Abstract
We report complete genome sequences of four blaNDM-1-harboring Gram-negative multidrug-resistant (MDR) isolates from Colombia. The blaNDM-1 genes were located on 193-kb Inc FIA, 178-kb Inc A/C2, and 47-kb (unknown Inc type) plasmids. Multilocus sequence typing (MLST) revealed that these isolates belong to sequence type 10 (ST10) (Escherichia coli), ST392 (Klebsiella pneumoniae), and ST322 and ST464 (Acinetobacter baumannii and Acinetobacter nosocomialis, respectively). Our analysis identified that the Inc A/C2 plasmid in E. coli contained a novel complex transposon (Tn125 and Tn5393 with three copies of blaNDM-1) and a recombination “hot spot” for the acquisition of new resistance determinants.
TEXT
At this time, blaNDM is recognized as a major global health threat. Guatemala and Colombia reported the first cases of blaNDM-1-harboring isolates in Latin America (1, 2). In both instances, blaNDM-1 was discovered in hospital-acquired, clonally related Klebsiella pneumoniae isolates that were recovered from patients who had not travelled recently. The molecular epidemiology of blaNDM carbapenemases in South America has been investigated only in a limited fashion, and data in Colombia are very scarce. In order to understand the dissemination of blaNDM-1 in Colombia, we performed a genomic analysis of four sentinel isolates, Acinetobacter baumannii, Acinetobacter nosocomialis, Escherichia coli, and K. pneumoniae collected in 2012, shortly after the first reported outbreak (1).
(Part of this work was presented at the 55th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], San Diego, CA, 17 to 21 September 2015.)
Phenotypic characterization revealed that two of the isolates (E. coli and A. nosocomialis) were resistant to all antibiotics tested, including polymyxin B and tigecycline (see Table S1 in the supplemental material). On the other hand, the A. baumannii isolate was also multidrug resistant but susceptible to polymyxin B, tigecycline, and ciprofloxacin. The K. pneumoniae isolate was susceptible to polymyxin B and exhibited relatively low susceptibility to carbapenems, highlighting once again the difficulty clinical microbiology laboratories have detecting carbapenemase genes that are expressed at low levels (3, 4). None of the isolates tested positive for carbapenemases using the modified Hodge test, but they tested positive (with the exception of the E. coli isolate) in the three-dimensional (3D) bioassay using an imipenem disk (5). Double-disk synergy testing using EDTA (DDST+EDTA) confirmed the presence of a metallo-β-lactamase for the members of the family Enterobacteriaceae, but not in the Acinetobacter species isolates.
In order to understand the genetic background of these early blaNDM-1-containing strains, the complete chromosome and plasmid sequences were obtained by assembly of Pacific Biosciences single-molecule real-time (SMRT) sequence data, with the exception of K. pneumoniae, where the chromosome was assembled into three ordered contigs (Table 1). Genome sequencing results showed that all isolates possessed multiple plasmids (Table 1) and revealed that blaNDM-1 was localized in one plasmid per strain, as confirmed by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) (6).
TABLE 1.
Accession number and resistome of blaNDM-1-harboring isolates
| Species | Parameter | Chromosome | Plasmids | ||
|---|---|---|---|---|---|
| A. baumannii | Accession no. | NZ_CP010397.1 | CP010398.1 | CP010399.1 | CP010400.1 |
| Size (bp) | 3,902,527 | 114,848 | 47,274 | 9,327 | |
| Resistance determinant(s) | blaADC-80 and blaOXA-94 | None | aph(3′)VIIa and blaNDM-1 | None | |
| A. nosocomialis | Accession no. | CP010368.1 | NZ_CP010369.1 | CP010903.1 | CP010370.2 |
| Size (bp) | 3,858,956 | 89,111 | 66,409 | 47,274 | |
| Resistance determinant(s) | blaADC-80 | None | None | aph(3′)VIIa and blaNDM-1 | |
| K. pneumoniae | Accession no. | NZ_JWRK01000001.1 | CP010390.1 | CP010391.1 | |
| Size (bp) | 5,329,244a | 198,371 | 178,193 | ||
| Resistance determinant(s) | blaCTXM-15, blaSHV-11, oqxA, oqxB, and fosA | strA, strB, aac(3′)IIa, aac(6′)lb-a, qnrB66, sul2, tetA, dfrA14, catB3, blaTEM-1, blaCTXM-15, and blaOXA-1 | aph3′VIa, aacA29, aadA2, mph(E), msr(E), catB3, cmlA1, sul2, sul1, blaNDM-1, and blaCARB-2 | ||
| E. coli | Accession no. | NZ_CP010371.1 | NZ_CP010373.2 | NZ_CP010372.1 | |
| Size (bp) | 4,761,012 | 193,908 | 151,583 | ||
| Resistance determinant(s) | sul1 | strA, strB, catA1, sul2, sul1, tetB, dfrA1, aadA16, and blaNDM-1 (3 times) | catA1, sul1, tetB, dfrA7, and blaTEM-1 | ||
The genome was not closed. The size is estimated based on the size of the three contigs.
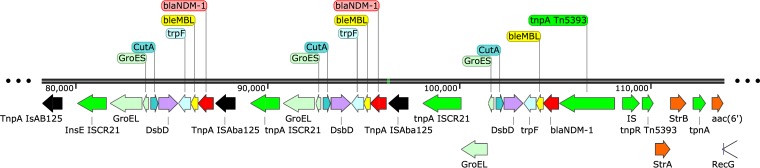
Multilocus sequence type (MLST) analysis revealed that Acinetobacter species isolates belonged to sequence type 322 (ST322) (A. baumannii) and ST464 (A. nosocomialis), none regarded as “high-risk” clones (7). Both A. baumannii and A. nosocomialis harbored three plasmids and carried the blaNDM-1 gene on a Tn125 backbone (Fig. 1) located on a 47,274-bp plasmid that was 99% similar to plasmid pNDM-BJ01 (GenBank accession no. JQ001791.1) reported in an Acinetobacter lwoffi isolate from China (8, 9). This plasmid also carried the aminoglycoside phosphotransferase aph(3′)VIIa gene, and a type IV secretion system (T4SS) gene cluster encoding a P-type T4SS that has been reported to encode a short, rigid pilus characteristic of broad-host-range conjugative plasmids (10).
FIG 1.
Organization of the blaNDM-1-containing element in E. coli. The first two copies of blaNDM-1 are contained within the Tn125 transposon (flanked by ISAba125 [shown in black]). The third copy lacks the right-flanking ISAba125 and is followed instead by a Tn5393-like element. Multiple transposable elements are found within this region, indicating that it may serve as a “hot spot” for the incorporation of new resistance determinants through homologous recombination via IS elements, site-specific recombination, or transposition.
We next discovered that a chromosomally encoded class C β-lactamase blaADC-80 was found in both Acinetobacter species isolates. This surprising finding is particularly interesting, as it highlights the possibility that blaADC evolved similarly in two different species of Acinetobacter. Consistent with the species identification, A. baumannii also harbored blaOXA-94 (a blaOXA-51-like derivative). None of the other plasmids contained additional resistance genes, and interestingly, A. baumannii resistance islands (AbaRI) were not found in the chromosome or on plasmids harbored by either of the Acinetobacter species isolates.
In contrast with Acinetobacter species isolates, both members of the Enterobacteriaceae contained multiple resistance genes in two large plasmids (151 to 198 kb) (Table 1), including blaCTXM-15, consistent with the previously documented predominance of that extended-spectrum β-lactamase (ESBL) in Colombia (11, 12). The K. pneumoniae isolate belonged to ST392, previously associated with the dissemination of blaKPC, blaOXA-48, and other ESBLs (13, 14). In addition, this isolate also harbored a chromosomal blaSHV-11 and blaCTX-M-15, the latter located downstream of ISEcp1, as previously reported in other Enterobacteriaceae isolates from Spain, Japan, Germany, the Netherlands, and the United Kingdom (15–18). The largest plasmid of K. pneumoniae (198 kb) was a multireplicon Inc FII/FIB type plasmid, and it carried antibiotic determinants conferring resistance to aminoglycosides [aph(3′)-Ib, aph(6′)-Id, aac(3)-IIa, and aac(6′)-Ib-cr], quinolones (qnrB66), sulfonamides (sul2), tetracycline [tet(A)], trimethoprim (dfrA14), chloramphenicol (catB3), and β-lactams (blaTEM-1, blaCTX-M-15, and blaOXA-1). In this case, the second copy of blaCTX-M-15 was found to be part of a previously reported structure: a Tn3-like transposon also carrying blaTEM-1 which has its tnpA gene disrupted by ISEcp1-blaCTX-M-15 (19). In this plasmid, this entire structure is also followed by an IS26 element, previously shown to have a critical role in the mobilization and reorganization of antibiotic resistance genes in Gram-negative bacteria (20, 21).
The blaNDM-1-bearing plasmid (178 kb) contained an Inc A/C2 replicon, extensively associated with antibiotic resistance in Gram-negative bacteria (22). The plasmid backbone shares similarity with other plasmids carrying blaNDM-1 and other β-lactamases in a variety of Gram-negative species (see Table S2 in the supplemental material). Additionally, this plasmid carried determinants conferring resistance to most antimicrobial classes, including β-lactams (blaCARB-2), aminoglycosides [aph(3′)-VIa, aacA29, and aadA2], chloramphenicol (catB3 and cmlA1), sulfonamides (sul2 and sul1), macrolides [mph(E)], streptogramin B (strB), and lincosamide [msr(E)].
The E. coli isolate belonged to ST10 (phylogroup A), which has been associated with ESBLs and hyperexpressed AmpC enzymes (7). E. coli harbored most of the resistance determinants in plasmids; only the sulfonamide resistance gene sul2 was present in the chromosome. The Inc FIA/FIB 151-kb plasmid carried blaTEM-1, catA1, sul1, tetB, and dfrA7, while the 193-kb Inc A/C2 plasmid harbored not only blaNDM-1 but also strA. strB, catA1, sul2, sul1, tetB, and dfrA1. It is noteworthy that there were three tandem repeats of blaNDM-1 in the 193-kb plasmid, two of them within a Tn125 structure and the last one lacking the right side copy of ISAba125 (Aba stands for A. baumannii) (Fig. 1). We interpret this to be a consequence of insertion of Tn125 within a Tn5393-like structure, as evidenced by the presence of tnpA, tnpR, strA, and strB, characteristic of this Tn3 transposon, originally reported for Erwinia amylovora (23), but now found in several Gram-negative species in clinical, ecological, and agricultural niches (24–27). This complex array of transposons is followed by an aminoglycoside resistance gene (aadA16) flanked by transposable genetic elements, indicating that this whole region could be serving as a “hot spot” for the incorporation of genetic determinants either by homologous recombination via IS elements, site-specific recombination, or transposition. A similar 20-kb resistance region is found on environmental plasmid pRSB101 which was originally isolated from bacterial populations residing in the activated sludge compartment of a wastewater treatment plant (28). Furthermore, one of those transposable elements was identified as IS26 and found not only flanking the above-mentioned region, but also next to the first copy of blaNDM-1 containing Tn125. This would reinforce the hypothesis of a “hot spot region,” given the replicative transposition mechanism of IS26 and its previously shown critical role in the mobilization and reorganization of antibiotic resistance genes in Gram-negative bacteria (20, 21). Most importantly, although blaNDM in tandem repeats has been observed before, this is to our knowledge the first report of such a structure in E. coli. In both previously reported cases, it occurred in K. pneumoniae isolates: the first from a Taiwanese patient with a hospitalization history in New Delhi, India (250-kb Inc FIB/FII type plasmid) (29) and the second from an outbreak in a neonatal unit in Nepal (304-kb Inc HIB/FIB type plasmid) (30).
All strains were nosocomially acquired and isolated from elderly patients with severe systemic infections, three patients, who presented several comorbidities, died (see Table S3 in the supplemental material). First, since evidence of international travel or travel to Bogota, Colombia (where the first Colombian blaNDM-1 was reported) could not be established for any of the patients or their families and second, given that they originally lived in rural areas or small cities, this emergence in a variety of species in two different geographic locations, is extremely worrisome. Colombia has often been among the first countries in the region to report the circulation of important resistance determinants, including CTX-M-15, KPC, and NMC-A, all of which have become widely disseminated, even becoming endemic, as is the case for KPC (31–34). Even though information regarding molecular epidemiology of blaNDM-1 in Colombia is still very limited, the National Institute of Health of Colombia is reporting increased numbers of patients infected with NDM-producing bacteria. Interestingly, K. pneumoniae and Providencia rettgeri are the most prevalent blaNDM-1-expressing Gram-negative bacterial species (35, 36). We hypothesize that the rapid spread of this resistance gene (blaNDM-1) is aided by the circulation of broad-host-range, transferable plasmids such as Inc A/C found in this study.
The widespread dissemination of blaNDM in Colombia portends a significant antibiotic resistance problem in Latin America (1). Colombia's situation may be only the “tip of the iceberg”; therefore, studies assessing the real prevalence of blaNDM, especially in countries where few reports are available, are warranted. It is of great importance that the findings of surveillance and genomic studies like the present one help inform new and more-effective infection control and stewardship programs that can be translated into appropriate national policies to prevent a situation where it becomes endemic.
Sequence accession numbers.
Sequences have been deposited in GenBank under the accession numbers given in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Ceron, A. Villareal, and M. Guerrero, Fundación Hospital San Pedro, Pasto, Colombia, and J. Osorio and E. Garcia, Hospital Universitario Hernando Moncaleano Perdomo, Neiva, Colombia.
Research reported in this publication was supported by the Genome Center for Infectious Diseases grant U19AI110819 to M.D.A. and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grants R01AI100560 and R01AI063517 to R.A.B. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Veterans Affairs Merit Review Program Award 1I01BX001974, and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B. This work was supported by Merck Sharp & Dohme, Janssen-Cilag SA, Pfizer SA, AstraZeneca Colombia SA, and Merck Colombia, which contributed to the formation of the Bacterial Resistance Surveillance Network.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding Statement
Research reported in this publication was supported by the Genome Center for Infectious Diseases grant U19AI110819 to M.D.A. Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI100560, R01AI063517, and R01AI072219 to R.A.B. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, award number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03072-15.
REFERENCES
- 1.Escobar Pérez JA, Olarte Escobar NM, Castro-Cardozo B, Valderrama Márquez IA, Garzón Aguilar MI, Martinez de la Barrera L, Barrero Barreto ER, Marquez-Ortiz R, Moncada Guayazán MV, Vanegas Gómez N. 2013. Outbreak of NDM-1-producing Klebsiella pneumoniae in a neonatal unit in Colombia. Antimicrob Agents Chemother 57:1957–1960. doi: 10.1128/AAC.01447-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasteran F, Albornoz E, Faccone D, Gomez S, Valenzuela C, Morales M, Estrada P, Valenzuela L, Matheu J, Guerriero L. 2012. Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J Antimicrob Chemother 67:1795–1797. doi: 10.1093/jac/dks101. [DOI] [PubMed] [Google Scholar]
- 3.Evans SR, Hujer AM, Jiang H, Hujer KM, Hall T, Marzan C, Jacobs MR, Sampath R, Ecker DJ, Manca C, Chavda K, Zhang P, Fernandez H, Chen L, Mediavilla JR, Hill CB, Perez F, Caliendo AM, Fowler VG Jr, Chambers HF, Kreiswirth BN, Bonomo RA, Antibacterial Resistance Leadership Group. 2016. Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis 62:181–189. doi: 10.1093/cid/civ837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viau RA, Hujer AM, Marshall SH, Perez F, Hujer KM, Briceño DF, Dul M, Jacobs MR, Grossberg R, Toltzis P, Bonomo RA. 2012. “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in northeast Ohio. Clin Infect Dis 54:1314–1321. doi: 10.1093/cid/cis036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coudron PE, Moland ES, Thomson KS. 2000. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol 38:1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 7.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 8.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 11.Martinez P, Garzón D, Mattar S. 2012. CTX-M-producing Escherichia coli and Klebsiella pneumoniae isolated from community-acquired urinary tract infections in Valledupar, Colombia. Braz J Infect Dis 16:420–425. doi: 10.1016/j.bjid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Villegas MV, Correa A, Perez F, Miranda MC, Zuluaga T, Quinn JP. 2004. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn Microbiol Infect Dis 49:217–222. doi: 10.1016/j.diagmicrobio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Potron A, Poirel L, Rondinaud E. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D. 2013. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 19:E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 15.Coelho A, González-López JJ, Miró E, Alonso-Tarrés C, Mirelis B, Larrosa MN, Bartolomé RM, Andreu A, Navarro F, Johnson JR, Prats G. 2010. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents 36:73–78. doi: 10.1016/j.ijantimicag.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Hirai I, Fukui N, Taguchi M, Yamauchi K, Nakamura T, Okano S, Yamamoto Y. 2013. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents 42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez I, Thomas K, Van Essen A, Schink AK, Day M, Chattaway M, Wu G, Mevius D, Helmuth R, Guerra B. 2014. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents 43:553–557. doi: 10.1016/j.ijantimicag.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Zhou K, Lokate M, Deurenberg RH, Arends J, Lo-Ten Foe J, Grundmann H, Rossen JWA, Friedrich AW. 2015. Characterization of a CTX-M-15 producing Klebsiella pneumoniae outbreak strain assigned to a novel sequence type (1427). Front Microbiol 6:1250. doi: 10.3389/fmicb.2015.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smet A, Van Nieuwerburgh F, Vandekerckhove TTM, Martel A, Deforce D, Butaye P, Haesebrouck F. 2010. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One 5:e11202. doi: 10.1371/journal.pone.0011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Chiou CS, Jones AL. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other Gram-negative bacteria. J Bacteriol 175:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi D, Jiang X, Sheng Z-K, Ngmenterebo D, Tai C, Wang M, Deng Z, Rajakumar K, Ou H-Y. 2015. Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J Antimicrob Chemother 70:2770–2774. doi: 10.1093/jac/dkv204. [DOI] [PubMed] [Google Scholar]
- 25.Harmer CJ, Holt KE, Hall RM. 2015. A type 2 A/C2 plasmid carrying the aacC4 apramycin resistance gene and the erm(42) erythromycin resistance gene recovered from two Salmonella enterica serovars. J Antimicrob Chemother 70:1021–1025. doi: 10.1093/jac/dku489. [DOI] [PubMed] [Google Scholar]
- 26.L'Abée-Lund TM, Sørum H. 2000. Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl Environ Microbiol 66:5533–5535. doi: 10.1128/AEM.66.12.5533-5535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantengoli E, Rossolini GM. 2005. Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum β-lactamase gene and other resistance determinants. Antimicrob Agents Chemother 49:3289–3296. doi: 10.1128/AAC.49.8.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, Schlüter A. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 29.Huang T-W, Chen T-L, Chen Y-T, Lauderdale T-L, Liao T-L, Lee Y-T, Chen C-P, Liu Y-M, Lin A-C, Chang Y-H, Wu K-M, Kirby R, Lai J-F, Tan M-C, Siu L-K, Chang C-M, Fung C-P, Tsai S-F. 2013. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One 8:e62774. doi: 10.1371/journal.pone.0062774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, Didelot X, Bashir A, Sebra R, Kasarskis A, Sthapit B, Shakya M, Kelly D, Pollard AJ, Peto TEA, Crook DW, Donnelly P, Thorson S, Amatya P, Joshi S. 2014. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco VM, Rojas LJ, De La Cadena E, Maya JJ, Camargo RD, Correa A, Quinn JP, Villegas MV. 2013. First report of a nonmetallocarbapenemase class A carbapenemase in an Enterobacter cloacae isolate from Colombia. Antimicrob Agents Chemother 57:3457. doi: 10.1128/AAC.02425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz SJ, Montealegre MC, Ruiz-Garbajosa P, Correa A, Briceño DF, Martinez E, Rosso F, Muñoz M, Quinn JP, Cantón R. 2011. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J Clin Microbiol 49:1993–1996. doi: 10.1128/JCM.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M, Quinn JP. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother 50:2880–2882. doi: 10.1128/AAC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Instituto Nacional de Salud. 2014. Circulación de carbapenemasas tipo Nueva Delhi metalo-β-lactamasa (NDM) en Colombia 2012-2014. Dirección de Redes en Salud Pública (DRSP), Subdirección Laboratorio Nacional de Referencia (SLNR), Laboratorio de Microbiología, Bogota, Colombia: http://www.ins.gov.co/tramites-y-servicios/examenes-de-interés-en-salud-publica/Microbiologa/Circulacion%20NDM%20Colombia%202012-2014.pdf. [Google Scholar]
- 36.Saavedra-Rojas S-Y, Duarte-Valderrama C, González-de-Arias M-N, Ovalle-Guerro MV. Emergencia de Providencia rettgeri NDM-1 en dos departamentos de Colombia, 2012-2013. Enferm Infecc Microbiol Clin, in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.