Abstract
Purpose
Multiple myeloma (MM) is the second most prevalent hematological malignancy and it remains incurable despite the introduction of several novel drugs. The discrepancy between preclinical and clinical outcomes can be attributed to the failure of classic two-dimensional (2D) culture models to accurately recapitulate the complex biology of MM and drug responses observed in patients.
Experimental design
We developed 3D tissue engineered bone marrow (3DTEBM) cultures derived from the BM supernatant of MM patients to incorporate different BM components including MM cells, stromal cells, and endothelial cells. Distribution and growth were analyzed by confocal imaging, and cell proliferation of cell lines and primary MM cells was tested by flow cytometry. Oxygen and drug gradients were evaluated by immunohistochemistry and flow cytometry, and drug resistance was studied by flow cytometry.
Results
3DTEBM cultures allowed proliferation of MM cells, recapitulated their interaction with the microenvironment, recreated 3D aspects observed in the bone marrow niche (such as oxygen and drug gradients), and induced drug resistance in MM cells more than 2D or commercial 3D tissue culture systems.
Conclusions
3DTEBM cultures not only provide a better model for investigating the pathophysiology of MM, but also serve as a tool for drug development and screening in MM. In the future, we will use the 3DTEBM cultures for developing personalized therapeutic strategies for individual MM patients.
Keywords: Multiple myeloma, 3D, tissue-engineering, drug resistance, tumor microenvironment, culture model
INTRODUCTION
Multiple myeloma (MM) is the second most prevalent hematological malignancy and remains incurable with a median survival time of 3-5 years [1, 2]. Despite the introduction of several novel drugs and their high efficacy in vitro, only about 60% of patients initially respond to therapy, and among relapsed patients more than 90% develop drug resistance [3-6].
The discrepancy between in vitro efficacy and clinical outcomes can be attributed to limitations of classic two-dimensional (2D) tissue culture and drug screening models. First, despite the fact that the interactions of MM cells with bone marrow (BM) microenvironment components was shown to induce resistance [7-10], most of the in vitro models use MM cell line mono-cultures and neglect the vital role of the microenvironment. Second, the BM niche is a three-dimensional (3D) structure which induces oxygen and drug concentration gradients as a function of distance from blood vessels known to significantly affect drug efficacy [11-14]. 2D tissue culture systems cannot reproduce the oxygen and drug gradients found in the BM niche, which limits the ability of 2D cultures to accurately predict drug sensitivity. Therefore, there is an urgent need to develop a model that addresses these limitations to investigate biological mechanisms and drug resistance in MM that are relevant and translatable to improved patient response.
Previous models have been developed to recreate the 3D microenvironment of the BM using collagen [15, 16], Matrigel [17], acrylic polymers [18], silk [19], hyaluronic acid [20], and ossified tissues [21]. These models have probed the importance of using 3D rather than 2D models to recreate myeloma growth; however, each has its limitations. For example, although hydrogel systems (such as collagen, Matrigel or synthetic polymers) [15-19] are simple and reproducible, these materials are not physiologically found in the BM and may cause significant changes in the culture milieu. Solid systems (such as ossified tissues) mimic BM physiological conditions[21, 22]; however, these are technically challenging due to reproducibility and adaptability problems, and rely on a normal BM microenvironment for the growth of MM cells, which was previously proven to be significantly different (in some cases opposite) from the effect of the MM microenvironment [23, 24].
In this study, we developed a 3D scaffold derived from the BM supernatant of MM patients to incorporate different BM components including MM cells, stromal cells, and endothelial cells. This model was defined as a 3D tissue engineered bone marrow (3DTEBM) culture, and we hypothesized that it will promote better growth of MM cells and provide a more patient relevant model for evaluating drug efficacy in MM (Figure 1A).
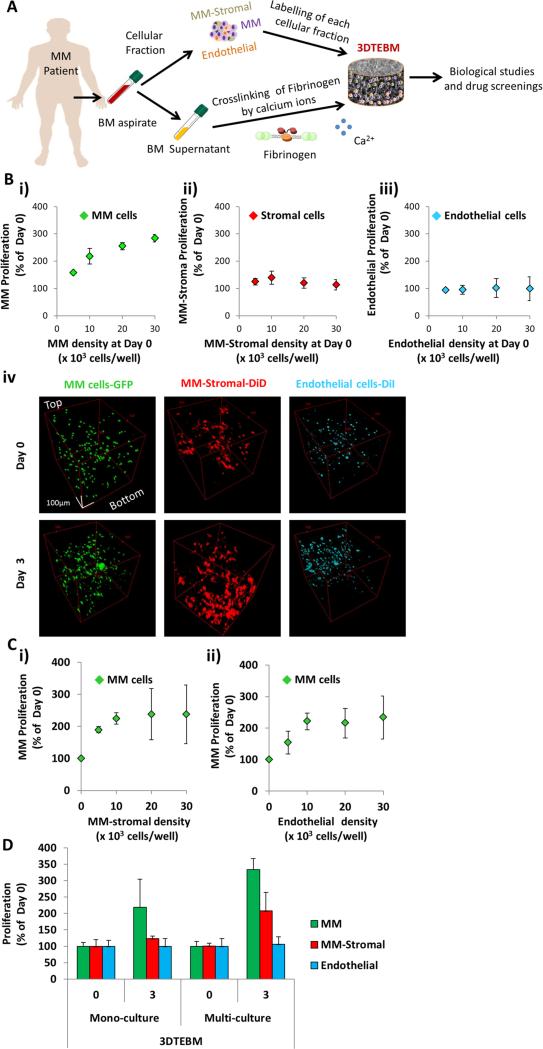
Figure 1. 3DTEBM cultures allow MM cell proliferation and interaction with accessory cells.
A) 3DTEBM cultures were developed through cross-linking of fibrinogen (naturally found in the plasma of BM supernatant) with calcium; numerous cellular components, including MM cells, MM-derived stromal cells, and endothelial cells, were pre-labeled and incorporated into the cultures. B) Effect of cell density (5 × 103 – 30 × 103 cells/well) on proliferation of i) MM, ii) MM-derived stroma, and iii) endothelial cells grown individually in 3DTEBM mono-cultures at day 3, and iv) confocal microscopy images of MM-GFP (green), MM-derived stroma-DiD (red), and endothelial cells-DiI (cyan) in mono-cultures inside 3DTEBM at days 0 and 3 represented by Z-Stack images from top to bottom in rotated view. Scale bar = 100 μm. C) Effect on MM cell proliferation in 3DTEBM (at day 3) when co-cultured with i) MM-derived stromal cells (0– 30 × 103 cells/well) or ii) endothelial cells (0– 30 × 103 cells/well). D) Summary of proliferation [MM cells (30 × 103 cells/well), MM-derived stromal cells (10 × 103 cells/well), and endothelial cells (10 × 103 cells/well)] in the 3DTEBM when cultured as mono-cultures or multi-cultures at day 3.
MATERIALS AND METHODS
Reagents
Calcium chloride (CaCl2), tranexamic acid, type I collagenase, dimethyl sulfoxide (DMSO), propidium iodide (PI, excitation, 488 nm; emission, 655 - 730 nm), and doxorubicin (excitation, 488 nm; emission, 585/40 nm) were purchased from Sigma-Aldrich (Saint Louis, MO). Cell trackers including DiO (excitation, 488 nm; emission, 525/50 nm), DiD (excitation, 635 nm; emission, 655 - 730 nm), DiI (excitation, 488 nm; emission, 585/40 nm) and Calcein violet (excitation, 405 nm; emission, 450/50 nm) were purchased from Invitrogen (Carlsbad, CA). Drugs including bortezomib and carfilzomib were purchased from Selleck Chemicals (Houston, TX).
Cell lines
The MM cell lines (MM1s, H929, RPMI8226, and MM1s-GFP-Luc) were a kind gift from Dr. Irene Ghobrial (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA). Human umbilical vein endothelial cells were purchased from Lonza (Walkersville, MD). All cells were cultured at 37°C, 5% CO2; MM cells in RPMI-1640 media (Corning CellGro, Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS, Gibco, Life technologies, Grand island, NY), 2 mmol/l of L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Corning CellGro), and endothelial cells in EGM-2 completed media (Lonza). Before experiments, MM cells and endothelial cells (1 × 106 cells/ml) were pre-labeled with Calcein violet (1 μg/ml) or DiI (10 μg/ml) for 1 hour, respectively.
Primary cells
Primary CD138+ and CD138− cells were isolated from BM aspirates of MM patients from the Siteman Cancer Center, Washington University in Saint Louis, by magnetic-bead sorting as previously described [7]. Informed consent was obtained from all patients with an approval from the Washington University Medical School IRB committee and in accord with the Declaration of Helsinki. For frozen samples, cells were viably frozen at −80°C in FBS with 10% DMSO (v/v). Plasma samples were extracted from peripheral blood (PB) and BM aspirates by centrifugation at 1620 g for 10 minutes, and finally frozen at −80°C. Primary CD138+ cells were cultured in RPMI-1640 media supplemented with 10% FBS, L-glutamine, penicillin, and streptomycin. Primary MM-derived stromal cells (CD138−) were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Corning CellGro) supplemented with 20% FBS, L-glutamine, penicillin, and streptomycin. MM-derived stromal cells were cultured for three weeks and monitored for the development of spindle-shaped cells. Before experiments CD138+ and MM-derived stromal cells (1 × 106 cells/ml) were pre-labeled with DiO (10 μg/ml) and DiD (10 μg/ml) for 1 hour, respectively.
Development of 3DTEBM cultures
3DTEBM cultures were developed through cross-linking of fibrinogen (naturally found in the plasma of the PB and BM supernatant) with CaCl2 concentrations ranging from 0 – 4 mg/ml, as previously described [25, 26]. Gelification time was measured and the CaCl2 concentration that induced the fastest gelification time was selected for further studies (Suppl Figure 1A). The effect of tranexamic acid addition at various concentrations (0 – 10 mg/ml) was studied for preventing the degradation of fibrin fibers and improving scaffold stability. The scaffold stability was measured at 3 weeks and assessed by comparing the tranexamic acid containing scaffold weight to the weight of non-stabilized scaffolds, and the tranexamic acid concentration that induced the maximal stabilization was selected for further studies (Suppl Figure 1B). The impact of tranexamic acid on the viability of myeloma cell lines was tested by MTT. No effect on viability was found at any concentration tested (1 – 10 mg/ml). The concentration of 4 mg/ml has no impact on MM cell lines (MM1s and H929) (Suppl Figure 1C). Briefly, 3DTEBM cultures were formed by mixing the following components: 40 μl of plasma (from PB and BM supernatants) diluted in RPMI-1640 completed media to form total volume of 100 μl, with a final concentration of 1 mg/ml CaCl2, and 4 mg/ml tranexamic acid. Scaffolds were allowed to gelify for 2 hours in a 96-well plate in incubator at 37°C, 5% CO2, before being covered with additional RPMI-1640 completed media. The medium was changed every 3 days during the culture period. Cells were added to the solution before fibrin clotting agents (CaCl2 and tranexamic acid). Figure 1A shows the strategy used for the development of 3DTEBM from MM patients. Morphology of the 3DTEBM scaffolds was analyzed with light microscopy (Axiovert 35, Zeiss) and size measurements were taken based on diameter and height (Image J software, NIH, Bethesda, MD) (Suppl Figure 1D). The structure of 3DTEBM with and without MM1s cells was studied using scanning electron microscopy (SEM) (Suppl Figure 1E), as previously described [26].
Confocal imaging
Distribution and growth of mono-cultures and multi-cultures of MM1s-GFP, endothelial cells-DiI, and MM-derived stromal cells-DiD through the 3DTEBM scaffolds were tested using confocal microcopy at days 1, 2, 3, 5 and 7. The 3DTEBM was glued to a glass slide. A ring was placed around the culture and filled with phenol red free DMEM. The cultures were imaged using a FV1000 confocal microscope with an XLUMPLFLN 20XW/1.0 immersion objective lens (Olympus, PA, USA). The cultures were excited at 488 nm (GFP/DiO), 543 nm (DiI), and 633 nm (DiD) and the emission light was collected at 500 – 530 nm, 555 – 625 nm, 650 long pass, for each channel respectively. Z-stack images of approximately 1 mm thickness were taken of each sample at 2 μm step sizes. Each frame consisted of a 520 × 520 pixel image, taken at a rate of 1 μs/pixel.
Cell proliferation using flow cytometry
Cell proliferation assays were performed by digestion of 3DTEBM cultures with type I collagenase (25 mg/ml for 2 - 3 hours at 37°C), and classic 2D cultures were washed with PBS and removed by pipetting or trypsinization. The effect of cell density (5 × 103 – 30 × 103 cells/well) of MM1s-GFP, MM-derived stromal cells-DiD, and endothelial cells-DiI on growth rate in mono-cultures at day 3 inside 3DTEBM was measured by flow cytometry. For all flow cytometry analysis, internal standard control cells labeled with Calcein violet were added, and a minimum of 5,000 events of these were acquired using BD FACS Aria (BD Biosciences) and DiVa v6.1.2 software. The data was analyzed using FlowJo program v10 (Ashland, OR). MM cells (cell lines and primary patient samples) were identified by gating cells with a high GFP/DiO signal (excitation, 488 nm; emission, 530/30 nm); MM-derived stromal cells were identified by gating cells with a high DiD signal (excitation, 638 nm; emission, 660/20 nm); endothelial cells were identified by gating cells with a high DiI signal (excitation, 488 nm; emission, 585/15 nm); and internal standard control cells were identified by gating cells with a high UV signal (excitation, 355 nm; emission, 450/50 nm). The effect of cellular interactions of MM-derived stromal cells and endothelial cells on proliferation of MM cells was tested at day 3.
We further tested the proliferation rate of MM cells in multi-culture within 3DTEBM cultures compared to classic 2D tissue culture and other 3D tissue culture systems such as PLGA microspheres, AlgiMatrix (a synthetic matrix), and Matrigel. Degradex PLGA microspheres 125 μm (Phosphorex Inc, MA) were used as previously described [27]; briefly, 100 μL of microspheres (0.66 mg/ml concentration) were transferred to 96-well plates and incubated for 2 hours at 37°C. Media was then aspirated carefully, leaving microparticles in the wells for cell seeding. Thereafter, 50 μl of a multi-culture cell suspension [MM1s-GFP (30 × 103 cells/well), MM-derived stromal cells-DiD (10 × 103 cells/well), and endothelial cells-DiI (10 × 103 cells/well)] was added to each well. After 3 hours of incubation, an additional 50 μl of media was added on the top of each well. At days 0, 3 and 7 media was removed; cells were detached by treating microparticles with 50 μL of 0.1 M citrate buffer for 1 hour at 37°C, followed by washing and flow cytometry analysis. Gibco AlgiMatrix 3D culture system (Life Technologies, CA) was prepared as previously described [28]; 100 μl of a multi-culture cell suspension was added to the top surface of a dry alginate sponge (pore size 50 - 200 μm). At days 0, 3 and 7 media was removed; the sponge was degraded by adding 100 μl of AlgiMatrix Dissolving buffer for 5 minutes, cells were washed and then analyzed by flow cytometry. Corning Matrigel Basement Membrane Matrix Growth Factor Reduced (Corning, NY) was prepared following manufacturer instructions for the thick gel method; a cell pellet of multi-culture cells was added to Matrigel diluted to 5 mg/ml. 50 μl of this mixture were added to each well in a 96-well plate, incubated 30 minutes at 37°C, and cultured media was added on top. At days 0, 3 and 7 media was removed; cells were recovered by adding 100 μl of cell recovery solution at 4°C for 2 hours, then cells were washed and analyzed by flow cytometry.
Frozen and fresh primary CD138+ cells from MM patients were cultured in multi-cultures in classic 2D culture system and in 3DTEBM cultures, and the number of viable MM cells was analyzed at days 3 and 7 by flow cytometry. In addition, fresh primary CD138+ cells from MM patients were cultured in multi-cultures in PLGA microspheres, AlgiMatrix, and Matrigel.
Cytokine secretion
The effect of 3DTEBM on cytokine secretion was tested by human cytokine antibody arrays. MM1s cells (30 × 103 cells/well) in mono-culture or multi-culture with MM-derived stromal cells (10 × 103 cells/well) and endothelial cells (10 × 103 cells/well) were grown in 3DTEBM or classic 2D cultures. As control, 3DTEBM and classic 2D cultures without cellular components (non-cultured) were included. After 3 days, 3DTEBM cultures were digested and media from classic 2D cultures was aspirated by pipetting. Then, the cell culture supernatants were analyzed with the C-Series Human Cytokine Antibody Array C5 (RayBiotech Inc., Norcross, GA) according to manufacturer's instructions. The chemiluminiscence signal of the membranes was captured and the intensity of each spot was measured by densitometric analysis (Image J Software). The value for each cytokine was determined by first subtracting negative controls and then normalizing to positive controls in each membrane.
Oxygen gradient and hypoxia in 3DTEBM cultures
Immunohistochemistry (IHC) analysis of the hypoxic and proliferative state of MM cells was performed in 3DTEBM cultures. MM cells were cultured 5 days in the 3DTEBM, fixed with 4% formaldehyde in PBS, dehydrated with ethanol, embedded in paraffin blocks, and longitudinally sectioned. Sections were stained with AlexaFluor488-anti-HIF1α (clone H1α67; Novus Biologicals) and AlexaFluor488-anti-Ki67 (MKI67; Novus Biologicals). Sections were imaged at 10X using a BX-51 epifluorescent microscope (Olympus). An emission cube was used for excitation at 465 - 505 nm and emission collection at 510 - 560 nm. Pictures were taken from five random areas from top and bottom, and fluorescence signal of HIF1α and Ki67 was quantified by Image J Software.
To further demonstrate the effect of depth on oxygen gradient in 3DTEBM cultures, MM1s-GFP cells were cultured for 48 hours in 3DTEBM scaffolds of various depths (made of 25 – 75 μl) or 2D cultures. Then, media was carefully removed and 100 μl of pimonidazole (PIM) solution (100 μg/ml, Hypoxyprobe, Burlington, MA) was added to each well, and incubated at 37°C for 24 hours. 3DTEBM cultures were digested and MM cells were retrieved from the classic 2D cultures by pipetting. MM cells from 2D and 3DTEBM cultures were fixed and permeabilized with 4% formalin and 90% cold methanol in PBS, washed with cold PBS, and blocked with 3% FBS in PBS. Cells were then stained with allophycocyanin(APC)-anti-PIM (excitation, 635 nm; emission, 655 - 730 nm) and analyzed by flow cytometry. MM cells were identified by gating cells with a high GFP signal; then mean fluorescence intensity (MFI) of PIM signal in the MM-GFP+ cells was measured.
Expression of CXCR4 and CD138
The expression of CD138 and CXCR4 in MM were previously reported to be affected by hypoxic conditions, therefore, we tested the effect of 3DTEBM cultures on the expression of CXCR4 and CD138 in MM cells. MM1s-GFP cells were cultured in 3DTEBM and classic 2D cultures, and after 3 days, 3DTEBM cultures were digested, and MM cells were retrieved from the classic 2D cultures by pipetting. Then, MM cells were washed with PBS and incubated with PerCP-Cy5.5-anti-CD138 (excitation, 488 nm; emission, 655-730 nm) and phycoerythrin (PE)-anti-CD184 (CXCR4) (excitation, 488 nm; emission, 585/40 nm) and isotype controls on ice for 1 hour, and analyzed by flow cytometry. MM cells were identified by gating cells with a high GFP signal, and then MFI of PE-CXCR4 and PerCP-Cy5.5-CD138 was detected and normalized to MFI of isotype controls.
Drug gradients in the 3DTEBM
To test the effect of the 3DTEBM on the development of drug gradients we tested the uptake of doxorubicin in MM cells cultured in 3DTEBM, classic 2D mono-cultures, PLGA microspheres, AlgiMatrix, and Matrigel. MM cells cultured in all the systems were treated with increasing concentrations (0 - 7.5 μg/ml) of doxorubicin for 6 hours. MM cells were retrieved from the different cultures and identified by gating cells with a high GFP/DiO signal; doxorubicin uptake was detected by MFI of PE signal.
To test the effect of depth of the 3DTEBM on drug gradients, MM1s-GFP cells (30 × 103 cells/well) were cultured in 3DTEBM scaffolds of various depths (made of 25 – 150 μl) and treated with doxorubicin (7.5 μg/ml) for 4 hours. Then, 3DTEBM's were digested, MM cells retrieved, and the uptake of doxorubicin was measured by flow cytometry, as described above.
Drug resistance in the 3DTEBM
The interaction of MM cells with the BM microenvironment, the degree of hypoxia and drug gradients are all known to affect drug efficacy; therefore, we tested the effect of the 3DTEBM on drug resistance in MM cells. MM1s-GFP cells (30 × 103 cells/well) were cultured in mono-culture or multi-culture with MM-derived stromal cells-DiD (10 × 103 cells/well), and endothelial cells-DiI (10 × 103 cells/well) in 3DTEBM and classic 2D cultures. Two hours after plating, cells were treated with increasing concentrations (0 – 30 nM) of bortezomib or carfilzomib (0, 5 nM) by adding drug solution on top of 2D cultures and 3DTEBM for 24 hours. Then, MM cells were retrieved from the different cultures, and cell survival was analyzed by flow cytometry against an internal standard.
In addition, we tested the effect of 3DTEBM on drug resistance in MM cells compared to 2D or other 3D tissue culture systems (PLGA microspheres, AlgiMatrix, and Matrigel). MM cells were cultured in these systems as described above and treated with bortezomib (0, 5 nM) or carfilzomib (0, 5 nM) for 24 hours. Then, MM cells were retrieved from the different cultures, and cell survival was analyzed by flow cytometry against an internal standard.
Finally, we sought to differentiate the effect of the 3DTEBM induced hypoxia and drug gradients on drug resistance. MM1s-GFP cells were treated with bortezomib (0, 5 nM) and carfilzomib (0, 5 nM) for 24 hours while incubated in classic 2D cultures (under normoxic conditions (21% O2) or hypoxic conditions (1% O2) in the hypoxic chamber from Coy, Grass Lake, MI), or in 3DTEBM cultures with drugs either uniformly mixed into the matrix or added only to the top surface. Then, MM cells were retrieved from the different cultures, and cell survival was analyzed by flow cytometry against an internal standard.
Statistical analysis
Experiments were performed in quintuplicates and repeated at least three times. Results were presented as mean ± standard deviation, and statistical significance was analyzed using student t-test or one-way ANOVA; a p value less than 0.05 was considered significant.
RESULTS
3DTEBM cultures allow MM cell proliferation and interaction with accessory cells
To optimize conditions for co-culturing MM cells with accessory cells, we first analyzed the effect of cell density (MM1s, MM-derived stromal cells, and endothelial cells individually) on their own growth rate when cultured alone in 3DTEBM. The optimal density for MM cell growth (30,000 cells/well) corresponded to a 275% increase in proliferation after 3 days (Figure 1Bi). A density of 10,000 cells/well induced highest growth in MM-derived stroma (Figure 1Bii), while cell density did not have an effect on the growth rate of endothelial cells (Figure 1Biii). Similarly, confocal images of MM1s-GFP (green) showed a homogeneous distribution pattern throughout the scaffold with a notable increase in MM cell number and clumping at day 3. MM-derived stromal cells-DiD (red) showed an increase in volume and 3D morphology (greater number of prolongations) with a preferential accumulation near the bottom of the 3DTEBM at day 3. In contrast, mono-cultures of endothelial cells-DiI (cyan) concentrated near the top of the 3DTEBM (Figure 1Biv).
Stromal cells derived from MM patients increased the growth of MM cells in the 3DTEBM with a positive correlation, in which 10,000 stromal cells increased the proliferation of MM cells by 250% compared to MM cells alone (Figure 1Ci). Similarly, endothelial cells provided the maximum increase to MM cell growth with a cell density of 10,000 cells per well (Figure 1Cii). No additional increase was observed at higher concentrations of stromal or endothelial cells. For the remaining multi-culture experiments described herein, an optimal MM (30,000 cells/ well), stromal (10,000 cells/ well), and endothelial (10,000 cells/ well) cell density was used.
We also tested the effect of multi-culture conditions (MM, stromal and endothelial cells all combined) in the 3DTEBM on the proliferation of each cell type. We found that MM cells proliferated about 200% when cultured alone, but in a multi-culture setting with both stromal and endothelial cells, MM cells proliferated about 350% (Figure 1D). When cultured alone, stromal cells showed a modest 125% increase in proliferation, but when cultured with MM and endothelial cells, proliferation increased to about 200%. No effect was observed on endothelial cell proliferation in either mono- or multi-culture.
3DTEBM cultures promote MM cell proliferation better than 2D and commercially available 3D systems
While MM1s proliferation increased about 200% and 300% at days 3 and 7 in classic 2D cultures, the 3DTEBM had significantly greater increases of 300% and 400% compared to day 0, respectively (Figure 2Ai). Similarly, H929 and RPMI showed significantly higher increases in proliferation in 3DTEBM compared to 2D tissue cultures (Figure 2Aii and iii, respectively).
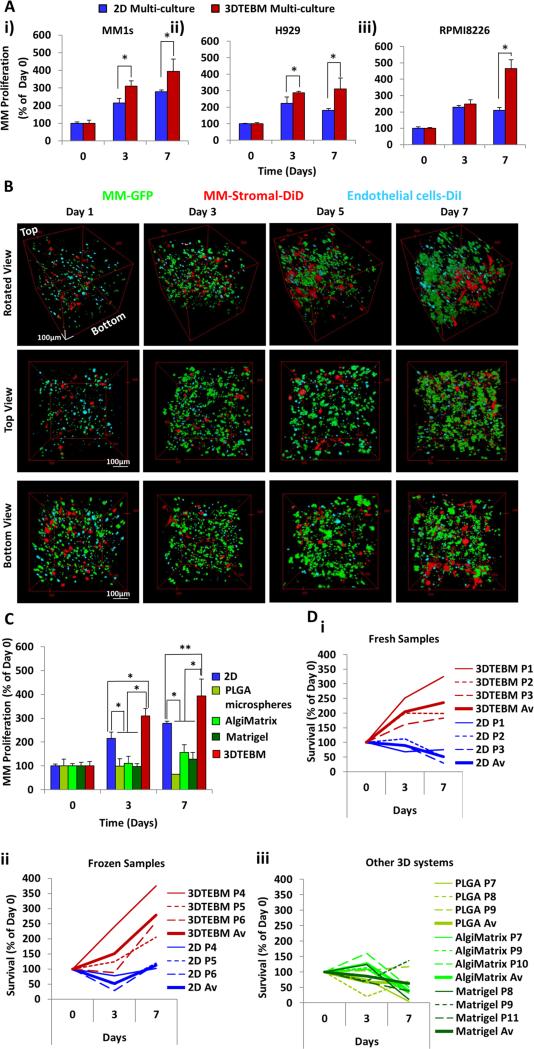
Figure 2. 3DTEBM cultures promote MM cell proliferation better than 2D and commercially available 3D systems.
A) Growth of MM cell lines i) MM1S, ii) H929, and iii) RPMI8226 in multi-culture with MM-derived stromal cells and endothelial cells in classic 2D cultures or in the 3DTEBM at days 3 and 7; (*) p< 0.02. B) Confocal microscopy images of multi-cultures of MM1s-GFP (green), MM-derived stroma-DiD (red) and endothelial cells-DiI (cyan) in the 3DTEBM at 1, 3, 5, and 7 days of culture, shown from 3 perspectives: Z-Stack rotated view, top down view, and bottom up view; Scale bar= 100 μm. C) Growth of MM1s in multi-culture with MM-derived stromal cells and endothelial cells in classic 2D cultures, PLGA microspheres, AlgiMatrix, Matrigel, and inside 3DTEBM at days 3 and 7 compared to day 0, (*) p< 0.01, (**) p< 0.05. D) Growth of i) primary fresh CD138+ plasma cells from three MM patients, ii) primary frozen CD138+ cells from three additional MM patients, in multi-culture conditions in classic 2D cultures (blue) or in the 3DTEBM (red), iii) and primary fresh in PLGA microspheres (green), AlgiMatrix (light green), and Matrigel (dark green) at days 3 and 7; bold line reflects average growth in the different conditions.
Confocal microscopy images indicated that MM1s-GFP cells were able to interact with accessory cells (endothelial cells-DiI and MM-derived stromal cells-DiD) in the 3DTEBM culture (Figure 2B). We found that MM cells (green) grew through the scaffold from top to bottom and their number increased with time to form a higher density and tumor bulk. Interestingly, we found that over time, the stromal cells (red) migrated towards the bottom of the scaffold and formed sheet-like structures; in contrast, endothelial cells (cyan) concentrated in the top part of the scaffold. MM cells direct interaction with stromal and endothelial was identified as a co-localization of green with red and cyan, respectively. Moreover, MM cells showed proliferation of about 200% and 300% at days 3 and 7 in classic 2D tissue cultures, they showed increased proliferation of 300% and 400% at days 3 and 7 in the 3DTEBM, respectively. In contrast, all the other commercially available 3D systems showed lower proliferation rates of MM cells compared to 2D cultures (Figure 2C).
We further evaluated the ability of the 3DTEBM to grow primary plasma cells from MM patients. 2D culture did not sustain the growth of fresh (Figure 2Di) or frozen (Figure 2Dii) primary MM cells. On the contrary, the 3DTEBM constantly increased the growth of fresh primary MM cells to 200% and 250% at days 3 and 7, respectively (Figure 2Di); as well as in three independent primary frozen primary cells, it increased the growth to 150% and 250% at 3 and 7 days, respectively (Figure 2Dii). In contrast, all the other 3D systems (PLGA microspheres, AlgiMatrix, and Matrigel) showed lower proliferation rates of primary MM cells compared to 3DTEBM, and were not able to sustain MM primary patient cell proliferation ex vivo (Figure 2Diii).
3DTEBM cultures induce changes in cytokine secretion in the MM environment
First, we tested the baseline profile of cytokines in the different cultures without the presence of cells. The cytokine profile of 3DTEBM non-cultured (BM supernatant) compared to 2D non-cultured (cell culture media) revealed that 23 of 40 detectable cytokines were enriched in the 3DTEBM with at least 3-fold increase compared to 2D tissue cultures (Figure 3A).
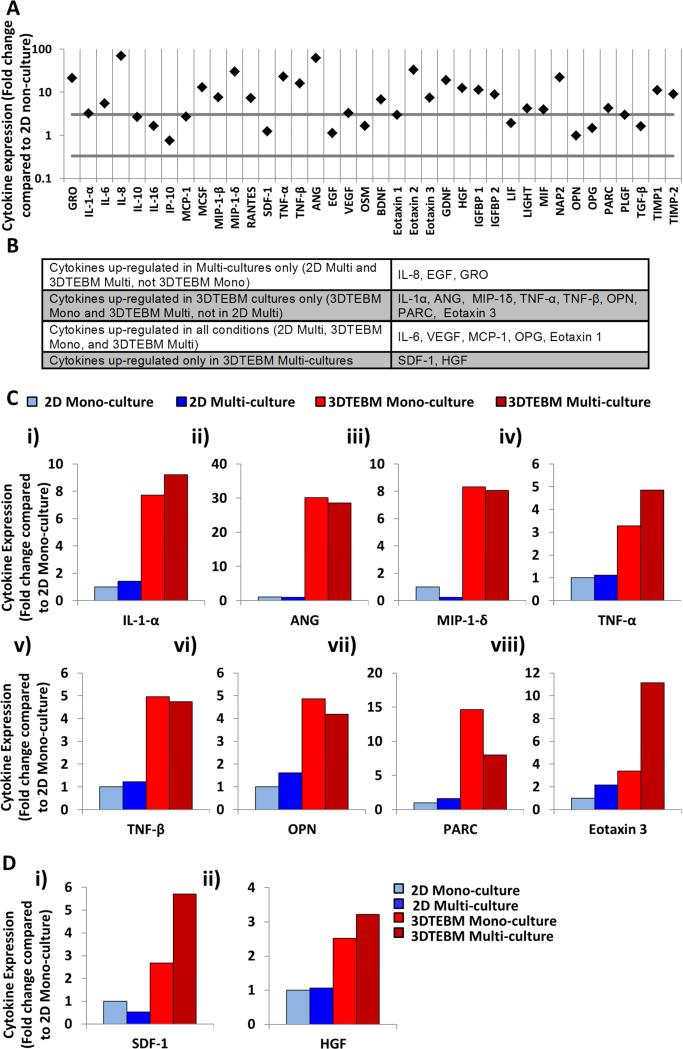
Figure 3. The 3DTEBM cultures induce changes in cytokine secretion in the MM environment.
A) Cytokines levels in non-cultured 3DTEBM (BM plasma) and 2D cultures (culture media). A significant change in expression was defined as a ≥ 3-fold increase or ≤0.3-fold decrease. B) Cytokines up-regulated due to multi-cultures, 3DTEBM cultures, or 3DTEBM multi-cultures. C) Cytokines up-regulated due to 3DTEBM including: i) Interleukin-1alpha (IL-1α), ii) Angiogenin (ANG), iii) Macrophage inflammatory protein-1delta (MIP-1δ), iv) Tumor necrosis factor-alpha (TNF-α), v) TNF-β, vi) Osteopontin (OPN), vii) Pulmonary and activation-regulated chemokine (PARC), and viii) Eotaxin 3. D) Cytokines up-regulated in multi-culture 3DTEBM included: i) Stromal cell-derived factor-1 (SDF-1), ii) Hepatocyte growth factor (HGF).
We then compared the cytokine secretion profile induced by multi-cultures only, by 3DTEBM cultures only, in all the conditions, and in 3DTEBM multi-cultures only. Cytokines levels were considered to be changed if there was ≥ 3-fold increase or ≤ 0.3-fold decrease, compared to 2D mono-cultured). Three cytokines were up-regulated in the multi-cultures only (IL-8, EGF, GRO) (Figure 3B); five cytokines were up-regulated in all the conditions tested including IL-6, VEGF, MCP-1, OPG, and Eotaxin 1 (Figure 3B); eight cytokines were up-regulated in the 3DTEBM cultures including IL-1α, ANG, MIP-1δ, TNF-α, TNF-β, OPN PARC, and Eotaxin 3 (Figure 3B and C). Only two cytokines were up-regulated only in multi-cultures in 3DTEBM including SDF-1, and HGF (Figure 3D).
3DTEBM cultures recapitulate oxygen gradients and hypoxia
While the top areas of the cultures showed few MM cells expressing HIF1α, deeper areas in the bottom of 3DTEBM showed an increased number of MM cells expressing HIF1α (Figure 4Ai). Quantifying the MFI of the HIF signal revealed a significant increase in hypoxia in the bottom layer compared to the top layer (Figure 4Aii). Furthermore, MM cells in the 3DTEBM exhibited higher binding of PIM (a hypoxia marker) compared to MM cells grown in classic 2D cultures, with in direct linear correlation with 3DTEBM depth (R2=0.9043) (Figure 4B). The presence of hypoxic microenvironment in the 3DTEBM was corroborated by decreased expression of CD138 and increased expression of CXCR4 in the 3DTEBM cultures (Figure 4Ci). Representative flow cytometry histograms of PerCP-Cy5.5-CD138 and PE-CXCR4 (Figure 4Cii and iii, respectively) signal compared to control MM cells cultured in 2D.
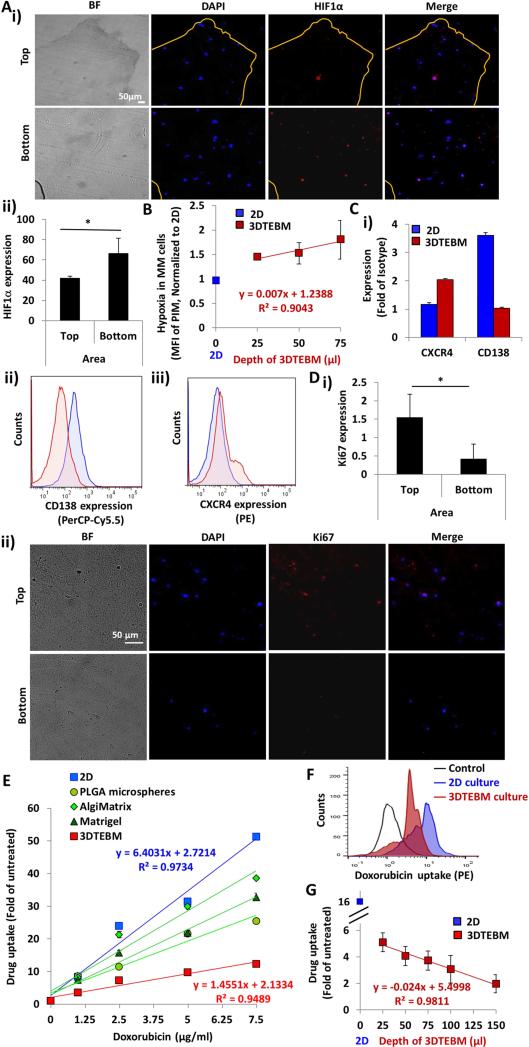
Figure 4. The 3DTEBM cultures recapitulate oxygen gradients, hypoxia and drug gradients.
A) i) Fluorescent imaging at day 5 of top and bottom sections of 3DTEBM cut longitudinally (Bright field (BF); DAPI, Blue; HIF1α, Red; Merge, Pink); Scale bar= 50 μm. ii) HIF1α expression quantified as MFI of AF488 expression in the top and bottom areas of 3DTEBM, (*) p< 0.05. B) Correlation between 3DTEBM depth and hypoxia in MM cells (MFI of PIM) compared to hypoxic state of MM cells in 2D cultures. Blue square shows drug uptake in 2D cultures, red squares show uptake in 3DTEBM. C) i) CD138 and CXCR4 expression measured as fold of MFI of PE-anti-CXCR4 and PerCPCy5.5-anti-CD138 to isotype controls in MM cells in the 3DTEBM vs 2D multi-cultures at day 3, and flow cytometry representative histogram of ii) CD138 (PerCP-Cy5.5) and iii) CXCR4 (PE). D) i) Ki67 expression measured as quantification of MFI AF488 expression in the top and bottom areas of the 3DTEBM, (*) p< 0.05. ii) Fluorescent imaging at day 5 of the top and bottom areas of 3DTEBM cut longitudinally (Bright field (BF); DAPI, Blue; Ki67, Red; Merge, Pink). Scale bar= 50 μm. E) Effect of increasing doxorubicin concentration (0 – 7.5 μg/ml) on MM cell uptake in 3DTEBM, classic 2D cultures, PLGA microspheres, AlgiMatrix, and Matrigel. F) Flow cytometry histogram representing doxorubicin uptake (PE signal) in MM cells grown in 3DTEBM and 2D cultures; Control: untreated MM cells. G) Correlation between various 3DTEBM and doxorubicin uptake of MM cells grown within. The blue square indicates doxorubicin uptake in 2D cultures (minimal depth) and red squares show uptake in various 3DTEBM scaffold depths).
Furthermore, quantification of the Ki67 signal revealed a significantly higher number of proliferating MM cells within the top layers of the 3DTEBM compared to the bottom layers (Figure 4Di). Quiescent MM cells were found predominantly in the bottom layers, while more proliferative cells (higher Ki67 expression) were found near the top layers of the 3DTEBM scaffold (Figure 4Dii).
3DTEBM cultures recapitulate drug gradients
Doxorubicin uptake linearly correlated with concentration in 2D and 3D systems; however, the uptake in 2D was higher than the 3DTEBM, in which the slope of the correlation in 2D (6.4) was 4.4-fold higher than the slope in the 3DTEBM (1.45). The other commercially available 3D systems showed lower uptake than in 2D, but higher uptake than in the 3DTEBM (Figure 4E). A representative flow cytometry histogram shows the reduced doxorubicin uptake in 3DTEBM compared to 2D mono-cultures (Figure 4F). Furthermore, the drug uptake of MM cells grown in 3DTEBM was inversely correlated with the depth of the scaffold (R2=0.9811) (Figure 4G).
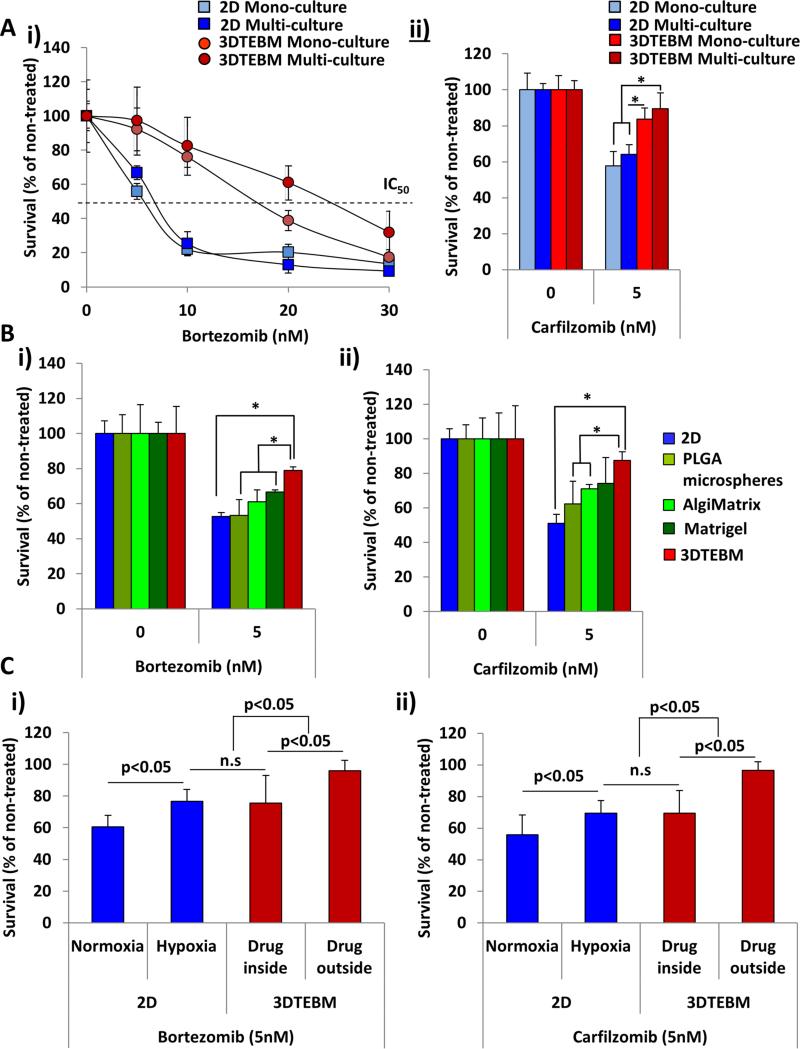
Effect of 3DTEBM cultures on drug resistance in MM
The inhibitory concentration 50 (IC50) of bortezomib in 2D mono-culture was approximately 5 nM, while the 2D multi-culture conditions demonstrated increased resistance (IC50 approximately 7 nM) (Figure 5Ai). Culturing MM cells in 3DTEBM as mono-culture induced a stronger resistance (IC50 = 17 nM), but culturing MM cells in 3DTEBM multi-culture showed the highest resistance (IC50 = 25 nM).
Figure 5. Effect of 3DTEBM cultures on drug resistance in MM.
A) Effect of i) increasing concentrations of bortezomib (24 hours) and ii) carfilzomib (5 nM, 24 hours) on MM survival when cultured in classic 2D (mono- or multi-culture) and 3DTEBM (mono- or multi-culture) systems, (*) p< 0.05. B) The effect of i) bortezomib (0, 5nM, 24 hours) and ii) carfilzomib (0, 5 nM, 24 hours) on MM cell multi-culture survival in 2D culture, PLGA microspheres, AlgiMatrix, Matrigel and 3DTEBM, (*) p< 0.01. C) The effect of i) bortezomib (0, 5nM, 24 hours), and ii) carfilzomib (0, 5 nM, 24 hours) on survival of MM cell mono-cultures in 2D cultures (normoxic or hypoxic conditions) compared to 3DTEBM cultures (drug added before or after 3DTEBM scaffold preparation), (*) p< 0.05.
Similar results were observed with carfilzomib (IC50 = 5 nM) (Figure 5Aii), in which classic 2D mono-culture induced about 40% killing of the MM cells, and 2D multi-culture modestly decreased the sensitivity of MM cells to carfilzomib. 3DTEBM cultures profoundly decreased the sensitivity of MM cells to carfilzomib, and multi-culture in the 3DTEBM induced the most profound resistance to carfilzomib by killing only 10% of the MM.
Furthermore, we found that bortezomib (Figure 5BEi) and carfilzomib (Figure 5Bii) induced about 50% killing in the 2D cultures, PLGA microspheres did not affect drug resistance, AlgiMatrix induced a modest increase in drug resistance, Matrigel induced a slightly higher drug resistance in MM cells, and the 3DTEBM induced the highest drug resistance among all other culture systems.
Finally, we tested the differential roles of 3D aspects of the 3DTEBM (hypoxia and drug gradients) on drug resistance. 2D cultures in normoxic conditions (no hypoxia and no drug gradients) showed 40 - 50% killing with bortezomib (Figure 5Ci) and carfilzomib (Figure 5Cii), hypoxic 2D cultures (no drug gradient but inducing hypoxia) showed significant drug resistance with only 25% killing. Similarly, 3DTEBM cultures with drug uniformly mixed during preparation (thereby eliminating drug gradient effects but maintaining hypoxia) showed comparable cell killing to the 2D hypoxic culture. However, when drugs were applied to the top surface of a preformed 3DTEBM (maintaining drug and hypoxia gradients), there was development of a profound resistance to treatment (less than 10% killing of MM cells).
DISCUSSION
The discrepancy between laboratory and clinical outcomes result from limitation of the current methods used to develop MM treatments including: (1) neglecting the vital role of the BM microenvironment in MM progression and drug resistance, and (2) the 3D structure with oxygen and drug concentration gradients which cannot be demonstrated in classic 2D cultures.
Several models are being used to study MM: 1) In vitro 2D culture of MM cell lines are widely used; but such models utilize MM cell lines alone, which clearly lack the effect of the BM microenvironment, the 3D aspects of the BM such as drug and oxygen gradients, and the personal heterogeneity between patients. In some cases, MM cells are used in co-culture with BM stromal cells [29], endothelial cells [30], and the extracellular matrix (ECM) [31]; which all add value to understanding the interaction of MM cells with the BM and its role in drug resistance, but still lack the 3D and the personal heterogeneity aspects of the disease. 2) In vitro 3D models made of acrylic polymers[18], Matrigel [17], or silk fibers [19] provide better alternatives to 2D cultures, but each has its own limitations. The acrylic polymer and the Matrigel based models use synthetic materials which are not naturally found in the BM, and may cause significant changes in the interaction between the different components of the culture and the matrix. Moreover, none of these models were shown to support proliferation of primary MM cells and they rely mainly on established MM cell lines. 3) In vivo models based on bone chips [32] use fetal bone chips implanted in SCID mice; after the fetal bones grow in the mouse, MM cells are injected into the bone. This model resembles BM physiological conditions more closely; however, it relies on normal BM microenvironment which we have different effects on the MM cell growth compared with the malignant microenvironment from MM [23]. In addition, this model is costly and labor intensive: it requires 4 weeks for the growth of the bone in mice and 4 more weeks for the growth of the MM cells in the bone (8 weeks in total), which adds more challenge to the technical feasibility of the model. 4) In vivo xenograft models of human MM cell lines in SCID mice are widely used. These xenografts are faster and less technically challenging than the bone chip models, however, this model demonstrates the interaction of human MM cells with normal-mouse microenvironment, which may have significant implications on the growth and drug resistance of MM cells. 5) C57BL/KaLwRij mice spontaneously develop MM at late age; this model mimics the slow pathophysiological development of MM and demonstrates the interaction of MM cells with malignant mouse-stroma. However, the disease presentation of mouse-MM afforded from this model differs significantly from the presentation of human-MM; such as absence of renal damage due to light chain deposition, which is considered one of the main complications of the human disease [33]. Moreover, this model cannot be used to represent heterogeneity between MM patients. In comparison, our new model is reproducible, fast and not technically challenging, incorporates the interactions of human MM cells with their malignant microenvironment, simulates 3D aspects of the BM niche such as drug gradients and hypoxia, takes into account the individual heterogeneity between patients, and contains minimal additions of exogenous materials to minimize their effect on the pathophysiological environment of MM. The main limitations of the 3DTEBM model are 1) unlike the in vivo systems, the 3DTEBM cannot be used to study spread and metastasis, 2) unlike the in vivo systems, the 3DTEBM lacks of shear flow components which recreates conditions similar to blood stream, and 3) unlike synthetic models, to create the 3DTEBM system it requires BM aspirates from patients which limits the availability of the model to laboratories without hospital collaborations.
In this study, unlike other 3D scaffolds developed from exogenous materials [17-19], we used fibrinogen naturally found in blood plasma and the BM supernatant to develop 3D scaffolds. The 3DTEBM included no exogenous polymer to produce the scaffold, which minimizes the manipulation of the natural pathophysiological environment of MM.
Fibrin-based scaffolds have been used before as 3D systems for cell culture and are characterized by safety, manageability, biocompatibility, and reproducibility [25, 26, 34-38]. Fibrin is an excellent scaffold for tissue engineering techniques (adipose, bone, cardiac, cartilage, muscle, ocular, skin, tendons, and vascular tissue engineering) and other strategies including delivery systems [38-45]. Fibrin-based scaffolds made from patient-derived plasma contains molecules that help cellular adhesion, such as fibronectin, and it is a reservoir for growth factors, platelets, cytokines and enzymes from the own patient [25, 37]. The 3D scaffold structure of fibrin for cell culture is composed of a network of interconnected pores, through which cells actively migrate, multiply and spread along the scaffold, the diffusion of nutrients and waste products is facilitated, and vascularization is promoted and developed [38]. In addition, cell-fibrin combinations have shown to be useful as bone graft substitutes due to their capacity to induce bone formation [38, 46, 47].
We have fine-tuned the conditions for developing 3DTEBM scaffolds using optimal concentration of calcium as a cross linker [48, 49] and tranexamic acid to reduce fibrin degradation, maintain structural integrity and stability, which it does not affect cell viability [25, 26, 38, 50, 51]. Furthermore, we optimized the density of MM, stromal, and endothelial cells at the time of preparation to promote the proliferation of MM cells in the 3DTEBM in a 96-well plate.
The 3D structure of 3DTEBM was shown to be a network of interconnecting pores, in which MM cells, stromal cells, and endothelial cells were able to interact with the fibers and between each other, and promote proliferation of the MM cells. We demonstrated that 3DTEBM recreated the cellular polarization in the 3D-axis similar to the BM niche, in which stromal cells migrated to the bottom of the scaffold and created a sheet-like structure, while endothelial cells migrated towards the top surface of the scaffold, and MM cells grew homogenously throughout the scaffold and showed interactions with both stromal and endothelial cells. The 3DTEBM scaffold promoted superior growth compared to classic 2D and other commercial 3D tissue culture systems. This is a proof of concept that different matrices have different effects on the proliferation of MM, and that the 3DTEBM derived from the BM supernatant of MM patients promotes more proliferation of MM cells.
Ex vivo culturing of primary MM cells has been a major challenge because of the lack of an in vitro technology capable of recreating the complicated bone marrow microenvironment which MM cells depend on for their survival. In this study, we have shown that the 3DTEBM promoted the progression of primary patient samples (fresh and frozen) for more than a week, while in 2D cultures or other commercially available 3D systems, the primary samples did not promote proliferation compared to Day 0. The higher proliferation rate observed in 3DTEBM cultures, in cell lines as well as in primary patient samples, in comparison to 2D and other 3D systems is clinically relevant due to this higher proliferation rate represent more closely the clinical manifestation of the disease. These results will have a significant impact in the field since it will allow validation of biological mechanisms and experimental procedures which need longer culture time in primary MM cells.
We have previously shown that indirect interactions such as secretion of cytokines and growth factors play a crucial role in the progression and drug resistance of MM cells [5-9, 14, 52, 53]. Therefore, we have tested the effect of culturing MM cells and other accessory cells in the 3DTEBM on the secretion of cytokines in the culture milieu. We found that the 3DTEBM itself served as a reservoir of cytokines and growth factors, since it was actually derived from the BM of MM patients. In addition, we found that culturing MM cells in the 3DTEBM increased the secretion of pro-angiogenic molecules (VEGF, IL-8 and OPN), pro-inflammatory (IL-1α, IL-6, TNF-α, and IL-8) and tumor cytokines involved in cell trafficking (VEGF, SDF-1, IL-8, GRO and MCP-1). In the last years, increased angiogenesis has been demonstrated in the BM microenvironment in hematologic malignancies, including MM, suggesting a potential pathophysiologic role for angiogenesis in MM [54]. MM patients with active disease have increased BM angiogenesis compared to patients with smoldering MM or early stage MM [55, 56]. Novel agents in MM, such as the immunomodulatory drug (IMiD) thalidomide, and the proteasome inhibitor (PI) bortezomib, have anti-angiogenic activity [57]. In addition, excessive tumor cell proliferation and the related hypoxia lead to overproduction of pro-angiogenic factors [58, 59]. Therefore, the development of a 3D culture model that recreates angiogenesis will be particularly attractive due to the importance of angiogenesis for tumor growth and drug response [58]. BM angiogenesis is regulated by an overbalance of pro-angiogenic factors by both myeloma cells and the microenvironment, such as vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), interleukin-6 (IL-6) and -8 (IL-8), angiopoietins, osteopontin, and hepatocyte growth factor (HGF) [60-62], and we found that secretion of most of these molecules (VEGF, IL-8, ANG, OPN, and HGF) were up-regulated by 3DTEBM cultures. In addition, the role of hypoxia and HIF-1α in the production of pro-angiogenic molecules (VEGF, IL-8 and OPN) by myeloma cells has been demonstrated [63]. Therefore, 3DTEBM cultures may provide a valuable tool to comprehend the role of microenvironment signals, such as angiogenesis or inflammation in MM cancer progression, as well as provide a means to study new potential targets in MM patients. IL-6, VEGF, AGN, OPN, MCP-1, SDF-1, and HGF have been previously reported to be increased in other 3D models [20, 32, 64-66]. The two cytokines up-regulated exclusively in our 3DTEBM multi-cultures were HGF and SDF-1. HGF is an angiogenic key signal in the evolution of MM acting as autocrine/paracrine growth factor and survival factor for MM cells [67]. HGF levels are dependent of the severity of MM suggesting that this cytokine may be useful for assessing disease progression and for predicting response to chemotherapy in MM patients [68, 69]. SDF-1 plays a critical role in supporting adhesion mediated-growth and cell trafficking of MM [70, 71]. Therefore, SDF-1 might be a key regulator to control disruption of adhesion and induction of mobilization, which could lead to increase sensitivity to therapeutic agents [8].
The BM niche is a 3D structure which creates drug concentration gradients that are inversely proportional to the distance from blood vessels [11, 12]. Moreover, we have previously found that oxygen gradients (hypoxia) developed in the BM during the progression of MM, which induced metastasis [14, 72] and drug-resistance in MM cells in vitro and in vivo [73, 74]. 2D and 3D models that grow MM cells on top of a polymer surface [18, 19] or bone chip [32] lack the development of drug gradients and hypoxia; therefore, these will not accurately depict proliferation and drug resistance of MM cells in the BM niche. We found that the 3DTEBM cultures recreated oxygen gradients throughout the depth of the scaffold, which was confirmed by increased endogenous (HIF1α) and exogenous (PIM) hypoxia markers; and other biological changes linked to hypoxic MM cells such as overexpression of CXCR4, down regulation of CD138 and decreased proliferation. These results show the recapitulation of the BM niche in MM, where the vascular niche (close to the blood vessels/top of the scaffold) presents high oxygenation levels and more proliferative cells, while the endosteal niche (close to the bone) is hypoxic, and includes less proliferative cells [11, 12, 14]. Similar to oxygen gradients the 3DTEBM recreated drug gradients through the depth of the tissue, and to achieve the same amount of drug uptake observed in 2D, concentrations of about 4-fold higher were needed in the 3DTEBM. The other commercially available 3D systems (PLGA microspheres, AlgiMatrix, and Matrigel) were not able to recreate the same drug gradient profile of the 3DTEBM, with lower uptake than in 2D, but higher uptake than in the 3DTEBM. The diffusion of drugs in the 3DTEBM was inversely correlated with the depth of the 3DTEBM. This is in agreement with our previous findings that treatment with bortezomib (3 weeks of 1mg/kg/week) in vivo was more effective in MM cells growing close to the vessels than in MM cells growing close to the bone [75].
The overall proliferation rate in the patient samples and cell lines in the 3DTEBM is higher than the 2D cultures and other commercially available 3D systems due to the fact that the microenvironment mimics better the BM microenvironment. However, in the 3DTEBM system we found areas with different rates of growth, while the upper (normoxic) layers are very highly proliferative, the bottom (hypoxic) layer is less proliferative compared to the upper layer.
We further demonstrated that multi-culture of MM cells with accessory cells in 2D (representing the role of the BM microenvironment), as well as culturing MM cells alone in 3DTEBM (representing 3D aspects and cytokines from the BM supernatant) decreased the sensitivity of MM cells to therapy. However, the most profound resistance of MM cells to therapy was observed when MM cells were multi-cultured with accessory cells in the 3DTEBM (representing the role of BM microenvironment, cytokines from the BM supernatant and the 3D aspects). MM cells in the 3DTEBM showed significantly higher resistance to bortezomib and carfilzomib compared to other 3D cultures.
Then, we sought to understand the differential roles that drug gradients and oxygen gradients play in the 3DTEBM and in the resistance mechanisms to these two proteasome inhibitors. We compared the drug resistance to bortezomib and carfilzomib in the 3DTEBM once when added them from outside (which reflects both drug gradient and hypoxia) or incorporated into the matrix (which rules out the role of drug gradient but maintains hypoxia); and we used the drugs in 2D cultures in hypoxia and normoxia (without drug gradient effects). It was shown that hypoxia alone in 2D culture induced resistance to bortezomib and carfilzomib; a comparable effect was observed in 3DTEBM when the drug was uniformly mixed in the 3DTEBM (representing only the effect of hypoxia but not drug gradient). However, a higher dug resistance was observed in the 3DTEBM when the drug was applied to the outside surface of the 3DTEBM (representing both he hypoxia and drug gradients). Therefore, we found that both two factors were important and contributed to the drug resistance to bortezomib and carfilzomib.
In conclusion, we developed 3DTEBM scaffolds by crosslinking the fibrinogen in BM supernatant preserving the natural pathophysiological environment; this is in contrast to other 3D culture methods that rely on exogenous polymer to produce the scaffold. Since the 3DTEBM is derived from the BM supernatant, it includes all the growth factors and cytokines naturally found in the MM microenvironment. MM cells were shown to proliferate in the 3DTEBM better than classic 2D and other commercial 3D tissue culture systems. Most importantly, primary MM cells (both fresh and frozen samples) were able to proliferate in the 3DTEBM but not in the 2D and other commercially available 3D systems, which may have significant implications on the field. The 3DTEBM recreated BM interactions and reproduced tissue-specific structural features, such as the endosteal and vascular niche. Moreover, the 3DTEBM recreated 3D aspects (such as oxygen and drug gradients) observed in the BM niche and, in turn, induced more drug resistance than 2D and commercial 3D tissue culture systems in MM cells (Figure 6). The 3DTEBM cultures not only provide a better model for investigating biological mechanisms of MM progression, but also it provides a paradigm shifting tool for drug development and screening in MM. In the future, we will develop 3DTEBM from individual MM patients (using autologous BM supernatant, MM cell and accessory BM cells from the same patient) to perform drug screens for each patient, and develop personalized therapeutic strategies for individual MM patients.
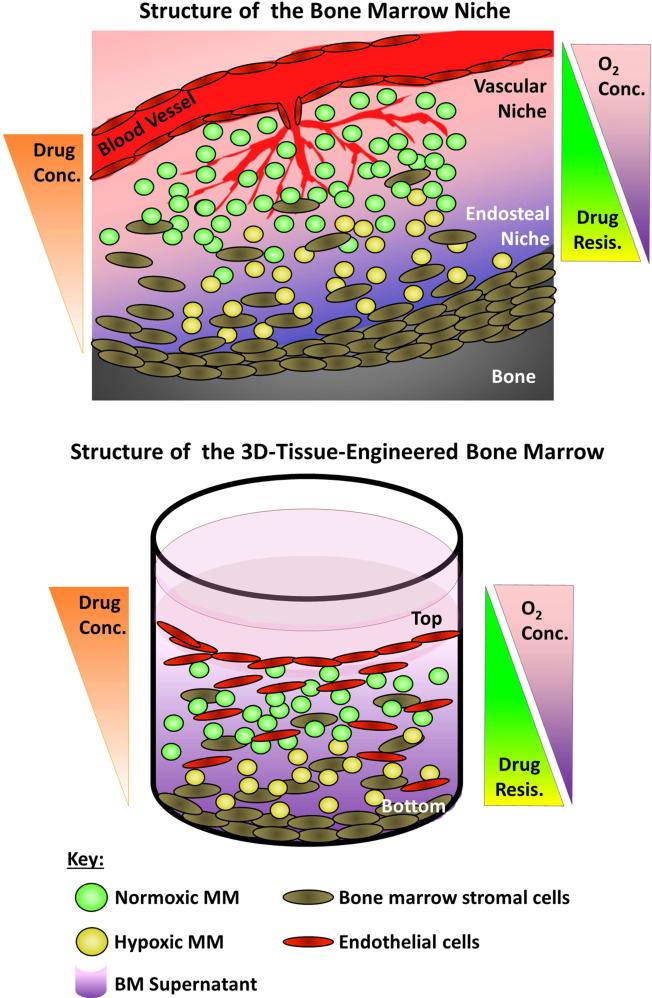
Figure 6. The 3DTEBM recapitulates the BM niche structure.
A) Cartoon of the BM niche with oxygen and drug concentration gradients as a function of the distance from blood vessels. The vascular niche (close to the blood vessels) provides higher levels of oxygenation and contains higher proliferating and more sensitive cells to therapy (green). The endosteal niche (close to the bone) is hypoxic, receives lower effective drug concentrations, and consists of less proliferative and more drug-resistant cells (yellow). B) Cartoon of the three-dimensional tissue engineered BM (3DTEBM) niche with oxygen and drug concentration gradients as a function of scaffold depth. The top of the 3DTEBM is enriched for endothelital cells, where MM cells are exposed to higher oxygen levels and drug concentrations, with more proliferative cells (green). The bottom of the 3DTEBM is hypoxic, receives lower concentration of drugs, and includes less proliferative and more drug-resistant cells (yellow). The 3DTEBM provides a tool for further studying the vascular and endosteal niches in ex-vivo experiments.
Further research is required to assess the role of other less characterized BM cellular components (osteoclasts, osteoblasts, and immune cells), as well as the cytokines and growth factors contained in the BM supernatant, on progression and drug resistance in the 3DTEBM. Additional work will be necessary using novel MM therapies including immunomodulatory drugs, proteasome inhibitors, monoclonal antibodies, cell signaling targeted therapies, and strategies targeting the tumor microenvironment. Moreover, we will introduce the use of the 3DTEBM in a bioreactor system with a shear-flow to study metastasis. Therefore, the 3DTEBM will serve as a promising model for evaluation of immunological events, metastasis, and prediction of therapeutic efficacy in patients. But most importantly, we will conduct prospective and retrospective studies between clinical response and the drug sensitivity/resistance in the 3DTEBM of individual patients.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank assistance from the Research Center for Auditory and Vestibular Studies, a P30 Research Core Center (P30 DC004665, Richard A. Chole, PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship: P.P.: Designed the study, performed research, analyzed and interpreted data, and wrote the manuscript. B.M., F.A., M.L, R.G. and S.A.: Performed research, analyzed and interpreted data. J.K. and R.V.: Provided primary MM samples, analyzed and interpreted data. A.K.A: Designed the study, analyzed and interpreted data, wrote the manuscript, and supervised the study.
CONFLICT OF INTEREST
Dr. Azab receives research support from Verastem, Selexys, Karyopharm and Cell Works, and is the founder and owner of Targeted Therapeutics LLC, and he has a provisional patent application on the method described in this manuscript. Dr. de la Puente has a provisional patent application on the method described in this manuscript. Other authors state no conflicts of interest.
REFERENCES
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. The New England journal of medicine. 2004;351:1860–73. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA: a cancer journal for clinicians. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Dimopoulos M, Spencer A, Attal M, Prince HM, Harousseau JL, Dmoszynska A, et al. Lenalidomide plus dexamethasone for relapsed or refractory multiple myeloma. The New England journal of medicine. 2007;357:2123–32. doi: 10.1056/NEJMoa070594. [DOI] [PubMed] [Google Scholar]
- 4.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. The New England journal of medicine. 2005;352:2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 5.de la Puente P, Azab AK. Contemporary drug therapies for multiple myeloma. Drugs Today (Barc) 2013;49:563–73. doi: 10.1358/dot.2013.49.9.2020941. [DOI] [PubMed] [Google Scholar]
- 6.de la Puente P, Muz B, Azab F, Luderer M, Azab AK. Molecularly targeted therapies in multiple myeloma. Leukemia research and treatment. 2014;2014:976567. doi: 10.1155/2014/976567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azab AK, Quang P, Azab F, Pitsillides C, Thompson B, Chonghaile T, et al. P-selectin glycoprotein ligand regulates the interaction of multiple myeloma cells with the bone marrow microenvironment. Blood. 2012;119:1468–78. doi: 10.1182/blood-2011-07-368050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–51. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azab AK, Azab F, Blotta S, Pitsillides CM, Thompson B, Runnels JM, et al. RhoA and Rac1 GTPases play major and differential roles in stromal cell-derived factor-1-induced cell adhesion and chemotaxis in multiple myeloma. Blood. 2009;114:619–29. doi: 10.1182/blood-2009-01-199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsiades CS, Mitsiades NS, Richardson PG, Munshi NC, Anderson KC. Multiple myeloma: a prototypic disease model for the characterization and therapeutic targeting of interactions between tumor cells and their local microenvironment. Journal of cellular biochemistry. 2007;101:950–68. doi: 10.1002/jcb.21213. [DOI] [PubMed] [Google Scholar]
- 11.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nature reviews Immunology. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 12.Burness ML, Sipkins DA. The stem cell niche in health and malignancy. Seminars in cancer biology. 2010;20:107–15. doi: 10.1016/j.semcancer.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Tunggal JK, Cowan DS, Shaikh H, Tannock IF. Penetration of anticancer drugs through solid tissue: a factor that limits the effectiveness of chemotherapy for solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 1999;5:1583–6. [PubMed] [Google Scholar]
- 14.Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood. 2012;119:5782–94. doi: 10.1182/blood-2011-09-380410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemeyer P, Krause U, Fellenberg J, Kasten P, Seckinger A, Ho AD, et al. Evaluation of mineralized collagen and alpha-tricalcium phosphate as scaffolds for tissue engineering of bone using human mesenchymal stem cells. Cells, tissues, organs. 2004;177:68–78. doi: 10.1159/000079182. [DOI] [PubMed] [Google Scholar]
- 16.Yang XB, Bhatnagar RS, Li S, Oreffo RO. Biomimetic collagen scaffolds for human bone cell growth and differentiation. Tissue engineering. 2004;10:1148–59. doi: 10.1089/ten.2004.10.1148. [DOI] [PubMed] [Google Scholar]
- 17.Kirshner J, Thulien KJ, Martin LD, Debes Marun C, Reiman T, Belch AR, et al. A unique three- dimensional model for evaluating the impact of therapy on multiple myeloma. Blood. 2008;112:2935–45. doi: 10.1182/blood-2008-02-142430. [DOI] [PubMed] [Google Scholar]
- 18.Calimeri T, Battista E, Conforti F, Neri P, Di Martino MT, Rossi M, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011;25:707–11. doi: 10.1038/leu.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reagan MR, Mishima Y, Glavey SV, Zhang Y, Manier S, Lu ZN, et al. Investigating osteogenic differentiation in multiple myeloma using a novel 3D bone marrow niche model2014. doi: 10.1182/blood-2014-02-558007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan NK, Duan B, Butcher JT, Mazumder A, Narayanan BA. Characterization of multiple myeloma clonal cell expansion and stromal Wnt/beta-catenin signaling in hyaluronic acid-based 3D hydrogel. In Vivo. 2014;28:67–73. [PubMed] [Google Scholar]
- 21.Zhang W, Lee WY, Siegel DS, Tolias P, Zilberberg J. Patient-Specific 3D Microfluidic Tissue Model for Multiple Myeloma. Tissue engineering Part C, Methods. 2014 doi: 10.1089/ten.TEC.2013.0490. [DOI] [PubMed] [Google Scholar]
- 22.Ferrarini M, Steimberg N, Ponzoni M, Belloni D, Berenzi A, Girlanda S, et al. Ex-vivo dynamic 3-D culture of human tissues in the RCCS bioreactor allows the study of Multiple Myeloma biology and response to therapy. PloS one. 2013;8:e71613. doi: 10.1371/journal.pone.0071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, et al. BM mesenchymal stromal cell- derived exosomes facilitate multiple myeloma progression. The Journal of clinical investigation. 2013;123:1542–55. doi: 10.1172/JCI66517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vacca A, Ria R, Semeraro F, Merchionne F, Coluccia M, Boccarelli A, et al. Endothelial cells in the bone marrow of patients with multiple myeloma. Blood. 2003;102:3340–8. doi: 10.1182/blood-2003-04-1338. [DOI] [PubMed] [Google Scholar]
- 25.de la Puente P, Ludena D, Fernandez A, Aranda JL, Varela G, Iglesias J. Autologous fibrin scaffolds cultured dermal fibroblasts and enriched with encapsulated bFGF for tissue engineering. Journal of biomedical materials research Part A. 2011;99:648–54. doi: 10.1002/jbm.a.33231. [DOI] [PubMed] [Google Scholar]
- 26.de la Puente P, Ludena D, Lopez M, Ramos J, Iglesias J. Differentiation within autologous fibrin scaffolds of porcine dermal cells with the mesenchymal stem cell phenotype. Experimental cell research. 2013;319:144–52. doi: 10.1016/j.yexcr.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Sahoo SK, Panda AK, Labhasetwar V. Characterization of porous PLGA/PLA microparticles as a scaffold for three dimensional growth of breast cancer cells. Biomacromolecules. 2005;6:1132–9. doi: 10.1021/bm0492632. [DOI] [PubMed] [Google Scholar]
- 28.Godugu C, Patel AR, Desai U, Andey T, Sams A, Singh M. AlgiMatrix based 3D cell culture system as an in-vitro tumor model for anticancer studies. PloS one. 2013;8:e53708. doi: 10.1371/journal.pone.0053708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azab AK, Runnels JM, Pitsillides C, Moreau A-S, Azab F, Leleu X, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–51. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azab AK, Quang P, Azab F, Pitsillides C, Thompson B, Chonghaile T, et al. P-selectin glycoprotein ligand regulates the interaction of multiple myeloma cells with the bone marrow microenvironment. Blood. 2012;119:1468–78. doi: 10.1182/blood-2011-07-368050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azab AK, Azab F, Blotta S, Pitsillides CM, Thompson B, Runnels JM, et al. RhoA and Rac1 GTPases play major and differential roles in stromal cell–derived factor-1–induced cell adhesion and chemotaxis in multiple myeloma. Blood. 2009;114:619–29. doi: 10.1182/blood-2009-01-199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urashima M, Chen BP, Chen S, Pinkus GS, Bronson RT, Dedera DA, et al. The development of a model for the homing of multiple myeloma cells to human bone marrow. Blood. 1997;90:754–65. [PubMed] [Google Scholar]
- 33.Libouban H, Onno C, Pascaretti-Grizon F, Gallois Y, Moreau MF, Basle MF, et al. Absence of renal lesions in C57BL/KaLwRij mice with advanced myeloma due to 5T2MM cells. Leukemia research. 2006;30:1371–5. doi: 10.1016/j.leukres.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Siedentop KH, Park JJ, Shah AN, Bhattacharyya TK, O'Grady KM. Safety and efficacy of currently available fibrin tissue adhesives. American Journal of Otolaryngology. 2001;22:230–5. doi: 10.1053/ajot.2001.24817. [DOI] [PubMed] [Google Scholar]
- 35.Linnes MP, Ratner BD, Giachelli CM. A fibrinogen-based precision microporous scaffold for tissue engineering. Biomaterials. 2007;28:5298–306. doi: 10.1016/j.biomaterials.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed TAE, Dare EV, Hincke M. Fibrin: A versatile scaffold for tissue engineering applications. Tissue Engineering - Part B: Reviews. 2008;14:199–215. doi: 10.1089/ten.teb.2007.0435. [DOI] [PubMed] [Google Scholar]
- 37.Clark RA. Fibrin is a many splendored thing. The Journal of investigative dermatology. 2003;121:xxi, xxii. doi: 10.1046/j.1523-1747.2003.12575.x. [DOI] [PubMed] [Google Scholar]
- 38.de la Puente P, Ludena D. Cell culture in autologous fibrin scaffolds for applications in tissue engineering. Experimental cell research. 2014;322:1–11. doi: 10.1016/j.yexcr.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Zhang YS, Gao JH, Lu F, Zhu M. [Adipose tissue engineering with human adipose-derived stem cells and fibrin glue injectable scaffold]. Zhonghua Yi Xue Za Zhi. 2008;88:2705–9. [PubMed] [Google Scholar]
- 40.Yamada Y, Boo JS, Ozawa R, Nagasaka T, Okazaki Y, Hata K, et al. Bone regeneration following injection of mesenchymal stem cells and fibrin glue with a biodegradable scaffold. J Craniomaxillofac Surg. 2003;31:27–33. doi: 10.1016/s1010-5182(02)00143-9. [DOI] [PubMed] [Google Scholar]
- 41.Jockenhoevel S, Zund G, Hoerstrup SP, Chalabi K, Sachweh JS, Demircan L, et al. Fibrin gel -- advantages of a new scaffold in cardiovascular tissue engineering. Eur J Cardiothorac Surg. 2001;19:424–30. doi: 10.1016/s1010-7940(01)00624-8. [DOI] [PubMed] [Google Scholar]
- 42.Eyrich D, Brandl F, Appel B, Wiese H, Maier G, Wenzel M, et al. Long-term stable fibrin gels for cartilage engineering. Biomaterials. 2007;28:55–65. doi: 10.1016/j.biomaterials.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 43.Huang YC, Dennis RG, Larkin L, Baar K. Rapid formation of functional muscle in vitro using fibrin gels. J Appl Physiol. 1985;98:706–13. doi: 10.1152/japplphysiol.00273.2004. [DOI] [PubMed] [Google Scholar]
- 44.Han B, Schwab IR, Madsen TK, Isseroff RR. A fibrin-based bioengineered ocular surface with human corneal epithelial stem cells. Cornea. 2002;21:505–10. doi: 10.1097/00003226-200207000-00013. [DOI] [PubMed] [Google Scholar]
- 45.Geer DJ, Swartz DD, Andreadis ST. Fibrin promotes migration in a three-dimensional in vitro model of wound regeneration. Tissue Eng. 2002;8:787–98. doi: 10.1089/10763270260424141. [DOI] [PubMed] [Google Scholar]
- 46.Yamada Y, Seong Boo J, Ozawa R, Nagasaka T, Okazaki Y, Hata K-i, et al. Bone regeneration following injection of mesenchymal stem cells and fibrin glue with a biodegradable scaffold. Journal of Cranio-Maxillofacial Surgery. 2003;31:27–33. doi: 10.1016/s1010-5182(02)00143-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhou H, Xu HHK. The fast release of stem cells from alginate-fibrin microbeads in injectable scaffolds for bone tissue engineering. Biomaterials. 2011;32:7503–13. doi: 10.1016/j.biomaterials.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Guehennec L, Goyenvalle E, Aguado E, Pilet P, Spaethe R, Daculsi G. Influence of calcium chloride and aprotinin in the in vivo biological performance of a composite combining biphasic calcium phosphate granules and fibrin sealant. Journal of materials science Materials in medicine. 2007;18:1489–95. doi: 10.1007/s10856-006-0086-x. [DOI] [PubMed] [Google Scholar]
- 49.Silver FH, Wang MC, Pins GD. Preparation of fibrin glue: a study of chemical and physical methods. Journal of applied biomaterials : an official journal of the Society for Biomaterials. 1995;6:175–83. doi: 10.1002/jab.770060306. [DOI] [PubMed] [Google Scholar]
- 50.Cholewinski E, Dietrich M, Flanagan TC, Schmitz-Rode T, Jockenhoevel S. Tranexamic acid--an alternative to aprotinin in fibrin-based cardiovascular tissue engineering. Tissue engineering Part A. 2009;15:3645–53. doi: 10.1089/ten.tea.2009.0235. [DOI] [PubMed] [Google Scholar]
- 51.Demol J, Eyckmans J, Roberts SJ, Luyten FP, Van Oosterwyck H. Does tranexamic acid stabilised fibrin support the osteogenic differentiation of human periosteum derived cells? European cells & materials. 2011;21:272–85. doi: 10.22203/ecm.v021a21. [DOI] [PubMed] [Google Scholar]
- 52.Azab AK, Azab F, Quang P, Maiso P, Ngo HT, Zimmermann J, et al. Dissecting the role of CXCR7 in Cell Trafficking of Endothelial-Cells and Endothelial-Progenitor-Cells in Multiple Myeloma. ASH Annual Meeting Abstracts. 2011;118:3934. [Google Scholar]
- 53.Azab F, Azab AK, Maiso P, Calimeri T, Flores L, Liu Y, et al. Eph-B2/Ephrin-B2 Interaction Plays a Major Role in the Adhesion and Proliferation of Waldenstrom's Macroglobulinemia. Clinical Cancer Research. 2012;18:91–104. doi: 10.1158/1078-0432.CCR-11-0111. [DOI] [PubMed] [Google Scholar]
- 54.Giuliani N, Storti P, Bolzoni M, Palma B, Bonomini S. Angiogenesis and Multiple Myeloma. Cancer Microenvironment. 2011;4:325–37. doi: 10.1007/s12307-011-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vacca A, Ribatti D, Roccaro AM, Frigeri A, Dammacco F. Bone marrow angiogenesis in patients with active multiple myeloma. Seminars in oncology. 2001;28:543–50. doi: 10.1016/s0093-7754(01)90022-3. [DOI] [PubMed] [Google Scholar]
- 56.Rajkumar SV, Mesa RA, Fonseca R, Schroeder G, Plevak MF, Dispenzieri A, et al. Bone marrow angiogenesis in 400 patients with monoclonal gammopathy of undetermined significance, multiple myeloma, and primary amyloidosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:2210–6. [PubMed] [Google Scholar]
- 57.Anargyrou K, Dimopoulos MA, Sezer O, Terpos E. Novel anti-myeloma agents and angiogenesis. Leukemia & lymphoma. 2008;49:677–89. doi: 10.1080/10428190701861686. [DOI] [PubMed] [Google Scholar]
- 58.Folkman J. Tumor angiogenesis: therapeutic implications. The New England journal of medicine. 1971;285:1182–6. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 59.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nature reviews Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 60.de la Puente P, Muz B, Azab F, Azab AK. Cell Trafficking of Endothelial Progenitor Cells in Tumor Progression. Clinical Cancer Research. 2013;19:3360–8. doi: 10.1158/1078-0432.CCR-13-0462. [DOI] [PubMed] [Google Scholar]
- 61.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nature reviews Cancer. 2002;2:727–39. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 62.Vacca A, Ribatti D. Bone marrow angiogenesis in multiple myeloma. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006;20:193–9. doi: 10.1038/sj.leu.2404067. [DOI] [PubMed] [Google Scholar]
- 63.Colla S, Storti P, Donofrio G, Todoerti K, Bolzoni M, Lazzaretti M, et al. Low bone marrow oxygen tension and hypoxia-inducible factor-1alpha overexpression characterize patients with multiple myeloma: role on the transcriptional and proangiogenic profiles of CD138(+) cells. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:1967–70. doi: 10.1038/leu.2010.193. [DOI] [PubMed] [Google Scholar]
- 64.Groen RWJ, Hideshima T, Cholujova D, Sedlak J, Mitsiades CS, Laubach JP, et al. Myeloma Patient- Derived Mesenchymal Stem Cells Grown In 3-D Culture Induce Primary Myeloma Cell Proliferation and Resistance To Therapy2013 [Google Scholar]
- 65.Zdzisinska B, Rolinski J, Piersiak T, Kandefer-Szerszen M. A comparison of cytokine production in 2- dimensional and 3-dimensional cultures of bone marrow stromal cells of multiple myeloma patients in response to RPMI8226 myeloma cells. Folia histochemica et cytobiologica / Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2009;47:69–74. doi: 10.2478/v10042-009-0015-1. [DOI] [PubMed] [Google Scholar]
- 66.Oswald J, Steudel C, Salchert K, Joergensen B, Thiede C, Ehninger G, et al. Gene-expression profiling of CD34+ hematopoietic cells expanded in a collagen I matrix. Stem Cells. 2006;24:494–500. doi: 10.1634/stemcells.2005-0276. [DOI] [PubMed] [Google Scholar]
- 67.Tjin EP, Derksen PW, Kataoka H, Spaargaren M, Pals ST. Multiple myeloma cells catalyze hepatocyte growth factor (HGF) activation by secreting the serine protease HGF-activator. Blood. 2004;104:2172–5. doi: 10.1182/blood-2003-12-4386. [DOI] [PubMed] [Google Scholar]
- 68.Iwasaki T, Hamano T, Ogata A, Hashimoto N, Kitano M, Kakishita E. Clinical significance of vascular endothelial growth factor and hepatocyte growth factor in multiple myeloma. British journal of haematology. 2002;116:796–802. doi: 10.1046/j.0007-1048.2002.03364.x. [DOI] [PubMed] [Google Scholar]
- 69.Sezer O, Jakob C, Eucker J, Niemoller K, Gatz F, Wernecke K, et al. Serum levels of the angiogenic cytokines basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) in multiple myeloma. European journal of haematology. 2001;66:83–8. doi: 10.1034/j.1600-0609.2001.00348.x. [DOI] [PubMed] [Google Scholar]
- 70.Hideshima T, Chauhan D, Hayashi T, Podar K, Akiyama M, Gupta D, et al. The biological sequelae of stromal cell-derived factor-1alpha in multiple myeloma. Molecular cancer therapeutics. 2002;1:539–44. [PubMed] [Google Scholar]
- 71.Sanz-Rodriguez F, Hidalgo A, Teixido J. Chemokine stromal cell-derived factor-1alpha modulates VLA-4 integrin-mediated multiple myeloma cell adhesion to CS-1/fibronectin and VCAM-1. Blood. 2001;97:346–51. doi: 10.1182/blood.v97.2.346. [DOI] [PubMed] [Google Scholar]
- 72.Azab A, Weisberg E, Sahin I, Liu F, Awwad R, Azab F, et al. The influence of hypoxia on CML trafficking through modulation of CXCR4 and E-cadherin expression. Leukemia. 2012 doi: 10.1038/leu.2012.353. [DOI] [PubMed] [Google Scholar]
- 73.Muz B, de la Puente P, Azab F, Luderer M, Azab AK. Hypoxia promotes stem cell-like phenotype in multiple myeloma cells. Blood Cancer J. 2014 Dec 5;4:e262. doi: 10.1038/bcj.2014.82. doi: 10.1038/bcj.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muz B, de la Puente P, Azab F, Luderer M, Azab AK. The Role of Hypoxia and Exploitation of the Hypoxic Environment in Hematologic Malignancies. Molecular Cancer Research. 2014;12:1347–54. doi: 10.1158/1541-7786.MCR-14-0028. [DOI] [PubMed] [Google Scholar]
- 75.Runnels JM, Carlson AL, Pitsillides C, Thompson B, Wu J, Spencer JA, et al. Optical techniques for tracking multiple myeloma engraftment, growth, and response to therapy. Journal of biomedical optics. 2011;16:011006. doi: 10.1117/1.3520571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.