Abstract
Background
Numerous genetic contributors to cardiovascular disease risk have been identified through genome-wide association studies (GWAS); however, identifying the molecular mechanism underlying these associations is not straightforward. The JUPITER trial of rosuvastatin users identified a sub-genome wide association of rs6924995, a SNP ~10kb downstream of MYLIP (aka IDOL, inducible degrader of LDLR), with LDL cholesterol statin response. Interestingly, though this signal was initially attributed to MYLIP, rs6924995 lies within RP1-13D10.2, an uncharacterized long noncoding RNA.
Methods and Results
Using simvastatin and sham incubated lymphoblastoid cell lines from participants of the Cholesterol and Pharmacogenetics simvastatin clinical trial, we found that statin induced change in RP1-13D10.2 levels differed between cell lines from the tails of the Caucasian and African American LDLC response distributions, while no difference in MYLIP was observed. RP1-13D10.2 overexpression in Huh7 and HepG2 increased LDLR transcript levels, increased LDL uptake, and decreased media levels of APOB. In addition, we found a trend of slight differences in the effects of RP1-13D10.2 overexpression on LDLR transcript levels between hepatoma cells transfected with the rs6924995 “A” vs. “G” allele, and a suggestion of an association between rs6924995 and RP1-10D13.2 expression levels in the CAP LCLs. Lastly, RP1-13D10.2 expression levels appear to be sterol regulated, consistent with its potential role as a novel lipid regulator.
Conclusions
RP1-13D10.2 is a long noncoding RNA that regulates LDLR and may contribute to LDLC response to statin treatment. These findings highlight the potential role of non-coding RNAs as determinants of inter-individual variation in drug response.
Keywords: low-density lipoprotein cholesterol, statin therapy, pharmacogenetics cholesterol, long-noncoding RNA
Introduction
Elevated plasma low-density lipoprotein cholesterol (LDLC) is a significant risk factor for cardiovascular disease (CVD), the leading cause of death in the world. Statins are the most widely prescribed class of drugs used to lower blood LDLC levels and reduce CVD risk. Specifically, statins competitively inhibit 3-hydroxy-3-methylglutaryl-Coenzyme A reductase (HMGCR), the rate limiting enzyme in the cholesterol biosynthesis pathway, and thus stimulate hepatic uptake of LDLC through up-regulation of the low density lipoprotein receptor (LDLR) 1. Although statin efficacy for reducing CVD mortality has been well-established, there is still substantial residual risk on treatment, and inter-individual response with regard to statin effects on cholesterol-lowering remains a concern 2.
While factors such as smoking status, race, and age have been reported to affect statin efficacy 3, the pharmacogenetics of statin response is an area of active study. Both clinical trial and population-based cohorts have identified variants in genes such as LPA, APOE, SORT1, HMGCR and LDLR that were associated with statin effects on LDLC lowering 4-10. To date, the largest genome-wide association study performed in a single statin clinical trial was reported in ~7000 participants of the JUPITER (Justification for the Use of Statins in Primary Prevention: An Intervention Trial Evaluating Rosuvastatin) clinical trial 5. Among the gene variants identified from this analysis, Chasman et al reported a sub-genome-wide significant association (p<1×10−6) between rs6924995 and statin-induced change in plasma LDLC. This particular association was notable as it was attributed to myosin regulatory light chain interacting protein (MYLIP; aka IDOL; inducible degrader of the low-density lipoprotein receptor), an E3 ubiquitin ligase that regulates LDLR stability, and thus activity, in response to changes in intracellular cholesterol levels 11. Although rs6924995 is located ~10kb downstream of MYLIP, to date, there is no evidence that rs6924995 impacts MYLIP expression levels or function.
Interestingly, rs6924995 is located within RP1-13D10.2, a processed pseudogene. Although RP1-13D10.2 has no known function, many pseudogenes have potential biological functions as noncoding RNAs 12. In addition, long noncoding RNAs (lncRNAs) have been implicated in cardiovascular disease. For example, the relationship between genetic variation at chromosome 9p21 associated with atherosclerotic risk has been attributed to expression changes in an antisense noncoding RNA 13, 14. Recently, the lncRNA RP5-833A20.1 was shown to modulate cholesterol homeostasis 15; however, the potential involvement of lncRNAs in statin response has not yet been assessed. Thus, here we sought to determine if RP1-13D10.2 acts as a novel lncRNA regulating cellular cholesterol metabolism, specifically hypothesizing that RP1-13D10.2 may mediate the association between rs6924995 and statin-induced change in LDLC.
Methods
Cell Culture
Lymphoblastoid cell lines (LCLs) from donors of the Cholesterol and Pharmacogenomics (CAP) population (ClinicalTrials.gov ID: NCT00451828) 3 with rs6924995 genotypes previously imputed 4, were grown in RPMI Medium 1640 supplemented with 10% fetal bovine serum (FBS; Hyclone), 500 U/mL penicillin/streptomycin and 2 nmol/L GlutaMAX. Cells were exposed to 2 μM activated simvastatin or sham buffer for 24 hr as described 16. HepG2 and Huh7 cell lines were maintained under standard conditions and grown in Eagle’s minimum essential medium (ATCC) supplemented with 10% FBS.
RNA-seq library preparation and data analyses
RNA-seq analyses were performed on 75 sham- and statin-exposed CAP LCLs chosen from the tails of the plasma LDLC statin response distribution (Table 1, Figure S1) that survived quality control criteria described below. These RNA-seq data form a subset of the RNA-seq data deposited in dbGaP under accession phs000481.v2.p1. Starting with 500 ng of total RNA, LabCorp (formerly Covance, Seattle, WA) made indexed, strand-specific, paired-end Illumina sequencing libraries, which were then sequenced at the University of Washington on Illumina HiSeq 2000 machines with a 101 bp read length. Paired-end fragments were aligned to the human (hg19) and EBV (NC_007605) genomes with the Ensembl v67 reference transcriptome using Tophatv2.0.4 17 allowing 4 mismatches. Suspected PCR duplicates were removed using Picard MarkDuplicates, and RNA genotypes in well transcribed (read depth >30) regions (that were also directly genotyped in the corresponding individuals’ genomic DNA) were estimated using allelic ratios derived from the SAMtools pileup command 18 and compared to genomic genotypes as an identity check. Samples of unknown identity, gender mismatches, sample mixtures, samples that were 5’->3’ bias outliers (evaluated using Picard CollectRnaSeqMetrics), and LCLs that did not have paired control and statin RNA-seq data were excluded from analyses. Aligned fragments in known Ensembl v67 genes were counted using HTSeq-count, adjusted for library size, and variance stabilized using DESeq 19. Statin-induced changes in gene expression (gene expression deltas) were calculated as statin-sham variance stabilized expression levels. Statistically significant differences between statin induced changes in gene expression between LDLC response groups (i.e. African American high versus low or Caucasian high versus low) was calculated from the variance stabilized deltas using a two-tailed t test.
Table 1.
Clinical characteristics of study participants split by race and LDLC statin response
| Caucasian American | African American | |||
|---|---|---|---|---|
| High | Low | High | Low | |
| N | 23 | 21 | 13 | 18 |
| Men | 48% | 62% | 85% | 44% |
| BMI | 27.9 ± 6.3 | 27.5 ± 4.5 | 32.1 ± 6.6 | 30.2 ± 5.6 |
| Age (yrs) | 50.6 ± 12.1 | 52.7 ± 9.7 | 50.8 ± 10.5 | 55.1 ± 15.0 |
| Before treatment LDLC level (mg/dl) | 136 ± 34 | 132 ± 28 | 141 ± 31 | 122 ± 42 |
| LDLC percent change after statin treatment (%) | -58.7 ± 4.1% | -22.0 ± 7.6% | -53.9 ± 5.7% | -26.7 ± 7.3% |
| LDLC level change after statin treatment (mg/dl) | -80.3 ± 22.4 | -29.3 ± 12.4 | -76.0 ± 18.3 | -33.6 ± 16.4 |
Data are presented as numbers, percentages or means ± SDs. None of these participants were smokers.
Transcript quantitation by qPCR
Cells were homogenized using QIAshredders (QIAGEN), total RNA was extracted using the PureLink RNA Mini Kit (Life Technologies), and RNA integrity was checked using an Agilent Bioanalyzer. Transcript levels were quantified using a qPCR SYBR Green assay with the following primers: RP1-13D10.2 (TGTGGCTCTATCACCCTCAA and AGGATGATTCGGAACACAGC), MYLIP (TCTCCTCTGCCACCTTGAAC and TCCATTGCCGACACAATCTG), RP1-13D10.3 (TGAAGCCAACAAAGCTGTCA and TGGATGCGAAACACTCAATT), and RP1-13D10.4 (CAGGAAGTGAGCCTGCTACC and TGTGGTTGAAGGATGGGTTT). HMGCR, LDLR, HMGCS1 and PCSK9 were quantified by qPCR with assays as previously reported 20. All reactions were performed in triplicate on an ABI PRISM 7900 Sequence Detection System. Given the lack of introns in RP1-13D10.2, RP1-13D10.3, and RP1-13D10.4, no reverse transcriptase (RT) controls were prepared for each sample, and transcript levels of these lncRNAs were calculated as the difference of the RT versus no RT sample to prevent detection of residual genomic DNA. All values were normalized to CLTPM1 as a loading control as previously described16.
RP1-13D10.2 overexpression construct
The pCMV6-Entry plasmid (OriGene), a mammalian expression vector, was used to overexpress RP1-13D10.2. A 512 bp fragment containing the RP1-13D10.2 gene was amplified from cDNA isolated from LCLs homozygous for either the rs6924995 A or G allele using the following primers: ATC GTC GCG ATC GCC TGG TGG GGC TCC TGC TCT G and ATC GTC ACG CGT TGG TCT GAG GTT GTC CGG GA. Fragments were cloned into the pCMV6-Entry Vector (OriGene) using SgfI and MluI, and plasmid sequences were confirmed by Sanger sequencing.
Cellular phenotyping
HepG2 and Huh7 plasmid transfections were performed with GenJet™ In Vitro DNA Transfection Reagent (Ver. II; SignaGen Laboratories), according to the manufacturer’s protocol. Twenty-four hours prior to transfection, HepG2 cells were seeded into 6-well plates at a concentration of 7.5 × 105 cells/well, and Huh7 cells were seeded into 6-well plates at a concentration of 5.0 × 105 cells/well. Cells were transfected for a total of 48 hours with either the pCMV6-entry plasmid (EV) or either allele of the RP1-13D10.2 overexpression plasmids, and cellular phenotypes were measured after 48 hours.
To measure LDL-uptake, cells transfected with the EV negative control or either allele of the RP1-13D10.2 expression constructs (rs6924995 ‘A’ or ‘G’) were incubated with 10 μg/mL of DiI-LDL (Biomedical Technologies Inc., BT-904) in EMEM media supplemented with 10% FBS for 2 hours. Cells were washed twice in PBS and scraped from wells. Levels of DiI-LDL uptake were quantified by fluorescence-activated cell sorting on a BD FACS Calibur Flow Cytometer, and values were calculated using the average of 10,000 gated events. Experiments were performed four times. Effects of RP1-13D10.2 overexpression on media levels of APOB were quantified by ELISA as previously described 21. To measure the effect of RP1-13D10.2 overexpression on LDLR transcript stability, Huh7 cells were transfected for 48 hours with either the pCMV6-entry plasmid or the RP1-13D10.2 overexpression plasmids, treated with 1 μg/mL actinomycin D and harvested over 6 hours. Transcript half-life was calculated as described previously 22.
SREBF1 and SREBF2 knock-down was achieved by 48 hour transfection of 1.5 × 105 Huh7 cells/well in 6-well plates using siPORT™ NeoFX™ transfection agent (Ambion) with either Silencer Select siRNA (Ambion) or non-targeting control, according to the manufacturer’s protocol. HepG2 cells were incubated with 1 μM GW3965 (Sigma Aldrich) dissolved in DMSO for 24 hr prior to collection for RNA isolation.
Cycloheximide treatment
Huh7 cells were transfected with RP1-13D10.2 “G” or “A” plasmid using GenJet (SignaGen Labs) according to manufacturer’s protocol. Cells were incubated with cycloheximide (20ug/ml) for 24 hours. To quantify total cellular protein, cells were dissolved in CellLytic M (Sigma-Aldrich) and centrifuged at 16,000xg. Supernatant was quantitated using the Bio-Rad Protein Assay kit (Bio Rad) and measured on a Synergy H1 microplate reader (Biotek).
Statistical Analyses
Differences between EV and RP1-13D10.2 overexpression (either “A” or “G” plasmid) on cellular phenotypes including gene expression, LDL uptake, and media APOB were identified using two-tailed t-tests in which samples within an experimental batch were paired. One-way ANOVA with post-hoc two-tailed paired t-tests was used to identify differences in gene expression levels after cellular incubation under various conditions. Unless otherwise stated, all statistical analyses were performed in GraphPad Prism 6.0.
Results
Statin-induced change in RP1-13D10.2 expression differs between high versus low LDLC statin responders
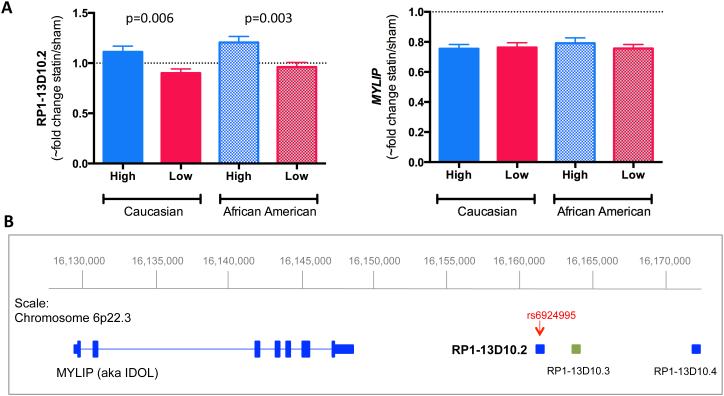
We first sought to determine if either RP1-13D10.2 or MYLIP gene expression was related to LDLC statin response using simvastatin and sham exposed lymphoblastoid cell lines (LCLs) from participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial with either high or low LDLC response (Table 1). Statin induced change in RP1-13D10.2 expression levels significantly differed between high and low LDLC responders in cell lines from both Caucasian (23 high responders vs. 21 low responders) and African American (13 high responders vs. 18 low responders) donors (Figure 1A). The statin induced change in RP1-13D10.2 expression in the high responders of both populations was greater than the low responders (1.11±0.06-fold vs. 0.90±0.04-fold respectively, p=0.006 in Caucasians, and 1.21±0.06-fold vs. 0.96±0.04-fold respectively, p=0.002 in African Americans). In contrast, we observed no relationship between statin response and change in MYLIP transcript levels (Figure 1A).
Figure 1.
A) Statin induced change in RP1-13D10.2 expression levels differs between high and low LDLC responders to statin treatment. CAP LCLs from the tails of the Caucasian and African American LDLC distribution were incubated with 2μM simvastatin or sham buffer for 24 hours, after which RP1-13D10.2 and MYLIP expression levels were quantified by RNA-seq. Since variance stabilization is approximately like a log2 transformation, the ~fold change was estimated as 2(variance stabilized sham-variance stabilized statin). Although fold changes (mean ± SE) are displayed for ease of interpretation, p-values were calculated from t-tests on the variance stabilized deltas. Transcripts on the + strand are indicated in blue, transcripts on the – strand are indicated in green. B) RP1-13D10.2 is located on chromosome 6 ~10 kb downstream of MYLIP and is proximal to two additional processed, uncharacterized pseudogenes, RP1-13D10.3 and RP1-13D10.4
RP1-13D10.2 transcript structure
RP1-13D10.2 is annotated to the + strand of chromosome 6. As shown in Figure 1B, it is located approximately 10 kb down-stream of MYLIP, and is adjacent to two processed pseudogenes, RP1-13D10.3 (annotated on the – strand) and MRPL42P2 (aka RP1-13D10.4, annotated on the + strand). Upon closer inspection of our RNA-seq data, we found evidence of a splice junction joining the annotated 3’ end of RP1-13D10.2 to another exon that partially overlaps RP1-13D10.3 in the anti-sense direction (Figure S2). There was no evidence of splicing of either pseudogene to RP1-13D10.4. We quantified RP1-13D10.2, RP1-13D10.3 and RP1-13D10.4 expression levels in immortalized lymphoblastoid cell lines (LCLs) (n=60) derived from participants of the Cholesterol and Pharmacogenetics (CAP) statin clinical trial after in vitro exposure to 2 μM simvastatin or sham buffer. Since these pseudogenes have no introns, we compared expression levels in cDNA versus no reverse transcriptase (RT) controls, prepared for each sample, to account for amplification from residual genomic DNA. RP1-13D10.2 expression was detected in both the statin and sham treated cells. In contrast, no expression of the annotated RP1-13D10.3 transcript or RP1-13D10.4 was detected in the statin or sham treated cells (Figure S3).
RP1-13D10.2 increases LDLR expression and stimulates LDL uptake
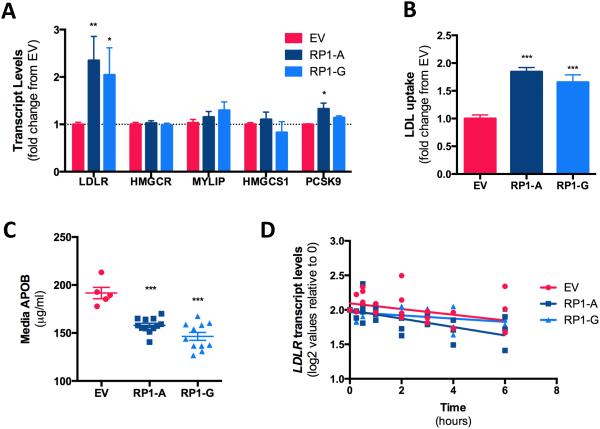
To determine if RP1-13D10.2 affects genes involved in cholesterol metabolism, we transiently transfected Huh7 cells with a plasmid overexpressing the RP1-13D10.2 gene containing either the rs6924995 “A” or “G” allele, or pCMV6-entry, a control plasmid that contained the empty vector backbone. Although we identified a novel splice variant of RP1-13D10.2, we chose to focus our functional investigation on the canonical RP1-13D10.2 transcript. After 48 hours, overexpression of RP1-13D10.2 was confirmed by qPCR (Figure S4A), and genes involved in cholesterol biosynthesis (HMGCR, HMGCS1) and cholesterol uptake (LDLR, MYLIP, and PCSK9) were quantified (Figure 2A). Overexpression of the RP1-13D10.2 containing the rs6924995 “A” allele of increased LDLR transcripts by 2.35±0.51 fold, p=0.002, while overexpression of the “G” allele increased LDLR transcripts by 2.04±0.57 fold, p=0.03. Notably, these effects appeared to be specific for LDLR as there were no consistent expression differences in any other genes, including MYLIP. Similar effects of RP1-13D10.2 overexpression were also observed in a second human hepatoma cell line, HepG2 (Figure S4). To verify the functional impact of RP1-13D10.2 overexpression, we tested the effect of overexpression on uptake of DiI-labeled LDL. Consistent with increased expression levels of LDLR, we found that RP1-13D10.2 overexpression increased DiI-LDL uptake with either the ‘A’ allele (1.85 ± 0.08 fold, p<0.0001) or ‘G’ allele (1.66 ± 0.13 fold, p=0.0002) (Figure 2B), as well as reduced media levels of APOB (Figure 2C). Finally, since LDLR transcript levels are known to be regulated at the level of transcription 23 as well as mRNA stability, we tested if RP1-13D10.2 altered the LDLR stability using incubation with Actinomycin D, but found no evidence of an effect (Figure 2D).
Figure 2.
RP1-13D10.2 overexpression increases LDLR expression and activity. Huh7 cells were transiently transfected with one of two different constructs expressing RP1-13D10.2 containing either the rs6924995 “A” (RP1-A) or “G” (RP1-G) allele, or an empty vector (EV) control for 48 hours. A) Gene expression levels were quantified by qPCR and normalized to CLPTM. N=12 B) Cells were incubated with DiI-LDL for 3 hours, and the mean fluorescence of 10,000 gated events was quantified to measure LDL uptake. N=8. C) Media APOB levels were measured by ELISA. N=5-12. D) Cells were incubated with 1 μg/mL actinomycin D after which aliquots of cells were harvested over 6 hours, and LDLR transcript levels were quantified by qPCR. N=4 For panels A and B, values shown are mean ± SE. *p<0.05, **p<0.01, ***p<0.001 by two tailed paired t-test.
RP1-13D10.2 impacts LDLR transcript in the absence of protein synthesis
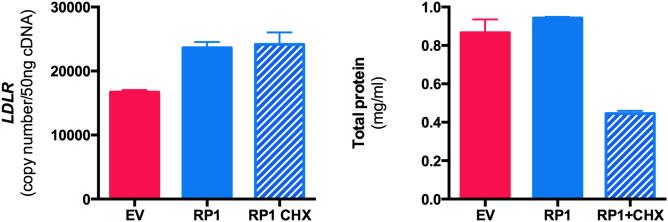
RP1-13D10.2 is currently annotated as a processed pseudogene by Gencode . However, recent findings have reported that some non-coding RNAs are translated and function as micropeptides rather than as non-coding RNAs 24. RP1-13D10.2 does have an open reading frame that, if translated, would encode a protein of 97 amino acids with the rs6924995 “G” allele (Figure S5). Notably, this putative open reading frame would be disrupted by the rs6924995 “A” allele, TGG (tryptophan) to TAG (stop), resulting in a protein of 82 amino acids. Thus, to test the possibility that RP1-13D10.2 functions as a protein, we tested if overexpression in the presence of cycloheximide, an inhibitor of protein synthesis, was able to stimulate increased expression levels of LDLR. While cycloheximide treatment reduced total cellular protein by ~50%, it had no effect on RP1-13D10.2 induction of LDLR expression levels (Figure 3).
Figure 3.
RP1-13D10.2 stimulation of LDLR does not require protein synthesis. Huh7 cells (n=3) were transfected with a construct containing either RP1-13D10.2 rs6924995 “G” alleles or an empty vector (EV) control in duplicate, after which one aliquot was treated with 20ug/ml cycloheximide for 24hrs and LDLR transcript was quantified by qPCR. Values shown are mean ± SE
To further evaluate the possibility that RP1-13D10.2 is translated, we used three in silico prediction programs, the coding-non-coding index (CNCI)25, the coding potential assessment tool (CPAT)26, and the coding potential calculator (CPC)27. Both the CNCI and CPAT analyses indicated that the RP1-13D10.2 transcript with either the “A” or “G” rs6924995 alleles was non-coding, while the CPC analysis indicated there was weak evidence that both “A” and “G” allele containing transcripts were coding.
Effects of rs6924995 on RP1-13D10.2 regulation of LDLR
We observed a consistent trend of slightly larger effect sizes of the “A” allele versus “G” allele overexpression on both LDLR expression and LDL uptake; however this difference was not statistically significant (p=0.10 for LDL uptake). Since this trend was observed in three different hepatoma cell lines (Figure S4C), we hypothesized that our model of extreme RP1-13D10.2 overexpression (~1010 fold increase) may obscure the potential differences between the two alleles. With reduced levels of RP1-13D10.2 overexpression (~107 fold) we continued to observe a similar trend of smaller increases in LDLR up-regulation with the “G” allele vs. “A” allele; however, this difference did not achieve statistical significance (p=0.06, Figure S4D and S4E). Further reductions in the degree of RP1-13D10.2 overexpression failed to produce consistent stimulation of LDLR transcript with either allele.
RP1-13D10.2 expression and rs6924995 genotype
Given the fact that rs6924995 is contained within RP1-13D10.2, we next sought to determine if this SNP was associated with expression levels of the pseudogene. We observed a suggestive association between rs6924995 genotype and RP1-13D10.2 expression in both the statin and sham treated cells, with trends toward greater RP1-13D10.2 expression observed in the GG homozygotes (Figure S6). In contrast, there was no relationship between rs6924995 and MYLIP expression levels (Figure S6). Closer examination of genomic region between MYLIP and RP1-13D10.2 found that rs6924995 is within a small block of linkage disequilibrium that contains RP1-13D10.2, but not MYLIP (Figure S7).
RP1-13D10.2 expression levels are sterol regulated
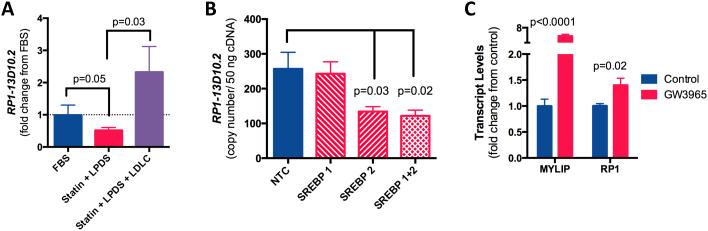
Many genes that impact cholesterol metabolism are themselves subject to sterol regulation. To test if RP1-13D10.2 transcript levels were impacted by changes in intracellular sterol content, we exposed HepG2 cells to conditions of extreme sterol depletion (2μM simvastatin + 10% lipoprotein deficient serum) for 24 hours, after which we added-back LDLC. Sterol depletion reduced RP1-13D10.2 expression levels 0.43±0.12 fold (Figure 4A). LDLC add-back not only reversed this effect but highly induced RP1-13D10.2 expression.
Figure 4.
RP1-13D10.2 expression levels are sterol regulated. A) RP1-13D10.2 transcript levels were quantified in HepG2 cells were exposed to four culture conditions: FBS (control), 48hr incubation with 2uM simvastatin + 10% lipoprotein deficient serum (LPDS), 24hr incubation with statin + LPDS after which LDLC or 25HC was added-back and incubated for an additional 24 hours. The experiment was repeated six times, with means ± standard error shown. Statistically significant differences in gene expression were assessed using one-way ANOVA (p=0.01), with two tailed paired t-tests used to identify differences between groups. B) RP1-13D10.2 transcript levels were quantified after SREBF1 and SREBF2 knock-down in Huh7 cells. C) RP1-13D10.2 and MYLIP transcript levels were quantified in HepG2 cells after 24 hour incubation with 1μM GW3965 (n=6).
SREBPs and LXR are well-known transcription factors that mediate cellular response to changes in intracellular sterols. RP1-13D10.2 expression levels were significantly reduced after SREBF2 knock-down in Huh7 cells (0.5±0.11-fold, p=0.03, Figure 4B and S8), no effect was observed with SREBF1 knock-down. We also found evidence that RP1-13D10.2 is regulated by LXR as incubation of HepG2 with GW3965, an LXR agonist, increased RP1-13D10.2 expression levels (1.4±0.13-fold, p=0.02, Figure 4C). Notably, these effects were much more modest compared to MYLIP, a known LXR target gene, which was increased 5.8±0.46-fold (p<0.0001).
Discussion
We have examined RP1-13D10.2, a long non-coding RNA containing a SNP, rs6924995, reported to have a sub-genome wide association with LDLC response to rosuvastatin 5. Here we report that RP1-13D10.2 overexpression increases transcript levels and activity of LDLR, which encodes the major receptor for uptake of plasma LDLC. RP1-13D10.2 expression appears to be sterol-regulated, and notably we observed a relationship between inter-individual variation in the magnitude of this regulation with statin-induced changes in LDLC from a panel of LCLs derived from participants of a statin clinical trial. In particular, statin incubation increased RP1-13D.10.2 expression levels in cell lines from both Caucasian and African Americans with high LDLC response to statin treatment, while either no change or reduced RP1-13D10.2 expression was detected in cell lines from donors with low LDLC response. Together, these findings support the identification of RP1-13D10.2 as a novel marker, and possibly determinant, of variation in statin efficacy for plasma LDLC lowering. Although lncRNAs have been well established to play a role in cardiovascular biology and disease 28, to our knowledge, RP1-13D10.2 is the first lncRNA that has been identified to play a role in statin response.
rs6924995 was first reported to be associated with LDLC response to statin in the JUPITER placebo-controlled trial of rosuvastatin (20 mg/day) response. The “A” allele was associated with both greater absolute (β=4.1, p=5.3E-07) and fractional (β=3.8, p=1.4E-06) LDLC reduction in individuals with genetically confirmed European ancestry. The Heart Protection Study, a five-year trial of 3895 self-reported Caucasians prescribed 40 mg simvastatin/day, failed to replicate this association with LDLC statin response 29. In addition, the largest genome-wide meta-analysis of LDLC response to statin treatment published to date, comprising 18,596 subjects from clinical trial and population based cohorts, did not identify this SNP 10; however, the report did not include a direct test for replication of this locus. This lack of replication may be due to the unique nature of the JUPITER study population. Statins are traditionally prescribed to individuals with hypercholesterolemia; in contrast JUPITER was comprised of participants with relatively normal levels of LDLC (< 130 mg/dL), but who had high measures of inflammation (C-reactive protein ≥2 mg/dL).
The association between rs6924995 and LDLC response to statin treatment was originally attributed to MYLIP (aka IDOL) 5, a known regulator of LDLR protein levels 11. Here, we failed to identify a relationship between rs6924995 genotype and MYLIP transcript levels in LCLs. In contrast, we found suggestions of an association between RP1-13D10.2, a lncRNA that contains rs6924995 and is located ~10kb downstream from MYLIP, expression levels with rs6924995 genotype. In addition, although we were unable to verify a genotype difference in RP1-13D10.2 effects on LDLR, the trend of slightly greater stimulation of LDLR transcript with the RP1 “A” allele constructs versus the “G” allele constructs is consistent with the GWAS association between the rs6924995 A allele and greater LDLC lowering upon statin treatment. Thus, additional study is necessary to ascertain the true relationship between rs6924995 and RP1-13D10.2 transcript levels, transcript structure, and/or activity.
We observed no direct effects of RP1-13D10.2 overexpression on MYLIP transcript levels. Although we did not test for an effect of RP1-13D10.2 on MYLIP protein levels, MYLIP is an E3-dependent ubiquitin ligase complex that mediates sterol-dependent degradation of LDLR protein 30, thus it is unlikely that the effects of RP1-13D10.2 on LDLR transcript levels are mediated by MYLIP. Although further study will be required to absolutely discount a relationship of either rs6924995 or RP1-13D10.2 to MYLIP, these findings suggest the intriguing possibility that RP1-13D10.2 and MYLIP may be mechanistically independent regulators of LDLR activity that happen to be in quite close proximity to one another, similar to other known clusters of lipid-related genes (i.e. APOC3 and APOA5, SREBF2 and mir33a).
We found that RP1-13D10.2 expression levels were increased with statin treatment in LCLs from donors with high LDLC response to statin treatment. Sterol response element binding protein 2 (SREBP2, gene name SREBF2) is a well-known transcription factor that is activated by conditions of sterol depletion, such as in vitro statin exposure. In human hepatoma cell lines, we found that SREBF2 knock-down reduced RP1-13D10.2 expression levels, consistent with the likelihood that RP1-13D10.2 may be an SREBF2 target gene. In addition, RP1-13D10.2 expression levels were also increased after incubation with an LXR agonist, suggesting that it may also be an LXR target gene. Notably, MYLIP is a well-known LXR target gene 11, and thus the close proximity between RP1-13D10.2 and MYLIP may allow for shared transcription factor regulatory sequences. Paradoxically, these two genes would be expected to oppose one another, as LXR-induced expression of RP1-13D10.2 would increase LDLR activity, while LXR-induced expression of MYLIP would stimulate LDLR decay. However, this is similar to the well-known phenomena in which SREBF2 both induces LDLR transcription, while stimulating expression of a factor, PCSK9, that promotes LDLR protein decay 31.
A question that remains is the precise mechanism by which RP1-13D10.2 specifically increases transcript levels of LDLR. Our observation that RP1-13D10.2 up-regulates LDLR after cycloheximide incubation supports the likelihood that RP1-13D10.2 functions as a lncRNA; however, since the cells were treated with cycloheximide 24 hr after transfection with the RP1 plasmid, these findings cannot exclude the possibility that RP1-13D10.2 acts as a highly stable protein that persists after inhibition of protein synthesis. However, when combined with our in silico analysis that does not support the coding potential of RP1-13D10.2, our findings strongly support the likelihood that RP1-13D10.2 functions as a non-coding RNA.
There are four major described functions of lncRNAs (recently reviewed by Uchida and Dimmeler28): 1) Imprinting – the lncRNA directly inhibits expression of a proximal locus. 2) Guide molecules – the lncRNA recruits functional proteins, often epigenetic or transcription factors, in a trans-acting manner. 3) Enhancer activation –the lncRNAs is transcribed from the site of an enhancer element and aids in enhancer activity. 4) Molecular sponges – the lncRNA binds miRNAs, disrupting miRNA inhibition of mRNAs. Endogenous levels of RP1-13D10.2 are quite low compared to LDLR, thus it is unlikely that RP1-13D10.2 functions as a molecular sponge, which often requires similar stoichiometry of the effector and target molecules 28. In addition, we found that RP1-13D10.2 overexpression increases LDLR transcript levels without affecting LDLR transcript stability, suggesting that RP1-13D10.2 enhances LDLR transcription. However, RP1-13D10.2 is located on chromosome 6, while LDLR is located on chromosome 19, thus RP1-13D10.2 does not likely impact LDLR through either imprinting or changes in LDLR enhancer activity. Thus, the most probable function of RP1-13D10.2 is as a guide molecule. Notably, neither moderate nor high expression is required for this function as the biological effects of even a very lowly expressed lncRNA may be amplified through a signaling cascade 28.
One of the major findings of the large transcriptomic projects of the past decade is the widespread transcription of the human genome, and recent estimates using RNA-seq data suggest that approximately 80% of the genome is transcribed, with many of these transcribed sequences representing ncRNAs 32, 33. Using a combination of gene expression and functional studies, here we identify the lncRNA RP1-13D10.2 as a novel marker, and possible determinant, of LDLC response to statin treatment that regulates LDLR. Thus, our findings illustrate the potential of non-coding regulatory RNA as a determinant of variability in drug response. In addition, as GWAS identified SNPs are most often annotated based on the their proximity to protein coding genes, these results demonstrate the importance of functional validation studies not only as an alternative approach for validating pharmacogenetic associations 34, but also for ensuring the correct annotation of GWAS findings.
Supplementary Material
Clinical Perspective.
Statins are among the most prescribed drugs in the United States, used to decrease LDL-cholesterol (LDLC) levels for the prevention and treatment of cardiovascular disease. However, response to statins is variable and many patients are left with insufficient LDLC-lowering despite treatment. Genome-wide association studies have identified a number of DNA variants that are associated with this inter-individual variation. Although simply identifying variants may be sufficient for the development of diagnostics, understanding the molecular mechanisms underlying these associations is essential for fully leveraging these findings into new biology that may inform advances in biomedical research. Previously, a sub-genome wide association was observed between rs6924995 and rosuvastatin response in the JUPITER clinical trial. Located downstream of MYLIP/IDOL, a gene known to regulate the major LDL receptor (LDLR), rs6924995 was originally annotated as the MYLIP/IDOL locus. However, rs6924995 is located within the uncharacterized long non-coding RNA (lncRNA), RP1-13D10.2, thus here we explored the contribution of this lncRNA to LDLC statin response. Using cell lines established from participants of a statin clinical trial with either high or low LDLC response, we found significant differences in statin-induced change in RP1-13D10.2 expression between the two groups, while no change was observed with MYLIP/IDOL expression levels. Furthermore, RP1-13D10.2 overexpression in hepatoma cell lines up-regulated the major LDL receptor (LDLR) and increased uptake of LDL. Our data support the hypothesis that RP1-13D10.2 is a novel marker, and possible determinant, of LDLC response to statin treatment, and highlight the importance of functional studies for annotation of GWAS identified loci.
Acknowledgments
We thank Jerome I. Rotter and Yi-D. Chen for creation of the CAP LCLs; and Ronald M. Krauss for his insightful discussions and critical review of this manuscript. This study would not have been possible without the contributions of the CAP participants.
Funding Sources: This work was supported by the NIH: U19 HL069757 (MWM, KM, AD, DN), R01 HL104133 (MWM, AD), P50 GM115318 (MWM) and NIH Pharmacogenomics Research Network RNA Sequencing Project (GM61390), as well as the American Heart Association 12POST10430005 (ET).
Footnotes
Journal Subject Terms: Lipids and Cholesterol; Functional Genomics; Gene Expression and Regulation; Genetics; Metabolism
Disclosures: None
References
- 1.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Sampson UK, Fazio S, Linton MF. Residual cardiovascular risk despite optimal LDL cholesterol reduction with statins: the evidence, etiology, and therapeutic challenges. Curr Atheroscler Rep. 2012;14:1–10. doi: 10.1007/s11883-011-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 4.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasman DI, Giulianini F, Macfadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic Determinants of Statin-Induced Low-Density Lipoprotein Cholesterol Reduction: The Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin (JUPITER) Trial. Circ Cardiovasc Genet. 2012;5:257–264. doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- 6.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr., Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 7.Deshmukh HA, Colhoun HM, Johnson T, McKeigue PM, Betteridge DJ, Durrington PN, et al. Genome-wide association study of genetic determinants of LDL-c response to atorvastatin therapy: importance of Lp(a) J Lipid Res. 2012;53:1000–1011. doi: 10.1194/jlr.P021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly LA, Doney AS, Dannfald J, Whitley AL, Lang CC, Morris AD, et al. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet Genomics. 2008;18:1021–1026. doi: 10.1097/FPC.0b013e3283106071. [DOI] [PubMed] [Google Scholar]
- 9.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, et al. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 10.Postmus I, Trompet S, Deshmukh HA, Barnes MR, Li X, Warren HR, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo X, Lin M, Rockowitz S, Lachman HM, Zheng D. Characterization of human pseudogene-derived non-coding RNAs for functional potential. PLoS One. 2014;9:e93972. doi: 10.1371/journal.pone.0093972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 14.Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR, Ma X, et al. RP5-833A20.1/miR-382-5p/NFIA-Dependent Signal Transduction Pathway Contributes to the Regulation of Cholesterol Homeostasis and Inflammatory Reaction. Arterioscler Thromb Vasc Biol. 2015;35:87–101. doi: 10.1161/ATVBAHA.114.304296. [DOI] [PubMed] [Google Scholar]
- 16.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina MW, Gao F, Naidoo D, Rudel LL, Temel RE, McDaniel AL, et al. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS One. 2011;6:e19420. doi: 10.1371/journal.pone.0019420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina MW, Bauzon F, Naidoo D, Theusch E, Stevens K, Schilde J, et al. Transmembrane Protein 55B Is a Novel Regulator of Cellular Cholesterol Metabolism. Arterioscler Thromb Vasc Biol. 2014;34:1917–1923. doi: 10.1161/ATVBAHA.113.302806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu CY, Theusch E, Lo K, Mangravite LM, Naidoo D, Kutilova M, et al. HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. Hum Mol Genet. 2014;23:319–332. doi: 10.1093/hmg/ddt422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida S, Dimmeler S. Long Noncoding RNAs in Cardiovascular Diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 29.Hopewell JC, Parish S, Offer A, Link E, Clarke R, Lathrop M, et al. Impact of common genetic variation on response to simvastatin therapy among 18 705 participants in the Heart Protection Study. Eur Heart J. 2013;34:982–992. doi: 10.1093/eurheartj/ehs344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Fairall L, Goult BT, Calkin AC, Hong C, Millard CJ, et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagace TA. PCSK9 and LDLR degradation: regulatory mechanisms in circulation and in cells. Curr Opin Lipidol. 2014;25:387–393. doi: 10.1097/MOL.0000000000000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aslibekyan S, Claas SA, Arnett DK. To replicate or not to replicate: the case of pharmacogenetic studies: Establishing validity of pharmacogenomic findings: from replication to triangulation. Circ Cardiovasc Genet. 2013;6:409–412. doi: 10.1161/CIRCGENETICS.112.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.