Abstract
The organization of the genome in the nucleus is believed to be crucial for different cellular functions. It is known that chromosomes fold into distinct territories, but little is known about the mechanisms that maintain these territories. To explore these mechanisms, we used various live-cell imaging methods, including single particle tracking to characterize the diffusion properties of different genomic regions in live cells. Chromatin diffusion is found to be slow and anomalous; in vast contrast, depletion of lamin A protein significantly increases chromatin motion, and the diffusion pattern of chromatin transforms from slow anomalous to fast normal. More than this, depletion of lamin A protein also affects the dynamics of nuclear bodies. Our findings indicate that chromatin motion is mediated by lamin A and we suggest that constrained chromatin mobility allows to maintain chromosome territories. Thus, the discovery of this function of nucleoplasmic lamin A proteins sheds light on the maintenance mechanism of chromosome territories in the interphase nucleus, which ensures the proper function of the genome.
Keywords: chromatin organization, diffusion, lamin A, nuclear structure, single particle tracking
Introduction
It is now well established that the organization of the genome in the nucleus of eukaryotic cells is not random. Chromosomes occupy well-defined chromosomal territories (CT) 1,2 and seem to fold in a fractal manner. Within each chromosome, Mbp-long strands called topological associated domains (TADs), are found to maintain increased number of internal contacts while lamina-associated domains (LADs) tend to interact with the lamina or other nuclear bodies.3 These organizational features were revealed to exist beyond the classical view of the local organization of the DNA which included the wrapping of chromatin around nucleosomes, and larger scale coiling and super-coiling.
During the last few years, functional effects of the nuclear organization were revealed.4 The regulation of gene expression is affected by the diffusion properties of transcription factors, chromatin condensation levels, the proximity of the chromosome regions to the lamina 5 or to the nucleus center,6 and the proximity of enhancers and other regulatory elements to the promoters within TADs, to name but a few. Other functions that are affected by the DNA organization include replication, the repair of DNA double strand breaks, chromosomal condensation during mitosis and more.
It is therefore clear that the maintenance of chromatin organization is critical for proper cell function, however, the mechanisms that are responsible for the organization and its maintenance are still unclear. Two conceptual methods have been used lately for exploring nuclear organization. First, there are fixed cell studies, based mainly on the chromosome conformation capture methods and fluorescence in situ hybridization (FISH). The second makes use of cell imaging techniques. These methods are complementary and both provide a vast amount of information.
Previous dynamic studies in the nucleus have shown that the motion of interphase chromatin is generally constrained.7 It was also shown that local chromatin diffusion is important in gene regulation,8 while the dynamic properties of chromatin seem to enhance the formation of chromosomal translocations.9 By following the dynamics of single genomic sites in the nucleus, we have recently shown 10 that lamin A plays an important role in the maintenance of chromatin organization. This was achieved by single particle tracking (SPT) of labeled telomeres and centromeres, and analysis of the measured data according to diffusion theories and biophysical modeling.11-13
SPT allows tracking the movement of individual particles and analyzing its dynamics. SPT is based on live-cell microscopy performed with a digital camera or a confocal setup combined with comprehensive image analysis, including corrections for cell drift and rotation and provides the spatio-temporal trajectories of single particles.14 For a detailed description, see.15
Here we show that lamin A affects not only the dynamics of telomeres and centromeres, but also the dynamics of nuclear bodies. This suggests that lamin A is important not only for the maintenance of chromatin organization but also for the positioning of general nuclear machineries and nuclear bodies. On the other hand, we show that lamin A does not affect the spatial distribution of chromatin in the nucleus, as shown below by image correlation spectroscopy.
Diffusion of genomic sites in the nucleus
Diffusion refers to the random motion of particles because of their kinetic (thermal) energy and interaction with the surrounding media. When following the dynamics of genetic sites in the nucleus, their diffusion pattern reflects on the structure of the chromatin and interactions with the nucleoplasm. Therefore, understanding the fine details of the diffusion pattern can lead to important insights.
There are different diffusion patterns that can be characterized from the experimental data. Most famous is normal diffusion resulting from a classical random walk process. It is characterized by the mean square displacement (MSD), , where nd is the spatial dimension of the motion and D is the diffusion coefficient. Thus, the average distance traveled by a particle is proportional to the square root of the travel-time.
Deviation from normal diffusion leads to anomalous diffusion where the MSD is not linear with time and gives , an anomalous exponent α that is different than 1. Anomalous diffusion can result from different mechanisms, each of them corresponds to a different biophysical phenomenon.16 When α<1 the process is called subdiffusion and can be interpreted in terms of interactions between the diffusing particles and other mobile or immobile structures. It was therefore proposed that the anomalous exponent can be used to quantify the crowding of the media at the molecular scale.17 Subdiffusing particles explore space slower than normally diffusing particles (Fig. 1) and have a higher chance of interacting with nearby targets while super-diffusing particles, where α>1, explore large distances and can reach far away targets faster.
Figure 1.
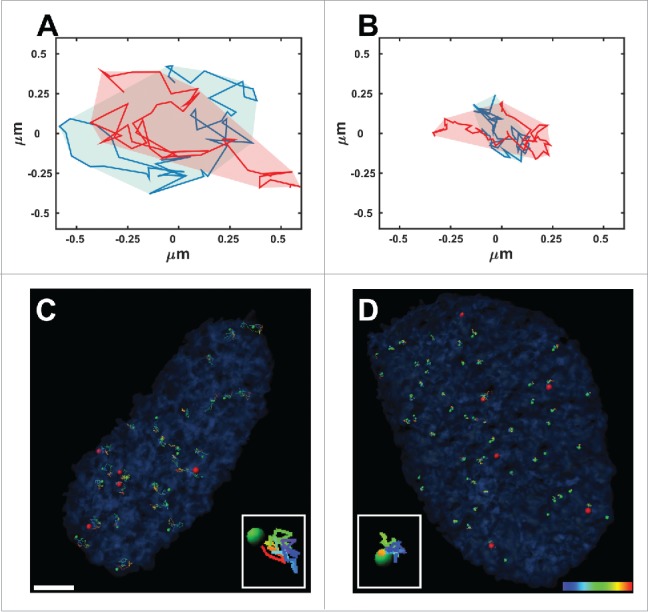
Normal and anomalous diffusion. Typical areas covered by 2 telomeres that undergo normal diffusion (A) and anomalous diffusion (B) over 50 time-points acquired during 925 seconds. (C, D) Trajectories of telomeres (green spots) and Cajal bodies (red spots) acquired from time-lapse fluorescence live-cell imaging over 925 seconds in Lmna−/− (C) and Lmna+/+ (D) cells. Note that the volume of movement in the Lmna−/− cells is much larger compared to that in Lmna+/+cells. Scale bar, 3µm.
The diffusion type can be identified from the plot of the MSD versus Δt, the latter being the time gap for which the MSD is calculated. It is common to present the plot on a log-log scale where the slope of the curve is equal to the anomalous coefficient α.10 An even better visual way to distinguish normal and anomalous diffusion is to plot MSD/Δt as a function of Δt, on a log-log scale, and in this case the slope is equal to . Therefore, normal diffusion translates to a zero slope and anomalous subdiffusion has a negative slope of .
Another tool that helps characterize the type of the dynamics is to observe the area scanned by a particle over a long time range e.g. imaging the cells for 15 minutes (Fig. 1A, B). This area depends on the diffusion type as well as on the actual value of the diffusion coefficient i.e. it depends both on the exponent value α and the preceding coefficient (D or A in the equations above). Therefore, the measured area directly emphasizes the level of order or chaos that may exist in the nucleus because of genetic modification.
Lamin proteins and nuclear organization
Lamin A and C are alternatively spliced proteins of a single gene, LMNA. They are type V intermediate filaments consisting of a short N-terminal non-helical head domain, an α-helical coiled-coil rod domain and a C-terminal non-helical tail domain containing a nuclear localization signal and an immunoglobulin-like domain.18 Lamin proteins are known to provide structural scaffolding to the nucleus. Lamin proteins form dimers by coiled-coil formation of their central rod domains and the dimers then assemble head to tail into polar polymers.19,20 It was shown that lamins incorporated into the lamina have little or no mobility, while a fraction of the nucleoplasmic lamin is mobile.21 When we followed different genomic sites, telomeres or centromeres, we could demonstrate that the loss of lamin A leads to a dramatic change of the diffusion type, as well as to significantly larger areas that are scanned by each entity at a given time.10
Genomic regions exhibit anomalous diffusion
To determine the diffusion pattern of chromatin, we used SPT for telomeres, centromeres and a single gene locus in living cells.10,22 There are normally dozens of telomeres or centromeres that can be simultaneously observed and measured in the same cell. Telomeres were fluorescently labeled by the specific telomeric proteins DsRed-TRF1 or GFP-TRF2. Centromeres were labeled through GFP-CENPA and the gene locus by RFP-LacI. The analysis procedures are described elsewhere 10 and typical particles paths are shown in Figure 1C, D.
All studied genomic loci exhibited slow anomalous diffusion in normal cells with a value of α in the range of 0.4 – 0.7.10 The genomic sites are part of a long DNA chain, and therefore it is expected that their diffusion will be constrained. The anomalous diffusion of telomeres was found in different cell types (Fig. 2A, B).
Figure 2.
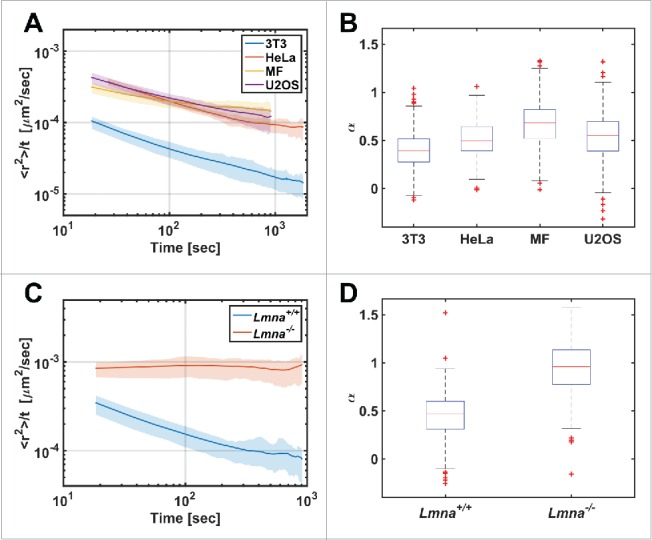
Diffusion patterns of telomeres in different cell types. (A) MSD/Δt vs Δt plot in log-log scale for different cell lines. (B) The distribution of α coefficient obtained from single telomeres trajectories (n=325, 166, 551, 958 for the 3T3, HeLa, MF, U2OS cell-lines respectively). (C) MSD/Δt vs Δt plot for Lmna+/+and Lmna−/− cells. (D) Box plot for the α coefficients distribution (n=223, 525 for the Lmna+/+ and Lmna−/− cells respectively). Note the significant difference showing anomalous diffusion in Lmna+/+ and normal diffusion in Lmna−/− cells.
It should be emphasized that similar slow anomalous diffusion is found for genomic sites that are located in the whole nucleoplasmic volume, both at the interior and the peripheral nuclear areas.
This reflects a very slow and localized motion that has major significance on the underlying biophysical mechanisms. The anomalous diffusion implies that the observed entities are not freely diffusing, but are rather interacting with their environment. Moreover, it will typically take a chromatin locus a long time (many hours) to diffuse through a chromosome territory that is typically larger than 1 µm. We suggest that the anomalous diffusion may be due to temporal chromatin-chromatin binding mediated by a certain protein. This will lead to very slow dynamics and necessarily to the maintenance of the chromosome territories in the nucleus, even though there are no membranes or other physical boundaries to keep them in place. We speculate that during mitosis, the binding mechanism can be switched off to allow for the major spatial reorganization of chromosomes.
Lamin A function in chromatin dynamics
In searching for a protein that may form chromatin-chromatin bonds, lamin A is a plausible candidate. It was shown before that lamin A interacts with chromatin through their non-α-helical C-terminal tail domain and the N- and C-terminal tail domains of core histones.23 Moreover, the effect of the loss of the lamin A protein on telomere distribution was previously shown.24 We therefore decided to measure the effect of lamin A on chromatin dynamics. Accordingly, we measured the diffusion of telomeres in lamin A depleted (Lmna−/−) cells and found that it changed from anomalous to normal diffusion (Fig. 2B, D). Moreover, each genomic site tested in the lamin A depleted cells tended to scan ∼10 times larger volumes compared to normal cells.
To further investigate the effect of lamin A proteins on chromatin dynamics, we measured telomere dynamics in Lmna−/− cells during stages of the cell cycle. For this purpose, we performed co-transfection of plasmids for telomere labeling and proteins that are expressed in the cell only during a specific stage of the cell cycle. To identify G1, S and late S-G2 stages we used PCNA-mCherry, hCDT1-mCherry and hGeminin-CFP proteins, respectively. PCNA proteins form distinct nuclear foci during the DNA synthesis phase of the cell cycle.25 The other 2 proteins are used in the FUCCI system 26 for following the cell cycle in living cells. hCDT1 levels peak in G1; as cells transition into S phase, hCDT1 levels fall and Geminin levels rise, remaining high until the cells are back in G1. We did not find significant differences in the movement volumes of telomere motion and α coefficient between different cell cycle stages. The average volume of movement of telomeres in the Lmna−/− cells during late S and G2 was V=0.12μm3, in comparison to V=0.10μm3 in early/mid S phase. For comparison, in Lmna+/+ cells the average volume of telomere movement was V=0.014μm3, an order of magnitude smaller.
The effect of lamin A depletion on nuclear body dynamics
Previous studies have shown that nuclear bodies such as mRNPs, Cajal and PML bodies move through the nucleoplasm.27,28 In light of our findings, it was important to explore whether the depletion of lamin A protein has an influence on the diffusion properties of nuclear bodies, even though they are not part of the DNA chains. We marked Cajal and PML bodies by transient transfection of DsRed-Colin and CFP-Sp100 plasmids, respectively. In each nucleus we observed 2-5 of each nuclear body, which is not enough to perform a correction of nucleus translation and rotation. Therefore, we imaged cells that had both telomeres and nuclear body staining and used the telomeres motion in order to produce the corrections of cell translation and rotation. We found that telomeres have the smallest volume of motion, while Cajal bodies are more mobile. In Lmna+/+ cells the average volume movement of telomeres during 925 sec was V=0.014μm3, while Cajal and PML bodies scanned an average volume of V=0.080 μm3 and V=0.024μm3, respectively. Lamin A depletion allowed both Cajal and PML bodies to scan larger volumes in the nucleus, although significantly less for Cajal than for PML bodies (Fig. 3). According to the corral model,29 nuclear body movement is influenced by chromatin motion and therefore large chromatin motion leads to higher mobility of nuclear bodies. The dynamics of the nuclear bodies themselves may be restricted as they are locked in between more dense chromatin regions 28,30 or they may be also connected to the chromatin network.
Figure 3.
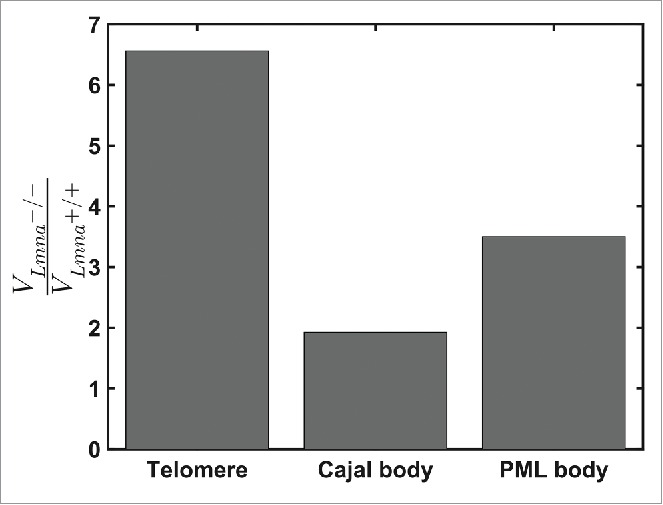
Comparison of the ratio of volumes of motions for telomeres, Cajal and PML bodies during 925 seconds in Lmna+/+ and Lmna−/− cells. For Lmna+/+ cells n=691, 74, 78 for telomeres, Cajal bodies and PML bodies, respectively. For Lmna−/− cells n=519, 109, 76 for telomeres, Cajal bodies and PML bodies, respectively. All entities explore smaller volumes in the Lmna+/+ compared to the Lmna−/− cells. The effect of lamin A depletion is more significant on chromatin dynamics (telomeres) compared to nuclear bodies.
TSA and osmotic pressure alter chromatin structure but not dynamics
We were interested in examining whether lamin A depletion leads to changes in spatial chromatin organization. For this purpose we used image correlation spectroscopy (ICS) and calculated the autocorrelation function g(r) of the nuclei of various cell types.31 Measurements and analysis were performed for chromatin labeled with Hoechst in control U2OS cells and following lamin A depletion by siRNA. In addition, we also used osmotic treatment that leads to chromatin condensation 30 and TSA incubation that induces a more open chromatin structure 32 in order to compare the effect of lamin A. The autocorrelation function, g(r), measures the spatial distribution of the chromatin in the nucleus and the obtained results show that lamin A depletion has no effect on chromatin compaction, while TSA and osmotic treatment both changed chromatin compaction levels.
TSA and osmotic treatments did not significantly change the type of chromatin diffusion i.e., the diffusion remained anomalous; while lamin A depletion changed the type of chromatin motion from anomalous to normal diffusion. These experiments provide evidence that the interaction between certain proteins and chromatin can govern chromatin dynamics while not changing its spatial distribution.
Summary
To shed light on the mechanisms of chromosome territory maintenance, we have used live cell imaging methods that are ideal for studying dynamic processes in living cells. We analyzed the diffusion of different genomic sites by measuring their trajectories through SPT. We compared the changes in the diffusion caused by different parameters such as genomic region, physical location inside the nucleus, chromatin condensation levels, as well as the effect caused by depletion of proteins that are known to interact with chromatin. We found that depletion of the lamin A protein has the strongest effect on chromatin diffusion properties. Diffusion analysis of chromatin motion in lamin A depleted cells strongly suggests that lamin A can bind chromatin thereby restricting its local motion and changes the diffusion pattern from anomalous to normal.
We propose that lamin A binds to chromatin and restricts its motion. Consequently, it dampens chromatin dynamics, which allows chromosomes to remain localized, each in its chromosome territory, during interphase (Fig. 4).
Figure 4.
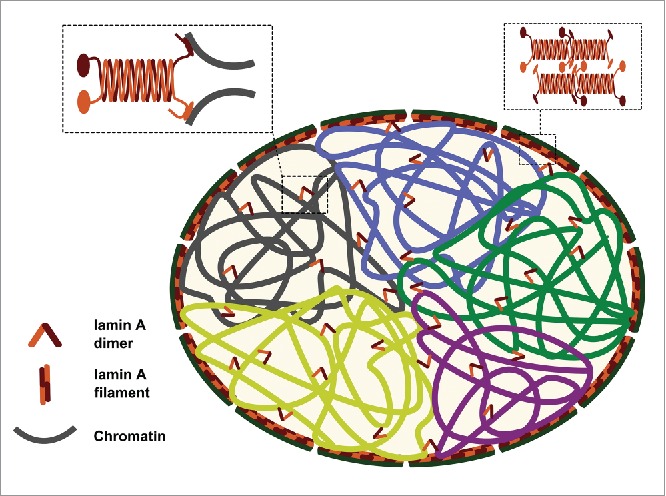
Suggested model explaining the role of lamin A in maintaining the intra-nuclear chromosomal structure. Lamin A serves as a “stapler” that connects adjacent chromosomal regions and helps in keeping chromosome territory structure in place during interphase.
By combining fluorescence microscopy, statistical analysis and live cell biological techniques, we were able to highlight the importance of lamin A on chromatin dynamics. Our experimental approach and data analysis enable the characterization of protein interactions with chromatin in living cells in an independent manner to biochemical assays. We are confident that future progress and implementation of such interdisciplinary approaches will enable the identification of new nuclear processes that take place in living cells.
Funding Statement
This work was supported in part by the Israel Centers of Research Excellence (ICORE) No. 1902/12 and Israel Science Foundation No. 51/12 (Y.G.) and by the European Research Council (Y.S.T).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Cremer C, Cremer T. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2001;2:292-301; PMID:11283701; http://dx.doi.org/ 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- 2.Lieberman-Aiden E, Berkum NLv, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; http://dx.doi.org/ 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickmore WA, Steensel BV. Genome architecture: domain organization of interphase chromosomes. Cell 2013; 152:1270-84; PMID:23498936; http://dx.doi.org/ 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 4.Woringer M, Darzacq X, Izeddin I. Geometry of the nucleus: a perspective on gene expression regulation. Curr Opin Chem Biol 2014; 20:112-9; PMID:24981829; http://dx.doi.org/ 10.1016/j.cbpa.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 5.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organization. Curr Opin Cell Biol 2007; 19:31-316; PMID:17184986; http://dx.doi.org/ 10.1016/j.ceb.2006.12.009 [DOI] [PubMed] [Google Scholar]
- 6.Kurz A, Lampel S, Nickolenko JE, Bradl J, Bermer A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol 1996; 135:1195-205; PMID:8947544; http://dx.doi.org/ 10.1083/jcb.135.5.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 1997; 7:930-9; PMID:9382846; http://dx.doi.org/ 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- 8.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al.. Chromatin architecture reorganization during stem cell differentiation. Nature 2015; 518:331-6; PMID:25693564; http://dx.doi.org/ 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soutoglou E, Misteli T. Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev 2007; 17:435-42; PMID:17905579; http://dx.doi.org/ 10.1016/j.gde.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronshtein I, Kepten E, Kanter I, Berezin S, Lindner M, Redwood AB, Mai S, Gonzalo S, Foisner R, Shav-Tal Y, et al.. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun 2015; 6:8044; PMID:26299252; http://dx.doi.org/ 10.1038/ncomms9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kepten E, Bronshtein I, Garini Y. Improved estimation of anomalous diffusion exponents in single-particle tracking experiments. Phys Rev E 2013; 87:052713; http://dx.doi.org/ 10.1103/PhysRevE.87.052713 [DOI] [PubMed] [Google Scholar]
- 12.Burnecki K, Kepten E, Janczura J, Bronshtein I, Garini Y, Weron A. Universal algorithm for identification of fractional Brownian motion. A case of telomere subdiffusion. Biophys J 2012; 103:1839-47; PMID:23199912; http://dx.doi.org/ 10.1016/j.bpj.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkai E, Garini Y, Metzler R. Strange kinetics of single molecules in living cells. Phys Today 2012; 65:29-35; http://dx.doi.org/ 10.1063/PT.3.1677 [DOI] [Google Scholar]
- 14.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J 2002; 82:2775-83; PMID:11964263; http://dx.doi.org/ 10.1016/S0006-3495(02)75618-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger IB, Kepten E, Garini Y Single-particle tracking for studying the dynamic properties of genomic regions in live cells. Methods Mol Biol 2013;1042:139–51. [DOI] [PubMed] [Google Scholar]
- 16.Metzler R, Jeon J-H, Cherstvy AG, Barkai E. Anomalous diffusion models and their properties: non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys Chem Chem Phys 2014; 16:24128-64; PMID:25297814; http://dx.doi.org/ 10.1039/C4CP03465A [DOI] [PubMed] [Google Scholar]
- 17.Weiss M, Elsner M, Kartberg F, Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys J 2004; 87:3518-24; PMID:15339818; http://dx.doi.org/ 10.1529/biophysj.104.044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22:832-53; PMID:18381888; http://dx.doi.org/ 10.1101/gad.1652708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CY, Lammerding J. Lamins at a glance. J Cell Sci 2012; 125:2087-93; PMID:22669459; http://dx.doi.org/ 10.1242/jcs.087288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruenbaum Y, Foisner R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 2015; 84:131-64; PMID:25747401; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034115 [DOI] [PubMed] [Google Scholar]
- 21.Broers JLV, Machiels BM, van Eys GJJM, Kuijpers HJH, Manders EMM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci 1999; 112:3463-75; PMID:10504295 [DOI] [PubMed] [Google Scholar]
- 22.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein E-M, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLOS Biology 2011; 9:e1000573; PMID:21264352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson PM, Lammerding J. Broken nuclei – lamins, nuclear mechanics, and disease. Trends Cell Biol 2013; 24:247-56; PMID:24309562; http://dx.doi.org/ 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, et al.. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J 2009; 28:2414-27; PMID:19629036; http://dx.doi.org/ 10.1038/emboj.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonardi E, Girlando S, Serio G, Mauri FA, Perrone G, Scampini S, Dalla Palma P, Barbareschi M. PCNA and Ki67 expression in breast carcinoma: Correlations with clinical and biological variables. J Clin Pathol 1992; 45:416-9; PMID:1350788; http://dx.doi.org/ 10.1136/jcp.45.5.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle -progression. Cell 2008; 132:487-98; PMID:18267078; http://dx.doi.org/ 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- 27.Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol 2002; 4:502-8; PMID:12068306; http://dx.doi.org/ 10.1038/ncb809 [DOI] [PubMed] [Google Scholar]
- 28.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science 2004; 304:1797-800; PMID:15205532; http://dx.doi.org/ 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Görisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci USA 2004; 101:13221-6; PMID:15331777; http://dx.doi.org/ 10.1073/pnas.0402958101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 2010; 12:543-52; PMID:20453848; http://dx.doi.org/ 10.1038/ncb2056 [DOI] [PubMed] [Google Scholar]
- 31.Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem Biophys 2007; 49:141-64; PMID:17952641; http://dx.doi.org/ 10.1007/s12013-007-9000-5 [DOI] [PubMed] [Google Scholar]
- 32.Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM–FRET. J Cell Biol 2009; 187:481-96; http://dx.doi.org/ 10.1083/jcb.200907029 [DOI] [PMC free article] [PubMed] [Google Scholar]


