Highlight
CHLI1 is indispensable for MgCh function in C. reinhardtii, while CHLI2 is not involved in MgCh activity. Inactivation of MgCh due to CHLI1 deficiency results in altered nuclear gene expression.
Key words: Chlamydomonas, CHLI, chloroplast retrograde signaling, Mg chelatase, protoporphyrin IX, tetrapyrrole biosynthesis pathway.
Abstract
The oligomeric Mg chelatase (MgCh), consisting of the subunits CHLH, CHLI, and CHLD, is located at the central site of chlorophyll synthesis, but is also thought to have an additional function in regulatory feedback control of the tetrapyrrole biosynthesis pathway and in chloroplast retrograde signaling. In Arabidopsis thaliana and Chlamydomonas reinhardtii, two genes have been proposed to encode the CHLI subunit of MgCh. While the role of CHLI1 in A. thaliana MgCh has been substantially elucidated, different reports provide inconsistent results with regard to the function of CHLI2 in Mg chelation and retrograde signaling. In the present report, the possible functions of both isoforms were analyzed in C. reinhardtii. Knockout of the CHLI1 gene resulted in complete loss of MgCh activity, absence of chlorophyll, acute light sensitivity, and, as a consequence, down-regulation of tetrapyrrole biosynthesis and photosynthesis-associated nuclear genes. These observations indicate a phenotypical resemblance of chli1 to the chlh and chld C. reinhardtii mutants previously reported. The key role of CHLI1 for MgCh reaction in comparison with the second isoform was confirmed by the rescue of chli1 with genomic CHLI1. Because CHLI2 in C. reinhardtii shows lower expression than CHLI1, strains overexpressing CHLI2 were produced in the chli1 background. However, no complementation of the chli1 phenotype was observed. Silencing of CHLI2 in the wild-type background did not result in any changes in the accumulation of tetrapyrrole intermediates or of chlorophyll. The results suggest that, unlike in A. thaliana, changes in CHLI2 content observed in the present studies do not affect formation and activity of MgCh in C. reinhardtii.
Introduction
The first committed enzyme in the chlorophyll branch of tetrapyrrole biosynthesis (TBS) in photosynthetic organisms is Mg chelatase (MgCh). MgCh is responsible for the catalysis of Mg2+ insertion into protoporphyrin IX (Proto), which results in formation of Mg protoporphyrin IX (MgProto). MgCh is a heterotrimeric complex consisting of least three separable proteins, CHLI (36–46kDa), CHLD (60–87kDa), and CHLH (120–155; Walker and Weinstein, 1991; Willows et al., 1996).
In the vascular plant Arabidopsis thaliana and in the green alga Chlamydomonas reinhardtii, one gene encodes the CHLD subunit, while two genes each for the CHLH and CHLI subunits have been identified. Based on the annotations and sequences obtained from the C. reinhardtii genome v5.5, Phytozome v10.2, expression of all these genes is supported by multiple expressed sequence tags (ESTs) in the dbEST of GenBank (http://www.ncbi.nlm.nih.gov/genbank, accessed 15 January 2016). The presence of two genes for CHLH and CHLI points to the possibility of different functions of the two isoforms of these proteins. However, despite the sizeable amount of research on MgCh, the exact role of each of its intrinsic subunits still remains elusive. CHLI contains Mg-ATP-binding motifs (Hansson et al., 2002), and the ATPase activity of CHLI was demonstrated in vitro (Sirijovski et al., 2006). CHLH was shown to bind Proto (Willows et al., 1996), while CHLD is thought to provide a platform for the assembly of the CHLI subunits (Sirijovski et al., 2006).
Previous analysis in A. thaliana indicated that the transcript levels of CHLI1 and CHLI2 are similar, but because CHLI2 protein could not be detected in the chli1 mutant, it was assumed that it undergoes a rapid post-translational turnover (Rissler et al., 2002). However, other studies in A. thaliana demonstrated that CHLI2 is expressed at a much lower level than CHLI1 (Brenner et al., 2000; Matsumoto et al., 2004; Huang and Li, 2009). It was also suggested that CHLI2 cannot be assembled into the hexameric ring because of changes in the residues of its C-terminus (Apchelimov et al., 2007). It was concluded that the function of CHLI2 cannot be determined due to its low expression level. Nevertheless, the recombinant CHLI2 of A. thaliana has an ATPase activity, although with a lower maximum reaction rate (V max) and higher Michaelis constant (K mATP) than CHLI1, and it shows thioredoxin-dependent reduction of a disulfide bond and thiol modulation of its ATPase activity (Kobayashi et al., 2008). However, in more recent work, it was demonstrated that overexpression of the native CHLI2 in A. thaliana can complement impairment caused by the mutation in CHLI1 (Huang and Li, 2009).
The chloroplast can exert changes on nuclear gene expression (Beck, 2001). A series of experiments led to the discovery of the genomes uncoupled (gun) mutants, which showed disturbed retrograde signaling, resulting in deregulation of nuclear gene expression (Susek et al., 1993). Inactivation of MgCh due to the mutation in CHLH was demonstrated in the A. thaliana gun5 mutant (genomes uncoupled 5), resulting in deregulated expression of the LHCB2 gene (Mochizuki et al., 2001). Interestingly, chlh and chld mutants characterized with deficient MgCh activity show plastid-mediated deregulation of selected nuclear genes (Chekunova et al., 2001; von Gromoff et al., 2008; Brzezowski et al., 2014).
Although it was reported that A. thaliana cs and ch42 mutants with defects in CHLI1 do not show the gun phenotype [modified gene expression in the presence of norflurazon (NF) in comparison with NF-treated wild-type seedlings (Mochizuki et al., 2001)], it was suggested that semi-dominant cs215/cs215 and the chli1/chli1 chli2/chli2 double mutant do, because they accumulate a higher level of LHCB1 transcript than the wild type upon NF treatment (Huang and Li, 2009).
Different studies correlated the product of MgCh activity with its function in chloroplast retrograde signaling, suggesting that MgProto is required for down-regulation of nuclear gene expression by means of transmitting the signal from the chloroplast (Johanningmeier and Howell, 1984; Susek et al., 1993; Oster et al., 1996; Kropat et al., 1997, 2000; La Rocca et al., 2001; Strand et al., 2003). Although the involvement of Mg porphyrins in chloroplast retrograde signaling was questioned in several studies (Mochizuki et al., 2008; Moulin et al., 2008; Brzezowski et al., 2014; Schlicke et al., 2014), it still seems to be a matter of debate (Zhang et al., 2011; Zhang et al., 2015).
The functions of the two CHLI isoforms in Mg chelation of Proto and the possible effect of disturbed tetrapyrrole biosynthesis on plastid-derived retrograde signaling were examined in the present study. The substantial role of CHLI1 in the MgCh reaction has been demonstrated, while overexpression or silencing of CHLI2 led to the conclusion that the second CHLI isoform in C. reinhardtii is apparently not necessary for MgCh activity. With regard to the effect of the chli1 knockout on chloroplast retrograde signaling, down-regulation of several tetrapyrrole biosynthesis-related and photosynthesis-associated nuclear (PhAN) genes was observed.
Materials and methods
Strains and culture conditions
Because of the acute light sensitivity (Fig. 2B; Supplementary Fig. S5 at JXB online) and a complete lack of chlorophyll (Supplementary Fig. S2A, B), the chli1/fdx3 mutant was maintained on agar-solidified TRIS-acetate-phosphate (TAP; Gorman and Levine, 1965; Harris, 1989) in the dark, at 23 °C. Genetic crosses were performed as described previously (Harris, 1989).
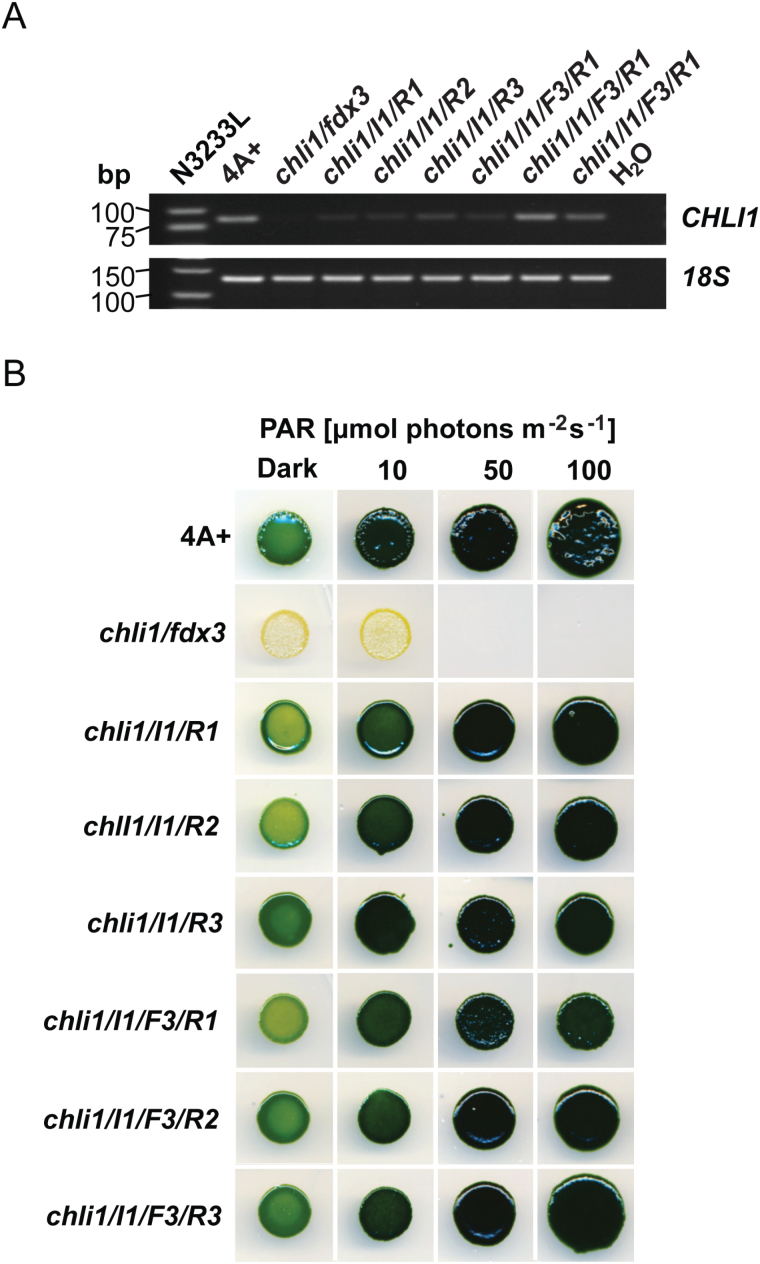
Fig. 2.
Analysis of chli1/fdx3 and strains obtained from the rescue transformation with CHLI1/pMS188 or FDX3/pMS586, or both plasmids simultaneously. (A) The RT–PCR examination of the presence of the CHLI1 transcripts; the low molecular weight DNA N3222L ladder (New England Biolabs GmbH, Frankfurt am Main, Germany) is shown. (B) General appearance, growth in the dark, and sensitivity to 10, 50, and 100 µmol photons m−2 s−1 of photosynthetically active radiation (PAR).
The CC-3395 cell wall-deficient (cwd) arginine auxotrophic (arg7-8), mating-type minus (mt–) strain was obtained from the Chlamydomonas Center (University of Minnesota, St. Paul, MN, USA). CC-3395 carries a mutation in argininosuccinate lyase (ARG7) and was maintained on agar-solidified TAP supplemented with 5 µg ml−1 arginine at 50 µmol photons m−2 s−1 and 23 °C. In the present study, both CC-3395 and 4A+ were referred to as the wild-type strains.
Analysis of tetrapyrrole intermediates and end-products
Samples from all the strains, normalized to contain 1.2×108 cells, were collected by centrifugation. Pellets were snap-frozen in LN2, followed by extraction with acetone/0.2M NH4OH (9/1, v/v) cooled to –20 °C to prevent chlorophyll degradation by chlorophyllase (Hu et al., 2013). The pellets obtained were used for the extraction of heme with acetone/HCl/DMSO (10/0.5/2, v/v/v) at room temperature. TBS intermediates and end-products were analyzed by HPLC (Czarnecki et al., 2011).
Preparation of nucleic acids and transcript analyses
The total DNA, used as a template for amplification of genomic CHLI1 and FDX3 for the rescue transformation of chli1/fdx3, was isolated from 4A+ as described previously (Brzezowski et al., 2012).
Total RNA was isolated using TRIzol Reagent (Life Technologies, http://www.lifetechnologies.com, accessed 15 January 2016) according to the manufacturer’s instruction. For both reverse transcription–PCR (RT–PCR) and quantitative RT–PCR (qRT–PCR), the constitutively expressed 18S rRNA was used as the control using PB69 and PB70 primers (Supplementary Table S2). Primers for RT–PCR and qRT–PCR were designed using PRIMER3PLUS (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi, accessed 15 January 2016; Supplementary Table S2). Transcripts for CHLI1, CHLI2, and CHLI1-neighboring genes were analyzed by RT–PCR in cells from liquid TAP media kept in the dark (Supplementary Fig. S1). Transcript analyses of the genes encoding enzymes of the TBS pathway and PhAN genes (Supplementary Fig. S3) were examined by qRT–PCR in the dark and following exposure to 20 µmol photons m−2 s−1 light, for 1, 2, 4, and 8h. The CHLI2 transcripts in strains overexpressing CHLI2 were examined in the dark, while transcripts of CHLI1, CHLI2, and GPX5 were analyzed in cells exposed to 50 µmol photons m−2 s−1 light.
Vector construction
Two vectors were constructed for the attempts to rescue chli1/fdx3. The pMS188 plasmid (Schroda et al., 2002) carries a bleomycin resistance cassette (BleR; Stevens et al., 1996). The 4282bp long genomic CHLI1 was PCR amplified using primers PB207 and PB208 with KpnI and PsiI restriction sites attached, respectively (Supplementary Table S2) and was introduced into pMS188 pre-digested with KpnI/PsiI to generate the CHLI1/pMS188 plasmid (Supplementary Fig. S4A). The primers PB209 and PB210, with PsiI and DraIII attached, respectively (Supplementary Table S2), were used to amplify a 1556bp genomic FDX3 fragment. The DNA was introduced into the pMS586 vector (pHyg3; obtained from Michael Schroda, University of Kaiserslautern, Germany) pre-digested with PsiI/DraIII, resulting in FDX3/pMS586 (Supplementary Fig. S4A). Transformation of chli1/fdx3 was performed with CHLI1/pMS188 or FDX3/pMS586, or both plasmids at the same time. Selection was conducted on 15 µg ml−1 zeocin, 10 µg ml−1 hygromycin, or both antibiotics, depending on the plasmid used. Following transformation and recovery in the dark, all plates were transferred to 50 μmol photons m−2 s−1 to increase the selection pressure.
The CHLI2 coding sequence (CDS) was amplified by PCR from cDNA using forward primers PB317 and PB313 with NdeI and NaeI restriction sites attached, respectively, and the reverse primer PB314 carrying an EcoRI site (Supplementary Table S2), followed by ligation into pGenD2 (Fischer and Rochaix, 2001) pre-digested with NdeI and EcoRI or NaeI and EcoRI, producing pCHLI2/GenD2 without and pCHLI2/GenD2-cTP with the CDS of the PSAD transit peptide (Supplementary Fig. S4B). Both vectors were used for co-transformation of the chli1/fdx3 strain with pMS188, and selection was conducted on TAP-agar plates with 15 µg ml−1 zeocin with illumination of 10 µmol photons m−2 s−1.
For the silencing of CHLI2, the MicroRNA Designer platform (WMD3, http://wmd3.weigelworld.org/cgi-bin/webapp.cgi, accessed 15 January 2016) was used to design suitable 90-mer oligonucleotides (PB449 and PB450; Supplementary Table S2) specific for targeting the region spanning exon 9 and the 3'-untranslated region (UTR) of CHLI2, carrying two mismatches corresponding to positions 1410 and 1427 of CHLI2 cDNA (genome v5.5, Phytozome v10.2; Supplementary Table S2). Following annealing of PB449 and PB450 (Supplementary Table S2), phosphorylation, and digestion with SpeI, the 90bp dsDNA was introduced into the SpeI site of the artificial miRNA (amiRNA) precursor cre-MIR1157 in the pChlamiRNA2 vector (Molnar et al., 2009), producing CHLI2/amiRNA-E9/3'UTR, which was used for CC-3395 transformation. Selection was based on the expression of ARG7 in pChlamiRNA2 on TAP plates without arginine, and at 10 µmol photons m−2 s−1, to avoid a possible phototoxic effect in potentially light-sensitive transformants. All C. reinhardtii transformations were performed by electroporation (Shimogawara et al., 1998).
Protein analyses
The total protein content was extracted in 400 µl of buffer containing 56mM Na2CO3, 56mM DTT, 2% SDS, 12% sucrose, and 2mM EDTA. The total protein amounts were determined using a Pierce BCA Protein Assay Kit (Life Technologies). For the analysis of the CHLI1 and CHLI2 content in chli1/fdx3 and strains overexpressing CHLI2, 50 µg of the total protein was loaded, while 10 µg was used for analysis in strains with silenced CHLI2. Proteins were separated by 12% SDS–PAGE, followed by transfer to a nitrocellulose membrane. The polyclonal CHLI1 antibody developed for A. thaliana was used, which recognized both CHLI1 and CHLI2 of C. reinhardtii. The chemiluminescence signal was detected using Stella 3200 (Raytest Isotopenmessgeräte GmbH, http://www.raytest.de, accessed 15 January 2016). Quantification of the signal was performed using AIDA/2D densitometry software (Raytest).
In silico analysis of the CHLI proteins
To analyze the homology between the CHLI proteins from different organisms, CHLI1 and CHLI2 sequences from A. thaliana and C. reinhardtii were used as queries within the Viridiplantae by the BLAST tool on Phytozome v10.2. The 10 highest scores for each BLAST search were selected, duplicates from different searches were removed, and transcript names were used for protein identification. Additionally, sequences for Synechocystis sp. PCC6803, Rhodobacter sphaeroides, and Nicotiana tabacum were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/protein/, accessed 15 January 2016), under the transcript names of WP_010871795.1, AAB97156.1, and O22436.1, respectively. The phylogenetic tree (Fig. 1) was constructed using DNAMAN software Version 6 (Lynnon Corporation, Pointe-Claire, QC, Canada). Bootstrap values were calculated from 1000 iterations; the alignment can be found in Supplementary Fig. S9.
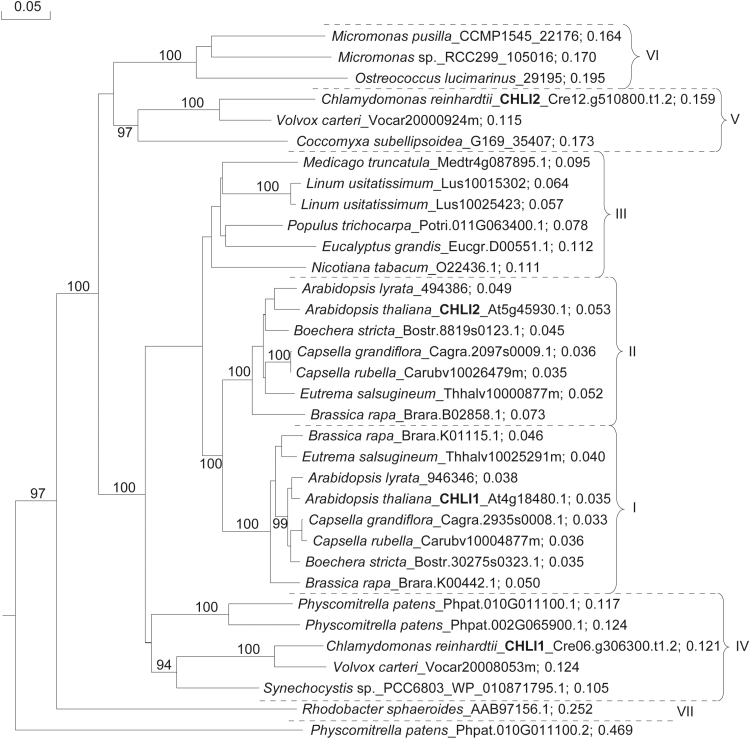
Fig. 1.
Phylogenetic tree constructed for comparative purposes, based on the alignment (Supplementary Fig. S9) of annotated CHLI amino acid sequences from selected organisms. Species, transcript names, and a sequence weight are indicated. The analysis was performed using DNAMAN, with quick alignment and based on observed divergence. Bootstrap values from 1000 iteration analysis are shown in italics; a detailed description is given in the text.
Results
Isolation of a mutant lacking the CHLI1 gene
CAL029C_08 is a light-sensitive brown mutant (Fig. 2B), which was isolated in a screen for C. reinhardtii mutants that are unable to grow photoautotrophically (Dent et al., 2015). The mutant was generated by transformation of the 4A+ wild-type strain with plasmid carrying the aphVIII cassette, which confers resistance to paromomycin (Sizova et al., 2001). Progeny of a cross between CAL029C_08 and the wild type showed a 2:2 pattern of segregation of the brown phenotype in complete tetrads (Supplementary Table S1), demonstrating that the phenotype is due to a single nuclear mutation. Altogether, a total of 73 progeny were obtained (including progeny from incomplete tetrads), of which 39 were wild type and 34 were mutants. All the mutants and none of the wild-type progeny were resistant to paromomycin (Supplementary Table S1), indicating that the insertion of the aphVIII cassette is tightly linked to the mutation causing the brown phenotype.
Isolation of the genomic DNA region adjacent to the insertion site in CAL029C_08 was conducted by RESDA-PCR (Gonzalez-Ballester et al., 2005), which yielded a 267bp flanking sequence on chromosome 6:8351310-8351576, upstream from CHLI1 (genome v5.5, Phytozome v10.2). CHLI1 mRNA was undetectable in RT–PCR analysis, demonstrating that CAL029C_08 carries a null mutation in the CHLI1 gene (chli1/fdx3; Fig. 2A; Supplementary Fig. S1A). However, because insertional mutagenesis in C. reinhardtii often results in deletions of relatively large fragments of genomic DNA (Tam and Lefebvre, 1993; Matsuo et al., 2008), the effect of the introduced exogenous DNA on CHLI1-neighboring genes was analyzed. Indeed, not only the mRNA of CHLI1, but also transcripts of several other genes located upstream or downstream of CHLI1 were missing in the CAL029C_08 mutant (Supplementary Fig. S1B, C). Among the genes affected by the insertion was FERREDOXIN 3 (FDX3, Cre06.g306350; genome v5.5, Phytozome v10.2; Supplementary Fig. S1B, C). Because no other gene affected by the insertion carried an annotation, the CAL029C_08 mutant strain was labeled chli1/fdx3.
The chli1/fdx3 mutant is affected in the chlorophyll branch of TBS
Analysis of the tetrapyrrole and pigment accumulation in chli1/fdx3 in the dark revealed a >100-fold increase in Proto compared with the wild type (Supplementary Fig. S2A; Table 1). Remarkably, there was no detectable MgProto and chlorophyll in chli1/fdx3. The MgProto steady-state level in the wild type was low, but detectable, probably due to the lower chlorophyll synthesis in the dark compared with light conditions (Supplementary Fig. S2). Moreover, lower levels of violaxanthin, antheraxanthin, neoxanthin, lutein, and β-carotene were detected (Supplementary Figs S2A, S6D–F, H, I), while higher levels of zeaxanthin were observed in the mutant compared with the wild type (Supplementary Figs S2A, B, S6G). Although, antheraxanthin was always lower in chli1/fdx3 in the dark and at 4h in the light, compared with the wild type (Supplementary Figs S2A, S6F), exposure to light for 24h caused its increase in chli1/fdx3, with values exceeding the levels observed in the wild type (Supplementary Fig. S2B).
Table 1.
Accumulation of TBS intermediates and chlorophyll in dark conditions, in chli1/fdx3 and the rescued lines, obtained from the transformation with CHLI1/pMS188 (I1/R-1, I1/R-2, and I1/R-3) or both CHLI1/pMS188 and FDX3/pMS586 (I1/F3/R-1, I1/F3/R-2, and I1/F3/R-3).
| Proto | Chl a | Chl b | |
|---|---|---|---|
| 4A+ | 0.004±0.001 (1.0) | 2440.5±27.6 (1.0) | 1344.0±19.6 (1.0) |
| chli1/fdx3 | 0.565±0.260 (145.2) | 0 | 0 |
| I1/R-1 | 0.002±0.002 (0.6) | 786.4±7.2 (0.32) | 448.8±6.3 (0.3) |
| I1/R-2 | 0.010±0.009 (2.5) | 81.6±108.0 (0.03) | 69.9±121.1 (0.1) |
| I1/R-3 | 0.096±.148 (24.8) | 317.5±6.4 (0.13) | 242.1±5.6 (0.2) |
| I1/F3/R-1 | 0.138±0.011 (35.4) | 615.1±2.7 (0.25) | 378.6±3.4 (0.3) |
| I1/F3/R-2 | 0.009±0.005 (2.3) | 844.8±24.4 (0.35) | 620.2±28.2 (0.5) |
| I1/F3/R-3 | 0.027±0.029 (6.9) | 332.7±25.3 (0.14) | 182.7±44.2 (0.1) |
The experiment was performed in biological triplicates; results are presented as mean values ±SD.
Amounts are given in pmol 10–6 cells.
Numbers in parentheses indicate relative fold difference compared with the wild-type levels (4A+).
Accumulation of selected tetrapyrrole intermediates was also analyzed after pre-growth in the dark and exposure to 20 μmol photons m−2 s−1 for 24h. It is noteworthy that almost a 15 000-fold increase in Proto was observed in chli1/fdx3 compared with the wild type (Supplementary Fig. S2B). The chli1/fdx3 strain also showed a complete lack of chlorophyll during illumination (Supplementary Fig. S2B). Interestingly, heme accumulated to almost 5.3 times higher levels in chli1/fdx3 compared with the wild type (Supplementary Fig. S2C), which could be indicative of a passive or active mechanism redirecting excess Proto towards the heme branch. Violaxanthin and β-carotene levels reached nearly 24% and 20% of the wild-type level, respectively (Supplementary Fig. S2B). Following 24h exposure to 20 µmol photons m−2 s−1, 2-fold and >5-fold increases were observed in antheraxanthin and zeaxanthin in chli1/fdx3 compared with the wild type, respectively (Supplementary Fig. S2B).
Expression of the selected genes encoding components of TBS in chli1/fdx3
Lack of the functional MgCh in C. reinhardtii chlh and chld mutants results in the absence of chlorophyll and altered HSP70A expression (Chekunova et al., 2001; von Gromoff et al., 2008). Thus, to elucidate further the consequences of abolished chlorophyll synthesis at the level of Mg chelation on TBS-derived retrograde signaling, the expression kinetics of selected genes were analyzed by qRT–PCR in chli1/fdx3 in the dark and upon transfer from the dark to 20 μmol photons m−2 s−1 light after 1, 2, 4, and 8h (Supplementary Fig. S3). All of the examined TBS genes, including the five genes for MgCh subunits, showed reduced light-induced expression in chli1/fdx3 compared with the wild type. Expression of two CHLI-encoding genes in the wild type revealed a >4-fold higher expression of CHLI1 than CHLI2 at the 4h time point in the light (Supplementary Fig. S3L). In the wild type, CHLI2 showed 4.4-fold lower transcript levels than CHLI1 after 4h of light exposure (Supplementary Fig. S3L). Notably, FeC encoding ferrochelatase (FeCh) showed an increased expression after 1h and 2h of illumination in chli1/fdx3 compared with the wild type (Supplementary Fig. S3O), which may explain the increased heme levels in chli1/fdx3 exposed to light (Supplementary Fig. S2C). Genes encoding proteins of the light-harvesting complexes (LHCs) showed a reduced expression in chli1/fdx3 compared with the wild type (Supplementary Fig. S3P–R), which indicates, as in A. thaliana, that knockout of CHLI1 results in retrograde signaling-dependent modulation of nuclear gene expression in response to a complete hindrance in Mg chelation and Proto accumulation.
The chli1 knockout is responsible for the chlorophyll deficiency phenotype in the chli1/fdx3 strain
To determine if the phenotypes of chli1/fdx3 are due to mutation of CHLI1 and/or FDX3, we performed transformations using genomic versions of wild-type CHLI1 and FDX3 in the CHLI1/pMS188 and FDX3/pMS586 vector, respectively (Supplementary Fig. S4A). Transformants that rescued the mutant phenotype were recovered when the CHLI1/pMS188 vector was used with or without FDX3/pMS586, whereas no rescued colonies were obtained from the transformation with FDX3/pMS586 alone. Thus, the CHLI1 deletion is responsible for the phenotype of chli1/fdx3. Three out of 258 colonies obtained from transformation with CHLI1/pMS188 were selected and labeled chli1/I1/R1, chli1/I1/R2, and chli1/I1/R3. Transformation with both CHLI1/pMS188 and FDX3/pMS586 yielded 166 colonies, from which three were labeled chli1/I1/F3/R1, chli1/I1/F3/R2, and chli1/I1/F3/R3. The RT–PCR confirmed the CHLI1 transcript present in all of the analyzed rescued lines, although with certain variations with regard to the accumulated mRNA levels (Fig. 2A).
Examination of growth and light sensitivity demonstrated that chli1/fdx3 cannot survive in light conditions ≥50 μmol photons m−2 s−1 on heterotrophic medium (TAP; Fig. 2B; Supplementary Fig. S5A) and on photoautotrophic medium regardless of the light conditions (TP; Supplementary Fig. S5B). All the rescued lines showed greening in the dark and light. Moreover, lack of light sensitivity, as observed in the chli1/fdx3 mutant, confirmed the successful rescue of the wild-type phenotype and the key role of CHLI1 in chlorophyll biosynthesis.
HPLC analysis of samples of the dark-incubated mutant and the wild-type strain revealed 145-fold higher levels of Proto in chli1/fdx3, while in the rescued strains accumulation of this tetrapyrrole decreased to 60% higher Proto levels compared with the wild type (Table 1). Chl a and b reached 3–35% and 10–50% of the wild-type level, respectively (Table 1).
Homology between the CHLI proteins from selected organisms
The presence of two CHLI isoforms in A. thaliana and C. reinhardtii prompted a wider scale analysis of the homology between CHLIs from different organisms, to examine their occurrence and any phylogenetic correlation between their possible functions and their evolutionary origin.
The analyzed CHLI homologs were grouped into seven major distinct branches (I–VII; Fig. 1). Group I includes CHLI1 (At4g18480.1, genome TAIR10) of A. thaliana, which shows high similarity to one of the isoforms of Arabidopsis lyrata (946346, genome v1.0) and to other representatives of the Brassicaceae family (Fig. 1).
It is worth mentioning that all of the examined plants clustering in group I seem to have a second CHLI isoform, which is gathered in group II (Fig. 1), together with B. rapa, which seems to have even a third CHLI within this group (Brara.B02858.1). Based on the complementation of CHLI1 by CHLI2 in A. thaliana (Huang and Li, 2009), it would be interesting to compare the expression of CHLIs gathered in groups I and II, and their ability for reciprocal complementation.
Group III contains two CHLIs (Lus10015302 and Lus10025423, genome v1.0) from Linum usitatissimum (Linaceae family), which differ from each other only in seven amino acid residues (Fig. 1; Supplementary Fig. S9). It is likely that both sequences constitute the same protein and the difference originates from alternative sequencing or indeed a whole-genome duplication event (Wang et al., 2012). However, the Nicotiana tabacum CHLI (NCBI: O22436.1) also belongs to this group, and its function in MgCh activity has been examined previously (Papenbrock et al., 2000b ).
Group IV includes CHLI1 of C. reinhardtii (Cre06.g306300.t1.2, genome v5.5), which shows high amino acid sequence similarity to Vocar20008053m (genome v2.0) of Volvox carteri (Fig. 1). Both organisms belong to the same order of Chlamydomonadales (Volvocales) of the class Chlorophyceae (green algae). Considering the evolutionary routes of the analyzed organisms, the presence of the Synechocystis sp. PCC6803 CHLI (WP_011243252.1) in group IV is not surprising, but such a high similarity might be indicative of a relatively conserved sequence of C. reinhardtii CHLI1. Still within group IV, two other CHLI isoforms of Physcomitrella patens (genome v3.0; Fig. 1) branch further away. However, locus Phpat.010G011100 seems to have two alternative transcripts, but the putative protein encoded by Phpat.010G011100.2 branches furthest away from all the analyzed putative CHLIs (Fig. 1). Therefore, it is likely that it might be assigned incorrectly. In the same line, the early release of P. patens genome v3.1 indicates only the Phpat.010G011100.2 transcript from this locus.
Group V contains CHLI2 (Cre12.g510800.t1.2) of C. reinhardtii, which shows high sequence similarity to the putative second isoform of CHLI from V. carteri (Vocar20000924m; Fig. 1). The third CHLI in this group originates from Coccomyxa subellipsoidea C-169 (35407, genome v2.0), which belongs to the same class of Chlorophyceae, but to the order of Chlorococcales. Although the CHLI domain is recognizable in these proteins (PFAM, PF01078; EC 6.6.1.1; KEGGORTH, K03405), the entire group branches further away from groups I, II, and III, and functions of these proteins other than in the MgCh complex cannot be excluded.
Putative CHLIs from Micromonas sp. (CCMP1545 and RCC299, genome v3.0) and Ostreococcus lucimarinus (29195, genome v2.0), which belong to the same family of Mamiellaceae, diverged from the same branch as group V, and were categorized separately in group VI. The only representative of group VII is Rhodobacter sphaeroides (NCBI: AAB97156.1), whose bacterial I subunit (BchI) seems to be the most distantly related to all of the CHLIs analyzed here.
Proteins from Micromonas pusilla CCMP1545 (22176), Micromonas sp. RCC299 (105016), O. lucimarinus (29195), C. reinhardtii (Cre12.g510800.t1.2), V. carteri (Vocar20 000924m), and C. subellipsoidea C-169 (35407) possess a C-terminal 46–53 amino acid extension and shorter N-termini compared with all the other analyzed CHLI-like proteins, except CHLI from Synechocystis sp. and BchI from R. sphaeroides (Supplementary Fig. S9).
Analysis by TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/, accessed 15 January 2016; Emanuelsson et al., 2007) indicated that all the proteins from groups I–IV, except Synechocystis sp. PCC6803, carry the N-terminal sequence of a chloroplast transit peptide (cTP). The C-terminal 46–53 amino acid extensions of the proteins from group V and VI have been examined using the InterProScan 5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/, accessed 15 January 2016), but no indication with regard to the family membership, domains, repeats, or biological function could be determined.
Overexpression of CHLI2 in C. reinhardtii has no effect on chlorophyll synthesis
The comparative analysis of the expression kinetics of CHLI1 and CHLI2 upon the shift from dark to light (Supplementary Fig. S3L) demonstrated lower expression of CHLI2 compared with CHLI1, which resembles expression of the two CHLI genes in A. thaliana (Brenner et al., 2000; Matsumoto et al., 2004). Surprisingly, even CHLI2 expression did not contribute to low levels of chlorophyll, and chli1/fdx3 showed a chlorophyll-free brown phenotype, which resembles the chlh and chld mutants. Therefore, the C. reinhardtii strains overexpressing CHLI2 in the chli1/fdx3 background were generated and examined for chlorophyll and TBS intermediate accumulation, and light sensitivity.
Analyses performed by PredSL (Petsalaki et al., 2006), TargetP 1.1 (Emanuelsson et al., 2007), and Predotar (Small et al., 2004) all indicated the presence of the cTP in a 417 amino acid sequence of CHLI1, while predictions of 425 amino acids of CHLI2 indicated possible targeting to a secretory pathway or mitochondria, but not to the chloroplast. Therefore, a dual approach was undertaken to overexpress the CHLI2-coding sequence driven by the PSAD promoter in the chli1/fdx3 mutant background. The CHLI2 sequence was fused with the sequence encoding the PSAD transit peptide, while the construct carrying exclusively the native CHLI2 targeting sequence was used independently as the control (Supplementary Fig. S4B). Following transformation with the pCHLI2/GenD2 plasmid without the cTP of PSAD, six colonies were randomly selected and labeled I2/oe1, I2/oe2, I2/oe3, I2/oe4, I2/oe5, and I2/oe6. Similarly, six colonies were selected following transformation with pCHLI2/GenD2-cTP and labeled cTP-I2/oe1, cTP-I2/oe2, cTP-I2/oe3, cTP-I2/oe4, cTP-I2/oe5, and cTP-I2/oe6. All selected transformants were examined for the overexpression of CHLI2, based on the transcript and protein levels compared with chli1/fdx3 (Fig. 3A, B), following pre-growth in the dark and exposure to 20 μmol photons m−2 s−1 light, for 4h. Most of the strains transformed either with pCHLI2/GenD2 or pCHLI2/GenD2-cTP showed higher levels of CHLI2 mRNA compared with chli1/fdx3 (Fig. 3A). Based on the amino acid sequence and online tools (http://proteome.gs.washington.edu/cgi-bin/aa_calc.pl, accessed 15 January 2016, and http://bioinformatics.org/sms/prot_mw.html, accessed 15 January 2016), the calculated masses of the mature C. reinhardtii CHLI1 and CHLI2 proteins are 40kDa and 45kDa, respectively. The 40kDa CHLI1 is absent in chli1/fdx3 and all of the CHLI2-overexpressing strains, but it is present in the wild type (Fig. 3B). The CHLI2-overexpressor lines accumulate a protein of ~45kDa, which corresponds to the expected molecular mass of the mature CHLI2. Strains with the highest CHLI2 accumulation were selected for further analyses.
Fig. 3.
Analysis of CHLI2 overexpression in six strains obtained from the transformation with pCHLI2/GenD2 and six strains from the transformation with pCHLI2/GenD2-cTP. (A) Transcript levels of CHLI2 analyzed by qRT–PCR; results represent the calculated fold expression (2−ΔΔCt) normalized to chli1/fdx3. Analyses were performed in biological triplicates, and error bars represent the SD. (B) Western blot showing the 40kDa CHLI1 and 45kDa CHLI2 protein levels in strains overexpressing CHLI2, compared with wild- type 4A+ and chli1/fdx3, respectively; the unspecific immunodetection of the protein with a molecular mass >45kDa is marked by an asterisk; strains selected for further analysis are marked with two asterisks; the lower panel shows the blot stained with Ponceau S used as a loading control. (This figure is available in colour at JXB online.)
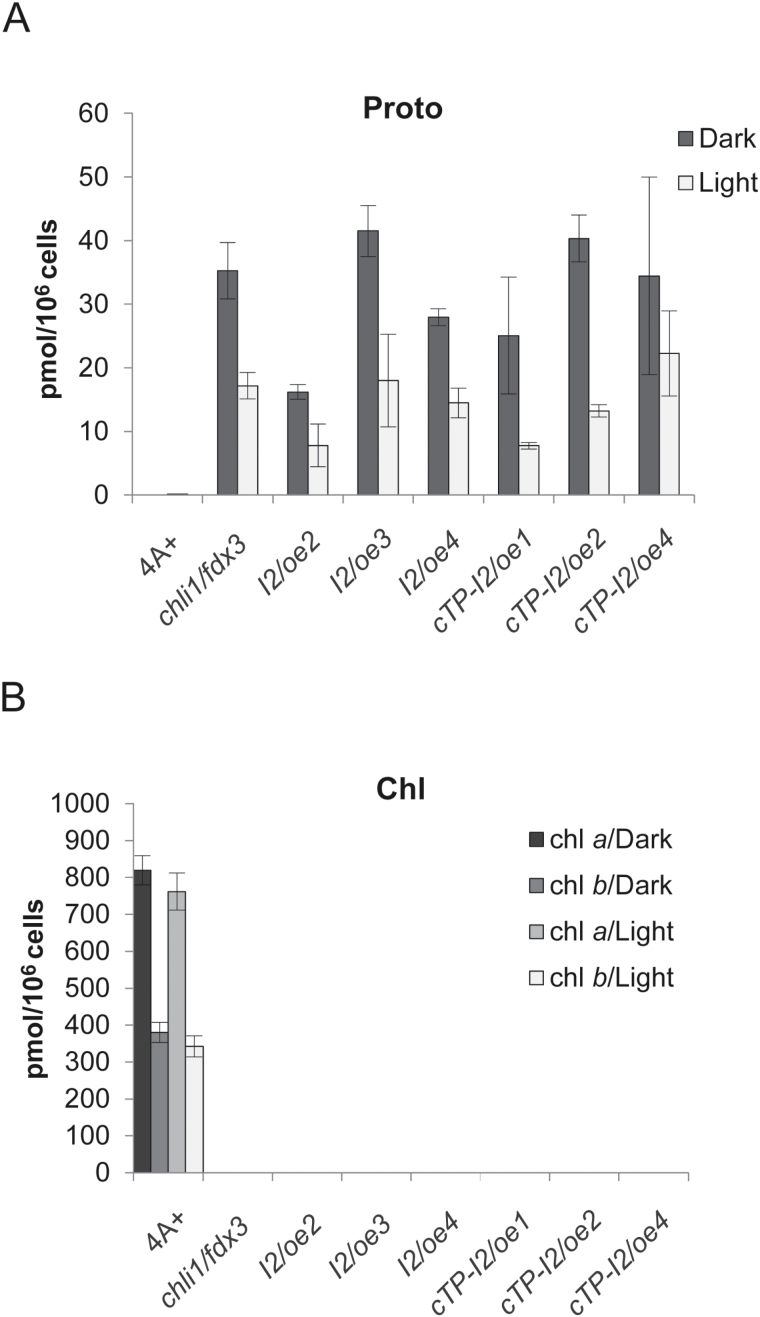
Analysis of the Proto content and pigments also did not show any differences between chli1/fdx3 and CHLI2-overexpressing strains. In all the strains, higher levels of Proto were observed compared with the wild type, both in the dark and upon 4h exposure to 20 µmol photons m−2 s−1, although certain variations in accumulation of this tetrapyrrole metabolite were observed (Fig. 4A). No PChlide, Chlide (Supplementary Fig. S6A, B), or even trace amounts of chlorophyll (Fig. 4B) could be detected in chli1/fdx3 and CHLI2-overexpressors, as compared with the wild type. Accumulation of Proto seems to be affecting heme biosynthesis, resulting in higher levels of this tetrapyrrole (Supplementary Fig. S2C; von Gromoff et al., 2008; Brzezowski et al., 2014). However, no differences in heme content correlating with CHLI2 levels in comparison with chli1/fdx3 were detected (Supplementary Fig. S6C). Additionally, carotenoid levels were determined in the dark and light, again showing no changes, which could be correlated with the CHLI2 levels (Supplementary Fig. S6D–I).
Fig. 4.
Analysis of the steady-state levels of Proto and chlorophyll in strains overexpressing CHLI2 with or without the cTP of PSAD in the dark and with 20 μmol photons m−2 s−1 compared with chli1/fdx3 and the wild type. The experiment was performed in biological triplicates; error bars represent the calculated SD. (A) Proto; (B) Chl a and Chl b content.
Examination of the light sensitivity in strains overexpressing CHLI2, regardless of the presence or absence of the PSAD chloroplast transit peptide, did not reveal any changes in chli1/fdx3 phenotype. While the growth of the wild type positively correlated with the light intensities, chli1/fdx3 and all the strains overexpressing CHLI2 were able to survive dark and light conditions in the examined range up to 30–40 µmol photons m−2 s−1, but 50 µmol photons m−2 s−1 light was always lethal.
Silencing of CHLI2 does not affect TBS or carotenoid biosynthesis
Because CHLI2 overexpression did not complement CHLI1 deficiency in chli1/fdx3, CHLI2 was silenced in the wild-type background to identify any possible function of CHLI2 specifically in the TBS pathway. The amiRNA silencing system was applied using the pChlamiRNA2 vector (Molnar et al., 2009). Following transformation of the CC-3395 strain, three colonies were selected and annotated as I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3. Additional transformation of cc-3395 was conducted with pChlamiRNA2 without the designed 90bp dsDNA. Such a control strain annotated as amiRNA2-E1 was necessary mainly to eliminate the need for arginine supplementation, which otherwise would cause differences in the rate of metabolism. Examination of the CHLI2 mRNA in I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3 showed a decrease to 50, 71, and 24% of the amiRNA2-E1 level, respectively, without a direct correlation with expression of CHLI1 and the GPX5 gene, which were used as the controls (Fig. 5A).
Fig. 5.
Analysis of the strains carrying CHLI2/amiRNA-E9/3'UTR plasmid for silencing of CHLI2. (A) Transcript levels of CHLI1, CHLI2, and GPX5 analyzed by qRT–PCR;the results represent calculated fold expression (2−ΔΔCt) normalized to amiRNA2-E1 transformed with the pChlamiRNA2 plasmid without the 90-mer dsDNA for amiRNA. Analyses were performed in biological triplicates; error bars represent the SD; the results are presented on a log2 scale. (B) Western blot analysis demonstrating silencing of CHLI2 in I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3 compared with amiRNA2-E1. CHLI1 is also visible, and unspecific immunodetection of the protein with molecular mass >45kDa is marked by an asterisk; the lower panel shows the blot stained with Ponceau S as a loading control. (This figure is available in colour at JXB online.)
Western blot analysis revealed that both the 40kDa and 45kDa proteins could be detected in amiRNA2-E1. The amount of CHLI1 did not differ in I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3 compared with amiRNA2-E1. However, the 45kDa band corresponding to CHLI2 could not be detected in I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3 (Fig. 5B), confirming the silencing event in these strains. As seen in Fig. 3B, a non-specific binding of the CHLI1 antibody was always detected (Fig. 5B). Analysis of the potential light sensitivity and growth pattern in 50, 100, 200, and 300 µmol photons m−2 s−1 light did not reveal differences between the strains with silenced CHLI2 and amiRNA2-E1 used as a control.
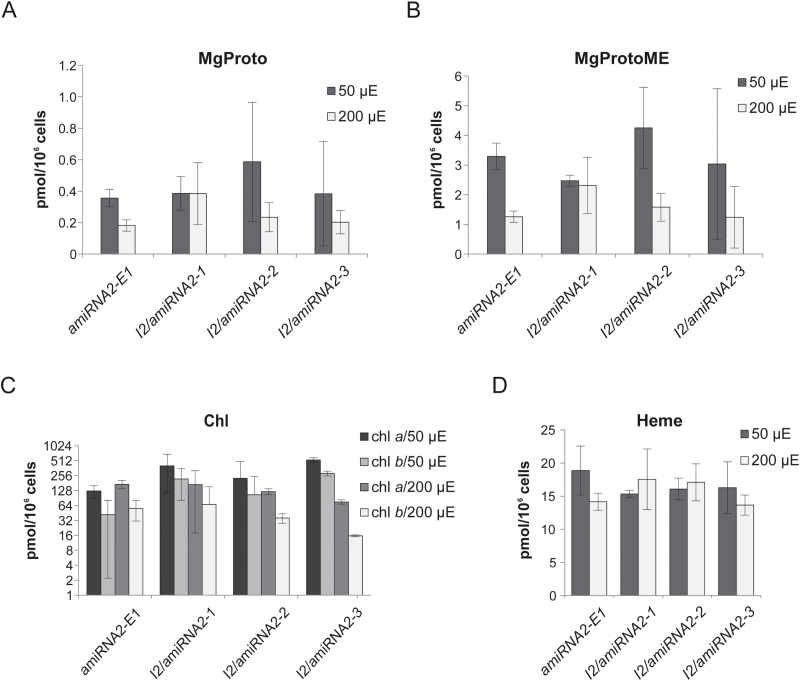
The steady-state levels of TBS pathway intermediates and end-products were analyzed in strains with silenced CHLI2 and compared with the levels observed in amiRNA2-E1 in 50 µmol photons m−2 s−1 and 200 µmol photons m−2 s−1 light. While Proto could not be detected under these light conditions, no correlation between MgCh activity and the reduced presence of CHLI2 could be determined, although a certain variation in MgProto levels was observed between biological replicates and strains (Fig. 6A). Also, accumulating MgProtoME, Chl a, and Chl b contents were not modified in I2/amiRNA2-1, I2/amiRNA2-1 and I2/amiRNA2-1 compared with amiRNA2-E1 (Fig. 6B, C). Additionally, PChlide, Chlide, and carotenoid levels did not differ in relation to the CHLI2 deficiency at either 50 µmol photons m−2 s−1 or 200 µmol photons m−2 s−1 (Supplementary Fig. S7A–H).
Fig. 6.
Analysis of the steady-state levels of selected TBS intermediates and end-products in I2/amiRNA2-1, I2/amiRNA2-2, and I2/amiRNA2-3 in comparison with amiRNA2-E1, following exposure to 50 μmol photons m−2 s−1 and 200 μmol photons m−2 s−1 light. Analyses were performed in biological triplicate; error bars represent the SD. (A) MgProto, (B) MgProtoME, (C) chlorophyll, with results presented on a log2 scale, and (D) heme.
Because Proto is a substrate for MgCh and FeCh, it was likely that any variations in Proto levels would affect not only the chlorophyll but also the heme biosynthesis in strains with silenced CHLI2, if it was involved in MgCh activity (Supplementary Figs S2C, S6C). However, the absence of CHLI2 did not result in different heme contents in I2/amiRNA2-1, I2/amiRNA2-1, and I2/amiRNA2-1 compared with amiRNA2-E1 (Fig. 6D).
Discussion
CHLI1 is indispensable for MgCh activity
Transformation of the chli1/fdx3 mutant with a genomic version of CHLI1 restored the wild-type phenotype (Fig. 2B; Table 1), pointing to a crucial role for this protein in MgCh activity. Although several genes neighboring CHLI1 were also affected by the insertion of the DNA during the mutagenesis (Supplementary Fig. S1), introduction of the wild-type CHLI1 was sufficient for the rescue of chlorophyll biosynthesis and recovery of the ability to grow photoautotrophically. In contrast, transformation with FDX3 had no effect on these phenotypes, confirming that deletion of CHLI1 is responsible for the defect in MgCh activity. Additionally, although FDX3 has been annotated as belonging to the FDX family in C. reinhardtii, it shows a high divergence compared with the other proteins from this group, and it clusters with FDXs with unknown function (Terauchi et al., 2009). More recently it was suggested that FDX3 of C. reinhardtii might be involved in nitrogen assimilation (Peden et al., 2013). Thus, it is unlikely that the mutation in FDX3 would have a direct effect on tetrapyrrole biosynthesis.
Lack of MgCh function due to the chli1 knockout results in modulated chloroplast retrograde signaling
Disturbance in chloroplast functions results in reduced expression of PhAN genes. Mutations affecting TBS in A. thaliana were reported to cause impairments in the chloroplast retrograde signaling in gun mutants upon NF treatment. Despite the dysfunction in tetrapyrrole biosynthesis, the chloroplast did not show severely reduced transcripts for PhAN genes in the presence of NF compared with the wild-type chloroplasts (Susek et al., 1993; Mochizuki et al., 2001; Cottage et al., 2007; Woodson et al., 2011). In the same line, inactivation of CHLI and CHLH expression by antisense RNA synthesis in tobacco caused modified nuclear gene expression of PhAN and TBS genes, as a result of impaired chlorophyll synthesis and consequently reduced assembly of photosynthetic complexes (Papenbrock et al., 2000a, b ). Different lines with disturbed retrograde signaling have been presented, such as mutants carrying defects in the CHLH subunit of MgCh, for example gun5 of A. thaliana (Mochizuki et al., 2001), or brs-1 and chl1 of C. reinhardtii (Kropat et al., 1997; Chekunova et al., 2001). It was also reported that the xantha-f, -g, and -h mutants of Hordeum vulgare L., with defects in CHLH, CHLD, and CHLI genes, respectively, are compromised in the normal plastid-derived control of nuclear gene expression (Gadjieva et al., 2005). The GUN4 protein was also shown to play a role in chloroplast retrograde signaling in A. thaliana (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003) and C. reinhardtii (Formighieri et al., 2012; Brzezowski et al., 2014).
It is relevant to refer to the current discussion on plastid-derived signaling, which can be summarized in two controversial conceptual ideas of tetrapyrrole-mediated signaling. Retrograde signaling is either induced in response to adverse environmental conditions and compromised chloroplast biogenesis, or it is a part of a continuous exchange of information between plastids and the nucleus to balance metabolic activities (Pogson et al., 2008).
In this line, it is worth mentioning that cs and ch42 mutants of A. thaliana, carrying defects in CHLI, do not show the NF-dependent gun phenotype (Mochizuki et al., 2001), but may compromise MgCh activity and chlorophyll supply, which both will hamper chloroplast function. In contrast, it was reported that the semi-dominant chli1 (cs215/cs215) mutant and the chli1/chli1 chli2/chli2 double mutant display a gun phenotype, because they accumulate a higher level of LHCB1 transcript upon NF treatment than the wild type (Huang and Li, 2009). Because the chli1 knockout mutant of A. thaliana does not display the gun phenotype, it was suggested that perhaps CHLI2 is sufficient for the signaling function in chli1 of A. thaliana (Nott et al., 2006). Analysis of the gene expression in chli1/fdx3 conducted in the present study demonstrated a severe effect of the CHLI1 knockout on PhAN and TBS genes, as the result of a plastid-mediated down-regulation (Supplementary Fig. S3). It is obvious that deficiency in chlorophyll synthesis is instantaneously communicated to alter nuclear gene expression, but in the light of the controversies on the contribution of metabolic products, it is worth emphasizing that in chli1 a direct participation of neither one of the other MgCh subunits nor of Proto and MgProto in signaling transfer to the nucleus can be deduced from these studies. This also seems reasonable when inability to carry out of photosynthesis and ROS-mediated signaling also down-regulate expression of TBS and PhAN genes.
The lack of the chlorophyll in the chli1 knockout affects carotenoid composition
The accumulation of Proto and the lack of chlorophyll cause a general disturbance in cell metabolism and also affect the carotenoid levels, both in the dark and in light (Supplementary Fig. S2A, B).
Compared with the wild type, an accumulation of zeaxanthin and lower levels of violaxanthin, antheraxanthin, and β-carotene were observed in the dark and in light in chli1/fdx3 (Supplementary Fig. S2A), while no change in the lutein content was detected either in dark or in light conditions (Supplementary S2A, B). The direct mechanism linking a defect in TBS with an alteration of carotenoid biosynthesis is unknown, and its elucidation would require more in-depth analysis. However, an interesting possibility is that the increased synthesis of heme in chli1/fdx3 perturbs the Fe- and heme-dependent enzymes involved in epoxidation of zeaxanthin and hydroxylation of β-carotene. Perturbed zeaxanthin epoxidase is a possible cause of zeaxanthin accumulation in chli1/fdx3 also because of the lower levels of antheraxanthin, violaxanthin, and neoxanthin in this strain compared with the wild type in the dark and after 4h exposure to light (Supplementary Figs S2A, S6E, F, H).
Zeaxanthin epoxidase uses reduced ferredoxin to epoxidize zeaxanthin (Bouvier et al., 1996), but no difference in zeaxanthin levels could be observed between strains transformed with CHLI1/pMS188 or with both CHLI1/pMS188 and FDX3/pMS586 (data not shown), which reduces the possibility that the second site mutation, fdx3, could be responsible for the zeaxanthin accumulation phenotype. However, involvement of FDX3 cannot be excluded, because these strains do not accumulate high levels of Proto observed in chli1/fdx3 (Table 1), and thus the fdx3 mutation effect may not be apparent.
Zeaxanthin is an efficient 1O2 quencher, and it was demonstrated that the increased levels of this pigment may reduce damage to membrane lipids caused by 1O2 (Havaux et al., 2007; Johnson et al., 2007). Increased levels of zeaxanthin may not be surprising in high light conditions in the wild type, but are puzzling in chli1/fdx3, considering a photosynthesizing effect of accumulating Proto and most probably increased generation of 1O2 in this strain in light (Supplementary Fig. S2B). Increased zeaxanthin is also observed in the dark (Supplementary Fig. S2A). Nevertheless, because the role of xanthophyll cycle pigments is mainly correlated with the energy dissipation at photosystem II (PSII; Demmig-Adams, 1990; Aro et al., 1993; Adams et al., 1995; Niyogi et al., 1998), a potential functionality of the xanthophyll cycle and accumulation of zeaxanthin in chli1/fdx3 remains to be elucidated. Indeed, zeaxanthin formation was demonstrated to be involved in a mechanism other than non-photochemical quenching (NPQ; Havaux and Niyogi, 1999). Additionally, xanthophylls have been demonstrated to function independently from binding to PSII antenna in the ch1 mutant of A. thaliana (Havaux et al., 2007). The ch1 mutant lacks Chl b, due to the null mutation in the gene encoding Chl a oxygenase, and does not assemble the PSII antenna complexes (Espineda et al., 1999). It shows a low capacity for NPQ (Havaux et al., 2007) and higher light sensitivity than the wild type (Havaux et al., 2004). However, the chli1/fdx3 mutant is not expected to assemble the functional photosystems at all, because of the complete lack of chlorophyll. This would be indicative of the other mechanism(s) governing the xanthophyll cycle, which are completely independent from the photosynthetic apparatus.
CHLI2 is not involved in MgCh activity and does not affect chlorophyll biosynthesis
In the chli1 knockout mutant of A. thaliana, chlorophyll is reduced to 10–17% of the wild-type level (Rissler et al., 2002). The C. reinhardtii chlh and chld mutants show complete lack of chlorophyll (Chekunova et al., 2001; von Gromoff et al., 2008), just like the chli1 knockout (Fig. 2; Supplementary Fig. S2A, B). Although the second isoform of the CHLH subunit also exists in C. reinhardtii, its function remains unknown. Previous experiments with the chlh mutant alleles indicated that the second C. reinhardtii CHLH cannot complement CHLH1 deficiency. In either case, results obtained in the present study indicate that the CHLI1 of C. reinhardtii might be the only isoform playing a role in MgCh formation.
CHLI2 of C. reinhardtii was overexpressed in the chli1/fdx3 background, to examine whether higher levels of CHLI2 can functionally complement the chli1 null mutation. Although higher levels of CHLI2 were observed in strains overexpressing the putative second CHLI isoform (Fig. 3), no correlation could be demonstrated between protein levels and MgCh function, or chlorophyll accumulation. Moreover, because of the presence of the light-independent protochlorophyllide reductase (DPOR) in C. reinhardtii and the ability to produce chlorophyll in the dark, the analyses were performed in both dark and light conditions, to examine the possibility of CHLI2 playing a role in any of these conditions. Nevertheless, based on the analysis of the TBS intermediates and end-products, no function in tetrapyrrole biosynthesis specifically could be assigned to CHLI2. However, it has to be mentioned that localization of the native CHLI2 should be examined in experiments involving cell fractionation, or localization studies involving green fluorescent protein (GFP)–CHLI2 or yellow fluorescent protein (YFP)–CHLI2 fusion proteins.
The potential functions of the CHLI2 protein were also examined in the CC-3395 strain with the silenced CHLI2 gene. The tetrapyrrole intermediates and end-products were analyzed in 50 µmol photons m−2 s−1 and 200 µmol photons m−2 s−1, but no difference could be observed between I2/amiRNA2-1, I2/amiRNA2-2, or I2/amiRNA2-3 and the amiRNA2-E1 strain used as the control. In the same line, it is not expected that the lower CHLI2 transcript levels correlate with modified nuclear gene expression for TBS and PhAN genes. Also, no difference could be determined between the strains with silenced CHLI2 and the control strain from examination of the general appearance and light sensitivity at 50, 100, 200, or 300 µmol photons m−2 s−1 light conditions. In summary, all the results combined suggest that Mg chelation does not depend on CHLI2 as a MgCh subunit, and that the CHLI1 isoform in C. reinhardtii is the indispensable protein involved in the formation of the functional enzymatic complex.
In silico analyses point to distinct roles for CHLI2 and CHLI1 in C. reinhardtii
Analysis of the 417 amino acid CHLI1 and 425 amino acid CHLI2 of C. reinhardtii using InterProScan 5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/, accessed 15 January 2016) and the NCBI database (http://www.ncbi.nlm.nih.gov, accessed 15 January 2016), based on the sequences obtained from Phytozome v10.2 (C. reinhardtii genome v5.5), showed that the same functional domains present in CHLI1 can also be identified in CHLI2, including a P-loop containing nucleoside triphosphate hydrolase (IPR027417) carrying an AAA+ ATPase domain (IPR003593), containing Walker A, Walker B, predicted phosphorylation sites, and the arginine finger motif (Supplementary Fig. S8A). However, the P-loop ATPase domain in CHLI2 seems to be incomplete, and its functionality as an ATPase has not been examined so far (Supplementary Fig. S8A). Alignments of the amino acid sequence using DNAMAN software indicated that CHLI2 shows only 48.7% similarity to CHLI1 in C. reinhardtii (Supplementary Fig. S8A). At present, no function can be assigned to CHLI2. Due to the low structural resemblance of both CHLI isoforms in C. reinhardtii, CHLI2 could act as a ‘surrogate protein’. This hypothesis is consistent with a replacement model, where both CHLI isoforms are synthesized in different amounts and the potential placeholder CHLI2 functions to complete the hexameric ring structure, which mainly consists of CHLI1 units, but lowers ATPase activity in conditions with attenuated chlorophyll synthesis. CHLI2 might be deposited when efficient MgCh activity is required.
In contrast, CHLI1 and CHLI2 in A. thaliana show 83% similarity in amino acid sequence (Supplementary Fig. S8B). Based on phylogenetic analysis (Fig. 1), these paralogs in A. thaliana resulted from gene duplication in the Brassicaceae. Perhaps that is why the overexpression of the native CHLI2 of A. thaliana can substitute for the CHLI1 deficiency in this organism (Huang and Li, 2009), but the same inter-relationship could not be observed in C. reinhardtii. Based on the phylogenetic analysis and the presence of A. thaliana CHLI2 (At5g45930.1) in group II (Fig. 1), together with the clustering pattern in group I and II, it can be hypothesized that group II gathers proteins whose expression might be lower compared with the major CHLI isoforms (group I), but which, according to previous results, might be essential under certain circumstances; for example, stress conditions (Moseley et al., 2000).
Based on the phenotypes of the chli1 knockout and the wild-type phenotype in strains rescued with the genomic version of the CHLI1 gene, it can be concluded that CHLI1 plays an essential role in MgCh activity, which cannot be substituted by CHLI2. The possible function of CHLI2 in tetrapyrrole biosynthesis has been examined by overexpressing the cDNA version of CHLI2 in the chli1/fdx3 background and by silencing of CHLI2 in the wild-type background. Based on the analysis of transcript levels, accumulation of TBS intermediates, TBS pathway end-products, carotenoid levels, light sensitivity, or general growth characteristics, no effect caused by higher accumulation of CHLI2 or absence of this protein could be observed.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Transcript analyses of the CHLI1-neighboring genes in the chli1/fdx3 mutant strain.
Figure S2. Analysis of the steady-state levels of intermediates and end-products of TBS, and carotenoids in chli1/fdx3 compared with the wild type.
Figure S3. Kinetics of the expression of selected genes upon shift from dark to 20 μmol photons m−2 s−1, examined at 1, 2, 4, and 8h time points.
Figure S4. Vectors used for C. reinhardtii transformations.
Figure S5. Examination of growth and sensitivity of chli1/fdx3 to light of 10, 50, and 100 μmol photons m−2 s−1 PAR.
Figure S6. Steady-state levels of TBS intermediates, heme, and pigments in strains overexpressing CHLI2 in the dark and after exposure to 20 µmol photons m−2 s−1 light for 4h, compared with the wild type and chli1/fdx3.
Figure S7. Steady-state levels of TBS intermediates and pigments in strains with silenced CHLI2 in 50 µmol photons m−2 s−1 and 200 µmol photons m−2 s−1 light, compared with the control strain amiRNA2-E1.
Figure S8. Alignment of the amino acid sequence of two CHLI isoforms.
Figure S9. Aligned amino acid sequences of the CHLI proteins used to construct the phylogenetic tree presented in Fig. 1.
Table S1. Results of crosses between CAL029C_08 (chli1/fdx3) and the wild type.
Table S2. Primers used in this study; underlined sequences indicate restriction sites.
Acknowledgements
We are grateful to Michael Schroda (University of Kaiserslautern, Germany) for providing pMS188 and pMS586 plasmids. The isolation, initial characterization, and genetic crossing of the chli1/fdx3 mutant was supported by funding from the US Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under Field Work Proposal number 449B granted to KKN. This project was supported by the Deutsche Forschungsgemeinschaft (DFG GR936/15-2) granted to BG.
References
- Adams WW, Demmig-Adams B, Verhoeven AS, Barker DH. 1995. ‘Photoinhibition’ during winter stress: involvement of sustained xanthophyll cycle-dependent energy dissipation. Australian Journal of Plant Physiology 22, 261–276. [Google Scholar]
- Apchelimov AA, Soldatova OP, Ezhova TA, Grimm B, Shestakov SV. 2007. The analysis of the ChlI 1 and ChlI 2 genes using acifluorfen-resistant mutant of Arabidopsis thaliana. Planta 225, 935–943. [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. 1993. Photoinhibition of photosystem-2—inactivation, protein damage and turnover. Biochimica et Biophysica Acta 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Beck CF. 2001. Signaling pathways in chloroplast-to-nucleus communication. Protist 152, 175–182. [DOI] [PubMed] [Google Scholar]
- Bouvier F, dHarlingue A, Hugueney P, Marin E, MarionPoll A, Camara B. 1996. Xanthophyll biosynthesis—cloning, expression, functional reconstitution, and regulation of beta-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). Journal of Biological Chemistry 271, 28861–28867. [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nature Biotechnology 18, 630–634. [DOI] [PubMed] [Google Scholar]
- Brzezowski P, Schlicke H, Richter A, Dent RM, Niyogi KK, Grimm B. 2014. The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii . The Plant Journal 79, 285–298. [DOI] [PubMed] [Google Scholar]
- Brzezowski P, Wilson KE, Gray GR. 2012. The PSBP2 protein of Chlamydomonas reinhardtii is required for singlet oxygen-dependent signaling. Planta 236, 1289–1303. [DOI] [PubMed] [Google Scholar]
- Chekunova E, Voronetskaya V, Papenbrock J, Grimm B, Beck CF. 2001. Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Molecular Genetics and Genomics 266, 363–373. [DOI] [PubMed] [Google Scholar]
- Cottage AJ, Mott EK, Wang JH, et al. 2007. GUN1 (GENOMES UNCOUPLED1) encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. Photosynthesis Research 91, PS1911. [Google Scholar]
- Czarnecki O, Peter E, Grimm B. 2011. Methods for analysis of photosynthetic pigments and steady-state levels of intermediates of tetrapyrrole biosynthesis. Methods in Molecular Biology 775, 357–385. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B. 1990. Carotenoids and photoprotection in plants: a role for the xanthophyll zeaxanthin. Biochimica et Biophysica Acta 1020, 1–24. [Google Scholar]
- Dent RM, Sharifi MN, Malnoe A, Haglund C, Calderon RH, Wakao S, Niyogi KK. 2015. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. The Plant Journal 82, 337–351. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Espineda CE, Linford AS, Devine D, Brusslan JA. 1999. The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 96, 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Rochaix JD. 2001. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Molecular Genetics and Genomics 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Formighieri C, Ceol M, Bonente G, Rochaix JD, Bassi R. 2012. Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii . Molecular Plant 5, 1242–1262. [DOI] [PubMed] [Google Scholar]
- Gadjieva R, Axelsson E, Olsson U, Hansson M. 2005. Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiology and Biochemistry 43, 901–908. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, de Montaigu A, Galvan A, Fernandez E. 2005. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Analytical Biochemistry 340, 330–335. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 54, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Willows RD, Roberts TH, Hansson M. 2002. Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA(+) hexamers. Proceedings of the National Academy of Sciences, USA 99, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use . San Diego: Academic Press. [DOI] [PubMed] [Google Scholar]
- Havaux M, Dall’Osto L, Bassi R. 2007. Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiology 145, 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Dall’Osto L, Cuine S, Giuliano G, Bassi R. 2004. The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. Journal of Biological Chemistry 279, 13878–13888. [DOI] [PubMed] [Google Scholar]
- Havaux M, Niyogi KK. 1999. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proceedings of the National Academy of Sciences, USA 96, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Tanaka A, Tanaka R. 2013. Simple extraction methods that prevent the artifactual conversion of chlorophyll to chlorophyllide during pigment isolation from leaf samples. Plant Methods 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Li HM. 2009. Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiology 150, 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanningmeier U, Howell SH. 1984. Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardii. Possible involvement of chlorophyll synthesis precursors. Journal of Biological Chemistry 259, 3541–3549. [PubMed] [Google Scholar]
- Johnson MP, Havaux M, Triantaphylides C, Ksas B, Pascal AA, Robert B, Davison PA, Ruban AV, Horton P. 2007. Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. Journal of Biological Chemistry 282, 22605–22618. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Mochizuki N, Yoshimura N, Motohashi K, Hisabori T, Masuda T. 2008. Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochemical and Photobiological Sciences 7, 1188–1195. [DOI] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rudiger W, Beck CF. 1997. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proceedings of the National Academy of Sciences, USA 94, 14168–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rudiger W, Beck CF. 2000. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. The Plant Journal 24, 523–531. [DOI] [PubMed] [Google Scholar]
- La Rocca N, Rascio N, Oster U, Rudiger W. 2001. Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213, 101–108. [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. [DOI] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T. 2004. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiology 135, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. 2008. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes and Development 22, 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98, 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. 2008. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA 105, 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. 2009. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. The Plant Journal 58, 165–174. [DOI] [PubMed] [Google Scholar]
- Moseley J, Quinn J, Eriksson M, Merchant S. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii . EMBO Journal 19, 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG. 2008. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proceedings of the National Academy of Sciences, USA 105, 15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Bjorkman O. 1998. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. The Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. 2006. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology 57, 739–759. [DOI] [PubMed] [Google Scholar]
- Oster U, Brunner H, Rudiger W. 1996. The greening process in cress seedlings. 5. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. Journal of Photochemistry and Photobiology B: Biology 36, 255–261. [Google Scholar]
- Papenbrock J, Mock HP, Tanaka R, Kruse E, Grimm B. 2000. a Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiology 122, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Pfundel E, Mock HP, Grimm B. 2000. b Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. The Plant Journal 22, 155–164. [DOI] [PubMed] [Google Scholar]
- Peden EA, Boehm M, Mulder DW, Davis R, Old WM, King PW, Ghirardi ML, Dubini A. 2013. Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. Journal of Biological Chemistry 288, 35192–35209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ. 2006. PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics, Proteomics and Bioinformatics 4, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Forster B, Small ID. 2008. Plastid signalling to the nucleus and beyond. Trends in Plant Science 13, 602–609. [DOI] [PubMed] [Google Scholar]
- Rissler HM, Collakova E, DellaPenna D, Whelan J, Pogson BJ. 2002. Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiology 128, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicke H, Hartwig AS, Firtzlaff V, Richter AS, Glaesser C, Maier K, Finkemeier I, Grimm B. 2014. Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole-mediated retrograde signaling. Molecular Plant 7, 1211–1227. [DOI] [PubMed] [Google Scholar]
- Schroda M, Beck CF, Vallon O. 2002. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. The Plant Journal 31, 445–455. [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirijovski N, Olsson U, Lundqvist J, Al-Karadaghi S, Willows RD, Hansson M. 2006. ATPase activity associated with the magnesium chelatase H-subunit of the chlorophyll biosynthetic pathway is an artefact. Biochemical Journal 400, 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221–229. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Stevens DR, Rochaix JD, Purton S. 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Molecular and General Genetics 251, 23–30. [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. [DOI] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi AM, Lu S-F, Zaffagnini M, et al. 2009. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. Journal of Biological Chemistry 284, 25867–25878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. 2008. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas . The Plant Cell 20, 552–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CJ, Weinstein JD. 1991. In vitro assay of the chlorophyll biosynthetic enzyme Mg-chelatase: resolution of the activity into soluble and membrane-bound fractions. Proceedings of the National Academy of Sciences, USA 88, 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hobson N, Galindo L, et al. 2012. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. The Plant Journal 72, 461–473. [DOI] [PubMed] [Google Scholar]
- Willows RD, Gibson LCD, Kanangara CG, Hunter CN, von Wettstein D. 1996. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. European Journal of Biochemistry 235, 438–443. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biology 21, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-W, Yuan S, Feng H, Xu F, Cheng J, Shang J, Zhang D-W, Lin H-H. 2011. Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs—new evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. Journal of Plant Physiology 168, 714–721. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Zhang GC, Zhu F, Zhang DW, Yuan S. 2015. The roles of tetrapyrroles in plastid retrograde signaling and tolerance to environmental stresses. Planta 242, 1263–1276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.