Abstract
The emergence and spread of Klebsiella pneumoniae carbapenemase (KPC) among Enterobacteriaceae presents a major public health threat to the world. Although not as common as in K. pneumoniae, KPC is also found in Escherichia coli strains. Here, we genetically characterized 9 carbapenem-resistant E. coli strains isolated from six hospitals in the United States and completely sequenced their blaKPC-harboring plasmids. The nine strains were isolated from different geographical locations and belonged to 8 different E. coli sequence types. Seven blaKPC-harboring plasmids belonged to four different known incompatibility groups (IncN, -FIA, -FIIK2, and -FIIK1) and ranged in size from ∼16 kb to ∼241 kb. In this analysis, we also identified two plasmids that have novel replicons: (i) pBK28610, which is similar to p34978-3 with an insertion of Tn4401b, and (ii) pBK31611, which does not have an apparent homologue in the GenBank database. Moreover, we report the emergence of a pKP048-like plasmid, pBK34397, in E. coli in the United States. Meanwhile, we also found examples of interspecies spread of blaKPC plasmids, as pBK34592 is identical to pBK30683, isolated from K. pneumoniae. In addition, we discovered examples of acquisition (pBK32602 acquired an ∼46-kb fragment including a novel replication gene, along with Tn4401b and other resistance genes) and/or loss (pKpQIL-Ec has a 14.5-kb deletion compared to pKpQIL-10 and pBK33689) of DNA, demonstrating the plasticity of these plasmids and their rapid evolution in the clinic. Overall, our study shows that the spread of blaKPC-producing E. coli is largely due to horizontal transfer of blaKPC-harboring plasmids and related mobile elements into diverse genetic backgrounds.
INTRODUCTION
Since the initial report of a Klebsiella pneumoniae strain producing K. pneumoniae carbapenemase (KPC) in 2001 (1), the rapid global spread of KPC and the movement of blaKPC among pathogenic Enterobacteriaceae have created a major public health crisis. KPC confers resistance to virtually all β-lactams, including carbapenems, the antibiotic of last resort. The blaKPC gene is harbored on a highly mobile Tn3-like transposon, Tn4401, and has been genotyped to numerous transferable plasmids belonging to various incompatibility groups (2–7).
Among the many medically significant Gram-negative bacteria (GNB), members of the family Enterobacteriaceae are the most common and important. Carbapenem resistance in Enterobacteriaceae (CRE) can occur through a number of different mechanisms, including the loss of porins, affecting the permeability of the outer membrane; upregulation of efflux systems in conjunction with hyperproduction of AmpC β-lactamases or extended-spectrum β-lactamases (ESBLs); or, more commonly, the acquisition and production of carbapenemases, such as KPC and New Delhi metallo-β-lactamase (NDM) (8, 9).
We have recently described the emergence and epidemic spread of carbapenem resistance among nosocomial K. pneumoniae in the New York-New Jersey region (8) and characterized the genetic structure of the successfully disseminated K. pneumoniae clone sequence type 258 (ST258). In addition, our study suggested that the dissemination of blaKPC in the northeastern United States is largely associated with some predominant blaKPC-harboring plasmids, pKpQIL-like plasmids (5), IncFIA plasmids (4), and IncI2 plasmids (10). Although not as common as in K. pneumoniae, carbapenem resistance has been identified in clinical Escherichia coli strains, the most common community-associated GNB. The movement of blaKPC plasmids into E. coli raises clinical concerns, as many strains are known pathogens and the frequent cause of urinary tract and intra-abdominal infections (11). The continued spread of resistance, by both plasmid transfer and the spread of primary clones, is further accelerated by the limited treatment options and our inability to reduce the disease burden (8).
blaKPC-harboring E. coli strains have been reported in different settings in the United States, Israel, China, Brazil, and European countries (12, 13). However, as a result of limited molecular data, it is unclear whether the spread of blaKPC in E. coli is attributable to the spread of a predominant strain, similar to the epidemic ST258 strain of K. pneumoniae (8), or the repeated acquisition of resistance plasmids in different E. coli strains. In this study, we genetically characterized carbapenem-resistant E. coli isolates and their blaKPC-harboring plasmids from 6 U.S. hospitals in 4 states during the period from 2009 to 2011 to explore whether the spread of KPC in E. coli is associated with a major genetic clone or certain predominant plasmids.
MATERIALS AND METHODS
Bacterial strains.
A total of 9 carbapenem-resistant E. coli isolates (BK28009, BK28610, BK28960, BK31611, BK32533, BK32602, BK33689, BK34397, and BK34592) were collected between 2009 and 2011 from 6 hospitals in 4 states—New Jersey, New York, Pennsylvania, and Illinois (Table 1)—as part of a retrospective study. The presence of the blaKPC gene was determined using a multiplex real-time PCR described previously (14). Multilocus sequence typing (MLST) analysis was performed using a method described previously (15).
TABLE 1.
Characteristics of KPC-producing E. coli isolates and their transformants
| Isolatea | ST | Location | Source | β-Lactamase(s) | Tn4401 | Inc | MIC (μg/ml)b |
Reference | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMI | ETP | MERO | DOR | CTX | CFP | ATM | SXT | TIC/CLA | GEN | TOB | CIP | LEV | TGC | CST | PMB | ||||||||
| BK28009 | 617 | NJ | Urine | KPC-3, TEM-1, SHV-2 | b | 4 | >4 | 8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | >8 | 8 | >2 | >8 | ≤0.25 | 0.5 | 1 | This study | |
| T-BK28009 | KPC-3, TEM-1 | b | N | 4 | >4 | >8 | >2 | 32 | >16 | >16 | >4/76 | >128/2 | ≤1 | 2 | >2 | >8 | 1 | 0.5 | 1 | ||||
| BK28610 | 457 | NY | Rectum | KPC-3 | b | 8 | >4 | 8 | >2 | >32 | >16 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | 2 | ≤0.25 | ≤1 | ≤0.25 | 0.5 | 0.5 | This study | |
| T-BK28610 | KPC-3 | b | New | 8 | 4 | 4 | 2 | >32 | >16 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | ≤1 | ≤0.25 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ||||
| BK28960 | 131 | NJ | Urine | KPC-2, TEM-1 | a | 8 | >4 | >8 | ≥4 | 32 | ≥32 | >16 | >4/76 | >128/2 | ≤1 | 2 | ≥4 | >8 | 0.5 | 1 | 1 | (4) | |
| T-BK28960 | KPC-2, TEM-1 | a | FIIK2 | 4 | 1 | 2 | 2 | 8 | 16 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | ≤1 | ≤0.25 | ≤1 | ≤0.25 | 0.5 | 1 | ||||
| BK31611 | 2287 | NJ | Urine | KPC-2, TEM-1 | b | >8 | >4 | >8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | 2 | 2 | >2 | >8 | 0.5 | 0.5 | 1 | This study | |
| T-BK31611 | KPC-2, TEM-1 | b | New | >8 | >4 | >8 | >2 | >32 | >16 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | ≤1 | ≤0.25 | ≤1 | ≤0.25 | 0.5 | 1 | ||||
| BK32533 | 2289 | NJ | Urine | KPC-3, TEM-1, OXA-9, FOX-5, PSE-1 | d | 4 | 4 | 8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | 2 | >8 | ≤0.25 | ≤1 | ≤0.25 | 0.5 | 0.5 | (1) | |
| T-BK32533 | KPC-3, TEM-1, OXA-9, FOX-5, PSE-1 | d | FIA, A/C | 2 | 1 | 2 | 1 | >32 | 8 | >16 | >4/76 | >128/2 | 2 | >8 | ≤0.25 | ≤1 | 0.5 | 0.25 | 0.5 | ||||
| BK32602 | 2288 | NY | Blood | KPC-3, TEM-1, OXA-9 | b | 4 | >4 | 8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | >8 | >8 | ≤0.25 | ≤1 | ≤0.25 | 0.5 | 0.5 | This study | |
| T-BK32602 | KPC-3, TEM-1, OXA-9 | b | N | 8 | >4 | 8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | >8 | >8 | ≤0.25 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ||||
| BK33689 | 167 | PA | Respiratory tract | KPC-2, TEM-1, SHV-12 | a | 4 | >4 | >8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | >8 | >8 | >2 | >8 | ≤0.25 | 0.5 | 0.5 | This study | |
| T-BK33689 | KPC-2, TEM-1 | a | FIIK1 | 8 | 4 | 4 | 2 | 16 | 8 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | ≤1 | ≤0.25 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ||||
| BK34397 | 167 | IL | Rectum | KPC-2, TEM-191 | NTEKPC | >8 | >4 | >8 | >2 | >32 | >16 | >16 | >4/76 | >128/2 | 4 | >8 | >2 | >8 | ≤0.25 | ≤0.25 | ≤0.25 | This study | |
| T-BK34397 | KPC-2 | NTEKPC | FII | 2 | 4 | 2 | 1 | 8 | 8 | >16 | ≤0.5/9.5 | >128/2 | ≤1 | ≤1 | ≤0.25 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ||||
| BK34592 | 1193 | NJ | Urine | KPC-3, TEM-1, OXA-9 | d | 2 | 4 | 4 | 2 | 32 | 16 | >16 | >4/76 | >128/2 | >8 | >8 | >2 | 4 | ≤0.25 | ≤0.25 | 0.5 | This study | |
| T-BK34592 | KPC-3, TEM-1, OXA-9 | d | FIA | 4 | 2 | 4 | 2 | 16 | 16 | >16 | >4/76 | >128/2 | >8 | >8 | ≤0.25 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ||||
T-, E. coli DH10B transformant.
MIC values were determined using the broth microdilution method. The values indicating resistance are in boldface. IMI, imipenem; ETP, ertapenem; MERO, meropenem; DOR, doripenem; CTX, cefotaxime; CFP, cefepime; ATM, aztreonam; SXT, trimethoprim-sulfamethoxazole; TIC/CLA, ticarcillin-clavulanate; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LEV, levofloxacin; TGC, tigecycline; CST, colistin; PMB, polymyxin B.
Characterization and manipulation of plasmids.
Plasmid DNA in these strains was extracted using a Qiagen Plasmid Midi kit (Qiagen, Valencia, CA) and electroporated into E. coli DH10B (Invitrogen) using a Gene Pulser II instrument (Bio-Rad Laboratories) as described previously (3, 16). E. coli DH10B transformants were selected on lysogeny broth (LB) agar plates with 100 μg/ml ampicillin. Conjugation experiments were performed using E. coli strain J53AzR as the recipient to examine the transfer abilities of blaKPC-harboring plasmids as described previously (3). Multiplex real-time PCR was used to screen for the presence of the blaKPC gene (14). S1 nuclease digestion of plasmid DNA followed by pulsed-field gel electrophoresis (S1-PFGE) was performed to estimate the number and size of plasmids (17). Plasmid incompatibility groups were determined using the PCR-based replicon-typing (PBRT) method of Carattoli et al. (18), as well as by the PCR assays previously developed in our laboratory for some of the common blaKPC-harboring plasmids (pKpQIL, pBK15692, pBK30683, and pBK32179) (4, 5, 10, 19).
Plasmid sequencing and bioinformatics.
Plasmid DNA was extracted from E. coli DH10B transformants of the nine strains described above using the Qiagen Plasmid Midi kit (Qiagen, Valencia, CA). The plasmid DNA was sequenced using an Illumina MiSeq system. The sequencing reads were de novo assembled, gaps between contigs were closed, open reading frames (ORFs) were predicted, and annotations were performed as described previously (3). Comparative genome alignment for different plasmids was performed using BLAST (http://blast.ncbi.nlm.nih.gov), Easyfig (20), and Mauve 2.4.0 (21).
Susceptibility testing.
The antimicrobial susceptibilities of the nine E. coli isolates and their E. coli DH10B transformants harboring a KPC-producing plasmid were determined by broth microdilution in cation-adjusted Mueller-Hinton broth (MHB) according to Clinical and Laboratory Standards Institute (CLSI) methods and interpretations (22, 23) using Sensititre GNX2F trays (Thermo Fisher Scientific, Waltham, MA) as described previously (3).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the plasmids from this study were deposited in GenBank with accession numbers KU295131 to KU295136.
RESULTS
Characteristics of clinical isolates.
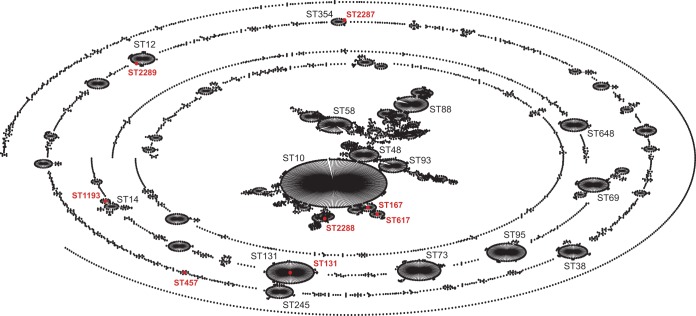
As part of a retrospective study, 9 carbapenem-resistant E. coli isolates from 4 different states (New Jersey, New York, Pennsylvania, and Illinois) were collected between 2009 and 2011. The isolates were recovered from urine samples (n = 5), rectum swabs (n = 2), blood (n = 1), and respiratory tract (n = 1) (Table 1). The genotyping results documented that they all harbored the blaKPC genes, including 5 blaKPC-3 (55.5%) and 4 blaKPC-2 (44.5%) variants. MLST analysis revealed that the isolates belonged to 8 different sequence types (STs): ST131 (n = 1), ST167 (n = 2), ST457 (n = 1), ST617 (n = 1), ST1193 (n = 1), ST2287 (n = 1), ST2288 (n = 1), and ST2289 (n = 1). Among them, endemic E. coli ST131 (MLST allele profile, 53-40-47-13-36-28-29) is a highly successful clone responsible for the dissemination of blaCTX-M-15 and blaKPC genes (6, 24). ST167 (10-11-4-8-8-13-2) and ST617 (10-11-4-8-8-13-73) belong to the ST10 complex, and ST457 (101-88-97-108-26-79-2) is unique (it does not belong to any clonal complex) (Fig. 1). In addition, ST1193 (14-14-10-200-17-7-10) is a single-locus variant (SLV) of the ST14 complex and is reported to be associated with ciprofloxacin resistance (25). Three STs, ST2287 (85-88-272-29-59-58-62), ST2288 (8-7-9-220-8-8-2), and ST2289 (13-13-5-13-16-10-9), are reported for the first time in this study. ST2288 is a triple-locus variant of the ST10 complex, while ST2287 and ST2289 are SLVs of the ST354 complex and the ST12 complex, respectively (Fig. 1).
FIG 1.
Population structure of KPC-producing E. coli. Shown is the population structure from the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) as of 5 January 2016, depicted graphically by eBURSTv3 (http://eburst.mlst.net). Major STs are in black. KPC-producing E. coli STs are highlighted by red dots.
Following electroporation, blaKPC-bearing plasmids were successfully transferred to E. coli DH10B. The E. coli DH10B transformants were analyzed by S1-PFGE analysis, which revealed that each transformant harbored a single plasmid ranging in size from 16 to 240 kb. Conjugation experiments showed that all blaKPC-harboring plasmids except pBK28610 and pBK31611 were successfully transferred to E. coli strain J53AzR, suggesting the plasmids harbor genes supporting conjugation (discussed below). The plasmid replicon PCR typing (4, 5, 10, 18, 19) revealed that the plasmids belonged to different incompatibility groups (Inc): IncFIA, -FIIK1, -FIIK2, and -N (Table 1). Two plasmids could not be typed by this PCR method, suggesting the presence of novel blaKPC-harboring plasmids.
Nine blaKPC-harboring plasmids from 8 different E. coli STs were fully sequenced. The data in Table 1 provide a comprehensive genotypic profile of the nine KPC-harboring E. coli strains. The nine parental clinical isolates and their corresponding E. coli DH10B transformants were tested for antibiotic susceptibility (the list of antimicrobial agents is shown in Table 1). All the parental isolates and their corresponding transformants were resistant to imipenem, ertapenem, and meropenem, consistent with the presence of a carbapenemase gene in the plasmid DNA. Moreover, all the isolates were resistant to cefotaxime, aztreonam, cefepime, and ticarcillin/clavulanate and susceptible to tigecycline, colistin and polymyxin B, with varying susceptibilities to gentamicin, tobramycin, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole (Table 1), a finding that suggests that the strains harbor different antimicrobial resistance genes. Resistance present in parental isolates that was not found in the E. coli DH10B transformants suggests that these resistance genes are linked to chromosomal genes or genes on different plasmids.
Novel replicon plasmids.
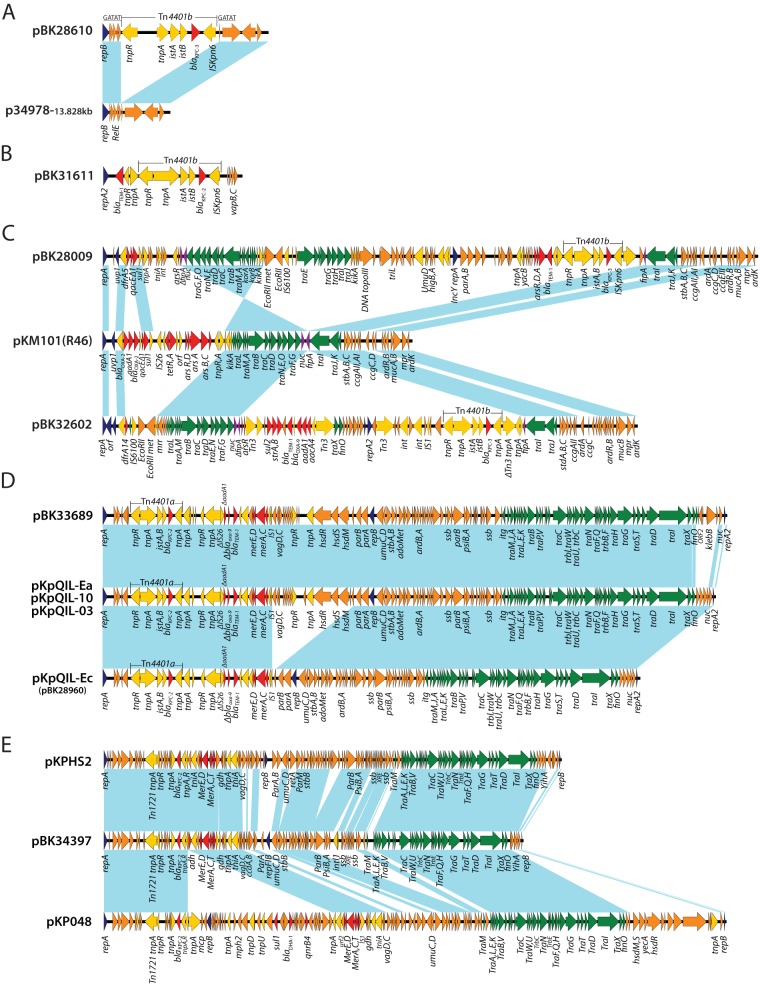
The nine plasmids in this study were completely sequenced, revealing diverse plasmid families and structures. Strain BK28610 (ST457) was isolated from a rectum swab from a New York patient in 2009 (Table 1). Plasmid pBK28610 is a circular molecule 16,926 bp in length with an average G+C content of 59.6% and harboring 16 predicted ORFs (Fig. 2A). It carries a blaKPC-3 gene on the Tn4401b transposon. pBK28610 carries a single novel replication gene, repB, with 100% sequence similarity to the replication gene of plasmid p34978-3 (GenBank accession no. CP010366.2; sequenced by J. Craig Venter Institute [JCVI] and our laboratory) identified in a KPC-harboring Enterobacter species. Full plasmid sequence BLAST against NCBI GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that pBK28610 has 40% query coverage with overall 99% nucleotide identity to p34978-3 (Fig. 2A). Close inspection of the sequence revealed that pBK28610 is identical to p34978-3 with an insertion of Tn4401b. The Tn3-like transposon created a 5-bp target site duplication (GATAT) upon insertion into a hypothetical protein-encoding gene (locus tag LI63_01490) (Fig. 2A). Our analysis suggests pBK28610 originates from the insertion of a blaKPC-3-harboring Tn4401b into the backbone of a p34978-3 like plasmid.
FIG 2.
Structures and comparative analysis of blaKPC-harboring plasmids from E. coli. The light-blue shading denotes shared regions of homology. ORFs are portrayed by arrows and colored based on the predicted gene function. The orange arrows indicate plasmid scaffold regions, the dark-blue arrows represent replication-associated genes, the green arrows represent genes associated with the tra locus, the red arrows represent antimicrobial resistance genes, and the yellow arrows represent accessory genes. The Tn4401 regions are marked, and the 5-bp Tn4401-adjacent sequences are marked and underlined.
Plasmid pBK31611 is harbored by strain BK31611 (ST2287), which was isolated from a urine culture from a New Jersey patient in 2010. Plasmid pBK31611 is 16,346 bp in length with an average G+C content of 56.1% and harbors 14 predicted ORFs (Fig. 2B). Plasmid sequence queries using PlasmidFinder (26) failed to find any significant match. pBK31611 carries a novel replication gene, repA, with 69% sequence similarity and 98% query coverage to plasmid pSG2 from Sodalis glossinidius (27). pBK31611 carries a blaTEM-1 gene upstream of a Tn4401b transposon harboring a blaKPC-2 gene. Moreover, pBK31611 harbors a type II toxin-antitoxin system (vapBC) downstream of Tn4401b (Fig. 2B). The vapBC loci play an important biological role in plasmid and gene stabilization, along with bacterial persistence (28). An element of ∼13.4 kb harboring a blaKPC-2 gene on a Tn4401b transposon, along with blaTEM-1, was inserted into an ∼2.8-kb region, creating 11-bp putative direct repeats (CGCCCTTGCAG) upon insertion. Interestingly, it appears that plasmid pBK31611, with a backbone of ∼3 kb, acquired this element, which is three times its size, containing both the blaTEM-1 gene and Tn4401b.
IncN-like chimera plasmids.
Plasmids pBK28009 and pBK32602 were isolated from strains BK28009 (ST617) and BK32602 (ST2288) from patients in New Jersey and New York hospitals in 2009 and 2011, respectively (Table 1). Sequence analysis of pBK28009 and pBK32602 revealed that they both carry plasmid incompatibility group N replicons. Plasmid pBK28009 is a circular molecule of 107,760 bp with an average G+C content of 53.6% harboring 135 predicted ORFs, while pBK32602 is 87,982 bp in length with an average G+C content of 53% and harbors 101 predicted ORFs (Fig. 2C). Both plasmids contain a blaKPC-3 gene on a Tn4401b transposon.
Full plasmid sequence queries of pBK28009 and pBK32602 against NCBI GenBank showed similarity to other IncN plasmids (like pYD626E [29] and pKPC-629 [30]), including the IncN prototype plasmid pKM101 (R46). pBK28009 has two β-lactamase genes, blaKPC-3 and blaTEM-1, and resistance genes, dfrA5 (trimethoprim resistance), sul1 (sulfonamide resistance), and arsR, arsD, and arsA (arsenic resistance). In comparison, pBK32602 contains dfrA14 (trimethoprim resistance), sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), and aadA1 and aacA4 (aminoglycoside resistance) and harbors three β-lactamase genes, blaKPC-3, blaOXA-9, and blaTEM-1 (Fig. 2C).
Overall, the core regions in pBK28009 and pBK32602 are similar to those of other IncN plasmids, with IncN replicons and the two regions involved in conjugation. The first region includes a truncated fipA gene, along with downstream tra genes (traG, traF, traO, traN, traE, traD, traC, traB, traM, traA, and traL), core genes (nuc, eex, korA, and korB), and EcoRII and EcoRII methyltransferase genes. Similar to pYD626E, the first region in pBK28009 is in an inverse orientation compared to most of the other IncN plasmids (pKM101 and pBK32602) (29, 31). The second regions in pBK28009 and pBK32602 are similar and include tra genes (traI, traJ, and traK), a second copy of a truncated fipA gene, core genes (stbA, stbB, stbC, ardAB, ardR, and ccgC and ccgD) and blaKPC-3 on a Tn4401b transposon.
However, these plasmids differ significantly in the number of acquired genes. pBK28009 acquired two distinct regions: an ∼11-kb fragment downstream of uvp1 and an ∼58-kb fragment located between the EcoRII gene and fipA, including a second replication gene (IncY), a second set of conjugation genes (tra), Tn4401b, and other core genes. This acquired region appears to belong to a novel plasmid with no significant match in NCBI GenBank (Fig. 2C). Similarly, pBK32602 acquired an ∼46-kb fragment downstream of nuc and upstream of fipA and carries a novel replication gene (repA2), Tn4401b, and a set of resistance genes (blaTEM-1, blaOXA-9, strAB, sul2, aadA1, and aacA4). Similar to most IncN plasmids, the first acquired region integrated downstream of the uvp1 gene and the second region was inserted between nuc and fipA in pBK32602 and between the EcoRII gene and fipA in pBK28009 (Fig. 2C). This suggests that the region upstream of fipA is a “hot spot” for integration of acquired regions with an IncN plasmid backbone. IncN plasmids typically carry the single replicon gene repA; however, we identified a second replicon gene in the acquired region, which suggests they have a mosaic plasmid structure.
pKpQIL-like plasmids.
pKpQIL, an IncFIIK2 group plasmid, is one of the most common blaKPC-harboring plasmids, reported in Israel, the United States, the United Kingdom, Colombia, Italy, and other countries (8). Our previous studies showed that pKpQIL-like plasmids accounted for ∼35% of KPC plasmids in the New Jersey-New York area (5). Here, we showed an example of their dissemination in E. coli. Strains BK33689 (ST167) and BK28960 (ST131) were recovered from patients in a Pennsylvania and a New Jersey hospital in 2011 and 2010, respectively. Interestingly, an IncF replicon sequence-typing scheme (32) using 88 bp of the copA region of the FII replicon showed plasmid pBK33689 belongs to incompatibility group IncFIIK1, which is different from the IncFIIK2 replicon type initially assigned for the pKpQIL plasmid identified in the K. pneumoniae ST258 strain from Israel (33).
Plasmid pBK33689 is 115,706 bp in length, with an average G+C content of 53.8%, and harbors 132 predicted ORFs (Fig. 2D). A BLAST search of the full plasmid sequence against the GenBank database showed a high degree of similarity to the previously published pKpQIL-like plasmid pKpQIL-10 (IncFIIK2) from a K. pneumoniae isolate (ST258) from a New York City patient (5), with 95% query coverage and overall 99% nucleotide identity. The major difference is the region between the finO and nuc genes in plasmid pBK33689, which shares no homology with pKpQIL-10 (Fig. 2D) (5). This ∼4-kb region carries 4 hypothetical protein-encoding genes, a klebicin B gene, and a gene encoding a colicin immunity protein.
pBK28960 is 99,142 bp in length with a G+C content of 54% and harbors 110 predicted ORFs. The complete sequence and comparative analysis for plasmid pBK28960 (pKpQIL-Ec; GenBank accession no. KJ146688) was published previously (5). Compared to plasmids spKpQIL-10 and pBK33689, plasmid pKpQIL-Ec has a 14.5-kb deletion downstream of the mer operon and upstream of the parAB operon (5).
IncFIA plasmids.
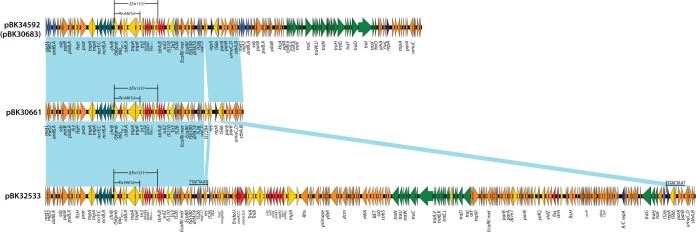
IncFIA plasmids are another example of successful plasmids that are widespread in the New Jersey-New York area and account for ∼20% of KPC-producing K. pneumoniae isolates (4). In our previous study, we identified plasmid pBK30683, an IncFIA plasmid with a cointegrate plasmid structure from K. pneumoniae (4). In this study, we found an identical plasmid, pBK34592, from E. coli isolate BK34592 from a patient in a New Jersey hospital in 2011. The sequence of pBK34592 is identical to that of pBK30683, suggesting the horizontal transfer of the same blaKPC-harboring IncFIA between different species. In addition, pBK32533, reported previously, is another example of a cointegrate IncFIA plasmid with 72-kb region, identical to the nonconjugative plasmid pBK30661, that acquired an additional 170-kb element containing genes for conjugative transfer (Fig. 3) (2). This suggests that the mobility of IncFIA plasmids across different species is facilitated by the formation of cointegrate plasmid structures.
FIG 3.
Plasmid structures of IncFIA plasmids pBK34592 (same as pBK30683), pBK30661, and pBK32533. The light-blue shading denotes shared regions. ORFs are represented by arrows and colored based on the predicted gene function. The orange arrows indicate plasmid scaffold regions. The genes associated with plasmid transfer are depicted by green arrows, and replication-associated genes are represented by dark-blue arrows. Antimicrobial resistance genes are indicated by red arrows. Other accessory genes are indicated by yellow arrows. The 8-bp target sequence duplications by IS26-mediated integration in pBK32533 are underlined.
Plasmids with blaKPC-bearing non-Tn4401 elements.
Plasmids harboring the blaKPC gene on non-Tn4401 mobile elements (NTEKPC) have been reported in China, Europe, and other regions (8). NTEKPC is usually in a Tn1722-blaKPC-2-Tn3 organization, with or without a truncated blaTEM integration upstream of blaKPC (8). To our knowledge, this is the first description of a plasmid with blaKPC on a non-Tn4401-like element from E. coli in the United States. Strain BK34397 is an ST167 strain isolated from a rectum swab from a patient in Illinois in 2011. pBK34397 is a circular molecule 101,915 bp in length with an average G+C content of 53% and harboring 127 predicted ORFs (Fig. 2E). pBK34397 belongs to incompatibility group IncFII and harbors a blaKPC-2 gene on a non-Tn4401-like element. A BLAST search of full plasmid sequences against the GenBank database showed an overall 90% query coverage with 98% sequence similarity to plasmid pKPHS2 from K. pneumoniae subsp. pneumoniae strain HS11286 (34). Moreover, it showed 79% query coverage with 99% sequence similarity to plasmid pKP048, isolated from K. pneumoniae strain KP048 (35). It showed that pBK34397 carries a backbone similar to those of the recently published plasmids pKPHS2 (34) and pKP048 (35) in China. Similarly, plasmid pBK34397 contained two replication regions, both belonging to the IncF group. Moreover, pBK34397 harbored a full complement of conjugative-transfer (tra) genes that were similar to those of plasmids pKP048 and pKPHS2. This region is able to facilitate the spread of non-Tn4401-like plasmids among different genera. Overall, pBK34397 has an IncF structure with modules for replication and stable inheritance, a mercury resistance operon, and conjugal transfer genes, similar to the backbone of pKPHS2 and pKP048 (Fig. 2E).
DISCUSSION
Among the Enterobacteriaceae, blaKPC has been primarily found in K. pneumoniae. In K. pneumoniae, KPC has predominantly spread worldwide in clonal group 258 (mainly ST258 and ST11) (8). In addition, KPC-harboring K. pneumoniae has been associated with several plasmids of different Inc groups, including IncFIA (4), IncFIIK2 (pKpQIL) (5), IncI2 (10), and IncN (3). In this study, we genetically characterized the plasmid content and genetic background of KPC-harboring E. coli to understand if similar plasmids have also spread in E. coli.
Our study revealed some important findings. In contrast to K. pneumoniae, where blaKPC is mainly found in certain Inc groups and is associated with an epidemic clone (CG258), our study identified a variety of blaKPC-harboring plasmids in different E. coli clones (STs). Nine carbapenem-resistant E. coli isolates were from 8 different STs (ST131, ST167, ST457, ST617, ST1193, ST2287, ST2288, and, ST2289). Among them, ST167 and ST617 belong to the ST10 complex, which is known to be associated with blaCTX-M-harboring E. coli isolates in Nigeria (36), Spain (37), France (38), and Mauritania (39). Moreover, ST457 has been previously associated with blaCTX-M-encoded β-lactamases in intestinal E. coli isolates from dogs in Korea (40), uropathogenic E. coli isolates from England (41), and fluoroquinolone-resistant E. coli isolates from canine feces in Australia (42). Our study demonstrated that blaKPC has emerged in these common ESBL-producing clones, raising concern that they will be able to continue to spread into diverse genetic backgrounds. In addition, a recent study on the global distribution of carbapenemase-producing E. coli reported ST131 as the genetic background responsible for the dispersal of blaKPC (63%) among E. coli isolates (43), and in our study, we have also identified carbapenem-resistant ST131 E. coli. Other genetic backgrounds identified in this study include ST1193, which is associated with ciprofloxacin resistance in human isolates from Australia (44) and Korea (25), and three strains with unique sequence types: ST2287 (a single-locus variant of the ST354 complex) and ST2288 and ST2289 (2) (single-locus variants of the ST12 complex). Our study suggests that, in contrast to the clonal spread of K. pneumoniae ST258, the spread of blaKPC in diverse E. coli genetic backgrounds is largely due to horizontal transfer of blaKPC-harboring plasmids and elements.
Among epidemic ST258 K. pneumoniae isolates, several major Inc plasmid groups are associated with the spread of blaKPC, including IncFII, -FIA, and -I2 (8). In the current study, we found that some of these predominant plasmids reported in K. pneumoniae are also found in E. coli. One example is a KPC-harboring pKpQIL-like plasmid, pBK33689. Using comparative plasmid genome analysis, pBK33689 is similar to pKPQIL-10 from K. pneumoniae (Fig. 2D) (5). Another example is plasmid pBK34592, an IncFIA plasmid from E. coli that is identical to pBK30683, previously reported from K. pneumoniae (ST963) (4). These findings strongly suggest the horizontal dissemination of these plasmids among strains of Enterobacteriaceae in the United States. Although the current epidemiology suggests that these carbapenemase-producing plasmids have not aggressively spread in E. coli, one wonders whether it is only a matter of time before we observe a plasmid-host pairing that will give rise to a successful multidrug-resistant clone.
The plasmid characterization in this study also identified novel plasmid groups and structures, such as the two small novel plasmids with unique replicons, pBK28610 and pBK31611. Both are ∼16 kb in size and harbor the blaKPC gene on a Tn4401b transposon. A search of the database revealed that plasmid p34978-3, found in Enterobacter cloacae, is the same plasmid without the Tn4401b insertion and that this acquisition generated a 5-bp target site duplication (Fig. 2B). The plasticity of the plasmid genome as a result of the acquisition and/or deletion of different elements is driving plasmid evolution. For example, a comparison of the IncN prototype plasmid pR46 and plasmid pBK32602 identified an ∼46-kb fragment upstream of traI and that includes a novel replication gene, Tn4401b, and a group of antibiotic resistance genes. Another example is the IncFIA cointegrate plasmids pBK32533 and pBK34592, which are similar to pBK30661 but have gained the ability to be mobilized by the acquisition of large insertions that harbor genes for conjugation. These examples highlight the rapid acquisition or loss of DNA and dramatically demonstrate how plasmids are able to adapt and evolve both antibiotic resistance and mobility. Complete plasmid sequencing and characterization have become necessary tools for genotyping and tracking plasmids and to identify key genetic features in intergenus and interspecies transfer associated with Tn4401 transposition.
Finally, we report the emergence of a pKP048-like plasmid (pBK34397) in the United States. This IncFIIK2 plasmid harbors blaKPC on a non-Tn4401-like element, similar to pKPHS2 (IncFIIK2) and pKP048 (IncFIIK5) from K. pneumoniae strains HS11286 and KP048, respectively, which were isolated from human sputum specimens from China (34). Although pKp048-like plasmids are the most predominant blaKPC-harboring plasmids in China and other regions, to our knowledge, this is the first report of this plasmid type in E. coli in the United States (39). It remains unclear whether the plasmid will spread further in the United States and become a predominant KPC-harboring plasmid.
Altogether, our study showed that, unlike in K. pneumoniae, blaKPC is harbored by different plasmid groups in E. coli strains with diverse genetic backgrounds. This finding suggests that the spread of KPC-producing E. coli is largely due to horizontal transfer of blaKPC-harboring plasmids and elements into diverse genetic backgrounds and that blaKPC-harboring plasmids continue to evolve by acquisition and/or deletion of different elements. This study provides insight into the molecular epidemiology and genetic composition of KPC-producing E. coli isolates.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (grant R01AI090155 to B.N.K. and R21AI117338 to L.C.). The research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI100560, R01AI063517, and R01AI072219 to R.A.B. The study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Veterans Affairs Merit Review Program award 1I01BX001974, and the Geriatric Research Education and Clinical Center grant VISN 10 to R.A.B.
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
Funding Statement
This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program Award 1I01BX001974, and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B.
REFERENCES
- 1.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chavda KD, Chen L, Jacobs MR, Rojtman AD, Bonomo RA, Kreiswirth BN. 2015. Complete sequence of a bla(KPC)-harboring cointegrate plasmid isolated from Escherichia coli. Antimicrob Agents Chemother 59:2956–2959. doi: 10.1128/AAC.00041-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Chavda KD, Melano RG, Hong T, Rojtman AD, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2289–2294. doi: 10.1128/AAC.02749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirth BN. 2014. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother 58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 57:5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratu S, Brooks S, Burney S, Kochar S, Gupta J, Landman D, Quale J. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin Infect Dis 44:972–975. doi: 10.1086/512370. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 13.Xu G, Jiang Y, An W, Wang H, Zhang X. 2015. Emergence of KPC-2-producing Escherichia coli isolates in an urban river in Harbin, China. World J Microbiol Biotechnol 31:1443–1450. doi: 10.1007/s11274-015-1897-z. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Wang ZQ, Liu B, Zhang XJ, Jiang F, Xiang JH. 2007. The study of optimal conditions of electroporation in Escherichia coli DH10B strain. Sheng Wu Gong Cheng Xue Bao 23:347–351. (In Chinese.) [PubMed] [Google Scholar]
- 17.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 18.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2013. Complete sequence of a bla(KPC-2)-harboring IncFII(K1) plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother 57:1542–1545. doi: 10.1128/AAC.02332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed (M07-A9) CLSI, Wayne, PA. [Google Scholar]
- 23.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement (M100-S24). CLSI, Wayne, PA. [Google Scholar]
- 24.Naas T, Cuzon G, Gaillot O, Courcol R, Nordmann P. 2011. When carbapenem-hydrolyzing β-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob Agents Chemother 55:4933–4934. doi: 10.1128/AAC.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang J, Yu J, Lee H, Ryu H, Park K, Park YJ. 2014. Prevalence and characteristics of lactose non-fermenting Escherichia coli in urinary isolates. J Infect Chemother 20:738–740. doi: 10.1016/j.jiac.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16:149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi Y, Inouye M. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog Mol Biol Transl Sci 85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- 29.Li JJ, Lee CS, Sheng JF, Doi Y. 2014. Complete sequence of a conjugative incn plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother 58:6974–6977. doi: 10.1128/AAC.03632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer PJ, Shanabruch WG, Walker GC. 1981. Functional organization of plasmid pKM101. J Bacteriol 145:1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 33.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou H-Y. 2012. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol 194:1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y. 2010. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother 54:3967–3969. doi: 10.1128/AAC.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. 2012. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect 18:E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x. [DOI] [PubMed] [Google Scholar]
- 37.Novais A, Viana D, Baquero F, Martinez-Botas J, Canton R, Coque TM. 2012. Contribution of IncFII and broad-host IncA/C and IncN plasmids to the local expansion and diversification of phylogroup B2 Escherichia coli ST131 clones carrying blaCTX-M-15 and qnrS1 genes. Antimicrob Agents Chemother 56:2763–2766. doi: 10.1128/AAC.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahmen S, Haenni M, Chatre P, Madec JY. 2013. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother 68:2797–2801. doi: 10.1093/jac/dkt291. [DOI] [PubMed] [Google Scholar]
- 39.Ben Sallem R, Ben Slama K, Estepa V, Cheikhna EO, Mohamed AM, Chairat S, Ruiz-Larrea F, Boudabous A, Torres C. 2015. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J Chemother 27:114–116. doi: 10.1179/1973947814Y.0000000172. [DOI] [PubMed] [Google Scholar]
- 40.Tamang MD, Nam HM, Jang GC, Kim SR, Chae MH, Jung SC, Byun JW, Park YH, Lim SK. 2012. Molecular characterization of extended-spectrum-beta-lactamase-producing and plasmid-mediated AmpC beta-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother 56:2705–2712. doi: 10.1128/AAC.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo S, Wakeham D, Brouwers HJ, Cobbold RN, Abraham S, Mollinger JL, Johnson JR, Chapman TA, Gordon DM, Barrs VR, Trott DJ. 2015. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect 17:266–274. doi: 10.1016/j.micinf.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. doi: 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platell JL, Trott DJ, Johnson JR, Heisig P, Heisig A, Clabots CR, Johnston B, Cobbold RN. 2012. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob Agents Chemother 56:3898–3904. doi: 10.1128/AAC.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]