Abstract
Single-molecule techniques can monitor the kinetics of transitions between enzyme open and closed conformations, but such methods usually lack the resolution to observe the underlying transition pathway or intermediate conformational dynamics. We have used a 1 MHz-bandwidth carbon nanotube transistor to electronically monitor single molecules of the enzyme T4 lysozyme as it processes substrate. An experimental resolution of 2 µs allowed the direct recording of lysozyme’s opening and closing transitions. Unexpectedly, both motions required 37 µs on average. The distribution of transition durations was also independent of the enzyme’s state, either catalytic or non-productive. The observation of smooth, continuous transitions suggests a concerted mechanism for glycoside hydrolysis with lysozyme’s two domains closing upon the polysaccharide substrate in its active site. We distinguish these smooth motions from a non-concerted mechanism, observed in approximately 10% of lysozyme openings and closings, in which the enzyme pauses for an additional 40 to 140 µs in an intermediate, partially closed conformation. During intermediate forming events, the number of rate limiting steps observed increases to four, consistent with four steps required in the step-wise, arrow-pushing mechanism. The formation of such intermediate conformations was again independent of the enzyme’s state. Taken together, the results suggest lysozyme operates as a Brownian motor. In this model, the enzyme traces a single pathway for closing and the reverse pathway for enzyme opening, regardless of its instantaneous catalytic productivity. The observed symmetry in enzyme opening and closing thus suggests that substrate translocation occurs while the enzyme is closed.
Graphical abstract
INTRODUCTION
Regulatory and catalytic functions of enzymes involve dynamic structural changes over a wide range of timescales. The mechanisms for protein transitions between conformational changes are often poorly understood, especially in the milli- to microsecond ranges where conventional experimental techniques and computational modeling are most limited.1 For example, only the most advanced molecular dynamics simulations can approach millisecond time-scales.2–4 Forster resonance energy transfer (FRET) experiments can monitor the motions of an enzyme,5–10 but the photon flux of the brightest FRET fluorophores limits the technique to observing movements on the timescale of 100 µs. State-of-the-art measurements using FRET and fluorescence correlation spectroscopy (FCS) monitoring many thousands of proteins have identified rare events that establish upper limits on the duration of time required for the folding of the protein α3D;11–14 however, such measurements fall short of continuous monitoring or direct observation.
Electronic single-molecule techniques using nanoscale transistors,15–19 tunnel junctions,20 or nanopores21, 22 provide opportunities to overcome some of the limitations of fluorescence-based techniques. For example, one electronic approach recently accomplished single-molecule monitoring of enzymatic catalysis by taking advantage of an enzyme’s electrostatic effects upon an underlying transistor channel.17, 19 In studies of T4 lysozyme, this electronic technique17–19 matched previous, single-molecule FRET experiments23–26 in measuring the timing and statistics of lysozyme’s open and closed conformations as it processed its peptidoglycan substrate. In principle, the higher temporal resolution possible with solid-state electronics promises improved recordings of much briefer single-molecule events, intermediate states, and information-rich fluctuations. This promise has been demonstrated using the fluctuations of an electronic tunnel junction to distinguish closely related amino acids and peptides.20
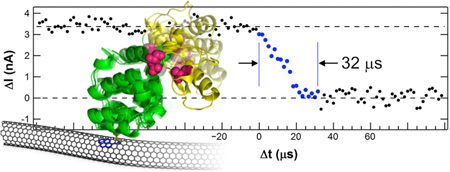
Here, we report electronic, single-molecule monitoring of T4 lysozyme with a resolution of 2 µs. This resolution allowed direct observation of lysozyme’s motions during transitions between its open and closed conformations. The mechanical motion from one conformation to the other required 37 µs on average. Furthermore, this transition time does not depend on the direction of motion nor on whether it occurred during catalysis or non-productive motions. Approximately 10% of the time, additional motions were observed in which long pauses during either closure or reopening extended the duration by 40 to 140 µs on average.
METHODS
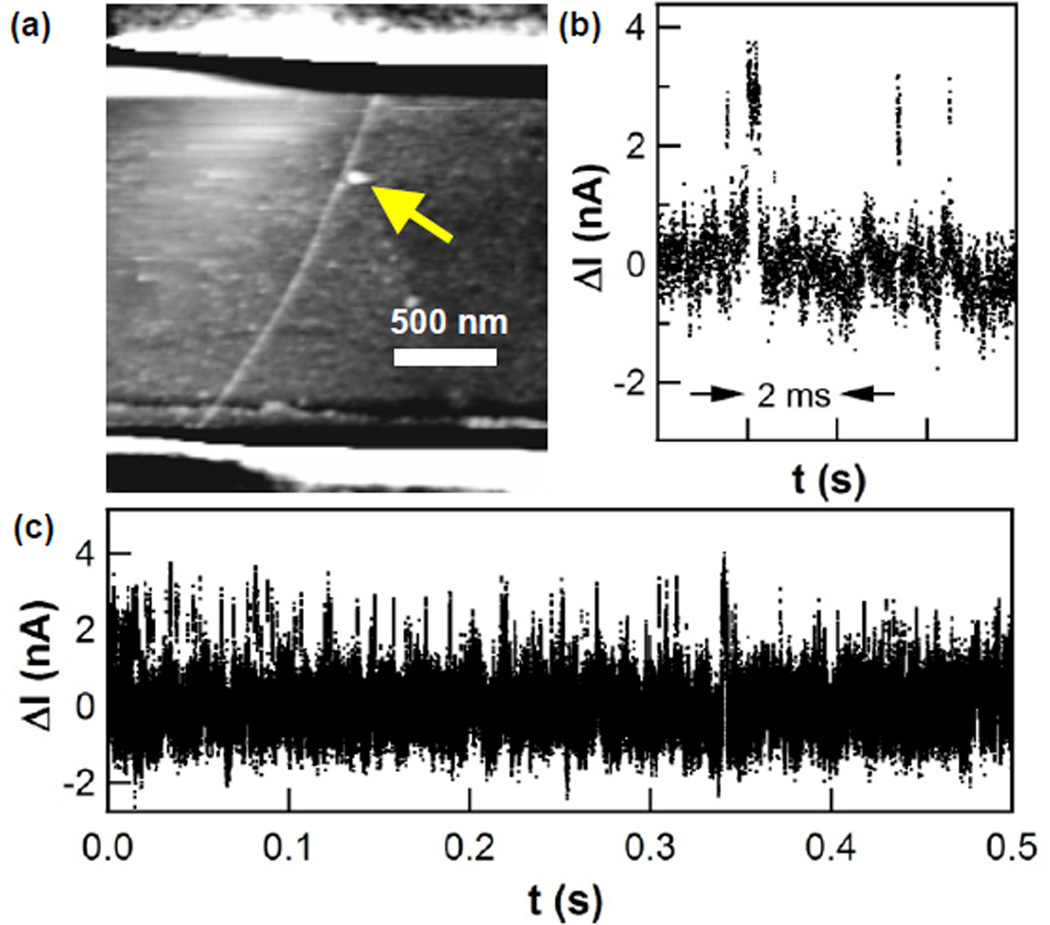
The methods of this experiment closely followed the protocols described in previous publications.17–19 Briefly, a pseudo wild-type variant of T4 lysozyme27 with the substitutions C54T/C97A/S90C, termed lysozyme here, was expressed in E. coli and then purified by cation exchange and size exclusion chromatography. The single cysteine substitution at residue 90 allowed facile and reproducible surface bioconjugation to the maleimide groups of N-(1-pyrenyl)maleimide linker molecules. The pyrene linkers adhered noncovalently to isolated single-walled carbon nanotubes (SWNTs)28 that were individually connected in field effect transistor (FET) configurations. The SWNT FET was first soaked in a solution of the pyrenemaleimide linkers (1 mM in ethanol, 30 min), rinsed to remove many of the linkers (5 min in ethanol with 0.1% Tween-20, then 5 min in deionized water), and then incubated for 120 min in a 54 µM solution of lysozyme in phosphate-buffered saline (PBS; 138 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.5). Finally, the device was rinsed with wash buffer (PBS with 0.05% Tween-20) to remove proteins nonspecifically adsorbed on the SiO2 surface. Figure 1a shows an example device imaged by atomic force microscopy, in which the single molecule attachment is clearly resolved. Fabrication details are described more completely in the Supporting Information (SI).
Figure 1.
Lysozyme-nanotube devices. (a) Atomic force micrograph of a single lysozyme protein attached to an electrically connected SWNT. Source and drain electrodes lay under a passivating polymer, seen as stripes across the top and bottom of the image. (b) Individual source-drain current fluctuations ΔI(t) observed when lysozyme is monitored in the presence of peptidoglycan, and (c) a representative stochastic series of such fluctuations.
Once bioconjugated, the lysozyme-SWNT devices were electrically monitored for 10 or 20 min in PBS solutions with and without peptidoglycan (Sigma-Aldrich).29 The electrolyte potential was held constant at a bias of −0.6 to −0.7 V (versus a Ag/AgCl reference electrode), which was chosen to maximize the signal-to-noise ratio for each device. 100 mV was applied across the source and drain terminals to drive a DC current I(t) in the SWNT channel. Fluctuations in this current were monitored using an externally mounted, 200 kHz preamplifier (Femto DLPCA-200) sampled at 600 kHz. A rise time of approximately 2 µs, which was 20 times shorter than reported in previous measurements,17–19 was realized by carefully limiting the system’s parasitic capacitances. The PBS solution was confined within a 100 µm wide microfluidic channel contained within a PDMS gasket and optically aligned over the SWNT active region, as previously developed for similar nanowire devices.30–32
RESULTS and DISCUSSION
Without peptidoglycan substrate in the surrounding electrolyte buffer, lysozyme remains in its open conformation.23–26 The SWNT current I(t) for a lysozyme-conjugated nanocircuit is featureless around a steady baseline. Furthermore, lysozyme variants with mutated active site residues known to abrogate enzymatic catalysis also result in a featureless I(t).15 After the addition of peptidoglycan, I(t) exhibits stochastic pulse trains of brief excursions to a higher current level. The excursions were analyzed by focusing on the excess current ΔI(t) above the baseline (Figures 1b,c). We observed these excursions to have all of the characteristics described previously.17–19 Specifically, excursions occurred at one of two average rates: either 20–50 s−1 corresponding to lysozyme’s processive hydrolysis of substrate, or else 200–400 s−1 corresponding to nonproductive motions in which lysozyme processivity and catalysis becomes blocked by peptidoglycan cross-links.18, 23, 25 We define these two possible ranges of activity as the catalytic state or the nonproductive state of lysozyme, respectively.
In either state, each transition of lysozyme to its closed conformation moved the positively charged sidechains Lys83 and Arg119 away from the SWNT. Immersed in an aqueous environment, the two charged sidechains, their appropriate counterions, and the liquid rearrange in sync with the protein’s motion. The FET records motions of this protein-ion system within the 1-nm Debye screening radius.19 Each time the enzyme closes, an 8 Å hinge motion brings the two lobes of the enzyme closer, swings the positively charged residues away from the SWNT, and electrostatically gates the electrical channel to produce the ΔI(t) signals analyzed here.
On average, such ΔI(t) increases were identical in magnitude, suggesting the enzyme sidechains moved the same distance each time it opened or closed upon substrate. Re-opening of lysozyme reversed the motions and returned I(t) to its baseline value. The mean duration of each excursion ΔI(t) was interpreted as the duration of the closed configuration, τclosed, and the pause between excursions was interpreted as the duration of the open configuration, τopen. Single-molecule distributions of τclosed and τopen followed Poisson statistics and had mean durations that were in excellent agreement with previous FRET studies of T4 lysozyme.23–26
Increasing the sampling rate of I(t) and the effective bandwidth of the SWNT device immediately resolved internal structure in the rising and falling transitions between open and closed conformations. With an experimental resolution increased to 2 µs, we indirectly observed observed the finite duration of transitions between the two conformations. These durations had a broad distribution that included transitions that occurred faster than 2 µs, transitions lasting over 100 µs, and transitions interrupted by long pauses in the middle of conformational changes. Here, we restrict attention to analysis of the transition durations, having analyzed τclosed and τopen in previous publications.17–19
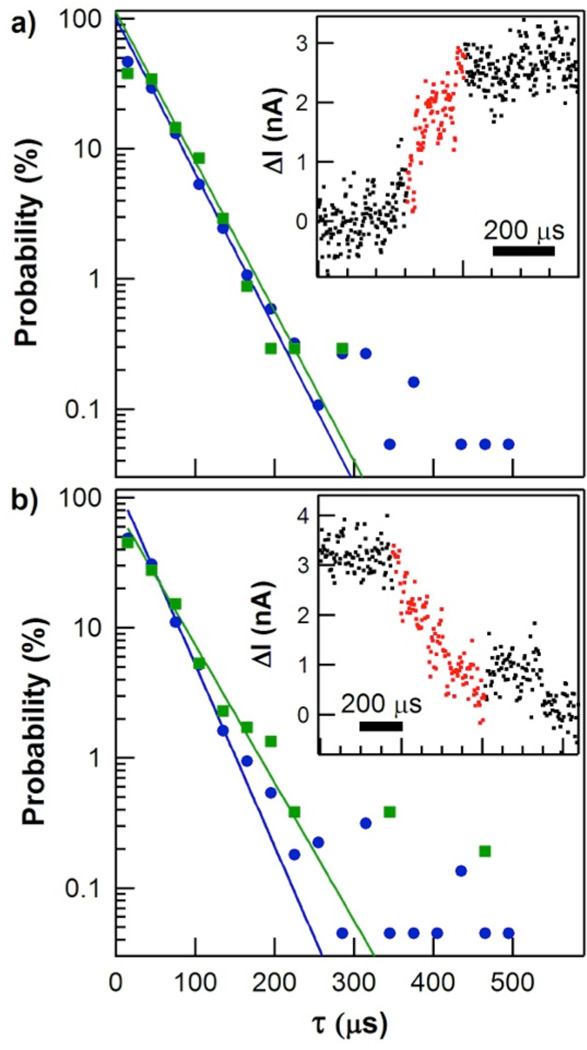
Figure 2a shows the probability distribution for the duration τclosing of transitions from the open to the closed conformation. One example closure event is shown in the inset (additional example events are plotted in the SI, Figure S1). We defined τclosing by defining local average values of ΔI(t) surrounding a single closure event and then measuring the duration for the signal to rise from 20% to 80% of this local average height. Similar measurements using 30% and 70% thresholds led to proportionally shorter τclosing values, whereas expanding the thresholds to 10% and 90% (the traditional criteria for defining rise and fall times in signal processing) led to very long values corrupted by the limited signal-to-noise ratio of the ΔI(t) signal. Other than the narrower thresholds used, the measurements of τclosing were reproduced by independent rise-time measurement algorithms. The τclosing distribution shown here results from semiautomated analysis of over 600 seconds of ΔI(t) records collected with three different lysozyme-SWNT devices, each of which gave the same results when analyzed independently.
Figure 2.
Probability distributions for the durations of hinge-bending motions. (a) Closing durations recorded during periods of catalytic processing (green) or nonproductive binding (blue). Inset shows an example ΔI(t) signal for a single closing event, with the portion used to calculate the transition’s duration highlighted in red. (b) Opening durations and example event. Solid lines depict single-exponential fits to each distribution. Note that examples shown are significantly longer than the distribution averages.
On average, lysozyme spent 8±3 consecutive seconds in either a slow, catalytically productive state or a fast, nonproductive state before switching states or becoming inactive.17, 18, 24 Consequently, a second-by-second average rate of closure events allowed us to separate individual events into two states of activity (catalytic or nonproductive) and to compare their probability distributions. As shown in Figure 2a, the τclosing distribution during catalytically productive closures (green) was statistically identical to the distribution for events during catalytically nonproductive, higher frequency closures (blue). Both distributions were semi-logarithmic with mean durations of approximately 37 µs. Table 1 gives precise values of each slope τ with its uncertainty (one standard deviation) as determined from a least-squares fit. Three lysozyme molecules measured with the new, high-bandwidth technique gave identical behaviors and were all consistent with lower-resolution measurements performed on dozens of single lysozyme devices.
Table 1.
Timing and energetics of T4 lysozyme closing and opening
| Conformation | Catalytically active | Nonproductive | |||
|---|---|---|---|---|---|
| τ (µs) | ΔE (kcal/mol) | τ (µs) | ΔE (kcal/mol) | ||
| open | 52,330 ± 2,290 | 0 | 2,670 ± 330 | 0 | |
| closinga | (no pause) | 38 ± 2 | 37 ± 1 | ||
| (with pause) | 81 ± 27 | 3.84 ± 0.20 | 111 ± 10 | 1.89 ± 0.09 | |
| closed | 589 ± 61 | 2.66 ± 0.07 | 176 ± 11 | 1.61 ± 0.08 | |
| openinga | (no pause) | 41 ± 5 | 31 ± 1 | ||
| (with pause) | 181 ± 54 | 3.36 ± 0.18 | 135 ± 24 | 1.77 ± 0.13 | |
As defined using 20% and 80% threshold criteria. Full transition times are up to 40% longer.
Figure 2b depicts the two probability distributions for the duration τopening of transitions from the closed conformation back to the open conformation. As with τclosing, the τopening distributions for the two enzyme conformations were statistically identical and did not depend on the enzyme’s state, either catalytic or non-productive. Furthermore, the mean value of τopening was the same as for τclosing. Inspection of the example transitions shown in the insets to Figure 2 and in Figure S2 indicate that the appearance and timing of lysozyme’s opening events were virtually identical to its closing events.
Thus, both opening and closing motions are driven by a common energy scale, and the speed of lysozyme’s motions is almost identical during high frequency nonproductive motions and less frequent catalytic ones. These findings are consistent with Kramers’ theory of transition kinetics,33 which expresses the duration of transition in terms of fundamental energy scales and implies that different transition paths should proceed at the same rate once initiated over a reaction barrier.34 In fact, proteins with vastly different folding rates can exhibit identical transition durations, a prediction experimentally verified by Chung et.al. using FRET.12, 13 In both protein folding and lysozyme closure, the duration is determined by dissipative interactions such as friction between sidechains surrounding lysozyme’s active site and the peptidoglycan substrate. These interactions manifest themselves in the frequency distributions of ΔI(t), but the approach described by Chung et. al.12 provides a simple method of estimating transition times directly from the values in Table 1. Taking τopen−1 as an upper bound for the Kramer’s rate of barrier crossing and requiring the energy barrier to be no less than the energy difference ΔE between open and closed conformations, theory predicts transition durations in the range of 10 to 200 µs, in good agreement with the experimental data.
Unlike protein folding, the duration of lysozyme’s motions is too transient to have been studied by ensemble methods like fluorescence or X-ray crystallography, and it is too long to be addressed by either NMR or typical first-principles simulations. For example, multiscale molecular dynamics modeling of lysozyme has proposed a <1 ns transition path through six protein conformations,23, 35 but this duration may reflect the limitations of simulation time more than true protein dynamics. The best comparable data comes from FCS, which arguably has the best time resolution among single-molecule techniques. Yirdaw and McHaourab applied FCS to T4 lysozyme and detected a 15 µs “relaxation time” between its open and closed conformations.14 This FCS measurement is in good agreement with the 37 µs average reported here, especially if one considers that FCS cannot resolve the frequent, short events or the very rare, long-lived events both contained in the full distributions (Figure 2). By directly observing both extremes, our electronic technique provides a much fuller representation of the range of lysozyme’s speeds. Nevertheless, we note that the most probable τclosing and τopening durations approach our experimental resolution of 2 µs. While the distributions in Figure 2 show hints of a roll-off at the shortest durations, example events in the SI demonstrate that further improvements are necessary to accurately resolve the shortest events and the true maximum speed of lysozyme’s motions.
The rare occurrence of long-duration events in both distributions led us to investigate possible correlations between τopening and τclosing, or between these transition durations and the times τclosed and τopen spent immediately before or after a transition. The four τ values were found to be independent of each other and uncorrelated in every way, at least within the statistical limits provided by a few thousand events. FCS suggests that much larger data sets, such as those obtained by monitoring many molecules, might be necessary to reveal weak correlations amongst rare events.14
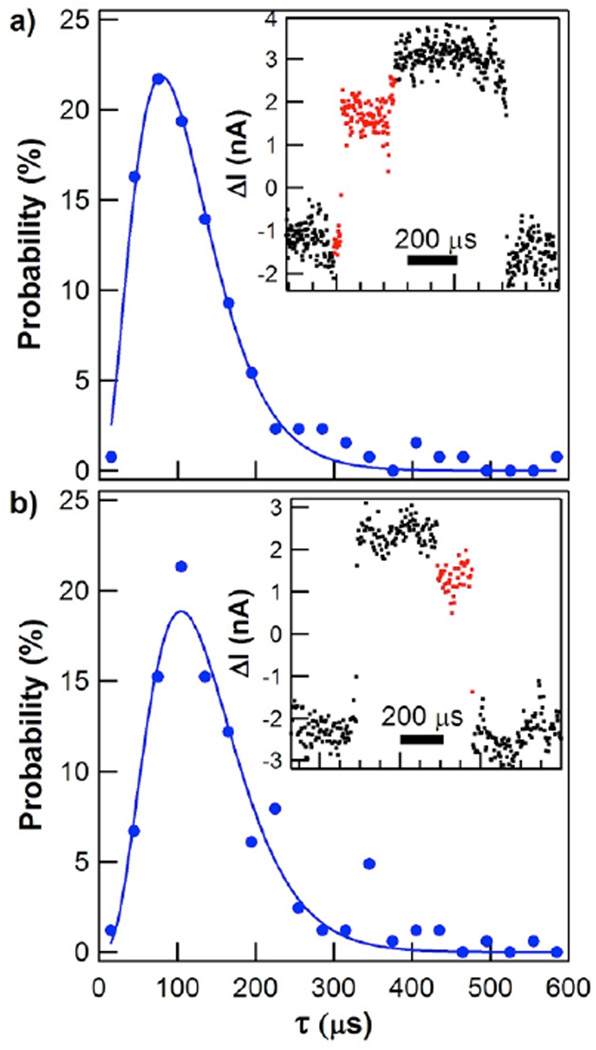
The smooth and continuous transitions analyzed in Figure 2 accounted for 90% of the transitions between open and closed conformations. The remaining 10% of events contained a single pause of 40 to 140 µs in the middle of the transition, leading to total transition times of 80 to 180 µs. Example transitions and their probability distributions are shown for closing (Figure 3a) and opening (Figure 3b). The transitions shown here are representative, but we note that the ΔI(t) levels of different pauses were observed to be randomly distributed throughout the entire 20–80% range of enzyme opening and closing. Because of the relative rarity of these events, their probability distributions had limited statistics. For clarity, the data sets in Figure 3 have not been separated into catalytic and nonproductive states, but an analysis of the catalytic and the nonproductive subsets is included in Table 1. When the closing transition was interrupted by a pause, the average duration increased from 37 µs to about 110 µs. The opening transitions paused even longer, with an average duration of 135 µs. Catalytically active cycles occurred 10 to 20 times less frequently than nonproductive ones, so the typical data set of catalytic motions with pauses contained fewer than 100 events. Within these limitations, catalytic activity did not significantly affect the pause duration compared to nonproductive motions.
Figure 3.
Probability distributions for the durations of hinge-bending motions when long-lived pauses occur. (a) Closing durations, with example signal for a single event (inset). (b) Opening durations and example event. Solid lines depict fits to gamma distributions.
Because the SWNT FET converts the motion of Lys83 and Arg119 sidechains to an electrical signal, the pauses indicate an interruption of these sidechains’ motions. Since these motions are allosterically linked to the entire protein structure, we interpret the interruptions to indicate that lysozyme is pausing in some conformation between its fully-open and fully-closed conformations.36 Furthermore, the durations of these pauses imply complex kinetics among multiple possible conformations. Solid lines in Figure 3 depict fits to the gamma probability distribution for N consecutive processes
where k is the shared rate of all N processes and Γ is the gamma function.37 Fitting the data to the gamma distribution set an upper bound on N, whereas the statistical variance σ2 of the same data determined a lower bound σ2=<τ>2/N.7, 38–41 In this manner, we determined closing to involve at least 2 but no more than 4 processes during those events which included an intermediate pause. Opening required 3 to 5 processes in series, an increase by one step that is consistent with the longer pauses during τopening. Bhabha et. al. have proposed10 that intermediate steps can play crucial roles in enzyme catalysis, but the intermediate step seen here seems to be independent of chemical activity. For comparison, variances of the smooth and continuous transitions analyzed in Figure 2 gave N=1.0±0.1, indicative of a single step or concerted process with no evidence of rate-limiting intermediates.
This statistical analysis is consistent with FCS observations of T4 lysozyme, which noted a range of brightnesses attributed to multiple intermediate states.14 However, the ΔI(t) signals corresponding to N>1 never contained two or more distinct levels. The long-lived pause in lysozyme’s motion was always associated with a single, intermediate configuration, even though statistical analysis indicated multiple consecutive processes. We hypothesize that the single plateau during the pause may represent a non-concerted mechanistic pathway during which protein is “stuck” in a partial conformation while waiting for the correct alignment of functional groups to complete a conformational change.
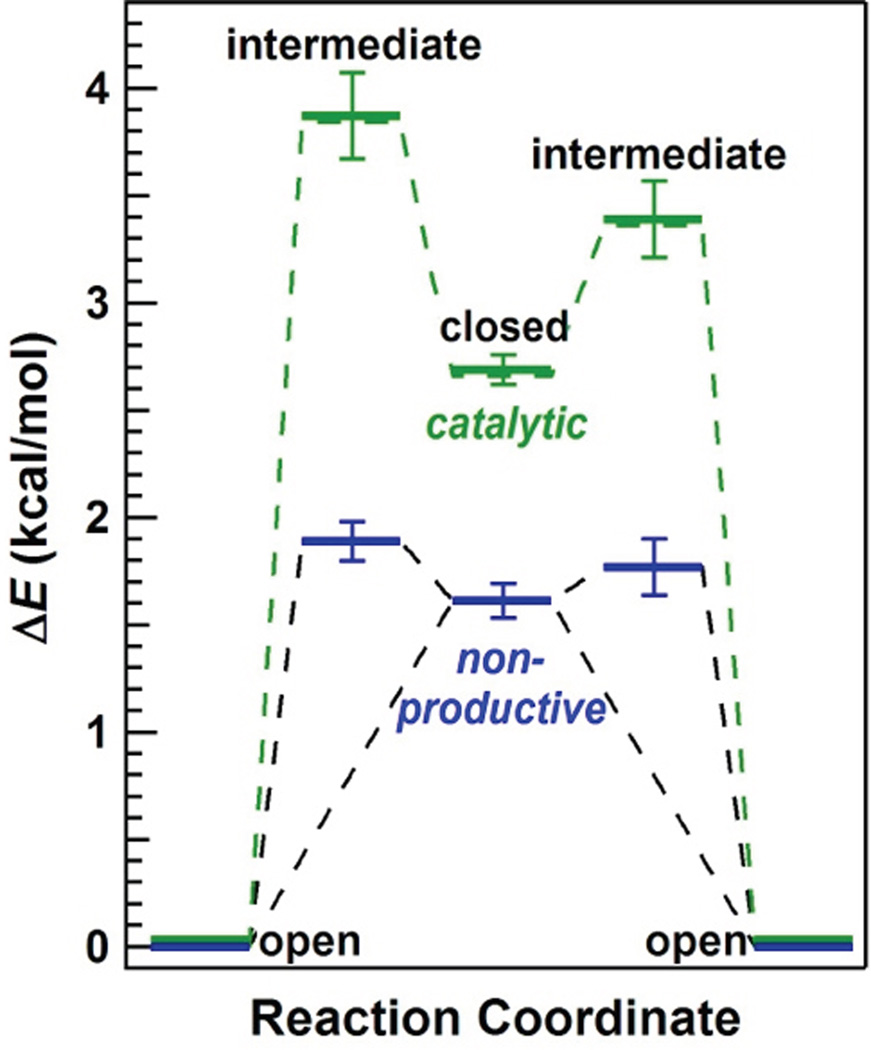
Figure 4 depicts a free-energy landscape for lysozyme’s primary hinge motions estimated from Boltzman statistics by comparing the durations of <τopen> to <τclosed>.7 Lysozyme’s nonproductive cycles occur during closures upon nonhydrolyzable cross-links in the peptidoglycan (blue), whereas catalytically active motions occur when lysozyme closes upon a hydrolyzable glycosidic bond between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) (green). Assuming that the open conformation provides a reference energy level that is insensitive to the peptidoglycan structure at the active site, the relative energy level ΔE of the closed conformation can be estimated using the Boltzman distribution of a two-state system, ΔE = kBT ln(<τclosed>/ <τopen>). As in the previous report, the closed configurations are at 1.6 and 2.6 kcal/mol for nonproductive and catalytic closures, respectively.17 The two closed conformations differ by 1.0 kcal/mol.17 Energetically, this difference is within the range expected for the formation of an additional hydrogen bond at the active site as a result of successful hydrolysis. The two- state model does not apply if lysozyme’s motions are limited by more substantial energy barriers; but FRET experiments observe lysozyme accessing a range of conformations, including the closed conformation, at high frequency and without any apparent stabilization when substrate is absent.26
Figure 4.
Free-energy landscape of T4 lysozyme. Single-molecule kinetics determine the energies of conformational states during catalytic processing (green) and nonproductive binding (blue). Direct transitions between the open and closed conformations occurred in 90% of events, but the remaining 10% of events paused in an intermediate, partially closed conformation. Energetic barriers exist between every conformational state but have been excluded for clarity.
An estimate for the energy level of intermediate configurations was added to this basic diagram using the mean pause durations observed in each type of cycle. Again, assumptions have been made that the observed waiting times are limited by thermodynamics rather than other, rate-limiting barriers. During nonproductive motions, the intermediate configuration was only slightly higher in energy than the closed configuration, by either 0.28±0.09 kcal/mol (closing) or 0.16±0.13 kcal/mol (opening). Essentially, the energy levels for these closing and opening pauses were statistically identical, suggesting a single unique conformation that was nearly isoenergetic with the nonproductive closed conformation. During catalytically productive cycles, the intermediate pauses occurred at much higher energies (3.4 to 3.8 kcal/mol) than the closed conformation.
The identical distributions of τopening and τclosing, independent of catalytic state, is unlikely to be coincidental. Taken together with a 10% propensity to form intermediates during opening and closing in both states, the results suggest the presence of symmetry in lysozyme’s hinge-bending motions. Such symmetry of motions has been invoked to explain the movements of Brownian ratchets, which rely on constrained motions and thermal energy to power their movement along a substrate.42, 43 In fact, lysozyme processively steps along a polysaccharide substrate to consecutively hydrolyze hundreds of glycosidic bonds,17 analogously to a Brownian motor or similar molecular machine.
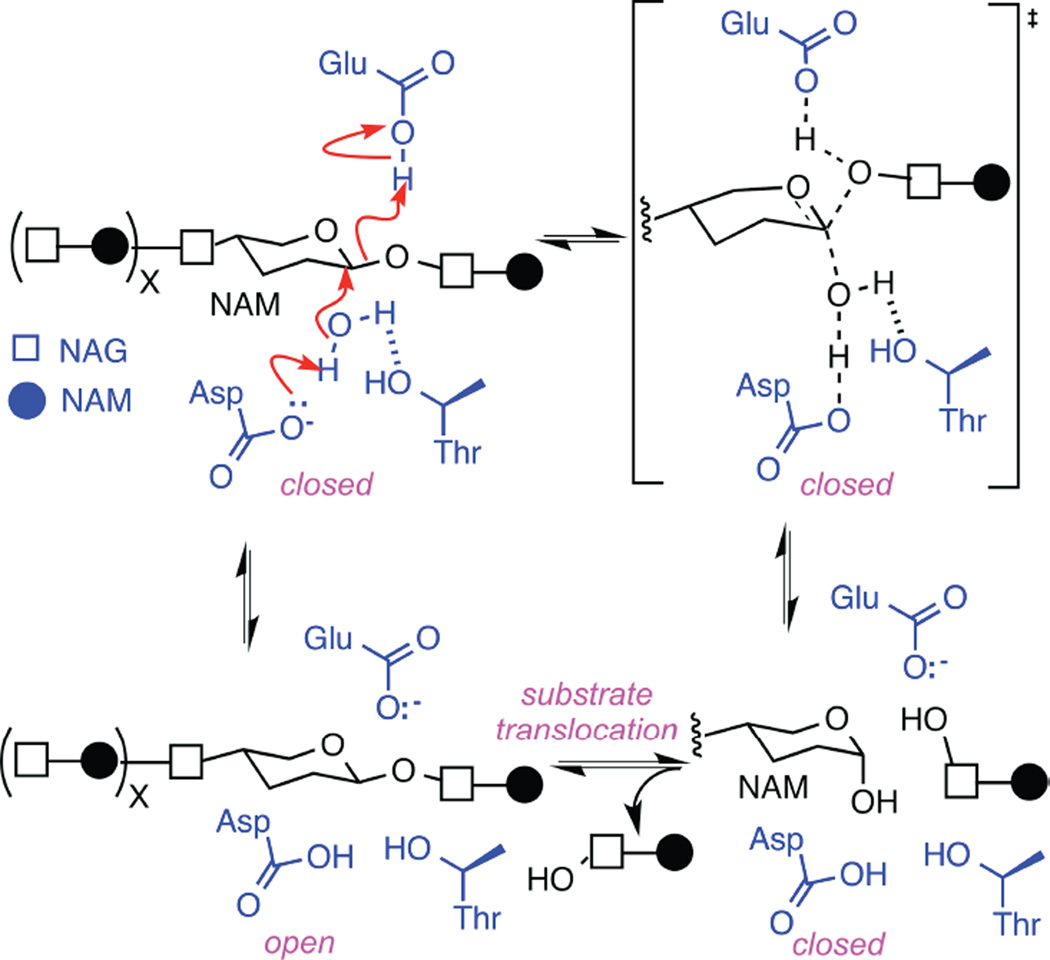
For this symmetry of motions to apply to lysozyme, the enzyme must open and close upon identical substrates. This condition is clearly satisfied by non-productive motions, during which the enzyme opens and closes upon a peptdioglycan cross-link without catalyzing any change in the substrate.15 Productive motions, on the other hand, hydrolyze one glycosidic bond during each closure. For the enzyme to open upon identical substrates after this chemical modification, the enzyme needs to move along the polysaccharide to the next glycosidic bond before reopening. Thus, the kinetic evidence supports a model in which the enzyme closes, hydrolyzes the substrate, translates to the next bond, and then opens. Consequently, we conclude that lysozyme operates as a Brownian motor. The 1.0 kcal/mol liberated in each catalytic cycle provides the energy for translocation along the polysaccharide. Thermal energy could be powering enzyme closing and opening along a constrained pathway. The fact that motions are disorganized until the enzyme binds its substrate23, 24 indicates that the substrate provides some essential structural feature, which could be identified by further modeling and structural studies. The catalytic mechanism of lysozyme is well established,44 and is depicted in Figure 5. The data described here suggests the enzyme closes and opens upon an identical glycosidic bond, which allows assignment of the enzyme open and closed conformations to this mechanism.
Figure 5.
Hydrolysis of glycosidic bonds catalyzed by T4 lysozyme. The glycosidic bond between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) subunits are depicted with one water molecule (black) surrounded by lysozyme’s key active-site functionalities Glu11, Asp20, and Thr26 (blue). The concerted mechanism shown with red arrows is observed in 90% of enzyme closures. Interruption of any step leads to the rarer, non-concerted mechanism, and causes the enzyme to pause in an intermediate, higher energy state. As described in the text, identical motions during re-opening suggest that the enzyme remains in the closed conformation during substrate translocation to an identical NAM-NAG glycosidic bond.
The formation of a rare intermediate conformation comprised 10% of enzyme openings and closings in both catalytic and non-productive states. Assuming this intermediate does result from heterogeneity in the polysaccharide substrate, the intermediate need not require an alternate conformational trajectory. Instead, we believe observations of the intermediate illustrates cases where the concerted mechanism of Figure 5 is converted into a multistep, non-concerted mechanism with higher transition state energies. The variable N matches the number of arrows drawn for the non-concerted reaction in the upper left of Figure 5. After any arrow, a pause to allow repositioning of sidechains or other reaction participants would result in the intermediate conformation being observed. The range of ΔI(t) heights for the observed intermediates suggests that no single step in the mechanism presents a special bottleneck. Furthermore, the symmetry of motions observed here could result from similar conformational pathways for enzyme closing and opening; thus, if the enzyme forms an intermediate 10% of the time while closing, the intermediate is also likely to occur during the constrained opening events. As noted above, we observe the intermediate occurring in both directions during both catalysis and non-productive motions, demonstrating the extent of conformational constraints for this Brownian motor.
CONCLUSION
In summary, we have demonstrated a generalizable approach to high-speed, single-molecule experiments. The faster time resolution directly resolved lysozyme’s motions and the presence of intermediate conformations interrupting these motions. Analysis of these motions strongly suggests that lysozyme is a Brownian motor. Other processive glycosidases (e.g., cellulase) could operate using similar mechanisms. The high temporal resolution experiments described here could guide the development of improved inhibitors, therapeutics, and engineered enzymes.
Supplementary Material
Acknowledgments
We thank J.F. Allard, J.S. Nowick, and S. Majumdar for helpful suggestions on this work. This research was supported financially by the NIH NCI (R01 CA133592-01), the NIH NIGMS (1R01GM106957-01) and the NSF (DMR-1104629 and ECCS-1231910).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Jensen MØ, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of Voltage Gating in Potassium Channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 3.Silva D-A, Weiss DR, Pardo Avila F, Da L-T, Levitt M, Wang D, Huang X. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma W, Schulten K. Mechanism of substrate translocation by a ring-shaped ATPase motor at millisecond resolution. J. Am. Chem. Soc. 2015 doi: 10.1021/ja512605w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha T, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. U.S.A. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu HP, Xun LY, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 7.Xie SN. Single-molecule approach to enzymology. Single Molecules. 2001;2:229–236. [Google Scholar]
- 8.Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Fluctuating Enzymes: Lessons from Single-Molecule Studies. Acc. Chem. Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 9.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. A Dynamic Knockout Reveals That Conformational Fluctuations Influence the Chemical Step of Enzyme Catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung HS, McHale K, Louis JM, Eaton WA. Single-Molecule Fluorescence Experiments Determine Protein Folding Transition Path Times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung HS, Eaton WA. Single-molecule fluorescence probes dynamics of barrier crossing. Nature. 2013;502:685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yirdaw RB, McHaourab HS. Direct Observation of T4 Lysozyme Hinge-Bending Motion by Fluorescence Correlation Spectroscopy. Biophys. J. 2012;103:1525–1536. doi: 10.1016/j.bpj.2012.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorgenfrei S, Chiu C-y, Gonzalez RL, Yu Y-J, Kim P, Nuckolls C, Shepard KL. Label-free single-molecule detection of DNA-hybridization kinetics with a carbon nanotube field-effect transistor. Nat. Nanotechnol. 2011;6:126–132. doi: 10.1038/nnano.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorgenfrei S, Chiu C-y, Johnston M, Nuckolls C, Shepard KL. Debye Screening in Single-Molecule Carbon Nanotube Field-Effect Sensors. Nano Lett. 2011;11:3739–3743. doi: 10.1021/nl201781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi Y, Moody IS, Sims PC, Hunt SR, Corso BL, Weiss GA, Collins PG. Single-Molecule Lysozyme Dynamics Monitored by an Electronic Circuit. Science. 2012;335:319–324. doi: 10.1126/science.1214824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y, Moody IS, Sims PC, Hunt SR, Corso BL, Seitz DE, Blaszcazk LC, Collins PG, Weiss GA. Single Molecule Dynamics of Lysozyme Processing Distinguishes Linear and Cross-linked Peptidoglycan Substrates. J. Am. Chem. Soc. 2012;134:2032–2035. doi: 10.1021/ja211540z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y, Olsen TJ, Sims PC, Moody IS, Corso BL, Dang MN, Weiss GA, Collins PG. Dissecting Single-Molecule Signal Transduction in Carbon Nanotube Circuits with Protein Engineering. Nano Lett. 2013;13:625–631. doi: 10.1021/nl304209p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Ashcroft B, Zhang P, Liu H, Sen S, Song W, Im J, Gyarfas B, Manna S, Biswas S, Borges C, Lindsay S. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat Nano. 2014;9:466–473. doi: 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, He J, Chang S, Zhang P, Liang F, Li S, Tuchband M, Fuhrmann A, Ros R, Lindsay S. Identifying single bases in a DNA oligomer with electron tunnelling. Nat. Nanotechnol. 2010;5:868–873. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutsui M, Rahong S, Iizumi Y, Okazaki T, Taniguchi M, Kawai T. Single-molecule sensing electrode embedded in-plane nanopore. Sci. Rep. 2011;1 doi: 10.1038/srep00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Hu DH, Vorpagel ER, Lu HP. Probing single-molecule T4 lysozyme conformational dynamics by intramolecular fluorescence energy transfer. J. Phys. Chem. B. 2003;107:7947–7956. [Google Scholar]
- 24.Hu D, Lu HP. Placing single-molecule T4 lysozyme enzymes on a bacterial cell surface: Toward probing single-molecule enzymatic reaction in living cells. Biophys. J. 2004;87:656–661. doi: 10.1529/biophysj.104.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu HP. Single-molecule spectroscopy studies of conformational change dynamics in enzymatic reactions. Current Pharmaceutical Biotechnology. 2004;5:261–269. doi: 10.2174/1389201043376887. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Lu HP. Bunching effect in single-molecule T4 lysozyme nonequilibrium conformational dynamics under enzymatic reactions. J. Phys. Chem. B. 2010;114:6669–6674. doi: 10.1021/jp1004506. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 28.Chen RJ, Zhan YG, Wang DW, Dai HJ. Noncovalent sidewall functionalization of single-walled carbon nanotubes for protein immobilization. J. Am. Chem. Soc. 2001;123:3838–3839. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- 29.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui Y, Wei QQ, Park HK, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 31.Patolsky F, Zheng G, Hayden O, Lakadamyali M, Zhuang X, Lieber CM. Electrical detection of single viruses. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xinjian Z, Moran-Mirabal JM, Craighead HG, McEuen PL. Supported lipid bilayer/carbon nanotube hybrids. Nat. Nanotechnol. 2007;2:185–190. doi: 10.1038/nnano.2007.34. [DOI] [PubMed] [Google Scholar]
- 33.Kramers HA. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7:284–304. [Google Scholar]
- 34.Hanggi P, Talkner P, Borkovec M. Reaction-rate theory - 50 years after Kramers. Reviews of Modern Physics. 1990;62:251–341. [Google Scholar]
- 35.de Groot BL, Hayward S, van Aalten DMF, Amadei A, Berendsen HJC. Domain motions in bacteriophage T4 lysozyme: A comparison between molecular dynamics and crystallographic data. Proteins-Structure Function and Genetics. 1998;31:116–127. doi: 10.1002/(sici)1097-0134(19980501)31:2<116::aid-prot2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Floyd DL, Harrison SC, van Oijen AM. Analysis of Kinetic Intermediates in Single-Particle Dwell-Time Distributions. Biophys. J. 2010;99:360–366. doi: 10.1016/j.bpj.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svoboda K, Mitra PP, Block SM. Fluctuation analysis of motor protein movement and single enzymekinetics. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnitzer MJ, Block SM. Statistical kinetics of processive enzymes. Cold Spring Harbor Symposia on Quantitative Biology. 1995;60:793–802. doi: 10.1101/sqb.1995.060.01.085. [DOI] [PubMed] [Google Scholar]
- 40.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 41.Xu WL, Kong JS, Chen P. Single-Molecule Kinetic Theory of Heterogeneous and Enzyme Catalysis. Journal of Physical Chemistry C. 2009;113:2393–2404. [Google Scholar]
- 42.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 43.Astumian RD. Microscopic reversibility as the organizing principle of molecular machines. Nat. Nanotechnol. 2012;7:684–688. doi: 10.1038/nnano.2012.188. [DOI] [PubMed] [Google Scholar]
- 44.Zechel DL, Withers SG. Glycosidase mechanisms: Anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000;33:11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.