---|---|---|---|
| Syndromic cleft palate and micrognathia | GTG-banded karyotyping, whole- genome sequencing | CAPZB | zebrafish | Z - exp, LOF (insertional mutant), GOF (mRNA) |
| Acrofacial Dysostosis - Cincinnati Type | Whole exome sequencing, Sanger sequencing | POLR1A | zebrafish | Z - exp, LOF (insertional mutant) |
| Mandibulofacial Dysostosis with Alopecia | Trio whole exome sequencing | EDNRA | zebrafish, mouse | Z - LOF (MO), GOF (mRNA); M - exp |
| Nonsyndromic cleft lip w/ or w/o cleft palate (NSCL/P) | Targeted whole-genome sequencing | FGFR2 (+others) | zebrafish | Z - NCE-exp |
| Isolated cleft lip and palate (CLP) | Non-coding enhancer analysis | FOXE1 | zebrafish, mouse | Z - NCE-exp; M - NCE-exp |
| Van der Woude syndrome | Family-based linkage, whole exome sequencing | GRHL3 | zebrafish, mouse | Z - GOF (dn-mRNA); M - LOF |
| X-Linked Cobalamin Disorder | Whole exome sequencing, Sanger sequencing | HCFC1 | zebrafish | Z - LOF (MO) |
| Noonan syndrome | Whole exome sequencing, Sanger sequencing | RIT1 | zebrafish | Z - GOF (mutant mRNA) |
| 3MC syndrome | SNP array - homozygosity mapping | COLEC11, MASP1 | mouse, zebrafish, quail | M - exp; Z - exp, LOF (MO), GOF; Q - GOF |
Note, bold examples are reviewed in more detail in the text. Abbreviations: dn, dominant-negative; exp - expression analysis; GOF, gain-of-function; LOF, loss-of-function; M, mouse; MO, morpholino; NCE, non-coding enhancer; Q, quail; Z, zebrafish.
Mutations affecting protein sequence
In the following three examples, we discuss how genome sequence analysis led to the identification of mutations in three different human craniofacial syndromes, 3MC, VWS2, and AFDCIN, as well as how animal models were used to tease apart the potential mechanism of action for each of the mutant alleles. 3MC syndrome (Carnevale, Mingarelli, Malpuech, and Michels syndrome) is a rare autosomal recessive disorder characterized by facial dysmorphism, including CL/P, hypertelorism, eyelid defects, and craniosynostosis, along with cognitive impairment, and hearing loss (Al Kaissi et al., 2007; Leal et al., 2008). SNP based arrays followed by target gene sequencing was used to identify homozygous mutations in COLEC11 and MASP1, two genes involved in the lectin complement pathway (Rooryck et al., 2011). Next, both zebrafish orthologs of COLEC11 and MASP1 were targeted using specific morpholinos and in each instance similar craniofacial defects were obtained. These morphant phenotypes included reduced mandibular length, malformed anterior neurocranium, and improper development of the ceratohyal cartilage, potentially mimicking some of the craniofacial defects observed in 3MC syndrome patients. Further analysis using specific markers revealed disrupted and disorganized streams of NC in morphant embryos, suggesting Colec11 and Masp1 may be involved in providing guidance cues for migrating NCCs. To test this hypothesis they applied beads soaked in Colec11 to either intact zebrafish or to quail neural tube explants and determined that NCCs preferentially migrated towards Colec11 beads but not control beads. Thus, they were able to use loss and gain of function assays in animal models to provide a rapid and feasible explanation for how the homozygous COLEC11 or MASP1 mutations would affect NCC migration and craniofacial development in 3MC Syndrome.
Unlike 3MC, both VWS2 (Van der Woude Syndrome 2) and AFDCIN (Acrofacial dysostosis – Cincinnati Type) are dominantly inherited craniofacial syndromes, but the animal studies accompanying the identification of the mutations in these two syndromes indicate very different mechanisms of action of the allelic variants. Van der Woude syndrome is characterized by a cleft lip and/or cleft palate, as well as hallmark pits and/or sinuses of the lower lip and the majority of cases result from mutations in one allele of IRF6 (Burdick et al., 1985; Kondo et al., 2002). Recently, exome sequencing was performed on a single large family that had VWS syndrome, but no detectable mutation at the IRF6 locus (Peyrard-Janvid et al., 2014). These studies, and follow up studies on additional families, revealed a series of mutations in the transcription factor GRHL3 that would result in amino acid substitutions or the expression of slightly truncated proteins. To assess the nature of the mutations, they next utilized a zebrafish periderm assay in which disruption of normal gene expression can lead to embryo rupture and lethality (Sabel et al., 2009). In this context, the zebrafish periderm is envisioned as a model for the ectoderm of the mammalian palatal shelves. Knockdown of either zebrafish irf6 or grhl-like genes leads to defective periderm and rupture, whereas the injection of RNA encoding wild-type human GRHL3 into the zebrafish embryo resulted in the presence of ectopic periderm suggesting that these genes are required for periderm development and integrity (de la Garza et al., 2013; Sabel et al., 2009). Zebrafish embryos were then injected with RNAs encoding the various mutant GRHL3 alleles found in VWS2 and in all instances resulted in periderm rupture, indicating that they inhibited the regulatory pathways required for integrity of this layer (Peyrard-Janvid et al., 2014). In the future, it would be interesting to generate the GRHL3 allele specific mutations in the mouse model to test whether similar craniofacial phenotypes are obtained as in human VWS2. However, the authors were able to show that Grhl3-null mice had an abnormal oral epithelium with oral adhesions and to a lesser extent, cleft palate. Overall, these studies indicate that GRHL3 acts as a second VWS locus (VWS2), and that mutations in this gene appear to have a dominant negative activity rather than the haploinsufficiency typical of IRF6 in VWS1 (Kondo et al., 2002).
Acrofacial dysostoses are characterized by down-slanted palpebral fissures, midface retrusion, micrognathia, and cleft palate alongside additional skeletal defects [reviewed in (Trainor and Andrews, 2013)]. There are a number of types of acrofacial dysostosis involving separate chromosomal loci and recent exome sequencing of an isolated individual identified a de novo heterozygous mutation in POLR1A, encoding a core component of RNA polymerase 1 (Weaver et al., 2015). Subsequent analysis of additional affected individuals also identified similar POLR1A mutations and the condition was named Acrofacial Dysostosis - Cincinnati Type (AFDCIN). Next, the authors performed a detailed examination of a zebrafish mutant, which contained a retroviral insertion in the 5′ UTR of polr1a, resulting in drastically reduced polr1a levels (Amsterdam et al., 1999; Weaver et al., 2015). Mutant embryos developed a number of defects including microophthalmia and jaw agenesis, which overlapped particular features of AFDCIN patients. Further analysis of the mutant zebrafish revealed significant reductions in rRNA levels, induction of p53, and greatly impaired NC survival. These findings are consistent with AFDCIN being a ribosomopathy caused by haploinsufficiency of POLR1A. In this context, multiple studies, in multiple animal model systems, have begun to link craniofacial defects with ribosomopathies, implying that facial development is particularly sensitive to defects in nucleolar function and ribosomal biogenesis (Dixon et al., 2006; Gazda et al., 2008; Griffin et al., 2015; Trainor and Andrews, 2013).
In sum, these three studies highlighted how animal model systems can be utilized in conjunction with loss-of-function, and gain-of-function assays to test the importance of a gene mutated in patients with a syndromic craniofacial anomaly.
Cis-regulatory enhancer mutations
A second class of genetic alterations that can drive human craniofacial disorders are those located within non-coding, cis-regulatory elements. Historically, these elements have been harder to detect compared to protein-coding regions, although new genome-wide techniques have facilitated their discovery (see above). More recently it has been appreciated that these elements may be the means of fine-tuning craniofacial morphogenesis between individuals and during evolution (Attanasio et al., 2013). Moreover, there are now several examples linking particular sequence variants at specific cis-acting motifs with human craniofacial defects (Rahimov et al., 2008). In one recent instance, Leslie and colleagues employed “targeted sequencing” to identify mutations associated with non-syndromic cleft lip with or without cleft palate (NSCL/P) using a case-parent trio design (Leslie et al., 2015b). Targeted loci chosen for sequencing (13 regions) were based on previous GWAS/genome-wide linkage studies (9/13) and candidate gene studies (4/13), encompassing ~6.3Mb of coding and non-coding sequence. This analysis detected a variety of both common and rare variants associated with NSCL/P as well as multiple de novo mutations. Of the 66 validated de novo mutations 63 were found to reside in non-coding sequence, with a subset (11/63) occurring within predicted regulatory elements.
As a proof-of-principle, one of these non-coding elements, located near FGFR2 (254.6Kb downstream of the transcription start site), was further analyzed using an in vivo, zebrafish based, reporter assay. This novel mutation was chosen given FGFR2’s known role in craniofacial development as well as the location of the mutation, residing within a region predicted to be an active NC enhancer based on chromatin marks (Rada-Iglesias et al., 2012; Riley et al., 2007; Rosenbloom et al., 2013; Stanier and Pauws, 2012). To test for functional activity of this putative enhancer, the human wild-type element was cloned upstream of a minimal promoter driving GFP, and introduced into zebrafish embryos to follow it’s ability to direct expression during development. GFP fluorescence was detected in the neural keel (where premigratory NCCs reside), brain, and emigrating NCCs, consistent with previous reports of Fgfr2 expression in zebrafish and mice (Orr-Urtreger et al., 1991). In contrast, when the mutant construct was used in the same assay, fewer embryos showed a similar expression pattern with the mutation present, consistent with the de novo mutation being a functional variant that disrupted NC expression. Given this analysis was done on the enhancer in isolation, it remains to be seen whether this alteration affects endogenous FGFR2 expression, a functional study more plausible with the advent of CRISPR/Cas technology. Nonetheless, this simple yet powerful type of in vivo reporter assay provides a convenient means to assess the functional potential of non-coding sequence and the effects of identified mutations on reporter activity.
A second example illustrating the potential influence of cis-acting sequences in human craniofacial pathology involves a locus at 8q24, which appears to be in a gene desert, and is associated with increased risk of human CL/P (Birnbaum et al., 2009; Ludwig et al., 2012). In an elegant series of genome manipulation analyses targeting the syntenic region of mouse chromosome 5, Uslu et al (Uslu et al., 2014) mapped a series of cis-acting elements that could drive LacZ reporter gene expression in the developing mouse face within this ~3Mb chromosomal interval. Deletion of a critical interval abolished LacZ expression in the face, and also decreased expression of the endogenous gene encoding Myc that lies close to this 8q24 interval. Moreover, homozygous deletion of this element resulted in mice with a low but significant incidence of CL/P as well as a more penetrant affect on post-natal skull shape in the survivors. These findings strongly suggest that the 8q24 region harbors cis-regulatory sequences controlling aspects of MYC expression in the developing human embryo and that the alteration of these sequences can result in craniofacial defects, including CL/P. In the future, it is likely that defining the importance of candidate mutations in cis-acting sequences will become a more common and important facet of human genetic analysis albeit one that may require detailed geometric morphometric analysis of resulting animal models (Attanasio et al., 2013).
Summary
In this review, we have highlighted forward and reverse genetic approaches that can be used to identify the regulatory hierarchy underlying craniofacial development. In addition, we have described recent approaches to test the importance of human mutations using various animal model systems. We further note that these studies illustrate that different animal model systems can often complement each other in analyzing the consequences of a given mutation. These types of studies are the first glimpse presaging an exciting new period in craniofacial research – one in which the identification of candidate human mutations resulting from genome-wide sequence analysis can be allied with the new gene-editing approaches and available mutant resources in animal models for the rapid expansion of our understanding and appreciation of this fascinating developmental system.
Highlights.
How animal model systems have been used to enhance our understanding of human craniofacial development
The use of animal models to test the function of candidate human disease variants
We provide a pipeline for the use of animal models in understanding human craniofacial development and disease
Acknowledgments
The authors would like to thank Dr. Tamim Shaikh for helpful discussions of human craniofacial syndromes and help with Figure 1. Work in the Artinger lab is supported by NIDCR (R01DE024034) and the Williams’ lab (NIDCR-F32DE023709, American Association of Anatomists, and Cleft Palate Foundation to EVO) and (U01DE024429 and 1R01HD081562 to TW). Finally, we would also like to acknowledge the many pertinent studies by our colleagues that due to space constraints we were unable to cover in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Developmental cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Ahlgren SC, Thakur V, Bronner-Fraser M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10476–10481. doi: 10.1073/pnas.162356199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Kaissi A, Klaushofer K, Safi H, Chehida FB, Ghachem MB, Chaabounni M, Hennekam RC. Asymmetrical skull, ptosis, hypertelorism, high nasal bridge, clefting, umbilical anomalies, and skeletal anomalies in sibs: is Carnevale syndrome a separate entity? American journal of medical genetics Part A. 2007;143:349–354. doi: 10.1002/ajmg.a.31610. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes & development. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva V, Connolly MH, Stewart-Swift C, Fraher D, Burt J, Cardarelli J, Yelick PC. Identification of adult mineralized tissue zebrafish mutants. Genesis. 2011;49:360–366. doi: 10.1002/dvg.20712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, Shao Y, Cho V, Kirk M, Singh M, Xia Y, Hager J, Winslade S, Sjollema G, Beutler B, Enders A, Goodnow CC. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open biology. 2012;2:120061. doi: 10.1098/rsob.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antin PB, Yatskievych TA, Davey S, Darnell DK. GEISHA: an evolving gene expression resource for the chicken embryo. Nucleic acids research. 2014;42:D933–937. doi: 10.1093/nar/gkt962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Banjo T, Okamoto N, Mizuno S, Kurosawa K, Ogata T, Takada F, Yano M, Ando T, Hoshika T, Barnett C, Ohashi H, Kawame H, Hasegawa T, Okutani T, Nagashima T, Hasegawa S, Funayama R, Nagashima T, Nakayama K, Inoue S, Watanabe Y, Ogura T, Matsubara Y. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. American journal of human genetics. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Ashe A, Butterfield NC, Town L, Courtney AD, Cooper AN, Ferguson C, Barry R, Olsson F, Liem KF, Jr, Parton RG, Wainwright BJ, Anderson KV, Whitelaw E, Wicking C. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Human molecular genetics. 2012;21:1808–1823. doi: 10.1093/hmg/ddr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA, Visel A. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric I, Perry MJ, Dear TN, Rodrigues Da Costa A, Salopek D, Marusic A, Hough T, Wells S, Hunter AJ, Cheeseman M, Brown SD. An ENU-induced mutation in the Ankrd11 gene results in an osteopenia-like phenotype in the mouse mutant Yoda. Physiol Genomics. 2008;32:311–321. doi: 10.1152/physiolgenomics.00116.2007. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 2004;131:1463–1477. doi: 10.1242/dev.01033. [DOI] [PubMed] [Google Scholar]
- Barron MJ, Brookes SJ, Draper CE, Garrod D, Kirkham J, Shore RC, Dixon MJ. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Human molecular genetics. 2008;17:3509–3520. doi: 10.1093/hmg/ddn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Gordon CT, Foster RG, Melin L, Abadie V, Baujat G, Vazquez MP, Amiel J, Lyonnet S, Heyningen V, Kleinjan DA. Functional assessment of disease-associated regulatory variants in vivo using a versatile dual colour transgenesis strategy in zebrafish. PLoS genetics. 2015;11:e1005193. doi: 10.1371/journal.pgen.1005193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee V, Mukhopadhyay P, Singh S, Johnson C, Philipose JT, Warner CP, Greene RM, Pisano MM. Neural crest and mesoderm lineage-dependent gene expression in orofacial development. Differentiation; research in biological diversity. 2007;75:463–477. doi: 10.1111/j.1432-0436.2006.00145.x. [DOI] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, Mangold E. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature genetics. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Human molecular genetics. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C. Morpholinos: Antisense and Sensibility. Developmental cell. 2015;35:145–149. doi: 10.1016/j.devcel.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, Nelson SF, Ababneh OH, Uz E, Guran T, Kayserili H, Hamamy H, Reversade B. Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nat Genet. 2012;44:709–713. doi: 10.1038/ng.2259. [DOI] [PubMed] [Google Scholar]
- Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, Perrin-Schmitt F. The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Hum Mol Genet. 1998;7:945–957. doi: 10.1093/hmg/7.6.945. [DOI] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, Bushell W, Collins FS, Desaintes C, Doe B, Economides A, Eppig JT, Finnell RH, Fletcher C, Fray M, Frendewey D, Friedel RH, Grosveld FG, Hansen J, Herault Y, Hicks G, Horlein A, Houghton R, Hrabe de Angelis M, Huylebroeck D, Iyer V, de Jong PJ, Kadin JA, Kaloff C, Kennedy K, Koutsourakis M, Lloyd KC, Marschall S, Mason J, McKerlie C, McLeod MP, von Melchner H, Moore M, Mujica AO, Nagy A, Nefedov M, Nutter LM, Pavlovic G, Peterson JL, Pollock J, Ramirez-Solis R, Rancourt DE, Raspa M, Remacle JE, Ringwald M, Rosen B, Rosenthal N, Rossant J, Ruiz Noppinger P, Ryder E, Schick JZ, Schnutgen F, Schofield P, Seisenberger C, Selloum M, Simpson EM, Skarnes WC, Smedley D, Stanford WL, Stewart AF, Stone K, Swan K, Tadepally H, Teboul L, Tocchini-Valentini GP, Valenzuela D, West AP, Yamamura K, Yoshinaga Y, Wurst W. The mammalian gene function resource: the International Knockout Mouse Consortium. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley JF, Borromeo C, Clarkson M, Cox TC, Cunningham MJ, Detwiler LT, Heike CL, Hochheiser H, Mejino JL, Travillian RS, Shapiro LG. The ontology of craniofacial development and malformation for translational craniofacial research. American journal of medical genetics Part C, Seminars in medical genetics. 2013a;163C:232–245. doi: 10.1002/ajmg.c.31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley JF, Mejino JL, Detwiler LT, Travillian RS, Clarkson M, Cox T, Heike C, Cunningham M, Hochheiser H, Shapiro LG. Towards understanding craniofacial abnormalities: the ontology of craniofacial development and malformation. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2013b;2013:20. [PubMed] [Google Scholar]
- Bronner ME, LeDouarin NM. Development and evolution of the neural crest: an overview. Developmental biology. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mammalian genome : official journal of the International Mammalian Genome Society. 2012a;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Disease models & mechanisms. 2012b;5:289–292. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Potter AS, Distasio A, Dexheimer P, Plassard A, Aronow BJ, Potter SS. A gene expression atlas of early craniofacial development. Developmental biology. 2014;391:133–146. doi: 10.1016/j.ydbio.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick AB, Bixler D, Puckett CL. Genetic analysis in families with van der Woude syndrome. Journal of craniofacial genetics and developmental biology. 1985;5:181–208. [PubMed] [Google Scholar]
- Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Lan Y, Jiang R. The cleft lip and palate defects in Dancer mutant mice result from gain of function of the Tbx10 gene. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7022–7027. doi: 10.1073/pnas.0401025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton P, Davey MG, Paton IR, Morrice DR, Francis-West PH, Burt DW, Tickle C. Craniofacial development in the talpid3 chicken mutant. Differentiation; research in biological diversity. 2004;72:348–362. doi: 10.1111/j.1432-0436.2004.07207006.x. [DOI] [PubMed] [Google Scholar]
- Caruana G, Farlie PG, Hart AH, Bagheri-Fam S, Wallace MJ, Dobbie MS, Gordon CT, Miller KA, Whittle B, Abud HE, Arkell RM, Cole TJ, Harley VR, Smyth IM, Bertram JF. Genome-wide ENU mutagenesis in combination with high density SNP analysis and exome sequencing provides rapid identification of novel mouse models of developmental disease. PloS one. 2013;8:e55429. doi: 10.1371/journal.pone.0055429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T. Phenotype-driven mouse ENU mutagenesis screens. Methods in enzymology. 2010;477:313–327. doi: 10.1016/S0076-6879(10)77016-6. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chang CF, Schock EN, O’Hare EA, Dodgson J, Cheng HH, Muir WM, Edelmann RE, Delany ME, Brugmann SA. The cellular and molecular etiology of the craniofacial defects in the avian ciliopathic mutant talpid2. Development. 2014;141:3003–3012. doi: 10.1242/dev.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HA, Medina-Ruiz S, Harland RM. Sp8 regulates inner ear development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6329–6334. doi: 10.1073/pnas.1319301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA. Cranial neural crest cells on the move: their roles in craniofacial development. American journal of medical genetics Part A. 2011;155A:270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Chatterjee B, Francis D, Yu Q, SanAgustin JT, Francis R, Tansey T, Henry C, Wang B, Lemley B, Pazour GJ, Lo CW. Disruption of Mks1 localization to the mother centriole causes cilia defects and developmental malformations in Meckel-Gruber syndrome. Disease models & mechanisms. 2011;4:43–56. doi: 10.1242/dmm.006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, Houston DW, Manak JR, Schutte BC, Wagner DS, Cornell RA. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. The Journal of investigative dermatology. 2013;133:68–77. doi: 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany ME. Genetic variants for chick biology research: from breeds to mutants. Mechanisms of development. 2004;121:1169–1177. doi: 10.1016/j.mod.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Dennis JF, Kurosaka H, Iulianella A, Pace J, Thomas N, Beckham S, Williams T, Trainor PA. Mutations in Hedgehog acyltransferase (Hhat) perturb Hedgehog signaling, resulting in severe acrania-holoprosencephaly-agnathia craniofacial defects. PLoS genetics. 2012;8:e1002927. doi: 10.1371/journal.pgen.1002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nature reviews Genetics. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmoto A, Shimizu K, Asada Y, Maeda T. Quantitative trait loci on chromosomes 10 and 11 influencing mandible size of SMXA RI mouse strains. Journal of dental research. 2002;81:501–504. doi: 10.1177/154405910208100714. [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Developmental cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM, Kimmel CB. FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC developmental biology. 2013;13:23. doi: 10.1186/1471-213X-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE Mouse Genome Database G. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic acids research. 2015;43:D726–736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Choi I, Clouthier DE, Niswander L, Williams T. The Ptch1(DL) mouse: a new model to study lambdoid craniosynostosis and basal cell nevus syndrome-associated skeletal defects. Genesis. 2013;51:677–689. doi: 10.1002/dvg.22416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Molecular and cellular biology. 2008;28:315–325. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, Phang T, Geraci M, Spritz RA, Hunter LE, Williams T. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PloS one. 2009;4:e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Halpern ME. Patterning the zebrafish axial skeleton requires early chordin function. Nature genetics. 1999;23:442–446. doi: 10.1038/70557. [DOI] [PubMed] [Google Scholar]
- Friedel RH, Seisenberger C, Kaloff C, Wurst W. EUCOMM--the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic. 2007;6:180–185. doi: 10.1093/bfgp/elm022. [DOI] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. American journal of human genetics. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoumid J, Drevillon L, Alavi-Naini SM, Bondurand N, Rio M, Briand-Suleau A, Nasser M, Goodwin L, Raymond P, Yanicostas C, Goossens M, Lyonnet S, Mowat D, Amiel J, Soussi-Yanicostas N, Giurgea I. ZEB2 zinc-finger missense mutations lead to hypomorphic alleles and a mild Mowat-Wilson syndrome. Hum Mol Genet. 2013;22:2652–2661. doi: 10.1093/hmg/ddt114. [DOI] [PubMed] [Google Scholar]
- Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS genetics. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Attanasio C, Bhatia S, Benko S, Ansari M, Tan TY, Munnich A, Pennacchio LA, Abadie V, Temple IK, Goldenberg A, van Heyningen V, Amiel J, FitzPatrick D, Kleinjan DA, Visel A, Lyonnet S. Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence. Hum Mutat. 2014;35:1011–1020. doi: 10.1002/humu.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Petit F, Kroisel PM, Jakobsen L, Zechi-Ceide RM, Oufadem M, Bole-Feysot C, Pruvost S, Masson C, Tores F, Hieu T, Nitschke P, Lindholm P, Pellerin P, Guion-Almeida ML, Kokitsu-Nakata NM, Vendramini-Pittoli S, Munnich A, Lyonnet S, Holder-Espinasse M, Amiel J. Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. American journal of human genetics. 2013;93:1118–1125. doi: 10.1016/j.ajhg.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Weaver KN, Zechi-Ceide RM, Madsen EC, Tavares AL, Oufadem M, Kurihara Y, Adameyko I, Picard A, Breton S, Pierrot S, Biosse-Duplan M, Voisin N, Masson C, Bole-Feysot C, Nitschke P, Delrue MA, Lacombe D, Guion-Almeida ML, Moura PP, Garib DG, Munnich A, Ernfors P, Hufnagel RB, Hopkin RJ, Kurihara H, Saal HM, Weaver DD, Katsanis N, Lyonnet S, Golzio C, Clouthier DE, Amiel J. Mutations in the endothelin receptor type A cause mandibulofacial dysostosis with alopecia. American journal of human genetics. 2015;96:519–531. doi: 10.1016/j.ajhg.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Governale LS. Craniosynostosis. Pediatr Neurol. 2015 doi: 10.1016/j.pediatrneurol.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder FB. Mutant Mouse Regional Resource Center Program: a resource for distribution of mouse models for biomedical research. Comp Med. 2002;52:203. [PubMed] [Google Scholar]
- Griffin JN, Sondalle SB, Del Viso F, Baserga SJ, Khokha MK. The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS genetics. 2015;11:e1005018. doi: 10.1371/journal.pgen.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Handschuh K, Feenstra J, Koss M, Ferretti E, Risolino M, Zewdu R, Sahai MA, Benazet JD, Peng XP, Depew MJ, Quintana L, Sharpe J, Wang B, Alcorn H, Rivi R, Butcher S, Manak JR, Vaccari T, Weinstein H, Anderson KV, Lacy E, Selleri L. ESCRT-II/Vps25 constrains digit number by endosome-mediated selective modulation of FGF-SHH signaling. Cell reports. 2014;9:674–687. doi: 10.1016/j.celrep.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Wicks MN, Davidson DR, Burger A, Ringwald M, Baldock RA. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Semantics. 2013;4:15. doi: 10.1186/2041-1480-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke K, Bowen ME, Harris MP. Identification of mutations in zebrafish using next-generation sequencing. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 7. Vol. 104. 2013a. p. 13. [DOI] [PubMed] [Google Scholar]
- Henke K, Bowen ME, Harris MP. Perspectives for identification of mutations in the zebrafish: making use of next-generation sequencing technologies for forward genetic approaches. Methods. 2013b;62:185–196. doi: 10.1016/j.ymeth.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Henken DB, Rasooly RS, Javois L, Hewitt AT National Institutes of Health Trans NIHZCC. The National Institutes of Health and the growth of the zebrafish as an experimental model organism. Zebrafish. 2004;1:105–110. doi: 10.1089/zeb.2004.1.105. [DOI] [PubMed] [Google Scholar]
- Hernandez-Porras I, Fabbiano S, Schuhmacher AJ, Aicher A, Canamero M, Camara JA, Cusso L, Desco M, Heeschen C, Mulero F, Bustelo XR, Guerra C, Barbacid M. K-RasV14I recapitulates Noonan syndrome in mice. Proc Natl Acad Sci U S A. 2014;111:16395–16400. doi: 10.1073/pnas.1418126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–497. doi: 10.1016/s0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- Hisano Y, Ota S, Kawahara A. Genome editing using artificial site-specific nucleases in zebrafish. Development, growth & differentiation. 2014;56:26–33. doi: 10.1111/dgd.12094. [DOI] [PubMed] [Google Scholar]
- Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S, Okamoto H, Yamamoto T, Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci Rep. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser H, Aronow BJ, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, Fraser SE, Hallgrimsson B, Iwata J, Klein O, Marazita ML, Murray JC, Murray S, de Villena FP, Postlethwait J, Potter S, Shapiro L, Spritz R, Visel A, Weinberg SM, Trainor PA. The FaceBase Consortium: a comprehensive program to facilitate craniofacial research. Developmental biology. 2011;355:175–182. doi: 10.1016/j.ydbio.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Verzi MP, Robinson AS, Tang PL, Hua LL, Xu SM, Kwok PY, Black BL. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Development. 2015;142:2775–2780. doi: 10.1242/dev.126391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla PH, Simoes-Costa M, Sanchez-Vasquez E, Bronner ME. DNA methyltransferase 3B regulates duration of neural crest production via repression of Sox10. Proc Natl Acad Sci U S A. 2014;111:17911–17916. doi: 10.1073/pnas.1318408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubler M, Volkert M, Kaser-Hotz B, Arnold S. Palliative irradiation of Scottish Fold osteochondrodysplasia. Veterinary radiology & ultrasound : the official journal of the American College of Veterinary Radiology and the International Veterinary Radiology Association. 2004;45:582–585. doi: 10.1111/j.1740-8261.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Irie K, Ishizaki H, Tanaka-Okamoto M, Morimoto K, Inoue E, Ohtsuka T, Miyoshi J, Takai Y. Roles of cell-adhesion molecules nectin 1 and nectin 3 in ciliary body development. Development. 2005;132:1525–1537. doi: 10.1242/dev.01697. [DOI] [PubMed] [Google Scholar]
- Jayasena CS, Bronner ME. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. The Journal of cell biology. 2012;199:453–466. doi: 10.1083/jcb.201204138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Human molecular genetics. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PloS one. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic acids research. 2015;43:D756–763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal KD, van Bokhoven H, Roscioli T, Carels CE, Zhou H. Genomic approaches for studying craniofacial disorders. American journal of medical genetics Part C, Seminars in medical genetics. 2013;163C:218–231. doi: 10.1002/ajmg.c.31379. [DOI] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nature genetics. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen CK, Easty R, Di Fenza A, Fiegel T, Grifiths M, Horne A, Karp NA, Kurbatova N, Mason JC, Matthews P, Oakley DJ, Qazi A, Regnart J, Retha A, Santos LA, Sneddon DJ, Warren J, Westerberg H, Wilson RJ, Melvin DG, Smedley D, Brown SD, Flicek P, Skarnes WC, Mallon AM, Parkinson H. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic acids research. 2014;42:D802–809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almen MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerova M, Rubin CJ, Wang C, Zamani N, Grant BR, Grant PR, Webster MT, Andersson L. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- Law SH, Sargent TD. The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PloS one. 2014;9:e100268. doi: 10.1371/journal.pone.0100268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Dupin E. The neural crest in vertebrate evolution. Current opinion in genetics & development. 2012;22:381–389. doi: 10.1016/j.gde.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Leal GF, Silva EO, Duarte AR, Campos JF. Blepharophimosis, blepharoptosis, defects of the anterior chamber of the eye, caudal appendage, radioulnar synostosis, hearing loss and umbilical anomalies in sibs: 3MC syndrome? American journal of medical genetics Part A. 2008;146A:1059–1062. doi: 10.1002/ajmg.a.32252. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Klingenberg CP, Sherratt E, Wolf JB, Cheverud JM. A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity. 2008;101:518–526. doi: 10.1038/hdy.2008.79. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Pomp D, Eisen EJ, Cheverud JM. Quantitative trait loci for directional but not fluctuating asymmetry of mandible characters in mice. Genetical research. 2000;76:27–40. doi: 10.1017/s0016672300004559. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Routman EJ, Cheverud JM. An epistatic genetic basis for fluctuating asymmetry of mandible size in mice. Evolution; international journal of organic evolution. 2002;56:642–653. doi: 10.1111/j.0014-3820.2002.tb01373.x. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Koboldt DC, Kang CJ, Ma L, Hecht JT, Wehby GL, Christensen K, Czeizel AE, Deleyiannis FW, Fulton RS, Wilson RK, Beaty TH, Schutte BC, Murray JC, Marazita ML. IRF6 mutation screening in non-syndromic orofacial clefting: analysis of 1521 families. Clin Genet. 2015a doi: 10.1111/cge.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, Carlson JC, Hetmanski JB, Wang H, Larson DE, Fulton RS, Kousa YA, Fakhouri WD, Naji A, Ruczinski I, Begum F, Parker MM, Busch T, Standley J, Rigdon J, Hecht JT, Scott AF, Wehby GL, Christensen K, Czeizel AE, Deleyiannis FW, Schutte BC, Wilson RK, Cornell RA, Lidral AC, Weinstock GM, Beaty TH, Marazita ML, Murray JC. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. American journal of human genetics. 2015b;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Liu H, Bullard SA, Bonde G, Machida J, Visel A, Uribe LM, Li X, Amendt B, Cornell RA. A single nucleotide polymorphism associated with isolated cleft lip and palate, thyroid cancer and hypothyroidism alters the activity of an oral epithelium and thyroid enhancer near FOXE1. Human molecular genetics. 2015;24:3895–3907. doi: 10.1093/hmg/ddv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KC. A knockout mouse resource for the biomedical research community. Annals of the New York Academy of Sciences. 2011;1245:24–26. doi: 10.1111/j.1749-6632.2011.06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KC, Meehan T, Beaudet A, Murray S, Svenson K, McKerlie C, West D, Morse I, Parkinson H, Brown S, Mallon AM, Moore M. Precision medicine: Look to the mice. Science. 2015;349:390. doi: 10.1126/science.349.6246.390-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, Bohmer AC, Mattheisen M, Alblas MA, Barth S, Kluck N, Lauster C, Braumann B, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Daratsianos N, Kreusch T, Murray JC, Marazita ML, Ruczinski I, Scott AF, Beaty TH, Kramer FJ, Wienker TF, Steegers-Theunissen RP, Rubini M, Mossey PA, Hoffmann P, Lange C, Cichon S, Propping P, Knapp M, Nothen MM. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nature genetics. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga AM, Navarro N, Cunningham ML, Cox TC. Quantitative trait loci affecting the 3D skull shape and size in mouse and prioritization of candidate genes in-silico. Frontiers in physiology. 2015;6:92. doi: 10.3389/fphys.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N, Eberhart JK. Gene-ethanol interactions underlying fetal alcohol spectrum disorders. Cell Mol Life Sci. 2014;71:2699–2706. doi: 10.1007/s00018-014-1578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N, Wetherill L, Lovely CB, Swartz ME, Foroud TM, Eberhart JK. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development. 2013;140:3254–3265. doi: 10.1242/dev.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH. Insertional mutation of ‘classical’ and novel genes in transgenic mice. Trends in genetics : TIG. 1992;8:341–344. doi: 10.1016/0168-9525(92)90278-c. [DOI] [PubMed] [Google Scholar]
- Melvin VS, Feng W, Hernandez-Lagunas L, Artinger KB, Williams T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:817–831. doi: 10.1002/dvdy.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller KA, Ah-Cann CJ, Welfare MF, Tan TY, Pope K, Caruana G, Freckmann ML, Savarirayan R, Bertram JF, Dobbie MS, Bateman JF, Farlie PG. Cauli: a mouse strain with an Ift140 mutation that results in a skeletal ciliopathy modelling Jeune syndrome. PLoS genetics. 2013;9:e1003746. doi: 10.1371/journal.pgen.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci U S A. 2013;110:549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Morriss-Kay GM. Derivation of the mammalian skull vault. Journal of anatomy. 2001;199:143–151. doi: 10.1046/j.1469-7580.2001.19910143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Ishii K, Pillalamarri V, Kammin T, Atkin JF, Hickey SE, Xi QJ, Gusella JF, Talkowski ME, Morton CC, Maas RL, Liao EC. Actin capping protein CAPZB regulates cell morphology, differentiation, and neural crest migration in craniofacial morphogenesis. Human molecular genetics. 2016 doi: 10.1093/hmg/ddw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P, Brock G, Pihur V, Webb C, Pisano MM, Greene RM. Developmental microRNA expression profiling of murine embryonic orofacial tissue. Birth defects research Part A, Clinical and molecular teratology. 2010;88:511–534. doi: 10.1002/bdra.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy J, Bhaskar L. Current concepts in genetics of nonsyndromic clefts. Indian journal of plastic surgery : official publication of the Association of Plastic Surgeons of India. 2009;42:68–81. doi: 10.4103/0970-0358.53004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Abdelilah S, Stainier DY, Driever W. Mutations affecting craniofacial development in zebrafish. Development. 1996;123:357–367. doi: 10.1242/dev.123.1.357. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140:2765–2775. doi: 10.1242/dev.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Amsterdam A, Hopkins N. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev Biol. 2006;6:28. doi: 10.1186/1471-213X-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. Journal of anatomy. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nature genetics. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Norris DP, Grimes DT. Mouse models of ciliopathies: the state of the art. Disease models & mechanisms. 2012;5:299–312. doi: 10.1242/dmm.009340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Developmental expression of two murine fibroblast growth factor receptors, flg and bek. Development. 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- Pan L, Shah AN, Phelps IG, Doherty D, Johnson EA, Moens CB. Rapid identification and recovery of ENU-induced mutations with next-generation sequencing and Paired-End Low-Error analysis. BMC Genomics. 2015;16:83. doi: 10.1186/s12864-015-1263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. Genesis. 2012;50:155–163. doi: 10.1002/dvg.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, Rautio J, Pegelow M, Karsten A, Basel-Vanagaite L, Gordon W, Andersen B, Svensson T, Murray JC, Cornell RA, Kere J, Schutte BC. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. American journal of human genetics. 2014;94:23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T, Ahn DG, Schilling TF, Nair S, Ruvinsky I, Geisler R, Rauch GJ, Haffter P, Zon LI, Zhou Y, Foott H, Dawid IB, Ho RK. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development. 2003;130:5043–5052. doi: 10.1242/dev.00704. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang YJ, Heisenberg CP, Beuchle D, Grandel H, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nature genetics. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J. Enhancer Divergence and cis-Regulatory Evolution in the Human and Chimp Neural Crest. Cell. 2015;163:68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Justice MJ. Mouse mutagenesis with the chemical supermutagen ENU. Methods in enzymology. 2010;477:297–312. doi: 10.1016/S0076-6879(10)77015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana AM, Geiger EA, Achilly N, Rosenblatt DS, Maclean KN, Stabler SP, Artinger KB, Appel B, Shaikh TH. Hcfc1b, a zebrafish ortholog of HCFC1, regulates craniofacial development by modulating mmachc expression. Developmental biology. 2014;396:94–106. doi: 10.1016/j.ydbio.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell stem cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nature genetics. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. High-Throughput Sequencing Technologies. Molecular cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Green GE, Park SS, Stamper BD, Gordon CT, Johnson JM, Cunniff CM, Smith JD, Emery SB, Lyonnet S, Amiel J, Holder M, Heggie AA, Bamshad MJ, Nickerson DA, Cox TC, Hing AV, Horst JA, Cunningham ML. A human homeotic transformation resulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome. American journal of human genetics. 2012;90:907–914. doi: 10.1016/j.ajhg.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M, Marazita ML, Murray JC. Impaired FGF signaling contributes to cleft lip and palate. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS genetics. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooryck C, Diaz-Font A, Osborn DP, Chabchoub E, Hernandez-Hernandez V, Shamseldin H, Kenny J, Waters A, Jenkins D, Kaissi AA, Leal GF, Dallapiccola B, Carnevale F, Bitner-Glindzicz M, Lees M, Hennekam R, Stanier P, Burns AJ, Peeters H, Alkuraya FS, Beales PL. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nature genetics. 2011;43:197–203. doi: 10.1038/ng.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Tam PPL. Mouse Development Patterning, Morphogenesis, and Organogenesis. Academic Press [Imprint] Elsevier Science & Technology Books; San Diego: 2002. p. 712. [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Ruzicka L, Bradford YM, Frazer K, Howe DG, Paddock H, Ramachandran S, Singer A, Toro S, Van Slyke CE, Eagle AE, Fashena D, Kalita P, Knight J, Mani P, Martin R, Moxon SA, Pich C, Schaper K, Shao X, Westerfield M. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabel JL, d’Alencon C, O’Brien EK, Van Otterloo E, Lutz K, Cuykendall TN, Schutte BC, Houston DW, Cornell RA. Maternal Interferon Regulatory Factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Developmental biology. 2009;325:249–262. doi: 10.1016/j.ydbio.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Iulianella A, Melton KR, Lynn M, Walker M, Inman KE, Bhatt S, Leroux-Berger M, Crawford M, Jones NC, Dennis JF, Trainor PA. A phenotype-driven ENU mutagenesis screen identifies novel alleles with functional roles in early mouse craniofacial development. Genesis. 2011;49:342–359. doi: 10.1002/dvg.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes & development. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nature reviews Neuroscience. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Piotrowski T, Grandel H, Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Hammerschmidt M, Kane DA, Mullins MC, van Eeden FJ, Kelsh RN, Furutani-Seiki M, Granato M, Haffter P, Odenthal J, Warga RM, Trowe T, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Schmitt SM, Gull M, Brandli AW. Engineering Xenopus embryos for phenotypic drug discovery screening. Advanced drug delivery reviews. 2014;69–70:225–246. doi: 10.1016/j.addr.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Ostrander EA. Insights into morphology and disease from the dog genome project. Annual review of cell and developmental biology. 2014;30:535–560. doi: 10.1146/annurev-cellbio-100913-012927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan-Rooney K, Swartz ME, Zhao F, Liu D, Eberhart JK. Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypoparathyroidism, sensorineural deafness and renal dysplasia (HDR) Disease models & mechanisms. 2013;6:1285–1291. doi: 10.1242/dmm.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, Brown-John M, Chand S, Charest D, Charters AM, Cullum R, Dhalla N, Featherstone R, Gerhard DS, Hoffman B, Holt RA, Hou J, Kuo BY, Lee LL, Lee S, Leung D, Ma K, Matsuo C, Mayo M, McDonald H, Prabhu AL, Pandoh P, Riggins GJ, de Algara TR, Rupert JL, Smailus D, Stott J, Tsai M, Varhol R, Vrljicak P, Wong D, Wu MK, Xie YY, Yang G, Zhang I, Hirst M, Jones SJ, Helgason CD, Simpson EM, Hoodless PA, Marra MA. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JMW. Essential developmental biology. 3. Wiley; Chichester, West Sussex ; Hoboken, NJ: 2013. [Google Scholar]
- Snider TN, Mishina Y. Cranial neural crest cell contribution to craniofacial formation, pathology, and future directions in tissue engineering. Birth defects research Part C, Embryo today : reviews. 2014;102:324–332. doi: 10.1002/bdrc.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Kontarakis Z, Rossi A. Making sense of anti-sense data. Developmental cell. 2015;32:7–8. doi: 10.1016/j.devcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Stanier P, Pauws E. Development of the lip and palate: FGF signalling. Frontiers of oral biology. 2012;16:71–80. doi: 10.1159/000337618. [DOI] [PubMed] [Google Scholar]
- Stottmann R, Beier D. ENU mutagenesis in the mouse. Current protocols in mouse biology. 2014;4:25–35. doi: 10.1002/9780470942390.mo140029. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Beier DR. Using ENU mutagenesis for phenotype-driven analysis of the mouse. Methods in enzymology. 2010;477:329–348. doi: 10.1016/S0076-6879(10)77017-8. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Bjork BC, Doyle JB, Beier DR. Identification of a Van der Woude syndrome mutation in the cleft palate 1 mutant mouse. Genesis. 2010;48:303–308. doi: 10.1002/dvg.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson ML, Field RA, Wheeler GN. Xenopus as a model organism in developmental chemical genetic screens. Molecular bioSystems. 2005;1:223–228. doi: 10.1039/b506103b. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Andrews BT. Facial dysostoses: Etiology, pathogenesis and management. American journal of medical genetics Part C, Seminars in medical genetics. 2013;163C:283–294. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh le A, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, Chow E, Gonzales C, Leung HY, Solomon I, Bronner-Fraser M, Megason SG, Fraser SE. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes & development. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki M, Nakane Y, Wada A. Hereditary multiple malformation in Japanese quail: a possible powerful animal model for morphogenetic studies. The Journal of heredity. 1998;89:24–31. doi: 10.1093/jhered/89.1.24. [DOI] [PubMed] [Google Scholar]
- Tucci V, Kleefstra T, Hardy A, Heise I, Maggi S, Willemsen MH, Hilton H, Esapa C, Simon M, Buenavista MT, McGuffin LJ, Vizor L, Dodero L, Tsaftaris S, Romero R, Nillesen WN, Vissers LE, Kempers MJ, Vulto-van Silfhout AT, Iqbal Z, Orlando M, Maccione A, Lassi G, Farisello P, Contestabile A, Tinarelli F, Nieus T, Raimondi A, Greco B, Cantatore D, Gasparini L, Berdondini L, Bifone A, Gozzi A, Wells S, Nolan PM. Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. The Journal of clinical investigation. 2014;124:1468–1482. doi: 10.1172/JCI70372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nature genetics. 2014;46:753–758. doi: 10.1038/ng.2971. [DOI] [PubMed] [Google Scholar]
- Varshney GK, Burgess SM. Mutagenesis and phenotyping resources in zebrafish for studying development and human disease. Briefings in functional genomics. 2014;13:82–94. doi: 10.1093/bfgp/elt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Huang H, Zhang S, Lu J, Gildea DE, Yang Z, Wolfsberg TG, Lin S, Burgess SM. The Zebrafish Insertion Collection (ZInC): a web based, searchable collection of zebrafish mutations generated by DNA insertion. Nucleic acids research. 2013a;41:D861–864. doi: 10.1093/nar/gks946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Lu J, Gildea DE, Huang H, Pei W, Yang Z, Huang SC, Schoenfeld D, Pho NH, Casero D, Hirase T, Mosbrook-Davis D, Zhang S, Jao LE, Zhang B, Woods IG, Zimmerman S, Schier AF, Wolfsberg TG, Pellegrini M, Burgess SM, Lin S. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome research. 2013b;23:727–735. doi: 10.1101/gr.151464.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, Harper U, Huang SC, Prakash A, Chen W, Sood R, Ledin J, Burgess SM. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome research. 2015 doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic acids research. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint rg: an atlas of gene expression patterns in the mouse embryo. Nucleic acids research. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Developmental cell. 2004;6:781–790. doi: 10.1016/j.devcel.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Developmental biology. 2007;304:194–207. doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao Y, Imsland F, Gu X, Feng C, Liu R, Song C, Tixier-Boichard M, Gourichon D, Li Q, Chen K, Li H, Andersson L, Hu X, Li N. The crest phenotype in chicken is associated with ectopic expression of HOXC8 in cranial skin. PloS one. 2012;7:e34012. doi: 10.1371/journal.pone.0034012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Yang F, Karim BO, Iacovelli AJ, Cai J, Lerner CP, Richtsmeier JT, Leszl JM, Hill CA, Yu K, Ornitz DM, Elisseeff J, Huso DL, Jabs EW. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development. 2005;132:3537–3548. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Niswander LA, Anderson KV. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Human molecular genetics. 2009;18:4565–4575. doi: 10.1093/hmg/ddp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KN, Watt KE, Hufnagel RB, Navajas Acedo J, Linscott LL, Sund KL, Bender PL, Konig R, Lourenco CM, Hehr U, Hopkin RJ, Lohmann DR, Trainor PA, Wieczorek D, Saal HM. Acrofacial Dysostosis, Cincinnati Type, a Mandibulofacial Dysostosis Syndrome with Limb Anomalies, Is Caused by POLR1A Dysfunction. American journal of human genetics. 2015;96:765–774. doi: 10.1016/j.ajhg.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DB, Pasumarthi RK, Baridon B, Djan E, Trainor A, Griffey SM, Engelhard EK, Rapp J, Li B, de Jong PJ, Lloyd KC. A lacZ reporter gene expression atlas for 313 adult KOMP mutant mouse lines. Genome research. 2015;25:598–607. doi: 10.1101/gr.184184.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijshake T, Baker DJ, van de Sluis B. Endonucleases: new tools to edit the mouse genome. Biochimica et biophysica acta. 2014;1842:1942–1950. doi: 10.1016/j.bbadis.2014.04.020. [DOI] [PubMed] [Google Scholar]
- Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, Cox TC, McHenry T, Narayan N, Feingold E, Wang X, Sliskovic S, Karmi N, Safra N, Sanchez C, Deleyiannis FW, Murray JC, Wade CM, Marazita ML, Bannasch DL. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS genetics. 2015;11:e1005059. doi: 10.1371/journal.pgen.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global strategies to reduce the health care burden of craniofacial anomalies: report of WHO meetings on international collaborative research on craniofacial anomalies. The Cleft palate-craniofacial journal : official publication of the American Cleft Palate-Craniofacial Association. 2004;41:238–243. doi: 10.1597/03-214.1. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charite J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yergeau DA, Kelley CM, Zhu H, Kuliyev E, Mead PE. Forward genetic screens in Xenopus using transposon-mediated insertional mutagenesis. Methods in molecular biology. 2012;917:111–127. doi: 10.1007/978-1-61779-992-1_6. [DOI] [PubMed] [Google Scholar]
- Yin L, Du X, Li C, Xu X, Chen Z, Su N, Zhao L, Qi H, Li F, Xue J, Yang J, Jin M, Deng C, Chen L. A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone. 2008;42:631–643. doi: 10.1016/j.bone.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Young NM, Wat S, Diewert VM, Browder LW, Hallgrimsson B. Comparative morphometrics of embryonic facial morphogenesis: implications for cleft-lip etiology. Anatomical record. 2007;290:123–139. doi: 10.1002/ar.20415. [DOI] [PubMed] [Google Scholar]
- Zhang D, Ighaniyan S, Stathopoulos L, Rollo B, Landman K, Hutson J, Newgreen D. The neural crest: a versatile organ system. Birth defects research Part C, Embryo today : reviews. 2014;102:275–298. doi: 10.1002/bdrc.21081. [DOI] [PubMed] [Google Scholar]
- Zhao C, Andreeva V, Gibert Y, LaBonty M, Lattanzi V, Prabhudesai S, Zhou Y, Zon L, McCann KL, Baserga S, Yelick PC. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS genetics. 2014;10:e1004074. doi: 10.1371/journal.pgen.1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]