abstract
An intricate machinery protects cells from the accumulation of misfolded, non-functional proteins and protein aggregates. Protein quality control pathways have been best described in the cytoplasm and the endoplasmic reticulum, however, recent findings indicate that the nucleus is also an important compartment for protein quality control. Several nuclear ubiquitinylation pathways target soluble and membrane proteins in the nucleus and mediate their degradation through nuclear proteasomes. In addition, emerging data suggest that nuclear envelope components are also degraded by autophagy, although the mechanisms by which cytoplasmic autophagy machineries get access to nuclear targets remain unclear. In this minireview we summarize the nuclear ubiquitin-proteasome pathways in yeast, focusing on pathways involved in the protein degradation at the inner nuclear membrane. In addition, we discuss potential mechanisms how nuclear targets at the nuclear envelope may be delivered to the cytoplasmic autophagy pathways in yeast and mammals.
Introduction
Misfolded and damaged proteins can be harmful for the cell. To eliminate these proteins and to maintain protein homeostasis, cells have developed an intricate protein quality control (PQC) system by which they assess the quality of proteins and take proper measures to either repair or eliminate the damaged components.1 Proteins can be targeted for proteasomal degradation by poly-ubiquitinylation, which is mediated by ubiquitin-activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin protein ligase (E3), the latter defining substrate specificity.2 E3 ligases recognize targets either directly or with the help of chaperones.3 While the proteasomes target predominantly soluble misfolded proteins, large insoluble protein aggregates are primarily degraded by autophagy.4 In macroautophagy, the cargo is sequestered within double membrane vesicles called autophagosomes, which fuse with the lysosome.5 Autophagosome formation requires yeast ubiquitin-like protein Atg8 or its mammalian homologues of the LC3 and GABARAP families, which become lipidated with phosphatidyl ethanolamine (PE) through ubiquitinylation-like reactions involving E1-like enzyme Atg7 and E2-like protein Atg3.6 In microautophagy, cargo is sequestered by invaginations of the lysosomal membrane, which then pinches off as small vesicles into the lysosome lumen7.
Degradation-mediated mechanisms in protein homeostasis have been best described in the cytoplasm and the endoplasmic reticulum (ER),8 but a number of recent studies identified PQC pathways also in the nucleus (Fig. 1).9 While proteasomes are long known to localize in the cytoplasm and the nucleus,10 PQC pathways and their targets in the nucleus have only been identified more recently.11 In particular, mechanisms mediating degradation of integral membrane proteins of the inner nuclear membrane (INM) have long remained elusive. In the cytoplasm, ER-associated degradation (ERAD) is the main pathway for the degradation-mediated PQC of membrane proteins. ERAD targets misfolded proteins, but also some correctly folded wild-type proteins to the proteasome.12 In yeast, 2 integral membrane proteins of the ER, Hrd1 and Doa10 are the core E3 ubiquitin ligases targeting ERAD substrates.13 While Hrd1 primarily targets proteins with lesions in domains oriented toward the ER lumen, Doa10 targets mainly proteins with lesions in their cytoplasmic or membrane regions.13 Hrd1 localizes exclusively to the ER, but Doa10 was also found in the INM14 and targets nuclear and INM proteins.14-16 In addition, novel E3 ligases were recently identified that enrich at the INM,17,18 suggesting that several proteasomal PQC pathways exist in the nucleus and target specific sets of proteins. Furthermore, increasing evidence suggests that nuclear proteins can also be degraded by autophagy, although the mechanisms remain largely unclear.19 In this minireview we summarize the recently identified protein degradation pathways at the INM and we discuss potential mechanisms how nuclear envelope (NE) proteins may be targeted by autophagic pathways.
Figure 1.
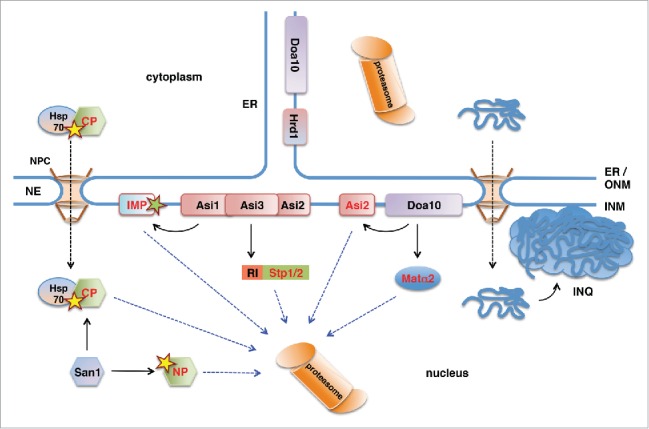
Ubiquitin-proteasome-dependent protein degradation pathways in the yeast nucleus. San1 is a nuclear E3 protein ubiquitin ligase that ubiquitinylates misfolded nuclear (NP) and cytoplasmic (CP) proteins. Delivery of CPs to nuclear San1 is assisted by Hsp70 chaperone Ssa1. Unlike the ER-membrane localized E3 ligase Hrd1, E3 ligase Doa10 localizes to both ER and the INM and targets INM protein Asi2 and transcriptional repressor Matα2 for proteasomal degradation. Asi1-Asi3 is an E3 ligase complex enriched in the INM that, together with Asi2, ubiquitinylates latent forms of transcription factors Stp1 and Stp2 via their RI degron. Asi1-Asi3 also ubiquitinylates misfolded or mislocalized integral membrane proteins (IMP) in the INM. The nucleus is also the compartment for the proteotoxic stress-induced deposit of misfolded cytoplasmic proteins and protein aggregates in the intranuclear quality control compartment (INQ).
Protein degradation in the cell nucleus
The NE consists of 2 membrane layers, the inner and the outer nuclear membrane, connected at the sites of nuclear pores.20 While the outer nuclear membrane (ONM) is an extension of the ER membrane, the INM has a protein composition different from that of the ONM and the ER. In metazoan cells the nuclear side of the INM is coated with a protein meshwork consisting of lamin intermediate filaments21,22 and lamin-interacting INM proteins and termed the nuclear lamina.23,24 Proteasomes are known to localize inside the nucleus in yeast and mammalian cells,25-31 but nuclear PQC pathways have not been described until recently (Fig. 1). A key pathway of nuclear PQC in yeast is mediated by the nuclear ubiquitin-protein ligase San1, which targets misfolded nuclear proteins for proteasomal degradation11 by recognizing their exposed hydrophobic regions.32,33 In addition, the ER integral membrane protein Doa10, which is a ubiquitin protein ligase involved in ERAD, localizes also to the INM, where it mediates degradation of transcription factor Matα214 and of the INM protein Asi2.15 Moreover, a novel ubiquitinylation machinery that specifically localizes at the yeast INM and targets several soluble and INM proteins for degradation has recently been discovered.17,18 Apart from the ubiquitin-proteasome-dependent protein degradation, portions of the nucleus can also be targeted by autophagy and be degraded by the vacuole/lysosome in both yeast and mammalian cells.34-39 Taken together, accumulating data show that the nucleus is an important compartment for protein degradation and quality control. This is further supported by findings that the stress-induced protein aggregate deposit in yeast, which has previously been described as “juxtanuclear quality control compartment,” surprisingly localizes to the nucleus.40
Asi1 and Asi3 - nuclear ubiquitin protein ligases involved in INM-associated degradation
INM proteins Asi1, Asi2 and Asi3 function as negative regulators of the amino acid induced Ssy1-Ptr3-Ssy5 (SPS) signaling pathway in yeast.41-43 In the absence of amino acids Asi proteins prevent promoter binding of latent transcription factors Stp1 and Stp2,44,45 however the molecular mechanism of this repression has not been clear so far. The first evidence that Asi proteins participate in the degradation of latent Stp1 and Stp2 in the nucleus came from findings that deletions in ASI genes stabilize specific forms of Stp1.46 Further studies showed that Asi1 and Asi3 function as ubiquitin protein ligases that ubiquitinylate latent forms of Stp1 and Stp2 in the nucleus, thereby targeting them for proteasomal degradation (Fig. 1).18
The role of Asi proteins is not limited to SPS-sensor signaling as additional substrates have been identified, including integral membrane proteins.17,18 For instance, membrane proteins Erg11 and Nsg1 involved in sterol synthesis were stabilized in asi1Δ and asi3Δ mutants,17 indicating that they are ubiquitinylation substrates of the Asi-complex. Protein levels of Erg11 were not affected by the levels of sterol metabolites,17 indicating that Asi1-mediated degradation of Erg11 is not involved in a homeostatic feedback mechanism. Rather, the purpose of Asi-mediated degradation of Erg11 is apparently to prevent accumulation of the ER-membrane protein Erg11 at the INM. The possibility that Asi-ubiquitin protein ligase has a role in clearance of mislocalized proteins at the INM is supported by the finding that vacuolar membrane proteins, which mislocalize to the ER/NE upon C-terminal epitope tagging, are also targeted by Asi1/3.18 In addition, Asi proteins may also recognize misfolded protein domains at the INM (Fig. 1), as the C-terminal epitope tagging of vacuolar proteins might also impair their proper folding. In support of this model a sec61-2 mutant that becomes misfolded at high temperature and is targeted to the INM when fused to a nuclear localization signal was degraded in an Asi1-dependent manner.17 Taken together, in addition to ensuring latency of transcription factors in the SPS-sensor signaling, and possibly in other pathways, Asi proteins may also be involved in the removal of misfolded and mislocalized integral membrane proteins from the INM.
Apart from misfolded sec61-2, which appears to be a common substrate of Hrd1 and Asi1, other substrates of the ERAD ubiquitin ligases Hrd1 and Doa10 were not affected by deletion of ASI1.17 Although the Asi-ubiquitinylation machinery works with E2 enzymes Ubc618 and Ubc7,17,18 which are also used in the ERAD pathway, Asi-mediated degradation is clearly distinct from ERAD, based on its predominant localization at the INM and its specific substrates and can thus be referred to as INM-associated degradation (INMAD).
INM protein Asi2 – a degradation mediator and a target
Ubiquitin ligases Asi1 and Asi3 form a complex with Asi2, an integral INM protein with 2 transmembrane regions and no apparent functional domains.17,43 Interestingly, Asi2 was required for degradation of some substrates of Asi1/Asi3, such as transcription factors Stp1/Stp2 and integral membrane proteins Erg11 and Nsg1, but degradation of misfolded sec61-217 and several other Asi1/Asi3 substrates18 did not require Asi2. Why Asi2 is necessary for degradation of some Asi-substrates, but not others is not clear, but Asi2 may recognize a specific type of degradation signal in a specific subset of Asi substrates. In the case of Stp1, the Asi-dependent degron has been defined as a 16 amino acids long sequence in the N-terminal region of Stp1, designated RI motif,46 but degron sequences in other substrates mediating Asi-dependent ubiquitinylation are not known.
Interestingly, we have recently shown that Asi2 itself is a degradation substrate of the ER/INM-localized ubiquitin ligase Doa10 (Fig. 1) and associated E2 enzymes Ubc6 and Ubc7.15 Like in most ubiquitinylated substrates, ubiquitinylation of Asi2 occurs predominantly on lysine residues.47 Ubiquitinylation of degradation substrates at alternative acceptor sites has been observed only in rare cases, such as in metazoan and virus-infected cells.48-55 Intriguingly, we found that a functional mutant of yeast Asi2 lacking all lysine residues is ubiquitinylated on atypical acceptor sites and targeted for proteasomal degradation in a Doa10-Ubc6-Ubc7 dependent manner,47 indicating that ubiquitinylation on alternative residues might be more prevalent than previously considered. The degradation signal that targets Asi2 for Doa10-dependent ubi-quitinylation may involve an amphipathic helix present close to the N-terminus of Asi2. Indeed, amphipathic helices are present in several other Doa10 substrates where Doa10 seems to recognize their exposed hydrophobic surfaces either directly or via chaperones.16,56,57 The degradation signal in Asi2 might become exposed upon changes in the molecular environment or upon loss of interaction partners. For instance, the loss of interaction between Asi2 and Asi1/Asi3 may uncover a region of Asi2 and affect Asi2 protein stability. In support of this possibility, we observed that degradation of Asi2 was faster in cells lacking Asi1 and Asi3.15 Our data suggest that the majority of Asi2 is ubi-quitinylation by Doa10 at the INM, although we could not experimentally exclude the possibility that Doa10 targets Asi2 also at the ER.15 Notably, as the inactivation of DOA10 does not completely abolish Asi2 degradation,15,47 additional pathways are likely involved in Asi2 degradation, which may function in parallel with Doa10 or upon inactivation of Doa10.
Autophagy of nuclear envelope proteins
In yeast, selective autophagy of nuclear material, also called nucleophagy, has been observed in a process called piecemeal microautophagy of the nucleus (PMN).34 In this process, which is induced by starvation, portions of the nuclear envelope form a direct physical interaction with the vacuolar membrane and form blebs that pinch off into the vacuole.34 A similar process that occurs after prolonged periods of nitrogen starvation was named late nucleophagy.35 A process similar to PMN has not been described in complex eukaryotes so far. However, an autophagy-mediated degradation of nuclear envelope proteins has recently been described in mammalian cells. Large perinuclear autophagosomes were observed in cells expressing muscular dystrophy-linked mutants of lamin A at the nuclear envelope or mutants of the INM protein emerin.58 Moreover, the lamin A mutant protein progerin, which is permanently farnesylated and thus tightly associated with the INM59-61 and causes premature aging Hutchinson-Gilford progeria syndrome (HGPS), was found to be degraded by the lysosomes in 2 recent studies (Fig. 2).37,38 Progerin-expressing cells exhibit abnormal nuclear shape, changes in heterochromatic marks, increased DNA damage and premature senescence,62 and treatment of HGPS cells with rapamycin, which is known to induce autophagy,63,64 ameliorated these phenotypes.37,38 These effects of rapamycin were linked to the increased rate of progerin degradation37 and reduced progerin levels in treated cells.37,38
Figure 2.
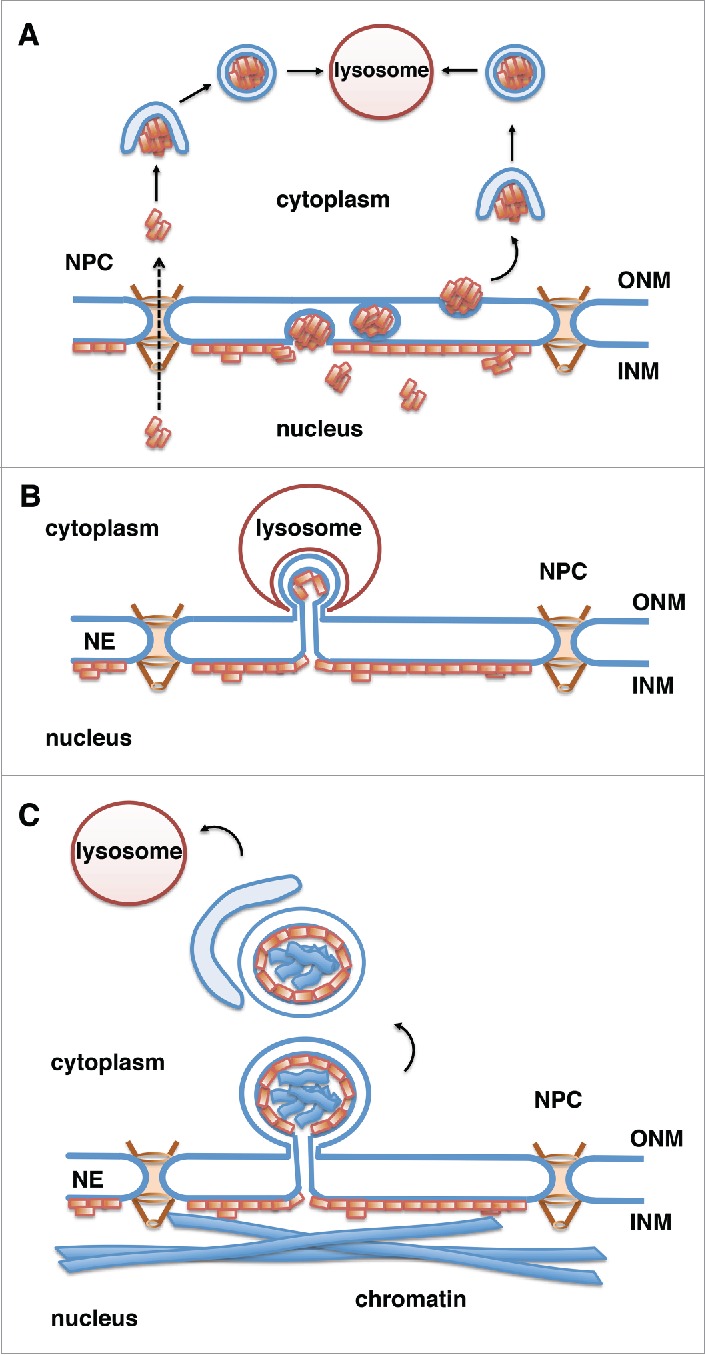
Potential mechanisms for lysosomal degradation of progerin by autophagy. Progerin (depicted by orange rectangles) is a lamin A mutant that associates with the INM. Progerin may become accessible for degradation by the cytoplasmic lysosomes in the following ways. (A) Progerin aggregates translocate to the cytoplasm through nuclear pores (NPCs) or vesicle-mediated transport through the double membrane of the NE (nuclear egress). Progerin aggregates are engulfed by the autophagosomal membrane in the cytoplasm and fuse with the lysosome.67 (B) Similar to piecemeal microautophagy of the nucleus in yeast, blebs of the nuclear envelope are engulfed by invaginations of the lysosomal membrane and then pinch off into the lysosomal lumen. (C) Vesicles or micronuclei bud off from the nuclear envelope, are engulfed by the growing isolation membrane producing autophagosomes, which fuse with lysosomes and deliver nuclear components to the lysosomal lumen.
The molecular details of the rapamycin-induced progerin degradation are not well understood. However, observations that inhibition of autophagosome formation by treatment of cells with 3-methyladenine or by knock-down of Atg 765,66 results in impaired progerin degradation37 suggested that progerin is degraded by macroautophagy. Since the autophagosomes and lysosomes are present in the cytoplasm, it is unclear how progerin at the nuclear envelope is delivered to the cytoplasm (Fig. 2). Potential routes could involve nuclear export through the nuclear pore complexes (NPC) or vesicle-mediated budding of larger progerin aggregates through the NE (nuclear egress, Fig. 2-A),67 similar to the transport of large ribonuclear particles in Drosophila68 or nuclear egress of herpesvirus.69 Alternatively, progerin might be degraded in a process similar to piecemeal microautophagy of the nucleus, in which blebs at the nuclear envelope are surrounded by invaginations of the lysosomal membrane and pinch off into the lysosmal lumen (Fig. 2-B), although this pathway has so far been described only in yeast.34
Interestingly, a recent study in yeast found that in response to rapamycin treatment portions of the NE are degraded by selective autophagy via budding of double membrane vesicles from the NE and their subsequent engulfment in autophagosomes (Fig. 2-C).36 Degradation of specific targets in the process of selective autophagy is achieved by receptor proteins that bind to degradation targets and to Atg8/LC3 on forming autophagosomal membranes, thereby facilitating cargo sequestration into the autophagosomes.70 Atg39 was identified as a novel Atg8-interacting autophagy receptor at the NE in yeast.36 Atg39 protein levels markedly increase upon rapamycin treatment and mediate the loading of NE-derived double-membrane vesicles into the autophagosomes.36 Atg39-mediated nucleophagy seems to be important for yeast cell survival under prolonged nutrient-limiting conditions but it is unclear which nuclear components are targeted. In view of these recent findings on Atg39-dependent nucleophagy in yeast, the rapamycin-induced degradation of progerin in mammalian cells may occur via a similar process, including formation of micronuclei encompassing nuclear material and their subsequent engulfment by autophagosomes (Fig. 2-C). Atg39-homologues were not found in complex eukaryotes yet, however other proteins may have similar functions in mammals.
In support of a vesicle-mediated delivery of nuclear material to cytoplasmic autophagosomes in mammals, a recent study found that lamin B1, a farnesylated and INM-associated protein, is targeted by macroautophagy via blebbing of the nuclear envelope, followed by sequestration of nuclear derived vesicles into autophagosomes.39 In contrast to degradation of progerin and Atg39-dependent nucleophagy in yeast, which were induced by rapamycin, lamin B1 autophagy was induced in response to oncogenic insult, such as activated RAS.39 Oncogene-induced autophagy selectively targets lamin B1, possibly because lamin B1, unlike other lamins, directly binds to LC3.39 Interaction of lamin B1 with LC3 is important for lamin B1 delivery to the cytoplasmic autophagosomes,39 although mechanistic details of this process remain unclear.
Both autophagy of lamin B1 in mammalian cells and Atg39-dependent nucleophagy in yeast involve the generation of vesicles from the NE and their engulfment by autophagosomes. Alternatively the autophagosomal membrane could also be derived from the nuclear envelope itself as described in macrophages and other cells infected with herpes simplex virus-1.72,73 In these studies 4-membrane layered autophagosome-like structures were found to emerge from the NE by coiling of the INM and ONM.72,73 This process was named NE-derived autophagy (NEDA).73 Taken together, although the molecular details of progerin and lamin B1 autophagy are not completely understood, available data clearly show that nuclear envelope components might be subject to autophagic degradation also in mammalian cells.
Concluding remarks
The nucleus is emerging as an important cellular compartment for protein degradation and PQC. Although PQC pathways in the nucleus have long been overlooked, recent discoveries revealed ubiquitin protein ligases that target PQC substrates in the nucleus and at the INM.11,14,17,18 Initially assumed that degradation of nuclear proteins relies entirely on the ubiquitin-proteasome system,10 recent findings show that nuclear material can also be subject to autophagic degradation both in yeast 34-36 and complex eukaryotic cells.37-39
Remarkably, nuclear pathways appear to be responsible for the quality control of not only nuclear, but also cytoplasmic proteins. Besides misfolded nuclear proteins, the nuclear ubiquitin protein ligase San1 also mediates degradation of misfolded cytoplasmic proteins (Fig. 1).74-76 Furthermore, it has recently been shown that a stress-induced protein aggregate deposit in yeast known as JUNQ (“juxtanuclear quality control compartment”) localizes inside the cell nucleus, and has accordingly been redefined as “intranuclear quality control compartment” (INQ, Fig. 1).40 Intriguingly, INQ serves as a deposit for both cytosolic and nuclear misfolded proteins.
Nuclear degradation-dependent PQC mechanisms are particularly important for proper function and survival of long-lived postmitotic cells, such as neurons. Unlike dividing cells, postmitotic cells cannot eliminate accumulated damage by asymmetric segregation or by dilution in cell divisions. Moreover, while in dividing cells the barrier between the nucleus and cytoplasm, the NE, breaks down during each cell division and consequently damaged proteins from the nucleus gain access to the cytoplasmic PQC pathways, postmitotic nuclei lack this possibility. Accordingly, several neurodegenerative diseases are associated with misfolding and aggregation of nuclear proteins, underscoring the importance of nuclear PQC.77-79
Abbreviations
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- HGPS
Hutchinson Gilford Progeria syndrome
- INQ
intranuclear quality control compartment
- INM
inner nuclear membrane
- INMAD
INM-associated degradation
- JUNQ
juxtanuclear quality control compartment
- NE
nuclear envelope
- NPC
nuclear pore complex
- ONM
outer nuclear membrane
- PQC
protein quality control
- PMN
piecemeal microautophagy of the nucleus
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work in the authors' laboratory was supported by grants from the Austrian Science Fund, grant number FWF P23805-B20 (to RF).
References
- 1.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell 2014; 157:52-64; PMID:24679526; http://dx.doi.org/ 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Intracellular protein degradation from a vague idea through the lysosome and the ubiquitin-proteasome system and on to human diseases and drug targeting: Nobel Lecture, December 8, 2004. Ann N Y Acad Sci 2007; 1116:1-28; PMID:18083918 [DOI] [PubMed] [Google Scholar]
- 3.Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J 2012; 279:532-42; PMID:22177318; http://dx.doi.org/ 10.1111/j.1742-4658.2011.08456.x [DOI] [PubMed] [Google Scholar]
- 4.Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett 2010; 584:2635-45; PMID:20412801; http://dx.doi.org/ 10.1016/j.febslet.2010.04.041 [DOI] [PubMed] [Google Scholar]
- 5.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118:7-18; PMID:15615779; http://dx.doi.org/ 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2:211-6; PMID:11265251; http://dx.doi.org/ 10.1038/35056522 [DOI] [PubMed] [Google Scholar]
- 7.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 2011; 7:673-82; PMID:21646866; http://dx.doi.org/ 10.4161/auto.7.7.14733 [DOI] [PubMed] [Google Scholar]
- 8.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 2010; 40:238-52; PMID:20965419; http://dx.doi.org/ 10.1016/j.molcel.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 9.Nielsen SV, Poulsen EG, Rebula CA, Hartmann-Petersen R. Protein quality control in the nucleus. Biomolecules 2014; 4:646-61; PMID:25010148; http://dx.doi.org/ 10.3390/biom4030646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury M, Enenkel C. Intracellular Dynamics of the Ubiquitin-Proteasome-System. F1000Research 2015; 4:367; PMID:26339477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell 2005; 120:803-15; PMID:15797381; http://dx.doi.org/ 10.1016/j.cell.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 12.Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol 2010; 22:1-10; PMID:20102790; http://dx.doi.org/ 10.1016/j.ceb.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 2008; 9:944-57; PMID:19002207; http://dx.doi.org/ 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 2006; 443:827-31; PMID:17051211; http://dx.doi.org/ 10.1038/nature05170 [DOI] [PubMed] [Google Scholar]
- 15.Boban M, Pantazopoulou M, Schick A, Ljungdahl PO, Foisner R. A nuclear ubiquitin-proteasome pathway targets the inner nuclear membrane protein Asi2 for degradation. J Cell Sci 2014; 127:3603-13; PMID:24928896; http://dx.doi.org/ 10.1242/jcs.153163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell 2011; 22:4726-39; PMID:21998200; http://dx.doi.org/ 10.1091/mbc.E11-05-0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science 2014; 346:751-5; PMID:25236469; http://dx.doi.org/ 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- 18.Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, et al.. Protein quality control at the inner nuclear membrane. Nature 2014; 516:410-3; PMID:25519137; http://dx.doi.org/ 10.1038/nature14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mijaljica D, Devenish RJ. Nucleophagy at a glance. J Cell Sci 2013; 126:4325-30; PMID:24013549; http://dx.doi.org/ 10.1242/jcs.133090 [DOI] [PubMed] [Google Scholar]
- 20.Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; PMID:20300205; http://dx.doi.org/ 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547; PMID:20826548; http://dx.doi.org/ 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 2015; 84:131-64; PMID:25747401; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034115 [DOI] [PubMed] [Google Scholar]
- 23.Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp Cell Res 2007; 313:2167-79; PMID:17451680; http://dx.doi.org/ 10.1016/j.yexcr.2007.03.012 [DOI] [PubMed] [Google Scholar]
- 24.Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci 2010; 123:1973-8; PMID:20519579; http://dx.doi.org/ 10.1242/jcs.019042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proceedings of the National Academy of Sciences of the United States of America 1993; 90:99-103; PMID:8380501; http://dx.doi.org/ 10.1073/pnas.90.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J 1998; 17:6144-54; PMID:9799224; http://dx.doi.org/ 10.1093/emboj/17.21.6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugle B, Kleinschmidt JA, Franke WW. The 22 S cylinder particles of Xenopus laevis. II. Immunological characterization and localization of their proteins in tissues and cultured cells. Eur J Cell Biol 1983; 32:157-63; PMID:6667692 [PubMed] [Google Scholar]
- 28.McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol 1997; 137:539-53; PMID:9151663; http://dx.doi.org/ 10.1083/jcb.137.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters JM, Franke WW, Kleinschmidt JA. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem 1994; 269:7709-18; PMID:8125997 [PubMed] [Google Scholar]
- 30.Russell SJ, Steger KA, Johnston SA. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem 1999; 274:21943-52; PMID:10419517; http://dx.doi.org/ 10.1074/jbc.274.31.21943 [DOI] [PubMed] [Google Scholar]
- 31.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol 2003; 35:579-89; PMID:12672451; http://dx.doi.org/ 10.1016/S1357-2725(02)00380-1 [DOI] [PubMed] [Google Scholar]
- 32.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell 2011; 22:2384-95; PMID:21551067; http://dx.doi.org/ 10.1091/mbc.E11-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al.. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 2011; 41:93-106; PMID:21211726; http://dx.doi.org/ 10.1016/j.molcel.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 2003; 14:129-41; PMID:12529432; http://dx.doi.org/ 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mijaljica D, Prescott M, Devenish RJ. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One 2012; 7:e40013; PMID:22768199; http://dx.doi.org/ 10.1371/journal.pone.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015; 522:359-62; PMID:26040717; http://dx.doi.org/ 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- 37.Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med 2011; 3:89ra58. [DOI] [PubMed] [Google Scholar]
- 38.Cenni V, Capanni C, Columbaro M, Ortolani M, D'Apice MR, Novelli G, Fini M, Marmiroli S, Scarano E, Maraldi NM, et al.. Autophagic degradation of farnesylated prelamin A as a therapeutic approach to lamin-linked progeria. Eur J Histochem 2011; 55:e36; PMID:22297442; http://dx.doi.org/ 10.4081/ejh.2011.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al.. Autophagy mediates degradation of nuclear lamina. Nature 2015; 527(7576):105-9; PMID: 26524528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, et al.. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J 2015; 34:778-97; PMID:25672362; http://dx.doi.org/ 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsberg H, Hammar M, Andréasson C, Moliner A, Ljungdahl PO. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 2001; 158:973-88; PMID:11454748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012; 190:885-929; PMID:22419079; http://dx.doi.org/ 10.1534/genetics.111.133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zargari A, Boban M, Heessen S, Andréasson C, Thyberg J, Ljungdahl PO. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J Biol Chem 2007; 282:594-605; PMID:17085444; http://dx.doi.org/ 10.1074/jbc.M609201200 [DOI] [PubMed] [Google Scholar]
- 44.Boban M, Ljungdahl PO. Dal81 enhances Stp1- and Stp2-dependent transcription necessitating negative modulation by inner nuclear membrane protein Asi1 in Saccharomyces cerevisiae. Genetics 2007; 176:2087-97; PMID:17603098; http://dx.doi.org/ 10.1534/genetics.107.075077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boban M, Zargari A, Andréasson C, Heessen S, Thyberg J, Ljungdahl PO. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J Cell Biol 2006; 173:695-707; PMID:16735580; http://dx.doi.org/ 10.1083/jcb.200601011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omnus DJ, Ljungdahl PO. Latency of transcription factor Stp1 depends on a modular regulatory motif that functions as cytoplasmic retention determinant and nuclear degron. Mol Biol Cell 2014; 25:3823-33; PMID:25253722; http://dx.doi.org/ 10.1091/mbc.E14-06-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boban M, Ljungdahl PO, Foisner R. Atypical ubiquitylation in yeast targets lysine-less Asi2 for proteasomal degradation. J Biol Chem 2015; 290:2489-95; PMID:25492870; http://dx.doi.org/ 10.1074/jbc.M114.600593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 2005; 309:127-30; PMID:15994556; http://dx.doi.org/ 10.1126/science.1110340 [DOI] [PubMed] [Google Scholar]
- 49.Carvalho AF, Pinto MP, Grou CP, Alencastre IS, Fransen M, Sa-Miranda C, Azevedo JE. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem 2007; 282:31267-72; PMID:17726030; http://dx.doi.org/ 10.1074/jbc.M706325200 [DOI] [PubMed] [Google Scholar]
- 50.Domingues C, Ryoo HD. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ 2012; 19:470-7; PMID:21886178; http://dx.doi.org/ 10.1038/cdd.2011.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem 2010; 285:23916-24; PMID:20519503; http://dx.doi.org/ 10.1074/jbc.M110.127936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci 2012; 125:539-48; PMID:22389393; http://dx.doi.org/ 10.1242/jcs.093567 [DOI] [PubMed] [Google Scholar]
- 53.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 2010; 40:917-26; PMID:21172657; http://dx.doi.org/ 10.1016/j.molcel.2010.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol 2007; 177:613-24; PMID:17502423; http://dx.doi.org/ 10.1083/jcb.200611063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 2007; 282:22534-43; PMID:17550898; http://dx.doi.org/ 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- 56.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 1998; 94:217-27; PMID:9695950; http://dx.doi.org/ 10.1016/S0092-8674(00)81421-X [DOI] [PubMed] [Google Scholar]
- 57.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 2006; 25:533-43; PMID:16437165; http://dx.doi.org/ 10.1038/sj.emboj.7600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy 2009; 5:795-804; PMID:19550147; http://dx.doi.org/ 10.4161/auto.8901 [DOI] [PubMed] [Google Scholar]
- 59.Candelario J, Borrego S, Reddy S, Comai L. Accumulation of distinct prelamin A variants in human diploid fibroblasts differentially affects cell homeostasis. Exp Cell Res 2011; 317:319-29; PMID:20974128; http://dx.doi.org/ 10.1016/j.yexcr.2010.10.014 [DOI] [PubMed] [Google Scholar]
- 60.Capanni C, Del Coco R, Squarzoni S, Columbaro M, Mattioli E, Camozzi D, Rocchi A, Scotlandi K, Maraldi N, Foisner R, et al.. Prelamin A is involved in early steps of muscle differentiation. Exp Cell Res 2008; 314:3628-37; PMID:18951892; http://dx.doi.org/ 10.1016/j.yexcr.2008.09.026 [DOI] [PubMed] [Google Scholar]
- 61.Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proceedings of the National Academy of Sciences of the United States of America 2007; 104:4955-60; PMID:17360326; http://dx.doi.org/ 10.1073/pnas.0700854104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prokocimer M, Barkan R, Gruenbaum Y. Hutchinson-Gilford progeria syndrome through the lens of transcription. Aging Cell 2013; 12:533-43; PMID:23496208; http://dx.doi.org/ 10.1111/acel.12070 [DOI] [PubMed] [Google Scholar]
- 63.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963-6; PMID:9461583; http://dx.doi.org/ 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- 64.Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol 1999; 155:135-42; PMID:10580846; http://dx.doi.org/ 10.1016/S0303-7207(99)00121-5 [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci 2011; 124:161-70; PMID:21187343; http://dx.doi.org/ 10.1242/jcs.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, et al.. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy 2011; 7:1273-94; PMID:21997368; http://dx.doi.org/ 10.4161/auto.7.11.17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rose A, Schlieker C. Alternative nuclear transport for cellular protein quality control. Trends Cell Biol 2012; 22:509-14; PMID:22858153; http://dx.doi.org/ 10.1016/j.tcb.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al.. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 2012; 149:832-46; PMID:22579286; http://dx.doi.org/ 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mettenleiter TC, Muller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 2013; 15:170-8; PMID:23057731; http://dx.doi.org/ 10.1111/cmi.12044 [DOI] [PubMed] [Google Scholar]
- 70.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16:495-501; PMID:24875736; http://dx.doi.org/ 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- 71.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 72.English L, Chemali M, Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy 2009; 5:1026-9; PMID:19556870; http://dx.doi.org/ 10.4161/auto.5.7.9163 [DOI] [PubMed] [Google Scholar]
- 73.Radtke K, English L, Rondeau C, Leib D, Lippe R, Desjardins M. Inhibition of the host translation shutoff response by herpes simplex virus 1 triggers nuclear envelope-derived autophagy. J Virol 2013; 87:3990-7; PMID:23365427; http://dx.doi.org/ 10.1128/JVI.02974-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proceedings of the National Academy of Sciences of the United States of America 2010; 107:1106-11; PMID:20080635; http://dx.doi.org/ 10.1073/pnas.0910591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 2010; 21:2117-27; PMID:20462951; http://dx.doi.org/ 10.1091/mbc.E10-02-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guerriero CJ, Weiberth KF, Brodsky JL. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem 2013; 288:18506-20; PMID:23653356; http://dx.doi.org/ 10.1074/jbc.M113.475905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 1998; 95:41-53; PMID:9778246; http://dx.doi.org/ 10.1016/S0092-8674(00)81781-X [DOI] [PubMed] [Google Scholar]
- 78.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 1997; 90:537-48; PMID:9267033; http://dx.doi.org/ 10.1016/S0092-8674(00)80513-9 [DOI] [PubMed] [Google Scholar]
- 79.Woulfe JM. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol Appl Neurobiol 2007; 33:2-42; PMID:17239006 [DOI] [PubMed] [Google Scholar]


