Abstract
We report on whole-exome sequencing (WES) of 213 melanomas. Our analysis established NF1, encoding a negative regulator of RAS, as the third most frequently mutated gene in melanoma, after BRAF and NRAS. Inactivating NF1 mutations were present in 46% of melanomas expressing wild-type BRAF and RAS, occurred in older patients and showed a distinct pattern of co-mutation with other RASopathy genes, particularly RASA2. Functional studies showed that NF1 suppression led to increased RAS activation in most, but not all, melanoma cases. In addition, loss of NF1 did not predict sensitivity to MEK or ERK inhibitors. The rebound pathway, as seen by the induction of phosphorylated MEK, occurred in cells both sensitive and resistant to the studied drugs. We conclude that NF1 is a key tumor suppressor lost in melanomas, and that concurrent RASopathy gene mutations may enhance its role in melanomagenesis.
Recent years have seen unprecedented growth in the understanding of genetic and genomic changes in melanoma. Much of the information is from next-generation sequencing (NGS), particularly exome sequencing, and has been instrumental in the discovery of new cancer-driver genes. These NGS studies have corroborated the previously identified frequent recurrent somatic mutations in BRAF and NRAS and revealed new melanoma mutations, including a recurrent mutation in RAC1 (RAC1P29S), the third most frequent activating mutation in sun-exposed melanoma after BRAF and NRAS, and the most frequent recurring mutation in the Rho GTPase family1–3. The mutant protein RAC1P29S is a fast-cycling GTPase favoring the GTP-bound state that accelerates cell proliferation and migration when expressed in normal melanocytes. Screens uncovered other mutations at recurrent positions in PPP6C that reduce the phosphatase’s catalytic activity, consequently dysregulating the kinase AURKA and causing chromosome instability4,5. Frequent inactivating mutations were also discovered in the tumor suppressors TP53, NF1 and ARID2, and less frequent mutations were found in TACC1, GRM3, MAP3K4 and MAP3K9 that are likely to enhance melanoma pathogenesis6,7. Furthermore, recent studies have shed light on variants in regulatory regions of the melanoma genome. Recurrent mutations in the TERT promoter, which alter a transcription factor–binding motif and possibly lead to increased expression of TERT, shield melanoma cells from senescence8,9. NGS has also fostered an increased understanding of the genetics of noncutaneous melanomas, with the discovery of frequent mutations in BAP1, EIF1AX and SF3B1 in uveal melanoma10,11.
We report here the results of WES analysis of 213 human melanoma samples, including samples from 109 patients that we studied previously3 (Supplementary Data). Matched normal DNA was sequenced and analyzed from 133 of the tumors. We also tested the response of melanoma cell lines to the MEK inhibitor selumetinib (AZD6244), currently in clinical trials, and to the ERK inhibitor SCH772984 and performed protein blot analyses to correlate the effects of specific mutations with drug response.
RESULTS
Identification of NF1-mutant melanoma subgroup
Nevi can harbor activating BRAF or NRAS mutations but remain in a growth-arrested state. In some melanomas, somatic mutations in PTEN, CDKN2A or PPP6C are likely to account for initiation of the proliferative state. To comprehensively understand the mutations that lead to malignant transformation, we analyzed genes for evidence of selection and significantly increased mutation burden. We applied the ‘20/20 rule’ to identify genes with nonsilent mutations at recurrent positions that constituted 20% or more of all observed mutations or genes with at least 20% inactivating mutations, that is, nonsense, splice-site variant or insertion-deletion (indel) mutations12. The top 40 ranked genes from this analysis are shown in Table 1 (details are also provided in the Supplementary Data). Among those, we identified 11 genes that exhibited statistically significant mutation counts above what was expected on the basis of a driver gene analysis by MutSigCV13 (Fig. 1 and Supplementary Data).
Table 1.
Top mutated genes across the Yale cohort (n = 213)
| Symbol | Samples with mutations at recurrent positions |
Incidence (%) |
Symbol | Samples with inactivating mutations |
Incidence (%) |
|---|---|---|---|---|---|
| BRAF | 93 | 43.7 | NF1 | 28 | 13.1 |
| NRAS | 62 | 29.1 | ARID2 | 20 | 9.4 |
| RAC1 | 13 | 6.1 | TP53 | 17 | 8.0 |
| PPP6C | 12 | 5.6 | CDKN2A | 11 | 5.2 |
| GABRA3 | 11 | 5.2 | KMT2B | 9 | 4.2 |
| ABCB5 | 9 | 4.2 | ATM | 6 | 2.8 |
| TRRAP | 8 | 3.8 | ASPM | 5 | 2.3 |
| CYP7B1 | 8 | 3.8 | ARID1A | 5 | 2.3 |
| PCDHGA1 | 7 | 3.3 | RB1 | 4 | 1.9 |
| DGKI | 7 | 3.3 | TNRC6B | 4 | 1.9 |
| KNSTRN | 6 | 2.8 | SETD2 | 4 | 1.9 |
| BCL2L12 | 5 | 2.3 | FAM58A | 4 | 1.9 |
| IDH1 | 5 | 2.3 | ITGA5 | 4 | 1.9 |
| PLCE1 | 5 | 2.3 | NIN | 4 | 1.9 |
| LTN1 | 5 | 2.3 | SPRED1 | 4 | 1.9 |
| MAP2K1 | 4 | 1.9 | OAS3 | 4 | 1.9 |
| RQCD1 | 4 | 1.9 | USP24 | 4 | 1.9 |
| PROS1 | 4 | 1.9 | PTEN | 3 | 1.4 |
| EZH2 | 4 | 1.9 | MME | 3 | 1.4 |
| FRMD4B | 4 | 1.9 | YLPM1 | 3 | 1.4 |
The list shows the top 40 expressed genes with high mutation burdens according to the 20/20 rule12. The rule identifies genes with mutations at recurrent positions that constitute 20% or more of all observed mutations as likely oncogenes (three leftmost columns) and genes with at least 20% inactivating mutations (i.e., premature termination, splice-site variant or indel) as likely tumor suppressors (three rightmost columns). Also shown is the number of samples with qualifying mutations in these genes.
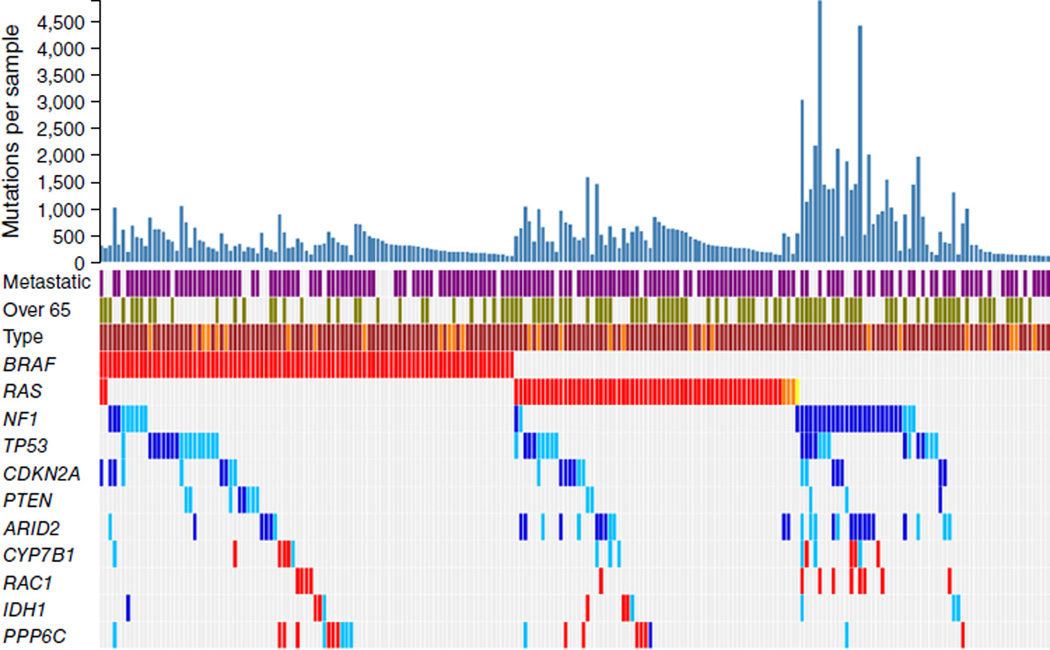
Figure 1.
Melanoma mutational landscape (n = 213). Top 11 melanoma-driver genes that reach genome-wide significance according to background mutation-frequency estimation. Purple, metastatic melanoma; green, patients over 65 years old; red, mutations at recurrent positions; dark blue, inactivating mutations (nonsense, splice, indel); light blue, predicted harmful mutations. Brown and darker orange represent sun-exposed tumors and tumors of unknown origin, respectively. Mutations in HRAS and KRAS are marked in light orange and yellow, respectively. Mutation counts correspond to novel mutations that are not found in repositories of common human variants.
Three genes were mutated with an incidence greater than 10%: BRAF and NRAS, with known recurrent activating mutations, and NF1, with a high number of inactivating mutations. In our cohort, we observed BRAFV600 mutations in 38.5% of melanomas (82 of 213) and RASQ61/G12/G13 mutations (58 NRAS and 3 HRAS) in 28.6% (61 of 213), and two melanomas were BRAF-NRAS double mutants (Table 2). Ninety percent of the NF1-mutant melanomas (38 of 42) harbored mutations of the inactivating or predicted damaging type (Supplementary Fig. 1 and Supplementary Data), with the majority (26 of 38) in melanomas expressing wild-type BRAF-RAS, as noted previously by our group3 and recently by The Cancer Genome Atlas (TCGA) consortium14; two NRASQ61R-mutant, one KRASG12I-mutant and nine BRAFV600-mutant melanomas had concurrent NF1 mutations. Among the tumors with no detectable BRAF or RAS mutation, a total of 46.4% (26 of 56) were NF1 mutants (Table 2). Of those, more than 80% (21 of 26) either showed loss of heterozygosity (LOH) across the NF1 locus or were compound heterozygotes harboring two NF1 mutations. Conversely, of the 12 BRAF-mutant or RAS-NF1 double-mutant melanomas, one-third (4 of 12) showed LOH or compound heterozygosity (Supplementary Data).
Table 2.
Mutational status of the Yale cohort (n = 213)
| RAS | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| BRAF | NRAS | HRAS | KRAS | NF1 | WT | ||
| BRAF | 82 (38.5%) | ||||||
| RAS | NRAS | 2 (0.9%) | 58 (27.2%) | ||||
| HRAS | 0 | 0 | 3 (1.4%) | ||||
| KRAS | 0 | 0 | 0 | 0 | |||
| NF1 | 9 (4.2%) | 2 (0.9%) | 0 | 1 (0.5%) | 26 (12.2%) | ||
| WT | 0 | 0 | 0 | 0 | 0 | 30 (14.1%) | |
Breakdown by activating mutations in BRAF (p.Val600) and RAS (p.Gln61/Gly12/Gly13) and inactivating mutations (stop mutations, splice-site variants, indels) and predicted damaging NF1 mutations. WT, triple–wild-type melanomas.
NF1-mutant melanomas harbored significantly more somatic mutations (P = 1.5 × 10−10) and occurred in significantly older patients (P = 0.017), but they were associated with similar overall patient survival compared to BRAF-mutant, RAS-mutant and ‘triple–wild-type’ (BRAF-RAS-NF1–wild-type) melanomas (Fig. 1 and Supplementary Fig. 2).
Mutations in known melanoma-driver genes affecting cell proliferation and survival, including RAC1, PTEN, CDKN2A, MAP2K1 and RB1, were also detected. This list included the X-chromosome gene FAM58A, which harbored several inactivating mutations. FAM58A (also known as cyclin M) negatively regulates RAF1 expression via interaction with CDK10, and mutations in FAM58A cause the inherited human developmental disease STAR syndrome15. Early-termination alterations in FAM58A were present in four tumors from male patients. All of the tumors were BRAF-RAS wild type; two were NF1-mutant and the other two were triple–wild type. The analysis also showed a recurrent R18W mutation in BCL2L12, which encodes an anti-apoptotic factor16. This mutation was adjacent to F17F, a recurrent silent mutation with increased inhibitory potential against UV-induced apoptosis due to increased BCL2L12 gene transcript and protein levels17.
Mutations in genes involved in chromatin modification or DNA repair were identified, some for the first time in melanoma. The list included two SWI/SNF family members, ARID1A (linked to gastric cancers18) and ARID2, reiterating the role of chromatin remodeling in malignancy. An additional gene in this category was the polycomb protein–encoding gene EZH2, which is upregulated in melanoma19; we noted recurrent EZH2Y646N mutations in four tumors. The mutation was located in the SET domain, which is involved in histone methylation20. A considerable number of inactivating mutations were in histone modifiers. Mutations were identified in KMT2B (MLL2) and SETD2, the latter of which is a recognized tumor suppressor in renal cell carcinoma21, as well as in TRRAP, involved in histone acetylation, which showed a previously described recurrent TRRAPS722F mutation22.
Finally, in the metabolism category, we found 22 mutations in CYP7B1, some of which occurred at recurrent positions (H286Y, E321K and P338S), suggesting an important role for cytochrome P450 in melanoma. Diminished expression of CYP7B1 is associated with breast cancer, possibly through modification of cholesterol metabolism23. We also observed a recurrent IDH1R132C mutation in four tumors. IDH1R132C, frequently present in glioma24 is a neomorphic mutation that causes production of D-2-hydroxyglutarate rather than the normal product, α-keto glutarate25.
NF1-mutant melanomas harbor co-mutations in RASopathy genes
A search for subtype-specific variants showed that damaging mutations in RASA2, which encodes a GTPase-activating protein that suppresses RAS function26, were exclusively present in NF1-mutant melanomas that were BRAF-RAS wild type (P = 0.006; Supplementary Data). Of the nine RASA2 mutations, two were nonsense and three were recurrent at position R551C. Four of the seven RASA2-mutant tumors showed LOH across the locus (all with a single RASA2 mutation), and two were compound heterozygotes. The recurrent RASA2R511C variant was recently described in the germline of a patient with Noonan syndrome27, a RASopathy otherwise linked to mutations in SOS1, PTPN11, RAF1 and other genes affecting RAS-MAPK signaling28. A search across these genes showed that four NF1-mutant, BRAF-RAS–wild-type melanomas harbored disease-causing mutations in PTPN11, with p.Tyr279Cys, p.Ala461Thr, p.Pro491Leu and p.Glu506Pro linked to the Noonan and LEOPARD syndromes29,30. TwoNF1-mutant, BRAF-RAS–wild-type melanomas harbored mutations in SOS1 linked to Noonan syndrome (p.Gly434Arg and p.Arg552Lys)31–33, and another two had mutations in RAF1 (p.Pro261Leu and p.Thr491Ile) that were also associated with Noonan syndrome34. NF1-mutant melanomas also featured mutations in SPRY and Sprouty-related genes known to encode negative regulators of MAPK signaling. In particular, three NF1-mutant melanomas harbored protein-coding alterations in SPRED1, including early-termination mutations. SPRED1 enhances NF1 activity by recruiting the protein to the plasma membrane35, and early-termination mutations in SPRED1 are linked to Legius syndrome (another RASopathy), which is characterized by neurofibromatosis- like skin features such as multiple café-au-lait macules, but not neurofibromas36.
In all, out of 26 NF1-mutant, BRAF-RAS–wild-type melanomas, ~60% (15) carried mutations in known RASopathy genes RASA2, PTPN11, SOS1, RAF1 and SPRED1, the majority of which (10 of 15) were documented disease-causing mutations (Table 3 and Supplementary Data).
Table 3.
Co-mutations in RASopathy genes in NF1-mutant, BRAF-RAS–wild-type melanomas in the Yale cohort
| Sample | NF1 mutations | RASopathy gene mutations |
|---|---|---|
| YUBER | p.Arg262Cys, p.Trp784* | SOS1 G434R |
| YUCAS | p.Ile679AspfsX21 | – |
| YUDAB | p.Phe1464Leu | PTPN11 Q506P |
| YUFIGUR | p.Tyr628LeufsX6, p.Gln2234* | – |
| YUHEF | p.Lys2552ThrfsX2, p.Gln853* |
SOS1G434V,R398H,P102S, RAF1P261L |
| YUKAT | p.Trp9*, p.Gln2239* | SOS1 R1067G |
| YUKAY | p.Arg1534* | – |
| YUKLAB | p.Ser2025Leu,
p.Arg1204Trp, p.Arg416* |
PTPN11P491L,Q510L,D425N, RASA2P530L |
| YULAN | NF1: c.6819+1G>A, p.Pro1084Ser | – |
| YUMUJ | p.Lys1632MetfsX8 | PTPN11 A461T |
| YUNEKI | p.Gln803* | – |
| YUOMEGA | p.Asn793LysfsX14, p.Leu844Phe | SPRED1 R117Q |
| YUOTHO | p.Gln1010*, pGln2239* | RASA2 S400F |
| YUPADI | p.Ser2093Phe, p.Arg1362* | SPRED1 Q353* |
| YUPAER | p.Gln912* | – |
| YUPAT | p.Leu844Phe | – |
| YUPROST | p.Arg440* | RASA2 R310* |
| YURAY | p.Arg2450* | – |
| YUROG | p.Gln1218* | – |
| YUROO | p.Gln282*, p.Arg440* p.Leu2661* | RASA2 F585I |
| YURUS | p.Gln2434* |
RASA2R511C,P843S, SPRED1Q205* |
| YUSOC | p.Glu337Lys, pTrp336* | RASA2R511C,Q500*, RAF1T491I |
| YUTAR | p.Val2653SerfsX26 | RASA2 R511C |
| YUTOGS | p.Leu466Phe, p.Lys2535* | – |
| YUWAND | p.Phe694_Trp696del | PTN11Y279C, SOS1R552K |
| YUZAN | p.Met108Ile | – |
Mutations in RASA2 (pArg511Cys), PTPN11 (p.Tyr279Cys, p.Ala461Thr, p.Pro491Leu, p.Gln506Pro), SOS1 (p.Gly434Arg, p.Arg552Lys) and RAF1 (p.Pro261Leu, p.Thr491Ile) have been previously shown to be disease causing in RASopathy syndromes (see text for details).
Although the occurrence of RASopathy gene mutations was significantly associated with NF1-mutant, BRAF-RAS–wild-type melanomas (P = 0.0004; Supplementary Data), 8.6% (16 of 187) of the remaining melanomas in our cohort also featured mutations in these genes (Supplementary Fig. 3). Mutations in three of these genes were thought to be disease causing: (1) two early-termination mutations in SPRED1 (in two different tumors, one with BRAFV600E and another with NRASQ61L mutation); (2) p.Asn58Ser in PTPN11, a known gain-of-function mutation in lung cancer37 that occurred in a melanoma with NRASQ61R; and (3) a p.Met269Lys SOS1 mutation in a triple–wild-type melanoma. The mutated residue in SOS1 (p.Met269) has documented associations with Noonan syndrome38.
Of interest, the pathogenic PTPN11P491L and MAPK1D321V, the latter a mutation in the docking site for DUSP-ERK interaction, were present in YUKLAB NF1H729Q/R1204W/R416* melanoma. The protein encoded by MAPK1D321V is similar to the known D321N, which impairs binding to DUSPs and reduces the sensitivity to inactivation by phosphatases39. Finally, two NF1-mutant melanomas (YUROG NF1Q1218* and YUCAS NF1FS-Indel) contained KRASA146T and KRASQ22K, respectively, known activating mutations in colorectal cancer40.
NF1 mutations, NF1 expression and RAS activation
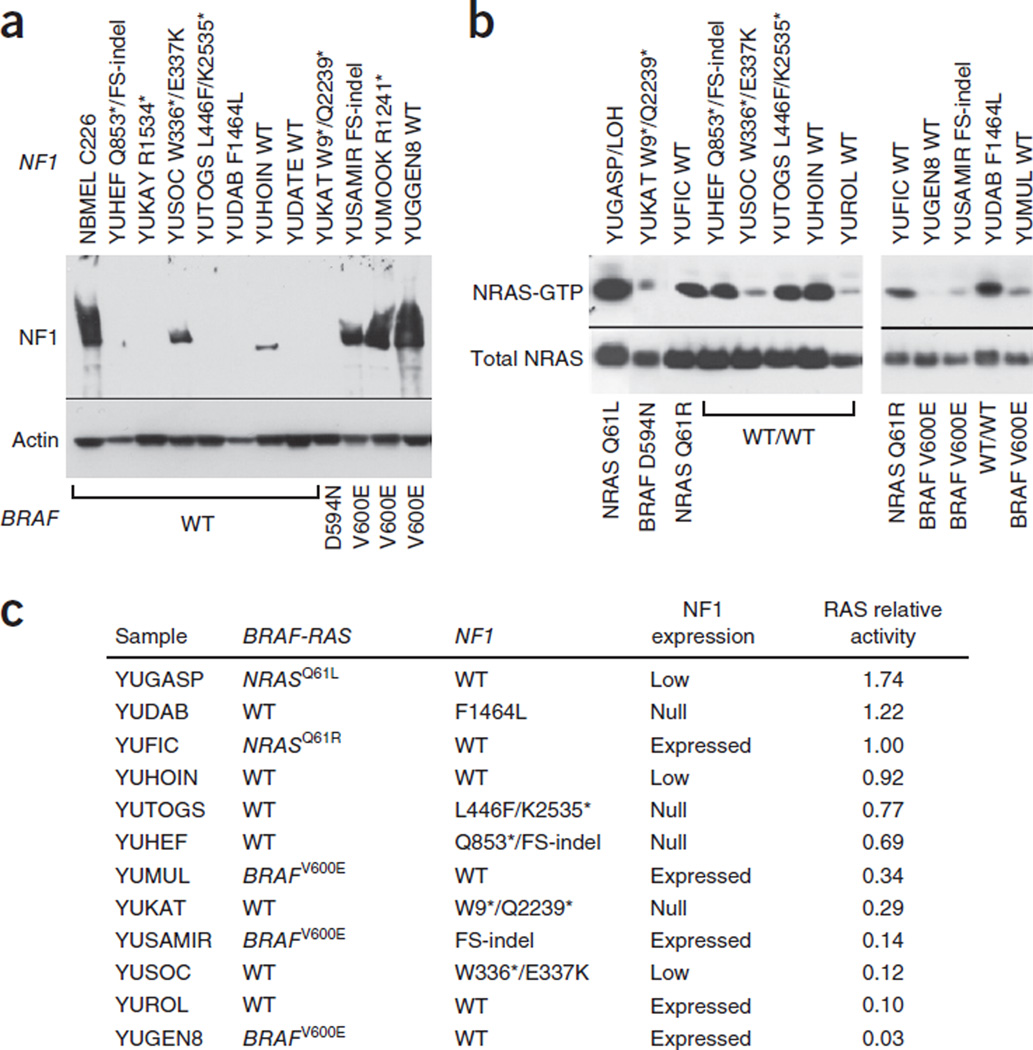
NF1 negatively regulates RAS by enhancing its RAS GTPase activity, converting active RAS-GTP to inactive RAS-GDP. We therefore investigated NF1 expression and NRAS activity in our exome-sequenced melanoma cell lines. Protein blot analysis showed that 75% of NF1-mutant melanomas (six out of ten), all BRAF-NRAS wild type, did not express NF1 or expressed it at low levels (Fig. 2a and Supplementary Fig. 4). One melanoma, YUDAB, homozygous for NF1F1464L mutation, did not express the protein, perhaps because of protein instability. Interestingly, the two NF1-mutant melanomas that expressed NF1 were also BRAFV600E mutants. One of them was from a neurofibromatosis patient with a germline NF1 frameshift mutation. This sample showed levels of NF1 expression about half those found in normal human melanocytes (YUSAMIR; Fig. 2a). Figure 2 also shows that two NF1–wild-type melanomas (YUHOIN and YUDATE) did not express NF1, and both showed LOH across the NF1 locus.
Figure 2.
NF1 expression and NRAS activity. (a) Protein blot showing NF1 expression in melanoma cells (YU designation) relative to that in normal human melanocytes (NBMEL) derived from a single newborn foreskin; β-actin was used as a loading control. NF1 mutations are indicated at the top, and BRAF mutations at the bottom. The wild-type NF1 (WT) melanomas YUHOIN and YUDATE displayed LOH. All the melanoma cell lines represented in this panel are NRAS wild type. The reproducibility data and protein expression in additional representative melanoma cell lines are presented in Supplementary Figure 4. (b) NRAS activity as determined by NRAS-GTP pulldown assay showing active NRAS-GTP and total NRAS detected in lysates. The same results were obtained with pan-RAS antibodies (data not shown). (c) Ranking of NRAS activity with BRAF, NRAS and NF1 mutation and expression status. We derived numbers by scanning the bands and quantitating the bands’ intensity relative to input NRAS using ImageJ and normalizing against the NRASQ61R YUFIC melanoma. FS, frame shift.
RAF pulldown assay showed that loss or low levels of NF1 were associated with the presence of activated GTP-bound RAS in most but not all cases (Fig. 2b,c), demonstrating the likely involvement of other RAS regulators.
NF1 expression and drug response
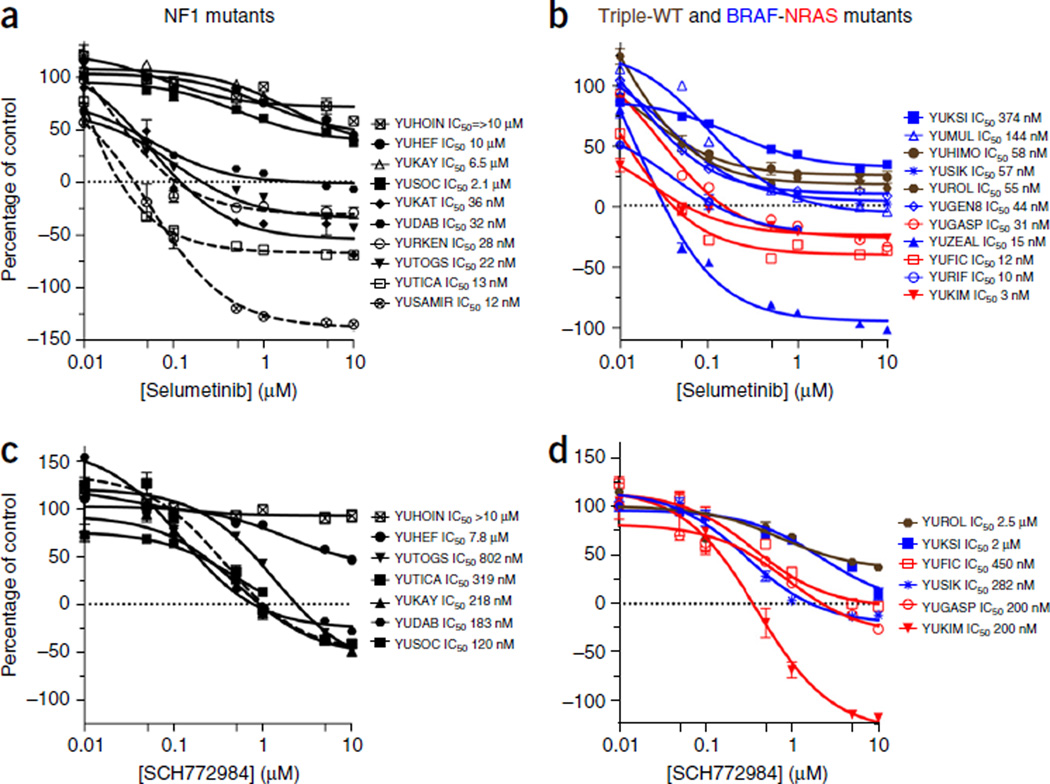
One of the main goals of identifying driver mutations in cancer is to make it easier to choose an appropriate targeted therapy, such as vemurafenib or dabrafenib, drugs that inhibit mutant BRAF melanomas, for the treatment of patients with BRAFV600-mutant tumors41,42. Loss of NF1 expression was suggested to mediate resistance to the MEK inhibitor selumetinib (AZD6244) in one report43 and to be a biomarker for high sensitivity to the MEK inhibitor PD0325901 in another44. A more recent study showed that NF1-negative and NF1-positive melanoma cell lines display the same sensitivity to the MEK inhibitor trametinib45. We therefore tested the effect of selumetinib, which is currently in clinical trials, on a panel of 21 melanoma cell lines, 10 NF1 mutants (or null by LOH) and 11 NF1 wild type, all characterized by exome sequencing (Fig. 3). The data showed that 60% of the NF1-mutant melanoma cell lines (six out of ten) were highly sensitive (IC50 of 0.036 µM or below), and four were highly resistant (IC50 of 2.1–10 µM) (Fig. 3a), indicating that suppression of NF1 is not always associated with sensitivity or resistance to a MEK inhibitor as was previously suggested43,44. Seven of the ten NF1-mutant, BRAF-NRAS–wild-type melanoma cell lines did not express NF1 or expressed it at extremely low levels (Fig. 2a). Of those, ~43% (three out of seven) were sensitive to MEK inhibition.
Figure 3.
Growth responses to selumetinib and SCH772984. (a–d) Growth of NF1-mutant (a,c) and of triple–wild-type (brown), BRAFV600-mutant (blue) and NRAFSQ61-mutant (red) melanoma cell lines (b,d). In a, the NF1 mutants YURKEN, YUTICA and YUSAMIR (dashed lines) are double-mutant cell lines (BRAFV600E, NRASQ61R and BRAFV600E, respectively) that express normal levels of NF1. The rest of the lines are null for NF1 (Fig. 2a). The selumetinib experiments were repeated at least twice. The data represent cell viability (CellTiter-Glo luminescent assay) as a percentage of the control at the end of 72 h of treatment. Each measurement is the average of triplicate or quadruplet wells. Error bars denote s.e.m.
All three double-mutant melanoma cell lines in this panel were sensitive to selumetinib. These included YURKEN, which carries NF1P228S-BRAFV600E mutation (IC50 = 0.028); YUTICA, which harbors NF1P166L-NRASQ61R mutation; and YUSAMIR, an NF1FS indel–BRAFV600E mutant from a person with neurofibromatosis (Fig. 3a). All three expressed high levels of NF1 (Fig. 2a). Also, YURKEN was highly resistant to the BRAF inhibitor vemurafenib, whereas YUSAMIR was highly sensitive to the drug (Supplementary Fig. 5). The results suggest that patients with double mutations should not be a priori excluded from vemurafenib treatment on the basis of expected resistance to the drug43,44.
Melanoma cell lines that did not carry an NF1 mutation showed diverse sensitivity to selumetinib, with an IC50 range of 0.003–0.374 µM, and ~55% (6 of 11) showed only a cytostatic response (Fig. 3b).
We also tested the response to SCH772984, a selective ATP-competitive ERK inhibitor that is in phase I clinical trials. Again, the highly selumetinib-resistant NF1-null YUHOIN and YUHEF melanoma cells were also the most resistant to this ERK inhibitor (Fig. 3c). However, in general, drug response followed a similar distribution in the NF1-null, triple–wild-type and BRAF-NRAS mutant melanoma cell lines (Fig. 3c,d).
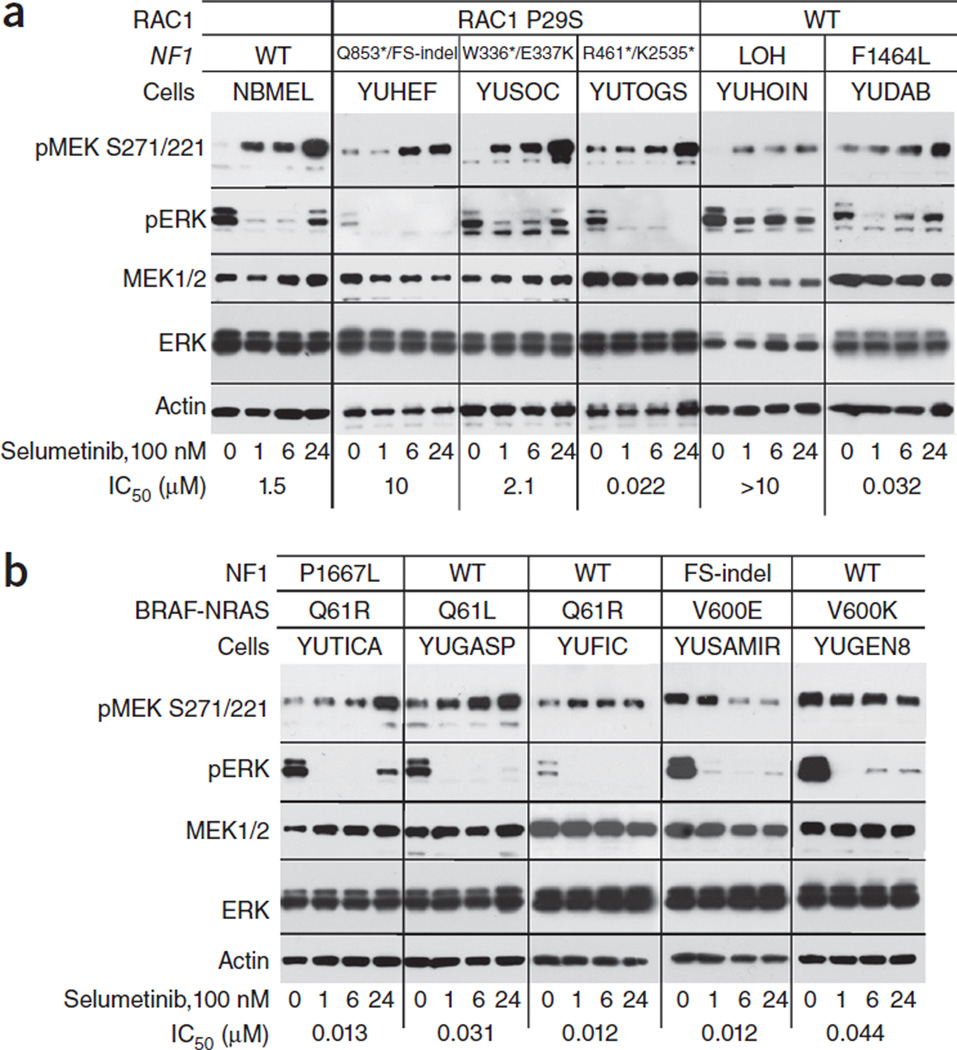
We explored the possibility that selumetinib induces upstream-pathway activation that is more prominent in NF1-null resistant cells than in sensitive cell lines, as this was suggested to characterize MEK resistance44. Protein blot analyses showed increased levels of phosphorylated MEK (pMEK) in all melanoma cells that did not carry the BRAFV600E/K mutation (i.e., NF1-null, NRASQ61R/L-mutant and even normal human melanocytes), regardless of the cells’ sensitivity to selumetinib in cell proliferation assays (Fig. 4a,b). Furthermore, although pERK was suppressed within an hour in response to selumetinib in all cell lines, its rebound at later time points was not indicative of resistance to the drug. For example, pERK did not recover in the resistant YUHEF cell line at the end of 24 h of treatment with 100 nM selumetinib, whereas it did recover in the resistant YUSOC and YUHOIN lines, as well as in the sensitive cell line YUDAB (Fig. 4a). Taken together, the data show that the feedback that leads to ERK activation in response to selumetinib in NF1-null melanoma cells is not necessarily associated with resistance to the drug, and that pERK phosphorylation is not required for selumetinib resistance.
Figure 4.
Changes in MEK1/2 and ERK phosphorylation in response to selumetinib. Normal human melanocytes (NBMEL) and melanoma cell lines were untreated (0) or treated with 100 nM selumetinib for 1, 6 and 24 h. The panels show protein blots probed with the indicated antibodies. (a) Normal melanocytes and NF1-mutant melanoma cell lines that are wild type for BRAF and NRAS. (b) Melanoma cell lines that are NF1, NRAS or BRAF mutants, as indicated. Numbers on the bottom show the IC50 in response to selumetinib (Fig. 3). The types of NF1 and RAC1 mutations are indicated for each cell line at the top. All the cell lines in b express wild-type RAC1. Figure 2a and supplementary Figure 4 show the corresponding levels of NF1 expression. The results represent one of two similar experiments.
Somatic copy-number alterations in triple–wild-type melanomas
We derived the genomic copy-number status and somatic copynumber alterations (SCNAs) on the basis of differences in read coverage between normal and matched tumor samples. Recent bioinformatics approaches allow for the long-range prediction of SCNAs by means of exome sequencing46. The corresponding SCNA segments can then be analyzed for broad and focal amplified and deleted regions with downstream programs such as GISTIC47. The GISTIC results for our matched melanoma samples are listed in the Supplementary Data and are presented graphically in Supplementary Figure 6. We observed known deletions in 9p21.3 (CDKN2A), 17p13.1 (TP53) and 10q23.31 (PTEN) and amplifications in 1q31.3 (ASPM) and 7q34 (BRAF). We identified a significant amplification signal in 7p22.1, near the PMS2 locus, and an amplification peak in 5p15.33, which harbors TERT. The latter supports the role of telomerase maintenance in melanoma based on recurrent somatic mutations in the TERT promoter8,9.
We also conducted GISTIC analysis with a focus on triple–wild-type melanomas, specifically, 19 matched melanomas that were wild type for BRAF, RAS and NF1. The analysis revealed a distinctive amplification peak at 4q12, harboring the KIT-KDR locus (Supplementary Fig. 7) that was present in two melanomas. KIT and KDR are receptor tyrosine kinases that activate the MAPK pathway48,49. Triple–wild-type melanomas carried additional characteristic copy-number alterations, including an amplification peak in MAP3K8 in YUROL that was not seen in any other melanoma specimen (Supplementary Fig. 8a,b). MAP3K8 expression has been implicated as a resistance mechanism to BRAF-inhibitor therapy in melanoma50. Another amplification peak was seen in YUDATE melanoma that included the MAPK1 (ERK2) locus (Supplementary Fig. 8c).
DISCUSSION
The genome-wide mutational analysis of melanoma reinforces the role of known frequent mutations in this devastating cancer and enhances our understanding of the processes that lead to melanomagenesis. This study showed that sun-exposed melanomas harbored a large number of missense mutations, but only a few were of the gain-of-function type characteristic of oncogenes such as BRAF, NRAS, RAC1, MAP2K1, EZH2 and IDH1 that are amenable to targeted therapy. In fact, mutations at recurrent positions can be of the inactivating type, as was demonstrated for PPP6C5. The majority of the loss-of-function changes affect processes involved in cell cycle progression; chromatin remodeling, DNA repair and/or transcription including histone methyltransferases (KMT2A, SETD2); and members of the SWI/SNF complex (ARID2 and ARID1A). Together these changes are likely to modify gene expression that releases normal cellular safeguards or constraints for which the design of inhibitors is more challenging.
The results of our large-scale exome sequencing highlight NF1 as a major player in melanomagenesis. In our cohort, 46% of BRAF-RAS–wild-type melanomas harbored NF1 mutations, which are often associated with LOH due to hemizygous deletions or copy-neutral gene conversions, or carried two or more single-nucleotide variants, suggesting biallelic mutation. NF1-BRAF and NF1-RAS double-mutants displayed considerably less LOH and fewer biallelic NF1 mutations (4 out of 12) than did NF1-mutant, BRAF-RAS–wild-type melanomas (21 of 26), and when tested they were found to express normal or high levels of NF1, suggesting a minor role for this tumor suppressor in these melanomas. In contrast, all of the NF1-mutant, BRAF-NRAS–wild-type melanomas did not express NF1. The loss of NF1 expression led to increased RAS activation in most but not all of these cases; RAS relative activity remained low in two out of the six NF1-null melanoma cell lines, probably because of the effects of other RAS-GTPase–activating proteins.
We found that NF1-null melanoma cell lines could be either sensitive or resistant to MEK or ERK inhibitors. We searched for additional mutations that may confer resistance to these inhibitors by activating alternative signaling pathways. Two of the three selumetinib-resistant NF1-mutant melanomas also carried the RAC1P29S mutation (YUHEF and YUSOC), in line with a recent publication indicating that RAC1mutant melanomas are relatively resistant to MEK inhibitors51. However, the YUTOGS melanoma is also a RAC1P29S-NF1 double-mutant but was highly sensitive to selumetinib (IC50 = 0.022 µM). We speculate that other damaging mutations could facilitate activation of the phosphoinositide 3–kinase (PI3K)-mTOR pathway. For example, YUHEF melanoma carries three mutations in the RASopathy gene SOS1 (p.Pro102Ser, p.Arg398Gly and p.Gly434Val) that may confer constitutive RAS activity, and PHLPP2 mutations are present in YUSOC (p.Pro142Ser) and YUHEF (p.Ser915Phe). PHLPP1 and PHLPP2 (similar to PTEN, which was not altered in these cells) are phosphatases that target AKT, PKC, S6K and RAF1, and their inactivation has been demonstrated in other cancers52. Furthermore, downregulation of PHLPP1 increases AKT activity, enhances the proliferation of melanoma cells and melanocytes and results in anchorage-independent growth of melanocytes53.
Neurofibromatosis is a genetic disorder that affects the nervous system and causes benign cutaneous neurofibromas and elevated risk for multiple Schwann cell tumors54. Epidemiological studies show that there is a general increase in the incidence of 13 other cancers among people with neurofibromatosis, albeit at a much lower rate, and melanoma is one of them55. Interestingly, melanocytes from café-au-lait macules of NF1 patients carry a somatic or second-hit mutation in NF1, indicating that a loss of NF1 provides a growth stimulus to normal melanocytes but does not accelerate their malignant transformation56. One possibility is that NF1 loss may be a more effective NRAS activator in the nervous system than in melanocytes57. Another likely explanation may relate to the timing of the mutation, with inherited loss of NF1 leading to the neurofibromatosis phenotype, and somatic mutations later in life, accompanied by additional alterations, leading to a variety of human cancers58. A similar observation has been made for retinoblastoma: although small-cell lung cancers show some of the highest somatic RB1 mutation rates, long-term retinoblastoma survivors with RB1 germline mutations have only a slightly increased incidence of secondary lung cancers59.
We identified documented disease-causing mutations in RASopathy genes RASA2, PTPN11, SOS1, RAF1 and SPRED predominantly in NF1-mutant melanomas, but also in BRAF-RAS mutant and triple– wild-type tumors, to a lesser degree. To our knowledge this is the first report to describe the presence of several known RASopathy gene mutations in melanoma, highlighting a possible role for these altered genes in cancer pathogenesis. Most of these mutations are found in NF1-mutant melanomas, and our functional studies showed that a loss of NF1 expression did not always lead to NRAS activation. These results raise the intriguing possibility that NF1-mutant melanomas may require additional changes for robust MAPK pathway activation. Indeed, clinically, concurrent NF1 and PTPN11 mutations cause severe or lethal forms of Noonan syndrome60,61. There is a case of a child with concurrent NF1 and PTPN11 mutation who developed bilateral optic nerve glioma, whereas the remaining family members, who carried only an NF1 mutation, displayed mild neurofibromatosis symptoms (café-au-lait spots)62. In addition, there is evidence that a loss of NF1 expression enhances the activity of noncanonical RAS mutations63, which is relevant to our observation of two cases of NF1-mutant melanomas, one with KRASA146T mutation and the other with KRASQ22K mutation.
ONLINE METHODS
Melanoma tumors and cell cultures
Normal human melanocytes (NBMEL) were cultured from foreskins of newborn males. The foreskins were incubated in dispase overnight at 4 °C, and epidermal melanocytes were further dissociated after 10 min of incubation in 0.25% trypsin-EDTA solution (GIBCO-Life Technologies). The cells were sedimented, and the pellets were incubated in OptiMEM (GIBCO), 5% FCS (Gemini Bio-Products), 1% Pen Strep (GIBCO) supplemented with 10 ng/ml basis fibroblast growth factor (ConnStem, F1001), 1 ng/ml heparin (Sigma, H3393), 0.1 mM dbcAMP (Sigma, D-0627) and 0.1 mM 3-isobutyl-1-methylxanthine (Sigma, I-5879). Melanoma tumors were excised to alleviate tumor burden, and cells were cultured in OptiMEM plus 5% FCS and Pen Strep. Specimens were collected by the Tissue Resource Core of the Yale SPORE in Skin Cancer with participants’ informed signed consent according to Health Insurance Portability and Accountability Act (HIPAA) regulations with a Human Investigative Committee protocol. The melanomas used for sequencing were from snap-frozen tumors or from short-term cell cultures64. The cell cultures were routinely checked for mycoplasma contamination and were discarded when they tested positive. Matching normal DNA was obtained from circulating lymphocytes or normal skin.
DNA extraction
The DNeasy purification kit (Qiagen, Valencia, CA) was used to extract DNA from cell pellets and freshly frozen tumors. The OneStep PCR Inhibitor Removal Kit (Zymo Research Corporation, Irvine, CA) was used to remove melanin from highly pigmented samples.
Exome sequencing data analysis
SNV and indel calling; somatic versus novel variants
We followed a previously described protocol for read alignment, single-nucleotide variant (SNV) and indel calling, data filtering, and somatic calling3. In short, we used the bwa program for read mapping and SAMtools for SNV calling65,66. SNVs were filtered according to the following quality criteria: (i) mutant allele frequency of ≥13%; (ii) SAMtools mapping score of ≥40; (iii) at least one forward and one backward read; and (iv) a minimum coverage of four mutant and eight total reads at the variant position. An SNV was called somatic in the absence of variant reads in the germline DNA samples, with one mutant read tolerated in the normal samples, and expecting a sufficient variant-to-total-read ratio in tumor and normal samples as assessed by Fisher’s exact test (P value threshold of 0.001). Generated data are with respect to Genome Reference Consortium human genome build 37 (GRCh37).
Somatic versus novel mutations
For our samples for which we had both tumor and matched germline DNA, we derived somatic SNVs, splice variants (SVs) and indels. For the entire cohort of matched and unmatched samples, we derived all SNVs, SVs and indels that were determined to be ‘novel’, i.e., not found in repositories of common human variation (phase 1 data from the 1,000 Genome Project and 2,577 non-cancer exomes sequenced at Yale) at a frequency of less than 0.05% and therefore enriched in somatic and private inherited mutations. We used the somatic mutations for the MutSigCV analysis and to calculate whether a gene had at least 20% somatic mutations at recurrent positions or at least 20% inactivating somatic mutations (integrating somatic data from the Broad Institute of MIT and Harvard and TCGA screens66). We used the list of mutations that were novel and predicted to be damaging for identifying paired and unpaired samples with mutations in driver genes, and for detecting gene mutations associated with the NF1 subgroup (redistribution analysis, discussed below). For all calculations that compare mutations counts across samples, we restricted the counts to genes with consistent coverage across samples.
Integrated mutation data from the Broad Institute of MIT and Harvard and TCGA
We integrated melanoma data from a recent publication by the Broad Institute2, and from the TCGA project66. For the TCGA project, we downloaded the automated somatic calls provided by the Baylor College of Medicine (4/17/2014) and Broad Institute (3/22/2014), available at the TCGA data portal. Both projects provided the data in MAF format. We removed samples that were in common between the data sets before integrating the data by calculating the overlap in somatic mutations between the cohorts and removing samples that shared a majority of mutations with another screen. After consolidation, we created an integrated data set of 397 melanomas from the Broad and TCGA cohorts.
Redistribution analysis for identifying co-mutated genes
To identify genes that were significantly co-mutated across the melanoma subgroups, we first applied the χ2 test to the equality of mutation counts across the four groups (NF1, BRAF, RAS and wild type). We additionally performed a redistribution analysis by adjusting for the total number of mutations (nt) per group. For each gene, we first computed the weighted mutation counts, or expected counts, for each group with weights n1/nt, n2/nt, n3/nt and n4/nt, respectively, where n1, n2, n3 and n4 are the total number of mutations per group and nt = n1 + n2 + n3 + n4. Then a scalar was multiplied by the weighted counts to ensure that the total number of counts for the gene stayed the same before and after redistribution. A χ2 test was then performed to determine whether the redistribution counts were equal across the four groups. For final selection, we filtered for genes with a redistribution analysis FDR of <0.2 (Supplementary Data). We performed a similar analysis to calculate whether the set of RASopathy genes were associated with the NF1 subgroup by tallying the mutation across the RASopathy genes and performing a redistribution analysis (Supplementary Data).
Gene mutation burden analysis: 20/20 rule
We calculated the top mutated and expressed genes on the basis of a heuristic rule12 that selects genes in which 20% or more of the observed somatic mutations are at recurrent sites, or in which 20% or more of the observed somatic mutations are inactivating (i.e., nonsense, splice-site, or indel). To increase power, we decided to integrate the somatic calls from the Yale cohort with the somatic calls from the integrated Broad-TCGA cohort for candidate gene identification. Genes with >20% somatic mutations and at least 1% frequency in the combined cohort were then ranked by the incidence of novel mutations in the entire Yale cohort (Table 1 and Supplementary Data).
Gene-mutation burden analysis using MutSigCV
The list of driver genes was identified using MutSigCV (Version 1.4)13, which distinguishes significantly mutated genes from others by taking into account mutational heterogeneity caused by replication timing, gene expression levels and chromatin state. We ran MutSigCV using the default input files as provided by the MutSigCV download page and prepared the mutational data using a MAF file comprising the somatic calls of expressed genes across the 133 matched samples of the Yale cohort. We selected significantly mutated genes using an FDR threshold of <0.2 among all genes that had at least one somatic mutation.
Survival analysis across the melanoma subgroups
We carried out an analysis to determine whether mutational status (NF1, BRAF, RAS and wild type) is associated with melanoma survival. The analysis was done for four mutational groups (NF1 mutant versus NF1 wild type, BRAFV600 versus BRAF wild type, RAS mutant versus RAS wild type, and BRAF-RAS wild type versus BRAF-RAS mutant). We considered two types of survival times by taking into account (1) the difference between age at last encounter and age of metastasis onset, and (2) the difference between age at last encounter and age of primary onset. The survival times and vital status were then used to obtain Kaplan-Meier curves. The analysis was done both with and without adjustment for age; for example, for the NF1 mutant–versus–wild type group analysis (age of metastasis onset), the P values with and without adjustment for age effects were 0.216 and 0.241, respectively.
CNV analysis
We used Excavator (Version 2.1)46 and GISTIC (Version 2.0.16)47 to calculate significantly amplified or deleted regions. Excavator was run with the default settings. The resulting chromosome-level segmentation data (genomic intervals), together with the location of the exome probes, were then used in GISTIC to generate the copy-number results. GISTIC was run with parameters of logR threshold for calling a segment amplified of ±0.25 and capping logR values at 2.
Tumor-purity calculation
We calculated sample purity in our paired samples by two approaches. The first approach was based on mutant allele frequency (MAF) estimation in regions of LOH. First, we established all LOH regions using the R module DNACopy. For each LOH region, we determined the median MAF for heterozygous inherited SNVs. We then used 2|0.5 – MAF| as a proxy for purity. Second, for samples with no LOH, we assumed that the majority of somatic mutations were heterozygous and used two times the median MAF of somatic mutations as a proxy for purity.
Automated tumor-purity calculation of our 133 paired samples showed a mean tumor-purity value of 0.63 ± 0.21, with 1.0 being the value of pure samples (Supplementary Data). Purities as low as 26% would theoretically be sufficient for calling heterozygous mutations with our mutation-calling thresholds and level of coverage. Review of selected samples and/or slides by a pathologist showed agreement with the automated analysis, with a mean purity of 71.7% across 24 paired and unpaired samples (Supplementary Data).
Sequencing statistics
For our tumor samples, we calculated a mean error rate of 0.2% ± 0.08% per called SNV, a mean coverage of 204 ± 76 independent reads per base, and coverage of at least eight independent reads across 96.5% ± 3% of the covered bases. Our matched normal samples were sequenced with a slightly smaller coverage of 141 ± 42 independent reads per base.
Variant-call precision
We Sanger validated 418 novel SNVs that were called using our variant-calling pipeline. We measured a precision of 93%, corresponding to 389 true positive and 29 false positive variant calls.
Protein blot analyses
Cell pellets were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors, and the levels of proteins were estimated with the Bio-Rad kit (Bio-Rad Laboratories). For NF1 analysis, cell extracts from different melanomas (20 µg/lane) were subjected to protein blot analysis with a 3%–8% Tris-acetate gel, Tris-acetate SDS running buffer and NuPAGE transfer buffer (Life Technologies, NP0006). The membranes were probed with anti-NF1 (Bethyl Laboratories, A300-140A) followed by anti–β-actin monoclonal antibody (clone AC-74, A5316, Sigma-Aldrich) as a control for protein loading. For MEK signaling pathway analysis, normal human melanocytes and melanoma cells were either untreated or treated with 100 nM selumetinib (Selleckchem Chemicals, S1008). Cells were harvested 1, 6, and 24 h later, and cell lysates were subjected to protein blot analysis in 4–12% Bis-Tris gels (Novex) as described64. The antibodies used were to phospho-MEK1/2 pSer217/221 (9121), MEK1/2 (4694), phospho-ERK2 pThr202/Tyr204 (9106), and ERK1/2 (4695), all from Cell Signaling Technology.
Analysis of intracellular RAS-GTP
We analyzed the RAS activation state with the Active RAS Pull-Down and Detection Kit from Thermo Scientific (16117) according to the manufacturer’s instructions. Cell lysates (400 µg protein) were incubated with GST-bound RAF1-RAS binding domain (GST-RAF1-RBD), and the levels of RAS-GTP were evaluated by protein blotting of bound material with anti-NRAS (or anti-panRAS). Whole-cell lysates (20 µg) were loaded in parallel to estimate the input levels of NRAS. The levels of active NRAS were estimated by band intensity relative to total NRAS, measured using the NIH software ImageJ 1.46r and normalized to NRASQ61R YUFIC melanoma.
Cell proliferation assays
Cell proliferation assays were performed in 96-well plates with the CellTiter-Glo Luminescent Cell Viability Assay in triplicate or quadruplet wells, measured at the end of 72 h of treatment with increasing concentrations of selumetinib, SCH772984 (Selleckchem Chemicals, S7101), or the BRAFV600 inhibitor vemurafenib (PLX4032, Plexxikon). The IC50 values (drug concentrations that reduced cell viability to 50% of the control) were calculated from the slope of the drug response generated in GraphPad Prism.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the Yale SPORE in Skin Cancer, funded by the National Cancer Institute, US National Institutes of Health, under award number 1 P50 CA121974 (R.H.); the Melanoma Research Alliance (Team award to R.H., M.B. and M.K.); Gilead Sciences, Inc. (J.S. and R.H.); the Howard Hughes Medical Institute (R.P.L.); the Department of Dermatology; and the Yale Comprehensive Cancer Center. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institutes of Health.
Footnotes
Accession codes. The entire data set has been deposited in dbGAP under accession number phs000933.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.K. and R.H. designed and performed the research, analyzed and interpreted the data, and wrote the manuscript. R.P.L. and J.S. designed the experiments. A.B., E.C., R.S. and M. Serin conducted the experiments. M.K., Y.K., P.E., J.P.M., S. Mane and N.P. analyzed the data from whole-exome sequencing. S. Ma and C.W. performed statistical analysis. M.B. evaluated the tumor percentage in the clinical specimens. S.A., D.N., M. Sznol and H.M.K. provided the clinical specimens and clinical annotation, as well as input in writing the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Davis MJ, et al. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc. Natl. Acad. Sci. USA. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold HL, et al. PP6C hotspot mutations in melanoma display sensitivity to Aurora kinase inhibition. Mol. Cancer Res. 2014;12:433–439. doi: 10.1158/1541-7786.MCR-13-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond D, et al. Melanoma-associated mutations in protein phosphatase 6 cause chromosome instability and DNA damage owing to dysregulated Aurora-A. J. Cell Sci. 2013;126:3429–3440. doi: 10.1242/jcs.128397. [DOI] [PubMed] [Google Scholar]
- 6.Prickett TD, et al. Exon capture analysis of G protein–coupled receptors identifies activating mutations in GRM3 in melanoma. Nat. Genet. 2011;43:1119–1126. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat. Genet. 2012;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 10.Martin M, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guen VJ, et al. CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proc. Natl. Acad. Sci. USA. 2013;110:19525–19530. doi: 10.1073/pnas.1306814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou CH, et al. GSK3β regulates Bcl2L12 and Bcl2L12A anti-apoptosis signaling in glioblastoma and is inhibited by LiCl. Cell Cycle. 2012;11:532–542. doi: 10.4161/cc.11.3.19051. [DOI] [PubMed] [Google Scholar]
- 17.Gartner JJ, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc. Natl. Acad. Sci. USA. 2013;110:13481–13486. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 19.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J. Cutan. Pathol. 2007;34:597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 20.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duns G, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5:637–645. doi: 10.1016/j.celrep.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin G, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarwood S, Bouyoucef-Cherchalli D, Cullen PJ, Kupzig S. The GAP1 family of GTPase-activating proteins: spatial and temporal regulators of small GTPase signalling. Biochem. Soc. Trans. 2006;34:846–850. doi: 10.1042/BST0340846. [DOI] [PubMed] [Google Scholar]
- 27.Chen PC, et al. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:11473–11478. doi: 10.1073/pnas.1324128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratner N, Miller SJA. RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 30.Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepri F, et al. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum. Mutat. 2011;32:760–772. doi: 10.1002/humu.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tartaglia M, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 33.Roberts AE, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 34.Pandit B, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 35.Stowe IB, et al. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brems H, et al. Review and update of SPRED1 mutations causing Legius syndrome. Hum. Mutat. 2012;33:1538–1546. doi: 10.1002/humu.22152. [DOI] [PubMed] [Google Scholar]
- 37.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 38.Ko JM, Kim JM, Kim GH, Yoo HW. PTPN11, SOS1, KRAS, and RAF1 gene analysis, and genotype-phenotype correlation in Korean patients with Noonan syndrome. J. Hum. Genet. 2008;53:999–1006. doi: 10.1007/s10038-008-0343-6. [DOI] [PubMed] [Google Scholar]
- 39.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 40.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson DB, et al. Combined BRAF (dabrafenib) and MEK inhibition (trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J. Clin. Oncol. 2014;32:3697–3704. doi: 10.1200/JCO.2014.57.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittaker SR, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nissan MH, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranzani M, et al. BRAF/NRAS wild-type melanoma, NF1 status and sensitivity to trametinib. Pigment Cell Melanoma Res. 2015;28:117–119. doi: 10.1111/pcmr.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magi A, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14:R120. doi: 10.1186/gb-2013-14-10-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji Z, Flaherty KT, Tsao H. Targeting the RAS pathway in melanoma. Trends Mol. Med. 2012;18:27–35. doi: 10.1016/j.molmed.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibuya M. VEGFR and type-V RTK activation and signaling. Cold Spring Harb. Perspect. Biol. 2013;5:a009092. doi: 10.1101/cshperspect.a009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson IR, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu. Rev. Pharmacol. Toxicol. 2014;54:537–558. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong L, et al. Oncogenic suppression of PHLPP1 in human melanoma. Oncogene. 2014;33:4756–4766. doi: 10.1038/onc.2013.420. [DOI] [PubMed] [Google Scholar]
- 54.Gallino G, et al. Association between cutaneous melanoma and neurofibromatosis type 1: analysis of three clinical cases and review of the literature. Tumori. 2000;86:70–74. doi: 10.1177/030089160008600113. [DOI] [PubMed] [Google Scholar]
- 55.Seminog OO, Goldacre MJ. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. Br. J. Cancer. 2013;108:193–198. doi: 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Schepper S, et al. Somatic mutation analysis in NF1 cafe au lait spots reveals two NF1 hits in the melanocytes. J. Invest. Dermatol. 2008;128:1050–1053. doi: 10.1038/sj.jid.5701095. [DOI] [PubMed] [Google Scholar]
- 57.Johnson MR, Look AT, DeClue JE, Valentine MB, Lowy DR. Inactivation of the NF1 gene in human melanoma and neuroblastoma cell lines without impaired regulation of GTP.Ras. Proc. Natl. Acad. Sci. USA. 1993;90:5539–5543. doi: 10.1073/pnas.90.12.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen LB, et al. Mutations in the neurofibromatosis 1 gene in sporadic malignant melanoma cell lines. Nat. Genet. 1993;3:118–121. doi: 10.1038/ng0293-118. [DOI] [PubMed] [Google Scholar]
- 59.Marees T, et al. Cancer mortality in long-term survivors of retinoblastoma. Eur. J. Cancer. 2009;45:3245–3253. doi: 10.1016/j.ejca.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Nyström AM, et al. A severe form of Noonan syndrome and autosomal dominant cafe-au-lait spots—evidence for different genetic origins. Acta Paediatr. 2009;98:693–698. doi: 10.1111/j.1651-2227.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- 61.Prada CE, et al. Lethal presentation of neurofibromatosis and Noonan syndrome. Am. J. Med. Genet. A. 2011;155A:1360–1366. doi: 10.1002/ajmg.a.33996. [DOI] [PubMed] [Google Scholar]
- 62.Thiel C, et al. Independent NF1 and PTPN11 mutations in a family with neurofibromatosis-Noonan syndrome. Am. J. Med. Genet. A. 2009;149A:1263–1267. doi: 10.1002/ajmg.a.32837. [DOI] [PubMed] [Google Scholar]
- 63.Stites EC, Trampont PC, Haney LB, Walk SF, Ravichandran KS. Cooperation between noncanonical Ras network mutations. Cell Rep. 2015;10:307–316. doi: 10.1016/j.celrep.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 64.Halaban R, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.