Abstract
Alzheimer’s disease (AD) is one of the most common neurodegenerative diseases, with increasing prevalence affecting millions of people worldwide. Currently, only autopsy is able to confirm the diagnosis with a 100% certainty, therefore, biomarkers from body fluids obtained by non-invasive means provide an attractive alternative for the diagnosis of Alzheimer`s disease. Global changes of the protein profile were examined by quantitative proteomics; firstly, electrophoresis and LC-MS/MS were used, thereafter, SRM-based targeted proteomics method was developed and applied to examine quantitative changes of tear proteins. Alterations in the tear flow rate, total tear protein concentration and composition of the chemical barrier specific to AD were demonstrated, and the combination of lipocalin-1, dermcidin, lysozyme-C and lacritin was shown to be a potential biomarker, with an 81% sensitivity and 77% specificity.
Introduction
Alzheimer’s disease (AD) is one of the most common forms of age-related dementia, affecting more than 25 million people worldwide, with the number of new cases being continuously on the rise, particularly in the developed and developing countries [1]. The etiology of AD is still unknown. The disease can be characterized by massive cognitive decline, occurring in either sporadic or familial forms. Evidence suggests that abnormal production and accumulation of misfolded, toxic proteins like β-amyloid (Aβ) peptides, the microtubule-associated protein tau [2], and the presynaptic protein α-synuclein are involved in the pathogenesis of AD [3]. The pathological hallmarks of AD are the appearance of senile plaques and neurofibrillary tangles in the brain tissue, in addition to the development of cerebral amyloid angiopathy, with depositions of Aβ peptides in the vessel walls [4].
Based on reported data, AD affects the entire visual system; AD-related changes have been observed in the eye, the visual pathway, as well as the visual cortex. Aβ depositions in the lens and retina were also detected, along with changes such as the reduction of the numbers of retinal ganglion cells, thinning of the nerve cell layer, optic atrophy and decline in the numbers of axons affecting mainly the large diameter axons in the optic nerve [5,6]. These changes result in the impairment of visual acuity and color vision, defects in fixation, delay in saccadic eye movement and changes in contrast sensitivity [7].
Multiple studies highlighted the role of Aβ and hyperphosphorylated tau [8] in the pathophysiology of the disease, however, new evidences suggest the involvement of inflammatory processes [9] and the increased oxidation of brain proteins [10], besides the amyloid depositions and neurofibrillary tangle formation. The oxidation of brain proteins is probably the consequence of higher reactive oxygen species (ROS) production that can be caused by inflammatory processes, or by mitochondrial dysfunction, which is often associated with the appearance of AD [11].
Several biomarkers for AD were identified, yet an ideal biomarker that is able to improve differential diagnosis, track disease progression and measure treatment efficacy has not been discovered. Despite advances in neuroimaging techniques [12,13], and the opportunities provided by the “omics” and systems biology studies [14], autopsy remains the only mean to provide diagnosis with a 100% certainty. Genetic markers; such as mutations in the gene of amyloid precursor protein (APP), presenilin 1 and 2 (PS1 and PS2) and tau, were identified for early onset AD, while apolipoprotein E (APOE) and clusterin gene (CLU) mutations were identified for the late onset form [15,16]. Disease-specific mutations indicating the alteration of fatty acid and amino acid metabolism, innate immune response, apoptosis, cell communication, and cell adhesion pathways in the brain of AD patients were also highlighted [17]. For biomarker analyses, brain tissue, cerebrospinal fluid (CSF), blood plasma and serum were used, and several candidate protein biomarkers were identified using advanced proteomics techniques, such as two dimensional gel electrophoresis, oxyblotting, mass spectrometry, and multiplex assays [14,18].
Selected reaction monitoring (SRM)-based targeted proteomics analysis is a versatile tool for biomarker studies. SRM is a special scan mode of the triple quadrupole-containing mass spectrometers, which allows for highly specific identification; and with the help of stable isotope-labeled (SIL) synthetic peptides, it enables the relative or absolute quantification of the analytes of interest [19,20]. Another advantage of SRM is the multiplex feature, multiple molecules can be analyzed simultaneously from the same sample, which allows for a more cost-effective analysis compared to the classical antibody-based quantification techniques [21].
With increasing AD prevalence, there is a high demand for novel diagnostic methods utilizing body fluids collected by non-invasive means, such biomarkers are of valuable importance. Tear fluid; therefore, provides a viable source, and with its relatively high protein content [22], it is a widely used candidate for biomarker studies [23–25]. Tear samples are easy to collect, thus, they provide excellent sample source for developing bedside diagnostic tests for pathological conditions affecting the ocular system, or the body as a whole [26,27]. The tear fluid creates a chemical barrier at the surface of the eye as part of the innate immune system, which provides protection against pathogens by secreting antibacterial and immunomodulatory proteins (AMPs) that inhibit bacterial growth [28]. The major tear proteins; such as lipocalin-1, lactotransferrin and lysozyme-C are involved in the immune and inflammatory processes and defense against pathogens [22]. The relatively high abundance of these proteins makes them the major AMPs of tears. Besides the major AMPs, tear fluid contains several prototypic AMPs, such as defensins, LL-37 cathelicidin in addition to dermcidin [22].
Tear fluid was used as a potential biomarker source in a study of neuroinflammation in Parkinson’s disease; a neurodegenerative condition associated with inflammation, and increased TNF-α levels were found in tears of PD patients [29].
Considering the extensive influence of AD on the visual system, in this pilot study, we wanted to examine the changes of tear production, and tear protein composition in patients with AD and in their age-matched controls. A two-step procedure was applied; firsty, gel-based LC-MS/MS was used for protein identification on a small sample size, thereafter, an SRM-based targeted proteomics method was developed for the identified proteins, in order to examine the AD-dependent alterations on a larger sample size, in order to identify possible tear biomarkers for AD.
Methods
Sample collection
In total, 23 donors were recruited in this study; 14 patients with AD and 9 age-matched controls. Sample collection complied with the guidelines of the Helsinki Declaration and ethical approval was obtained from the University of Debrecen Ethics Committee (2980–2009), while the subjects gave informed written consent. In the case of AD patients, the consent was acquired in the presence of a caregiver, although none of them were placed under guardianship. All of the donors were patients of the University of Debrecen, Faculty of Medicine, and the assessment of AD was done by a psychiatrist. Diagnosis of Alzheimer’s disease was based on the NINCDS-ADRDA [30] and the DSM-IV-TR [31] criteria. Besides psychiatric and neurological assessment, patients underwent general medical examination, basic laboratory testing (blood chemistry analysis, complete blood cell count, hepatic and renal function test, vitamin B12 and folate level determination, thyroid function test) and CT or MR imaging of the brain to rule out other causes of dementia. Patients with a history of sudden onset, early extrapyramidal signs, early behavioral changes or focal neurological features were excluded. The clinical evaluation of the age-matched control subjects consisted of a structured interview, demographic information, medical history, current medication, history of alcohol consumption and a subjective assessment of memory problems using the Mini-Mental State Examination [32]. Only controls without any signs of cognitive impairment were included in the study. Patients with autoimmune disorders, systemic inflammation or ophthalmological disorders were excluded. In case of AD group, the women to men ratio was approximately 1:2 and the mean age was 77 years, whereas in the control group, the women to men ratio was approximately 1:1 and the mean age was 72 years. After assessment of the anterior ocular status of each patient, tear was collected from the inferior meniscus of one or both eyes. The non-traumatic tear collection was carried out without topical anesthesia using standard capillary collection technique [33], thereafter, collected samples were frozen and stored at -70°C until analysis.
SDS-PAGE
The protein concentration of each tear sample was measured using the Bradford method [34]. 20μg tear protein from three randomly selected AD patients and two controls were subjected to SDS-PAGE analysis on a 10% SDS polyacrylamide gel. Electrophoresis was carried out in a Bio-Rad mini tetra cell (Bio-Rad) on 100 A constant current for one hour. The protein bands were stained using Coomassie PageBlue (Fermentas) solution and scanned with a Pharos FX Plus laser scanner (Bio-Rad). Image analysis was done using the QuantityOne software (Bio-Rad). Band intensities in each case were determined and statistically analyzed by Mann–Whitney U test using SigmaPlot 12.0 software (Systat Software Inc.).
LC-MS/MS analysis
Bands with significantly different intensities between AD and control samples were excised, followed by in-gel digestion with trypsin. Initially, reduction was performed using 20mM dithiothreitol for one hour at 56°C, followed by alkylation with 55mM iodoacetamide for 45 minutes. Overnight trypsin digestion was carried out using stabilized MS grade TPCK-treated bovine trypsin (ABSciex) at 37°C, thereafter; the digested peptides were extracted and lyophilized. The peptides were re-dissolved in 10μl 1% formic acid before LC-MS/MS analysis.
Prior to mass spectrometric analysis, peptides were separated using a 90 minute water/acetonitrile gradient, with an increase in acetonitrile concentration from 0 to 100% in a 60 minute time interval, using an EasynLCII (Bruker) nano HPLC. The peptide mixture was desalted on a Zorbax 300SB-C18 in-line trap column (5 x 0.3 mm, 5 μm pore size, Agilent), followed by separation on a Zorbax 300SB-C18 analytical column (150 mm x 75 μm 3.5 μm pore size, Agilent). Solvent A was 0.1% formic acid in LC water, solvent B was LC acetonitrile containing 0.1% formic acid, while the flow rate was 300nl/min.
Positive mode LC-MS/MS scans were performed on a 4000 QTRAP (ABSciex) mass spectrometer using a NanoSpray II MicroIon source, controlled by the Analyst 1.4.2 software (ABSciex). The spray voltage was 2800V, the ion source gas was 50 psi, the curtain gas was 20 psi and the source temperature was 70°C. Information Dependent Acquisition method was utilized; after the first mass scan (mass range 400–1700 amu), an enhanced resolution experiment was carried out to establish the charge state of the precursor ions. The MS/MS spectra of the two most intensive ions were recorded (mass range 100–1900 amu) in Enhanced Product Ion mode at a scan rate of 4000 amu/s, and rolling collision energy was applied with the maximum of 90eV.
The acquired LC-MS/MS data were used for protein identification with the help of ProteinPilot 4.0 (ABSciex) search engine searching the SwissProt database (release: 2015.07, 548872 sequence entries), and using the biological modification table included in the ProteinPilot 4.0 software. A minimum of two peptide sequences with ≥95% confidence were used for protein identification.
Development of SRM-based targeted proteomics experiment
The amino acid sequences of lipocalin-1, lactotransferrin, extracellular glycoprotein lacritin, lysozyme-C, lipophilin A, Ig λ-chain C region, prolactin inducible protein, Zn-α2 glycoprotein, galectin 3 binding protein and dermcidin were retrieved from the UniProt database (P31025, P02788, Q9GZZ8, P61626, P60201, P0CG04, P12273, P25311, Q08380, P81605, respectively), in silico trypsin digestions were then carried out. In order to determine the unique tryptic sequences, BLASTp analyses were performed (http://blast.ncbi.nlm.nih.gov), searching the NCBI non-redundant protein sequence database for unique, protein-specific sequences. The protein specific sequences were subjected to SRM transition design using the Skyline software (www.brendanx-uw1.gs.washington.edu). Stable isotope-labeled (SIL) synthetic crude peptides were obtained from JPT Peptide Technologies GmbH, Germany, and the SRM spectra of all singly charged “y” ions were registered in case of all peptides. The best two transitions for each peptide were selected for further analyses.
SRM-based targeted mass spectrometry analysis
37 tear samples originating from 14 AD patients and 9 age-matched controls were denatured using 6 M urea for 30 mins at room temperature, followed by a digestion procedure similar to that used for LC-MS/MS sample preparation described above. In the denatured, reduced, and alkylated samples, the urea concentration was diluted with 25mM ammonium bicarbonate to 1M immediately before trypsin digestion. Samples were desalted with C18 PierceTips (Thermo Scientific), lyophilized and re-dissolved in 1% formic acid.
SRM-based analyses of digested tear samples were carried out in triplicates on the 4000 QTRAP mass spectrometer using the same voltage, gas, and temperature, as described above in section LC-MS/MS analysis. The declustering potential (DP) and collision energy (CE) values were optimized, and the cycle time was set to 2.5 sec. Chromatographic separation was done on the EasynLC II, using a 30 min acetonitrile/water gradient with an increase in acetonitrile concentration from 0 to 100% during a 15 minute time interval.
Sample blocking was carried out before analyzes; a randomly selected AD sample was paired with a randomly selected control sample and analyzed one after the other using the same conditions. SIL peptides were added to the samples immediately before the analyses. Data were evaluated using the Skyline software, spectra were examined manually and the AUC values were calculated by the software. The SRM data were uploaded to the Panorama website: (https://panoramaweb.org/labkey/project/__r1225/begin.view?) and are publicly available.
The primary AUC data were transformed to MSstats R-package format [35] by an in-house developed software. After the normalization based on the SIL standard peptides, and log2 transformation of data, group differences were examined by a mixed-effect variance analysis [36]. Groups were modeled as fixed effect, while the subject level variances were modeled as random effects. After analysis, the raw p-values were adjusted by the Benjamini and Hochberg type false discovery rate method for multiple testing purposes [37]. Besides the adjusted p-values, the log2 fold change, the standard error and the t-values were also examined. Receiver-operating characteristic (ROC) analyses [38] were carried out by the pROC software [39] and Confidence Interval (CI) values were calculated.
Results
Tear protein profile changes in AD
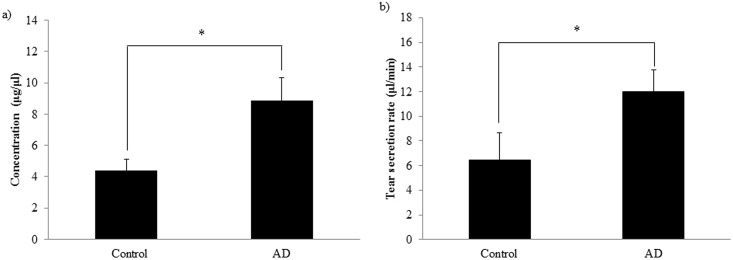
The amount of proteins in biological fluids; especially in tear, can provide information of diagnostic relevance. The typical protein concentration in tear fluid is 5–7 μg/μl [40], and the tear production rate is approximately 2 μl/min. Monitoring the tear production rate and tear protein concentration during sample collection (Table 1), significant differences were observed in case of AD patients compared to controls. The flow rate of 6 ± 2 μl/min observed in controls was significantly increased in AD patients (12±2 μl/min), along with a significant increase in tear protein concentration from 4.4 ± 1.4 μg/μl in controls to 8.8 ± 2.9 μg/μl in AD patients (Fig 1).
Table 1. Patient data and tear collection parameters.
| Patient Nr. | Age (years) | Gender | Tear flow rate (μl/min) | Tear protein concentration (μg/μl) |
|---|---|---|---|---|
| AD 1 | 67 | M | OD: 10.0 | OD:7.1 |
| AD 2 | 77 | M | OD: 12.4 OS: 16.7 |
OD:3.6 OS:8.3 |
| AD 3 | 80 | F | OD: 14.5 OS: 16.7 |
OD:4.1 OS:3.9 |
| AD 4 | 80 | M | OD: 5.0 | OD:6.9 |
| AD 5 | 80 | M | OS: 5.0 | OS:11.5 |
| AD 6 | 72 | F | OD: 15.9 OS: 7.7 |
OD:12.1 OS:14.4 |
| AD 7 | 90 | F | OD: 5.6 OS: 13.0 |
OD:4.8 OS:5.0 |
| AD 8 | 76 | F | OD: 11.8 | OD:12.1 |
| AD 9 | 82 | M | OD: 10.7 OS: 12.7 |
OD:17.0 OS:15.9 |
| AD 10 | 81 | M | OS: 9.6 | OS: 9.6 |
| AD 11 | 65 | F | OD: 18.1 OS: 18.7 |
OD:9.4 OS:9.6 |
| AD 12 | 66 | M | OD: 10.0 OS: 17.0 |
OD: 4.7 OS:9.4 |
| AD 13 | 76 | M | OD: 14.1 OS: 17.7 |
OD:9.1 OS:7.5 |
| AD 14 | 86 | M | OD: 10.0 | OD:8.8 |
| Control 1 | 73 | M | OD: 5.0 OS: 0.8 |
OD:3.5 OS:3.2 |
| Control 2 | 62 | M | OD: 10.4 OS: 2.6 |
OD:4.7 OS:4.7 |
| Control 3 | 71 | M | OD: 7.0 OS: 1.2 |
OD:7.4 OS:7.0 |
| Control 4 | 71 | F | OS: 4.6 | OS: 3.6 |
| Control 5 | 81 | M | OD: 1.2 OS: 8.4 |
OD:4.4 OS:4.4 |
| Control 6 | 89 | F | OD: 3.5 OS: 3.0 |
OD:2.9 OS:2.3 |
| Control 7 | 68 | F | OD: 17.7 OS: 4.6 |
OD:7.4 OS:6.3 |
| Control 8 | 66 | F | OD: 3.2 | OD:1.6 |
| Control 9 | 65 | F | OD: 6.6 | OD:1.4 |
The eye from which the tear was collected is indicated. OD: Oculus dexter (right eye), OS: Oculus sinister (left eye).
Fig 1. Protein concentration and flow rate of collected tear.
The bars represent mean values with the standard error of mean of a) total protein concentration measured by Bradford method b) tear secretion rate. * indicates p <0.05.
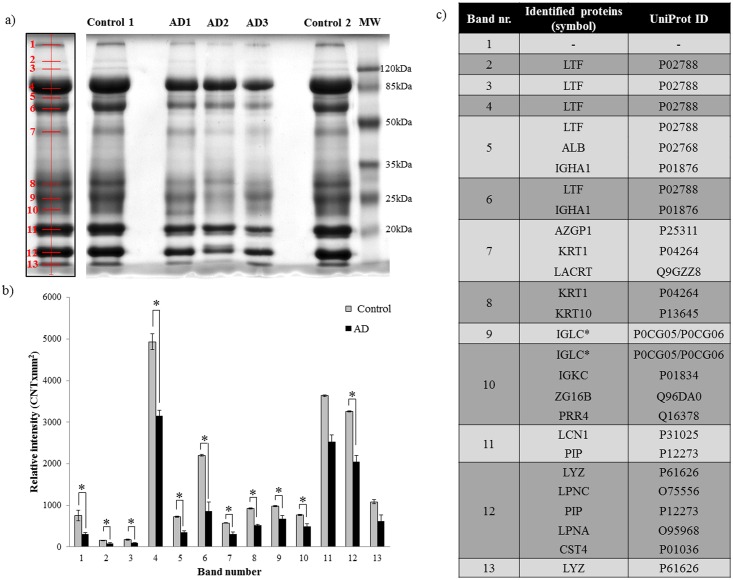
Due to the limited amount of tear fluid; initially, tear profile changes were studied from a few randomly selected samples, thereafter, validation of the observed differences was carried out on a larger sample size. In order to monitor global tear protein changes characteristic to AD, equal amounts of tear proteins from three randomly selected AD patients and two controls were analyzed by electrophoresis. After visualization of protein bands and carrying out densitometric evaluation, 13 bands were observed in each case (Fig 2a), out of which 11 bands showed a significant decrease in band intensity in AD samples (Fig 2b). The bands were excised and digested using trypsin, followed by LC-MS/MS based protein identification (Fig 2c, S1 Table). Only those proteins; of which at least two peptides with ≥95% confidence could be detected, were accepted as present (S1 Table). Examining the differentially expressed proteins, we have identified them as being involved in the host defense, and are components of the chemical barrier of the eye. These data suggest that AD can alter the composition of the chemical barrier, this is in accordance with previous experiments showing alterations of the chemical barrier by different stimuli and pathological conditions [41–43].
Fig 2. Changes in the tear proteome in AD.
a) Gel image of tear proteins from three AD and two control subjects. The left panel shows the representative band distribution detected with QuantityOne. b) Densitometric analysis of gel bands. The bars show mean values with the standard error of mean, grey bars indicate control group while the black bars indicate the AD group. * indicates p <0.05. c) List of identified proteins with the corresponding UniProt reference numbers. * indicates the presence of Ig λ, though the Ig λ-2 chain C region and Ig λ-3 chain C region of the identified peptides were undistinguishable. LTF: lactotransferrin, ALB: serum albumin, IGHA1: Ig α-1 chain C region, AZGP1: Zn-α2 glycoprotein, KRT1: keratin, type II cytoskeletal 1, LACRT: extracellular glycoprotein lacritin, KRT10: Keratin, type I cytoskeletal 10, IGLC: Ig λ chain C region, IGKC: Ig κ chain C region, ZG16B: zymogen granule protein 16 homolog B, PRR4: proline-rich protein 4, LCN1: lipocalin-1, PIP: prolactin-inducible protein, LYZ: lysozyme-C, LPNC: lipophilin C, LPNA: lipophilin A, CST4: cystatin S.
SRM-based quantitative proteomic method development for tear proteins
In order to validate changes of the chemical barrier in the tears of AD patients on a larger sample size, an SRM-based targeted proteomics approach was developed. The SRM scan mode provides a highly selective and sensitive method to monitor the level of multiple molecules in a single analysis; however, this method requires the proteins under examination to be defined priorly. For SRM assay design, lipocalin-1, lactotransferrin, lysozyme-C, extracellular glycoprotein lacritin, Ig λ, Zn α2 glycoprotein, prolactin inducible protein and lipophilin A proteins identified in bands showing significantly different intensity in AD samples as compared to controls were chosen. It is well known that the major proteins of sweat and tears play a role in host defense against potential pathogens [22,44], thus, based on previous experiments, galectin-3 binding protein and dermcidin; considered as proteins with an important role in host defense and immunomodulation [45,46], were also included into the panel of analyzed tear proteins.
SRM-based quantitative proteomic experiments were designed and optimized for the ten selected proteins (S2 Table). In order to enhance the specificity of the method, stable isotope-labeled (SIL) synthetic counterparts of the studied peptides were purchased and used in the analyses as an internal standard (S1 Fig). The sensitivity of the method was assessed by analyzing increasing amounts of SIL peptides added to the tear in a range of 10000 fold– 5 fold dilution. Most of the peptides were detectable in 10000 fold dilution (approximately 7.5 fmol), and the SRM signal intensity was proportional to the amount of peptides introduced in a broad range of dilutions (S3 Table). Based on our results, it appears that the developed SRM method is sensitive enough, and has a broad dynamic range enabling it to be a useful tool to monitor alterations of tear protein amounts.
Changes in the tear chemical barrier`s composition in AD
The composition of the chemical barrier constantly changes upon different stimuli, as a response to mechanical and microbial challenges, in order to protect the human organism efficiently from potentially invading pathogens [47]. Moreover, systemic conditions can also affect the production and secretion of antimicrobial peptides, leading to changes in the composition of this antimicrobial peptide cocktail [47]. Tear fluid contains a variety of antimicrobial and immunomodulatory proteins as part of the first line defense of the innate immune system [22].
In order to study changes in the tear`s chemical barrier in AD, level of the ten selected proteins was analyzed using the developed SRM method in 37 tear samples from 14 patients with AD, and 9 controls in triplicates (S4 table). In line with results obtained from electrophoresis, the levels of lipocalin-1, lactotransferrin, extracellular glycoprotein lacritin, lysozyme-C, and prolactin inducible protein were significantly decreased, while the level of dermcidin was significantly elevated in AD tears, as compared to those of the controls (Fig 3,Table 2). The down-regulated proteins are expressed by the lacrimal glands, indicating lacrimal gland dysfunction in AD.
Fig 3. Quantitative analysis of tear proteins by SRM.
The log2 fold change of the studied proteins in tears of patients with AD compared to controls. * indicate p≤0.05. AZGP1: Zn-α2 glycoprotein, PIP: prolactin-inducible protein, LYZ: lysozyme-C, LTF: lactotransferrin, LPNA: lipophilin A, LCN1: lipocalin-1, LACRT: extracellular glycoprotein lacritin, IGLC: Ig λ chain C region, GAL3BP: galectin 3-binding protein, DCD: dermcidin.
Table 2. Result of the mixed effect variance analysis.
| Protein | Log2FC | SE | Tvalue | Adjusted p value |
|---|---|---|---|---|
| Zn-α2 glycoprotein | -0.25 | 0.17 | -1.43 | 0.26 |
| Prolactin-inducible protein | -0.66 | 0.14 | -4.75 | <0.0001 |
| Lysozyme-C | -1.11 | 0.15 | -7.29 | <0.0001 |
| Lipophilin A | 0.09 | 0.23 | 0.39 | 0.76 |
| Lipocalin-1 | -0.76 | 0.13 | -5.79 | <0.0001 |
| Lactotransferrin | -1.16 | 0.17 | -6.66 | <0.0001 |
| Ig λ chain C region | -0.27 | 0.23 | -1.19 | 0.35 |
| Galectin 3-binding protein | 0.13 | 0.19 | 0.68 | 0.59 |
| Extracellular glycoprotein lacritin | -2.04 | 0.18 | -11.53 | <0.0001 |
| Dermcidin | 0.85 | 0.23 | 3.64 | 0.0006 |
The log2 fold change (log2FC) standard error (SE), T values (Tvalue) and the FDR corrected p-values are shown.
Identification of potential AD-specific tear biomarkers
Proteins differentially expressed in the tears of AD patients were subjected to further analyses, in order to examine their potential use as future predictive biomarkers for AD. A receiver operator characteristic (ROC) analysis was carried out for each significantly expressed protein, and multivariate ROC curves were constructed to test the different combinations of potential biomarkers. The sensitivity, specificity, positive and negative predictive values and the area under the ROC curve (AUC) were calculated in each setting (Table 3). An AUC value close to 1.0 indicates a well-performing biomarker; whereas values close to 0.5 suggest a biomarker performing no better than random.
Table 3. ROC analysis of possible tear biomarkers for AD.
| Proteins | Sensitivity (%) | Specificity (%) | PV+ (%) | PV- (%) | AUC | Accuracy | 95% CI |
|---|---|---|---|---|---|---|---|
| LCN1 | 53.1 | 79.7 | 22.7 | 43.3 | 0.68 | 0.72 | 0.56–0.80 |
| LTF | 78.1 | 51.6 | 17.5 | 55.4 | 0.67 | 0.73 | 0.55–0.78 |
| LYZ | 90.6 | 50 | 8.6 | 52.5 | 0.68 | 0.67 | 0.57–0.79 |
| PIP | 71.9 | 56.2 | 20 | 54.9 | 0.6 | 0.67 | 0.48–0.72 |
| LACRT | 90.6 | 57.8 | 7.5 | 48.2 | 0.71 | 0.65 | 0.59–0.82 |
| DCD | 62.5 | 76.6 | 19.7 | 42.9 | 0.7 | 0.70 | 0.59–0.82 |
| LCN1+DCD | 53.1 | 89.1 | 20.8 | 29.2 | 0.74 | 0.76 | 0.63–0.85 |
| LYZ+LACRT | 90.6 | 59.4 | 7.3 | 47.3 | 0.72 | 0.64 | 0.61–0.83 |
| LCN1+DCD+ LACRT | 90.6 | 65.6 | 6.7 | 43.1 | 0.8 | 0.69 | 0.71–0.89 |
| LCN1+DCD+LTF | 53.1 | 89.1 | 20.8 | 29.2 | 0.74 | 0.76 | 0.62–0.84 |
| LCN1+DCD+LYZ | 59.4 | 82.8 | 19.7 | 36.7 | 0.75 | 0.76 | 0.64–0.86 |
| LCN1+LTF+LYZ | 87.5 | 50 | 11.1 | 53.3 | 0.72 | 0.71 | 0.61–0.83 |
| LYZ+LACRT+ DCD | 90.6 | 65.6 | 6.7 | 43.1 | 0.79 | 0.71 | 0.69–0.89 |
| LYZ+LACRT+ LCN1 | 87.5 | 59.4 | 9.5 | 48.1 | 0.72 | 0.67 | 0.62–0.83 |
| PIP+LACRT+DCD | 93.8 | 60.9 | 4.9 | 45.5 | 0.79 | 0.68 | 0.69–0.97 |
| DCD+LCN1+LTF+PIP | 62.5 | 76.6 | 19.7 | 42.9 | 0.74 | 0.72 | 0.64–0.84 |
| LCN1+DCD+LYZ+LACRT | 81.2 | 76.6 | 10.9 | 36.6 | 0.8 | 0.72 | 0.71–0.89 |
In all cases, the AUC value was above 0.6, and when different combinations of two or more proteins were analyzed, AUC value was above 0.7, indicating an additive effect of the different proteins in improving the performance of the test. The highest sensitivity was obtained by combination of lysozyme-C and extracellular glycoprotein lacritin; yielding 91% sensitivity. The most balanced performance was achieved when lipocalin-1, dermcidin, lysozyme-C and extracellular glycoprotein lacritin were combined. In this case, the AUC was 0.80, the sensitivity was 81% and the specificity was 77%. Considering these values, it seems that a test combining these four potential biomarkers may therefore be used as a first screen in the diagnosis of AD.
Discussion
Tear fluid is an excellent candidate for biomarker studies, given the facts that samples may be collected non-invasively, in addition to providing information not only on local ocular conditions; such as dry-eye disease and diabetic retinopathy, but also on systemic pathophysiological processes [22,25,29]. The presence of amyloid plaques was demonstrated in the retina and lens of patients with Alzheimer’s disease, and animal model studies indicated correlation in amyloid depositions between the retina and the brain [5,6]. Changes in the retinal vasculature and retinal morphology were detected in the eyes of patients with AD, resulting in the reduced visual performance observed in AD patients [13]. We have hypothesized that changes in retinal morphology and blood flow related to AD may alter the microenvironment of the eye, and this alteration may therefore be reflected at the level of tear proteins.
In our study, a significant increase in the flow rate and protein concentration, along with a significant difference in the amount of the studied tear proteins was observed in samples obtained from AD patients. This is in good agreement with previously published data suggesting extensive ocular alterations related to AD [5–7]. The protein level changes observed with electrophoretic analysis of five samples were validated by targeted proteomics analyses on 37 samples. The only increase in tear proteins characteristic to AD has been observed in case of dermcidin, which is produced by epithelial cells, [47] and is the main sweat antimicrobial peptide with a broad range antimicrobial activity [44]. The proteins with reduced amount were involved in first line defense of the eye, produced by the lacrimal gland.
The altered composition of the chemical barrier, along with the reduced level of studied defense proteins might imply an increased risk of ocular infections, yet there has been no reported increase of ocular infections in patients with AD according to the scientific literature. However, reduced corneal sensitivity and abnormal tear functions were reported in AD patients as compared to controls, in addition to other neurodegenerative diseases [48]. Dermcidin; with its broad antimicrobial spectrum, appears to be a plausible factor responsible for limiting bacterial over-growth and; therefore, possible infections.
Given the easy, non-invasive collection possibility of tears, the prospect of a bedside test development is worth exploring. Patients with an increased tear flow rate (above 5 μl/min according to our data), along with increased tear protein concentration, and who also show altered level of extracellular glycoprotein lacritin, lipocalin-1, lysozyme-C and dermcidin, may be subjected to further imaging, neuropsychological testing (CT, MRI, PET) and cerebrospinal fluid analyses (Aβ42, total tau and p-tau levels). Considering the small sample size analyzed in this pilot study, more studies carried out on a large population scale are required, in order to evaluate the applicability of the proposed biomarkers. After validation, tear analysis, as an easily executable test, can be used in population screening by general practitioners, and patients with a positive test may be further evaluated by clinical centers for the establishment of the diagnosis. If early diagnosis and treatment are promptly provided, the quality of life can be improved for AD patients and their caregivers, which will undoubtedly aid in decreasing the socio-economic burden of the disease.
Supporting Information
The y axis shows the intensity while the x axis shows the retention time. The blue line refers for the synthetic, stabile isotope labeled peptide, while the red line for the endogenous counterparts.
(PPTX)
The name and accession numbers of the identified proteins in each band are presented along with the protein identification data. The sequence coverage (%Cov), the identified peptide sequences, the confidence of sequence identification (Conf), post-translational modifications (Modifications), cleavages, theoretical molecular weights and m/z values, the recorded precursor mass and m/z values, the delta mass (dMass) values are indicated along with the ID of spectrum used for sequence identification and retention time.
(XLSX)
Bold amino acids represent carbamidomethylated cysteines while * indicates the stable isotope-labeled amino acids. DP: de-clustering potential, CE: collision energy.
(DOCX)
The values represent the dilution range where the amount of peptide introduced into the mass spectrometer is proportional with the signal intensity.
(DOCX)
The values of three independent replicates are indicated.
(XLS)
Acknowledgments
This research was supported by TÁMOP-4.2.2.D-15/1/KONV-2015-0016, TÁMOP 4.2.4.A/2-11-1-2012-0001, TÁMOP-4.2.2.B-15/1/KONV-2015-0001, TÁMOP 4.2.2.A-11/1/KONV-2012-0045 and National Brain Research Program KTIA_13_NAP-A-II/3. Adrienne Csutak is receiver of Janos Bolyai and Lajos Szodoray Postdoctoral Fellowships. We thank Dr. Mohamed Mahdi for the English proofreading.
Gergő Kalló was supported by Astellas Pharma Ltd. in form of support for chemicals and consumables which does not alter our adherence to PLOS ONE policies on sharing data and materials.
Data Availability
The SRM data are publicly available at the Panorama website: (https://panoramaweb.org/labkey/project/__r1225/begin.view?)
Funding Statement
This work was supported by the New Széchenyi Plan TÁMOP 4.2.4.A/2-11-1-2012-0001 to GK, New Széchenyi Plan TÁMOP 4.2.2.A-11/1/KONV-2012-0045 to: GK AC ÉC, New Széchenyi Plan TÁMOP-4.2.2.D-15/1/KONV-2015-0016 to: JT ÉC GK, New Széchenyi Plan TÁMOP-4.2.2.B-15/1/KONV-2015-0001 to GK, National Brain Research Program KTIA_13_NAP-A-II/3 to ME, Astellas Pharma Ltd. Fellowship to GK, Janos Bolyai and Lajos Szodoray Postdoctoral Fellowships to AC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Qiu C, Kivipelto M, von Strauss E. Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci. 2009;11: 111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bali J, Ben Halima S, Felmy B, Goodger Z, Zurbriggen S, Rajendran L. Cellular basis of Alzheimer’s disease. Ann Indian Acad Neurol. 2010;13: S89–93. 10.4103/0972-2327.74251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crews L, Tsigelny I, Hashimoto M, Masliah E. Role of synucleins in Alzheimer’s disease. Neurotox Res. 2009;16: 306–317. 10.1007/s12640-009-9073-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love S. Contribution of cerebral amyloid angiopathy to Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2004;75: 1–4. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1757461&tool=pmcentrez&rendertype=abstract [PMC free article] [PubMed] [Google Scholar]
- 5.Koronyo-Hamaoui M, Koronyo Y, Ljubimov A V., Miller C a., Ko MK, Black KL, et al. Identification of amyloid plaques in retinas from Alzheimer’s patients and noninvasive in vivo optical imaging of retinal plaques in a mouse model. Neuroimage. Elsevier Inc.; 2011;54: S204–S217. 10.1016/j.neuroimage.2010.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein LE, Muffat JA, Cherny RA, Moir RD, Ericsson MH, Huang X, et al. Cytosolic beta-amyloid deposition and supranuclear cataracts in lenses from people with Alzheimer’s disease. Lancet (London, England). 2003;361: 1258–65. 10.1016/S0140-6736(03)12981-9 [DOI] [PubMed] [Google Scholar]
- 7.Armstrong RA. Alzheimer’s Disease and the Eye. J Optom. Elsevier; 2009;2: 103–111. 10.3921/joptom.2009.103 [DOI] [Google Scholar]
- 8.Prvulovic D, Hampel H. Amyloid β (Aβ) and phospho-tau (p-tau) as diagnostic biomarkers in Alzheimer’s disease. Clin Chem Lab Med. 2011;49: 367–374. 10.1515/CCLM.2011.087 [DOI] [PubMed] [Google Scholar]
- 9.Wyss-coray T, Rogers J. Inflammation in Alzheimer Disease—A Brief Review of the Basic Science and Clinical Literature. Cold Spring Harb Perspect Biol. 2012; 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korolainen M a, Goldsteins G, Nyman T a, Alafuzoff I, Koistinaho J, Pirttilä T. Oxidative modification of proteins in the frontal cortex of Alzheimer’s disease brain. Neurobiol Aging. 2006;27: 42–53. 10.1016/j.neurobiolaging.2004.11.010 [DOI] [PubMed] [Google Scholar]
- 11.Takuma K, Yan SS, Stern DM, Yamada K. Mitochondrial dysfunction, endoplasmic reticulum stress, and apoptosis in Alzheimer’s disease. J Pharmacol Sci. 2005;97: 312–316. 10.1254/jphs.CPJ04006X [DOI] [PubMed] [Google Scholar]
- 12.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005;1: 55–66. 10.1016/j.jalz.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laske C, Sohrabi HR, Frost SM, López-de-Ipiña K, Garrard P, Buscema M, et al. Innovative diagnostic tools for early detection of Alzheimer’s disease. Alzheimers Dement. 2015;11: 561–78. 10.1016/j.jalz.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 14.Korolainen M a., Nyman T a., Aittokallio T, Pirttilä T. An update on clinical proteomics in Alzheimer’s research. J Neurochem. 2010;112: 1386–1414. 10.1111/j.1471-4159.2009.06558.x [DOI] [PubMed] [Google Scholar]
- 15.Kim DH, Yeo SH, Park J-M, Choi JY, Lee T-H, Park SY, et al. Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene. Elsevier B.V.; 2014;545: 185–193. 10.1016/j.gene.2014.05.031 [DOI] [PubMed] [Google Scholar]
- 16.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41: 1088–1093. 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podtelezhnikov AA, Tanis KQ, Nebozhyn M, Ray WJ, Stone DJ, Loboda AP. Molecular insights into the pathogenesis of Alzheimer’s disease and its relationship to normal aging. PLoS One. 2011;6: e29610 10.1371/journal.pone.0029610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korolainen M a, Nyman T a, Nyyssönen P, Hartikainen ES, Pirttilä T. Multiplexed proteomic analysis of oxidation and concentrations of cerebrospinal fluid proteins in Alzheimer disease. Clin Chem. 2007;53: 657–65. 10.1373/clinchem.2006.078014 [DOI] [PubMed] [Google Scholar]
- 19.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9: 555–66. 10.1038/nmeth.2015 [DOI] [PubMed] [Google Scholar]
- 20.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4: 222 10.1038/msb.2008.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalló G, Chatterjee A, Tóth M, Rajnavölgyi É, Csutak A, Tőzsér J, et al. Relative quantification of human β-defensins by a proteomics approach based on selected reaction monitoring. Rapid Commun Mass Spectrom. 2015;29: 1623–1631. 10.1002/rcm.7259 [DOI] [PubMed] [Google Scholar]
- 22.Zhou L, Zhao SZ, Koh SK, Chen L, Vaz C, Tanavde V, et al. In-depth analysis of the human tear proteome. J Proteomics. Elsevier B.V.; 2012;75: 3877–3885. 10.1016/j.jprot.2012.04.053 [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Beuerman RW, Chan CM, Zhao SZ, Li XR, Yang H, et al. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J Proteome Res. 2009;8: 4889–905. 10.1021/pr900686s [DOI] [PubMed] [Google Scholar]
- 24.Herber S, Grus FH, Sabuncuo P, Augustin AJ. Two-dimensional analysis of tear protein patterns of diabetic patients. Electrophoresis. 2001;22: 1838–44. [DOI] [PubMed] [Google Scholar]
- 25.Csősz É, Boross P, Csutak A, Berta A, Tóth F, Póliska S, et al. Quantitative analysis of proteins in the tear fluid of patients with diabetic retinopathy. J Proteomics. 2012;75: 2196–2204. 10.1016/j.jprot.2012.01.019 [DOI] [PubMed] [Google Scholar]
- 26.An HJ, Ninonuevo M, Aguilan J, Liu H, Lebrilla CB, Alvarenga LS, et al. Glycomics analyses of tear fluid for the diagnostic detection of ocular rosacea. J Proteome Res. 4: 1981–7. 10.1021/pr0501620 [DOI] [PubMed] [Google Scholar]
- 27.Haeckel R, Hänecke P. The application of saliva, sweat and tear fluid for diagnostic purposes. Ann Biol Clin (Paris). 1993;51: 903–10. Available: http://www.ncbi.nlm.nih.gov/pubmed/8210068 [PubMed] [Google Scholar]
- 28.Sperandio B, Fischer N, Sansonetti PJ. Mucosal physical and chemical innate barriers: Lessons from microbial evasion strategies. Semin Immunol. Elsevier Ltd; 2015;27: 111–118. 10.1016/j.smim.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 29.Çomoğlu SS, Güven H, Acar M, Öztürk G, Koçer B. Tear levels of tumor necrosis factor-alpha in patients with Parkinson’s disease. Neurosci Lett. 2013;553: 63–67. 10.1016/j.neulet.2013.08.019 [DOI] [PubMed] [Google Scholar]
- 30.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34: 939–44. Available: http://www.ncbi.nlm.nih.gov/pubmed/6610841 [DOI] [PubMed] [Google Scholar]
- 31.Diagnostic and Statistical Manual of Mental Disorders Source Information [Internet]. U.S. National Library of Medicine; Available: https://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/DSM4/
- 32.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12: 189–198. 0022-3956(75)90026-6 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Berta A. Collection of tear samples with or without stimulation. Am J Ophthalmol. 1983;96: 115–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/6869470 [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72: 248–54. Available: http://www.ncbi.nlm.nih.gov/pubmed/942051 [DOI] [PubMed] [Google Scholar]
- 35.Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30: 2524–6. 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- 36.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw. 2015;67 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 37.Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing When researchers tend to select pursuing multiple the (statistically) and support of conclusions. An unguarded use in a greatly results of single-inference inc. 2015;57: 289–300. [Google Scholar]
- 38.Shultz EK. Multivariate receiver-operating characteristic curve analysis: prostate cancer screening as an example. Clin Chem. 1995;41: 1248–55. Available: http://www.ncbi.nlm.nih.gov/pubmed/7628115 [PubMed] [Google Scholar]
- 39.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12: 77 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fullard RJ, Snyder C. Protein levels in nonstimulated and stimulated tears of normal human subjects. Invest Ophthalmol Vis Sci. 1990;31: 1119–1126. [PubMed] [Google Scholar]
- 41.Sugi K, Saitoh O, Hirata I, Katsu K. Fecal lactoferrin as a marker for disease activity in inflammatory bowel disease: comparison with other neutrophil-derived proteins. Am J Gastroenterol. 1996;91: 927–34. Available: http://www.ncbi.nlm.nih.gov/pubmed/8633583 [PubMed] [Google Scholar]
- 42.Rieg S, Steffen H, Seeber S, Humeny A, Kalbacher H, Dietz K, et al. Deficiency of dermcidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J Immunol. 2005;174: 8003–8010. 10.1016/S0093-3619(08)70018-2 [DOI] [PubMed] [Google Scholar]
- 43.Mumcu G, Cimilli H, Karacayli U, Inanc N, Ture-Ozdemir F, Eksioglu-Demiralp E, et al. Salivary levels of antimicrobial peptides Hnp 1–3, Ll-37 and S100 in Behcet’s disease. Arch Oral Biol. Elsevier Ltd; 2012;57: 642–6. [DOI] [PubMed] [Google Scholar]
- 44.Csősz É, Emri G, Kalló G, Tsaprailis G, Tőzsér J. Highly abundant defense proteins in human sweat asrevealed by targeted proteomics and label-free quantification mass spectrometry. J Eur Acad Dermatology Venereol. 2015;29: 2024–2031. 10.1111/jdv.13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullrich A, Sures I, D’Egidio M, Jallal B, Powell TJ, Herbst R, et al. The secreted tumor-associated antigen 90K is a potent immune stimulator. J Biol Chem. 1994;269: 18401–18407. [PubMed] [Google Scholar]
- 46.Schittek B. The multiple facets of dermcidin in cell survival and host defense. J Innate Immun. 2012;4: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1: 440–464. 10.4161/viru.1.5.12983 [DOI] [PubMed] [Google Scholar]
- 48.Örnek N, Dağ E, Örnek K. Corneal Sensitivity and Tear Function in Neurodegenerative Diseases. Curr Eye Res. 2015;40: 423–428. 10.3109/02713683.2014.930154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The y axis shows the intensity while the x axis shows the retention time. The blue line refers for the synthetic, stabile isotope labeled peptide, while the red line for the endogenous counterparts.
(PPTX)
The name and accession numbers of the identified proteins in each band are presented along with the protein identification data. The sequence coverage (%Cov), the identified peptide sequences, the confidence of sequence identification (Conf), post-translational modifications (Modifications), cleavages, theoretical molecular weights and m/z values, the recorded precursor mass and m/z values, the delta mass (dMass) values are indicated along with the ID of spectrum used for sequence identification and retention time.
(XLSX)
Bold amino acids represent carbamidomethylated cysteines while * indicates the stable isotope-labeled amino acids. DP: de-clustering potential, CE: collision energy.
(DOCX)
The values represent the dilution range where the amount of peptide introduced into the mass spectrometer is proportional with the signal intensity.
(DOCX)
The values of three independent replicates are indicated.
(XLS)
Data Availability Statement
The SRM data are publicly available at the Panorama website: (https://panoramaweb.org/labkey/project/__r1225/begin.view?)