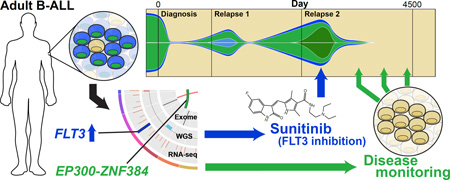
Abstract
The genomic events responsible for the pathogenesis of relapsed adult B lymphoblastic leukemia (B-ALL) are not yet clear. We performed integrative analysis of whole genome, exome, custom capture, RNA-seq, and locus-specific genomic assays across nine time points from a patient with primary de novo B-ALL. Comprehensive genome and transcriptome characterization revealed a dramatic tumor evolution during progression, yielding a tumor with complex clonal architecture at second relapse. We observed and validated point mutations in EP300 and NF1, a highly expressed EP300-ZNF384 gene fusion, a microdeletion in IKZF1, a focal deletion affecting SETD2, and large deletions affecting RB1, PAX5, NF1, and ETV6. While the genome analysis revealed events of potential biological relevance, no clinically actionable treatment options were evident at the time of the second relapse. However, transcriptome analysis identified aberrant overexpression of the targetable protein kinase encoded by the FLT3 gene. Although the patient had refractory disease after salvage therapy for the second relapse, treatment with the FLT3 inhibitor sunitinib rapidly induced a near complete molecular response, permitting the patient to proceed to a matched unrelated donor stem cell transplant. The patient remains in complete remission more than 4 years later. Analysis of this patient’s relapse genome revealed an unexpected, actionable therapeutic target that led to a specific therapy associated with a rapid clinical response. For some patients with relapsed or refractory cancers, this approach may indicate novel therapeutic interventions that could alter patient outcomes.
Graphical Abstract
INTRODUCTION
B lymphoblastic leukemia (B-ALL) is the most common acute leukemia occurring in children, but it is more rare in adults [1]. It is generally a sporadic, non-inherited disease in children, though recent genome-wide association studies have identified susceptibility loci including GATA3, ARID5B, CEBPE, ETV6, and IKZF1 [2–4]. Additional studies have implicated rare TP53 [5], IKZF1 [6] and PAX5 [7] germline mutations that can confer susceptibility to B-ALL. Early genomic studies of B-ALL using cytogenetics or array-based technologies established a high frequency of chromosomal alterations, ranging from focal copy number alterations (e.g., IKZF1) to large-scale inter-chromosomal rearrangements (e.g., BCR-ABL, ETV6-RUNX1) and genome-wide aneuploidy, which have been used to classify and risk-stratify patients [8, 9]. More recent next-generation sequencing-based approaches have identified additional recurrent mutations and fusion events involved in the pathogenesis of childhood B-ALL [10, 11]. Taken together, specific pathways have emerged as recurrently altered in B-ALL implicating impaired lymphoid development (PAX5, EBF1, IKZF1), altered cell cycle maintenance (CDKN2A, CDKN2B, RB1) and activated kinase signaling (JAK2, ABL1, IL7R, FLT3) [6]. Analysis of relapsed ALL cases has identified mutations in TP53, NT5C2 and CREBBP that could influence treatment response and prognosis [12–15]. These studies have provided valuable insights into the molecular pathogenesis of B-ALL, however, they have primarily focused on pediatric patients. The frequency of genomic alterations and corresponding tumor complexity increase with age, and treatment outcomes in adults remain distinctly inferior to those in children [16]. To date, no pilot studies or case studies have been performed to assess the clinical impact of comprehensive exome, whole genome, and RNA-seq in patients with relapsed adult B-ALL.
METHODS
Study design, sequencing, and data analysis
Bone marrow biopsy specimens and normal skin samples were obtained from a single patient at nine distinct time points spanning 4,024 days from diagnosis. The subject provided written informed consent on a form that contained specific language authorizing whole genome sequencing and data sharing. All genomic testing was done in a research environment, since no CLIA-approved facility was available at the time of relapse (mid 2011) to create or validate results. However, the consent document allowed for the return of results to the patient and treating physician if they were thought to be clinically actionable. This consent procedure was approved by the Washington University School of Medicine Human Research Protection Office (HRPO) on 10/23/06 and renewed annually thereafter. Since the second relapse sample occurred after an allograft from a sibling donor, we needed to flow enrich tumor cells from the “blast gate” to reduce contamination from the allograft donor, which would have confounded the sequencing results (Supplementary Methods). Whole genome, exome and transcriptome sequencing (RNA-seq) were performed using library construction and capture hybridization methods previously described [17, 18]. Alignment of whole genome and exome data was performed using the Genome Modeling System (GMS) essentially as described in Griffith et al. [18]. Germline and somatic variant detection was performed using multiple tools for each type of variant, including single nucleotide variants, small insertions and deletions, large amplifications and deletions, and structural variants. Validation of somatic variants and deep sequencing assessment at all time points was performed by custom hybrid capture sequencing using a NimbleGen (Roche) probe design. This reagent targeted 5,628 regions of interest identified by whole genome sequencing of the patient’s second relapse. Analysis of RNA-seq data including read alignment, estimation of transcript abundance, and fusion detection were conducted essentially as previously described [19].
RESULTS
Case report
A 25-year-old white male presented with a two-month history of fatigue, bone pain, night sweats and frequent upper respiratory infections. There was no family history of cancer or leukemia. A complete blood count revealed a white blood cell count of 2,700 cells/microliter, a hemoglobin level of 9.8 g/dl, and a platelet count of 269,000/microliter. A bone marrow biopsy showed >95% cellular marrow. The aspirate revealed >95% blasts. Flow cytometry was notable for co-expression of CD10/19/33/34/79a, HLA-DR and TdT, with absent expression of myeloperoxidase. CD20 was negative. A diagnosis of acute B Lymphoblastic Leukemia was made. Cytogenetics showed a 46,XY,del(12)(p13) karyotype in 4/25 metaphases (Figure S1A). FISH for KMT2A (MLL) deletions, ETV6-RUNX1 (TEL/AML) fusion, and BCR-ABL1 fusion were all negative. Cerebrospinal fluid (CSF) analysis demonstrated no evidence of central nervous system involvement. He was treated with a multidrug induction regimen according to CALGB protocol 8811, which yielded a complete remission was documented. He went on to receive post-remission intensification, and subsequently completed maintenance chemotherapy for a total of two years of chemotherapy. Central nervous system prophylaxis consisted of 8 doses of intrathecal chemotherapy with methotrexate.
His first relapse was documented in the bone marrow 64 months after the initial diagnosis, when the patient presented with bone pain and fatigue. CSF studies were again negative. The blast immunophenotype was similar to his initial presentation, with the addition of partial (dim) CD52 expression. Cytogenetics showed a 46,XY karyotype in 20/20 metaphases. He was treated with hyper-CVAD chemotherapy for 4 cycles (B → A → B → A) and intrathecal chemoprophylaxis (4 doses of methotrexate), and subsequently underwent matched (12/12) sibling donor bone marrow transplantation while in a second complete remission, following conditioning with total body irradiation (1,320 cGy), etoposide (30 mg/kg), and high dose cyclophosphamide (120 mg/kg). He was treated with standard methotrexate and tacrolimus for GVHD prophylaxis. The post-transplant course was complicated by acute skin GVHD, successfully treated with corticosteroids and etanercept. Subsequently, his tacrolimus was weaned, and the patient was maintained on sirolimus for two years post-transplant.
The patient’s second relapse was documented in the bone marrow 102 months after initial diagnosis, when the patient presented with mild pancytopenia and bone pain; CSF studies were negative for malignant cells. The immunophenotype was again positive for CD10 (partial)/19/34/79a. CD20 and TdT were negative. CD33 and CD52 were not tested. Cytogenetics demonstrated the 46,XY,del(12)(p13) lesion in 5/20 metaphases (Figure S1B). FISH with ETV6 break apart probes revealed a 12p deletion in 92/200 nuclei. ETV6-RUNX1 (TEL/AML1) FISH using break-apart probes suggested that this deletion did not correspond to an ETV6-RUNX1 (TEL-AML1) fusion (Figure S1C). He was treated with ifosfamide and etoposide with concurrent filgrastim, followed by a donor lymphocyte infusion, and four doses of intrathecal methotrexate. He developed severe pancytopenia, and developed grade IV enteritis complicated by sepsis. A repeat marrow examination one month after recovery from salvage revealed 7% lymphoblasts (based on both morphology and flow cytometry), and 3/20 metaphases were positive for del(12)(p13). FISH with the ETV6 break-apart probes revealed a 12p deletion in 21/200 nuclei, consistent with persistent, refractory disease. Molecular chimerism studies demonstrated 81% donor and 19% recipient cells. He was started on a low intensity maintenance regimen of 6-mercaptopurine, sirolimus, weekly methotrexate, monthly vincristine (only one dose was administered), and dexamethasone (days 1–4/28). No other viable treatment options were available, and the patient experienced clinical deterioration with progressive pancytopenia, transfusion dependence, and fatigue.
Genome analysis
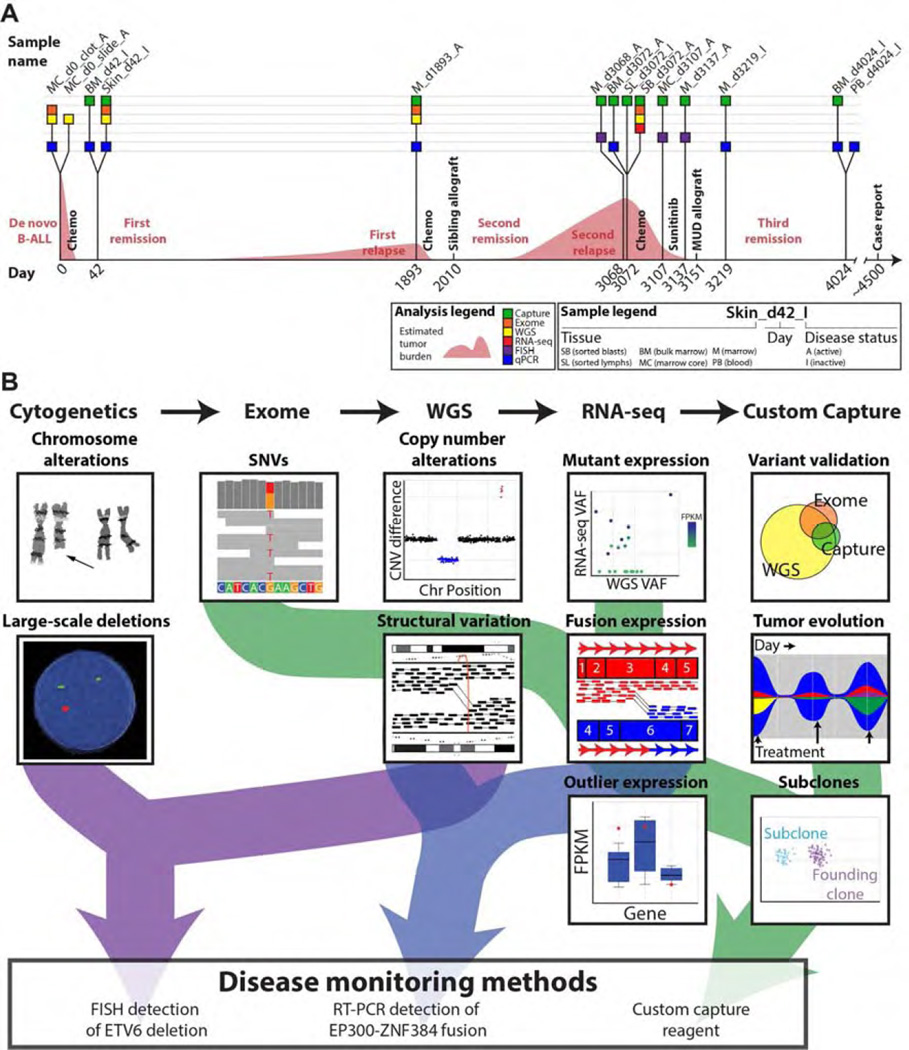
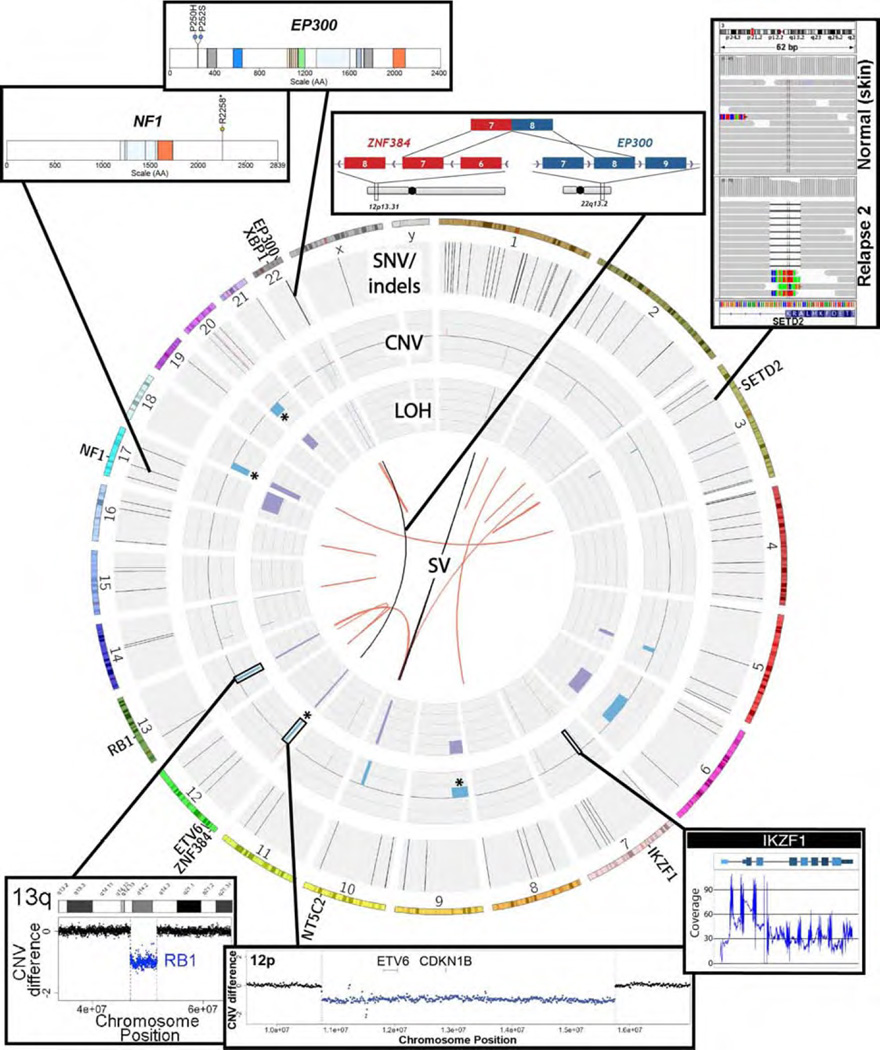
The patient (hereafter referred to as ALL1) was consented for whole genome sequencing on an IRB-approved protocol. We performed a combination of genotype microarray analysis, expression microarray analysis, whole genome sequencing (WGS), whole exome sequencing, transcriptome sequencing, custom capture sequencing, qPCR, and quantitative interphase FISH. Although the immediate analysis focused on the second relapse sample, we were able to ultimately obtain 18 samples from nine time points throughout disease progression to better understand the evolution of this tumor over time (Figure 1A, Table S1). The specific advantages of each technology were leveraged to create a comprehensive combinatorial analysis of the tumor, leading to the development of custom disease monitoring methods (Figure 1B, Figure S2–S12, Table S2–S4). Analysis began with the search for clinically relevant somatic mutations in the whole genome and exome data of the second relapse. Variant candidates identified in these data were used to design a custom capture reagent (Methods). This capture reagent was used to perform deep (~800–900x) validation sequencing for variants discovered by WGS or exome, and later, to assess the presence of each variant at additional samples/time points not previously sequenced (Table S2). We identified events that persisted throughout the course of the disease, suggesting that they were present in the original founding clone. We also observed variants that were lost during disease progression, and variants that were acquired or enriched during progression (Figure S11–S12). Based on these observations, we inferred that at least six distinct subclones existed at some point during the disease. While the data obtained were insufficient to create a complete model of clonal evolution [17], we have sufficient data to conclude that a dramatic shift in clonal architecture occurred between the initial presentation and the second relapse. Potentially relevant somatic variants were identified in the second relapse by integrated analysis of all data using multiple algorithms for detection, annotation, and visualization of each type of variant (Table S3 and Methods). All notable somatic variants are summarized in Table 1 and extensive additional details of the genome analysis are provided in the Supplementary Results and Figures S13–S40, and Tables S3–S7. Germline variant analysis was also performed, but did not reveal any established candidates for susceptibility to B-ALL. While the genome analysis led to the identification of several events relevant to the biology of this tumor, none suggested specific treatment strategies once the patient was determined to have refractory disease following salvage therapy.
Figure 1. Study overview.
(A) Timeline depicting sample collection during the course of disease treatment and progression. Each sample profiled by one of five approaches is indicated as a box placed along a timeline representing the patient’s disease course. Time points are indicated in days with day 0 representing the day of B-ALL diagnosis. Coloring of each box depicts the type of molecular profiling applied to that sample. Tissue details are encoded in the abbreviated sample names. Use of ‘A’ or ‘I’ in sample names indicates expected disease status (Active or Inactive). The most comprehensively profiled sample used for much of the mutation discovery described in this work was ‘SB_d3072_A’, a sorted blast sample (CD45−/CD19+/CD34+) obtained from marrow at the time of second relapse on day 3,072. At the time of publication the patient had achieved a complete response and has been disease free for ~4 years. For additional details on each sample and data type generated, refer to Table S1. (B) Overview of the integrated analysis approach used in this case along with simplified depictions of key analysis activities.
Table 1.
Notable variants identified in the second relapse
| Variant | Variant info | Description |
|---|---|---|
| EP300 p.P250H | Missense mutation in third exon (NM_001429). 22:41521887(C/A) |
This mutation was detected in WGS, exome, and custom capture data and was validated by cDNA cloning and Sanger sequencing. This mutation is observed at high VAF in the primary as well as relapse 1 and relapse 2. The high VAF suggests that this mutation is heterozygous and is an early/initiating event present in all tumor cells. Expression of this mutation was observed in the RNA-seq data for relapse 2. |
| EP300 p.P252S | Missense mutation in third exon (NM_001429). 22:41521892(C/T) |
The features of this mutation very closely mirror those of the p.P250H mutation described above. Both EP300 mutations are in linkage with each other and with the EP300-ZNF384 fusion event described below. |
| NF1 p.R2258* | Nonsense mutation in the 45th exon (NM_001042492). 17:29665110(C/T) |
This mutation is predicted to result in loss of function by introduction of a stop codon. This position falls within an ‘Armadillo-type fold domain’ (IPR016024). Observed at 65% VAF in relapse 2, this variant is heterozygous but a subclone exhibits a copy neutral LOH event leading to >50% VAF (see NF1 LOH event below). This mutation is not detected in the primary and appears to have been acquired (or selected from a rare subclone) between primary and relapse 1. Expression of this mutation was observed in the RNA-seq data for relapse 2. |
| NF1 LOH | 17q copy neutral LOH resulting in increased frequency of NF1 p.R2258*. 17:26000000–81165572 |
The q arm of chromosome 17 exhibits copy neutral loss of heterozygosity affecting the NF1 locus as well as many others. Based on the magnitude of LOH observed in the WGS data, this LOH event is likely subclonal, affecting ~30% of cells in the second relapse. RNA-seq data shows preferential expression of the mutant copy of NF1 (p.R2258*). |
| SETD2 p.R2510fs | 14 bp deletion affecting the 20th exon (NM_014159). 3:47059121–47059134 (GGAGTACCTTGCGA/-) |
This mutation is a predicted to result in loss of function by introducing a frameshift. The 14 bp deletion spans the donor splice site of SETD2 exon 20. This position falls within an ‘SRI, Set2 Rpb1 interacting’ domain (IPR013257). This mutation is only observed in relapse 2 and was not detected in primary or relapse 1. |
|
SETD2 focal deletion (3p21.31) |
2.36 Mb deletion. 3:45750000–48110000 |
A deletion affecting SETD2 was observed by CNV analysis of WGS coverage data. LOH is observed in heterozygous SNPs from the region. Both CNV and LOH analyses suggest that this event is subclonal in relapse 2. |
|
IKZF1 (Ikaros) focal deletion |
80 kb deletion affecting the last 5 exons of IKZF1. 7:50400000–50480000 |
This mutation is predicted to result in loss of function by deleting 5 of 8 exons of IKZF1. WGS CNV and LOH analyses suggest that this deletion is a heterozygous loss present in all tumor cells in primary and second relapse. |
|
RB1 large deletion (13q14.13-13q14.3) |
4.80 Mb deletion. 13:46900000–51700000 |
This mutation is predicted to result in loss of function by deletion of the entire RB1 locus. WGS CNV and LOH analyses suggest that this deletion is a heterozygous loss present in all tumor cells at relapse 2. |
|
ETV6 large deletion (12p13.2-12p12.3) |
4.99 Mb deletion. Affecting ETV6 (TEL) and CDKN1B. 12:10760000–15750000 |
This mutation is predicted to result in loss of function by deletion of the entire ETV6 locus. WGS CNV and LOH analyses suggest that this deletion is a heterozygous loss present in all tumor cells at relapse 2. |
|
10q24.1-10q24.33 large complex deletion |
7.82 Mb complex deletion. Affecting TLX1, NFKB2, SUFU, and NT5C2. 10:97420000–105240000 |
This region contains complex deletions and rearrangements with multiple breakpoints. Some portions of the region appear to exhibit heterozygous loss while others exhibit homozygous loss. These events appear to be present in all tumor cells at relapse 2. This region also results in a SUFU-TBX19 translocation and ADCY10- CC2D2B translocation. |
| 9p deletion (PAX5) | 38.50 Mb deletion. 9:200000–38700000 |
This large deletion affects multiple cancer genes including some tumor suppressors. Both CNV and LOH analyses suggest that this event is subclonal in relapse 2. Cancer genes affected*: CD274, CDKN2A, FANCG, JAK2, MLLT3, NFIB, PAX5, PDCD1LG2, and PSIP1. |
|
EP300-ZNF384 fusion |
Interchromosomal translocation t(12;22)(p13;q13). Breakpoints: 22:41535555–41536555 12:6784786–6785786 |
A DNA translocation within the introns of EP300 and ZNF384 results in a fusion gene that was confirmed as expressed in relapse 2 by RNA-seq. The fusion gene cDNA sequence was validated by full length cloning and Sanger sequencing. qPCR established this fusion as an early initiating event present in the dominant clone of primary, relapse 1 and relapse 2 disease. Full length cDNA sequencing confirmed that the open reading frame is maintained. A fusion protein consisting of the C- terminal and N-terminal portions of EP300 and ZNF384 is predicted. The fusion sequence also contained both EP300 missense mutations described above. |
|
FLT3 overexpression |
Aberrantly high RNA expression. 13:28577410–28674729 |
High expression of FLT3 was observed within the RNA- seq data of relapse 2. Comparison to additional RNA-seq data from sorted blood lineage cells from healthy donors also suggested high expression in the patient. Comparisons to microarray data from ~200 B-ALLs suggested that FLT3 expression was also high compared to other tumors. |
| XBP1 focal deletion | 13 kb deletion affecting the 1st exon of XBP1. 22:29195159–29207958 |
This focal deletion was identified by SV analysis with apparent read support in WGS and exome data. Competitive realignment provided evidence that this deletion is heterozygous and somatic as no deletion- spanning reads aligned to the reference allele. |
Visualizations and additional supporting data are available for all notable events in the Supplementary Materials (Figure S13–S40, Table S3–S7).
Cancer genes defined by the Cancer Gene Consensus.
Transcriptome analysis
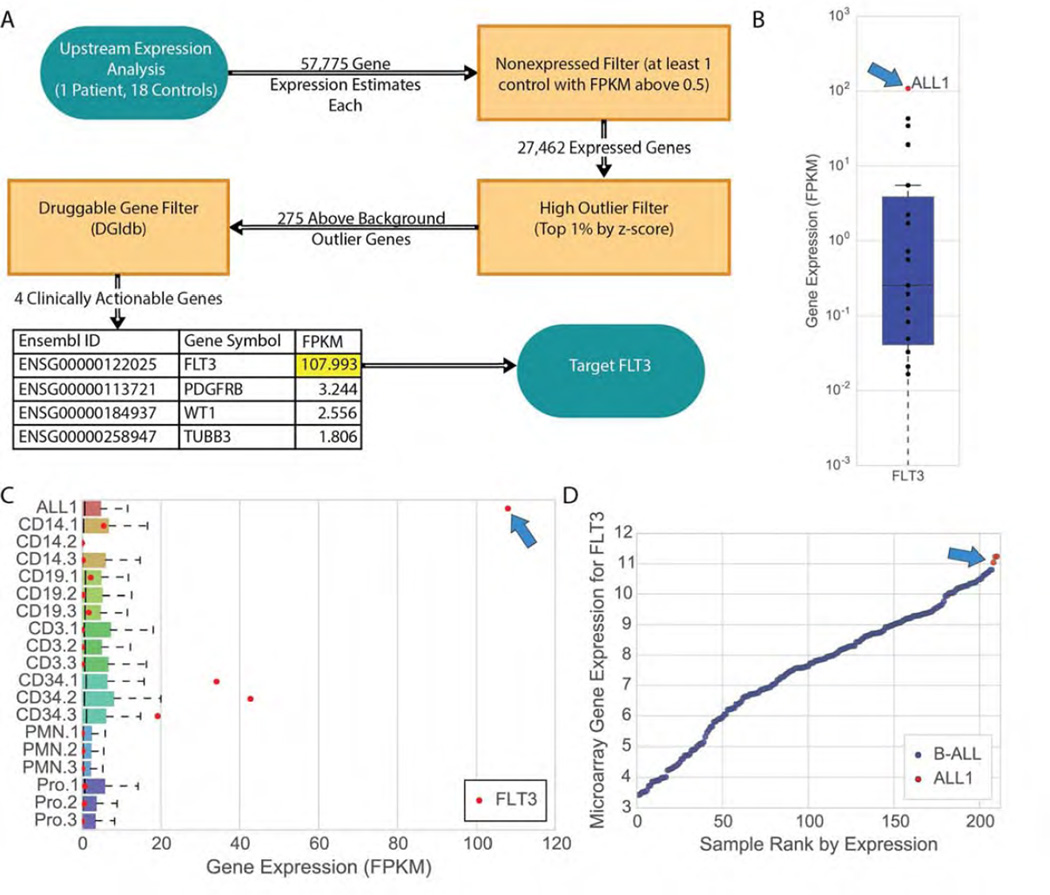
Several transcriptome analyses and methods for integration of RNA-seq data with WGS and exome data were developed during this study (Supplementary Materials). Comparison of expression data from the second relapse to several B-ALL sample cohorts was used to identify possible outlier expression of genes (Methods, Figure 3). Sample cohorts for comparison consisted of sorted blood cells from healthy donors, and 207 additional B-ALL tumors from pediatric patients. Using predictive analysis of microarrays and outlier clustering of microarray gene expression data performed in triplicate, this case was not “Ph-like”, but clustered strongly with the “R5” subset of ALL samples [10, 20, 21]. To identify expression outliers, a multiple-step filter approach was taken, selecting for differentially expressed, clinically actionable, and druggable genes using a combination of statistical thresholds and the Drug-Gene Interaction database [22, 23] (Methods). The fms-related tyrosine kinase, FLT3, had an estimated transcript abundance that was several orders of magnitude above the other three druggable candidates that were identified (PDGFRB, WT1, and TUBB3; Figure 3A). Evaluation of the RNA expression estimates of FLT3 in ALL1 and controls revealed an aberrant overexpression of this gene in the second relapse sample of ALL1 (Figure 3B). FLT3 was highly expressed in the second relapse in both the absolute sense (i.e. when compared to all genes expressed in that tumor, see ‘ALL1’ in Figure 3C), as well as in the relative sense (i.e. when compared to other B-ALL samples; Figure 3D). In the second relapse sample, FLT3 had an estimated expression level (FPKM of 108.0) that placed it within the top 0.51% of all genes (Table S5). FLT3 expression in the second relapse was also an outlier compared to blood cell types sorted from healthy donors, including hematopoietic stem and progenitor-enriched CD34+ fractions (Figure 3B, 3C) as well as additional B-ALL samples from Kang et al. 2010 (Figure 3D) [24]. The overexpression of FLT3 was confirmed orthogonally by evaluation on an exon array platform (in triplicate), and was compared directly to exon array data from other studies (Methods). Finally, immunohistochemical analysis of the bone marrow biopsy obtained at the second relapse confirmed high level expression of the FLT3 protein (CD135) in lymphoblasts (Figure S41).
Figure 3. Transcriptome analysis of second relapse reveals aberrant expression of FLT3.
(A) Analysis of the ALL1 transcriptome compared to sorted blood cells from 18 healthy individuals (Methods) revealed FLT3 as an important and clinically actionable target. (B) Evaluation of the expression levels of FLT3 in ALL1 versus the healthy control-derived blood cells reflects a difference of several orders of magnitude. (C) FLT3 expression is an outlier across all expressed genes in ALL1, a statistical anomaly otherwise observed only in hematopoietic progenitor (CD34+) cells, and at a magnitude several times higher than in those cells. Legend for control samples: CD14+ (monocytes), CD19+ (B-cells; CD33−/CD19+), CD3+ (T-cells; CD33−/CD3+), CD34+ (hematopoietic progenitors), PMN (neutrophils; CD33−/CD15+/CD16+), and Pro (promyelocytes; CD14−/CD15+/CD16low/−). (D) Microarray evaluation of ALL1 and 207 childhood B-ALL samples reveals that FLT3 is more highly expressed in ALL1 than in any other B-ALL sample in the pediatric study. The three red points shown for ALL1 represent technical replicates run on the same microarray expression platform as the 207 B-ALLs (Affymetrix Human Genome U133 Plus 2.0 Array, Methods).
We investigated whether cis-acting regulatory mutations might be increasing FLT3 transcription levels (Supplementary Materials). To test this, we made use of a heterozygous C/T single nucleotide polymorphism (SNP) in exon 2 (Methods). We found that at each time point—diagnosis, first relapse, and second relapse—the relative contribution of each allele to total FLT3 mRNA was equivalent, suggesting that cis-acting mutations did cause FLT3 overexpression. These data indicate that both FLT3 alleles were massively overexpressed in the second relapse sample, suggesting that the gene was activated in trans by an unknown mechanism. PAX5 is a negative regulator of FLT3 expression in mice [25, 26], and its heterozygous deletion in this tumor (Table 1, Figure S21) could potentially be related to the increased expression of FLT3.
Clinical course after sunitinib therapy was initiated
The initial sequencing of this case took five weeks from receipt of the blast cell DNA sample to return of a detailed report summarizing the whole genome, exome, and transcriptome results (day 3,107, Figure 4A). Based on the observation of aberrant FLT3 expression, and a lack of viable alternative therapeutic options, the patient was offered sunitinib on a compassionate need basis, because of it ability to inhibit signaling from the wild type FLT3 receptor. Sunitinib is a receptor tyrosine kinase inhibitor approved by the FDA for treatment of renal cell carcinoma. In addition to FLT3, sunitinib is reported to inhibit KIT, KDR, FLT4, PDGFRB, FLT1, PDGFRA, and RET [22, 23]. While it is possible that the “off-target” activity of this drug could have been relevant for this tumor, the expression of FLT3 (FPKM of 108.0) was substantially higher than any of the other known targets of the inhibitor. Of these seven additional targets, only two are expressed above background levels: PDGFRB (FPKM of 3.2) and FLT4 (FPKM of 2.8) (Table S5).
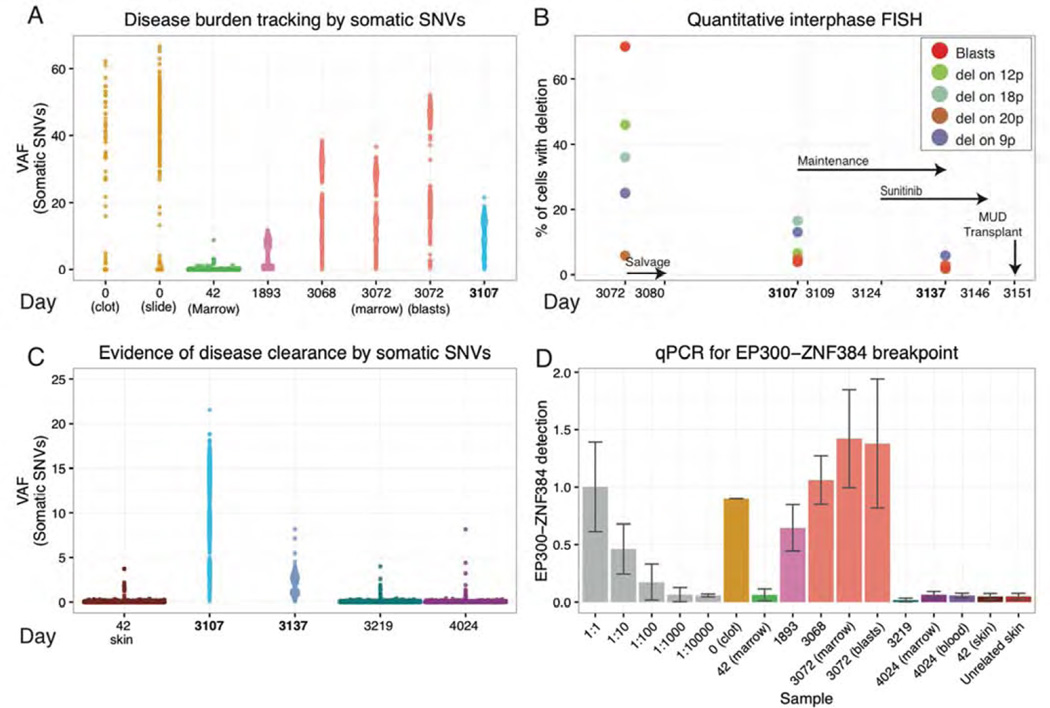
Figure 4. Personalized genomic assays for assessment of tumor burden and response to treatment.
Disease burden and evidence for disease clearance prior to the final MUD allograft were assessed by three orthogonal approaches: VAFs from deep custom capture sequencing for validated somatic SNVs, quantitative FISH for four somatic large-scale deletions, and qPCR of a somatic EP300-ZNF384 translocation breakpoint found to be a possible early/initiating event in this tumor. Time points assayed range from the day of initial diagnosis (day 0) to the patient’s most recent follow-up biopsy obtained ~2.5 years into his current sustained remission (day 4,024). (A) VAFs of 1,588 somatic variants are displayed for eight samples across six time points from diagnosis of the primary B-ALL (day 0), first relapse (day 1,893), and second relapse (day 3,068/3,072). A 40× coverage filter was applied to the primary samples further limiting variants to approximately 581 and 270 for the slide and clot, respectively (Methods). (B) Results from quantitative FISH (Methods) are displayed for four structural deletions utilized to assess tumor burden at the second relapse and following weeks. Refer to Table S3 for details on each deletion assayed. Key treatment time points are indicated with black arrows. (C) VAFs of somatic variants used to assess disease clearance at day 3,219 and day 4,024 are contrasted with their VAFs during refractory disease post salvage therapy (day 3,107) and in the normal skin sample obtained during the first remission (day 42). The sample from day 3,137 was obtained 13 days after initiation of sunitinib, and demonstrates a highly significant reduction in average VAFs compared to that of day 3,107 (p-value < 2.2e-16; n=1,588; Wilcox signed-rank test). (D) qPCR data representing abundance of the EP300-ZNF384 breakpoint in genomic DNA is displayed for a dilution series of the day 3,072 relapse sample with an unrelated skin sample (grey), six disease samples designated by the day of collection, three samples obtained after the MUD transplant, a matched skin normal obtained in first remission (day 42), and a skin sample from an unrelated donor who did not have this translocation (used as a reference to measure assay “noise”) Note the non-linear, square root transformed values used for display purposes (Methods).
Sunitinib administration (50 mg daily) began on day 3,124. Within 3 days, the patient had a notable improvement in his sense of well-being. Quantitative interphase FISH for large somatic deletions, morphology (blast counts), and deep sequencing of somatic SNVs were used to track response to this therapy. By day 3,137 (day 13 of 22 days of sunitinib) a significant reduction in disease burden was apparent (Figure 4B–C). At the same time point, the VAFs of somatic SNVs dropped from 11.1% at day 3,107, to 2.0% at day 3,137 (p-value < 2.2×10−16, n=1,588, paired, two-tailed Wilcoxon test) (Figure 4C). Sunitinib was continued for nine more days, and then the patient received conditioning (busulfan and fludarabine) for a second allograft from a matched unrelated donor (MUD) on day 3,151. No samples were obtained between the end of sunitinib treatment (day 3,146) and the MUD allograft. However, a bone marrow biopsy obtained at day 3,219 (68 days post-MUD allograft) showed complete tumor clearance by VAF assessment of somatic SNVs; mutation clearance persisted at day 4,024 (873 days post-MUD allograft; Figure 4C). The distribution of VAFs for somatic SNVs at days 3,219 and 4,024 were indistinguishable from the normal skin sample obtained during the first remission (day 42) (Figure 4C). VAFs of variants detected at day 3,219 and 4,024 were not significantly different from what would be expected from sequencing errors alone [17]. Finally, a DNA-based qPCR assay was developed for the EP300-ZNF384 fusion (Table 1), a possible initiating event in this tumor [27, 28]. We used this assay to monitor levels of the fusion in bone marrow, blood, and skin samples obtained at many time points during the disease (Figure 4D). This assay was quantitative, and revealed a strong correlation with the mutation burden measured by somatic SNV VAFs. Importantly, there was no statistical difference in the levels of the EP300-ZNF384 fusion detected between an unrelated skin sample, and any of the bone marrow or blood samples obtained after the MUD transplant. Flow cytometry for minimal residual disease in blood and bone marrow samples obtained at day 4,024 showed no evidence of persistent disease. Taken together, these results suggested that at day 4,024, the patient was in a complete molecular remission. The patient remains in a remission at this writing (4+ years post-MUD allograft).
All sequence data described in this publication including the whole genome, exome, RNA-seq and custom capture data have been deposited in the Database of Genotypes and Phenotypes (dbGAP) [29] (accession: phs001066.v1.p1; Table S8 for details).
DISCUSSION
In this study, we used an integrated genomic approach to evaluate a patient with refractory disease after a second relapse of adult B-ALL. The analysis suggested a novel therapy for this patient, which resulted in a clinical response that served as a bridge to definitive therapy with an unrelated donor stem cell allotransplant. The whole genome data and transcriptome data added considerably to what would have been identified with exome sequencing, or an assay panel of cancer genes. Extensive interpretation by an experienced team of analysts allowed all of these data to be rapidly mined for potential clinical relevance.
This tumor contained a large number of possible driver mutations. Potentially relevant loss-of-function events included multiple mutations involving NF1 and SETD2, as well as heterozygous losses affecting RB1, ETV6, PAX5, and IKZF1. Possible gain-of-function events included a complex mutation involving missense mutations of EP300, combined with a fusion event between EP300 and ZNF384. Although none of these mutations are therapeutic targets, they were valuable for tracking disease progression and response to treatment. Finally, a likely gain-of-function event in the form of aberrant overexpression of FLT3 suggested a druggable target, and allowed for the rational use of an inhibitor of wild type FLT3 that would not otherwise have been used in this setting. We were unable to quantify the reduction of disease burden that could be definitively attributed to sunitinib treatment or salvage chemotherapy. Similarly, while the rapid response of the tumor to sunitinib suggested the tumor might be hyperresponsive to FLT3 signaling, we can not quantify how much this response was related to FLT3 overexpression or possibly other factors that may influence the function of FLT3. While a previous report suggested that FLT3 inhibition can selectively kill childhood ALL cells with high levels of FLT3 expression [30], the use of sunitinib in an adult B-ALL case has not been previously reported. Some preliminary studies have suggested a possible relationship between FLT3 expression and response to FLT3 tyrosine kinase inhibitors in childhood ALL, and have suggested that FLT3 overexpression is associated specifically with the MLL (KMT2A) rearranged subset of childhood ALL [30–32]. Additional studies in adult ALL have reported high expression of FLT3 in MLL rearranged [33, 34] or cytogenetically normal patients [35]. We observed no evidence of rearrangements involving the MLL region in this tumor (Figure S23); however, Figure 3D includes 21 MLL rearranged pediatric B-ALL cases with lower FLT3 expression than observed in this case. Additional studies will need to be performed in order to determine whether sunitinib and the predictive value of FLT3 expression may play a role in the treatment of other relapsed adult B-ALL patients and identify which subset of patients would be most likely to respond.
Comprehensive analysis of massively parallel sequencing data from a tumor genome in ‘real time’ remains a great challenge [36]. Lack of optimization in pipelines that perform automated analysis, and a lack of tools for report generation, make a fast and clinically relevant turnaround time difficult to achieve. Even with a sophisticated analysis pipeline, many features of genomic analysis still require ad hoc approaches, manual review, and considerable effort from analysis experts. This tumor was characterized mostly by mutations that caused loss-of-function events that are generally not druggable. In some cases, these losses may lead to the activation of a pathway that may be targeted therapeutically, but algorithms that make these predictions are not yet available. Furthermore, some types of somatic alterations are still difficult to detect with current sequencing platforms and analytical approaches (e.g. indel detection from WGS/exome data, structural variant detection from WGS data, fusion detection from RNA-seq data, the identification of aberrant splicing events, and the detection of integrated viruses). Each of these limitations results in false negative results that may be biologically significant.
Despite these caveats, the comprehensive approach described for this case may serve as a guide for future clinical cancer sequencing for patients with limited or no treatment options, using integrated analysis of whole genome and transcriptome data. The data generated for this case can serve as a reference for future method development, and as an example of a successful application of these principles.
Supplementary Material
Figure 2. An overview of genomic findings in the second relapse.
Circos plot of the somatic events identified in the second relapse. The outermost ring displays an ideogram highlighting genes outlined in Table 1. The first data track identifies single nucleotide variants (black) and small insertions (red) and deletions (blue). The next innermost track displays copy number gains (red) and losses (blue) relative to the normal sample. Deletions targeted for FISH analysis are indicated with an asterisk (*). The innermost track displays loss of heterozygosity as the change in variant allele frequency from normal to tumor cells at germline heterozygous sites. The center of the ring displays structural variants identified in the DNA from two algorithms (Breakdancer = black, Manta = red).
Acknowledgments
We are grateful to the ALL1 patient for his gracious consent to share his identity and story with the world [36]. We thank Drs. Richard Harvey and Charles Mullighan for their assistance with analysis of the RNA array data. We thank the Siteman Cancer Center Flow Cytometry Core for cell sorting services. We also thank the McDonnell Genome Institute’s LIMS, Analysis Pipeline, and Systems groups for developing and maintaining the automated sequence analysis pipelines used in this work.
Funding
Supported by grants from the National Institutes of Health (NIH) National Cancer Institute (NCI) (PO1 CA101937, P30 CA91842) with support for analysis from NIH National Human Genome Research Institute (NHGRI) (U54 HG003079). Malachi Griffith was supported by a NIH NHGRI grant (K99 HG007940). Obi L. Griffith was supported by a NCI grant (K22 CA188163). Christopher A. Maher was supported by NCI grants (R21 CA185983 and R00 CA149182).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Andreu V, Roberts KG, Xu H, et al. A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood. 2015;125:680–686. doi: 10.1182/blood-2014-09-595744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 4.Moriyama T, Metzger ML, Wu G, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol. 2015;16:1659–1666. doi: 10.1016/S1470-2045(15)00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol. 2015;12:344–357. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 7.Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 10.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, Rapado I, Li Y, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hof J, Krentz S, van Schewick C, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol. 2011;29:3185–3193. doi: 10.1200/JCO.2011.34.8144. [DOI] [PubMed] [Google Scholar]
- 13.Inthal A, Zeitlhofer P, Zeginigg M, et al. CREBBP HAT domain mutations prevail in relapse cases of high hyperdiploid childhood acute lymphoblastic leukemia. Leukemia. 2012;26:1797–1803. doi: 10.1038/leu.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzoneva G, Perez-Garcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 2012;26:123–135. doi: 10.1016/j.blre.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Griffith M, Miller CA, Griffith OL, et al. Optimizing cancer genome sequencing and analysis. Cell Syst. 2015;1:210–223. doi: 10.1016/j.cels.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith M, Griffith OL, Smith SM, et al. Genome Modeling System: A Knowledge Management Platform for Genomics. PLoS Comput Biol. 2015;11:e1004274. doi: 10.1371/journal.pcbi.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith M, Walker JR, Spies NC, Ainscough BJ, Griffith OL. Informatics for RNA Sequencing: A Web Resource for Analysis on the Cloud. PLoS Comput Biol. 2015;11:e1004393. doi: 10.1371/journal.pcbi.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner AH, Coffman AC, Ainscough BJ, et al. DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res. 2016;44:D1036–D1044. doi: 10.1093/nar/gkv1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang H, Chen IM, Wilson CS, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20:933–938. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang J, Wei L, de Ridder J, et al. PAX5 is a tumor suppressor in mouse mutagenesis models of acute lymphoblastic leukemia. Blood. 2015;125:3609–3617. doi: 10.1182/blood-2015-02-626127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gocho Y, Kiyokawa N, Ichikawa H, et al. A novel recurrent EP300-ZNF384 gene fusion in B-cell precursor acute lymphoblastic leukemia. Leukemia. 2015;29:2445–2448. doi: 10.1038/leu.2015.111. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda T, Tsuzuki S, Kawazu M, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016 doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- 29.Tryka KA, Hao L, Sturcke A, et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 31.Stam RW, den Boer ML, Schneider P, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106:2484–2490. doi: 10.1182/blood-2004-09-3667. [DOI] [PubMed] [Google Scholar]
- 32.Brown P, Levis M, McIntyre E, Griesemer M, Small D. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL-rearranged ALL cells in a sequence-dependent manner. Leukemia. 2006;20:1368–1376. doi: 10.1038/sj.leu.2404277. [DOI] [PubMed] [Google Scholar]
- 33.Kohlmann A, Schoch C, Schnittger S, et al. Pediatric acute lymphoblastic leukemia (ALL) gene expression signatures classify an independent cohort of adult ALL patients. Leukemia. 2004;18:63–71. doi: 10.1038/sj.leu.2403167. [DOI] [PubMed] [Google Scholar]
- 34.Torelli GF, Guarini A, Porzia A, et al. FLT3 inhibition in t(4;11)+ adult acute lymphoid leukaemia. Br J Haematol. 2005;130:43–50. doi: 10.1111/j.1365-2141.2005.05556.x. [DOI] [PubMed] [Google Scholar]
- 35.Chiaretti S, Li X, Gentleman R, et al. Gene expression profiles of B-lineage adult acute lymphocytic leukemia reveal genetic patterns that identify lineage derivation and distinct mechanisms of transformation. Clin Cancer Res. 2005;11:7209–7219. doi: 10.1158/1078-0432.CCR-04-2165. [DOI] [PubMed] [Google Scholar]
- 36.Wartman LD. A case of me: clinical cancer sequencing and the future of precision medicine. Molecular Case Studies. 2015;1:a000349. doi: 10.1101/mcs.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.