Abstract
Rhabdomyosarcoma (RMS) is the most frequent soft tissue sarcoma in children that shares many features of developing skeletal muscle. TBX2, a T-box family member, is highly up regulated in tumor cells of both major RMS subtypes where it functions as an oncogene. TBX2 is a repressor that is often over expressed in cancer cells and functions in bypassing cell growth control, including the repression of the cell cycle regulators p14 and p21. We have found that TBX2 directly represses the tumor suppressor PTEN in both RMS and normal muscle. Exogenous expression of TBX2 in normal muscle cells down regulates PTEN, and depletion or interference with TBX2 in RMS cells up regulates PTEN. Human RMS tumors show high levels of TBX2 and correspondingly low levels of PTEN. The expression of PTEN in clinical RMS samples is relatively uncharacterized and we establish that suppression of PTEN is a frequent event in both subtypes of RMS. TBX2 represses PTEN by directly binding to the promoter and recruiting the histone deacetylase, HDAC1. RMS cells have high levels of activated AKT due to the deregulation of PI3K signaling, and depletion or interference with TBX2, which up regulates PTEN, results in a reduction of phospho-AKT. We have also found that the highly related T-box family member TBX3 does not repress PTEN in the muscle lineage. This work suggests that TBX2 is a central component of the PTEN/PI3K/AKT signaling pathway deregulation in RMS cells and that targeting TBX2 in RMS tumors may offer a novel therapeutic approach for RMS.
Keywords: TBX2, PTEN, RMS, myogenesis
Introduction
Phosphatase and tensin homolog (PTEN) is a well known tumor suppressor and loss or inactivation of PTEN has been implicated in many types of cancer 4. PTEN plays an essential role in normal development, physiology and tumor suppression. Homozygous deletion of Pten causes embryonic lethality, suggesting that PTEN is essential for embryonic development 8. Heterozygous deletion of Pten promotes tumorigenesis of several cancers including medulloblastoma 3, intestinal tumors 41 and prostate cancer 9. In medulloblastoma, patients whose tumor express a low to absent level of PTEN show a worse survival ratio 3 and in prostate cancer PTEN level inversely correlates with occurrence of invasive prostate cancer 9. Germline mutation of PTEN causes multiple disease syndromes, including Cowden disease, Bannayan-Riley-Ruvalcaba syndrome and Lhermitte-Duclos syndrome 4.
PTEN is known to function at the cytoplasmic membrane to antagonize the PI3K signaling pathway by dephosphorylating phosphatidylinositol-3,4,5-triphosphate PIP3, the important secondary-messenger molecule of PI3K pathways 16. Inactivation of PTEN results in activation of the PI3K/AKT pathway and subsequent increase in cell cycle progression, migration and survival 5,17. PTEN also functions in the nucleus where PTEN is indicated to have multiple roles including cell cycle control 52,51 and stabilizing chromosomes 42.
In the cytoplasm, PTEN prefers PIP3 as the major biological phosphoprotein substrate for dephosphorylation and converts PIP3 to PIP2 25. PIP3 is absent or very low in quiescent cells, but is rapidly up regulated by PI3K in response to growth factors or extracellular signaling. PIP3 is the major activator of AKT. AKT is recruited via PIP3 to the plasma membrane, where AKT can then be fully activated by phosphorylation.
In muscle, activation of PI3K/AKT pathway induced by serum starvation is crucial for myoblast differentiation in vitro. Loss of PTEN significantly promotes AKT phosphorylation and enhances myoblast differentiation 27. Surprisingly, a muscle specific depletion of Pten driven by muscle creatine kinase (MCK) promoter was found to protect mice from insulin resistance and did not grossly affect muscle histology or induce tumor development 53.
In the nucleus, PTEN regulates cell cycle progression by down regulating transcriptional expression and protein stability of cyclin D1, as well as inhibiting its nuclear localization 32. Besides cyclin D1, PTEN also is shown to potentially repress cyclin D2 13 and cyclin D3 55 to arrest the cell cycle at G1. PTEN is also been shown to modulate the cell cycle by up regulating the CDK inhibitor p27 46.
The status of PTEN in rhabdomyosarcoma has not been extensively studied. A recent genome wide mutational analysis revealed that mutations in the receptor tyrosine kinase/RAS/PIK3CA genetic axis are common in RMS 43. In 147 human tumors analyzed in this study, only one homozygous mutation in PTEN was identified 43. This work established that mutation of PTEN is not a frequent event in RMS cells, but the expression of PTEN in clinical RMS samples has not been characterized. In RMS cells, the fusion protein PAX3-FOXO1 has been shown to contribute to repression of PTEN 18. Depletion of PAX3-FOXO1 in RMS cells up regulated PTEN and exogenous expression of PAX3 in C2C12 cells down regulated PTEN 18. In both C2C12 normal myoblasts and RMS cells, the level of PTEN has been shown to be inversely correlated with AKT serine 473 phosphorylation 50, which is mediated by the rapamycin-insensitive mTOR complex (mTORC2) 39 and required for full activation of AKT 47. It has also been shown that microRNA miR-183 functions as an oncogene in RMS cells by targeting the transcription factor EGR1, which is an activator of PTEN 40.
Here, we show that TBX2 directly represses PTEN in RMS cells. The repression is mediated at least in part through recruitment of the histone deacetylase, HDAC1 to the PTEN promoter. TBX2 expression and PTEN expression are inversely correlated in both RMS cell lines and human RMS tumor samples representing both ERMS and ARMS cells. We show that PTEN expression is suppressed in a majority of clinical RMS samples representing both subtypes, suggesting that the repression of PTEN is a frequent event in RMS. Depletion or interference with TBX2 in RMS cells up regulates PTEN and reduces phospho-AKT. A highly related factor, TBX3, which has been shown to repress PTEN in other cell types, does not repress PTEN in the muscle linage. These data suggest that TBX2 represents an important new target for inhibiting the PI3K pathway implicated in RMS proliferation and progression.
Results
PTEN is induced upon muscle cell differentiation
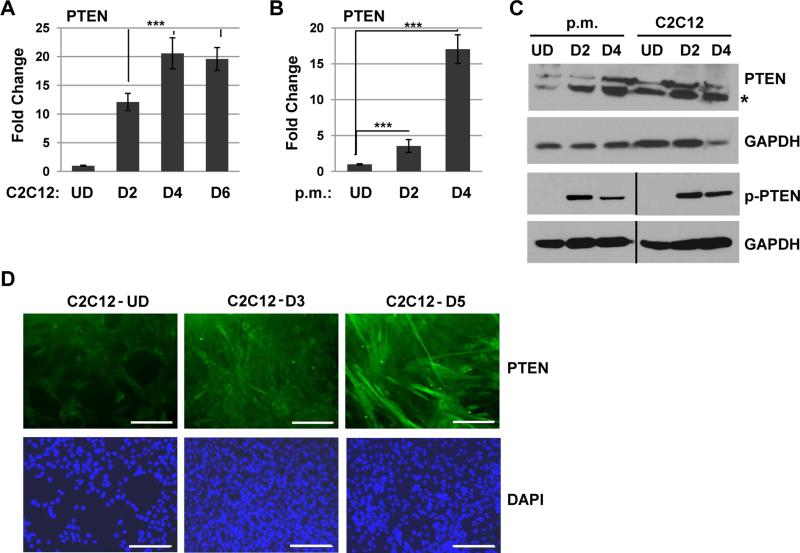
To understand the expression pattern of PTEN during normal skeletal muscle cell differentiation, we first assayed for the expression of PTEN in proliferating and differentiated C2C12 cells, a= murine cell line commonly used as a model of myogenesis. We found that PTEN was up regulated at the RNA level during differentiation (Figure 1A). To confirm this result, the experiment was repeated in primary murine myoblasts and we found that PTEN was strongly up regulated upon differentiation in these cells as well (Figure 1B). The up regulation of PTEN was also observed at the protein level in both differentiated primary myoblasts and C2C12 cells (Figure 1C). PTEN activity is also regulated by phosphorylation of the C-terminus, which inactivates the protein 37. Using an antibody specific to the phosphorylation of Ser380/Thr392/383 on PTEN, we found that p-PTEN could be detected in differentiated C2C12 cells, indicating that both the expression and phosphorylation of PTEN is regulated in normal muscle (Figure 1C). Immunocytochemistry with specific antibodies against PTEN displayed a robust increase of PTEN protein upon C2C12 differentiation in both the cytoplasm and nucleus (Figure 1D). These data demonstrate that PTEN is up regulated upon differentiation and regulated by phosphorylation in normal skeletal muscle cells.
Figure 1.
PTEN is up regulated upon muscle differentiation. A. PTEN mRNA is up regulated upon C2C12 differentiation. C2C12 cells were assayed for PTEN expression by qRT-PCR while proliferating (UD) and after 2 days (D2), 4 days (D4) and 6 days (D6) of differentiation. Bars, SD. ***, P<0.001. B. PTEN mRNA is up regulated upon primary myoblast (p.m.) differentiation. Primary myoblasts were assayed as in A. C. PTEN is up regulated and phosphorylated at the protein level upon differentiation in both primary myoblasts and C2C12 cells. Western blots were probed with antibodies as indicated. The asterisk indicates the predicted size of PTEN. D. PTEN is up regulated upon C2C12 cell differentiation. C2C12 cells were immunostained with antibodies against PTEN and with DAPI. Images were taken at 200X magnification and scale bars represent 10 μm.
PTEN is not expressed in the majority of RMS tumors
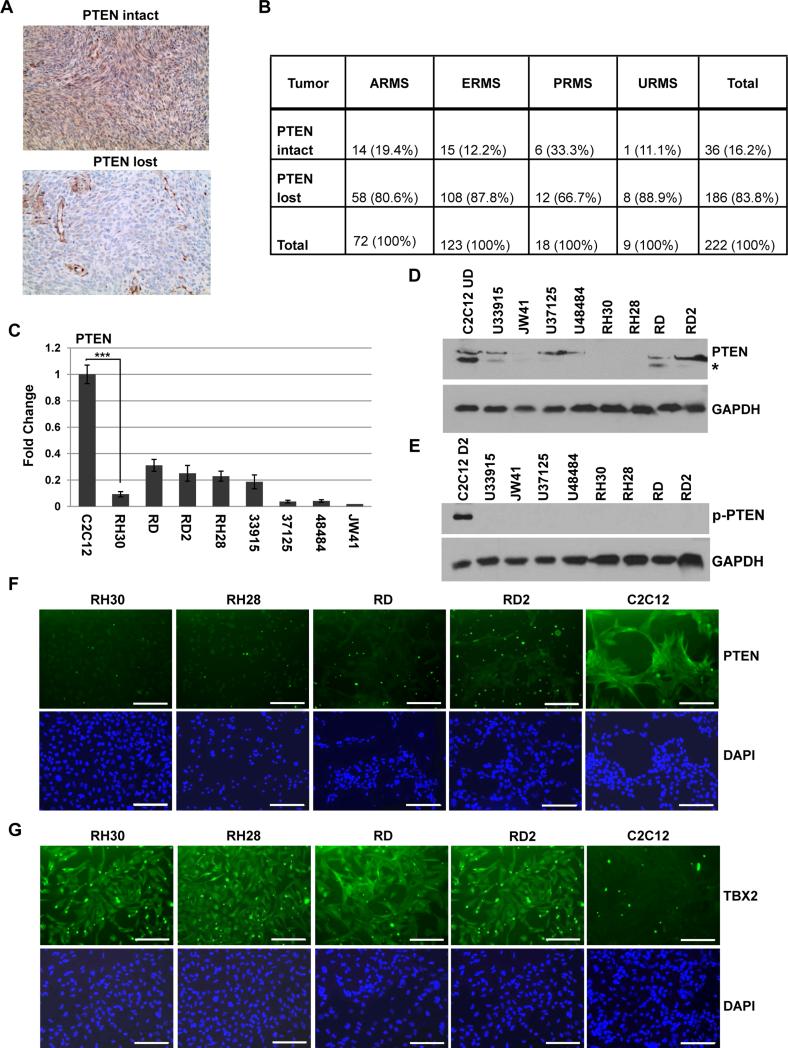
To determine the frequency of the loss of PTEN expression in clinical RMS cases, we identified 222 archival cases of RMS tumor specimens. These cases were previously diagnosed as ARMS (n = 72), ERMS (n = 123), PRMS (n = 18), and URMS (n = 9) (alveolar, embryonal, pleomorphic, and undifferentiated subtypes, respectively). Representative sections were immunostained for PTEN and the signal graded as intact or lost (Figure 2A). We found that PTEN expression was lost in the majority of both ERMS and ARMS cases (Figure 2B). 87.8% of ERMS tumors and 80.6% of ARMS tumors did not express PTEN (Figure 2B). A similar trend was noted in URMS, with PTEN loss detected at 88.9% (Figure 2B). PTEN expression was more retained in PRMS, with the loss of PTEN detected in only 66.7% of cases (Figure 2B). One limitation of the study is the lack of molecular genetic data for FOXO1 in early archival cases for ARMS, thus, the status of the PAX-FOXO1 fusion protein, which has been shown to repress PTEN 18, is not known in these samples. The data show that the absence of PTEN is a highly frequent event in human RMS tumors, including ERMS, which do not contain the PAX-FOXO1 fusion protein.
Figure 2.
PTEN expression is repressed in RMS. A-B. PTEN is repressed in a majority of RMS tumors. Archived tissue sections were immunostained for PTEN and images were taken at 100X magnification. A representative section is shown for the signal grading of “intact” or “lost” (A.). Results are summarized according to signal grading (B.). ARMS, ERMS, PRMS, and URMS represent alveolar, embryonal, pleomorphic, and undifferentiated subtypes, respectively. C. PTEN mRNA is down regulated in RMS cell lines. RD (ERMS), RD2 (ERMS), RH28 (ARMS), RH30 (ARMS) are human RMS cell lines and JW41 (ERMS), U33915 (ERMS), U37125 (ERMS), U48484 (ARMS) are primary murine RMS cells. qRT-PCR was used with primers specific to human and murine PTEN. Bars, SD. ***, P<0.001. D. PTEN is down regulated at the protein level in RMS cells. Samples from cells as described in C. were prepared for western blot probed with antibodies against PTEN and GAPDH. The asterisk indicates the predicted size of PTEN. E. P-PTEN is not detectable in RMS cells. Western blot was probed with antibodies against p-PTEN and GAPDH. The black line demarcates the two blots used for the analysis. F. PTEN is suppressed in human RMS cells. C2C12 cells and human RMS cells were immunostained with antibodies against PTEN. Images were taken at 200X magnification and scale bars represent 10 μm. The punctate staining in the images is a non specific artifact present in all controls, including samples immunostained without primary antibody. G. TBX2 is over expressed in RMS cells. Cells described in C. were immunostained with antibodies against TBX2. Images were as described in F.
PTEN expression is decreased in RMS cells
Next, we sought to validate our finding in representative RMS cell lines to provide appropriate cell models for functional studies defining the mechanism of repression of PTEN. The expression of PTEN was assayed in both RMS cell lines and primary tumor cells. We found that the PTEN transcript was reduced when compared to proliferating C2C12 cells in RH30, RD, RD2 and RH28 cell lines as well as in primary cells derived from mouse tumor models of RMS that represent both major tumor subtypes (Figure 2C). The protein expression of PTEN was detected with antibodies against PTEN that recognize both murine and human proteins. We found that PTEN protein expression was reduced in all tested RMS cells when compared to proliferating C2C12 cells (Figure 2D). We also examined p-PTEN, but, corresponding to the low expression of PTEN, p-TEN levels were not detectable in RMS cells (Figure 2E) Immunostaining for PTEN confirmed the low level of PTEN expression in RMS cell lines (Figure 2F). To correlate the expression of PTEN with TBX2 expression in these cell lines, we also performed immunocytochemistry for TBX2 (Figure 2G). Consistent with our previously published western blot analysis 54, we found that TBX2 was robustly up regulated in RMS cells and correlated inversely with PTEN expression.
PTEN expression is inversely correlated with TBX2 expression in human tumors
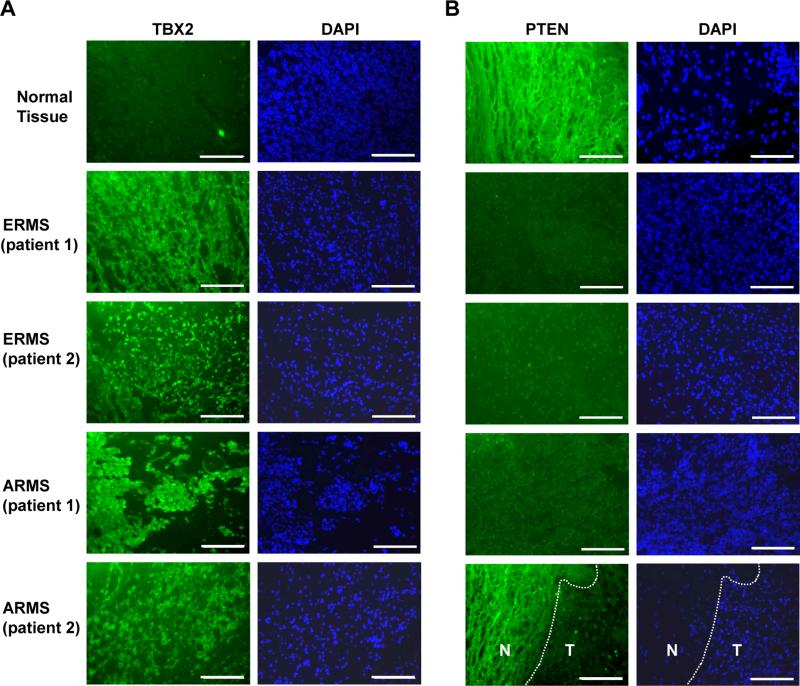
To determine if TBX2 was also inversely correlated with PTEN in human RMS tumors, we performed immunohistochemistry for TBX2 and PTEN in human clinical RMS tumor samples which included both ERMS and ARMS tumor samples (Figure 3A). We found that TBX2 was highly expressed in human tumor samples of both ERMS and ARMS subtypes when compared to normal human tissue. We then performed immunohistochemistry for PTEN in the same human clinical RMS tumor samples and found that RMS tumors displayed low levels of PTEN protein when compared with normal muscle tissue (Figure 3B). The result agrees with our previous results in RMS cell lines and primary tumor cells from engineered RMS mice. The inverse expression pattern of PTEN and TBX2 suggests that PTEN is a potential downstream target gene of TBX2 in RMS.
Figure 3.
PTEN expression is inversely related with TBX2 expression in human RMS tumors. A. TBX2 is up regulated in RMS tumor tissue. Immunostaining of primary RMS tumor and normal skeletal muscle sections using TBX2 specific antibodies is shown. Images were taken at 100X magnification and scale bars represent 20 μm. B. PTEN is down regulated in human tumor tissue. Immunostaining of samples shown in A. using PTEN specific antibodies is shown. Images were taken at 100X magnification and scale bars represent 20 μm. The section shown in the lower right panel contained both normal tissue (N) and tumor (T), and the demarcation of the tumor boundary is indicated with a dashed line.
PTEN expression is repressed by TBX2
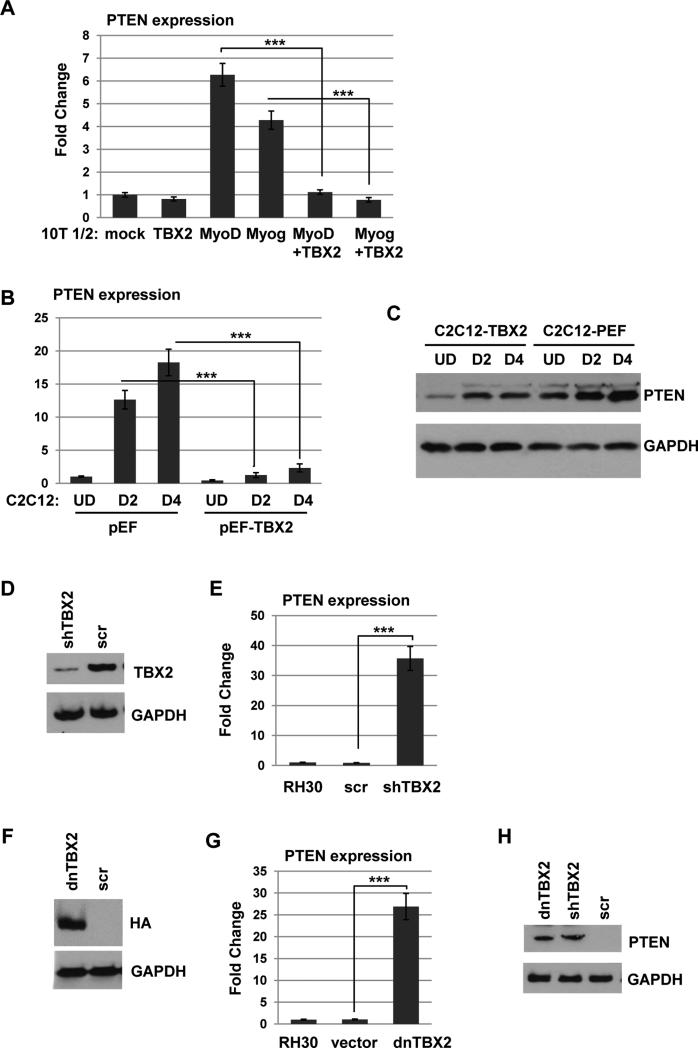
To determine if PTEN expression is repressed by TBX2, 10T1/2 cells, a fibroblast cell line which can be induced to express muscle specific genes upon transfection with MRFs 7, were transfected with an expression construct for TBX2 in combination with expression constructs for myogenin or MyoD and gene expression changes were determined for PTEN. When 10T1/2 cells were transfected with myogenin or MyoD to induce skeletal muscle differentiation, PTEN expression was up regulated, but co-transfection with TBX2 significantly inhibited PTEN expression, suggesting that TBX2 does repress PTEN (Figure 4A). To confirm this result, a stable C2C12 cell line over expressing TBX2 was assayed for PTEN expression. We found that ectopically expressed TBX2 markedly reduced PTEN at both the RNA (Figure 4B) and protein (Figure 4C) level in C2C12 cells. To determine if the high level of TBX2 in RMS cells blocks PTEN expression, we stably introduced TBX2 shRNA constructs (shTBX2) into RH30 cells. The depletion of TBX2 was confirmed by western blot analysis (Figure 4D). Depletion of TBX2 strongly elevated PTEN expression at the level of RNA (Figure 4E). We also used a dominant negative construct of TBX2 (dnTBX2) which lacks the C-terminal domain of TBX2 necessary for transcriptional repression and interaction with HDAC1, but retains amino acids 1-301 of the T-box DNA binding activity of TBX2, thus blocking the function of endogenous TBX219. The expression of dnTBX2 was confirmed by western blot analysis (Figure 4F). We found that stable expression of dnTBX2 in RH30 cells also led to an up regulation of PTEN expression (Figure 4G). The up regulation of PTEN by either depletion of TBX2 or expression of dnTBX2 was confirmed by western blot analysis (Figure 4H). The western blot was also probed for p-PTEN, but p-PTEN was not detected in the presence of shTBX2 or dnTBX2 (data not shown). Taken together, these results suggest that TBX2 represses PTEN expression in both normal muscle cells and RMS cells.
Figure 4.
PTEN expression is repressed by TBX2. A. TBX2 represses PTEN expression in 10T1/2 cells. 10T1/2 cells were transiently transfected with expression constructs for myogenin, MyoD and TBX2 as indicated and harvested for RNA 48 hours post transfection. Gene expression was assayed by qRT-PCR. B. Stable C2C12 cell lines over expressing TBX2 (pEFTBX2) or a vector control (pEF) were assayed for PTEN expression by qRT-PCR. C. Stable C2C12 cell lines as in B. were assayed for PTEN expression by western blot analysis with the indicated antibodies. D-E. Depletion of TBX2 up regulates PTEN. Stable RH30 cell lines expressing shRNA constructs against TBX2 (shTBX2) or the scrambled control (scr) were confirmed by western blot analysis (D.) and assayed for PTEN mRNA expression by qRT-PCR (E.). F-G. Interference with TBX2 up regulates PTEN. Stable RH30 cell lines expressing HA tagged dominant negative TBX2 (dnTBX2) or a vector control pcDNA3.1 (vector) were confirmed by western blot analysis (F.) and assayed for PTEN mRNA expression by qRTPCR(G). H. The up regulation of PTEN by depletion or interference with TBX2 was confirmed at the protein level by western blot analysis.
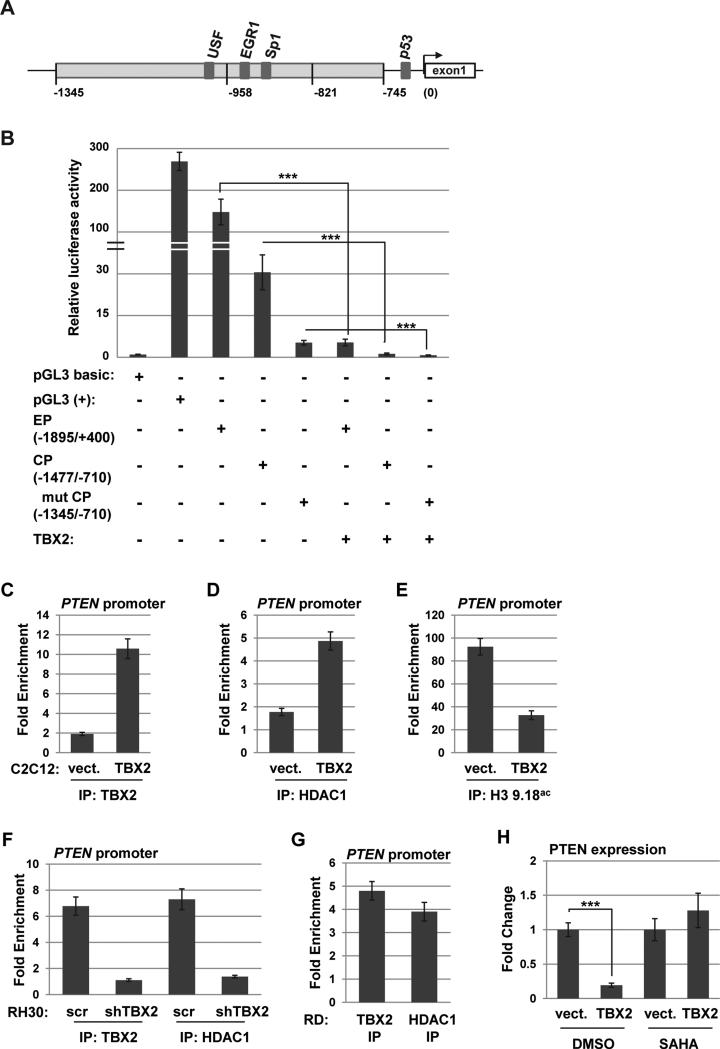
TBX2 inhibits PTEN by recruiting HDAC1 to the promoter
We used PTEN promoter specific luciferase constructs to determine promoter elements of PTEN that were required for TBX2 dependent repression. The promoter proximal elements of the PTEN promoter are diagrammed in Figure 5A. The constructs contain the extended promoter (EP-PTEN), covering the PTEN promoter region between positions −1895 to + 400, the core promoter region (CP-PTEN) lying between −1477 and −710, and the deletion mutant (Δmut CP-PTEN), which contains positions −1345 and −710. 10T1/2 cells were transfected with individual promoter constructs in the presence or absence of TBX2. All three promoter constructs were robustly inhibited by TBX2 (Figure 5B). The result suggests that the DNA region responsive to TBX2 is located between positions −1345 and −710.
Figure 5.
TBX2 directly represses PTEN. A. Diagram of the PTEN promoter. B. TBX2 represses PTEN promoter activity. 10T1/2 cells were transfected with the indicated constructs. EP represents EP-PTEN (−1895/+400); CP represents CP-PTEN (−1477/−710) and Δmut represents Δmut CP-PTEN (−1345/−710). pGL3 basic, which lacks a promoter, represents a negative control and pGL3 (+), which contains the SV40 promoter, represents a positive control. C-E. TBX2 and HDAC1 bind to the PTEN promoter and block histone acetylation. ChIP assays were performed on proliferating stable C2C12 cell lines over expressing TBX2 or a vector control (vect.) with antibodies against TBX2 (C.), HDAC1 (D.) and histone H3, lysine 9,18ac (H3 K9,18ac, E.) and PTEN promoter specific primers. All values were normalized against the IgG antibody sample. F. ChIP assays were performed with antibodies against TBX2 and HDAC1 in stable RH30 cell lines expressing shRNA constructs against TBX2 (shTBX2) or the scrambled control (scr). G. ChIP assays were performed on RD cells for TBX2 and HDAC1. H. Histone deacetylase inhibitors (HDACi) block TBX2 repression of PTEN. C2C12 cells expressing exogenous TBX2 or vector control were treated with vehicle control (DMSO) or SAHA for 24 hours while differentiating, and were then assayed for PTEN expression by qRT-PCR. Data are presented with respect to the vector control value, which was set to one.
To investigate the molecular mechanism of TBX2 repression on the PTEN promoter, we performed chromatin immunoprecipitation (ChIP) assays for TBX2 and the class I histone deacetylase, HDAC1, on the PTEN promoter as TBX2 has been shown to recruit HDAC1 to target gene promoters 48. ChIP assays were also used to assay for the acetylation status of histones by examining the acetylation of lysine 9 and 18 on histone H3 (H3K9,18ac). We found that in C2C12 cells expressing exogenous TBX2, TBX2 (Figure 5C) and HDAC1 (Figure 5D) were bound to the PTEN promoter. In C2C12 cells expressing vector, this binding was not observed (Figure 5C and D). The binding of TBX2 and HDAC1 was concomitant with a reduction in histone H3 K9,18 acetylation at the PTEN promoter (Figure 5E).
To determine if TBX2 and HDAC1 bound to the PTEN promoter in RMS cells, ChIP assays for TBX2 and HDAC1 were repeated in RD and RH30 cells, which express high amounts of endogenous TBX2. We found that both TBX2 and HDAC1 bound to the PTEN promoter in both RH30 (Figure 5F) and RD (Figure 5G) cells. To determine if the binding of HDAC1 was dependent on TBX2, TBX2 was depleted with shRNA construct in RH30 cells and we found that the occupancy of both TBX2 and HDAC1 was reduced in TBX2 depleted RH30 cells (Figure 5F). To determine if HDACs were required for the repression mediated by TBX2, C2C12 cells expressing TBX2 or a vector control were treated with the class I and class II HDAC inhibitor SAHA while differentiating. We found that the addition of SAHA blocked the repression of PTEN by TBX2 (Figure 5H).
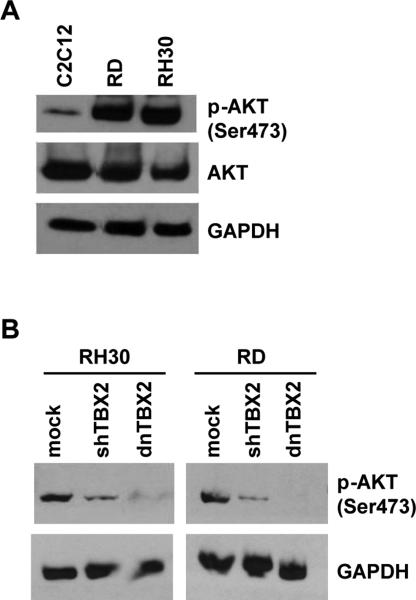
Low level of PTEN is related with high activity of AKT
PTEN acts as a negative regulator of the PI3K/AKT signaling pathway. To correlate AKT activity with PTEN expression in normal myoblasts and RMS cells, we assayed for activated AKT by western blot assays with antibodies against both phosphorylated (pAKT) and total AKT. The phosphorylation status of Ser473 on AKT, the target of mTOR2, was used to detect activated AKT. We found that while the total amount of AKT protein was very similar in RMS cells (RH30 and RD) compared with C2C12 cells, the RMS cell lines contained remarkably elevated pAKT (Figure 6A). It has been shown that transfection of wild-type PTEN into RMS and C2C12 cells results in reduced AKT phosphorylation 50. As we show here that TBX2 represses PTEN, we asked if depletion or interference with TBX2 would result in reduced phosphorylation of AKT. RH30 and RD cells were transfected with shTBX2 or dnTBX2 and assayed for the phosphorylation of Ser473. We found that either depletion or interference with TBX2 results in reduced pAKT (Figure 6B). These results suggest that the high activity of PI3K/AKT signaling is modulated by decreased PTEN expression in RMS mediated by TBX2.
Figure 6.
TBX2 expression is correlated with AKT activation in RMS cells. A. RMS cell lines have high levels of activated AKT. The level of pAKT (Ser 473) and total AKT were assayed in proliferating C2C12, RH30 and RD cells by western blot analysis. B. Depletion or interference of TBX2 reduces p-AKT. Stable RH30 or RD cell lines expressing shTBX2 or dnTBX2 were assayed for pAKT by western blot analysis.
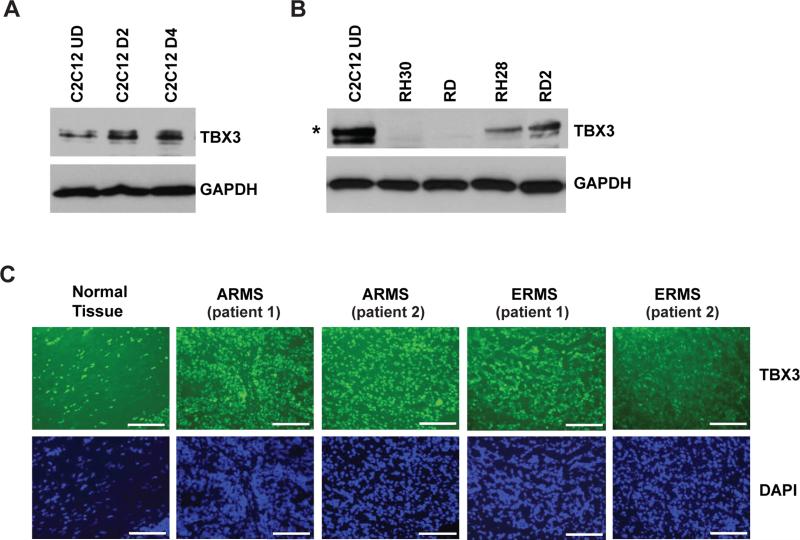
TBX3 does not repress PTEN in skeletal muscle cells
TBX2 and TBX3 are highly related members of the T-box family which both function as repressors. In several cancer types, TBX2 and TBX3 function together to regulate cancer proliferation, metastasis and invasion 10, 30. TBX3 has previously been shown to be both a repressor of myogenesis in C2C12 cells 2 and a direct repressor of PTEN in head and neck squamous cell carcinoma 1. Thus, we sought to determine if TBX3 played a similar role to TBX2 in skeletal muscle cells and RMS. We have shown that TBX2 is sharply down regulated upon C2C12 differentiation 54 so we assayed for the expression of TBX3 during C2C12 differentiation. We found that TBX3 was robustly expressed in C2C12 cells and that the expression did not decrease upon differentiation (Figure 7A). We next assayed RMS cells for TBX3 and found that TBX3 was reduced in expression relative to the level seen in C2C12 cells (Figure 7B). While reduced in expression, TBX3 was detectable in the RH28 and RD2 cell lines but was not detectable in RH30 or RD cell lines. We also assayed for the expression of TBX3 in human RMS tumor samples by immunostaining for TBX3, and found that TBX3 was expressed in normal tissue as well as in RMS tumor tissue (Figure 7C). TBX3 did not appear to be up regulated or down regulated with respect to normal tissue. Taken together, these data show that TBX3 expression patterns are entirely different from that of TBX2 in both normal skeletal muscle and RMS cells, thus suggesting that TBX3 has an independent function in the muscle lineage.
Figure 7.
TBX3 is expressed in normal skeletal muscle and not over expressed in RMS cells. A. TBX3 is expressed in C2C12 cells throughout differentiation. C2C12 cells were differentiated for the indicted number of days and analyzed by western blot analysis. B. TBX3 is not over expressed in RMS cells. Western blot was probed with specific antibodies against TBX3 and GAPDH. Asterisk indicates the predicted size of TBX3. C. TBX3 is expressed in both normal tissue and primary RMS tumors. Immunofluorescence staining of primary RMS tumor and normal skeletal muscle sections using TBX3 specific antibodies is shown. Images were taken at 100X magnification and scale bars represent 20 μm.
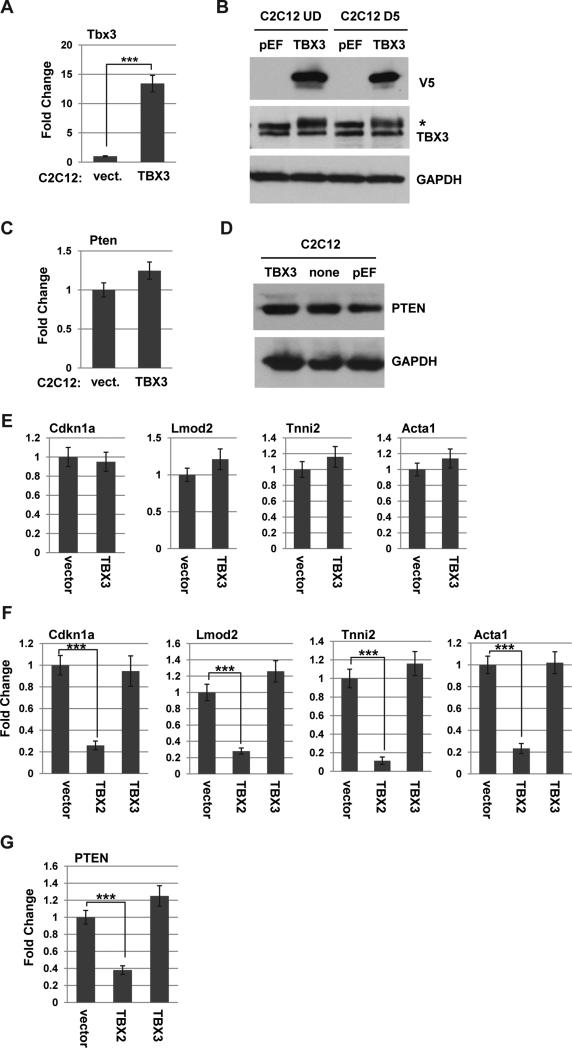
To determine if TBX3 represses PTEN in muscle, an expression construct for TBX3 was stably expressed in C2C12 cells. The exogenous expression of TBX3 was confirmed by RNA (Figure 8A) and protein analysis (Figure 8B). We next assayed for PTEN expression and found that no repression of PTEN was observed at the RNA (Figure 8C) or protein (Figure 8D) level. Thus, TBX3 does not repress PTEN in C2C12 cells. The results suggest that TBX2, not TBX3, is the major regulator of PTEN in the muscle linage, including RMS cells. To determine if TBX3 repressed myogenesis as we have shown for TBX2, we also assayed for muscle specific gene expression in the TBX3 expressing cell line described above. C2C12 cells expressing exogenous TBX3 or vector control were differentiated for two days and examined for gene expression of genes normally induced upon differentiation including actin (Acta1), leiomodin2 (Lmod2), troponin 1, type 2 (Tnni2) and p21 (Cdkn1a). We have previously shown that exogenous expression of TBX2 represses each of these genes54. We found that TBX3 expression had no effect on the expression of any of these genes (Figure 8E). To confirm our results, expression constructs for both TBX2 and TBX3 were transiently transfected into C2C12 cells and cells were assayed for gene expression after one day of differentiation. We found that TBX2 robustly repressed the expression of Acta1, Tnni2, Lmod2 and Cdkn1a while TBX3 had no significant effect on the expression of these genes (Figure 8F). The expression of PTEN was also assayed and we found that TBX2, but not TBX3, repressed PTEN (Figure 8G).
Figure 8.
TBX3 does not repress PTEN in C2C12 cells. A-B. Stable C2C12 cell lines over expressing V5 tagged TBX3 (TBX3) or a vector control (pEF) were assayed for expression of TBX3 by qRT-PCR (A.) and western blot analysis with indicated antibodies (B.). C-D. Over expression of TBX3 does not repress PTEN. The expression of PTEN was assayed in the cell lines described in A. by qRT-PCR (C.) and western blot analysis (D.) E. Over expression of TBX3 does not inhibit myogenic differentiation. Cell lines described in A. were differentiated for 2 days and gene expression for the indicated genes was analyzed by qRT-PCR. F. TBX2, not TBX3, inhibits myogenesis. C2C12 cells were transiently transfected with expression constructs for vector control, TBX2 or TBX3 and differentiated for 24 hours prior to RNA isolation 48 hours post transfection. Gene expression for the indicated genes was analyzed by qRT-PCR. G. TBX3 does not regulate PTEN in C2C12 cells. Cells described in F. were analyzed for PTEN expression by qRT-PCR.
Discussion
The role of altered PI3K signaling in RMS origin and progression is well known. We show here that the tumor suppressor PTEN, which is a negative regulator of the PI3K pathway, is down regulated in a majority of RMS tumors and that TBX2, a novel oncogene in RMS, is a direct regulator of PTEN. Thus, TBX2 represents an important new target for targeting aberrant PI3K activity in RMS cells. It has been shown that both the PI3K/AKT/mTOR pathway and the RAS/RAF/MEK/ERK pathway are active in RMS cells, leading to resistance in treating either pathway alone. However, co-inhibition of both pathways has reduced tumor growth in xenograft models 35. As p21 can be induced by MEK/ERK pathway inhibition in RMS cells 6 and we have previously shown that TBX2 regulates p21 and p14 54, this suggests that targeting TBX2 might block downstream targets of both the PI3K/AKT/mTOR pathway and the RAS/RAF/MEK/ERK pathway.
It was surprising that the highly related T-box family member TBX3 did not regulate PTEN in our experiments. TBX3 is a known regulator of PTEN in head and neck carcinoma and was shown to repress PTEN in both HeLa and HEK293 cells 1. However, the distinct roles of TBX2 and TBX3 in the muscle lineage are not entirely surprising. TBX2 and TBX3 have been shown to be over expressed in a number of cancers, including breast, pancreatic, ovarian, liver, cervical and melanoma 14, 15, 21, 31. However, TBX2 and TBX3 have also been shown to exhibit mutually exclusive expression in melanoma cell lines 36 and the homologues have distinct functions in embryonic development and cancer 10, 11, 14, 24, 26, 31, 38, 44, 48. TBX2 and TBX3 have distinct roles in cancer progression, with TBX2 functioning as a pro-proliferation factor and TBX3 promoting migration and invasion 29, 33. Recent work has shown that the anti-proliferative function of TGF-β1 is mediated through repression of TBX2 by TBX3 19. We do not currently understand the function of TBX3 in skeletal muscle, but it is expressed throughout differentiation, suggesting that it does play a role in myogenesis. However, overexpression of TBX3, unlike TBX2, is not sufficient to alter myogenesis.
PAX3 and the PAX3/FOXO1 fusion found in ARMS tumors have also been shown to repress PTEN 18. Depletion of PAX3/FOXO1 in ARMS cells lines or exogenous expression of PAX3 in C2C12 cells repressed PTEN 18. PAX3 was shown to bind to the PTEN promoter, but the repression mechanism is unclear as PAX3 and PAX3/FOXO1 are known transcriptional activators 34. PAX3 does interact with negative modulators such as DAXX, but the PAX3-FOXO1 fusion protein is unresponsive to DAXX 12. PAX3 has also been shown to activate TBX2 in melanoma cells 20, suggesting that PAX3 might regulate TBX2 in RMS cells as well. Thus, the negative regulation of PTEN by PAX3 may be mediated through TBX2.
PTEN is also known to be regulated in several cell types by the transcription factor EGR128, 49. EGR1 binds to the PTEN promoter and activates expression 49. Intriguingly, TBX2 has been shown to repress the tumor suppressor NDRG1 through an interaction with EGR1 33. TBX2 requires EGR1 to target the promoter and repress NDRG1 33. These results suggest that TBX2 may repress the EGR1 dependent gene program, including PTEN. EGR1 also regulates a number of other key tumor suppressor genes, thus, the repression of the EGR1 program by TBX2 has important implications for growth control and oncogenesis. Understanding whether this mechanism is active in RMS cells will be an important future direction for our studies.
The regulation of several key signaling pathway components and cell cycle regulators by TBX2 strongly suggests that TBX2 is an important target for therapeutic development. We have shown the depletion or interference with TBX2 in the ARMS cell line RH30 completely blocks tumor formation in vivo 54. Targeting TBX2 in RMS tumors may offer a novel therapeutic approach for controlling the proliferation and progression of this disease. In addition, TBX2 has been shown to be an oncogene in several cancers, suggesting that therapies designed to target TBX2 may be useful in the treatment of many cancers.
Materials and Methods
Cell Culture
RD and RH30 cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone) according to standard protocols. RD2 and RH28 were obtained from Denis Guttridge, Ohio State University, and grown as described above. All cell lines were authenticated by Bio-Synthesis (Lewisville, TX) using STR analysis on September 14, 2011. JW41 cells, isolated from an ERMS tumor from a p53−/−/cfos−/− mouse 45, were the gift of Charlotte Peterson, University of Kentucky. U33915 and U37125 represent ERMS-type tumors derived from the Pax7CreER, Ptch1, p53 conditional model of spindle ERMS/UPS8 (Rubin BP, Nishijo, 2011) and U48484 was derived from the Myf6Cre, Pax3:Fkhr, p53 conditional mouse model of ARMS (Nishijo K, Chen, 2009). Proliferating C2C12 myoblasts (ATCC), 10T1/2 cells (ATCC) and HEK293 cells (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (Hyclone). To induce differentiation of C2C12 myoblasts into myotubes, cells were grown to 90% confluence and the media switched to DMEM supplemented with 2% horse serum (Hyclone). C2C12 cells were grown in differentiation medium for the number of days indicated in each experiment. For treatment with the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, SCBT, Dallas, TX), cells were grown to 100% confluence, switched to differentiation media and incubated with 5 μM SAHA for 24 hr. Toxicity curves were generated for SAHA before examining the effects on gene expression. All assays were performed in triplicate.
Cloning
To clone murine genes, RNA from C2C12 cells was extracted with Trizol (Invitrogen) and used to generate cDNA through the use of the Superscript III First Strand Synthesis System for RT-PCR kit with oligo (dT) primers (Invitrogen). Murine TBX3 was amplified from cDNA generated from undifferentiated C2C12 cells using primers TBX3 ORF F 5′ ATGAGCCTCTCCATGAGAGATC 3′ and R 5′ AGGGGACCCGCTGCA 3′. The resulting PCR product was TOPO cloned using the pEF6/V5-His-TOPO TA Expression Kit (Invitrogen). All resulting clones were confirmed by sequencing.
Cell transfections
Cells were transfected with calcium phosphate according to standard protocols or Turbofect transfection reagent (Thermo Scientific). Transient transfections were harvested 48 hours post transfection. Stable cell lines were made by transfecting cells with linearized plasmids and selecting for drug resistant colonies. Individual clones were isolated and propagated.
Luciferase assays
10T1/2 cells were transiently transfected with calcium phosphate. The PTEN promoter reporter plasmid constructs were generously provided by Dr. Ugur Yavuzer, Department of Physiology, School of Medicine, Akdeniz University. The constructs included the extended promoter (EP-PTEN) covering PTEN promoter region between positions −1895 to + 400, the core promoter region (CP-PTEN) lying between −1477 and −710 and the deletion mutant (Δmut CP-PTEN) between positions −1345 and −710 1. Plasmid EMSV-myogenin was a gift of Dr. D. Edmondson, U.T. Medical School at Houston and the MyoD expression construct of pEMCIIs was provided by Dr. Andrew Lassar, Harvard Medical School. The expression construct for TBX2 (pEF TBX2) was described previously 54. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). 10T1/2 cells were seeded at a density of 5×103 cell per well in 96 well plates and transfected with 0.3 ug of DNA. Transfections were normalized to Renilla luciferase. Transfections were performed in triplicate and all data sets were repeated three times.
Quantitative real time PCR
RNA was isolated from cells by Trizol extractions (Invitrogen). Following treatment with DNase (Promega), two micrograms of total RNA was reversed transcribed with MultiScribe™ MuLV reverse transcriptase (Applied Biosystems). cDNA equivalent to 40 ng was used for quantitative polymerase chain reaction (qPCR) amplification (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems). Samples in which no reverse transcriptase was added (no RT) were included for each RNA sample. The relative levels of expression of genes were normalized according to those of HPRT1. qPCR data were calculated using the comparative Ct method (Applied Biosystems). Standard deviations from the mean of the [Δ] Ct values were calculated from three independent RNA samples. PTEN and TBX3 were amplified using primers pTEN m/h F 5′ GCTATGGGATTTCCTGCAG 3′, R 5′ CTAGCTGTGGTGGGTTATGG 3′ and TBX3 m/h F 5′ TGATGGACATTATAGCTGCTGA 3′, R 5′ AGTGACGACTTTGGACATCC 3′. Primers against LMOD2, TNNI2, ACTA1 and HPRT were previously described 23, as were primers against CDKN1A 54. Where possible, intron spanning primers were used. All quantitative PCR was performed in triplicate and three independent RNA samples were assayed for each time point. For measurements of relative gene expression (fold stimulation), a fold change was calculated for each sample pair and then normalized to the fold change observed at HPRT.
Western Blot Analysis
Cell extracts were made by lysing PBS washed cell pellets in radio-immunoprecipitation assay buffer (RIPA) supplemented with protease inhibitors (Complete protease inhibitor, Roche Diagnostics). Following incubation on ice, clear lysates were obtained by centrifugation. Protein concentrations were determined by Bradford's assay (Bio-Rad). For each sample, 30 μg of protein was loaded on each gel. Proteins were transferred onto a PVDF membrane using a tank blotter (Bio-Rad). The membranes were then blocked with 5% milk and 1X Tris buffered saline plus Tween 20 (TBST) and incubated with primary antibody overnight at 4°C. Membranes were then washed with 1X TBST and incubated with the corresponding secondary antibody. Membranes were again washed with 1X TBST, incubated with chemiluminescent substrate according to manufacturer's protocol (SuperSignal, Pierce) and visualized by autoradiography. The antibodies used include anti-PTEN (559600, BD Pharmingen), anti-phospho PTEN (9554, Cell Signaling), anti-TBX2 (C-17, Santa Cruz Biotechnology), anti-TBX2 (gift of C. Goding, University of Oxford), anti-TBX3 (A-20, Santa Cruz Biotechnology), anti-AKT (pan, C67E7, Cell Signaling), anti-phospho-AKT (Ser 473 D9E, Cell Signaling) and anti-GAPDH (6C5, Millipore). At least three biological replicates were performed for each experiment.
Chromatin immunoprecipitation assays
ChIP assays were performed and quantified as described previously 22 with the following modifications: 1×107 cells were used for each immunoprecipitation and protein A agarose beads (Invitrogen) were used to immunoprecipitate the antibody:antigen complexes. The following antibodies were used: anti-TBX2 (C-17, Santa Cruz Biotechnology), anti-HDAC1 (10E2, Cell Signaling) and anti-H3K9, 18ac (gift of Dr. S. Dent, U.T.M.D. Anderson Cancer Center). Rabbit IgG (Santa Cruz Biotechnology) was used as a non-specific control. Primers against the PTEN promoters were PTEN E2 F 5′ GGAATTTGGAAAGTTCCCC 3′ and R 5′ GTACGGAACGGTAGGAAGCT 3′. Primers against a gene desert region of Chr. 19 were previously described 54. The real time PCR was performed in triplicate. Values of [Δ][Δ] Ct were calculated using the following formula based on the comparative Ct method: [Δ] Ct, template (antibody) - [Δ] Ct, template (IgG) = [Δ][Δ] Ct. Fold enrichments were determined using the formula : 2 −[Δ] [Δ] Ct. (experimental)/2 −[Δ] [Δ] Ct (reference, Chr. 19). Standard error from the mean was calculated from replicate [Δ][Δ] Ct values. All ChIP assays shown are representative of at least three individual experiments.
Immunocytochemistry
Cells were grown on cover slips, fixed with paraformaldehyde, incubated with goat serum and 1.0 % NP-40 for one hour and washed with PBS. Primary antibodies anti-PTEN (559600, BD Pharmingen) and anti-TBX2 (gift of C. Goding, University of Oxford) were incubated overnight at 4°C, washed with PBS and detected by Alexa Fluor-488 goat anti-mouse antibody (1:500, Invitrogen). Cell nuclei were then stained by incubating with DAPI (1 μM, Invitrogen) for 5 min. At least three biological replicates were performed for each experiment.
Primary human RMS tumor samples on slides and accompanying pathology reports with no patient information except age and sex were obtained from the Nationwide Children's Hospital Biopathology Center, Colombus, Ohio. Samples were fixed by 3% H2O2 in methanol for 20 minutes at room temperature, washed in PBS and blocked by 0.1% Triton X-100 TBS solution supplemented with 1.5% Normal Goat Serum (1.5% Normal Donkey Serum for TBX3 primary antibodies) overnight at 4°C. Primary antibodies included anti-PTEN (559600, BD Pharmingen), anti-TBX2 (gift of C. Goding, University of Oxford) and anti-TBX3 (A-20, Santa Cruz Biotechnology). Primary antibodies were incubated for 2 hours at room temperature, and then washed with 0.1% Triton X-100 TBS before adding the corresponding secondary antibodies Alexa Fluor-488 goat anti-mouse or Alexa Fluor-488 donkey anti-goat antibody (1:500, Invitrogen) for incubation for 1 hour at room temperature. Mounting medium including DAPI was used for cell nuclei staining. Ten patient samples, with at least two slides per patient, were used for each antibody. Representative slides are shown.
The 222 archival samples derived from RMS specimens submitted for surgical pathology diagnosis. The cases were previously diagnosed as alveolar, embryonal, pleomorphic and undifferentiated subtypes. Formalin fixed paraffin embedded sections were stained with antibody clone PTEN D4.3 (Cell Signaling Technology, Danvers MA) at a 1:100 dilution after CC1 mild retrieval on a Benchmark Ultra instrument (Ventana, Tucson AZ).
Statistics
Data are presented as means ± standard deviation (SD). Statistical comparisons were performed using unpaired two-tailed Student's t tests, with a probability value of <0.05 taken to indicate significance.
Acknowledgements
We thank Terry Morgan, M.D. Ph.D., Oregon Health and Sciences University, for help with the statistical analysis of the archival RMS tumor samples. We thank Steven Verhulst, Ph.D., Southern Illinois University School of Medicine, for help with the statistical analysis of the primary RMS tumor samples. This work was supported by grant 159609 from the American Cancer Society, Illinois Division to J.D. and by grant RAR 060017A from the National Institute of Health, NIAMS division.
Footnotes
Conflict of Interest
The authors have no competing financial interests in relation to the work described.
References
- 1.Burgucu D, Guney K, Sahinturk D, Ozbudak IH, Ozel D, Ozbilim G, et al. Tbx3 represses PTEN and is over-expressed in head and neck squamous cell carcinoma. BMC cancer. 2012;12:481. doi: 10.1186/1471-2407-12-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ. Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation. Oncogene. 2002;21:3827–3835. doi: 10.1038/sj.onc.1205476. [DOI] [PubMed] [Google Scholar]
- 3.Castellino RC, Barwick BG, Schniederjan M, Buss MC, Becher O, Hambardzumyan D, et al. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PloS one. 2010;5:e10849. doi: 10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annual review of pathology. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung JH, Eng C. Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer research. 2005;65:8096–8100. doi: 10.1158/0008-5472.CAN-05-1888. [DOI] [PubMed] [Google Scholar]
- 6.Ciccarelli C, Marampon F, Scoglio A, Mauro A, Giacinti C, De Cesaris P, et al. p21WAF1 expression induced by MEK/ERK pathway activation or inhibition correlates with growth arrest, myogenic differentiation and onco-phenotype reversal in rhabdomyosarcoma cells. Molecular cancer. 2005;4:41. doi: 10.1186/1476-4598-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 8.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nature genetics. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 9.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nature genetics. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 10.Douglas NC, Papaioannou VE. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. Journal of mammary gland biology and neoplasia. 2013;18:143–147. doi: 10.1007/s10911-013-9282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer research. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- 12.Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. The EMBO journal. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Chang HY, Fei T, Wu H, Chen YG. GSK3 beta mediates suppression of cyclin D2 expression by tumor suppressor PTEN. Oncogene. 2007;26:2471–2482. doi: 10.1038/sj.onc.1210033. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nature genetics. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson G, Dahl C, Staaf J, Sandberg T, Bendahl PO, Ringner M, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26:4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 16.Leevers SJ, Vanhaesebroeck B, Waterfield MD. Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Current opinion in cell biology. 1999;11:219–225. doi: 10.1016/s0955-0674(99)80029-5. [DOI] [PubMed] [Google Scholar]
- 17.Li DM, Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li HG, Wang Q, Li HM, Kumar S, Parker C, Slevin M, et al. PAX3 and PAX3-FKHR promote rhabdomyosarcoma cell survival through downregulation of PTEN. Cancer letters. 2007;253:215–223. doi: 10.1016/j.canlet.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, et al. The Anti-proliferative Function of the TGF-beta1 Signaling Pathway Involves the Repression of the Oncogenic TBX2 by Its Homologue TBX3. The Journal of biological chemistry. 2014;289:35633–35643. doi: 10.1074/jbc.M114.596411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Cao J, Lv J, Dong L, Pier E, Xu GX, et al. TBX2 expression is regulated by PAX3 in the melanocyte lineage. Pigment cell & melanoma research. 2013;26:67–77. doi: 10.1111/pcmr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomnytska M, Dubrovska A, Hellman U, Volodko N, Souchelnytskyi S. Increased expression of cSHMT, Tbx3 and utrophin in plasma of ovarian and breast cancer patients. International journal of cancer. 2006;118:412–421. doi: 10.1002/ijc.21332. [DOI] [PubMed] [Google Scholar]
- 22.Londhe P, Davie JK. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet Muscle. 2011;1:14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Londhe P, Davie JK. Gamma Interferon Modulates Myogenesis through the Major Histocompatibility Complex Class II Transactivator, CIITA. Molecular and cellular biology. 2011;31:2854–2866. doi: 10.1128/MCB.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyng H, Brovig RS, Svendsrud DH, Holm R, Kaalhus O, Knutstad K, et al. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC genomics. 2006;7:268. doi: 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 26.Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, Karhu R, et al. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes, chromosomes & cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 27.Mandl A, Sarkes D, Carricaburu V, Jung V, Rameh L. Serum withdrawal-induced accumulation of phosphoinositide 3-kinase lipids in differentiating 3T3-L6 myoblasts: distinct roles for Ship2 and PTEN. Molecular and cellular biology. 2007;27:8098–8112. doi: 10.1128/MCB.00756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamura H, Yoshida K, Morimoto H, Haneji T. PTEN expression elicited by EGR-1 transcription factor in calyculin A-induced apoptotic cells. Journal of cellular biochemistry. 2005;94:117–125. doi: 10.1002/jcb.20283. [DOI] [PubMed] [Google Scholar]
- 29.Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, et al. The Highly Homologous T-Box Transcription Factors, TBX2 and TBX3, Have Distinct Roles in the Oncogenic Process. Genes & cancer. 2010;1:272–282. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peres J, Prince S. The T-box transcription factor, TBX3, is sufficient to promote melanoma formation and invasion. Molecular cancer. 2013;12:117. doi: 10.1186/1476-4598-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prince S, Carreira S, Vance KW, Abrahams A, Goding CR. Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer research. 2004;64:1669–1674. doi: 10.1158/0008-5472.can-03-3286. [DOI] [PubMed] [Google Scholar]
- 32.Radu A, Neubauer V, Akagi T, Hanafusa H, Georgescu MM. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Molecular and cellular biology. 2003;23:6139–6149. doi: 10.1128/MCB.23.17.6139-6149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redmond KL, Crawford NT, Farmer H, D'Costa ZC, O'Brien GJ, Buckley NE, et al. T-box 2 represses NDRG1 through an EGR1-dependent mechanism to drive the proliferation of breast cancer cells. Oncogene. 2010;29:3252–3262. doi: 10.1038/onc.2010.84. [DOI] [PubMed] [Google Scholar]
- 34.Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes & development. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renshaw J, Taylor KR, Bishop R, Valenti M, De Haven Brandon A, Gowan S, et al. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2013;19:5940–5951. doi: 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer research. 2008;68:7872–7881. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- 37.Ross AH, Gericke A. Phosphorylation keeps PTEN phosphatase closed for business. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1297–1298. doi: 10.1073/pnas.0812473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowley M, Grothey E, Couch FJ. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. Journal of mammary gland biology and neoplasia. 2004;9:109–118. doi: 10.1023/B:JOMG.0000037156.64331.3f. [DOI] [PubMed] [Google Scholar]
- 39.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York, NY. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 40.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer research. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 41.Shao J, Washington MK, Saxena R, Sheng H. Heterozygous disruption of the PTEN promotes intestinal neoplasia in APCmin/+ mouse: roles of osteopontin. Carcinogenesis. 2007;28:2476–2483. doi: 10.1093/carcin/bgm186. [DOI] [PubMed] [Google Scholar]
- 42.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 43.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer discovery. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinclair CS, Adem C, Naderi A, Soderberg CL, Johnson M, Wu K, et al. TBX2 is preferentially amplified in BRCA1- and BRCA2-related breast tumors. Cancer research. 2002;62:3587–3591. [PubMed] [Google Scholar]
- 45.Singh S, Vinson C, Gurley CM, Nolen GT, Beggs ML, Nagarajan R, et al. Impaired Wnt signaling in embryonal rhabdomyosarcoma cells from p53/c-fos double mutant mice. The American journal of pathology. 2010;177:2055–2066. doi: 10.2353/ajpath.2010.091195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toker A, Newton AC. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. The Journal of biological chemistry. 2000;275:8271–8274. doi: 10.1074/jbc.275.12.8271. [DOI] [PubMed] [Google Scholar]
- 48.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer research. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 49.Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nature cell biology. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 50.Wan X, Helman LJ. Levels of PTEN protein modulate Akt phosphorylation on serine 473, but not on threonine 308, in IGF-II-overexpressing rhabdomyosarcomas cells. Oncogene. 2003;22:8205–8211. doi: 10.1038/sj.onc.1206878. [DOI] [PubMed] [Google Scholar]
- 51.Weng LP, Brown JL, Eng C. PTEN coordinates G(1) arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Human molecular genetics. 2001;10:599–604. doi: 10.1093/hmg/10.6.599. [DOI] [PubMed] [Google Scholar]
- 52.Weng LP, Brown JL, Baker KM, Ostrowski MC, Eng C. PTEN blocks insulin-mediated ETS-2 phosphorylation through MAP kinase, independently of the phosphoinositide 3-kinase pathway. Human molecular genetics. 2002;11:1687–1696. doi: 10.1093/hmg/11.15.1687. [DOI] [PubMed] [Google Scholar]
- 53.Wijesekara N, Konrad D, Eweida M, Jefferies C, Liadis N, Giacca A, et al. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Molecular and cellular biology. 2005;25:1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu B, Zhang M, Byrum SD, Tackett AJ, Davie JK. TBX2 blocks myogenesis and promotes proliferation in rhabdomyosarcoma cells. International journal of cancer. 2014 doi: 10.1002/ijc.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu X, Kwon CH, Schlosshauer PW, Ellenson LH, Baker SJ. PTEN induces G(1) cell cycle arrest and decreases cyclin D3 levels in endometrial carcinoma cells. Cancer research. 2001;61:4569–4575. [PubMed] [Google Scholar]