Abstract
Despite the global economic and ecological importance of forest trees, the genomic basis of differential adaptation and speciation in tree species is still poorly understood. Populus tremula and Populus tremuloides are two of the most widespread tree species in the Northern Hemisphere. Using whole-genome re-sequencing data of 24 P. tremula and 22 P. tremuloides individuals, we find that the two species diverged ∼2.2–3.1 million years ago, coinciding with the severing of the Bering land bridge and the onset of dramatic climatic oscillations during the Pleistocene. Both species have experienced substantial population expansions following long-term declines after species divergence. We detect widespread and heterogeneous genomic differentiation between species, and in accordance with the expectation of allopatric speciation, coalescent simulations suggest that neutral evolutionary processes can account for most of the observed patterns of genetic differentiation. However, there is an excess of regions exhibiting extreme differentiation relative to those expected under demographic simulations, which is indicative of the action of natural selection. Overall genetic differentiation is negatively associated with recombination rate in both species, providing strong support for a role of linked selection in generating the heterogeneous genomic landscape of differentiation between species. Finally, we identify a number of candidate regions and genes that may have been subject to positive and/or balancing selection during the speciation process.
Keywords: Populus tremula, Populus tremuloides, whole-genome re-sequencing, demographic histories, heterogeneous genomic differentiation, linked selection, recombination
Introduction
Understanding how genomes diverge during the process of speciation has received a great deal of attention in the evolutionary genetics literature in recent years (Nosil et al. 2009; Strasburg et al. 2012; Seehausen et al. 2014). Under strict neutrality, differentiation is expected to accumulate as a result of the stochastic fixation of polymorphisms by genetic drift (Coyne and Orr 2004). Demographic processes, such as population bottlenecks or expansions, can accelerate or decelerate the rate of differentiation through changes in the effective population sizes of nascent daughter species (Avise 2000). Random genetic drift and demographic processes are both expected to affect the entire genome (Luikart et al. 2003). Natural selection, however, only influence loci involved in ecological specialization and/or reproductive isolation, resulting in patterns of polymorphisms and divergence that deviate from neutral predictions (Luikart et al. 2003; Via 2009). The functional architectures of genomes, for example, mutation and recombination rates, are also important factors in determining genomic landscape of differentiation (Noor and Bennett 2009; Nachman and Payseur 2012; Renaut et al. 2013). For example, suppressed recombination could increase genetic differentiation either by limiting inter-species gene flow to prevent the break-up of co-adapted alleles, or through the diversity-reducing effects of linked selection (Noor and Bennett 2009). However, disentangling the relative importance of these evolutionary forces when interpreting patterns of genomic divergence remains a challenge in speciation genetics.
With the advance of next generation sequencing (NGS) technologies, a growing number of studies have found highly heterogeneous patterns of genomic differentiation between recently diverged species (Turner et al. 2005; Ellegren et al. 2012; Renaut et al. 2013; Carneiro et al. 2014; Feulner et al. 2015). A common explanation for these patterns is that levels of gene flow between species differ across the genome. Increased genetic divergence is usually observed in a small number of regions containing loci involved in reproductive isolation (‘speciation islands’), whereas the remainder of the genome is still permeable to ongoing gene flow and therefore shows lower levels of differentiation (Nosil et al. 2009; Sousa and Hey 2013). However, some recent studies have argued that highly differentiated regions represent ‘incidental islands’ that are not tied to the speciation processes per se. Rather they are seen simply as a result of the diversity-reducing effects of linked selection that accelerate lineage sorting of ancestral variation and increase interspecific differentiation, especially in regions of reduced recombination (Turner and Hahn 2010; Cruickshank and Hahn 2014). In addition, long-term balancing selection is supposed to maintain stable trans-species polymorphisms and leave signatures of unusually low genetic differentiation between species (Charlesworth 2006). Under these scenarios, natural selection alone is sufficient to generate patterns of heterogeneous genomic differentiation even under complete allopatry (Noor and Bennett 2009; Turner and Hahn 2010). Finally, strictly neutral forces, such as stochastic genetic drift and complex demographic processes, can also create heterogeneous genomic divergence and generate patterns of divergence and polymorphism that mimic the effects of selection (Nosil et al. 2009; Campagna et al. 2015). In general, the three hypotheses listed above are not mutually exclusive and exhaustive examination of these hypotheses requires detailed information on the speciation process, such as the timing of speciation, the geographic and demographic context in which it occurred (Nosil and Feder 2012).
Although largely understudied compared with other model species, forest trees represent a promising system to understand the genomic basis of species divergence and adaptive evolution; as a group they have developed diverse strategies to adapt and thrive across a wide range of climates and environments (Neale and Kremer 2011). Populus tremula (European aspen) and Populus tremuloides (American aspen) are two of the most ecologically important and geographically widespread tree species of the Northern Hemisphere (fig. 1A). Both are keystone species, display rapid growth, with high tolerance to environmental stresses and long-distance pollen and seed dispersal via wind (Eckenwalder 1996; Müller et al. 2012). In addition, they both harbor among the highest level of intraspecific genetic diversity reported in plant species so far (Ingvarsson 2008; Callahan et al. 2013; Wang et al. 2016). Based on their morphological similarity and close phylogenetic relationship, they are considered to be sister species, or less commonly, conspecific subspecies (Eckenwalder 1996; Wang et al. 2013). They can readily cross and artificial hybrids usually show high heterosis (Hamzeh and Dayanandan 2004; Tullus et al. 2012). A recent study based on a handful of nuclear and chloroplast loci suggests that the first opening of the Bering land bridge may have driven the allopatric speciation of the two species (Du et al. 2015).
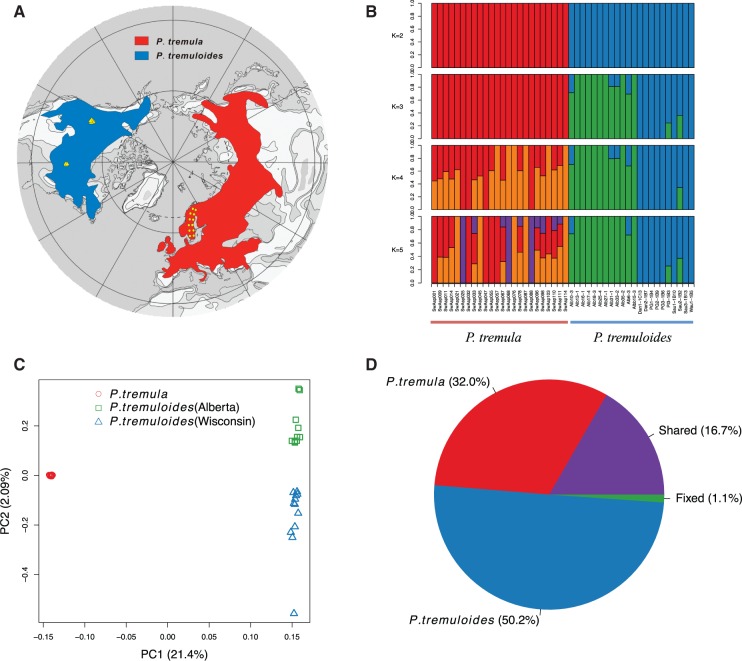
Fig. 1.
Geographic distribution and genetic structure of 24 P. tremula and 22 P. tremuloides. (A) Map showing the current geographic distribution of P. tremula (red) and P. tremuloides (blue). Yellow circles and triangles indicate the locations where the 24 individuals of P. tremula and 22 individuals of P. tremuloides were sampled. (B) Genetic structure of the two species inferred using NGSadmix. The y-axis quantifies subgroup membership, and the x-axis shows the sample ID for each individual. (C) PCA plot based on genetic covariance among all individuals of P. tremula (red circle) and P. tremuloides (green square and blue triangle). The first two principle components (PCs) are shown, with PC1 explaining 21.04% (P = 2.51 × 10−19, Tacey–Widom test) of the overall genetic variation and separating the two species, and PC2 explaining 2.09% (P = 9.65 ××10−4, Tracy–Widom test) of the overall variation and separating samples from Wisconsin (blue triangle) and Alberta (green square) in P. tremuloides. (D) Pie chart summarizing the proportion of fixed, shared, and exclusive polymorphisms of the two species.
Due to their continent-wide distributions, extraordinary levels of genetic and phenotypic diversity, along with the availability of a high-quality reference genome in the congener, Populus trichocarpa (Tuskan et al. 2006), P. tremula and P. tremuloides represent a promising system for evaluating how various evolutionary processes have shaped the patterns of genomic divergence during speciation. In this study, we use whole-genome re-sequencing data from both species to estimate and infer their divergence time and historical demographic processes. Explicit characterizations of the demographic history not only allow us to estimate historical population size fluctuations in both species, but also increase the accuracy of identifying regions or genes that have been under natural selection. By incorporating the inferred demographic scenarios into the null model, we investigate the extent to which demographic and selective events have contributed to the overall patterns of genomic differentiation between the two species. We also identify a number of outlier regions and genes that likely have evolved in response to positive and/or balancing selection during the speciation process.
Results
We generated whole-genome re-sequencing data for 24 P. tremula and 22 P. tremuloides. The high extent of conserved synteny between the genomes of aspen and P. trichocarpa (Pakull et al. 2009; Robinson et al. 2014) allowed us to map most of the sequenced reads (>88%, supplementary table S1, Supplementary Material online) from the two aspen species to the P. trichocarpa reference genome (v3.0) (Tuskan et al. 2006) following adapter removal and quality trimming (see “Materials and Methods” section). The mean coverage of uniquely mapped reads per site was 25.1 and 22.5 in samples of P. tremula and P. tremuloides, respectively (supplementary table S1, Supplementary Material online). Two complementary bioinformatics approaches were used in this study (supplementary fig. S1, Supplementary Material online): (1) For those population genetic statistics that relied on inferred site-frequency-spectrum (SFS), estimation was performed directly from genotype likelihoods without calling genotypes (Nielsen et al. 2011) as implemented in ANGSD (Korneliussen et al. 2014). (2) For those estimations that required accurate genotype calls, single nucleotide polymorphisms (SNPs) and genotypes were called with HaplotypeCaller in GATK (Danecek et al. 2011). In total, we identified 5,894,205 and 6,281,924 SNPs passing filtering criteria (see “Materials and Methods” section ) across the 24 P. tremula samples and 22 P. tremuloides samples, respectively.
Population Structure
We used NGSadmix (Skotte et al. 2013) to infer individual ancestry based on genotype likelihoods, which takes the uncertainty of genotype calling into account. It clearly sub-divided all sampled individuals into two species-specific groups when the number of clusters (K) was 2 (fig. 1B). When K = 3, there was evidence for further population sub-structuring in P. tremuloides, where individuals from populations of Alberta and Wisconsin clustered into two subgroups. With K = 4, most individuals of P. tremula were inferred to be a mixture of two genetic components, showing slight clinal variation with latitude. No further structure was found when K = 5 (fig. 1B). A principal component analysis (PCA) further supported these results (fig. 1C). Only the first two components were significant based on the Tracy–Widom test (supplementary table S2, Supplementary Material online), which explained 21.4% and 2.1% of total genetic variance, respectively (fig. 1C). Among the total number of polymorphisms in the two species, fixed differences between P. tremula and P. tremuloides accounted for 1.1%, whereas 16.7% of polymorphisms were shared between species, with the remaining polymorphic sites being private in either of the two species (fig. 1D).
To further examine the extent of population subdivision in P. tremuloides, we measured FST and dxy between the two subpopulations (Alberta and Wisconsin) along individual chromosomes (supplementary table S3, Supplementary Material online). We found low levels of genetic differentiation (average FST: 0.0443 ± 0.0325) between the two subpopulations (supplementary table S3, Supplementary Material online). Total sequence differentiation in the inter-population comparison (mean dxy = 0.0165 ± 0.0083) was similar to mean sequence differences in intra-population comparisons (πAlberta: 0.0161 ± 0.0081; πWisconsin: 0.0157 ± 0.0080, supplementary table S3, Supplementary Material online), indicating that individuals of the two populations were genetically not more different from each other than individuals within each population. Based on the summaries of site frequency spectrum (Tajima’s D and Fay & Wu’s H), both populations exhibited strong skews toward low-frequency variants (negative D) and intermediate skews toward high frequency-derived variants (negative H) (supplementary table S3, Supplementary Material online), suggesting that they likely experienced similar species-wide demographic events.
Demographic Histories
We used fastsimcoal2 (Excoffier et al. 2013), a coalescent simulation-based method, to infer the past demographic histories of P. tremula and P. tremuloides from the joint SFS. Eighteen divergence models were evaluated (supplementary fig. S2 and table S4, Supplementary Material online), and all models began with the split of the ancestral population into two derived populations and differed in terms of (1) whether post-divergence gene flow was present or not, (2) levels and patterns of gene flow between the two species, and (3) how population size changes occurred, either at the time of species divergence or afterwards (supplementary fig. S2, Supplementary Material online). The best-fitting model was a simple isolation-with-migration model where, after the two species diverged, P. tremuloides experienced exponential growth and whereas a stepwise population size change occurred in P. tremula (fig. 2A). The exact parameter estimates of divergence time, gene flow, effective population sizes and their associated 95% confidence intervals (CIs) are given in table 1. The estimated divergence time between P. tremula and P. tremuloides (TDIV) was ∼2.3 million years ago (Mya) (bootstrap range [BR]: 2.2–3.1 Mya). The contemporary effective population sizes (Ne) of P. tremula (Ne-P.tremula) and P. tremuloides (Ne-P.tremuloides) were 102,814 (BR: 93,688–105,671) and 309,500 (BR: 247,321–310,105) respectively, with both being larger than the effective population size of their common ancestor (Ne-ANC = 56,235 [48,012–69,492]). Both the per generation migration rate (m) from P. tremula to P. tremuloides (5.14 × 10−7 [5.56 × 10−7–1.11 × 10−6]), and that from P. tremuloides to P. tremula [6.53 × 10−7 [6.31 × 10−7–1.21 × 10−6]) were fairly low, which is not unexpected given the large geographical distance and disjunct distributions between the two species.
Fig. 2.
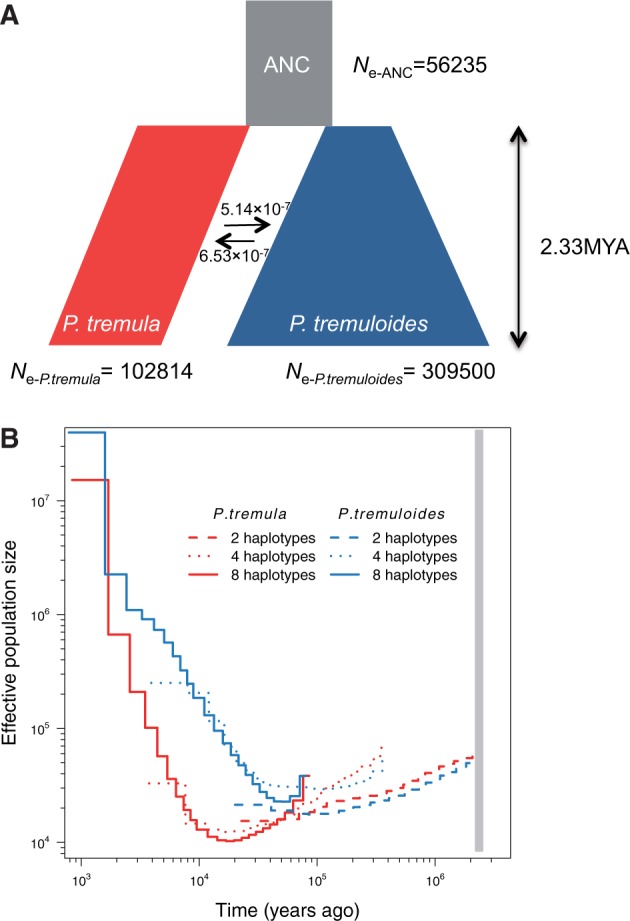
Demographic history of P. tremula and P. tremuloides. (A) Simplified graphical summary of the best-fitting demographic model inferred by fastsimcoal2. The ancestral population is in grey, P. tremula in red, P. tremuloides in blue, and their widths represent the relative effective population sizes (Ne). The arrows indicate the per generation migration rate (m) between P. tremula and P. tremuloides. The inferred demographic parameters are described in the text and shown in table 1 as well. (B) MSMC estimates of the effective population size (Ne) changes for P. tremula (red line) and P. tremuloides (blue line) based on the inference from two (dashed), four (dotted), and eight (solid) phased haplotypes in each species. Time scale on the x-axis is calculated assuming a neutral mutation rate per generation (μ) = 3.75 × 10−8 and generation time (g) = 15 years. The grey bar indicates the speciation time inferred by fastsimcoal2.
Table 1.
Inferred Demographic Parameters of the Best-Fitting Demographic Model Shown in figure 2A.
| Parameters | Point estimation | 95% CIa |
|
|---|---|---|---|
| Lower bound | Upper bound | ||
| Ne-ANC | 56,235 | 48,012 | 69,492 |
| Ne-P.tremula | 102,814 | 93,688 | 105,671 |
| Ne-P.tremuloides | 309,500 | 247,321 | 310,105 |
| mP.tremuloides->P.tremula | 6.53 × 10−7 | 6.31 × 10−7 | 1.21 × 10−6 |
| mP.tremula->P.tremuloides | 5.14 × 10−7 | 5.56 × 10−7 | 1.11 × 10−6 |
| TDIV | 2,332,410 | 218,6760 | 3,113,520 |
Note.—Parameters are defined in figure 2A. Ne-P.tremula, Ne-P.tremuloides, Ne-ANC indicate the effective population sizes of P. tremula, P. tremuloides and their ancestral population respectively, mP.tremuloides->P.tremula indicates the per generation migration rate from P. tremuloides to P. tremula, mP.tremula->P.tremuloides indicates the per generation migration rate from P. tremula to P. tremuloides, TDIV indicates the estimated divergence time between the two species obtained from fastsimcoal2.
aParametric bootstrap estimates obtained by parameter estimation from 100 datasets simulated according to the overall maximum composite likelihood estimates shown in point estimation columns. Estimations were obtained from 100,000 simulations per likelihood.
We employed the multiple sequential Markovian coalescent (MSMC) method to investigate changes of Ne over time based on inferring the time to the first coalescence between pairs of haplotypes (Schiffels and Durbin 2014). Higher resolution of recent population size changes is expected when more haplotypes are used (Schiffels and Durbin 2014). We therefore applied MSMC to phased whole-genome sequences from one (two haplotypes), two (four haplotypes), and four (eight haplotypes) individuals in each species, respectively. We did not include more haplotypes because of the high computational cost of larger samples. The MSMC-based estimates of Ne for both P. tremula (60,796) and P. tremuloides (49,701) at the beginning of species divergence (∼2.3 Mya) were very similar to the fastsimcoal2-based estimates of Ne for their ancestral population (fig. 2). The two species experienced similar magnitudes of population decline following initial divergence (fig. 2B). Population expansion in P. tremuloides occurred around 50,000–70,000 years ago and continued up to the present (fig. 2B), whereas P. tremula experienced a population expansion following a substantially longer periods of bottleneck (fig. 2B).
To assess the possible confounding effects of population subdivision and biased sampling scheme on demographic inferences in both species, we first applied MSMC analysis for P. tremuloides individuals originating from populations in Alberta and Wisconsin separately and compared them with the result obtained from the pooled samples (supplementary fig. S3, Supplementary Material online). Although the Wisconsin population was found to have undergone a decline in population size during the last 2000–3000 years ago (supplementary fig. S3, Supplementary Material online), both local populations of P. tremuloides experienced a longer period of species-wide expansion compared with P. tremula, which were in accordance with the results observed from the pooled samples. Second, demographic inferences of both species were also supported by the summary statistics based on the nucleotide diversity (θπ) and SFS (Tajima’s D and Fay & Wu’s H) (supplementary fig. S4, Supplementary Material online). The θπ in the two subpopulations of P. tremuloides were all marginally higher than in P. tremula, suggesting that the large effective population size found in P. tremuloides was not influenced by the presence of intraspecific population subdivision (supplementary fig. S4A, Supplementary Material online). In addition, the signal of more negative values of Tajima’s D in both local and pooled samples of P. tremuloides (supplementary fig. S4B, Supplementary Material online) suggests that it may have gone through a more pronounced and/or longer period of population expansion compared to P. tremula. The lower values of the genome-wide Fay &Wu’s H in P. tremula (supplementary fig. S4C, Supplementary Material online), on the other hand, might reflect the relatively longer period of low population size during the bottleneck. Taken together, these results suggest that population subdivision of P. tremuloides and the unbalanced sampling schemes between the two species have negligible effects on our demographic inferences.
Genome-Wide Patterns of Differentiation and Identification of Outlier Regions against the Best-Fitting Demographic Model
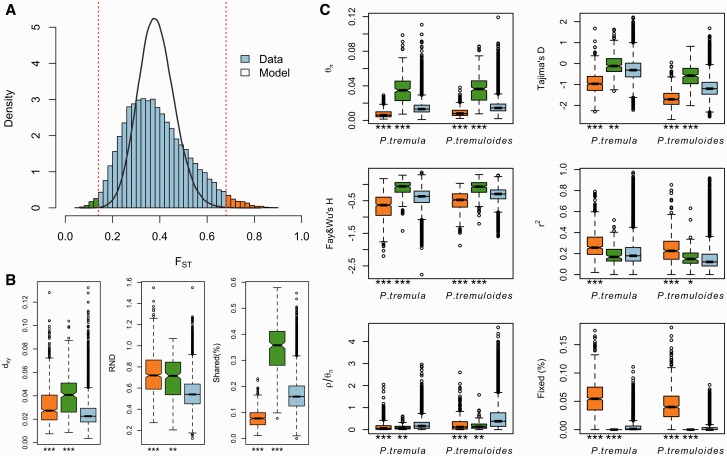
To investigate patterns of interspecific genetic differentiation across the genome, we calculated the standard measure of genetic divergence, FST, between P. tremula and P. tremuloides over non-overlapping 10 kbp windows (fig. 3). Levels of genetic differentiation varied greatly throughout the genome, with the majority of windows showing high genetic differentiation (mean FST = 0.386 ± 0.134) between species (fig. 3).
Fig. 3.
Genome-wide divergence. Chromosomal distribution of genetic differentiation (FST) between P. tremula and P. tremuloides. The small, light blue dots indicate FST values estimated over 10 kbp non-overlapping windows. Grey lines indicate FST values estimated over 100 kbp non-overlapping windows. Locations for windows displaying extreme differentiation relative to demographic simulations are highlighted with colored bars above the plot. Among them, outlier windows displaying significantly high differentiation (orange bars) are located on the topside; outlier windows displaying significantly low differentiation (green bars) are located at the bottom.
In order to test the extent to which historical demographic events can explain the observed patterns of genetic divergence between the two species. We used coalescent simulations performed in msms (Ewing and Hermisson 2010) to compare the observed patterns of differentiation to that expected under three demographic models (supplementary fig. S5, Supplementary Material online). The demographic scenario in model 1 was same as the best-fitting model inferred by fastsimcoal2 (supplementary fig. S5A and table S5, Supplementary Material online). In another two models, we incorporated the population subdivision of P. tremuloides into the best-fitting demographic model. In model 2 (supplementary fig. S5B and table S5, Supplementary Material online), we assume that there was no gene flow between the two subpopulations of P. tremuloides and explored different values of their divergence time until the simulated FST values between the two subpopulations matched those observed (supplementary fig. S6, Supplementary Material online). The same procedure was applied to model 3 (supplementary fig. S5C and table S5, Supplementary Material online), except that we there assume the per-generation gene flow between the two subpopulations of P. tremuloides (4Nem) was equal to 10 and increased their divergence time in tandem with gene flow. To assess the fit of these models, we compared two summary statistics, θπ and Tajima’s D, between the simulated and observed data for both species. As can be seen from supplementary figures S7 and S8, Supplementary Material online, there was generally good agreement between observed and simulated datasets for all three models. In addition, the above three models showed consistent distributions of simulated FST values between P. tremula and P. tremuloides, indicating that the presence of population subdivision in P. tremuloides has little effect on the overall patterns of genomic divergence that we observe between the two species (supplementary fig. S9, Supplementary Material online).
Comparing the empirical distribution of inter-specific FST with that obtained from simulations based on the best-fitting demographic model, we found that the empirical distribution was flatter and contained greater proportions of regions falling in the extremes of distribution (fig. 4A). We identified 674 and 262 outlier windows exhibiting significantly (False Discovery Rate (FDR) <0.01) high and low inter-specific FST compared with the expected null distribution obtained from the coalescent simulations (fig. 4A). These outlier windows likely have been subject to natural selection, although the coalescent simulations performed here are obvious simplifications of the true demographic histories of the two aspen species (Akey 2009). After examining the genomic distribution, physical sizes and overlaps of these outlier windows, we found that both highly and lowly differentiated regions were randomly distributed across the genome (fig. 3) and that the sizes of these regions appeared to be rather small, with the majority occurring on a physical scale smaller than 10 kbp (supplementary fig. S10, Supplementary Material online).
Fig. 4.
Identification of outlier windows that are candidates for being affected by natural selection. (A) Distribution of genetic differentiation (FST) between P. tremula and P. tremuloides from the observed (blue bar) and simulated datasets (black line). The dashed lines indicate the thresholds for determining significantly (FDR <1%) high (orange bars) and low (green bars) inter-specific differentiation based on coalescent simulations. (B) Comparisons of dxy, RND, and the proportion of inter-specific shared polymorphisms among regions displaying significantly high (orange boxes) and low (green boxes) differentiation versus the genomic background (blue boxes). (C) Comparisons of multiple population genetic statistics, nucleotide diversity (θπ), Tajima’s D, Fay & Wu’s H, LD (r2), recombination rate (ρ/θπ), the proportion of fixed differences caused by derived alleles fixed in either P. tremula or P. tremuloides, among regions displaying significantly high (orange boxes) and low differentiation (green boxes) versus the genomic background (blue boxes). Asterisks designate significant differences between outlier windows and the rest of genomic regions by Mann–Whitney U-test (* P value <0.05; ** P value <1e−4; ***P value <2.2e−16).
Signatures of Selection in Outlier Regions
As FST is a relative measure of differentiation and is thus sensitive to any processes that alter intra-species genetic variation (Charlesworth 1998; Cruickshank and Hahn 2014), we quantified and compared inter-specific genetic differentiation between two unions of outlier windows and the rest of the genome using three additional approaches: (1) pairwise nucleotide divergence between species (dxy), which is a measure that is independent of within-species diversity (Nei 1987); (2) relative node depth (RND) (Feder et al. 2005), which takes into account possible variation in the mutation rate among genomic regions by dividing dxy of the two aspen species with dxy between aspens and a third more distantly related species (P. trichocarpa); and (3) the proportion of inter-specific shared polymorphisms. Compared with the genomic background averages, both dxy and RND revealed significantly greater divergence between the two species in regions of high differentiation (fig. 4B; supplementary table S6, Supplementary Material online) and, in accordance with these patterns, the proportion of inter-specific shared polymorphisms was significantly lower in these regions (fig. 4B; supplementary table S6, Supplementary Material online). In addition, these regions are characterized by multiple signatures of positive selection within both species (Nielsen 2005), including significantly reduced levels of polymorphism (θπ), skewed allele frequency spectrum toward rare alleles (more negative Tajima’s D), increased high-frequency derived alleles (more negative Fay & Wu’s H), and stronger signals of linkage disequilibrium (LD) (higher squared correlation coefficients, r2, between pairs of SNPs) (P < 0.001, Mann–Whitney U test) (fig. 4C; supplementary table S6, Supplementary Material online). Relative to genome-wide averages, these regions also contained significantly higher proportions of fixed differences that were caused by derived alleles fixed in either species (fig. 4C; supplementary table S6, Supplementary Material online).
In contrast to patterns found in regions of high differentiation, regions of low differentiation showed significantly higher levels of polymorphism, excesses of intermediate-frequency alleles (higher Tajima’s D and Fay & Wu’s H values), higher proportions of inter-specific shared polymorphisms and negligible proportions of fixed differences between species compared to the genomic background (fig. 4B,C; supplementary table S6, Supplementary Material online). It is therefore likely that some of these regions have been targets of long-term balancing selection in both species (Charlesworth 2006). Consistent with this prediction, we found slightly lower or comparable levels of LD in these regions (fig. 4C; supplementary table S6, Supplementary Material online), which is likely due to the long-term effects of recombination on old balanced polymorphisms (Leffler et al. 2013). The higher dxy and RND values we observe in these regions may, however, be a consequence of the higher levels of ancestral polymorphisms that were maintained predating the split of the two species (fig. 4B; supplementary table S6, Supplementary Material online) (Cruickshank and Hahn 2014).
Impact of Recombination Rate on Patterns of Genetic Differentiation
We examined relationships between the scaled recombination rates (ρ = 4Nec) and levels of inter-species divergence over non-overlapping 10 kbp windows across the genome (supplementary fig. S11, Supplementary Material online). We found a significant negative correlation between relative divergence, measured as FST that depends on genetic diversity within species, and recombination rates in both P. tremula (Spearman’s ρ = −0.121, P value <0.001) and P. tremuloides (Spearman’s ρ = −0.157, P value <0.001) (supplementary fig. S11a, Supplementary Material online). In contrast to FST, we observed significantly positive correlations between absolute divergence dxy and recombination rates in both species (P. tremula: Spearman’s ρ = 0.199, P value <0.001; P. tremuloides: Spearman’s ρ = 0.140, P value <0.001) (supplementary fig. S11B, Supplementary Material online).
Because ρ = 4Nec, where c is the per-generation recombination rate and Ne is the effective population size, a reduction of Ne in regions linked to selection will lower local estimates of ρ even if local c is identical to other regions in the genome. In order to account for such effects and to obtain a measure of recombination that is independent of local Ne, we compared ρ/θπ between regions with extreme genetic differentiation and the remainder of the genome. Relative to the genomic background, our results showed significantly suppressed recombination in outlier regions displaying either exceptionally high or low inter-specific differentiation (fig. 4C).
Genes under Selection
The availability of the annotated P. trichocarpa genome enabled functional analyses of candidate target genes within regions that were likely under selection. In total, 722 and 391 genes were located in outlier windows displaying exceptionally high and low differentiation (supplementary tables S7 and S8, Supplementary Material online), respectively. Compared with the genome overall, we did not find significantly higher gene density in these outlier windows (P > 0.05, Mann–Whitney U test; supplementary fig. S12, Supplementary Material online). We used the Gene Ontology (GO) assignments of those candidate genes to assess whether specific GO terms were significantly over-represented. After accounting for multiple comparisons, we did not detect over-representation of any functional category among the candidate genes within regions of high differentiation. However, we identified 60 significantly overrepresented GO terms for genes located within regions showing significantly low genetic differentiation and most of these GO categories were associated with immune and defense responses, signal transduction or apoptosis (supplementary table S9, Supplementary Material online). Although multiple signatures of long-term balancing selection were found in these regions as shown above, some caution should still be applied when interpreting candidate genes as targets of selection. In particular, a skewed pattern of low coverage breadth was observed in lowly differentiated regions compared with either the genomic background or to those highly differentiated regions (supplementary fig. S13, Supplementary Material online). Such unequal coverage breadth most likely results from the inherent technical hurdle of short-read sequencing technologies (Brandt et al. 2015), where the high polymorphism in regions potentially under balancing selection may not only prevent the mapping of short sequence reads to the reference genome but also may result in more reads and informative sites to be discarded after the stringent quality filtering procedures (see “Materials and Methods” section). Therefore, future studies, incorporating a combination of careful experimental design and long-read sequencing technologies, are needed to explore the evolutionary history of these candidate selection genes in greater detail.
Discussion
We use a population genomic approach to resolve the evolutionary histories of two widespread and closely related forest tree species, and to highlight how genome-wide patterns of differentiation have been influenced by a variety of evolutionary processes. Our simulation-based analyses indicate that P. tremula and P. tremuloides diverged around 2.2–3.1 Mya during the Late Pliocene and/or Early Pleistocene. This timing corresponds closely with the first opening of the Bering Strait, which occurred 3.1–5.5 Mya and broke up the overland intercontinental migration route of terrestrial floras between Eurasia and North America (Marincovich and Gladenkov 1999; Gladenkov et al. 2002). This may have been less of an immediate barrier to wind-dispersed Populus than some other tree species, but the severing of the Bering land bridge associated with the onset of dramatic climatic oscillations throughout the Pleistocene were likely the principal drivers for initial divergence between P. tremula and P. tremuloides (Comes and Kadereit 1998; Milne and Abbott 2002; Du et al. 2015). Given the modern-day geographic isolation, disjunct distribution and extremely low rates of gene flow, our results support an allopatric model of speciation for these two aspen species (Morjan and Rieseberg 2004). The coalescent-based, intra-specific demographic analyses using MSMC demonstrate that both species have experienced substantial population expansions following long-term declines after species divergence. The population expansion of P. tremuloides has occurred over the last 50,000–70,000 years, following the retreat of the penultimate glaciation and has continued up to the present (Kaufman and Manley 2004). P. tremula, in contrast, experienced a more extended population contraction and, consistent with many other forest trees in Europe, the initiation of the population expansion in P. tremula coincided with the end of the Last Glacial Maximum (Hewitt 2000, 2004).
Consistent with the expectation for allopatric speciation, where the absence of gene flow allowed for the accumulation of inter-specific differentiation due to stochastic genetic drift (Coyne and Orr 2004), we detect widespread genomic differentiation between the two species. Although neutral processes can largely explain the observed patterns of genomic differentiation between the two species (Coyne and Orr 2004; Strasburg et al. 2012), a number of regions exhibit more extreme genetic differentiation compared with expectations based on demographic simulations and show multiple evidences of the action of natural selection (Nielsen et al. 2009). If natural selection has truly been one of the dominant evolutionary forces shaping patterns of genetic differentiation between the two species, regions of low recombination would be expected to show increased FST values, but not increased dxy values (Noor and Bennett 2009; Cruickshank and Hahn 2014). This occurs because natural selection (through either selective sweeps and/or background selection) removes neutral variation over longer distances in regions of low recombination (Begun and Aquadro 1992). As a consequence, relative measures of divergence (e.g., FST) that rely on within-species diversity are expected to be higher in regions with restricted recombination (Noor and Bennett 2009; Nachman and Payseur 2012). Increased absolute divergence (e.g., dxy), however, is only expected if reduced gene flow occurred in regions of low recombination (Nachman and Payseur 2012). Contrary to expectations of heterogeneous gene flow, we observe a significant negative relationship between population-scaled recombination rates (ρ) and FST, but not with dxy, in both species (Noor and Bennett 2009; Keinan et al. 2010). Our findings thus highlight significant effects of linked selection and recombination in generating the heterogeneous differentiation landscape we observe between the two Populus species (Turner and Hahn 2010; Cruickshank and Hahn 2014; Burri et al. 2015).
Rather than being physically clustered into just a few large, discrete genomic ‘islands’ as expected when species diverge in the presence of gene flow (Turner et al. 2005), differentiation islands in our study system are distributed throughout the genome, being narrowly defined and mostly located in regions with substantially suppressed recombination. Because linked selection occurred either in the form of positive selection for advantageous mutations (genetic hitchhiking) or purifying selection against weakly deleterious mutations (background selection) could be involved in the evolution of differentiation islands (Noor and Bennett 2009; Cruickshank and Hahn 2014), we assessed multiple population genetic parameters to disentangle the mechanisms generating the observed pattern of exceptional differentiation in both species (see “Results” section). We find that high differentiation regions exhibit some selective signatures that are unique to hitchhiking under positive selection, such as an excess of high-frequency derived variants, stronger LD between pairs of SNPs and increased absolute interspecific divergence (higher values of dxy and RND) (Nielsen 2005). Therefore, although a contribution of background selection to the observed patterns cannot be completely discounted, the independent action of positive selection in both P. tremula and P. tremuloides is expected to be the dominant driver for the evolution of reduced diversity and increased differentiation in most islands of differentiation. Moreover, the lack of functional over-representation for candidate genes located in regions of exceptional differentiation further suggests that a wide range of genes and functional categories may have been involved in the adaptive evolution of the two species after they became geographically isolated (Wolf et al. 2010).
In addition to the highly differentiated regions that show signs of species-specific positive selection, we also identify a number of lowly differentiated regions that are candidates for being affected by long-term balancing selection in both species (Charlesworth 2006). Apart from low inter-specific divergence and high intra-specific diversity, these regions contain an excess of sites at intermediate frequencies, a greater proportion of shared polymorphisms between the two species and lack of fixed inter-specific differences. Genes located within these regions are enriched for functional categories of immune and defense response, signal transduction, and apoptosis. Our findings thus indicate that long-term balancing selection, likely mediated by ‘trench warfare’ (Stahl et al. 1999) or ‘recycling polymorphism’ (Holub 2001) of co-evolutionary interaction between hosts and natural enemies, may have maintained functional genetic diversity at immunity and defense-related genes over long periods of time (Tiffin and Moeller 2006; Salvaudon et al. 2008). That said, because of the difficulty for accurate assessment of variation in highly polymorphic regions, more caution is required when interpreting the functional properties of the candidate genes identified here. Future studies of these candidate genes are clearly needed to better assess the adaptive genetic potential of these widespread forest tree species to current and future climate change.
A number of factors may have influenced our demographic inferences and the detection of natural selection in the two species. First, the presence of within-species population subdivision could have magnified the inference of demographic expansion in P. tremuloides, because pooling samples from populations of the Alberta and Wisconsin skews the SFS toward low-frequency polymorphism (more negative Tajima’s D) (supplementary fig. S4, Supplementary Material online) and results in larger estimates of effective population sizes than estimates obtained from local samples (supplementary fig. S3, Supplementary Material online). However, all our analyses suggest that this confounding effect is very weak (see “Results” section) and the divergence between the two subpopulations of P. tremuloides likely is too recent to have any major effects on the demographic reconstruction and tests of selection in these species (Chikhi et al. 2010). Another caveat concerns the sampling scheme used in the two species. Local samples in P. tremula may not adequately reflect species-wide demography compared with the pooled samples in P. tremuloides. However, the extent to which this might influence the estimates of inter-specific FST deserves further study. More generally, sampling should likely be more extensive in both species to capture a greater proportion of the species-wide diversity, although local sampling is expected to only have small effects in species with high gene flow like Populus (Wakeley 2000). Third, inter-specific hybridization in either species could potentially bias our results. However, there are no other species of Populus occurring in the regions from where the P. tremula individuals were sampled. For P. tremuloides, naturally occurring hybridization is only known to occur with P. grandidentata in central and eastern North America where the two species co-occur (Pregitzer and Barnes 1980). Therefore, any possible hybridization in this study would be limited to samples from the Wisconsin population of P. tremuloides but, as noted above, we did not detect any major differences in patterns of genetic variation between the two subpopulations, suggesting little or no effect of hybridization. Finally, it should be noted that the coalescent simulations performed here are probably too simplistic to recapitulate the complex demographic perturbations of real populations, and considerable caution is therefore needed when interpreting outlier regions as targets of selection.
Conclusion
Here, we provide insights into the speciation and recent evolutionary histories of two closely related forest tree species, P. tremula and P. tremuloides. This study supports an allopatric model of speciation for the two species, which are estimated to have diverged around 2.2–3.1 Mya as a result of the first opening of Bering Strait. Coalescent simulations suggest that stochastic genetic drift and historical demographic processes can largely explain the genome-wide patterns of differentiation between species. However, there is an excess of regions displaying extreme inter-specific genetic differentiation in the observed data compared with demographic simulations. We infer that heterogeneous genomic divergence is strongly driven by linked selection and variation in recombination rate in the two species. Instead of being clustered into a few large genomic “islands” as is expected under a model of speciation with gene flow, regions of pronounced differentiation are characterized by multiple signatures of positive selection in both species, and are distributed throughout the genome at many small, independent locations. Regions displaying exceptionally low differentiation are likely candidate targets of long-term balancing selection, which are strongly enriched for genes involved in immune and defense response, signal transduction, and apoptosis, suggesting a possible link to long-term co-evolutionary interactions of plant–herbivore or plant–pathogen. Our study highlights that future work should integrate more information on the natural histories of speciation, such as divergence time, geographical context, magnitudes of gene flow, demographic histories, and sources of adaptation, when interpreting the meaning of observed patterns of genomic divergence between closely related species.
Materials and Methods
Population Samples, Sequencing, Quality Control, and Mapping
A total of 24 individuals of P. tremula and 22 individuals of P. tremuloides were collected and sequenced (fig. 1a; supplementary table S1, Supplementary Material online). For each individual, genomic DNA was extracted from leaf samples and paired-end sequencing libraries with an insert size of 600 bp were constructed according to the Illumina library preparation protocol. Sequencing was carried out on the Illumina HiSeq 2000 platform at the Science for Life Laboratory in Stockholm, Sweden. All samples were sequenced to a target coverage of 25×. The sequencing data have been deposited in the Short Read Archive at NCBI under accession numbers SRP065057 and SRP065065 for samples of P. tremula and P. tremuloides, respectively.
For raw sequencing reads (Wang et al. 2015), we used Trimmomatic (Lohse et al. 2012) to remove adapter sequences and cut off bases from either the start or the end of reads when the base quality was <20. Reads were completely discarded if there were fewer than 36 bases remaining after trimming. We then mapped all reads to the P. trichocarpa reference genome (v3.0) (Tuskan et al. 2006) with default parameters implemented in bwa-0.7.10 using the BWA-MEM algorithm (Li H, unpublished data, http://arxiv.org/abs/1303.3997, last accessed May 26, 2013). Local realignment was performed to correct for the mis-alignment of bases in regions around insertions and/or deletions (indels) using RealignerTargetCreator and IndelRealigner in GATK v3.2.2 (DePristo et al. 2011). In order to account for the occurrence of PCR duplicates introduced during library construction, we used MarkDuplicates in Picard (http://picard.sourceforge.net) to remove reads with identical external coordinates and insert lengths. Only the read with the highest summed base quality was kept for downstream analyses.
Filtering Sites
Prior to variant and genotype calling, we employed several filtering steps to exclude potential errors caused by paralogous or repetitive DNA sequences. First, after investigating the empirical distribution, we removed sites showing extremely low (<100 reads across all samples per species) or high (>1200 reads across all samples per species) read coverage. Second, as a mapping quality score of 0 is assigned for reads that could be equally mapped to multiple genomic locations, we removed sites containing >20 such reads among all samples in each species. Third, we removed sites that overlapped with known repeat elements as identified by RepeatMasker (Tarailo-Graovac and Chen 2009). After all filtering steps, there were 42.8% of sites across the genome left for downstream analyses. Among them, 54.9% were found within gene boundaries, and the remainder (45.1%) was located in intergenic regions.
SNP and Genotype Calling
We employed two complementary approaches for SNP and genotype calling (supplementary fig. S1, Supplementary Material online): (1) Direct estimation without calling genotypes was implemented in the software ANGSD v0.602 (Korneliussen et al. 2014). Only reads with a minimal mapping quality of 30 and bases with a minimal quality score of 20 were considered. For all filtered sites in both species, we defined the alleles that were the same as those found in the P. trichocarpa reference genome as the ancestral allelic state. We used the −doSaf implementation to calculate the site allele frequency likelihood based on the SAMTools genotype likelihood model at all sites (Li et al. 2009), and then used the −realSFS implementation to obtain a maximum likelihood estimate of the unfolded SFS using the Expectation Maximization (EM) algorithm (Kim et al. 2011). Several population genetic statistics were then calculated based on the global SFS (supplementary fig. S1, AX). (2) Multi-sample SNP and genotype calling was implemented in GATK v3.2.2 with HaplotypeCaller and GenotypeGVCFs (supplementary fig. S1, Supplementary Material online) (DePristo et al. 2011). A number of filtering steps were performed to reduce false positives from SNP and genotype calling: (1) Remove SNPs that were located in regions not passing all previous filtering criteria; (2) Remove SNPs with more than two alleles in both species; (3) Remove SNPs at or within 5 bp from any indels; (4) Assign genotypes as missing if their quality scores (GQ) were <10, and then remove SNPs with more than two missing genotypes in each species; (5) Remove SNPs showing significant deviation from Hardy–Weinberg Equilibrium (P < 0.001) in each species.
Population Structure
Population genetic structure was inferred using the program NGSadmix (Skotte et al. 2013), with only sites containing <10% of missing data being used. We used the SAMTools model (Li et al. 2009) in ANGSD to estimate genotype likelihoods and then generated a beagle file for the subset of the genome that was determined as being variable using a likelihood ratio test (P value <10−6) (Kim et al. 2011). We predefined the number of genetic clusters K from 2 to 5, and the maximum iteration of the EM algorithm was set to 10,000.
As another method to visualize the genetic relationships among individuals, we performed PCA using ngsTools that accounted for sequencing errors and uncertainty in genotype calls (Fumagalli et al. 2014). The expected covariance matrix across pairs of individuals in both species was computed based on the genotype posterior probabilities across all filtered sites. Eigenvectors and eigenvalues from the covariance matrix were generated with the R function eigen, and significance levels were determined using the Tracy–Widom test as implemented in EIGENSOFT version 4.2 (Patterson et al. 2006).
Demographic History
We inferred demographic histories associated with speciation for P. tremula and P. tremuloides using a coalescent simulation-based method implemented in fastsimcoal 2.5.1 (Excoffier et al. 2013). Two-dimensional joint SFS (2D-SFS) was constructed from posterior probabilities of sample allele frequencies by ngsTools (Fumagalli et al. 2014). A total of 100,000 coalescent simulations were used for the estimation of the expected 2D-SFS and log-likelihood for a set of demographic parameters in each model. Global maximum likelihood estimates for each model were obtained from 50 independent runs, with 10–40 conditional maximization algorithm cycles. Model comparison was based on the maximum value of likelihood over the 50 independent runs using the Akaike information criterion and Akaike’s weight of evidence (Excoffier et al. 2013). The model with the maximum Akaike’s weight value was chosen as the optimal one. We assumed a mutation rate of 2.5 × 10−9 per site per year and a generation time of 15 years in Populus (Koch et al. 2000) when converting estimates to units of years and individuals. Parameter confidence intervals of the best model were obtained by 100 parametric bootstraps, with 50 independent runs in each bootstrap.
We then employed MSMC method to estimate variation of scaled population sizes (Ne) over historical time in both species (Schiffels and Durbin 2014), which is an extension of a pairwise sequential Markovian coalescent method (Li and Durbin 2011). Prior to the analysis, all segregating sites within each species were phased and imputed using fastPHASE v1.4.0 (Scheet and Stephens 2006). A generation time of 15 years and a rate of 2.5 × 10−9 mutations per nucleotide per year (Koch et al. 2000) were used to convert the scaled times and population sizes into real times and sizes.
Genome-Wide Patterns of Differentiation
We have previously shown that LD decays within 10 kbp in both P. tremula and P. tremuloides (Wang et al. 2016), and we thus divided the genome into 39,406 non-overlapping windows of 10 kbp in size to investigate patterns of genomic differentiation between species. For a window to be included in the downstream analyses, there should be at least 1,000 bases left after all above filtering steps. Levels of genetic differentiation between species at each site were estimated using method-of-moments FST estimators implemented in ngsFST from the ngsTools package (Fumagalli et al. 2014), which calculates indices of the expected genetic variance between and within species from posterior probabilities of sample allele frequencies, without relying on SNP or genotype calling (Fumagalli et al. 2013). We then averaged FST values of all sites within each 10 kbp non-overlapping window.
Coalescent Simulations for Detecting Outlier Windows
In order to examine thresholds for detection of outlier windows that may have been targets of natural selection, we conducted coalescent simulations to compare observed patterns of genetic differentiation (FST) to those expected under different demographic models (see “Results” section). All simulations were performed using the program msms v3.2rc (Ewing and Hermisson 2010) based on demographic parameters derived from the best-fitting model inferred by fastsimcoal2.5.1 (Excoffier et al. 2013). Population-scaled recombination rates (ρ) were assumed to be between 1 and 5 kbp−1 given the large variation we found in both species (Wang et al. 2016). We simulated genotypes corresponding to a 10 kbp region with the same sample size as the real data for 100,000 replications, from where we simulate genotype likelihoods using the program msToGlf in ANGSD (Korneliussen et al. 2014) by assuming a mean sequencing depth of 20× and an error rate of 0.5%. We estimated two summary statistics, nucleotide diversity (θπ), and Tajima’s D, from sample allele frequency likelihoods in ANGSD for all simulation replicates to test whether the simulated data match the observed data. To assess whether any of the observed windows display FST values deviating significantly from neutral expectations, we determined the conditional probability (P value) of observing more extreme inter-specific FST values among simulated datasets than among the observed data. The determination of significance was based on running 500,000 coalescent simulations of the most acceptable demographic null model (see “Results” section). We then corrected for multiple testing by using the FDR adjustment, and only windows with FDR <1% were considered as candidate regions affected by selection (Storey 2002).
Molecular Signatures of Selection in Outlier Regions
To assess the occurrence of selection in outlier windows displaying either exceptionally high or low differentiation, we compared the two unions of outlier windows with the remaining portion of the genome by a variety of additional population genetic statistics in both species. First, θπ, Tajima’s D (Tajima 1989), and Fay & Wu’s H (Fay and Wu 2000) were calculated from sample allele frequency likelihoods in ANGSD over non-overlapping 10 kbp windows. Second, levels of LD and population-scaled recombination rates (ρ) were estimated and compared. To evaluate levels of LD within each 10 kbp window, the correlation coefficients (r2) between SNPs with pairwise distances larger than 1 kbp were calculated using VCFtools v0.1.12b (Danecek et al. 2011). Population-scaled recombination rates (ρ) were estimated using the Interval program of LDhat 2.2 (McVean et al. 2004) with 1,000,000 MCMC iterations sampling every 2,000 iterations and a block penalty parameter of five. The first 100,000 iterations of the MCMC iterations were discarded as burn-in. Although no experimental recombination map is available for any of the two aspen species at the moment, a previous study on P. trichocarpa found a high correlation between recombination rates estimated indirectly from the re-sequencing data and that estimated directly from low-resolution genetic maps (Spearman’s ρ = 0.50, P < 10−10) (Slavov et al. 2012). This suggests that the population-scaled recombination rate we used in this article can largely reflect the true patterns of recombination events in these species. Resulting estimates of r2 and ρ were then averaged over each 10 kbp window. In both species, windows were discarded in the estimation of r2 and ρ if there were <3 kbp and/or ten SNPs left from previous filtering steps. Finally, we used the program ngsStat (Fumagalli et al. 2014) to calculate several additional measures of genetic differentiation: (1) with P. trichocarpa as an outgroup, the proportion of fixed differences that is caused by derived alleles fixed in either P. tremula or P. tremuloides among all segregating sites; (2) the proportion of inter-specific shared polymorphisms among all segregating sites; (3) dxy, which was calculated from sample allele frequency posterior probabilities at each site and was then averaged over each 10 kbp window; and (4) the RND, which was calculated by dividing the dxy of the two aspen species with dxy between aspen (represented by 24 samples of P. tremula in this study) and P. trichocarpa (24 samples; see Wang et al. 2016). Significance of the differences between outlier windows and the genome-wide averages for all above-mentioned population genetic statistics were examined using one-sided Wilcoxon ranked-sum tests.
GO Enrichment
To determine whether any functional classes of genes were overrepresented among regions that were candidates for being under selection, we performed functional enrichment analysis of GO using Fisher’s exact test by agriGO’s Term Enrichment tool (http://bioinfo.cau.edu.cn/agriGO/index.php) (Du et al. 2010). GO groups with fewer than two outlier genes were excluded from this analysis. P values of Fisher’s exact test were further corrected for multiple testing with Benjamini–Hochberg FDR (Benjamini and Hochberg 1995). GO terms with a corrected P value <0.05 were considered to be significantly enriched.
Supplementary Material
Supplementary figures S1–S13 and Supplementary tables S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Rick Lindroth for providing access to the samples of P. tremuloides used in this study. We thank Carin Olofsson for extracting DNA for all samples used in this study. We thank both the editor and two anonymous referees for their useful comments on the manuscript. The research has been funded through grants from Vetenskapsrådet and a Young Researcher Award from Umeå University to P.K.I. J.W. was supported by a scholarship from the Chinese Scholarship Council. The authors also would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure (NGI), and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
References
- Akey JM. 2009. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 19:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Begun DJ, Aquadro CF. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356:519–520. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 57:289–300. [Google Scholar]
- Brandt DY, Aguiar VR, Bitarello BD, Nunes K, Goudet J, Meyer D. 2015. Mapping bias overestimates reference allele frequencies at the Hla genes in the 1000 Genomes Project Phase I Data. G3 5:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bureš S, Garamszegi LZ, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25:1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan CM, Rowe CA, Ryel RJ, Shaw JD, Madritch MD, Mock KE. 2013. Continental‐scale assessment of genetic diversity and population structure in quaking aspen (Populus tremuloides). J Biogeogr. 40:1780–1791. [Google Scholar]
- Campagna L, Gronau I, Silveira LF, Siepel A, Lovette IJ. 2015. Distinguishing noise from signal in patterns of genomic divergence in a highly polymorphic avian radiation. Mol Ecol. 24:4238–4251. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Albert F, Afonso S, Pereira R, Burbano H, Campos R, Melo-Ferreira J, Blanco-Aguiar J, Villafuerte R, Nachman M, et al. 2014. The genomic architecture of population divergence between subspecies of the European Rabbit. PLoS Genet. 10:e1003519.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 15:538–543. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2:e64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. 2010. The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186:983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comes HP, Kadereit JW. 1998. The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 3:432–438. [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Cruickshank TE, Hahn MW. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23:3133–3157. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Wang Z, Ingvarsson PK, Wang D, Wang J, Wu Z, Tembrock LR, Zhang J. 2015. Multilocus analysis of nucleotide variation and speciation in three closely related Populus (Salicaceae) species. Mol Ecol. 24:4994–5005. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38:W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenwalder JE. 1996. Systematics and evolution of Populus In: Stettler RF, Bradshaw HD, Heilman PE, Hinckley TM, editors. Biology of populus and its implications for management and conservation. Ottawa: NRC Research Press; p. 7–32. [Google Scholar]
- Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491:756–760. [DOI] [PubMed] [Google Scholar]
- Ewing G, Hermisson J. 2010. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26:2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa V, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9:e1003905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. 2000. Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Xie X, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M. 2005. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci USA. 102:6573–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feulner P, Chain F, Panchal M, Huang Y, Eizaguirre C, Kalbe M, Lenz T, Samonte I, Stoll M, Bornberg-Bauer E, et al. 2015. Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genet. 11:e1004966.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sánchez E, Albrechtsen A, Nielsen R. 2013. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195:979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T, Nielsen R. 2014. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30:1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladenkov AY, Oleinik AE, Marincovich L, Barinov KB. 2002. A refined age for the earliest opening of Bering Strait. Palaeogeogr Palaeoclimatol Palaeoecol. 183:321–328. [Google Scholar]
- Hamzeh M, Dayanandan S. 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am J Bot. 91:1398–1408. [DOI] [PubMed] [Google Scholar]
- Hewitt G. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci. 359:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. [DOI] [PubMed] [Google Scholar]
- Holub EB. 2001. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2:516–527. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. 2008. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman D, Manley W. 2004. Quaternary glaciations—extent and chronology Part II: North America In: Ehlers J, Gibbard PL. editors. Developments in quaternary science. Amsterdam: Elsevier; p. 1–440. [Google Scholar]
- Keinan A, Reich D, Begun DJ. 2010. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 6:e1000886.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lohmueller KE, Albrechtsen A, Li Y, Korneliussen T, Tian G, Grarup N, Jiang T, Andersen G, Witte D, et al. 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12:231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol. 17:1483–1498. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Ségurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, et al. 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339:1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger A, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40:W622–W627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. 2003. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 4:981–994. [DOI] [PubMed] [Google Scholar]
- Marincovich L, Gladenkov AY. 1999. Evidence for an early opening of the Bering Strait. Nature 397:149–151. [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304:581–584. [DOI] [PubMed] [Google Scholar]
- Milne RI, Abbott RJ. 2002. The origin and evolution of Tertiary relict floras. Adv Bot Res. 38:281–314. [Google Scholar]
- Morjan CL, Rieseberg LH. 2004. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 13:1341–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Leuschner C, Horna V, Zhang C. 2012. Photosynthetic characteristics and growth performance of closely related aspen taxa: on the systematic relatedness of the Eurasian Populus tremula and the North American P. tremuloides. Flora 207:87–95. [Google Scholar]
- Nachman MW, Payseur BA. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond B Biol Sci. 367:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Kremer A. 2011. Forest tree genomics: growing resources and applications. Nat Rev Genet. 12:111–122. [DOI] [PubMed] [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 39:197–218. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andrés AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, et al. 2009. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 19:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. 2011. SNP calling, genotype calling, and sample allele frequency estimation from New-Generation Sequencing data. PloS One 7:e37558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Bennett SM. 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Feder JL. 2012. Genomic divergence during speciation: causes and consequences. Philos Trans R Soc Lond B Biol Sci. 367:332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz‐Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Pakull B, Groppe K, Meyer M, Markussen T, Fladung M. 2009. Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet Genomes 5:505–515. [Google Scholar]
- Patterson N, Price A, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregitzer KS, Barnes BV. 1980. Flowering phenology of Populus tremuloides and P. grandidentata and the potential for hybridization. Can J Forest Res. 10:218–223. [Google Scholar]
- Renaut S, Grassa C, Yeaman S, Moyers B, Lai Z, Kane N, Bowers J, Burke J, Rieseberg L. 2013. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 4:1827.. [DOI] [PubMed] [Google Scholar]
- Robinson KM, Delhomme N, Mähler N, Schiffthaler B, Önskog J, Albrectsen BR, Ingvarsson PK, Hvidsten TR, Jansson S, Street NR. 2014. Populus tremula (European aspen) shows no evidence of sexual dimorphism. BMC Plant Biol. 14:276.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaudon L, Giraud T, Shykoff JA. 2008. Genetic diversity in natural populations: a fundamental component of plant–microbe interactions. Curr Opin Plant Biol. 11:135–143. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre G-P, Bank C, Brännström Å, et al. 2014. Genomics and the origin of species. Nat Rev Genet. 15:176–192. [DOI] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, Rodgers‐Melnick E, Lipphardt MF, Pennacchio CP, Hellsten U, Pennacchio LA, et al. 2012. Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol. 196:713–725. [DOI] [PubMed] [Google Scholar]
- Sousa V, Hey J. 2013. Understanding the origin of species with genome-scale data: modelling gene flow. Nat Rev Genet. 14:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400:667–671. [DOI] [PubMed] [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 64:479–498. [Google Scholar]
- Strasburg JL, Sherman NA, Wright KM, Moyle LC, Willis JH, Rieseberg LH. 2012. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans R Soc Lond B Biol Sci. 367:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using repeatmasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 4:1–4.. [DOI] [PubMed] [Google Scholar]
- Tiffin P, Moeller DA. 2006. Molecular evolution of plant immune system genes. Trends Genet. 22:662–670. [DOI] [PubMed] [Google Scholar]
- Tullus A, Rytter L, Tullus T, Weih M, Tullus H. 2012. Short-rotation forestry with hybrid aspen (Populus tremula L.× P. tremuloides Michx.) in Northern Europe. Scand J Forest Res. 27:10–29. [Google Scholar]
- Turner T, Hahn M, Nuzhdin S. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3:e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. 2010. Genomic islands of speciation or genomic islands and speciation? Mol Ecol. 19:848–850. [DOI] [PubMed] [Google Scholar]
- Tuskan G, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604. [DOI] [PubMed] [Google Scholar]
- Via S. 2009. Natural selection in action during speciation. Proc Natl Acad Sci USA. 106:9939–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. 2000. The effects of subdivision on the genetic divergence of populations and species. Evolution 54:1092–1101. [DOI] [PubMed] [Google Scholar]
- Wang J, Scofield D, Street N, Ingvarsson P. 2015. Variant calling using NGS data in European aspen (Populus tremula) In: Sablok G, Kumar S, Ueno S, Kuo J, Varotto C, editors. Advances in the understanding of biological sciences using next generation sequencing (NGS) approaches. New York: Springer; p. 43–61. [Google Scholar]
- Wang J, Street NR, Scofield DG, Ingvarsson PK. 2016. Natural selection and recombination rate variation shape nucleotide polymorphism across the genomes of three related Populus species. Genetics 202:1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Du S, Dayanandan S, Wang D, Zeng Y, Zhang J. 2013. Phylogeny reconstruction and hybrid analysis of Populus (salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PloS One 9:e103645.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Lindell J, Backström N. 2010. Speciation genetics: current status and evolving approaches. Philos Trans R Soc Lond B Biol Sci. 365:1717–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.