Abstract
Diphtheria toxin translocation (T) domain undergoes conformational changes in acidic solution and associates with the lipid membranes, followed by refolding and transmembrane insertion of two nonpolar helices. This process is an essential step in delivery of the toxic catalytic domain of the diphtheria toxin to the infected cell, yet its molecular determinants are poorly characterized and understood. Therefore, an atomistic model of the T-domain–membrane interaction is needed to help characterize factors responsible for such association. In this work, we present atomistic model structures of T-domain membrane-bound conformations and investigate structural factors responsible for T-domain affinity with the lipid bilayer in acidic solution using all-atom molecular dynamics (MD) simulations. The initial models of the protein conformations and protein–membrane association that serve as starting points in the present work were developed using atomistic simulations of partial unfolding of the T-domain in acidic solution (Kurnikov, I. V.; et al. J. Mol. Biol. 2013, 425, 2752–2764), and coarse-grained simulations of the T-domain association with the membranes of various compositions (Flores-Canales, J. C.; et al. J. Membr. Biol. 2015, 248, 529–543). In this work we present atomistic level modeling of two distinct configurations of the T-domain in association with the anionic lipid bilayer. In microsecond-long MD simulations both conformations retain their compact structure and gradually penetrate deeper into the bilayer interface. One membrane-bound conformation is stabilized by the protein contacts with the lipid hydrophobic core. The second modeled conformation is initially inserted less deeply and forms multiple contacts with the lipid at the interface (headgroup) region. Such contacts are formed by the charged and hydrophilic groups of partially unfolded terminal helixes and loops. Neutralization of the acidic residues at the membrane interface allows for deeper insertion of the protein and reorientation of the protein at the membrane interface, which corroborates that acidic residue protonation as well as presence of the anionic lipids may play a role in the membrane association and further membrane insertion of the T-domain as implicated in experiments. All simulations reported in this work were performed using AMBER force-field on Anton supercomputer. To perform these reported simulations, we developed and carefully tested a force-field for the anionic 1-palmitoyl-2-oleoyl-phosphatidyl-glycerol (POPG) lipid, compatible with the Amber 99SB force-field and stable in microsecond-long MD simulations in isothermal–isobaric ensemble.
Graphical abstract
1. INTRODUCTION
Diphtheria toxin, a soluble protein at neutral pH, binds to a cell surface receptor and blocks protein synthesis of eukaryotic cells by delivering an enzymatic fragment (catalytic domain) into cytoplasm by utilizing the cell’s endocytic pathway.1 Endosomal acidification triggers conformational changes in the diphtheria toxin translocation (T) domain facilitating its membrane insertion and translocation of its N-terminus attached to the catalytic domain.1 The membrane insertion and function of the stand-alone T-domain have been extensively studied. Different experimental conditions have suggested a diverse set of membrane-bound and inserted states of the T-domain.2–12 A better understanding of the unassisted pH-dependent membrane insertion of the T-domain at the molecular level of resolution may facilitate development of improved drug delivery methods for therapeutic applications such as oncogenic treatment.13
A stand-alone T-domain peptide adopts a monomeric all α-helical structure in a neutral pH solution (see Figure 1A), consistent with the structure of the full toxin protein.14,15 The T-domain protein structure undergoes conformational rearrangements in response to solution acidification, which facilitates its subsequent membrane association followed by the membrane insertion, possibly, of a hydrophobic hairpin (helices TH8–9).6–9 The latter is followed by rearrangement of the remaining helices and the translocation of the catalytic domain into a cytoplasm.16–18 In a series of experiments, free energy calculations, and atomistic molecular dynamics (MD) simulations of a T-domain in solution, we have shown that the protein structure unfolds partially upon protonation of histidines19,20 (see Figure 1B); in addition, previous work showed the role of histidines in the destabilization of the T-domain in solution.21 This partially unfolded state of the T-domain termed a membrane-competent state is characterized by refolded N-terminal helices and increased solvent exposure of some hydrophobic and charged residues.19 Early work showed pH-dependent membrane interactions of the T-domain, which suggested that protonation of acidic residues (at pH 4) facilitates interfacial interactions of the N-terminal helices.10
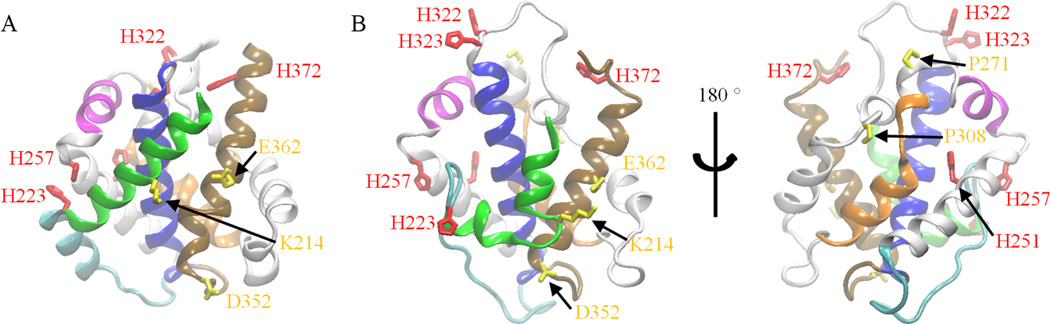
Figure 1.
(A) X-ray structure [PDB ID code 1F0L]. (B) Atomistic model of the partially unfolded T-domain in two different views. The model was obtained from the last MD snapshot of a 6.8 µs MD simulation of the T-domain with all histidines protonated. All histidines are represented in red licorice. Residues K214, P271, P308, D352, and E362 are shown in yellow licorice. Helices TH1, TH2, TH4, TH5, TH8, and TH9 are represented by green, cyan, magenta, orange, blue, and brown ribbons. Other helices are shown in gray ribbons.
A recent kinetics study of the T-domain insertion into anionic lipid bilayers aimed at distinguishing formation of a membrane-competent state and an insertion-competent state in mildly acidic solutions.6 It was shown by experiments that the pH-dependent formation of the insertion-competent state is affected by the increase of anionic lipid content. Namely, in the presence of lipid bilayers with higher ratio of the anionic lipids there was an increase in an apparent transition pKa value from the membrane-bound to the inserted state of the protein. It was also determined that the rate of transmembrane insertion of the T-domain is accelerated in solution with lower pH.6 Both these results suggest that protonation of acidic residues occurs in the membrane interface. Furthermore, this kinetics analysis suggested that the observed coexistence of a water-soluble and a transmembrane inserted state of the T-domain is due to the shift of pKa and, thus, protonation of acidic residues at the membrane interface. No experimentally determined structural information exists about multiple postulated conformational states of the T-domain at the membrane interfaces.
To gain insight into possible mechanisms and structures of the T-domain–protein association we have recently performed extensive coarse-grained MD simulations of the initial stages of the membrane association of the T-domain.22 Using a coarse-grained model allowed us to perform free unrestrained simulations of the T-domain association with membranes and to demonstrate that presence of anionic lipid in the membrane increased the lifetime of the protein–membrane complexes. Two distinctive membrane-bound conformations were identified in extensive equilibrium unrestrained simulations of the protein with the membrane, and characterized using umbrella sampling (US) simulations.22 Both predicted membrane-bound conformations showed helices TH8–TH9 with a near parallel orientation relative to the membrane plane, which was in agreement with low resolution information reported by solid state nuclear magnetic resonance (NMR).11 However, only limited characterization of the preferred modes of the protein binding with the lipid is possible using the coarse-grained approaches. Among numerous limitations of the coarse-grained models are their inability to maintain the protein in a specific conformation and the lack of individual pairwise interaction between atoms.
In this work, we employ extensive all-atom molecular dynamics simulations of the T-domain in association with the anionic lipid bilayer (POPG) to characterize in detail the proposed structure of the initial complexes of the T-domain with the membrane, and to explore possible structural implications of the acidic residue protonation at the membrane interface. Our microsecond-long simulations show that membrane-bound conformations proposed using the coarse-grained approach mainly retained the structure of the protein at the bilayer–water interface, but both conformations inserted deeply in the membrane interface. The simulations also allow us to suggest which protein regions interact most likely with the lipid headgroups or the acyl-chains. Protonation of the acidic residues facilitates reorientation and deeper insertion of a membrane-bound conformation of the protein into the membrane.
This work utilizes all-atom molecular dynamics simulations using AMBER FF99SB force-field for protein23 and a lipid force-field developed for POPG lipid in this work. Our force-field is compatible with AMBER and retains an improved structure in unrestrained constant pressure and temperature simulations compared to results from previous models (GaffLipid, Lipid11).24,25 It does not require specialized ensemble simulations unavailable in the Anton supercomputer. All microsecond-long simulations reported in this paper were performed using Anton supercomputer. The development and testing of the POPG force-field are also described in the Methods section and in the Supporting Information to greater extent.
2. METHODS
2.1. Lipid Force-Field Development
Atomistic MD simulations of proteins using Amber FF99SB force-field have resulted in good agreement with the NMR experimental data, folding of small α-helical or β-sheet proteins, and limited agreement with temperature-dependent helix–coil transitions.26–30 Moreover, we have recently reported the formation of a membrane-competent state in solution of the T-domain using FF99SB, which has features in good agreement with the fluorescence experiments.12,14 However, development of the force-field parameters for anionic lipids compatible with both tensionless MD simulations and Amber force-fields is still an ongoing process.23,24 To study the T-domain association to lipid bilayers composed of the anionic 1-palmitoyl-2-oleoyl-phosphatidyl-glycerol (POPG), POPG force-field parameters are described below. A schematic representation of POPG is shown in Figure S1. Recently, two related lipids force-fields have been reported, which are based on the general Amber force-field (GAFF).31 One of them, Lipid11, provides a systematic charge derivation methodology compatible with the Amber RESP charge fitting philosophy.25,32 However, to successfully reproduce essential properties and structure of the lipid bilayer Lipid11 requires an external surface tension NPT ensemble. To circumvent the use of the external surface tension NPT ensemble, which is not implemented in Anton supercomputer and, more generally, is not a desirable ensemble in biomolecular simulations, GAFF parameters of the acylchains have been reparameterized by fitting the torsion and Lennard-Jones parameters to reproduce properties of the pentadecane liquid (GaffLipid).24 Thus, GaffLipid successfully reproduced a range of properties including the area per lipid of the 1-palmitoyl-2-oleoyl-phosphatidyl-choline (POPC) lipid bilayers.24 However, there is no model of the POPG lipid for this recent force-field. An attempt to reuse the majority of the GaffLipid parameters to model POPG lipids was unsuccessful. Namely, the average area per lipid is around 55 Å2 while the experimental value is 67.3 Å2 for the POPG with modified GaffLipid at T = 310 K (see Figure S2).33 In order to alleviate the problem, we used Lennard-Jones parameters of headgroup atoms developed for POPG CHARMM36 force-field.34 Partial charges of the lipid headgroup atoms were fitted using the multiconformer RESP method,35 which used 19 different lipids conformations. All lipid conformations were optimized in gas phase using Hartree–Fock self-consistent field (HF-SCF) with a basis set HF/6-31G* and Gaussian03.36 The force-field modifications are shown in Table S1 (see Supporting Information). Atomic charges for the lipids tails were the same as described in Lipid11 (neutral residues palmitoyl and oleoyl). The capping strategy employed in LIPID11 was used to derive the partial charges of the headgroup atoms with the constrained charges of the capping groups (the total charge of the headgroup was −1). The test system consisted of 72 POPG lipids solvated by approximately 3600 TIP3P water molecules,37 and sodium counterions were added to neutralize the system.38
The MD simulations using the thus developed force-field yielded bilayers with the area per lipid around 60 Å2, which is an improvement relative to that obtained using GaffLipid parameters (see Figure S2). The order parameters were computed as SCD = 〈3 cos2 θ − 1〉/2, where θ is the angle between the vector joining the lipid tails C–H and the membrane normal. Large values of SCD represent ordered lipid tails, and smaller ones represent less ordered lipid tails. Lipid tails are slightly more ordered at a lower temperature (T = 310 K) than at T = 323 K (see Figure S3). Figure S4 shows the X-ray and neutron scattering form factors for MD simulations and experiments at temperatures T = 303 and 323 K. The estimated disagreement parameter χ between simulations and experiments of the X-ray scattering plots are significant for the abovementioned temperatures (see Figure S4).39 This indicates the need for further parametrization of POPG lipids, such as optimization of the dihedral angles of the headgroup and phosphate group atoms.
The density profile of pure POPG bilayers at two different temperatures (see Figure S5) shows that sodium counterions reside mostly in the phosphate group region, which is in agreement with previous MD simulations.40,41 However, to neutralize the protein-bilayer system (see details below) over 192 Na+ ions were added, which results in a concentration three times (ca. 450 mM) the physiological ion concentration. Recent work by Vila-Vicosa et al. reported on the negative effects of counterions added to neutralize the system on the structure of charged lipid bilayers.42 Neutralizing counterions located in the negatively charged headgroups induce an ordered state of the charged lipid bilayer. Future modeling of T-domain membrane binding could benefit from improved models of anionic lipid bilayers and ionic strength.
2.2. Coarse-Grained Model to the All-Atom Representation Conversion
A previous study of the membrane association of the low pH T-domain model predicted two membrane-bound conformations denominated B1 and B2, which were determined by unrestrained MD and umbrella sampling simulations of coarse-grained models of the protein and lipid bilayers.22 In this study, a representative snapshot from each bound conformation was extracted from an umbrella window simulation corresponding to the lowest free energy (see22). Each coarse-grained model of the T-domain bound to a bilayer was converted into an all-atom model using the conversion protocol of Cojocaru et al.43 The original number of phospholipids in the coarse-grained model was 512, which was reduced to 192 lipids in the atomistic model. Also, the coarse-grained model contained a mixture of POPC:POPG with a molar ratio of 1:3, which was changed to only POPG lipids in our atomistic models. The coarse-grained model of the protein was converted to all-atom by aligning the last frame of an MD simulation of a low pH T-domain model.19 The latter model was generated by a 6.8 µs MD simulation of the protein in explicit solvent with all histidines protonated and all other acidic side-chains in their standard state (see Figure 1B).19 It was shown that protonation of histidines triggers partial unfolding of the protein N-terminal in explicit solvent, which was in good agreement with experiments.19
2.3. System Preparation and Equilibration Protocols
Hydrogen atoms were added to the initial low pH T-domain structures (residues 201–380) using tLeap available in AMBER12 package.44 All histidines were protonated, and all acidic side-chains were set to their standard protonation states (unless otherwise noted). Note that this serial multiscale approach models the initial membrane binding of the protein. The simulation boxes of the membrane-bound states of the low pH T-domain model were created by adding approximately 14800 TIP3P explicit water molecules such that the distance between the protein/bilayer and the simulation box edge was 12.0 Å. The total number of atoms is approximately 72 000 atoms. The size of the simulation boxes were 120 Å × 121 Å × 116 Å and 123 Å × 112 Å × 122 Å for the membrane-bound conformations B1 and B2, respectively. Sodium counterions were added to neutralize the protein/membrane charge. Equilibration simulations were performed using PMEMD on GPU cards with the FF99SB-ILDN force-field.45–48 The simulation time step was 2 fs, and all hydrogen bonds were constrained via SHAKE.49 Periodic boundary conditions were set up, cutoff radius was set to 10 Å, and electrostatic calculations were performed using Particle Mesh Ewald (PME) method.50 Each system was equilibrated as follows. The protein and lipid heavy atoms were restrained, and the solvent was minimized for 250 steps of the steepest descent minimization method followed by 750 steps of conjugate gradient descent. Then, the solvent was restrained, and the protein was minimized by a total number of 5000 steps with the first 2500 steps of steepest descent. Each system was slowly heated in five stages of 5 ps from T = 0.1 K up to T = 310 K with NVT ensemble using Langevin thermostat. On each stage restraints were applied as follows: (a) Harmonic restraints with a force constant of 10 kcal mol−1 Å−2 were applied to protein and lipid atoms. (b) Harmonic restraints with a force constant of 5 kcal mol−1 Å−2 were applied to protein and lipid atoms. (c) Harmonic restraints with a force constant of 2.5 kcal mol−1 Å−2 were applied to protein atoms. Harmonic restraints with a force constant of 1.0 kcal mol−1 Å−2 were applied to protein atoms over the last two heating stages. Anisotropic NPT ensemble equilibrations using Berendsen barostat at 1 atm were carried out in three stages. In the first stage protein atoms were restrained with a force constant of 0.5 kcal mol−1 Å2 over 250 ps; the following stage has restraints on backbone atoms with a force constant of 0.25 kcal mol−1 Å2 over 250 ps, and all restraints were removed over the last stage of 170 ps. Then, each system was equilibrated over 200 ns using the NPT ensemble with semi-isotropic scaling available in GPU accelerated PMEMD.
2.4. Production Protocols on Specialized Computer Anton
The final structures of equilibration MD simulations were used for production simulations. The force-field parameters used for the equilibration simulations were also used for MD simulations on the MD specialized hardware Anton.51 In Anton simulations, the Gaussian Split Ewald method52 with a grid size of 64 × 64 × 64 was used with a cutoff radius of 12.36 Å for the van der Waals and short-range electrostatic interactions. A reversible multiple-time step algorithm was used with a time step of 2 fs for bonded and short-range nonbonded interactions and 6 fs for the long-range nonbonded forces.53 All bonded hydrogens were constrained with M-SHAKE algorithm.54 X and Y dimensions were coupled, and Z was free to change in the semiisotropic NPT conditions applied to the system. The following multigrator approach was used: a Noose–Hoover thermostat with an interval of 24 steps and a Martyna–Tobias–Klein barostat with an interval of 240 steps.55,56 The temperature was set to 310 K and pressure to 1.0 atm. MD simulations of conformation B1 and B2 were carried out over 3541 and 810 ns, respectively. An extended MD simulation of conformation B1 at T = 323 K was carried out over 2124 ns. A second extended MD simulation of conformation B1 at T = 323 K and with all acidic side-chains neutralized (residues Glu and Asp) was carried out over 3459 ns.
2.5. Analysis
Trajectory analysis was carried out with AmberTools1344 and in-house tcl scripts using VMD 1.9.1.57 The latter was also used for molecular visualization and preparation of figures. Protein and lipid heavy atoms were considered to be in contact if the distance was lower than 5 Å. Cα RMSD per residue were calculated after aligning the protein MD generated structures to the X-ray structure using Cα atoms of residues 206–375. Secondary structure content was calculated using the DSSP program available in AmberTools.58 Helicity profiles versus time were smoothed using an average window of 12 ns. Scattering form factors were calculated using the software SIMtoEXP.39 Number density profiles were calculated using VMD, and the experimental data was retrieved from Pan et al.33
3. RESULTS
3.1. Simulated Models
Multiple coarse-grained MD simulations of the T-domain and lipid bilayers of different compositions have suggested two distinctive membrane-associated conformations (denominated B1 and B2) of the low pH T-domain model.22 Conformation B1 was frequently observed during coarse-grained simulations, while B2 was observed in a single simulation and has the lower free energy of binding.22 In this work, we refine representative snapshots of these two membrane-bound conformations by using atomistic MD simulations. Conformation B1 is extensively studied because of its structural changes observed on a microsecond time-scale and its predicted high occurrence by coarse-grained simulations. To model the lipid bilayer, a pure anionic bilayer (POPG) is used because negatively charged lipids promote faster membrane binding in our coarse-grained MD simulations and facilitate faster transmembrane insertion of T-domain in experiments.6,22
3.2. MD Simulations
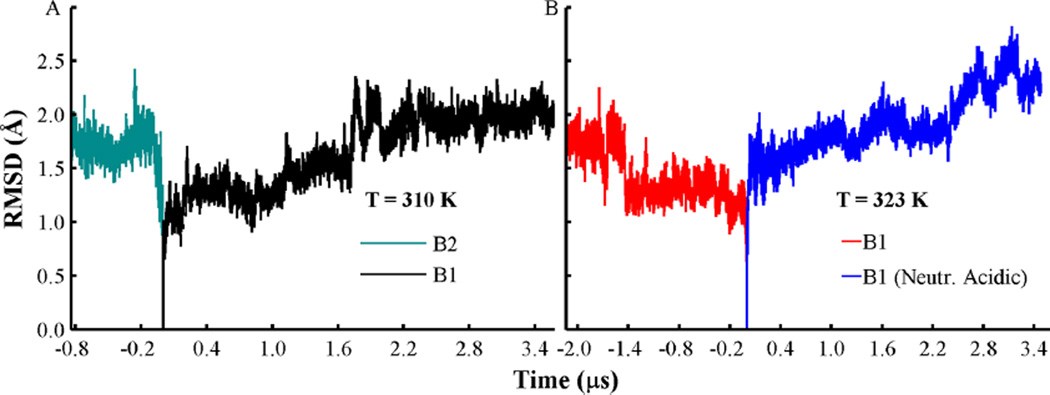
Coarse-grained conformations B1 and B2 were converted to atomistic representation followed by equilibration simulations (described in the Methods section). Production simulations are performed over 3.5 and 0.8 µs for each orientation B1 and B2 at T = 310 K, respectively. Structural rearrangements of the T-domain in our atomistic simulations are roughly estimated by the root-mean-square deviation (RMSD) of Cα atoms relative to their initial position in helices TH1–9, TH5′ (see Figure 2). This is calculated after translation and overlay of each MD frame relative to the initial protein structure. RMSD gradually increases to an average value of 2.0 Å over the initial 2.0 µs of the MD trajectory of conformation B1 (see Figure 2A), while conformation B2 shows an abrupt change of RMSD to 1.7 Å after the first 100 ns (see Figure 2A). In order to observe possible refolding of the less deeply inserted state B1, we extended the simulation of this conformation at a slightly higher temperature of T = 323 K. The temperature was increased to accelerate the sampling of protein conformational changes at membrane interfaces. The extended MD simulation of conformation B1 shows a slight increment of RMSD to 1.7 Å over the last 500 ns (see Figure 2B). Partial unfolding of the solvent exposed helix TH5 is observed over this segment. To explore the possible role of neutralization of acidic side-chains in the protein conformational changes, we retrieve an MD frame from the simulation of conformation B1 at T = 310 K at simulation time ca. 1.5 µs. All acidic side-chains are neutralized, while histidines are kept in their protonated state. Simulation of the neutralized conformation B1 results in an increase of RMSD to 2.4 Å over the last 500 ns. Helix TH5 is partially unfolded over this segment (see Figure 2B). These MD simulations show that the protein retains its compact structure, while it remains bound to the bilayer interface.
Figure 2.
Changes of the Cα root mean squared deviation (RMSD) from helices TH1–9, with TH5′ relative to the initial protein coordinates. (A) RMSD traces obtained from orientation B1 (black line) and orientation B2 (cyan). Simulations were performed at T = 310 K. (B) RMSD curves obtained from orientation B1 (red) and with acidic side-chains neutralized (blue lines). Simulations were performed at T = 323 K. All histidines are set in their protonated state in all simulations, and other acidic residues are set in their standard state, unless indicated.
To characterize structural changes observed in our simulations, we calculated the RMSD per residue and changes in the α-helical content of each helix. At T = 310 K, conformation B2 has significant displacement of the solvent exposed helices TH1–2, while B1 shows displacement of the Cα atoms in helices TH2, TH5′, the interhelical loops TH7–8, TH8–9, and the C-terminal of TH9 (see Figure S6). MD simulation of B1 at T = 323 K resulted in unfolding of helix TH4, which was not observed in other simulations. Extended MD simulation of B1 (T = 323 K) with neutralized acidic side-chains produced a similar RMSD profile than B1 at T = 310 K. Analysis of secondary structure content shows that helices TH2, TH5′, and TH7 are unfolded, while helices TH1, TH4, TH5, and TH9 are partially unfolded (see Figure S7). Among them, helix TH4 unfolds completely in the MD simulation of B1 with only histidines protonated at T = 323 K.
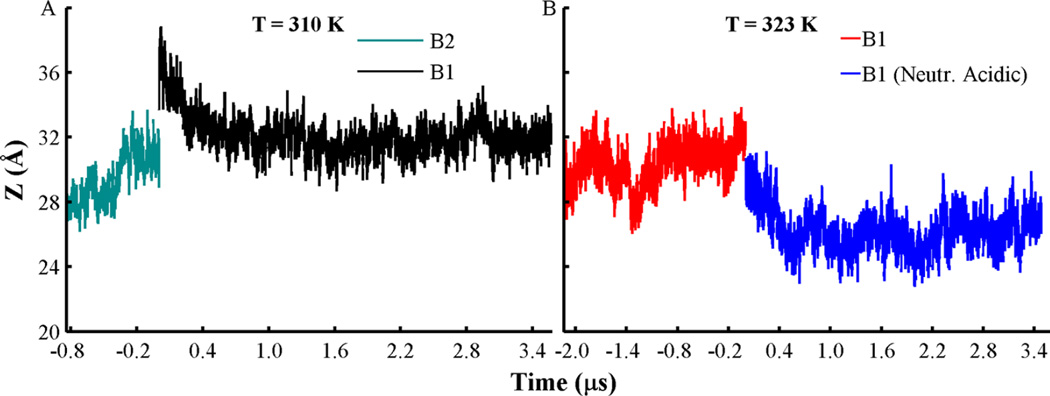
To monitor the degree of insertion of the T-domain in the membrane interface, we calculated the center of mass (COM) distance (Z) between the protein and the bilayer along the bilayer normal axis as a function of time (see Figure 3). For example, Figure 3A shows that the membrane-bound conformation B2 penetrates deeper in the bilayer interface, which is initially placed at ca. Z = 32 Å and then rapidly moves to a position near Z = 28 Å. In contrast, membrane-bound conformation B1 is initially positioned at ca. Z = 37 Å and gradually adopts a stable inserted state around Z = 32 Å over the last 1.0 µs of simulation time. At a higher temperature (T = 323 K), conformation B1 penetrates the membrane interface slightly deeper to an average Z = 30 Å (see Figure 3B). Further neutralization of acidic side-chains induces deeper insertion of B1 to an average Z = 26 Å.
Figure 3.
Changes of the center of mass (COM) distance (Z) between the low pH T-domain models and lipid bilayers along the bilayer normal axis. (A) COM distances obtained from orientation B1 (black line) and orientation B2 (cyan). Simulations were performed at T = 310 K. (B) COM distances obtained from orientation B1 (red) and acidic side-chains neutralized (blue lines). Simulations were performed at T = 323 K. All histidines are set in their protonated state in all simulations, and other acidic residues are set in their standard state, unless indicated.
3.3. Equilibrated Conformations B1 and B2
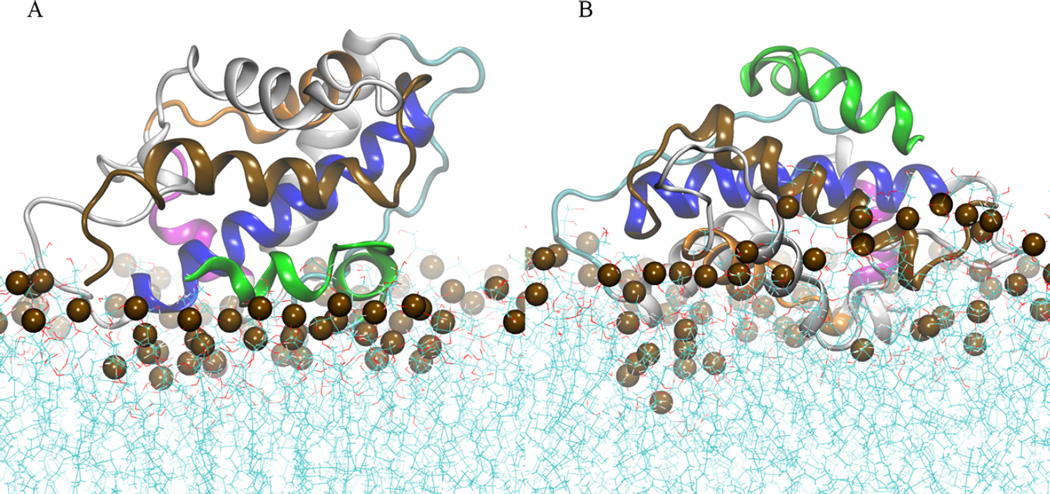
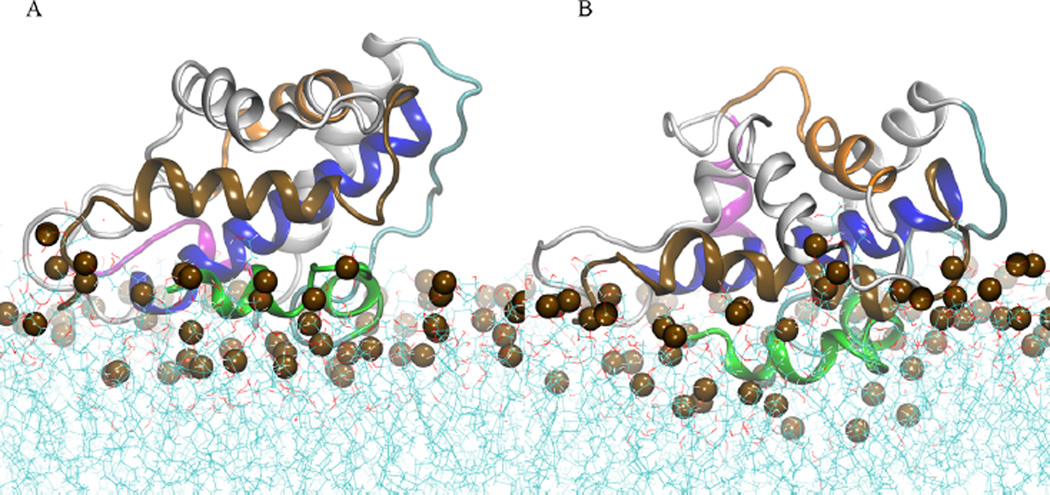
Figure 4A shows that the conformation B1 has protein–membrane contacts localized at residues in helices TH1–2, the loop TH3–TH4, the N-terminus of TH8, and C-terminus of TH9. In contrast, conformation B2 forms membrane contacts on residues of the loop TH2–TH3, TH3, TH5–6, the N-terminus of TH8, and C-terminus of TH9 (see Figure 4B). The latter orientation penetrates deeper in the bilayer–water interface than orientation B1 at simulation temperature T = 310 K. Increase of temperature in the extended simulations and neutralization of acidic side-chain facilitates deeper insertion of conformation B1 in the membrane interface, as shown in Figure 5. This shows the last MD frames of simulations of conformation B1 with different protonation states of acidic side-chains. Figure 5B shows that neutralization of acidic side-chains induced protein reorientation of conformation B1, which was not observed in the simulation with acidic residues in their standard state (see Figure 5A). As a result, the refolded helix TH1 penetrates the phosphate and the lipid acyl regions. The protein reorientation is accompanied by the formation of protein–membrane contacts in loops TH5′–TH6 and TH8–TH9.
Figure 4.
Final MD frames of atomistic simulations of low pH T-domain with fixed protonation state for histidines and standard state for the acidic side-chains: (A) orientation B1 and (B) orientation B2. Helices TH1, TH2, TH4, TH5, TH8, and TH9 are shown in green, cyan, magenta, orange, blue, and brown ribbon representation, respectively. Other helices are shown in gray ribbons. Phosphorus atoms are shown in dark brown space filled representation. Water molecules are not shown.
Figure 5.
Final MD frames of atomistic simulations of membrane-bound conformation B1 with fixed protonated state for all histidines. (A) Low pH T-domain structure with acidic residues set in their standard protonation state. (B) Low pH T-domain structure with acidic residues set in their neutralized state. Helices TH1, TH2, TH4, TH5, TH8, and TH9 are shown in green, cyan, magenta, orange, blue, and brown ribbon representation, respectively. Other helices are shown in gray ribbons. Phosphorus atoms are shown in dark brown space filled representation. Water molecules are not shown.
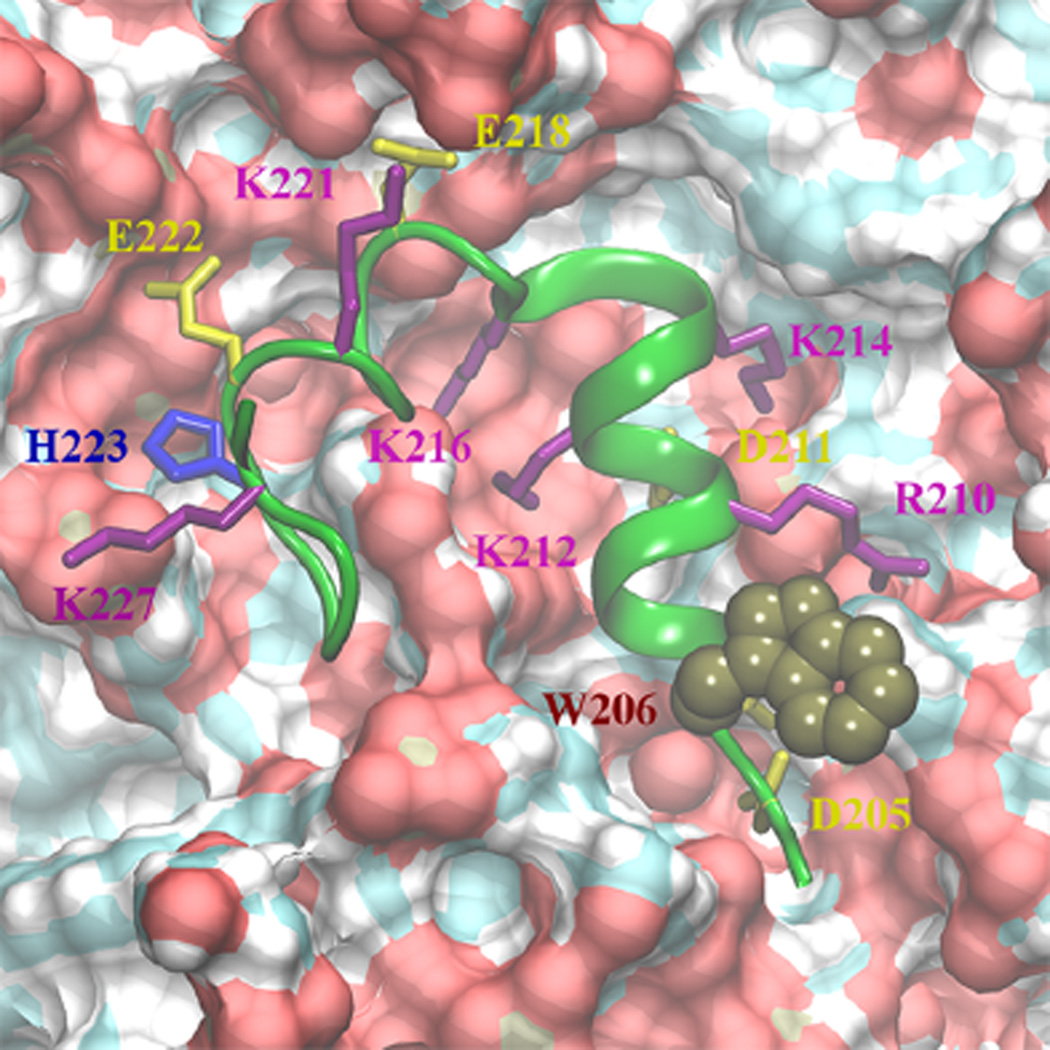
To gain structural insight on the membrane-bound state of TH1 with neutralized acidic side-chains, Figure 6 displays the top view of TH1 in contact with the membrane interface. After membrane binding and neutralization of acidic side-chains in conformation B1, helix TH1 exhibits two different patterns of membrane–protein interactions. At the N-terminus, neutralized side-chains of residues D205, D207, and D211 are buried by TH1 and lateral side-chains of R210, K212, K214, and K216 form contacts with the polar headgroups and acyl-chains. In the C-terminus, cationic side-chains are not facing the membrane interface, while neutralized glutamates and H223 interact with the lipid headgroups. Helix TH1 remains inserted in the membrane as shown by COM distance plots (Figure 3B). MD simulations of conformation B1 suggest that acidic neutralization allows for deeper insertion of the refolded helix TH1 at the membrane interface and that cationic residues are stabilized by the negatively charged headgroups.
Figure 6.
Top view of helix TH1 obtained from the last MD frame of B1 with all acidic residues in neutral state (residues 205–228 in green ribbon representation). The surface of the membrane is shown in red, cyan, and white corresponding to oxygen, carbon, and hydrogen atoms. Cationic residues R210, K212, K214, K216, K221, and K227 are shown in magenta licorice representation. H223 is in blue, and residues D205, D207 (not labeled), D211, E218, and E222 are in yellow licorice representation. W206 is shown in brown space filling representation.
3.4. Protein Reorientation in the Membrane Interface
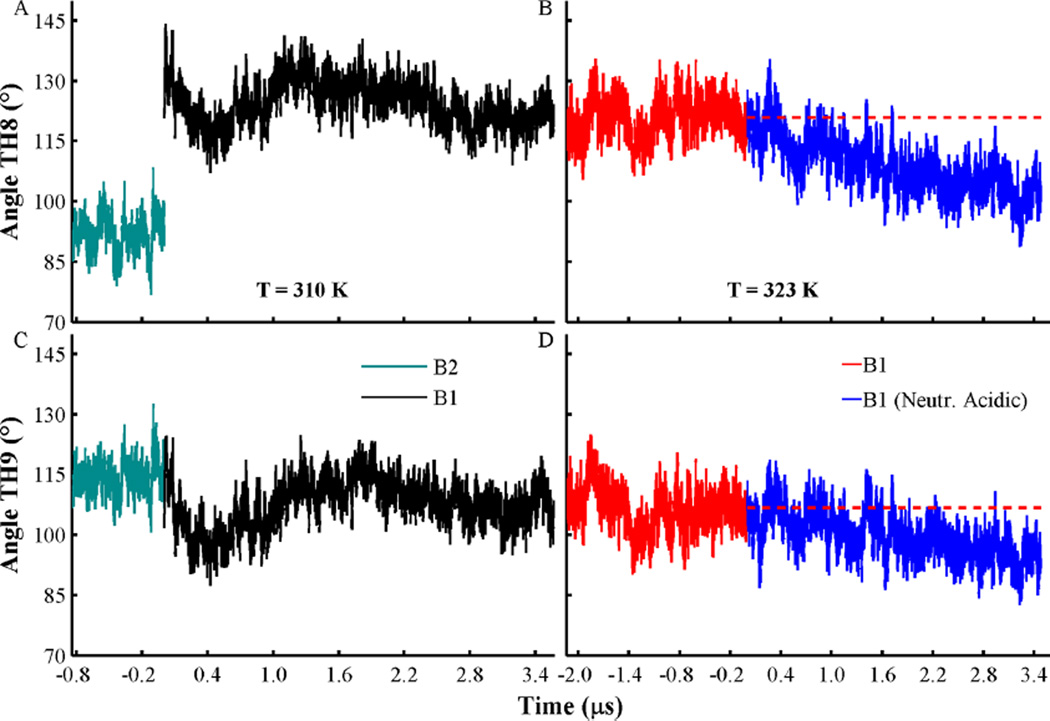
To determine changes in the overall orientation of the protein, we find it useful to compute the orientation of hydrophobic helices TH8 and TH9 relative to the membrane normal axis (see Figure 7). Figure 7A,C shows that both membrane-bound conformations B1 and B2 retain their overall orientation, in which helices TH8–TH9 exhibit oblique orientation relative to the membrane plane. MD simulation of conformation B1 at a slightly higher temperature of T = 323 K displays similar oblique conformations of helices TH8–TH9; however, neutralization of acidic side-chains drives changes of the orientation of helices TH8–TH9. These changes result in a near parallel orientation of these helices relative to the membrane plane (see Figure 7B,D).
Figure 7.
Changes of the angles formed by the axes of helices TH8 (A, B) and TH9 (C, D) relative to the membrane normal axis. Angle traces obtained from membrane-bound conformations B1 and B2, at T = 310 K, are shown in cyan and black, respectively. Simulations at T = 323 K of membrane-bound conformation B1 with acidic side-chains in their standard and neutralized state are shown in red and blue, respectively. The average values for helices TH8 (121°) and TH9 (107°) obtained from B1 are shown in red broken lines. A value of 90° corresponds to a parallel orientation of a helix relative to the membrane plane. All histidines are set in their protonated state in all simulations, and other acidic residues are set in their standard state, unless indicated.
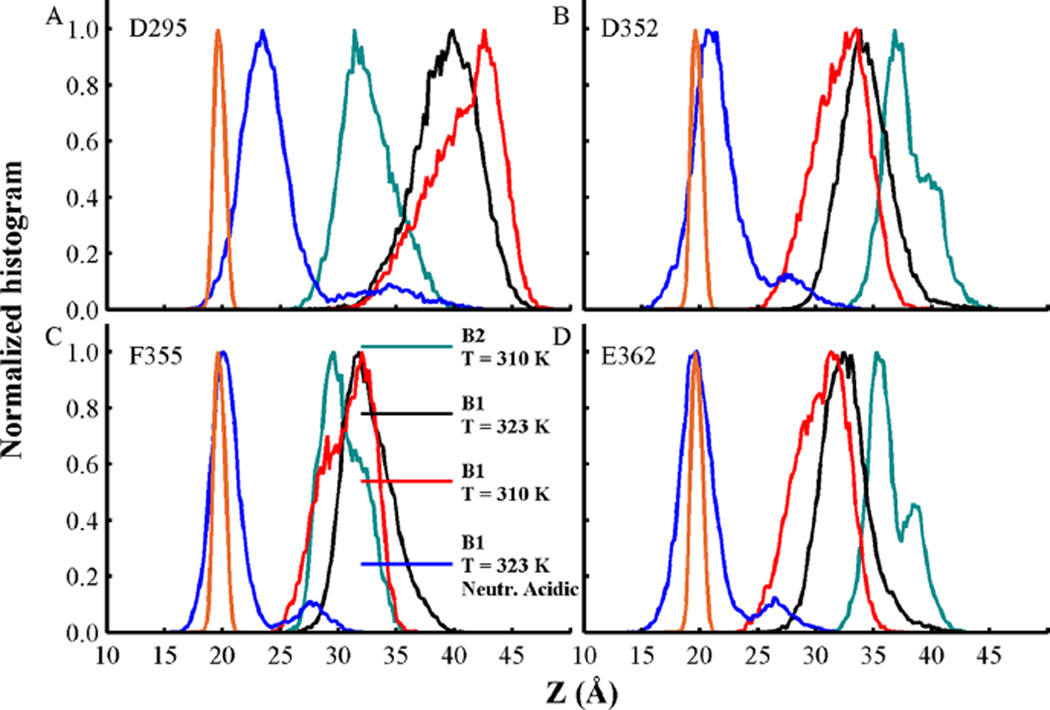
To analyze specific differences of the membrane-bound conformations of the T-domain in the simulations, the normalized histograms of protein and membrane interface contacts are calculated. Figure S8 shows the patterns of protein–membrane contacts obtained from the last 500 ns of all MD simulations. In general, conformations B1 and B2 have different patterns of protein–membrane interactions (see Figure S8A,B). Conformation B1 forms long-lived protein–interface contacts in residues of helices TH12 and in the loop TH3–TH4, while conformation B2 forms stable contacts at residues in the loop TH2–TH3, helix TH3, along TH4–6, and loop TH5′–TH6. Notice that both conformations show similar contacts in the N-terminus of TH8 and the C-terminus in helix TH9. An extended MD simulation of B1 (at T = 323 K) shows that the protein retains most of the protein–membrane contacts (see Figure S8C). A second extended MD simulation of B1 with neutralized acidic side-chains results in the formation of new protein–interface contacts in residues located in the unfolded helix TH5′, loop TH5′–TH6, loop TH8– TH9, and helix TH9 (see Figure S8D). This reorganization of protein–interface of contacts can be depicted by the increased bilayer insertion of residues D295, N296 located in the loop between helices TH5′–TH6, as shown in Figure 8A. Furthermore, residues D352, F355 located in the loop between TH8–TH9 and E362 in TH9 penetrate into the region of lipid acyl-chains (see Figure 8B–D).
Figure 8.
Normalized density of insertion depth of Cα atoms from residues (A) D295, (B) D352, (C) F355, and (D) E362. The distance Z is measured from the bilayer center located at Z = 0 along the bilayer normal axis. Histograms from membrane-bound conformations B1 and B2 at T = 310 K are shown in black and cyan, respectively. Simulations at T = 323 K of membrane-bound conformation B1 with acidic side-chains in their standard and neutralized state are shown in red and blue, respectively. Phosphate group density computed over all MD simulations is shown as a reference in orange lines. Data was obtained from the last 500 ns of each simulation.
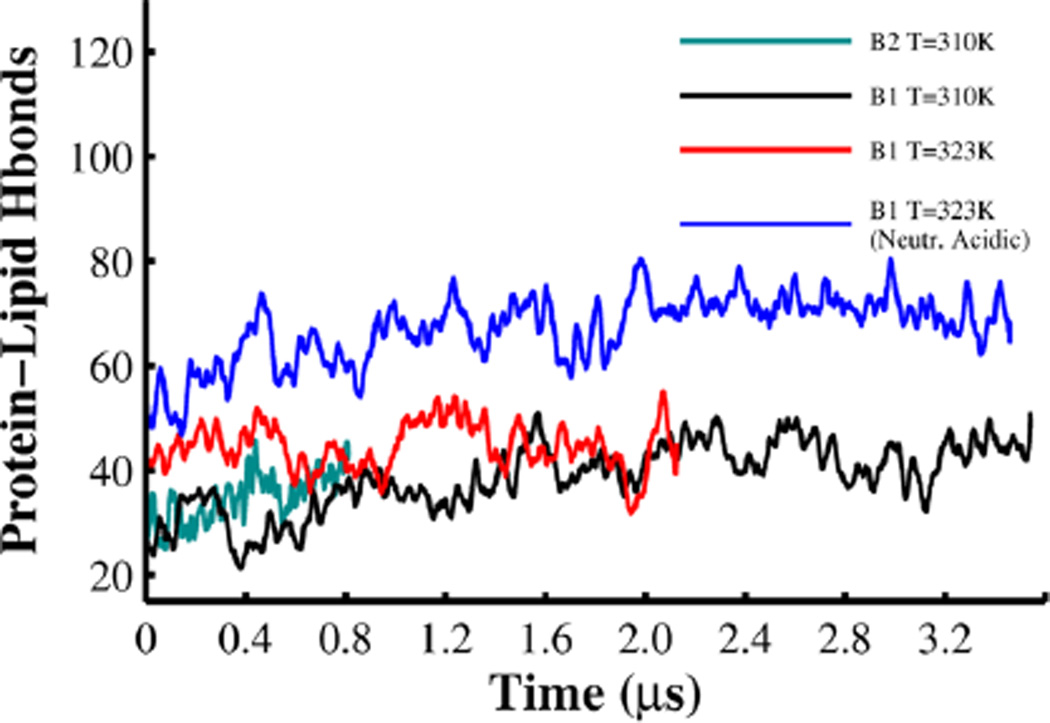
The change in the orientation of the protein and further insertion in the membrane interface of B1 also resulted in the increase of the number of hydrogen bonds between the protein and lipid headgroups (see Figure 9). This figure shows that neutralization of acidic side-chains in B1 allows for a larger average number of protein–lipid hydrogen bonds (70 over the last 500 ns). MD simulations of B1 at T = 310 K and T = 323 K with standard state of protonation of acidic side-chains resulting in a similar average number of hydrogen bonds (43 over the last 500 ns). Conformation B2 has the lowest average number of hydrogen bonds (40 over the last 500 ns). Changes in the orientation and deeper insertion of B1 with neutralized acidic side-chains are stabilized by the formation of an additional 27 hydrogen bonds between the protein and the lipids.
Figure 9.
Total number of protein–lipids hydrogen bonds (Hbonds) obtained from B1 and B2 at T = 310 K in black and cyan lines. B1 with acidic residues in standard and neutral state at T = 323 K in red and blue line, respectively. The curves were smoothed by a moving window of 24 ns.
Further inspection of the deeply inserted state of B1 reveals the formation of long-lived hydrogen bonds of residues located in the N-terminus of TH1 and helix TH9 (see Table S3). For example side-chains of W206, E362, Q369, and H372 form hydrogen bonds with lipids not observed in all other simulations. Notice that neutralization of side-chains in B1 results in the disruption of protein–lipid hydrogen bonds in helix TH1, which are stable in the other two simulations of B1 (see Table S3). Conformation B2 has fewer long-lived hydrogen bonds than those observed in B1.
3.5. T-Domain Insertion in the Bilayer Interface
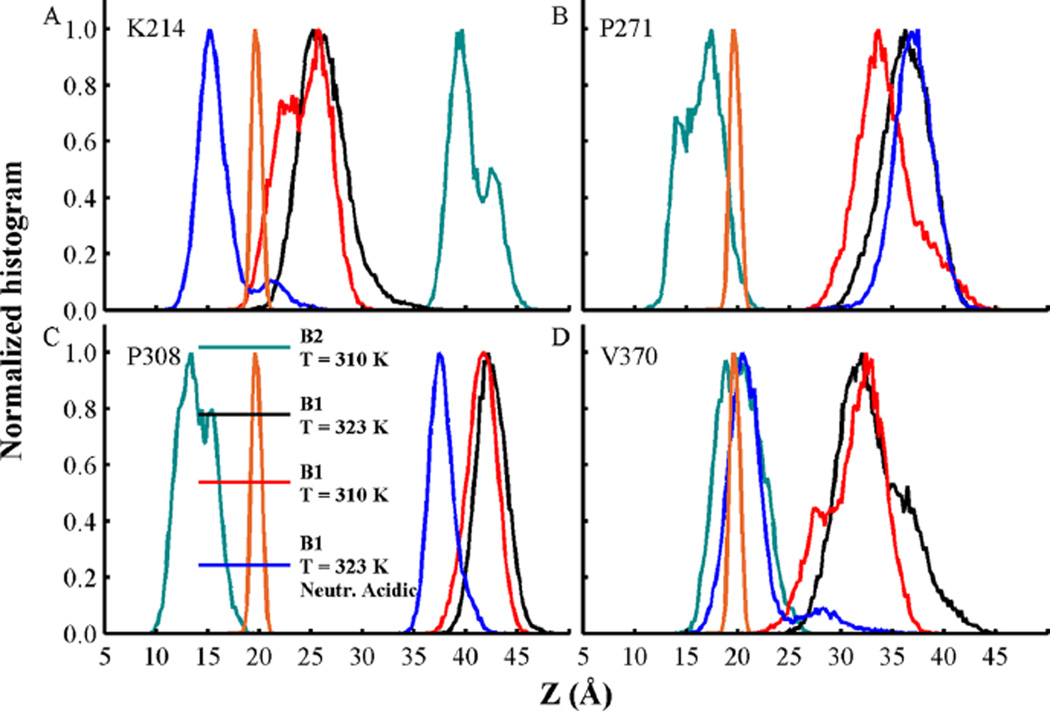
Conformations B1 and B2 display different extents of contacts with lipid acyl-chains, which are shown in histograms obtained from the last 500 ns of all simulations. Note that contact patterns are a rough characterization of protein–membrane interactions. Figure S9A shows that conformation B2 forms stable contacts at residues in the loop TH4–TH5 (residue P271), helix TH6 (residues E298, A302), loop TH6–TH7 (residues S305, I306, P308), and TH9 (residues N366, L367, V370, V371). In contrast, conformation B1 forms few contacts with the lipid acyl-chains (see Figures S9B,C). However, neutralization of acidic side-chains in conformation B1 promotes larger number of contacts between the protein and the bilayer hydrophobic core (see Figure S9D). The positively charged helix TH1 (+5 e−) forms long-lived protein–acyl-chain contacts (residues K214, T215) as well as residues in the loop between TH8–TH9 (residues D352, G354, F355), and in helix TH9 (Y358, Y375). The increased insertion of K214 in the bilayer interior is shown by its normalized density profile over the bilayer normal axis (see Figure 10A). Note that the insertion of positively charged residues is facilitated by interactions with the polar headgroups (see Figure 6) and formation of hydrogen bonds (Figure 9). The degree of insertion of K214 in the hydrophobic core region is similar to that observed for residues P271 and P308 in conformation B2 (see Figure 10B,C). Neutralization of acidic side-chains in conformation B1 induces further insertion of V370 in helix TH9, which is as similar to that observed in conformation B2 (see Figure 10D).
Figure 10.
Normalized density of insertion depth of Cα atoms from residues (A) K214, (B) P271, (C) P308, and (D) V370. The distance Z is measured from the center of the bilayer at Z = 0 along the bilayer normal axis. Histograms from membrane-bound conformations B1 and B2 at T = 310 K are shown in black and cyan, respectively. Simulations at T = 323 K of membrane-bound conformation B1 with acidic side-chains in their standard and neutralized state are shown in red and blue, respectively. Data was obtained from the last 500 ns of each simulation. Phosphate group density computed over all MD simulations is shown as a reference in orange lines.
3.6. Changes of Bilayers Properties
The membrane association of the T-domain can induce changes in the behavior of the membrane. Figure S10 shows the distance of the average positions of phosphate atoms (DPP) in the upper and lower leaflets as a function of time. Conformations B1 and B2 induce no or small decrease of the average thickness compared to that of a pure bilayer at T = 310 K, respectively (see Figure S10A). In particular, B2 induces a decrease of ΔDPP = −2 Å relative to a pure bilayer. MD simulation of conformation B1 at T = 323 K reduces the average thickness of the bilayer (ΔDPP = −1 Å), as shown in Figure S10B. The bilayer thickness is further decreased upon neutralization of acidic side-chains (ΔDPP = −2 Å) accompanied by further insertion of the protein (see Figure S10B).
One of the most important properties of lipid bilayers is the area per lipid. MD simulations of conformations B2 and B1 (with neutralized acidic side-chains) resulted in areas per lipid similar to those calculated for free lipid bilayers (see Figure S11A,B). In contrast, MD simulations of B1 with only protonated histidines resulted in a decrease of the area per lipid compared to the area per lipid calculated for a free lipid bilayer (see Figure S11A,B). The observed reduction in the area per lipid corresponds to an increase of order in the lipid tails, which is shown by the increase of the order parameter SCD of lipid acyl-chains (see Figure S12).
4. DISCUSSION
Previously, we reported on MD simulations of the T-domain in aqueous solution in conjunction with spectroscopic experiments, which demonstrated a key role of the histidine residues in partial protein unfolding in acidic conditions, as well as predicted formation of a membrane-competent state of the T-domain.19 The model of the partially unfolded state resulted from such MD simulations is characterized by exposed hydrophobic and charged sites on the protein surface that were assumed to facilitate subsequent membrane binding.19,22 Furthermore, the coarse-grained MD simulations suggested that the partially unfolded model of the T-domain initially associates to the membrane through two distinctive conformations (B1 and B2).22 In this study, atomistic MD simulations at T = 310 K showed that conformations B1 and B2 retained their overall structure, orientation, and membrane contacts over several hundreds of nanoseconds. Conformation B1 adopts a shallowly inserted state in the membrane interface with membrane contacts located in protein regions that underwent conformational changes upon protonation of histidines in explicit solvent.19 These regions involve partially unfolded helices TH1–TH2, the loop between TH3–TH4, and N-terminal residues in TH8 and C-terminus of helix TH9. In contrast, conformation B2 is deeply inserted in the bilayer–water interface and partially penetrates the hydrophobic core of the lipid bilayer. B2 insertion is facilitated by persistent protein–lipid acyl-chain interactions at residues in the loop TH4–TH5, helix TH6, the loop TH6–TH7, and helix TH9. The resulting final structures of the predicted membrane-bound conformations B1 and B2 show an oblique orientation of C-terminal helices TH8–TH9 relative to the membrane plane.
It has been strongly suggested on the basis of experimental data that, in addition to histidine residue protonation, acidic residues are also protonated upon T-domain association with the membranes.4,5,59–61 In the present work, we modeled the implication of such protonation of the acidic residues onto membrane–protein conformation by setting all acidic residues in the T-domain to their neutralized states in our simulations. Extended MD simulations of the shallowly bound conformation B1, at T = 323 K, resulted in partial unfolding of the solvent exposed helix TH5 and deeper insertion in the membrane interface. However, the pattern of protein–membrane contacts remained similar to those observed in the MD simulation at T = 310 K. Neutralization of acidic side-chains favors deeper insertion of the protein and triggers an overall change in the orientation of helices TH8–TH9 relative to the membrane normal axis (see Figures 3 and 6). Note that neutralization of acidic side-chains increases the net positive charge of helix TH1 from +1 to +5, which favors deep insertion of helix TH1 in the bilayer interface. Previous fluorescence experiments have reported changes of the environment surrounding the N-terminal helix TH1 of the membrane-bound protein as a function of pH.10 Interestingly, the authors also showed that peptides containing the amino acid sequence of TH1 and TH3 have stronger interaction with membranes than peptides resembling helices TH2 and TH4. It was hypothesized that protonation of acidic residues and the net positive charge of helix TH1 allows for stabilizing interactions with anionic lipid headgroups.10 Our MD simulations are the first to study the effects of protonation of acidic residues in the context of the complete T-domain bound to lipid bilayers.
The removal of negative charges in the loops between TH5′–TH6 and TH8–TH9 facilitates the insertion of these loops in the early stages of insertion. Experimentally, neutralization of anionic residues located in the loop TH8–TH9 (E349 and D352) and in TH9 (E362) has been suggested to facilitate the protein–bilayer interactions (Figure 2 in Ghatak et al.).62 Neutralization of E362 was also shown to participate in the transmembrane insertion of the hydrophobic hairpin TH8–9 (Figure 8B in Ghatak et al.).62 In general, changes in the protonation state of residues at the membrane interface have been suggested to occur during the membrane interface binding of the T-domain and peptides.6,11,63
Experiments performed at different conditions have shown at least two different states of the T-domain bound to membranes: a noninserted and a transmembrane state of helices TH8–9.6,11 The noninserted state was found to be stabilized by low anionic content of lipid bilayers. Also, T-domain associated with bilayers of low anionic content and NMR experiments showed that helices TH8–9 were stabilized in a near parallel orientation to the membrane plane.11 Other variables, such as protein concentration and thickness of the lipid bilayer, were also shown to affect the stability of a shallow or a deeply inserted state of helices TH8–9.3 States B1 and B2 obtained through MD simulations have a nearly parallel orientation of helices TH8–9 similar to the states experimentally characterized by Chenal et al.11 However, the simulation time-scale is still limited to microseconds, which precludes sampling of further structural changes of the T-domain bound to the membrane and the transmembrane insertion of helices TH8–9.
5. CONCLUSIONS
In summary, atomistic MD simulations of the predicted membrane-bound conformations B1 and B2 showed that the protein remains stable in the anionic bilayer interface. MD simulations showed that the deep insertion of conformation B2 is facilitated by long-lived contacts between protein hydrophobic sites and the lipid acyl-chains, while the N-terminal helices TH1–TH2 are exposed to the solvent. Additional MD simulations suggested that neutralization of acidic residues in conformation B1 induces changes in its insertion depth and overall orientation relative to the membrane plane. A similar feature of both conformations is the oblique or nearly parallel orientation relative to the membrane plane of the hydrophobic helices TH8–TH9.6,7,17,55 Therefore, both membrane-bound conformations have been hypothesized to be part of the initial stages of the insertion folding pathway of T-domain.22 Future studies addressing the refolding of the protein on the membrane will require the use of advanced sampling techniques such as accelerated molecular dynamics and replica exchange methods.
Supplementary Material
Acknowledgments
We are grateful to Prof. Alexey S. Ladokhin for fruitful discussions of this work, and to Mr. Mauricio Vargas-Uribe for careful reading of the manuscript and suggestions during the editing of this manuscript. The authors would like to thank the anonymous reviewers for their valuable suggestions. This research was supported by the National Institutes of Health Grant GM-069783. Anton computer time was provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center (PSC) through Grant RC2GM093307 from the National Institutes of Health. The Anton machine at PSC was generously made available by D. E. Shaw Research.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Several tables, including tables of force-field and lipid parameters, and several plots, including plots of electron densities and atom distances (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Murphy JR. Mechanism of Diphtheria Toxin Catalytic Domain Delivery to the Eukaryotic Cell Cytosol and the Cellular Factors That Directly Participate in the Process. Toxins. 2011;3:294–308. doi: 10.3390/toxins3030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senzel L, Gordon M, Blaustein RO, Oh KJ, Collier RJ, Finkelstein A. Topography of Diphtheria Toxin’s T Domain in the Open Channel State. J. Gen. Physiol. 2000;115:421–434. doi: 10.1085/jgp.115.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Malenbaum SE, Kachel K, Zhan H, Collier RJ, London E. Identification of Shallow and Deep Membrane-Penetrating Forms of Diphtheria Toxin T Domain That Are Regulated by Protein Concentration and Bilayer Width. J. Biol. Chem. 1997;272:25091–25098. doi: 10.1074/jbc.272.40.25091. [DOI] [PubMed] [Google Scholar]
- 4.Chenal A, Savarin P, Nizard P, Guillain F, Gillet D, Forge V. Membrane Protein Insertion Regulated by Bringing Electrostatic and Hydrophobic Interactions into Play. A Case Study with the Translocation Domain of Diphtheria Toxin. J. Biol. Chem. 2002;277:43425–43432. doi: 10.1074/jbc.M204148200. [DOI] [PubMed] [Google Scholar]
- 5.Ladokhin AS, Legmann R, Collier RJ, White SH. Reversible Refolding of the Diphtheria Toxin T-Domain on Lipid Membranes. Biochemistry. 2004;43:7451–7458. doi: 10.1021/bi036157w. [DOI] [PubMed] [Google Scholar]
- 6.Kyrychenko A, Posokhov YO, Rodnin MV, Ladokhin AS. Kinetic Intermediate Reveals Staggered Ph-Dependent Transitions Along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain. Biochemistry. 2009;48:7584–7594. doi: 10.1021/bi9009264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh KJ, Zhan H, Cui C, Hideg K, Collier RJ, Hubbell WL. Organization of Diphtheria Toxin T Domain in Bilayers: A Site- Directed Spin Labeling Study. Science. 1996;273:810–812. doi: 10.1126/science.273.5276.810. [DOI] [PubMed] [Google Scholar]
- 8.Oh KJ, Zhan H, Cui C, Altenbach C, Hubbell WL, Collier RJ. Conformation of the Diphtheria Toxin T Domain in Membranes: A Site-Directed Spin-Labeling Study of the Th8 Helix and Tl5 Loop. Biochemistry. 1999;38:10336–10343. doi: 10.1021/bi990520a. [DOI] [PubMed] [Google Scholar]
- 9.Kachel K, Ren J, Collier RJ, London E. Identifying Transmembrane States and Defining the Membrane Insertion Boundaries of Hydrophobic Helices in Membrane-Inserted Diphtheria Toxin T Domain. J. Biol. Chem. 1998;273:22950–22956. doi: 10.1074/jbc.273.36.22950. [DOI] [PubMed] [Google Scholar]
- 10.Montagner C, Perier A, Pichard S, Vernier G, Menez A, Gillet D, Forge V, Chenal A. Behavior of the N-Terminal Helices of the Diphtheria Toxin T Domain During the Successive Steps of Membrane Interaction. Biochemistry. 2007;46:1878–1887. doi: 10.1021/bi602381z. [DOI] [PubMed] [Google Scholar]
- 11.Chenal A, Prongidi-Fix L, Perier A, Aisenbrey C, Vernier G, Lambotte S, Haertlein M, Dauvergne MT, Fragneto G, Bechinger B, et al. Deciphering Membrane Insertion of the Diphtheria Toxin T Domain by Specular Neutron Reflectometry and Solid-State Nmr Spectroscopy. J. Mol. Biol. 2009;391:872–883. doi: 10.1016/j.jmb.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 12.Malenbaum SE, Collier RJ, London E. Membrane Topography of the T Domain of Diphtheria Toxin Probed with Single Tryptophan Mutants. Biochemistry. 1998;37:17915–17922. doi: 10.1021/bi981230h. [DOI] [PubMed] [Google Scholar]
- 13.Prince HM, Duvic M, Martin A, Sterry W, Assaf C, Sun Y, Straus D, Acosta M, Negro-Vilar A. Phase III Placebo-Controlled Trial of Denileukin Diftitox for Patients with Cutaneous T-Cell Lymphoma. J. Clin. Oncol. 2010;28:1870–1877. doi: 10.1200/JCO.2009.26.2386. [DOI] [PubMed] [Google Scholar]
- 14.Steere B. Ph. D. Dissertation. Los Angeles, CA: University of California; 2001. Characterization of High-Order Oligomerization and Energetics in Diptheria Toxin. [Google Scholar]
- 15.Li J, Rodnin MV, Ladokhin AS, Gross ML. Hydrogen-Deuterium Exchange and Mass Spectrometry Reveal the pH-Dependent Conformational Changes of Diphtheria Toxin T Domain. Biochemistry. 2014;53:6849–6856. doi: 10.1021/bi500893y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senzel L, Huynh PD, Jakes KS, Collier RJ, Finkelstein A. The Diphtheria Toxin Channel-Forming T Domain Translocates Its Own NH2-Terminal Region across Planar Bilayers. J. Gen. Physiol. 1998;112:317–324. doi: 10.1085/jgp.112.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh KJ, Senzel L, Collier RJ, Finkelstein A. Translocation of the Catalytic Domain of Diphtheria Toxin across Planar Phospholipid Bilayers by Its Own T Domain. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8467–8470. doi: 10.1073/pnas.96.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodnin MV, Kyrychenko A, Kienker P, Sharma O, Vargas-Uribe M, Collier RJ, Finkelstein A, Ladokhin AS. Replacement of C-Terminal Histidines Uncouples Membrane Insertion and Translocation in Diphtheria Toxin T-Domain. Biophys. J. 2011;101:L41–L43. doi: 10.1016/j.bpj.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurnikov IV, Kyrychenko A, Flores-Canales JC, Rodnin MV, Simakov N, Vargas-Uribe M, Posokhov YO, Kurnikova M, Ladokhin AS. pH-Triggered Conformational Switching of the Diphtheria Toxin T-Domain: The Roles of N-Terminal Histidines. J. Mol. Biol. 2013;425:2752–2764. doi: 10.1016/j.jmb.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores-Canales JC, Simakov NA, Kurnikova M. Proteins: Struct., Funct., Bioinf. Submitted to. [Google Scholar]
- 21.Perier A, Chassaing A, Raffestin S, Pichard S, Masella M, Menez A, Forge V, Chenal A, Gillet D. Concerted Protonation of Key Histidines Triggers Membrane Interaction of the Diphtheria Toxin T Domain. J. Biol. Chem. 2007;282:24239–24245. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 22.Flores-Canales JC, Vargas-Uribe M, Ladokhin AS, Kurnikova M. Membrane Association of the Diphtheria Toxin Translocation Domain Studied by Coarse-Grained Simulations and Experiment. J. Membr. Biol. 2015;248:529–543. doi: 10.1007/s00232-015-9771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins: Struct., Funct., Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson CJ, Rosso L, Betz RM, Walker RC, Gould IR. Gafflipid: A General Amber Force Field for the Accurate Molecular Dynamics Simulation of Phospholipid. Soft Matter. 2012;8:9617–9627. [Google Scholar]
- 25.Skjevik AA, Madej BD, Walker RC, Teigen K. Lipid11: A Modular Framework for Lipid Simulations Using Amber. J. Phys. Chem. B. 2012;116:11124–11136. doi: 10.1021/jp3059992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fawzi NL, Phillips AH, Ruscio JZ, Doucleff M, Wemmer DE, Head-Gordon T. Structure and Dynamics of the Aβ21–30 Peptide from the Interplay of NMR Experiments and Molecular Simulations (Vol 130, Pg 6145, 2008) J. Am. Chem. Soc. 2011;133:11816–11816. doi: 10.1021/ja710366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Showalter SA, Bruschweiler R. Validation of Molecular Dynamics Simulations of Biomolecules Using NMR Spin Relaxation as Benchmarks: Application to the AMBER99SB Force Field. J. Chem. Theory Comput. 2007;3:961–975. doi: 10.1021/ct7000045. [DOI] [PubMed] [Google Scholar]
- 28.Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic Validation of Protein Force Fields against Experimental Data. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan YB, et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 30.Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 32.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the RESP Model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 33.Pan JJ, Heberle FA, Tristram-Nagle S, Szymanski M, Koepfinger M, Katsaras J, Kucerka N. Molecular Structures of Fluid Phase Phosphatidylglycerol Bilayers as Determined by Small Angle Neutron and X-Ray Scattering. Biochim. Biophys. Acta, Biomembr. 2012;1818:2135–2148. doi: 10.1016/j.bbamem.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the Charmm All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupradeau FY, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. Tools: Advances in RESP and ESP Charge Derivation and Force Field Library Building. Phys. Chem. Chem. Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Rob MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, et al. Gaussian 03. Wallingford, CT: Gaussian, Inc.; 2003. [Google Scholar]
- 37.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Joung IS, Cheatham TE. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kucerka N, Katsaras J, Nagle JF. Comparing Membrane Simulations to Scattering Experiments: Introducing the SIMtoEXP Software. J. Membr. Biol. 2010;235:43–50. doi: 10.1007/s00232-010-9254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henin J, Shinoda W, Klein ML. Models for Phosphatidylglycerol Lipids Put to a Structural Test. J. Phys. Chem. B. 2009;113:6958–6963. doi: 10.1021/jp900645z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolokh IS, Vivcharuk V, Tomberli B, Gray CG. Binding Free Energy and Counterion Release for Adsorption of the Antimicrobial Peptide Lactoferricin B on a POPG Membrane. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2009;80:031911. doi: 10.1103/PhysRevE.80.031911. [DOI] [PubMed] [Google Scholar]
- 42.Vila-Vicosa D, Teixeira VH, Santos HAF, Baptista AM, Machuqueiro M. Treatment of Ionic Strength in Biomolecular Simulations of Charged Lipid Bilayers. J. Chem. Theory Comput. 2014;10:5483–5492. doi: 10.1021/ct500680q. [DOI] [PubMed] [Google Scholar]
- 43.Cojocaru V, Balali-Mood K, Sansom MS, Wade RC. Structure and Dynamics of the Membrane-Bound Cytochrome P450 2C9. PLoS Comput. Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, et al. Amber 12. San Francisco, CA: University of California; 2012. [Google Scholar]
- 45.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 46.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved Side-Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins: Struct., Funct., Bioinf. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Grand S, Gotz AW, Walker RC. SPFP: Speed without Compromise-A Mixed Precision Model for GPU Accelerated Molecular Dynamics Simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 49.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical Integration of Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 50.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 51.Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, et al. Millisecond-Scale Molecular Dynamics Simulations on Anton; Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis (SC09); Portland, Oregon. Nov 14–20, 2009; New York, NY: ACM; 2009. pp. 1–11. [Google Scholar]
- 52.Shan YB, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian Split Ewald: A Fast Ewald Mesh Method for Molecular Simulation. J. Chem. Phys. 2005;122:054101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 53.Tuckerman M, Berne BJ, Martyna GJ. Reversible Multiple Time Scale Molecular-Dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 54.Lippert RA, Bowers KJ, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Shaw DE. A Common, Avoidable Source of Error in Molecular Dynamics Integrators. J. Chem. Phys. 2007;126:046101. doi: 10.1063/1.2431176. [DOI] [PubMed] [Google Scholar]
- 55.Martyna GJ, Klein ML, Tuckerman M. Nose-Hoover Chains - the Canonical Ensemble Via Continuous Dynamics. J. Chem. Phys. 1992;97:2635–2643. [Google Scholar]
- 56.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular-Dynamics Algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 58.Kabsch W, Sander C. Dictionary of Protein Secondary Structure - Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 59.Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D. The Crystal Structure of Diphtheria Toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 60.Montecucco C, Schiavo G, Tomasi M. Ph-Dependence of the Phospholipid Interaction of Diphtheria-Toxin Fragments. Biochem. J. 1985;231:123–128. doi: 10.1042/bj2310123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silverman JA, Mindell JA, Finkelstein A, Shen WH, Collier RJ. Mutational Analysis of the Helical Hairpin Region of Diphtheria Toxin Transmembrane Domain. J. Biol. Chem. 1994;269:22524–22532. [PubMed] [Google Scholar]
- 62.Ghatak C, Rodnin MV, Vargas-Uribe M, McCluskey AJ, Flores-Canales JC, Kurnikova M, Ladokhin AS. Role of Acidic Residues in Helices TH8–TH9 in Membrane Interactions of the Diphtheria Toxin T Domain. Toxins. 2015;7:1303–1323. doi: 10.3390/toxins7041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fendos J, Barrera FN, Engelman DM. Aspartate Embedding Depth Affects pHLIP’s Insertion pKa. Biochemistry. 2013;52:4595–4604. doi: 10.1021/bi400252k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.