Abstract
Voltage-gated ion channels respond to changes in membrane potential with conformational shifts that either facilitate or stem the movement of charged ions across the cell membrane. This controlled movement of ions is particularly important for the action potentials of excitable cells such as cardiac myocytes, and therefore essential for timely beating of the heart. Inherited mutations in ion channel genes and in the genes encoding proteins that regulate them can cause lethal cardiac arrhythmias either by direct channel disruption or by altering interactions with therapeutic drugs, the best-understood example of both these scenarios being Long QT Syndrome (LQTS). Unsurprisingly, mutations in the genes encoding ion channel pore-forming α subunits underlie the large majority (~90%) of identified cases of inherited LQTS. Given that inherited LQTS is comparatively rare in itself (~0.04% of the US population), is pursuing study of the remaining known and unknown LQTS-associated genes subject to the law of diminishing returns? Here, with a particular focus on the KCNE family of single transmembrane domain K+ channel ancillary subunits, the significance to cardiac pharmacogenetics of ion channel regulatory subunits is discussed.
Keywords: voltage-gated potassium channel, KCNQ1, hERG, ancillary subunit, long QT syndrome, atrial fibrillation
Background
Pharmacogenetics – the interplay between natural human gene sequence variation and therapeutic or recreational drugs – represents at once a major challenge to the pharmaceutical industry, a major opportunity for drug companies willing to consider genetic variability when developing drugs, and both a substrate and raison d’être for basic and translational scientists aiming to understand human biology and disease in the genomic era. The pharmacogenetics of human cardiac arrhythmias is a particularly engrossing and clinically important field, primarily for two reasons. First, adverse drug-gene interactions can cause sudden cardiac death in a manner often both unpredictable and reversible only with defibrillation within minutes of onset. Second, a specific cardiac voltage-gated potassium (Kv) channel, the human ether-à-go-go related gene product (hERG, a.k.a. KCNH2) exhibits an alarming combination of properties that have made it the nemesis of the modern pharmaceutical industry: its efficient function is essential for healthy human cardiac rhythm [1,2], its protein folding and trafficking are exquisitely sensitive to genetic or pharmacologic perturbation [3,4], and it is the single most susceptible potassium channel to inhibition by a wide range of therapeutic drugs [5,6].
Voltage-gated sodium (Nav) and Kv channels are the dominant proteins controlling electrical activity in cardiac myocytes of the four chambers of the heart, and their disruption is therefore the most common culprit in electrically-based inherited cardiac arrhythmias [7]. As in neurons and other electrically excitable cells, the upstroke (depolarization phase) of the action potential of atrial and ventricular cardiomyocytes is controlled by Nav channels, whereas the repolarization phase is primarily controlled by Kv channels. Voltage-gated calcium (Cav) channels also influence the atrial and ventricular myocyte repolarization time, and in specialized pacemaking cells such as those in the sinoatrial node, are thought to dominate the depolarization, rendering it slower than in the majority of cardiac myocytes (although the role of Nav channels in sinoatrial node cells is perhaps greater than originally considered [8,9]). Focusing primarily here on the ventricles, for which the inherited basis of electrical abnormalities is somewhat better understood than the atria, increased depolarizing current through Nav channels, or decreased repolarizing current through Kv channels, delays repolarization – translating into an extended QT interval on the surface electrocardiogram (ECG), hence Long QT Syndrome (LQTS) (Figure 1A) [7].
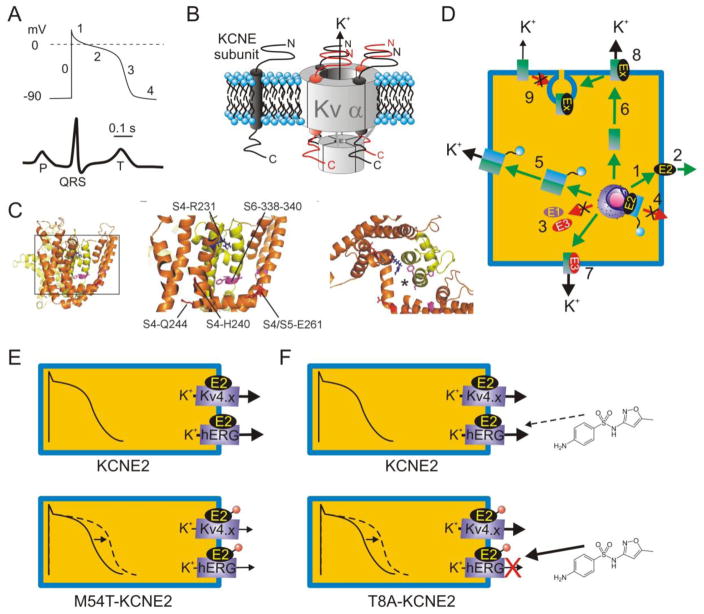
Figure 1.
A. Upper, generic ventricular myocyte action potential indicating phases 0–4; lower, generic surface ECG with time-course and scale-bar corresponding to action potential above.
B. Left, KCNE topology in cell membrane; right, possible stoichiometries of KCNE subunits in complex with a tetramer of Kv α subunits.
C. Putative positioning of KCNE subunits within the Kv channel complex. Left, lateral view of open-state model of KCNQ1 based on Rosetta modeling after the Kv1.2 structure [153–155]. Boxed region is expanded in the center panel, which shows the S6 region known to interact with KCNE subunits and S4/S4–5 linker residues important to KCNQ1 function in the presence and/or absence of KCNE subunits. Right, top view of region in center panel, with asterisk indicating one putative position of KCNE transmembrane region based on structure-function studies.
D. KCNE subunits are extremely versatile, regulating many aspects of K+ channel biology. For example, KCNE2 can reach the surface alone (1) and is also putatively secreted extracellularly (2); KCNE1 and KCNE3 probably require α subunits for surface expression (3). (4) KCNE2 suppresses N-type α subunit surface expression unless they are rescued by same-subfamily non-N-type α subunits (5). (6) KCNE2 and KCNE3 direct contrasting polarized trafficking of α subunits (7), and KCNEs regulate multiple facets of α subunit function (8). KCNE1 and KCNE2 also mediate α subunit endocytosis (9). Ex = KCNE subunit. For detailed review see [80].
E. KCNE2 is thought to form complexes with hERG and Kv4.x channels α subunits in some ventricular cardiomyocytes. Inherited human KCNE2 loss-of-function mutations (red circle) delay myocyte repolarization (dashed line) and are associated with inherited LQTS. One example, KCNE2-M54T, quickens hERG-KCNE2 deactivation [33].
F. The KCNE2-T8A polymorphism is “silent” in the absence of drugs but increases sensitivity to sulfamethoxazole, and can predispose to drug-induced LQTS [99].
The apparent simplicity of the yin-yang of Na+ and K+ movement (with Ca2+ also contributing to the membrane potential, and more importantly as a signaling entity in excitation-contraction coupling) masks the reality that in myocytes, as in all cell types, each ion channel is a signaling hub involving perhaps hundreds of different interacting proteins, with a pore-forming α subunit (or subunits) at the core [10]. How can we make sense of the pharmacogenetics of these complex systems, and is the best strategy to focus on the main players, and ignore the rest? Mutations in KCNE genes underlie a relatively small fraction of identified monogenic inherited cardiac arrhythmias (see below), but does it necessarily follow that ongoing efforts to examine the role of KCNE genes in cardiac arrhythmias (500+ publications to date) constitute “much ado about nothing”?
Inherited cardiac arrhythmias by numbers
Inherited LQTS is one of the currently best-understood groups of disorders in which relatively rare inherited or sporadic point mutations have been linked to, or associated with, diseases arising from dysfunction of an ion channel (“channelopathies”), and certainly the one for which most is understood vis-à-vis KCNE-associated pathology. There are at time of writing thirteen formally recognized genetic types of inherited LQTS (LQT1-13) (Table 1). LQT1 is caused by mutations in the KCNQ1 gene encoding the KCNQ1 Kv α subunit [11,12], LQT2 by mutations in KCNH2 encoding the hERG Kv α subunit [1,2]. These two forms account for ~80% of known genotyped cases of inherited LQTS, while mutations in Nav α subunit gene SCN5A explain another ~10% [13]. The remaining ~10% are divided between genes encoding other channel α subunits (e.g., CACNA1c), ion channel integral ancillary subunits (KCNE1, KCNE2, SCN4B), and other regulatory proteins (e.g., AKAP9); other genes may underlie some of the as-yet non-genotyped LQTS cases. KCNE1 mutations account for approximately 1% of inherited LQTS, and KCNE2 mutations are thought to underlie <1% of cases in this disorder. Perhaps 25% of apparently inherited LQTS cases have no identified gene association; percentages for identified and non-identified forms are estimates and vary depending upon the source [14,15]. There is one report in the literature of KCNE3 mutations potentially associated with prolonged QT with/without drug interaction [16] but this has not yet been recognized as a discrete genetic subtype, and to date there is no LQTS association for KCNE4 or 5, the remaining KCNE family genes (Table 2). It is important to note that these numbers, and the following estimates, are derived from the available databases and registries, which comprise primarily data from northern European Caucasians; thus they may not be as applicable to other ethnicities.
Table 1. Genes in designated forms of inherited LQTS and the proteins they encode.
Note that in addition, a recently postulated novel genetic form of LQT (LQT14) has been tentatively associated with Ryanodine Receptor 2 (RYR2) sequence variants [28].
| Syndrome | Gene | Protein | Protein type | References |
|---|---|---|---|---|
| LQT1 | KCNQ1 | KCNQ1 (Kv7.1) | Kv α subunit | [12] |
| LQT2 | KCNH2 | hERG (Kv11.1) | Kv α subunit | [2] |
| LQT3 | SCN5A | Nav1.5 | Nav α subunit | [142] |
| LQT4 | ANK2 | Ankyrin B | Cytoskeleton-associated adaptor protein | [143] |
| LQT5 | KCNE1 | KCNE1 (MinK) | ancillary subunit for KCNQ1, hERG, etc | [144] |
| LQT6 | KCNE2 | KCNE2 (MiRP1) | ancillary subunit for KCNQ1, hERG, etc | [33] |
| LQT7 | KCNJ2 | Kir2.1 | Kir α subunit | [145] |
| LQT8 | CACNA1c | Cav1.2 | Cav α subunit | [146] |
| LQT9 | CAV3 | Caveolin 3 | Structural component of caveolae | [147] |
| LQT10 | SCN4B | Navβ4 | ancillary subunit for Nav1.5 | [148] |
| LQT11 | AKAP9 | AKAP9 (Yotiao) | A-kinase anchoring protein | [149] |
| LQT12 | SNTA1 | α-Syntrophin | Cytoskeletal protein, regulates Nav1.5 | [150] |
| LQT13 | KCNJ5 | Kir3.4 | Kir α subunit | [151] |
Table 2.
Genetic evidence for the requirement of KCNEs in mammalian cardiac physiology.
| Gene (other names) | Disease Association | Favored mechanism | Additional possible mechanisms | Genetic evidence [references] |
|---|---|---|---|---|
|
| ||||
| KCNE1 (MinK) | LQT5 | ↓ IKs | ↓ IKr | H [144] |
| diTdP | ↓ IKs | ↓ IKr | H [117] | |
| AF | ↑ IKs | ↑ IKr | H [19], M [152] | |
|
| ||||
| KCNE2 (MiRP1) | LQT6 | ↓ IKr | ↓ IKCNQ1-KCNE2 | H [33], M [46] |
| diTdP | ↓ IKr | ↓ IKCNQ1-KCNE2 | H [33] M [46] | |
| AF | ↑ IQ1-E2 | ↑ IKur | H [20] | |
| Early-onset MI | ? | ? | H [102] | |
| Cardiac hypertrophy | hypothyroidism | Direct cardiac effect | M [104] | |
|
| ||||
| KCNE3 (MiRP2) | LQTS | ↓ IQ1-E3 | ↓ IKr | H [16] |
| AF | ↑ IQ1-E3 | ? | H [22] | |
| BrS6 | ↑ Ito | ? | H [17] | |
|
| ||||
| KCNE4 (MiRP3) | AF | ↑ IQ1-E4 | ? | H [23] |
|
| ||||
| KCNE5 (KCNE1L, MiRP4) | AF | ↑ IKs | ? | H [24] |
| IVF | ↑ Ito | ? | H [18] | |
| BrS11 | ↑ Ito | ? | H [18] | |
Abbreviations: AF, atrial fibrillation; BrS, Brugada Syndrome; diTdP, drug-induced Torsades de Pointes; Ex, KCNEx; H, human; IVF, idiopathic ventricular fibrillation; LQTS, Long QT Syndrome; M, mouse; MI, myocardial infarction; Q1, KCNQ1.
It is extremely difficult to generate accurate frequency estimates in rare diseases such as LQTS, especially as it is likely be underdiagnosed, particularly when sub-clinical, but for the sake of argument we will discuss some hypothetical values. The incidence of inherited LQTS in the US population has been estimated at 1 in 2–3000 (~0.04%) [14]. Therefore, given the available evidence, the proportion of the US population harboring an LQTS-associated KCNE gene mutation is likely to be in the order of at least 1 in 100,000 (0.001%) – roughly 3000 people. This compares to an estimated 80,000 US citizens harboring inherited LQTS-associated mutations in the genes encoding the two principle ventricular myocyte-repolarizing Kv channel α subunits, KCNQ1 or KCNH2 (calculated based on existing data). Therefore, KCNE gene mutations unquestionably represent a relatively minor genetic component of inherited “monogenic” LQTS, and a relatively unimportant one in terms of the impact of inherited monogenic LQTS on the population health of the US (and by extension other countries). However, an alternative method to compare the significance of KCNEs versus other genes in inherited LQTS is to normalize the incidence of pathologic point mutations to the number of bases per gene. The entire gene size (introns and exons) varies dramatically among KCNE genes, being for example 65,585 bases for KCNE1 compared to just 7,118 bases for KCNE2 – despite their protein products being similar in size (129 vs 123 amino acids, respectively). This is compared to 101,617 bases for SCN5A. Assuming KCNE1 and KCNE2 gene variants each account for 1% of inherited LQTS, this represents 1/66 or 0.015% of inherited LQTS cases per KCNE1 kilobase, versus 1/7 or 0.14% inherited LQTS cases per KCNE2 kilobase. This compares to 10/102 or 0.098% per SCN5A kilobase using a similar calculation. Thus, KCNE2 is of more pathologic significance base-for-base than SCN5A, assuming KCNE2 accounts for 1% of LQTS cases, which is an approximation at this stage.
As the available data for protein-coding regions is far more developed than our knowledge of potential intronic sequence variant effects on LQTS, in particular for the KCNE genes, an alternative comparison utilizing solely protein-coding sequences is also worth assessing. For KCNE1, with 387 bases coding for protein, assuming 1% of inherited LQTS cases are caused by KCNE1 coding region mutations gives 1/387 or 0.0026% of inherited LQTS cases per KCNE1 base. KCNE2 gives a similar value of 1/369 or 0.0027%. In contrast, SCN5A, which is linked to 10% of inherited LQTS cases, numbers 6048 protein-coding bases - constituting only 0.0017% of inherited LQTS cases per SCN5A base. By this (admittedly unorthodox) calculation, base-for-base both the KCNE1 and KCNE2 genes are of more pathologic significance with respect to inherited LQTS than SCN5A.
Aside from LQTS, sporadic, rare KCNE gene mutations have been identified and suggested to be causative in patients with Brugada Syndrome (KCNE3 and KCNE5) [17,18] and lone atrial fibrillation (AF) (KCNE1-5) [19–24]. A KCNE5 polymorphism is also suggested to confer protection against AF [25] and another, rs697829, was associated with prolonged QT interval and survival in acute coronary syndromes patients [26]. Brugada Syndrome is exemplified by loss-of-function mutations in SCN5A that impair phase 0 depolarization and manifest as coved-type ST-segment elevation in atypical right-bundle branch block (leads V1 to V3 on body-surface electrocardiogram). This associates with a predisposition for polymorphic ventricular tachyarrhythmias and sudden cardiac death [27]. The twelve other genes associated with Brugada Syndrome account for less than 5% of cases combined, although it is important to note that only in ~25% of clinical Brugada cases has a potentially causative mutation been identified [28]. KCNE3 and KCNE5 mutations may mimic SCN5A loss-of-function or perturb phase 0 depolarization by increasing Ito, the transient outward K+ current that is generated primarily by Kv4 family α subunits (in human ventricles) and rapidly follows and counteracts phase 0 depolarization.
Unlike LQTS, the dominant causative factor in most AF cases is structural heart disease leading to areas of non-conduction; however, rare “lone AF”, i.e., in patients lacking identifiable structural defects, has been associated with ion channel gene mutation. Sporadic KCNE mutations almost certainly constitute an extremely rare cause of AF per se; a recent KCNE1-targeted screen of 209 unrelated early-onset AF patients uncovered two patients with KCNE1 mutations that increased IKs density in vitro [19]. Also in contrast to LQTS, AF appears to associate most frequently with a shortening of the atrial effective refractory period, therefore increased KCNQ1 current density is one potential mechanism, and most if not all lone AF-associated KCNE gene mutations increase KCNQ1 current density when co-expressed with this α subunit in heterologous expression studies [20,21]. However, it is important to note that while KCNE gene variants for the most part represent a tiny fraction of AF cases, there are an estimated 2.3 million AF sufferers in the United States and so an understanding of even relatively rare genetic forms is still warranted from both a public health standpoint and in the interests of learning more about fundamental mechanisms of AF etiology.
What is understood of the mechanisms of inherited KCNE-associated cardiac arrhythmias?
Shakespeare’s comedy Much Ado About Nothing is centered around two couples: Claudio and Hero are besotted with one another yet Claudio is convinced into rejecting Hero; Benedick and Beatrice seem anything but amorous toward each other and yet are fooled into announcing their love. Ultimately, all ends well as the source of the scandalous “noting” that caused all the trouble is revealed [29]. Most of the research into human ventricular repolarization has also centered around two couples (KCNQ1-KCNE1 and hERG-KCNE2), and other parallels with the famous play are apparent.
The KCNQ1-KCNE1 pairing has until recently largely been viewed as set in stone as the molecular correlate of IKs because the unusual activation kinetics of this pairing match the established native IKs kinetics (and other functional attributes) [30,31] and because KCNE1 stood as the only known KCNE subunit for a decade [32], until we and others discovered its relatives [33,34]. More recently, however, scientists have started to acknowledge that KCNQ1 probably also dallies with other KCNE partners in the heart [35,36], even forming complexes with more than one KCNE at the same time [37,38]. Particular attention has been paid to the potential role of KCNQ1-KCNE2 in the atria, with AF-associated KCNE2 and KCNQ1 mutations increasing KCNQ1-KCNE2 current, a potential mechanism for AF [20,39].
Conversely, the hERG-KCNE2 partnership was greeted with skepticism, with difficulties reported in repeating elements of our original findings [33,40], belief hampered by the relatively subtle effects of KCNE2 upon hERG, and difficulties in detecting KCNE2 expression in the ventricles [41]. More recently, however, native cardiac ERG-KCNE2 complexes have been detected [42], potential roles for KCNE2 in several cardiac pathologies have been uncovered [20,43], and ventricular KCNE2 expression convincingly confirmed (and shown to be higher in ventricles than atria) [44].
Nevertheless, KCNE2 regulates a number of different Kv α subunits in mammalian heart (and may do so in man), including Kv4.2/3 [45,46], Kv1.5 [46], Kv2.1 [47] and probably HCN subunits [48,49] and KCNQ1 [36]. KCNE1 also regulates hERG [50,51] and probably others in the heart as well [47]. While the two couples of KCNQ1-KCNE1 and hERG-KCNE2 have attracted the most attention, they most likely represent just two of many KCNE-α complexes to be found at different stages of development, disease, and location in the myocardium of humans and other species [52,53]. Of the five known human KCNE proteins (KCNE1-5), all form complexes with and functionally modulate the KCNQ1 α subunit, at least in heterologous expression studies [30,31,54–56]. However, each KCNE subunit is also known to have the capacity to regulate other Kv α subunits, and KCNE functional interactions with the HCN “pacemaker” α subunits that do not select between Na+ and K+ [48,49], and with the Ca2+-activated BK channel α subunit [57], have also been reported. Uncertainty surrounding the cardiac role of KCNE2 (and KCNE3-5) probably arises from the complexity of their roles as much as the magnitude of the impact upon cardiac health of their disruption [58].
In addition to their promiscuity, this complexity arises from other aspects of the functional versatility of KCNE subunits. KCNE proteins are single-transmembrane (TM) domain subunits that co-assemble with Kv α subunits to form heteromeric channel complexes with altered functional properties [59]. The debate continues about whether 2, 4, or a variable number of KCNE subunits join the Kv channel α subunit tetramer in functional native channels [60–64] (Figure 1B). Based mostly on site-directed mutagenesis structure-function studies, it is thought that KCNEs sit close to the Kv α subunit voltage sensor, pore-lining S6, the S4–S5 loop that connects the voltage sensor to the channel gate, and the C-terminus [65–73] (Figure 1C) – although it is important to recognize that almost all KCNE structure-function studies pertain to KCNE1, 2 and 3 interactions with the KCNQ1 α subunit, and some or all of these may not be directly extrapolated to other KCNE-α subunit complexes. This location affords KCNE subunits the capacity to control channel voltage dependence, gating kinetics, ion selectivity, conductance, regulation by other proteins, and pharmacology (for review, see [53]). KCNE subunits have also been found to direct multiple aspects of α subunit trafficking in vitro [74–83] and in vivo [46,84,85] (Figure 1D).
As mentioned, the best-understood complex is KCNQ1-KCNE1 – it is primarily responsible for human cardiac IKs, a slow-activating K+ current important for ventricular myocyte repolarization, particularly at high heart-rates or when other repolarizing currents are compromised [31,86]. KCNE1 increases KCNQ1 conductance [61], slows its activation 5–10-fold [30,31], eliminates its inactivation [87], alters its ion selectivity [88] and pharmacology [89], changes its regulation by other proteins [78] and lipids [90,91], and mediates its internalization from the plasma membrane by clathrin-mediated endocytosis [76]. Most pathologic KCNE1 mutations identified to date are those that underlie inherited LQTS, thought primarily to be by reducing IKs density (loss-of-function mutations) [13]. The autosomal dominant form of KCNE1-associated LQTS is referred to as Romano-Ward syndrome, while the predominantly autosomal recessive form, Jervell and Lange-Nielsen syndrome, also impairs hearing due to loss-of-function of KCNQ1-KCNE1 current in the inner ear, where this channel mediates K+ secretion from the endolymph [92,93]. The potential complexity of KCNE1-associated LQTS becomes apparent, however, when one appreciates that KCNE1 can also regulate the hERG α subunit, which generates the crucial human ventricular IKr repolarization current (KCNE1 increases hERG current 2-fold in vitro) [50]. Furthermore, not only have KCNE1-hERG complexes been identified in vivo, but so have KCNQ1-hERG complexes; in fact, KCNQ1 mutations can potentially cause LQTS by impairing function of the hERG α subunit [94,95]. We do not yet know whether KCNQ1-hERG-KCNE1 complexes exist in vivo, but it is a distinct possibility.
KCNE2, mutations in which are also associated with inherited LQTS (Table 1, 2), was – together with KCNE3,4 and 5 – cloned a decade after KCNE1 [33,34]. KCNE2 modulates hERG in vitro (gating kinetics, conductance, regulation by other proteins, pharmacology) [58], forms cardiac complexes with it in vivo, and loss-of-function KCNE2 mutations may underlie LQT6 by disrupting IKr [33,34]. However, KCNE2 can also modulate KCNQ1, largely stripping it of its voltage dependence and reducing its conductance such that it forms a “background” K+-selective leak that may provide an ever-present repolarizing force in some cells [54]. While little is known of potential KCNQ1-KCNE2 cardiac relevance, it potentially impacts formation of KCNQ1-KCNE1 complexes [36,38], or possibly contributes to “steady-state” current (Iss) that can be observed in cellular electrophysiology experiments after the transient components have inactivated. The importance of KCNQ1-KCNE2 in epithelial tissues has been extensively studied (see below). KCNE2 also regulates a host of other Kv α subunits and its disruption in mice delays ventricular repolarization by reduction of IKur and Ito (generated, respectively, by Kv1.5 and Kv4.2/3) [46] rather than IKr and IKs, which are essentially absent in adult mouse ventricular myocytes [96].
KCNE1 and KCNE2 gene polymorphisms: drug-SNP interactions and beyond
In contrast to the rarity of KCNE gene mutations underlying purely inherited forms of cardiac arrhythmia, more common KCNE gene variants bring into sharp focus the necessity of considering KCNEs in the cardiac health and treatment of the general human population (Table 2). In the original report of KCNE2 cloning and association with inherited LQTS [33], we also described a new phenomenon: a pathologic gene variant in KCNE2 actually increased sensitivity of hERG-KCNE2 channels to inhibition by the macrolide antiobiotic clarithromycin, for which the timing of therapeutic administration made this drug a likely contributing factor to the QT prolongation of the patient involved. The gene variant (resulting in a Q9E substitution in the extracellular region of KCNE2) was later discovered to be a polymorphism (typically a gene variant earns this recognition when present in a specific population at a prevalence of ≥ 1%) present in 3% of African-Americans (and apparently absent in U.S. Caucasians) [97]. At time of writing, data from the National Heart, Lung and Blood Institute Exome Variant Server (NHLBI EVS) (http://evs.gs.washington.edu/EVS/) still indicate absence of the rarer G allele from European Americans (n = 8600 alleles), whereas it is present in 1.7% of alleles from African Americans (n = 4406) in the United States. The threefold-increased clarithromycin sensitivity of hERG-Q9E-KCNE2 channels suggests against use of high-dose clarithromycin (and other macrolide antibiotics) in certain populations, and certainly once routine genetic testing can be used to quickly identify such polymorphisms before specific drugs are indicated. The Q9E polymorphism was also subsequently found in one case during post-mortem testing for Sudden Infant Death Syndrome (SIDS) [98].
KCNE2 G/A polymorphism rs2234916, with the rarer G allele encoding a T8A substitution, is found in 0.7% of European Americans (n = 8600), but only 0.1% of African Americans (n = 4406) (NHLBI EVS). The G allele was originally identified in a man who developed prolonged QT after taking the antibiotic Bactrim [99]. The sulfamethoxazole component of Bactrim was found to block hERG-T8A-KCNE2 channels more effectively than T8-containing channels, thus demonstrating a second example of a KCNE2 polymorphism mediating genetic predisposition to drug-induced LQTS. A 0.7% incidence is significant enough to consider avoiding administration of Bactrim or sulfamethoxazole-related compounds to patients harboring T8A once genetic testing of patients becomes routine. Interestingly, the T8A variant was found to eliminate a glycosylation site that apparently shields the hERG-KCNE2 channel from sulfamethoxazole block [100]. While Q9E both reduces hERG-KCNE2 current pre-drug, and also increases drug sensitivity, T8A has no noticeable pre-drug effects (in cellular electrophysiology experiments) and therefore is likely to be undetectable in a patient’s ECG. Other KCNE2 variants that were identified in adverse drug interactions thus far were found not to alter drug sensitivity of hERG-KCNE2, but reduced its current at baseline and were likely pathogenic in the drug-induced LQTS patient cohort in which they were identified because mutation + drug effects were additive [99] and sufficient to impact the “repolarization reserve”, the repolarizing current density we need in order to maintain normal cardiac rhythm [101] (Figure 1E, F).
While hERG’s unique drug sensitivity has meant that the pharmacology of KCNE2 polymorphisms has focused on hERG-KCNE2, it is entirely possible that other channels are involved. To give one example, Kcne2 knockout predisposes adult mice to sevoflurane-induced LQTS, even though mERG is not important in adult mouse ventricular repolarization [46]. In terms of KCNEs in general, it is important to recognize that they can radically alter channel pharmacology, and therefore in cases where their α subunit partner is ubiquitously expressed but their KCNE partner’s expression is more confined, KCNEs could help provide therapeutic windows of drug specificity. For example, KCNE3 greatly increases the sensitivity of KCNQ1 to block by chromanols, compared to homomeric KCNQ1 or heteromeric KCNQ1-KCNE1 channels [89].
A polymorphism near the KCNE2 locus has also been associated with early-onset myocardial infarction, suggesting a possible link between KCNE2 disruption and structural abnormalities, although other genes in the 21q22 region harboring the variant (MRPS6 and SLC5A3) are equally likely to be involved (or none of the three) [102]; interestingly, ventricular KCNE2 expression is elevated by acute myocardial infarction [103]. Kcne2 deletion in mice causes hypothyroidism because basolateral KCNQ1-KCNE2 channels in thyrocytes are required for adequate iodide uptake through the thyroid sodium iodide symporter (NIS). This impacts cardiac structure in mice (contributing to hypertrophy and impaired cardiac contractility in the offspring of Kcne2−/− dams and in aging Kcne2−/− mice) but these recent observations have yet to be extended in human populations [104]. KCNQ1-KCNE2 channels feature heavily in epithelial cells, providing a constitutively active K+ current. In gastric parietal cells, KCNQ1-KCNE2 provides a K+ recycling pathway to facilitate gastric acid secretion by the apical H+/K+ATPase. Accordingly, recent reports demonstrate that Jervell-Lange-Nielsen Syndrome can manifest as hypergastrinemia and gastric hyperplasia in addition to LQTS and deafness [105,106], reflecting previous gastric findings in rodents [105,107–113]. KCNE2 is also expressed in the choroid plexus, where it regulates KCNQ1 and Kv1.3 (KCNA3) [85], and we recently found that Kcne2 deletion in mice reduces HCN current and protein expression in some neurons, by an as yet undefined mechanism [114]. The neural consequences of human KCNE2 polymorphisms are yet to be assessed, but the potential exists for a cardiocerebral phenotype in which KCNE2-associated seizures could precipitate arrhythmias [115], similar to the mechanism suggested for a mouse model of sudden unexplained death in epilepsy (SUDEP) manipulated to induce CNS expression of a human KCNQ1 mutant [116].
The KCNE1 D85N polymorphism is found in 1.22% of European American alleles (n = 8600) but only 0.2% of African American alleles (n = 4406) (NHLBI EVS). D85N was also recently linked to drug-induced torsades de pointes (TdP), a dangerous ventricular arrhythmia that can arise from QT prolongation. A D85N allele was found in 8.6% of drug-induced TdP cases, versus 2.9% of drug-exposed controls and 1.8% of population controls in a total cohort of 207 patients with QT lengthening after drug exposure and 837 controls [117]. The D85N polymorphism presumably works by impairing IKs current such that it is less likely to compensate for drug-inhibited IKr, but as covered earlier, the potential mechanistic explanations are legion and extremely difficult to distinguish. Another KCNE1 polymorphism of potential pathological significance is the rs1805127 C/T polymorphism, which results in either a glycine or serine at KCNE1 residue 38. The rarer T allele, encoding serine, is represented in 36% of European American alleles (n = 8600) compared to 29% of African American alleles (n = 4406) (NHLBI EVS). Variation at the S38G site in KCNE1 has previously been implicated in both general and gender-specific inherited LQTS predisposition; predisposition to sudden cardiac death, heart failure, and AF; and exacerbation of LQT2 severity [118–128].
Postoperative AF (POAF), which is incompletely understood but appears to stem at least in part from postoperative oxidative stress and inflammation, affects as many as two-thirds of patients shortly after cardiac surgery and a third after other forms of thoracic surgery, and significantly increases postoperative hospital stays, morbidity and mortality [129–133]. Adopting a transcriptomic approach to identify potential transcriptional changes in a porcine model of susceptibility to POAF, we recently found that left atrial KCNE1 transcript downregulation was evident in pigs 3 days following lung lobectomy [134]. This is the typical time window during which patients undergoing thoracic surgery are susceptible to onset of POAF, and suggests KCNE1 involvement in either the etiology of, or early compensatory remodeling in response to, POAF. As a two-fold reduction in KCNE1 expression would be predicted to mimic the effects of the KCNE1 38G polymorphism which is associated with lone AF [128], future studies could be directed toward determining whether or not there is a causal relationship between KCNE1 38G (or other human polymorphisms) and increased POAF susceptibility. The ultimate goal of this research would be to establish whether or not routine, targeted genetic testing could potentially improve management and outcomes of cardiothoracic surgery patients.
Beyond KCNE1 and KCNE2, few sequence variants in the KCNE3, 4 and 5 genes have been functionally characterized with cellular electrophysiology, and this will be an important endeavor particularly as the number of identified human disease-associated variants in these subunits increases as expected. The challenge is these cases is to determine which Kv α subunits are functionally affected by the normal and mutant KCNE proteins in vivo in the various tissues in which they are expressed. A nice example is the BrS-linked rs121908441 mutation that results in R99H-KCNE3. Although KCNE3 has profound effects on KCNQ1 function, the R99H mutation does not impact this, but instead increases current through Kv4.3 (KCND3) channels. Together with data showing KCNE3-Kv4.3 co-immunoprecipitation from human atria, these functional effects strongly support a role for KCNE3 in regulating Ito in human heart, and in R99H in pathologic disruption of Ito [17]. Similar effects on Kv4.3 were subsequently discovered for KCNE5 variants associated with BrS and IVF [18].
While an arrhythmia-associated mutation in cardiac Kv channel α subunit KCNQ1 was found to reduce sensitivity of KCNQ1-KCNE1 to potentially therapeutic activation by the benzodiazepine R-L3 [135], little is known about the impact of KCNE sequence variants on the efficacy of antiarrhythmic drugs. Thus, known KCNE SNP-drug interactions typically fall into the category of increased sensitivity to block by proarrhythmic medications given for noncardiac indications. However, one group has exploited this latter phenomenon in proof-of-concept studies in which they introduced macrolide-hypersensitive Q9E into the atria of pigs, to increase susceptibility of native atrial pERG channels to block by clarithromycin. They then used this approach to therapeutically lengthen the atrial effective refractory period (a common and effective antiarrhythmic strategy to combat atrial fibrillation) in a dose window that did not delay ventricular repolarization (because the wild-type KCNE2-pERG channels there were less sensitive to clarithromycin block) [136,137]. This approach could have potential as a future therapeutic strategy in human patients if and when gene therapy safety and affordability issues are resolved.
In a study of another form of drug-gene interaction, the antiarrhythmic drug amidarone – a highly effective drug in both ventricular tachyarrhythmias and atrial fibrillation – was administered to mice for 6 weeks at various doses and then ion channel gene microarrays were probed for potential gene expression changes in response to drug treatment. In addition to alterations in a variety of voltage-gated ion channel α subunit genes, Kcne2 and Kcne3 expression was upregulated whereas Kcne1 expression was unchanged [138]. Given the promiscuity and substantial functional effects of these subunits on multiple Kv channels, it would be fascinating to determine if this result is reproducible in human studies, and whether it contributes to the therapeutic action (or otherwise) of amiodarone.
Expert Commentary
KCNE gene mutations represent a relatively uncommon fraction of the recognized inherited LQTS genetic variants, but more common KCNE polymorphic variants contribute significantly to predisposition to drug-induced LQTS, general prolonged QT interval, and potentially AF. The promiscuity of KCNE subunits, combined with their wide range of functional effects on ion channels both inside and outside the heart, have made understanding of their place in cardiac physiology and disease a challenging goal and very much a work in progress. In contrast their relatively small size has made them highly amenable to genetic testing. Study of this intriguing family of proteins has taught us much about ion channel function and regulation and the genetics of channelopathies, and has a role to play in improving human health by identifying genetic predisposition to several potentially lethal disorders.
Five-year view
From a clinical point of view, ongoing advances in the speed and reduction of the cost of genetic testing will generate within the next 5 years a fuller picture of the landscape of KCNE-associated diseases and facilitate avoidance of unwanted drug interactions when warranted, or proactive intervention where appropriate. As most clinicians and scientists have understandably focused on the more common causes of disease, routine whole-genome sequencing will have an even greater impact on our understanding of the less common forms of genetic disease or predisposition to adverse drug interactions. From a basic science viewpoint, the single most important breakthrough in our understanding of KCNEs will be a high-resolution structure of a mammalian KCNE-α subunit channel complex, as has already been achieved for cytoplasmic Kv β subunits [139,140]. This breakthrough is likely to happen within the next 5 years, and will not only enhance our understanding of fundamental mechanisms of KCNE function, but will provide a 3D view of the positioning of, for example, drug interaction susceptibility “hotspots”. This, in turn, will facilitate future avoidance once widespread adoption of channel structure-based drug design is a reality – which will likely take longer. While most studies to date have focused on point mutations, ever-increasing sequencing power and larger patient cohorts will facilitate more of an appreciation of the role of other sequence variants, including indels, fusions, copy number variation and inversions in the genetics and pharmacogenetics of KCNE-associated diseases. Interestingly, while KCNE1 and KCNE2 are both on chromosome 21 and therefore overrepresented in individuals with trisomy 21, we do not yet know their potential role in cardiac defects and other features of Down Syndrome. Similarly, KCNE5 lies in a stretch of genomic DNA deleted in AMME contiguous gene syndrome but its potential role in cardiac or other abnormalities in this disorder is unknown [34]. Finally, with burgeoning clinical, pharmacological and genetic data, online pharmacogenomics databases (for a recent review see [141]) will become an increasingly widely used tool for clinicians and basic scientists in both academia and industry in the drive for avoidance of adverse drug-SNP interactions and ultimately (but not to any great extent within the next 5 years) bespoke, genotype-specific prescriptions.
Key Issues (bullet points).
Discovery of rare gene variants discovered in familial forms of disease such as the KCNE-associated arrhythmias can pave the way for large-scale population studies and reveal predispositions associated with more common gene polymorphisms.
Because of their promiscuity, versatility of function and widespread distribution in the body, identification of all the mechanistic underpinnings of KCNE-associated genetic disorders is challenging.
Despite their small size and the availability of high-resolution structural data for their larger α subunit partners, we still lack direct structural information on KCNE-α complexes, and even their stoichiometry is still debated.
The small size of KCNE genes (~400 bases coding region) makes them highly amenable to large-scale genetic screening.
Mouse models of KCNE dysfunction are currently being used to identify novel roles for both KCNEs and their α subunit partners, paving the way for future human genetic studies of disease linkage and future drug development.
KCNEs (both wild-type and mutant) can radically alter channel pharmacology and therefore can provide challenges but also opportunities for specificity in drug development.
Acknowledgments
G.W.A. is grateful for financial support from US National Institutes of Health HL079275 and HL101190.
References
Papers of special interest have been highlighted as
* of interest
**of special interest
- 1.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81(2):299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 2.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80(5):795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 3.Anderson CL, Delisle BP, Anson BD, et al. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006;113(3):365–373. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 4.Rajamani S, Eckhardt LL, Valdivia CR, et al. Drug-induced long QT syndrome: hERG K+ channel block and disruption of protein trafficking by fluoxetine and norfluoxetine. British journal of pharmacology. 2006;149(5):481–489. doi: 10.1038/sj.bjp.0706892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown AM. HERG block, QT liability and sudden cardiac death. Novartis Foundation symposium. 2005;266:118–131. doi: 10.1007/978-1-59259-884-7_4. discussion 131–115, 155–118. [DOI] [PubMed] [Google Scholar]
- 6.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440(7083):463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 7.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 8.Smits JP, Koopmann TT, Wilders R, et al. A mutation in the human cardiac sodium channel (E161K) contributes to sick sinus syndrome, conduction disease and Brugada syndrome in two families. Journal of molecular and cellular cardiology. 2005;38(6):969–981. doi: 10.1016/j.yjmcc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Veldkamp MW, Wilders R, Baartscheer A, Zegers JG, Bezzina CR, Wilde AA. Contribution of sodium channel mutations to bradycardia and sinus node dysfunction in LQT3 families. Circulation research. 2003;92(9):976–983. doi: 10.1161/01.RES.0000069689.09869.A8. [DOI] [PubMed] [Google Scholar]
- 10.Panaghie G, Abbott GW. The impact of ancillary subunits on small-molecule interactions with voltage-gated potassium channels. Current pharmaceutical design. 2006;12(18):2285–2302. doi: 10.2174/138161206777585175. [DOI] [PubMed] [Google Scholar]
- 11.Splawski I, Timothy KW, Vincent GM, Atkinson DL, Keating MT. Molecular basis of the long-QT syndrome associated with deafness. The New England journal of medicine. 1997;336(22):1562–1567. doi: 10.1056/NEJM199705293362204. [DOI] [PubMed] [Google Scholar]
- 12.Neyroud N, Tesson F, Denjoy I, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nature genetics. 1997;15(2):186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 13.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart rhythm : the official journal of the Heart Rhythm Society. 2005;2(5):507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Lehnart SE, Ackerman MJ, Benson DW, Jr, et al. Inherited arrhythmias: a National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116(20):2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 15.Prystowsky EN, Padanilam BJ, Joshi S, Fogel RI. Ventricular arrhythmias in the absence of structural heart disease. Journal of the American College of Cardiology. 2012;59(20):1733–1744. doi: 10.1016/j.jacc.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 16.Ohno S, Toyoda F, Zankov DP, et al. Novel KCNE3 mutation reduces repolarizing potassium current and associated with long QT syndrome. Human mutation. 2009;30(4):557–563. doi: 10.1002/humu.20834. [DOI] [PubMed] [Google Scholar]
- 17.Delpon E, Cordeiro JM, Nunez L, et al. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circulation Arrhythmia and electrophysiology. 2008;1(3):209–218. doi: 10.1161/CIRCEP.107.748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno S, Zankov DP, Ding WG, et al. KCNE5 (KCNE1L) variants are novel modulators of Brugada syndrome and idiopathic ventricular fibrillation. Circulation Arrhythmia and electrophysiology. 2011;4(3):352–361. doi: 10.1161/CIRCEP.110.959619. [DOI] [PubMed] [Google Scholar]
- 19.Olesen MS, Bentzen BH, Nielsen JB, et al. Mutations in the potassium channel subunit KCNE1 are associated with early-onset familial atrial fibrillation. BMC medical genetics. 2012;13(1):24. doi: 10.1186/1471-2350-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Xia M, Jin Q, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. American journal of human genetics. 2004;75(5):899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundby A, Ravn LS, Svendsen JH, Hauns S, Olesen SP, Schmitt N. KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2008;21(1–3):47–54. doi: 10.1159/000113746. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DF, Liang B, Lin J, Liu B, Zhou QS, Yang YQ. [KCNE3 R53H substitution in familial atrial fibrillation] Chinese medical journal. 2005;118(20):1735–1738. [PubMed] [Google Scholar]
- 23.Ma KJ, Li N, Teng SY, et al. Modulation of KCNQ1 current by atrial fibrillation-associated KCNE4 (145E/D) gene polymorphism. Chinese medical journal. 2007;120(2):150–154. [PubMed] [Google Scholar]
- 24.Ravn LS, Aizawa Y, Pollevick GD, et al. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart rhythm : the official journal of the Heart Rhythm Society. 2008;5(3):427–435. doi: 10.1016/j.hrthm.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravn LS, Hofman-Bang J, Dixen U, et al. Relation of 97T polymorphism in KCNE5 to risk of atrial fibrillation. The American journal of cardiology. 2005;96(3):405–407. doi: 10.1016/j.amjcard.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 26.Palmer BR, Frampton CM, Skelton L, et al. KCNE5 polymorphism rs697829 is associated with QT interval and survival in acute coronary syndromes patients. Journal of cardiovascular electrophysiology. 2012;23(3):319–324. doi: 10.1111/j.1540-8167.2011.02192.x. [DOI] [PubMed] [Google Scholar]
- 27.Brugada P, Brugada J, Brugada R. The Brugada syndrome. Cardiac electrophysiology review. 2002;6(1–2):45–48. doi: 10.1023/a:1017978903909. [DOI] [PubMed] [Google Scholar]
- 28.Sarquella-Brugada G, Campuzano O, Iglesias A, et al. Genetics of sudden cardiac death in children and young athletes. Cardiology in the young. 2012:1–15. doi: 10.1017/S1047951112001138. [DOI] [PubMed] [Google Scholar]
- 29.Shakespeare W. Much Ado About Nothing. Wise and Aspley; London: 1600. [Google Scholar]
- 30.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384(6604):78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 31.Sanguinetti MC, Curran ME, Zou A, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384(6604):80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 32.Takumi T, Ohkubo H, Nakanishi S. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science. 1988;242(4881):1042–1045. doi: 10.1126/science.3194754. [DOI] [PubMed] [Google Scholar]
- 33.Abbott GW, Sesti F, Splawski I, et al. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97(2):175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 34.Piccini M, Vitelli F, Seri M, et al. KCNE1-like gene is deleted in AMME contiguous gene syndrome: identification and characterization of the human and mouse homologs. Genomics. 1999;60(3):251–257. doi: 10.1006/geno.1999.5904. [DOI] [PubMed] [Google Scholar]
- 35.Bendahhou S, Marionneau C, Haurogne K, et al. In vitro molecular interactions and distribution of KCNE family with KCNQ1 in the human heart. Cardiovascular research. 2005;67(3):529–538. doi: 10.1016/j.cardiores.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Wu DM, Jiang M, Zhang M, Liu XS, Korolkova YV, Tseng GN. KCNE2 is colocalized with KCNQ1 and KCNE1 in cardiac myocytes and may function as a negative modulator of I(Ks) current amplitude in the heart. Heart rhythm : the official journal of the Heart Rhythm Society. 2006;3(12):1469–1480. doi: 10.1016/j.hrthm.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Toyoda F, Ueyama H, Ding WG, Matsuura H. Modulation of functional properties of KCNQ1 channel by association of KCNE1 and KCNE2. Biochemical and biophysical research communications. 2006;344(3):814–820. doi: 10.1016/j.bbrc.2006.03.213. [DOI] [PubMed] [Google Scholar]
- 38.Jiang M, Xu X, Wang Y, et al. Dynamic partnership between KCNQ1 and KCNE1 and influence on cardiac IKs current amplitude by KCNE2. The Journal of biological chemistry. 2009;284(24):16452–16462. doi: 10.1074/jbc.M808262200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299(5604):251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 40.Weerapura M, Nattel S, Chartier D, Caballero R, Hebert TE. A comparison of currents carried by HERG, with and without coexpression of MiRP1, and the native rapid delayed rectifier current. Is MiRP1 the missing link? The Journal of physiology. 2002;540(Pt 1):15–27. doi: 10.1113/jphysiol.2001.013296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pourrier M, Zicha S, Ehrlich J, Han W, Nattel S. Canine ventricular KCNE2 expression resides predominantly in Purkinje fibers. Circulation research. 2003;93(3):189–191. doi: 10.1161/01.RES.0000084851.60947.B5. [DOI] [PubMed] [Google Scholar]
- 42.Anantharam A, Abbott GW. Does hERG coassemble with a beta subunit? Evidence for roles of MinK and MiRP1. Novartis Foundation symposium. 2005;266:100–112. discussion 112–107, 155–108. [PubMed] [Google Scholar]
- 43.Jiang M, Zhang M, Tang DG, et al. KCNE2 protein is expressed in ventricles of different species, and changes in its expression contribute to electrical remodeling in diseased hearts. Circulation. 2004;109(14):1783–1788. doi: 10.1161/01.CIR.0000124225.43852.50. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Wang Y, Jiang M, et al. KCNE2 protein is more abundant in ventricles than in atria and can accelerate hERG protein degradation in a phosphorylation-dependent manner. American journal of physiology. Heart and circulatory physiology. 2011;302 (4):H910–922. doi: 10.1152/ajpheart.00691.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Jiang M, Tseng GN. minK-related peptide 1 associates with Kv4.2 and modulates its gating function: potential role as beta subunit of cardiac transient outward channel? Circulation research. 2001;88(10):1012–1019. doi: 10.1161/hh1001.090839. [DOI] [PubMed] [Google Scholar]
- 46.Roepke TK, Kontogeorgis A, Ovanez C, et al. Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f) The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22(10):3648–3660. doi: 10.1096/fj.08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mccrossan ZA, Roepke TK, Lewis A, Panaghie G, Abbott GW. Regulation of the Kv2.1 potassium channel by MinK and MiRP1. The Journal of membrane biology. 2009;228(1):1–14. doi: 10.1007/s00232-009-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu J, Kryukova Y, Potapova IA, et al. MiRP1 modulates HCN2 channel expression and gating in cardiac myocytes. The Journal of biological chemistry. 2004;279(42):43497–43502. doi: 10.1074/jbc.M405018200. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Wu J, Potapova I, et al. MinK-related peptide 1: A beta subunit for the HCN ion channel subunit family enhances expression and speeds activation. Circulation research. 2001;88(12):E84–87. doi: 10.1161/hh1201.093511. [DOI] [PubMed] [Google Scholar]
- 50.Mcdonald TV, Yu Z, Ming Z, et al. A minK-HERG complex regulates the cardiac potassium current I(Kr) Nature. 1997;388(6639):289–292. doi: 10.1038/40882. [DOI] [PubMed] [Google Scholar]
- 51.Finley MR, Li Y, Hua F, et al. Expression and coassociation of ERG1, KCNQ1, and KCNE1 potassium channel proteins in horse heart. American journal of physiology. Heart and circulatory physiology. 2002;283(1):H126–138. doi: 10.1152/ajpheart.00622.2001. [DOI] [PubMed] [Google Scholar]
- 52.Franco D, Demolombe S, Kupershmidt S, et al. Divergent expression of delayed rectifier K(+) channel subunits during mouse heart development. Cardiovascular research. 2001;52(1):65–75. doi: 10.1016/s0008-6363(01)00349-2. [DOI] [PubMed] [Google Scholar]
- 53.Mccrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47(6):787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 54.Tinel N, Diochot S, Borsotto M, Lazdunski M, Barhanin J. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. The EMBO journal. 2000;19(23):6326–6330. doi: 10.1093/emboj/19.23.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schroeder BC, Waldegger S, Fehr S, et al. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403(6766):196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- 56.Grunnet M, Rasmussen HB, Hay-Schmidt A, et al. KCNE4 is an inhibitory subunit to Kv1.1 and Kv1.3 potassium channels. Biophysical journal. 2003;85(3):1525–1537. doi: 10.1016/S0006-3495(03)74585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy DI, Wanderling S, Biemesderfer D, Goldstein SA. MiRP3 acts as an accessory subunit with the BK potassium channel. American journal of physiology. Renal physiology. 2008;295(2):F380–387. doi: 10.1152/ajprenal.00598.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbott GW. KCNE2 and the K (+) channel: The tail wagging the dog. Channels (Austin) 2012;6(1) doi: 10.4161/chan.19126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abbott GW, Goldstein SA. A superfamily of small potassium channel subunits: form and function of the MinK-related peptides (MiRPs) Quarterly reviews of biophysics. 1998;31(4):357–398. doi: 10.1017/s0033583599003467. [DOI] [PubMed] [Google Scholar]
- 60.Chen H, Kim LA, Rajan S, Xu S, Goldstein SA. Charybdotoxin binding in the I(Ks) pore demonstrates two MinK subunits in each channel complex. Neuron. 2003;40(1):15–23. doi: 10.1016/s0896-6273(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 61.Sesti F, Goldstein SA. Single-channel characteristics of wild-type IKs channels and channels formed with two minK mutants that cause long QT syndrome. The Journal of general physiology. 1998;112(6):651–663. doi: 10.1085/jgp.112.6.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang KW, Goldstein SA. Subunit composition of minK potassium channels. Neuron. 1995;14(6):1303–1309. doi: 10.1016/0896-6273(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 63.Morin TJ, Kobertz WR. Counting membrane-embedded KCNE beta-subunits in functioning K+ channel complexes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1478–1482. doi: 10.1073/pnas.0710366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakajo K, Ulbrich MH, Kubo Y, Isacoff EY. Stoichiometry of the KCNQ1 - KCNE1 ion channel complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(44):18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Zheng R, Melman YF, Mcdonald TV. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PloS one. 2009;4(4):e5143. doi: 10.1371/journal.pone.0005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melman YF, Domenech A, De La Luna S, Mcdonald TV. Structural determinants of KvLQT1 control by the KCNE family of proteins. The Journal of biological chemistry. 2001;276(9):6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- 67.Melman YF, Krumerman A, Mcdonald TV. A single transmembrane site in the KCNE-encoded proteins controls the specificity of KvLQT1 channel gating. The Journal of biological chemistry. 2002;277(28):25187–25194. doi: 10.1074/jbc.M200564200. [DOI] [PubMed] [Google Scholar]
- 68.Melman YF, Krummerman A, Mcdonald TV. KCNE regulation of KvLQT1 channels: structure-function correlates. Trends in cardiovascular medicine. 2002;12(4):182–187. doi: 10.1016/s1050-1738(02)00158-5. [DOI] [PubMed] [Google Scholar]
- 69.Melman YF, Um SY, Krumerman A, Kagan A, Mcdonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42(6):927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Panaghie G, Abbott GW. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. The Journal of general physiology. 2007;129(2):121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panaghie G, Purtell K, Tai KK, Abbott GW. Voltage-dependent C-type inactivation in a constitutively open K+ channel. Biophysical journal. 2008;95(6):2759–2778. doi: 10.1529/biophysj.108.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. The Journal of physiology. 2006;570(Pt 3):455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng R, Thompson K, Obeng-Gyimah E, et al. Analysis of the interactions between the C-terminal cytoplasmic domains of KCNQ1 and KCNE1 channel subunits. The Biochemical journal. 2010;428(1):75–84. doi: 10.1042/BJ20090977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bas T, Gao GY, Lvov A, Chandrasekhar KD, Gilmore R, Kobertz WR. Post-translational N-Glycosylation of Type I Transmembrane KCNE1 Peptides: IMPLICATIONS FOR MEMBRANE PROTEIN BIOGENESIS AND DISEASE. The Journal of biological chemistry. 2011;286(32):28150–28159. doi: 10.1074/jbc.M111.235168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bianchi L, Shen Z, Dennis AT, et al. Cellular dysfunction of LQT5-minK mutants: abnormalities of IKs, IKr and trafficking in long QT syndrome. Human molecular genetics. 1999;8(8):1499–1507. doi: 10.1093/hmg/8.8.1499. [DOI] [PubMed] [Google Scholar]
- 76.Xu X, Kanda VA, Choi E, et al. MinK-dependent internalization of the IKs potassium channel. Cardiovascular research. 2009;82(3):430–438. doi: 10.1093/cvr/cvp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biliczki P, Girmatsion Z, Brandes RP, et al. Trafficking-deficient long QT syndrome mutation KCNQ1-T587M confers severe clinical phenotype by impairment of KCNH2 membrane localization: evidence for clinically significant IKr-IKs alpha-subunit interaction. Heart rhythm : the official journal of the Heart Rhythm Society. 2009;6(12):1792–1801. doi: 10.1016/j.hrthm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Kanda VA, Purtell K, Abbott GW. Protein kinase C downregulates I(Ks) by stimulating KCNQ1-KCNE1 potassium channel endocytosis. Heart rhythm : the official journal of the Heart Rhythm Society. 2011;8(10):1641–1647. doi: 10.1016/j.hrthm.2011.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanda VA, Lewis A, Xu X, Abbott GW. KCNE1 and KCNE2 inhibit forward trafficking of homomeric N-type voltage-gated potassium channels. Biophysical journal. 2011 doi: 10.1016/j.bpj.2011.08.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanda VA, Abbott GW. KCNE Regulation of K(+) Channel Trafficking - a Sisyphean Task? Frontiers in physiology. 2012;3:231. doi: 10.3389/fphys.2012.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanda VA, Lewis A, Xu X, Abbott GW. KCNE1 and KCNE2 provide a checkpoint governing voltage-gated potassium channel alpha-subunit composition. Biophysical journal. 2011;101(6):1364–1375. doi: 10.1016/j.bpj.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roura-Ferrer M, Sole L, Oliveras A, et al. Impact of KCNE subunits on KCNQ1 (Kv7.1) channel membrane surface targeting. Journal of cellular physiology. 2010;225(3):692–700. doi: 10.1002/jcp.22265. [DOI] [PubMed] [Google Scholar]
- 83.Um SY, Mcdonald TV. Differential association between HERG and KCNE1 or KCNE2. PloS one. 2007;2(9):e933. doi: 10.1371/journal.pone.0000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roepke TK, King EC, Purtell K, Kanda VA, Lerner DJ, Abbott GW. Genetic dissection reveals unexpected influence of beta subunits on KCNQ1 K+ channel polarized trafficking in vivo. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25(2):727–736. doi: 10.1096/fj.10-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roepke TK, Kanda VA, Purtell K, King EC, Lerner DJ, Abbott GW. KCNE2 forms potassium channels with KCNA3 and KCNQ1 in the choroid plexus epithelium. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25(12):4264–4273. doi: 10.1096/fj.11-187609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silva J, Rudy Y. Subunit interaction determines IKs participation in cardiac repolarization and repolarization reserve. Circulation. 2005;112(10):1384–1391. doi: 10.1161/CIRCULATIONAHA.105.543306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tristani-Firouzi M, Sanguinetti MC. Voltage-dependent inactivation of the human K+ channel KvLQT1 is eliminated by association with minimal K+ channel (minK) subunits. The Journal of physiology. 1998;510 (Pt 1):37–45. doi: 10.1111/j.1469-7793.1998.037bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldstein SA, Miller C. Site-specific mutations in a minimal voltage-dependent K+ channel alter ion selectivity and open-channel block. Neuron. 1991;7(3):403–408. doi: 10.1016/0896-6273(91)90292-8. [DOI] [PubMed] [Google Scholar]
- 89.Bett GC, Morales MJ, Beahm DL, Duffey ME, Rasmusson RL. Ancillary subunits and stimulation frequency determine the potency of chromanol 293B block of the KCNQ1 potassium channel. The Journal of physiology. 2006;576(Pt 3):755–767. doi: 10.1113/jphysiol.2006.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Zaydman MA, Wu D, et al. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9095–9100. doi: 10.1073/pnas.1100872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loussouarn G, Park KH, Bellocq C, Baro I, Charpentier F, Escande D. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. The EMBO journal. 2003;22(20):5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chorbachi R, Graham JM, Ford J, Raine CH. Cochlear implantation in Jervell and Lange-Nielsen syndrome. International journal of pediatric otorhinolaryngology. 2002;66(3):213–221. doi: 10.1016/s0165-5876(02)00181-7. [DOI] [PubMed] [Google Scholar]
- 93.Schulze-Bahr E, Wang Q, Wedekind H, et al. KCNE1 mutations cause jervell and Lange-Nielsen syndrome. Nature genetics. 1997;17(3):267–268. doi: 10.1038/ng1197-267. [DOI] [PubMed] [Google Scholar]
- 94.Ehrlich JR, Pourrier M, Weerapura M, et al. KvLQT1 modulates the distribution and biophysical properties of HERG. A novel alpha-subunit interaction between delayed rectifier currents. The Journal of biological chemistry. 2004;279(2):1233–1241. doi: 10.1074/jbc.M309087200. [DOI] [PubMed] [Google Scholar]
- 95.Ren XQ, Liu GX, Organ-Darling LE, et al. Pore mutants of HERG and KvLQT1 downregulate the reciprocal currents in stable cell lines. American journal of physiology. Heart and circulatory physiology. 2010;299(5):H1525–1534. doi: 10.1152/ajpheart.00479.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Feng ZP, Kondo CS, Sheldon RS, Duff HJ. Developmental changes in the delayed rectifier K+ channels in mouse heart. Circulation research. 1996;79(1):79–85. doi: 10.1161/01.res.79.1.79. [DOI] [PubMed] [Google Scholar]
- 97.Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clinic proceedings. Mayo Clinic. 2003;78(12):1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 98.Arnestad M, Crotti L, Rognum TO, et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115(3):361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 99.Sesti F, Abbott GW, Wei J, et al. A common polymorphism associated with antibiotic-induced cardiac arrhythmia. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10613–10618. doi: 10.1073/pnas.180223197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park KH, Kwok SM, Sharon C, Baerga R, Sesti F. N-Glycosylation-dependent block is a novel mechanism for drug-induced cardiac arrhythmia. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17(15):2308–2309. doi: 10.1096/fj.03-0577fje. [DOI] [PubMed] [Google Scholar]
- 101.Kannankeril PJ, Roden DM. Drug-induced long QT and torsade de pointes: recent advances. Current opinion in cardiology. 2007;22(1):39–43. doi: 10.1097/HCO.0b013e32801129eb. [DOI] [PubMed] [Google Scholar]
- 102.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41(3):334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia S, Wang Y, Zhang Y, et al. Dynamic changes in HCN2, HCN4, KCNE1, and KCNE2 expression in ventricular cells from acute myocardial infarction rat hearts. Biochemical and biophysical research communications. 2010;395(3):330–335. doi: 10.1016/j.bbrc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 104.Roepke TK, King EC, Reyna-Neyra A, et al. Kcne2 deletion uncovers its crucial role in thyroid hormone biosynthesis. Nature medicine. 2009;15(10):1186–1194. doi: 10.1038/nm.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rice KS, Dickson G, Lane M, et al. Elevated serum gastrin levels in Jervell and Lange-Nielsen syndrome: a marker of severe KCNQ1 dysfunction? Heart rhythm : the official journal of the Heart Rhythm Society. 2011;8(4):551–554. doi: 10.1016/j.hrthm.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 106.Winbo A, Sandstrom O, Palmqvist R, Rydberg A. Iron-deficiency anaemia, gastric hyperplasia, and elevated gastrin levels due to potassium channel dysfunction in the Jervell and Lange-Nielsen Syndrome. Cardiology in the young. 2012:1–10. doi: 10.1017/S1047951112001060. [DOI] [PubMed] [Google Scholar]
- 107.Grahammer F, Herling AW, Lang HJ, et al. The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology. 2001;120(6):1363–1371. doi: 10.1053/gast.2001.24053. [DOI] [PubMed] [Google Scholar]
- 108.Kuwamura M, Okajima R, Yamate J, Kotani T, Kuramoto T, Serikawa T. Pancreatic metaplasia in the gastro-achlorhydria in WTC-dfk rat, a potassium channel Kcnq1 mutant. Veterinary pathology. 2008;45(4):586–591. doi: 10.1354/vp.45-4-586. [DOI] [PubMed] [Google Scholar]
- 109.Lee MP, Ravenel JD, Hu RJ, et al. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. The Journal of clinical investigation. 2000;106(12):1447–1455. doi: 10.1172/JCI10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takagi T, Nishio H, Yagi T, et al. Phenotypic analysis of vertigo 2 Jackson mice with a Kcnq1 potassium channel mutation. Experimental animals/Japanese Association for Laboratory Animal Science. 2007;56(4):295–300. doi: 10.1538/expanim.56.295. [DOI] [PubMed] [Google Scholar]
- 111.Vallon V, Grahammer F, Volkl H, et al. KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(49):17864–17869. doi: 10.1073/pnas.0505860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roepke TK, Anantharam A, Kirchhoff P, et al. The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. The Journal of biological chemistry. 2006;281(33):23740–23747. doi: 10.1074/jbc.M604155200. [DOI] [PubMed] [Google Scholar]
- 113.Roepke TK, Purtell K, King EC, La Perle KM, Lerner DJ, Abbott GW. Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PloS one. 2010;5(7):e11451. doi: 10.1371/journal.pone.0011451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ying SW, Kanda VA, Hu Z, et al. Targeted Deletion of Kcne2 Impairs HCN Channel Function in Mouse Thalamocortical Circuits. PloS one. 2012;7(8):e42756. doi: 10.1371/journal.pone.0042756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Heron SE, Hernandez M, Edwards C, et al. Neonatal seizures and long QT syndrome: a cardiocerebral channelopathy? Epilepsia. 2010;51(2):293–296. doi: 10.1111/j.1528-1167.2009.02317.x. [DOI] [PubMed] [Google Scholar]
- 116.Goldman AM, Glasscock E, Yoo J, Chen TT, Klassen TL, Noebels JL. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Science translational medicine. 2009;1(2):2ra6. doi: 10.1126/scitranslmed.3000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaab S, Crawford DC, Sinner MF, et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circulation. Cardiovascular genetics. 2012;5(1):91–99. doi: 10.1161/CIRCGENETICS.111.960930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gouas L, Nicaud V, Berthet M, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. European journal of human genetics : EJHG. 2005;13(11):1213–1222. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- 119.Lahtinen AM, Marjamaa A, Swan H, Kontula K. KCNE1 D85N polymorphism--a sex-specific modifier in type 1 long QT syndrome? BMC medical genetics. 2011;12:11. doi: 10.1186/1471-2350-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marjamaa A, Newton-Cheh C, Porthan K, et al. Common candidate gene variants are associated with QT interval duration in the general population. Journal of internal medicine. 2009;265(4):448–458. doi: 10.1111/j.1365-2796.2008.02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakajima T, Kaneko Y, Manita M, Iso T, Kurabayashi M. Aborted cardiac arrest in a patient carrying KCNE1 D85N variant during the postpartum period. Intern Med. 2010;49(17):1875–1878. doi: 10.2169/internalmedicine.49.3859. [DOI] [PubMed] [Google Scholar]
- 122.Nishio Y, Makiyama T, Itoh H, et al. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. Journal of the American College of Cardiology. 2009;54(9):812–819. doi: 10.1016/j.jacc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 123.Nof E, Barajas-Martinez H, Eldar M, et al. LQT5 masquerading as LQT2: a dominant negative effect of KCNE1-D85N rare polymorphism on KCNH2 current. Europace : European pacing, arrhythmias, and cardiac electrophysiology : journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology. 2011;13(10):1478–1483. doi: 10.1093/europace/eur184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paulussen AD, Gilissen RA, Armstrong M, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med (Berl) 2004;82(3):182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 125.Porthan K, Marjamaa A, Viitasalo M, et al. Relationship of common candidate gene variants to electrocardiographic T-wave peak to T-wave end interval and T-wave morphology parameters. Heart rhythm : the official journal of the Heart Rhythm Society. 2010;7(7):898–903. doi: 10.1016/j.hrthm.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zeng Z, Tan C, Teng S, et al. The single nucleotide polymorphisms of I(Ks) potassium channel genes and their association with atrial fibrillation in a Chinese population. Cardiology. 2007;108(2):97–103. doi: 10.1159/000095943. [DOI] [PubMed] [Google Scholar]
- 127.Zeng ZY, Pu JL, Tan C, et al. [The association of single nucleotide polymorphism of slow delayed rectifier K+ channel genes with atrial fibrillation in Han nationality Chinese] Zhonghua xin xue guan bing za zhi [Chinese journal of cardiovascular diseases] 2005;33(11):987–991. [PubMed] [Google Scholar]
- 128.Ehrlich JR, Zicha S, Coutu P, Hebert TE, Nattel S. Atrial fibrillation-associated minK38G/S polymorphism modulates delayed rectifier current and membrane localization. Cardiovascular research. 2005;67(3):520–528. doi: 10.1016/j.cardiores.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 129.Elahi MM, Flatman S, Matata BM. Tracing the origins of postoperative atrial fibrillation: the concept of oxidative stress-mediated myocardial injury phenomenon. European journal of cardiovascular prevention and rehabilitation : official journal of the European Society of Cardiology, Working Groups on Epidemiology & Prevention and Cardiac Rehabilitation and Exercise Physiology. 2008;15(6):735–741. doi: 10.1097/HJR.0b013e328317f38a. [DOI] [PubMed] [Google Scholar]
- 130.Filardo G, Hamilton C, Hebeler RF, Jr, Hamman B, Grayburn P. New-onset postoperative atrial fibrillation after isolated coronary artery bypass graft surgery and long-term survival. Circulation. Cardiovascular quality and outcomes. 2009;2(3):164–169. doi: 10.1161/CIRCOUTCOMES.108.816843. [DOI] [PubMed] [Google Scholar]
- 131.Girerd N, Magne J, Pibarot P, Voisine P, Dagenais F, Mathieu P. Postoperative atrial fibrillation predicts long-term survival after aortic-valve surgery but not after mitral-valve surgery: a retrospective study. BMJ open. 2011;1(2):e000385. doi: 10.1136/bmjopen-2011-000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Imazio M, Brucato A, Ferrazzi P, et al. Colchicine reduces postoperative atrial fibrillation: results of the Colchicine for the Prevention of the Postpericardiotomy Syndrome (COPPS) atrial fibrillation substudy. Circulation. 2011;124(21):2290–2295. doi: 10.1161/CIRCULATIONAHA.111.026153. [DOI] [PubMed] [Google Scholar]
- 133.Mann CJ, Kendall S, Lip GY. Acute management of atrial fibrillation with acute haemodynamic instability and in the postoperative setting. Heart. 2007;93(1):45–47. doi: 10.1136/hrt.2006.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heerdt PM, Kant R, Hu Z, et al. Transcriptomic analysis reveals atrial KCNE1 down-regulation following lung lobectomy. Journal of molecular and cellular cardiology. 2012 doi: 10.1016/j.yjmcc.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Seebohm G, Pusch M, Chen J, Sanguinetti MC. Pharmacological activation of normal and arrhythmia-associated mutant KCNQ1 potassium channels. Circulation research. 2003;93(10):941–947. doi: 10.1161/01.RES.0000102866.67863.2B. [DOI] [PubMed] [Google Scholar]
- 136.Burton DY, Song C, Fishbein I, et al. The incorporation of an ion channel gene mutation associated with the long QT syndrome (Q9E-hMiRP1) in a plasmid vector for site-specific arrhythmia gene therapy: in vitro and in vivo feasibility studies. Human gene therapy. 2003;14(9):907–922. doi: 10.1089/104303403765701196. [DOI] [PubMed] [Google Scholar]
- 137.Perlstein I, Burton DY, Ryan K, et al. Posttranslational control of a cardiac ion channel transgene in vivo: clarithromycin-hMiRP1-Q9E interactions. Human gene therapy. 2005;16(7):906–910. doi: 10.1089/hum.2005.16.906. [DOI] [PubMed] [Google Scholar]
- 138.Le Bouter S, El Harchi A, Marionneau C, et al. Long-term amiodarone administration remodels expression of ion channel transcripts in the mouse heart. Circulation. 2004;110(19):3028–3035. doi: 10.1161/01.CIR.0000147187.78162.AC. [DOI] [PubMed] [Google Scholar]
- 139.Gulbis JM, Mann S, Mackinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97(7):943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 140.Gulbis JM, Zhou M, Mann S, Mackinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science. 2000;289(5476):123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- 141.Lagoumintzis G, Poulas K, Patrinos GP. Genetic databases and their potential in pharmacogenomics. Current pharmaceutical design. 2010;16(20):2224–2231. doi: 10.2174/138161210791792804. [DOI] [PubMed] [Google Scholar]
- 142.Wang Q, Shen J, Splawski I, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80(5):805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 143.Mohler PJ, Schott JJ, Gramolini AO, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421(6923):634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 144.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nature genetics. 1997;17(3):338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 145.Plaster NM, Tawil R, Tristani-Firouzi M, et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell. 2001;105(4):511–519. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 146.Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 147.Vatta M, Ackerman MJ, Ye B, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114(20):2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 148.Medeiros-Domingo A, Kaku T, Tester DJ, et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116(2):134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(52):20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ueda K, Valdivia C, Medeiros-Domingo A, et al. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(27):9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang Y, Liang B, Liu J, et al. Identification of a Kir3.4 Mutation in Congenital Long QT Syndrome. American journal of human genetics. 2010 doi: 10.1016/j.ajhg.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Temple J, Frias P, Rottman J, et al. Atrial fibrillation in KCNE1-null mice. Circulation research. 2005;97(1):62–69. doi: 10.1161/01.RES.0000173047.42236.88. [DOI] [PubMed] [Google Scholar]
- 153.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 154.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309(5736):903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 155.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(19):7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]