Summary
T help (Th) 17 cells are prominently featured in multiple autoimmune diseases, but the regulatory mechanisms that control Th17 cell responses are poorly defined. Here we found that stimulation of OX40 triggered a robust chromatin remodeling response and produced a “closed” chromatin structure at interleukin-17 (IL-17) locus to inhibit Th17 cell function. OX40 activated the NF-κB family member RelB, and RelB recruited the histone methyltransferases G9a and SETDB1 to the Il17 locus to deposit “repressive” chromatin marks at H3K9 sites, and consequently repressing IL-17 expression. Unlike its transcriptional activities, RelB acted independently of both p52 and p50 in the suppression of IL-17. In an experimental autoimmune encephalomyelitis (EAE) disease model, we found that OX40 stimulation inhibited IL-17 and reduced EAE. Conversely, RelB-deficient CD4+ T cells showed enhanced IL-17 induction and exacerbated the disease. Our data uncover a mechanism in the control of Th17 cells that may have important clinic implications.
Introduction
Signals from T cell costimulatory molecules are critical to the activation of naïve CD4+ T cells, and together with those from the T cell receptor (TCR) and cytokine receptors, they activate diverse signaling pathways that control the fate as well as the function of activated T cells (Sharpe, 2009). CD4+ T cells also have the capacity to differentiate into distinct T helper (Th) subsets (i.e., Th1, Th2, Th9, Th17, Tfh), as defined by differences in the cytokines they produce (Dong, 2008). This process is transcriptionally regulated and involves the induction of lineage-specific transcription factors (Li et al., 2014). Furthermore, complex chromatin remodeling responses that control the accessibility of transcription factors to their target genes provide another regulatory mechanism in Th cell differentiation (Falvo et al., 2013). As compared to other aspects of Th cell induction, signals and pathways that trigger either “permissive” or “repressive” chromatin remodeling responses during Th cell generation remain poorly defined.
Th17 cells are important in multiple autoimmune diseases (Korn et al., 2009). Induction of Th17 cells is best achieved in vitro with a combination of TGF-β and IL-6 (Mangan et al., 2006); these cytokines signal through SMAD2 and SMAD3, and STAT3, respectively, and converge on the induction of RORγt, a lineage-specific transcription factor for Th17 cell induction (Ivanov et al., 2006). Of note, other inflammatory cytokines, especially IL-1, TNF-α, IL-21, IL-23, and additional transcription factors (e.g., STAT3, RORα, BATF, c-Rel) also facilitate Th17 cell induction under certain ex vivo conditions (Dong, 2008). Once induced, Th17 cells produce copious IL-17A, IL-17F, IL-21, and through recruiting inflammatory cells, Th17 cells trigger robust tissue inflammation (Patel and Kuchroo, 2015). Thus, Th17 cells have been implicated in multiple autoimmune diseases, including colitis (Fantini et al., 2007), multiple sclerosis (Kebir et al., 2007), psoriasis (Ma et al., 2008), as well as in tumor immunity (Coursey et al., 2011) and transplant rejection (Yuan et al., 2008).
OX40 is a T cell costimulatory molecule in the Tumor necrosis factor receptor (TNFR) superfamily (Watts, 2005). One outstanding feature of OX40 is that it is highly expressed by activated T cells, but not naive T cells (Sugamura et al., 2004). As a member in the TNFR superfamily, OX40 signals through the NF-κB pathway, and under certain conditions, OX40 also triggers the activation of PI3K-AKT pathway, as well as the NFAT pathway (So et al., 2011a; So et al., 2011b). These signaling pathways exert a broad impact on T cell survival and proliferation. Furthermore, OX40 also regulates the fate and the functional attributes of activated T cells. In certain models, OX40 promotes the induction of Th1 cells (Demirci et al., 2004), whereas in others it is a powerful inducer of Th2 cell responses (Ito et al., 2005). We and others showed that OX40 potently inhibits Foxp3+ Treg cells, while strongly boosts the induction of Th9 cells, which results in prominent airway inflammation (Piconese et al., 2008; Xiao et al., 2012a). However, the role of OX40 in the induction of Th17 cells remains contested. In models of uveitis and intestinal inflammation, OX40 supports Th17 cells in vivo (Xin et al., 2014; Zhang et al., 2010), whereas in other models, OX40 engagement inhibits Th17 cell induction (Xiao et al., 2012a). Studies in humans also revealed an inhibitory effect of the OX40-OX40L pathway in Th17 cell induction, which can be reversed by neutralizing IFN-γ (Li et al., 2008). A key point from these studies is that OX40 and the cytokine signaling are critical determinants of Th cell differentiation programs, but the underlying mechanisms of how OX40 controls Th17 cells remain largely unresolved.
In the present study, we used multiple in vivo and in vitro models to examine the role of OX40 in Th17 cell function, and found that OX40 triggered a robust chromatin remodeling pathway through activation of the histone methyltransferases G9a and SETDB1. These histone methyltransferases were recruited to the Il17 locus by OX40-mediated induction of RelB and deposited “repressive” chromatin marks by catalyzing histone 3 lysine 9 (H3K9) trimethylation, resulting in the closure of Il17 locus, and consequently, inhibition of Th17 cell function and Th17 cell-mediated autoimmunity.
Results
OX40 stimulation inhibits IL-17 induction in vivo and in vitro
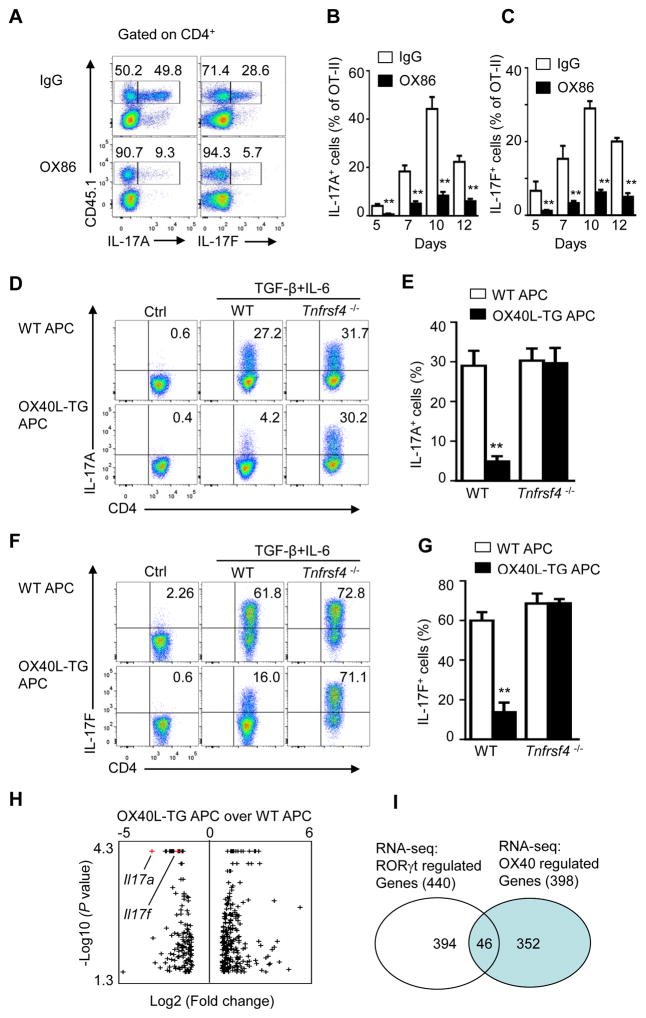
OX40 is best known for survival and proliferation of CD4+ T cells (Rogers et al., 2001). To determine whether OX40 also impacts IL-17 and Th17 cell generation, we adoptively transferred OT-II cells (CD45.1+) into C57BL/6 mice (CD45.2+) and immunized them with the cognate antigen OVA in complete Freund’s adjuvant. Groups of mice were also treated with an agonist anti-OX40 mAb OX86 to trigger OX40 signaling or a control IgG. At different time points (5 to 12 days), we examined IL-17A and IL-17F expression in the draining lymph nodes of immunized mice. We observed that in control IgG treated mice ~50% of the CD45.1+ OT-II T cells became IL-17A producing cells, and ~30% were IL-17F producing cells 10 days after OVA immunization (Figure 1A). Treatment of host mice with OX86 drastically inhibited the expression of IL-17A and IL-17F from OT-II cells (down to 5~9%). This inhibition of IL-17A and IL-17F was consistently observed at all time points examined in the immunized mice (5 to 12 days) (Figures 1B and 1C).
Figure 1. OX40 inhibits induction of IL-17 in vivo and in vitro.
(A) CD4+Foxp3− OT-II cells (CD45.1+) were transferred into wild-type B6 recipients (CD45.2+), and mice were immunized with OVA(323–339) in CFA a day later. Groups of mice were given OX86 or IgG on days 0, 1, and 2 post immunization. CD4+ T cells in draining LN were analyzed on day 10 for IL-17A and IL-17F expression by flow cytometry. The FACS plots shown are cells gated on CD4+CD45.1+ OT-II cells.
(B and C) The percentage of IL-17A and IL-17F producing cells among gated CD45.1+ OT-II cells analyzed on days 5, 7, 10 and 12 post immunization (n = 3 per group; mean ± SD). ** p <0.01
(D, F) Naive CD4+ T cells sorted from WT Foxp3gfp and Tnfrsf4 −/− Foxp3gfp reporter mice were activated with anti-CD3 plus WT APCs or OX40L-TG APCs under Th17-polarizing conditions or non-polarizing conditions (Controls or Ctrl) for 3 days, and induction of IL-17A(D) and IL-17F (F) producing cells was analyzed and shown.
(E, G) The tabulated percentage of IL-17A (E) and IL-17F (G) producing cells from all experiments performed, and the graph depicts Mean ± SD of 5 experiments with triplicate cultures. ** p <0.01
(H) Volcano plot of RNA-seq data from CD4+ T cells activated with anti-CD3 plus WT-APC (WT) or OX40LTG-APC (OX40L) under Th17 conditions, concentrating on OX40 regulated genes. Black dots indicate differentially expressed genes (minimum two fold changes with a P <0.05), and Il17a and Il17f are shown in Red.
(I) Venn diagram of private and overlapping genes induced by RORγt and OX40, based on RNA-seq data.
To further address the role of OX40 in suppression of IL-17, we sorted naïve CD4+Foxp3− T cells from Foxp3gfp reporter mice (B6 background) by flow cytometry, activated them in vitro with anti-CD3 and antigen presenting cells (APCs) under Th17 polarizing conditions (with TGF-β and IL-6), and examined the induction of IL-17A and IL-17F. To stimulate OX40 on activated CD4+ T cells, we included OX40Ltg APCs in the culture in which the ligand for OX40 is constitutively expressed (Sato et al., 2002). As shown in Figures 1D and 1E, naïve CD4+ T cells activated under Th17 conditions expressed high amounts of IL-17A. As compared to those activated without TGF-β and IL-6, ~30% of the CD4+ T cells became IL-17A producing cells 3 days later. Furthermore, induction of IL-17F was even more dramatic, and ~60% of the CD4+ T cells expressed IL-17F (Figures 1F and 1G). However, OX40 stimulation consistently inhibited the induction of IL-17A and IL-17F, and few CD4+ T cells became IL-17A or IL-17F producing cells following OX40 stimulation (~4% for IL-17A, and ~14% for IL-17F). The most striking effect of OX40 is the suppression of T cells producing both IL-17A and IL-17F (Figure S1). This inhibition was OX40 specific, as induction of IL-17A and IL-17F from OX40-deficient CD4+ T cells (Tnfrsf4−/− CD4+ T) was not affected, as comparable amounts of IL-17A and IL-17F were induced from Tnfrsf4−/− CD4+ T cells regardless of OX40 stimulation (Figures 1E and 1G).
To better appreciate the global effects of OX40 on IL-17 and Th17 cells, we performed RNA-seq on CD4+ T cells activated under Th17 conditions with or without OX40 stimulation. As show in Figure 1H, among the 398 genes that were differentially regulated by OX40, based on a 2 fold cutoff threshold with p<0.05, during Th17 induction, 144 were down-regulated and 254 up-regulated. Such genes were largely non-overlapping with those regulated by the RORγt (Figure 1I). A complete list of genes that were mostly affected by OX40 was shown in Table S1. Of the reported 440 genes regulated by RORγt (Ciofani et al., 2012), only 46 genes were affected by OX40, and Il17a and Il17f were prominently featured in the suppressed gene category (Figure 1H, red dot). These data suggest that OX40 does not affect Th17 commitment, but most likely impaired their effector differentiation. Because IL-17A and IL-17F are defining Th17 cytokines (Korn et al., 2009), their suppression suggests that OX40 inhibits effector functions of Th17 cells.
RORγt is induced by OX40 stimulation but functionally disabled
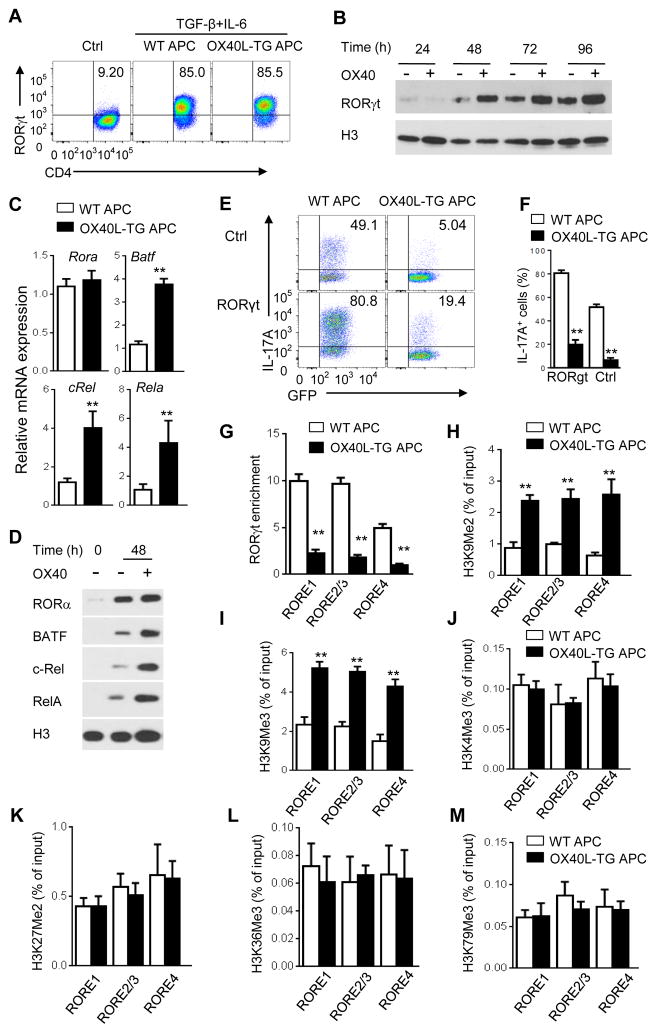
As induction of IL-17 and Th17 cells requires the transcription factor RORγt (Ivanov et al., 2006), we determined whether OX40 is capable of altering RORγt activities. We activated naïve CD4+ T cells under Th17 conditions with or without OX40 stimulation, and examined RORγt expression by flow cytometry. As compared to CD4+ T cells activated without polarizing cytokines, which did not express RORγt (Figure 2A), T cells activated under Th17 conditions expressed high amounts of RORγt, and its expression was similar in OX40 stimulated CD4+ T cells (Figure 2A). We also performed immunoblotting assays, which revealed abundant nuclear localization of RORγt in OX40 stimulated T cells (Figure 2B). In addition, other transcription factors that facilitate Th17 induction, including RORα, BATF, c-Rel, and RelA (Dong, 2006) were upregulated by OX40 stimulation, both at the transcript level (real-time PCR) and at the protein level (Western blot), (Figures 2C and 2D). Thus, induction of Th17-associated transcription factors is not perturbed by OX40 costimulation.
Figure 2. OX40 induces RORγt expression but impairs its transcriptional activities in CD4+ T cells.
(A) Flow cytometry plot showing expression of RORγt by WT CD4+ T cells activated under neutral control- or Th17-polarizing conditions for 3 days.
(B) Immunoblot analysis of RORγt in the nucleus of naive CD4+ T cells activated with anti-CD3 plus WT APCs (OX40 −) or OX40L-TG APCs (OX40 +) under Th17-polarizing conditions for 24–96 h.
(C) Real-time PCR quantification of Rora, Batf, Rel, and Rela mRNA in naive WT CD4+ T cells activated under Th17-polarizing conditions for 48 h with anti-CD3 plus WT APC or OX40L-TG APCs. Results are relative expression normalized to GAPDH mRNA (glyceraldehyde phosphate dehydrogenase).
(D) Immunoblot analysis of the induction of RORa, Batf, c-Rel, and RelA in the nucleus of naive CD4+ T cells activated for 48 h with or without OX40 stimulation.
(E and F) Flow cytometry analysis of the induction of Th17 cells from WT naive CD4+ T cells transduced with empty retroviral vector (Ctrl) or retrovirus expressing RORγt, cultured Th17-polarizing conditions with WT APCs or OX40L-TG APCs for 3 days (E). Graphs in (F) depict Mean ± SEM of 3 experiments with triplicate cultures. Data are presented as mean ± SD (C) and are representative of at least three individual experiments in (A–E). (G) ChIP analysis of the enrichment of RORγt at Il17a promoter and CNS5 regions in WT CD4+ T cells activated for 48 h as in Figure 2D. Values are presented as relative binding based on normalization to control IgG. Data are presented as mean ± SD (n=4). (H–M) ChIP analysis of H3K9me2 (H), H3K9Me3 (I), H3K4Me3 (J), H3K27Me2 (K), H3K36Me3 (L) and H3K79Me3 (M) at the Il17a promoter (RORE1, RORE2/3) and CNS5 (RORE4) regions in WT CD4+ T cells activated under Th17-polarizing conditions for 2 days. Data are presented as mean ± SD (n=3). ** p <0.01.
In a different set of experiments, we used a retroviral mediated gene transfer approach to overexpress RORγt in CD4+ T cells, followed by culturing such cells under Th17-polarizing conditions. Compared to control virus transduced T cells, overexpression of RORγt in CD4+ T cells led to an enhanced induction of IL-17 (~80% versus 49% IL-17A+ cells), but similar to control retroviral transduced cells, RORγ overexpression failed to rescue IL-17 from OX40 mediated suppression (Figures 2E and 2F). Thus, the RORγt function, not its expression and nuclear localization, seems to be impaired by OX40.
OX40 ligation induces a “closed” chromatin structure at Il17 locus
We focused on the prototypic Th17 cell cytokine IL-17A and performed extensive ChIP analysis to examine the binding of RORγt to the Il17a locus in OX40 stimulated CD4+ T cells. The Il17a locus contains at least 4 RORγt responsive elements (RORE1-4) that are potential RORγt binding sites (Figure S2). As expected, in CD4+ T cells activated under Th17 conditions, RORγt was highly enriched at RORE1-4 at Il17a locus (Figure 2G), which was correlated with robust induction of IL-17 producing cells (Figures 1A and 1D). OX40 stimulation led to markedly reduced RORγt enrichment at all RORE sites at Il17a locus (Figure 2G), suggesting that under Th17 conditions, OX40 signaling impairs RORγt binding to the Il17a locus.
We then examined the role of OX40 signaling in chromatin modifications at the Il17a locus in activated CD4+ T cells. As the promoter and conserved non-coding sequence (CNS) regions of Il17a locus are devoid of CpG islands (Yang et al., 2013), sites where epigenetic DNA modification occurs (Lal and Bromberg, 2009), we focused on changes in histone modifications in response to OX40 stimulation. As shown in Figures 2H and 2I, the most striking effect of OX40 stimulation in CD4+ T cells under Th17 conditions was the markedly enhanced H3K9 di- and tri-methylation (H3K9Me2, H3K9Me3) at RORE1-4 regions, whereas the methylation status of H3K4, H3K27, H3K36, and H3K79 did not show significant changes with or without OX40 stimulation (Figures 2J–2M), thus identifying H3K9 as a focal point of chromatin modification in response to OX40 stimulation. We also examined other time points (up to 120 hr after Th17 polarization), and observed similar changes in histone modification at Il17a locus (Figure S3). Because H3K9me2 and H3K9me3 are well-known “repressive” chromatin marks in gene transcription (Tachibana et al., 2007), we reasoned that H3K9 methylation at Il17a locus is likely involved in suppression of IL-17 and Th17 cells.
OX40 requires the NF-κB family member RelB to inhibit IL-17
Consistent with our previous report (Xiao et al., 2012a), stimulation of OX40 on CD4+ T cells under Th17-polarizing conditions activated both the canonical (p50-RelA) and the non-canonical (p52-RelB) NF-kB pathways, as shown by immunoblotting assays (Figure S4).
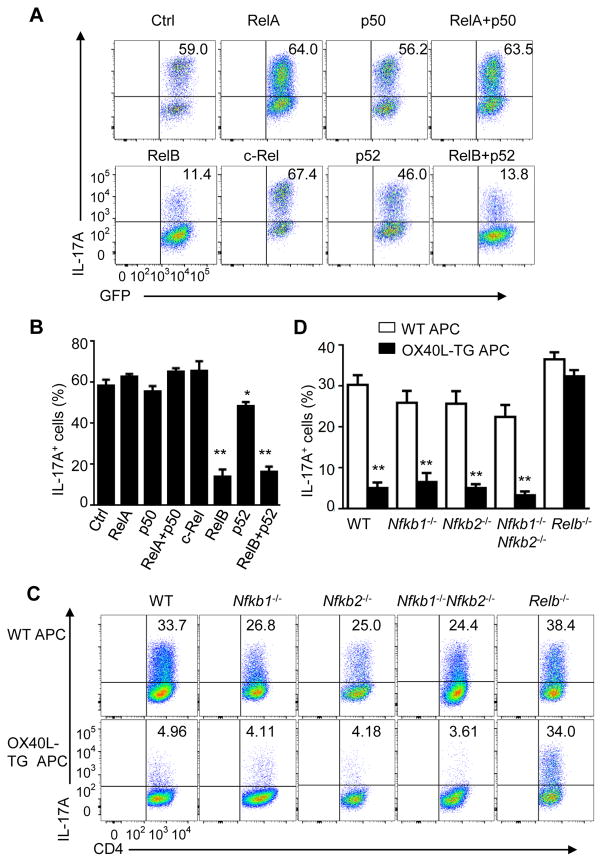
To examine the role of NF-kB family members in IL-17 expression, we firstly used a “gain of function” approach where we overexpressed each NF-κB family member in activated CD4+ T cells using the retrovirus-mediated gene transfer method, cultured them under Th17 conditions, and then examined the induction of IL-17. As shown in Figure 3A, in control retroviral transduced CD4+ T cells (marked by the expression of GFP), IL-17A producing cells could be readily induced, and ~60% of GFP+CD4+ T cells were IL-17-producing cells 3 days later (Figure 3B). Overexpression of RelA, p50 (encoded by Nfkb1), c-Rel, or p52 (encoded by Nfkb2), alone or a combination of RelA and p50 failed to show any effects on IL-17 expression, and similar numbers of IL-17A producing cells were induced under all conditions (~50% to 60% IL-17A+ cells) (Figure 3B). In contrast, overexpression of RelB or RelB plus p52 in CD4+ T cells led to a marked inhibition of IL-17 (down to ~10%). A combination of RelB and p52 did not lead to further reduction in IL-17 producing cells (Figure 3B), suggesting that RelB is a critical mediator in suppression of IL-17.
Figure 3. Critical role of RelB in suppression of Th17 cells by OX40.
(A and B) Induction of Th17 cells from WT CD4+ T cells transduced with empty vector encoding GFP alone (Ctrl) or with retrovirus expressing p50, RelA, p52, c-Rel, or RelB, and cultured for 3 days under Th17-polarizing conditions (A). Graphs in (B) depict Mean ± SEM of 5 experiments with triplicate cultures. * p <0.05; ** p <0.01.
(C and D) Induction of Th17 cells from WT B6, Nfkb1−/−, Nfkb2−/−, −/−, Nfkb1−/−Nfkb2−/− DKO, and Relb−/− CD4+ T cells activated for 3 days as in Figure 1D(C). Graphs in (D) depict Mean ± SEM of 5 experiments with triplicate cultures. ** p <0.01.
We also used a “loss of function” approach where we sorted naïve CD4+ T cells from WT B6, Nfkb1−/−, Nfkb2−/−, Nfkb1−/−Nfkb2−/−, as well as Relb−/− mice, activated them under Th17-culturing conditions with or without OX40 stimulation, and assessed the induction of IL-17. As shown in Figure 3C, comparable numbers of IL-17 producing cells were induced from WT, Nfkb1−/−, Nfkb2−/−, Nfkb1−/−Nfkb2−/−, and Relb−/− CD4+ T cells (~30%) by TGF-β and IL-6 (Figure 3D). Interestingly, induction of IL-17 producing cells from WT B6, Nfkb1−/−, Nfkb2−/−, Nfkb1−/−Nfkb2−/−, CD4+ T cells was strongly inhibited by OX40 stimulation. In contrast, deficiency of RelB (Relb−/− CD4+ T cells) completely abolished the effect of OX40 in suppression of IL-17, and similar proportion of Relb−/− CD4+ T cells became IL-17 producing cells (~35%) regardless of OX40 stimulation (Figures 3C and 3D). This is not due to the lack of OX40 expression on activated Relb−/− CD4+ T cells, as they express the same amounts of the OX40 on the cell surface as activated WT B6 CD4+ T cells (Figure S5). Together, these data suggest that RelB is indispensable in suppression of IL-17 and Th17 cells by OX40 stimulation.
RelB inhibits Th17 cells independent of p50, p52, or its transactivation domain
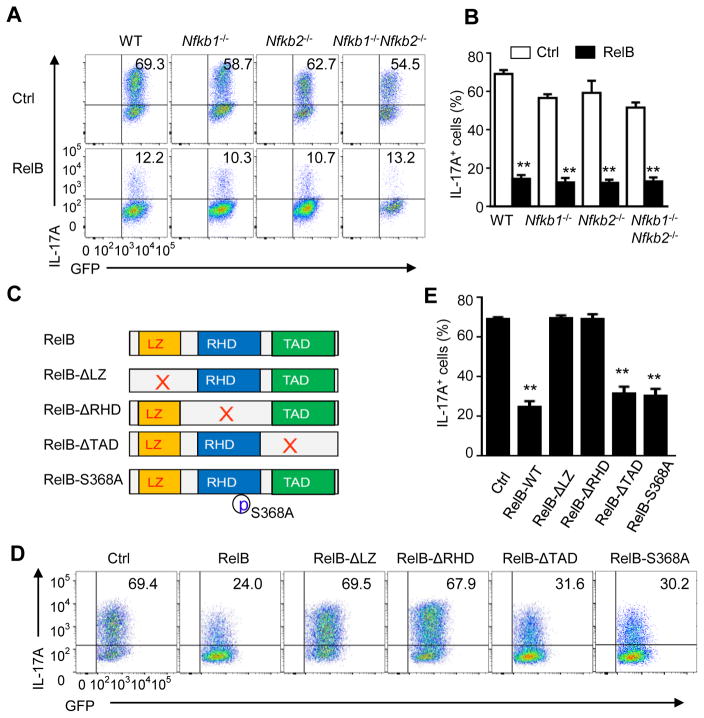
RelB usually dimerizes with p52 to form heterodimers to drive expression of NF-κB responsive genes (Vallabhapurapu and Karin, 2009). To determine how RelB inhibits IL-17 and Th17 cells, we again used the retrovirus-mediated gene transfer approach to overexpress RelB in WT B6, Nfkb1−/−, Nfkb2−/−, Nfkb1−/−Nfkb2−/− CD4+ T cells, and the transduced cells were then cultured under Th17-polarizing condition. IL-17 expression was determined 3 days later. As shown in Figure 4A, while IL-17 was readily induced from control vector transduced CD4+ T cells (~65%) (Figures 4A and 4B), and forced expression of RelB in Nfkb1−/− or Nfkb2−/− CD4+ cells strongly inhibited the induction of IL-17 (down to ~10%). Interestingly, overexpression of RelB in Nfkb1−/−Nfkb2−/− CD4+ T cells also strongly inhibited the induction of IL-17 producing cells (Figures 4A and 4B), suggesting that RelB inhibits IL-17 independent of both p50 and p52. These data suggest that, in contrast to its transcriptional activities, RelB controls IL-17 and Th17 cells without dimerizing with p50 and p52.
Figure 4. RelB inhibits Th17 cells independent of p50, p52, and its transactivation domain.
(A and B) Induction of Th17 cells from WT B6, Nfkb1−/−, Nfkb2−/−, −/−, Nfkb1−/−Nfkb2−/− DKO CD4+ T cells transduced with empty vector (Ctrl) or with retrovirus expressing RelB and cultured for 3 days under Th17-polarizing conditions (A). Graphs in (B) depict Mean ± SEM of 5 experiments. ** p <0.01.
(C) Schematic representation of RelB structure and RelB mutants showing deleted leucine zipper domain (LZ), Rel homology domain (RHD), transactivation domain (TAD), respectively, as well as a serine 368 to alanine point mutation (RelB-S368A).
(D and E) Induction of Th17 cells from WT CD4+ T cells transduced with empty vector (Ctrl) or with retroviruses expressing full length RelB (WT-RelB) or RelB mutants (as in C) and cultured for 3 d under Th17-polarizing conditions (D). Graphs in (E) depict Mean ± SEM of 3 independent experiments with triplicate cultures. ** p <0.01.
We also made a series of RelB mutants to further address how RelB inhibits IL-17 expression. Structurally, RelB has a leucine zipper domain (LZ), a Rel homology domain (RHD), and a transactivation domain (TAD). These domains are required for protein dimerization, DNA binding, and transcriptional activation of target genes (Vallabhapurapu and Karin, 2009). The RelB mutants we created are depicted in Figure 4C, with the goal of pinpointing specific domain that is essential in suppression of Th17 cells. We transfected such RelB mutants in frame with a GFP reporter, into activated WT B6 CD4+ T cells. The transduced T cells were cultured under Th17 conditions, and induction of IL-17 producing cells was assessed. As shown in Figure 4D, IL-17 producing cells were highly induced from control vector transduced CD4+ T cells (marked by GFP expression) 3 days later, and overexpression the full-length RelB inhibited IL-17 expression (Figures 4D and 4E). Deletion of the TAD domain did not affect RelB in suppression of IL-17. Similarly, a point mutation in the RHD domain that destroyed its dimerization with p52 or p50 also failed to affect RelB in suppression of IL-17 expression (Figures 4D and 4E), which is consistent with the findings using Nfkb1−/−Nfkb2−/− CD4+ T cells (Fig 4A). However, deletion of the LZ domain or the RHD domain, domains required for protein-protein interactions and DNA binding (Vallabhapurapu and Karin, 2009), abolished the effects of RelB in suppression of IL-17 expression (Figures 4D and 4E). These data suggest that suppression of IL-17 and Th17 cells by RelB most likely involves new mechanisms that are different from its transcription activities.
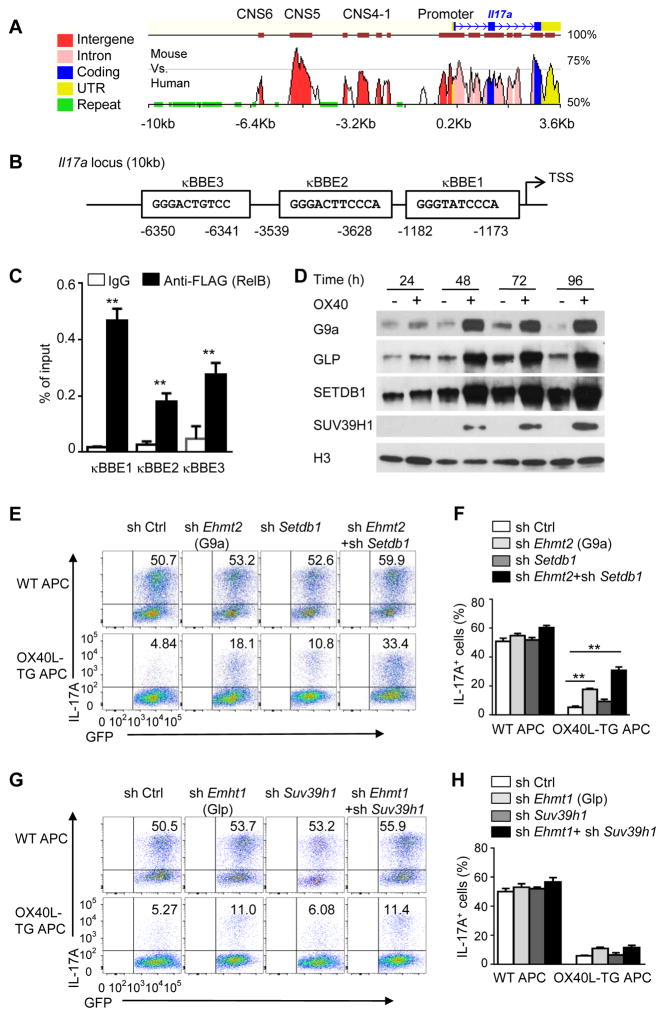
RelB recruits G9a and SETDB1 to Il17 locus to deposit repressive chromatin marks
Sequence analysis of Il17a locus revealed that there are at least 3 putative κB binding elements (κBBE) in the promoter and the CNS regions (Figures 5A and 5B); these sites are in close proximity to the RORγt sites RORE1-4 (Figure S6). We introduced a FLAG tag to the N-terminus of RelB, and transduced the FLAG-tagged RelB into activated CD4+ T cells, so that RelB can be immunoprecipitated using an anti-FLAG mAb (no ChIP grade anti-RelB available). The transduced T cells were cultured under Th17 conditions for 24 hrs and ChIP assay was performed to determine RelB binding to the Il17a locus. As shown in Figure 5C, RelB was highly enriched at the kB sites of Il17a locus in OX40 stimulated CD4+ T cells. It should be noted that binding of RelB to the Il17a locus is inversely correlated with that of the RORγt (Figure 2G).
Figure 5. RelB recruits histone methyltransferases G9a and SETDB1 to the IL-17 locus.
(A) Vista analysis showing CNS regions at Il17a locus (mouse vs. human). Positions are assigned relative to the transcriptional start site (TSS) of mouse Il17a (horizontal axis).
(B) The consensus sequences and positions of NF-κB binding sites in the region of 10 kb upstream of mouse Il17a, numbers below the diagram indicate relative position to the transcriptional start site (TSS).
(C) ChIP analysis of the enrichment of RelB at the Il17a locus in WT CD4+ T cells transduced with retrovirus expressing FLAG-RelB and cultured for 24 h under Th17-polarizing conditions. ** p <0.01
(D) Immunoblot analysis of the expression of G9a, GLP, SETDB1, and SUV39H1 in the nucleus of WT CD4+ T cells activated as in Figure 2B. Data are representative of three independent experiments.
(E and F) Induction of Th17 cells from WT CD4+ T cells transduced with empty vector (sh Ctrl) or with retroviruses expressing shRNA oligos targeting Ehmt2 (G9a), Setdb1 and cultured for 3 d under Th17-polarizing conditions (E). Graphs in (F) depict Mean ± SD of 3 experiments. ** p <0.01
(G and H) Induction of Th17 cells from WT CD4+ T cells transduced with empty vector (sh Ctrl) or with retroviruses expressing shRNA oligos targeting Ehmt1 (Glp), Suv39h1 and cultured for 3 d under Th17-polarizing conditions (G). Graphs in (H) depict Mean ± SD of 3 experiments. ** p <0.01
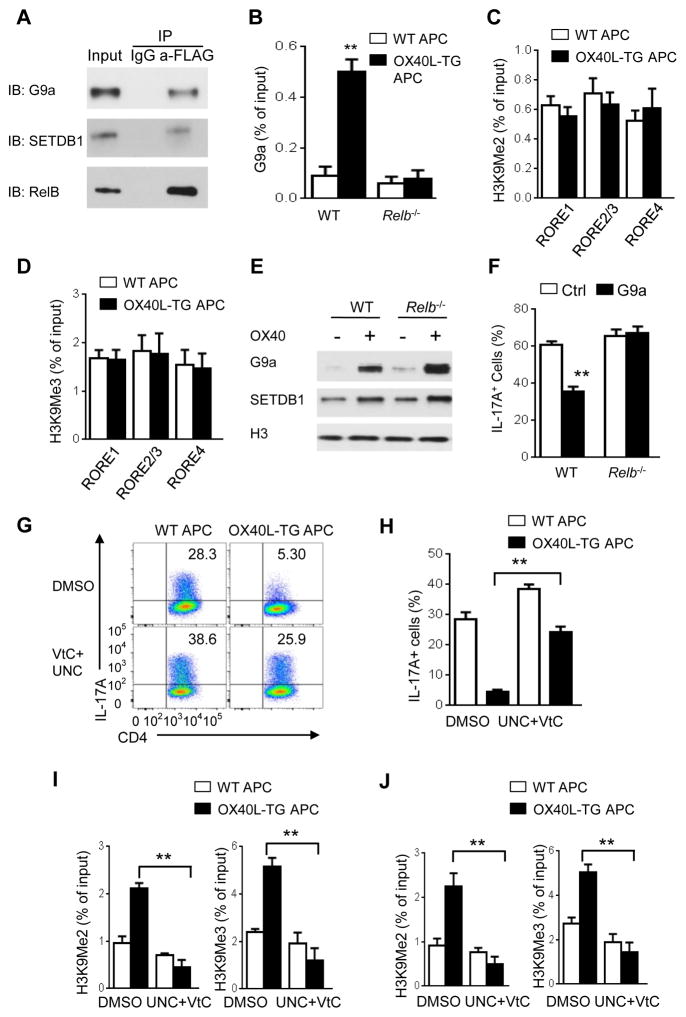
We then explore how RelB binding led to H3K9 trimethylation at Il17 locus. RelB itself does not have any histone methyltransferase activity, but other studies suggest that H3K9 dimethylation is catalyzed by the methyltransferases G9a and GLP, while H3K9 trimethylation involves SETDB1 and SUV39H1 (Shinkai and Tachibana, 2011). Immunoblotting assays showed that G9a, GLP, SETDB1, and SUV39H1 were induced by OX40 stimulation in CD4+ T cells, as compared to those without OX40 stimulation (Figure 5D). Such molecules also showed prominent nuclear localization, which was sustained for up to 4 days following OX40 stimulation (Figure 5D). To determine their relative contributions to suppression of IL-17, we designed gene-specific shRNA to specifically knockdown each molecule in activated CD4+ T cells (Figure S7A), and induction of IL-17 expression with or without OX40 stimulation was then examined. Interestingly, knockdown of both G9a and SETDB1 substantially rescued IL-17 expression from OX40-mediated suppression (Figure 5E, 5F), whereas knockdown of GLP and SUV39H1 showed minimal effects (Figures 5G and 5H), suggesting a predominant role for G9a and SETDB1 in suppression of IL-17 and Th17 cells by OX40. Furthermore, using FLAG-tagged RelB, we showed that in OX40 stimulated CD4+ T cells, RelB co-immunoprecipitated with G9a and SETDB1, and G9a and SETDB1 were readily detected in anti-FLAG (RelB) immunoprecipitates (Figure 6A), whereas GLP and SUV39H1 were not detectable in the RelB immunoprecipitates (data not shown), suggesting that RelB can physically interact with G9a and SETDB1. We confirmed in ChIP assays that G9a was enriched at Il17a locus in WT CD4+ T cells, but absent in Relb−/− CD4+ T cells activated under Th17 conditions in the presence of OX40 stimulation (Figure 6B). Moreover, in Relb−/− CD4+ T cells activated under Th17 conditions, the methylation status of H3K9 (i.e., H3K9me2, H3K9Me3) at the Il17a locus was not altered regardless of OX40 stimulation (Figures 6C and 6D), suggesting a critical role for RelB in G9a and SETDB1-mediated methylation of Il17 locus.
Figure 6. Inhibition of G9a and SETDB1 rescues Th17 cells from OX40-mediated suppression.
(A) Immunoblot (IB) analysis of G9a and SETDB1 in cell lysate immunoprecipitated with anti-FLAG (RelB) or IgG from WT CD4+ T cells transduced with retrovirus expressing FLAG-RelB and cultured for 48 h under Th17-polarizing conditions. Data are representative of three independent experiments.
(B) ChIP analysis of the enrichment of G9a in the Il17a promoter region in WT and Relb−/− CD4+ T cells activated as in Figure 6C.
(C and D) ChIP assays of H3K9me2 (C) and H3K9Me3 (D) at the Il17a locus by ChIP–qPCR in Relb−/− CD4+ T cells activated under Th17-polarizing conditions for 3 days. Data are presented as mean ± SD (n=3).
(E) Immunoblot analysis off G9a and SETDB1expression in the nucleus of WT or Relb−/− CD4+ T cells activated as in Figure 2B for 48 h. Data are representative of two independent experiments.
(F) Induction of Th17 cells from WT or Relb−/− CD4+ T cells transduced with retrovirus expressing G9a and cultured with WT APCs under Th17-polarizing conditions for 3 days. Graphs depict Mean ± SEM of 4 experiments with triplicate cultures. ** p <0.01.
(G and H) Induction of Th17 cells from WT naive CD4+ T cells activated for 3 days under Th17-polarizing conditions and treated with G9a inhibitor UNC0646 (500 nM) and Vitamin C (50ug/ml) or DMSO (G). Graphs in (H) depict Mean ± SEM of 5 experiments with triplicate cultures. ** p <0.01.
(I and J) ChIP analysis of H3K9Me2 and H3K9Me3 at RORγt binding sites RORE1 (I) and RORE2/3 (J) in the Il17a promoter region in WT CD4+ T cells activated and treated as in (C) for 3 days. Data are presented as mean ± SD (n=3). ** p <0.01.
In Relb−/− CD4+ T cells, OX40 stimulation also led to marked upregulation of G9a and SETDB1 (Figure 6E), suggesting that G9a and SETDB1 enrichment at Il17a locus, not their expression, is dependent on RelB in activated CD4+ T cells. Indeed, overexpression of G9a (using the retroviral approach) in Relb−/− CD4+ T cells failed to inhibit IL-17 expression when cultured under Th17 conditions (Figure 6F). To further address the relationship of RelB and G9a and SETDB1 in suppression of IL-17, we used pharmacological inhibitors that are known to block H3K9 methylation to examine the induction of IL-17 expression. It has been shown that UNC0646 (UNC) is a G9a specific inhibitor (Bittencourt et al., 2012), and Vitamin C (VtC) strongly inhibits H3K9 tri-methylation (Chen et al., 2013). We activated WT CD4+ T cells under Th17 conditions with or without OX40 stimulation, and performed extensive titrations of UNC and VtC in the cultures. As shown in Figure S7B, addition of UNC or VtC alone in the cultures partially rescued IL-17 from OX40-induced suppression. However, addition of both UNC and VtC in the cultures substantially abrogated the suppressive effects of OX40 in IL-17 expression (Figures 6G and 6H). Additionally, the reversal of IL-17 suppression was associated with markedly reduced H3K9 methylation at the promoter and the CNS region of Il17a locus (Figures 6I and 6J).
Critical role of the OX40-RelB pathway in suppression of Th17 cell-mediated autoimmunity in vivo
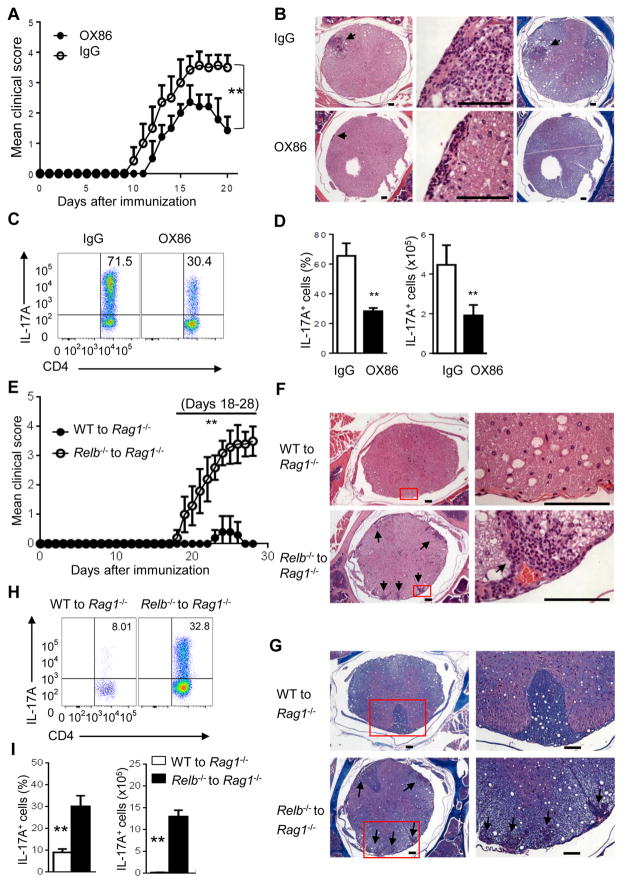
To examine the role of the OX40-RelB pathway in regulation of Th17-mediated autoimmunity in vivo, we used an experimental autoimmune encephalomyelitis (EAE) disease model in which the disease is mediated by Th1 and Th17 cells (Langrish et al., 2005). To specifically address the effect of IL-17 and Th17 cells, we used Ifng−/− mice as hosts, which are known to develop more aggressive EAE that requires Th17 cells (Korn et al., 2009). As shown in Figure 7A, immunization of Ifng−/− mice with MOG in complete Freund’s adjuvant induced aggressive EAE, and all mice were immobile by day 20 and required euthanasia. Treatment of host mice with the agonist anti-OX40 mAb OX86 (0.25mg i.p. on days 0, 1, 3, 6 in relation to MOG immunization) delayed the onset of EAE and markedly reduced the clinical scores. Also, the treated mice showed early signs of remission than the control IgG treated mice. Tissue histology revealed that OX86 treatment markedly reduced cellular infiltration and tissue damage, as well reduced demyelination in the spinal cord (Figure 7B). Furthermore, flow cytometry analysis showed substantially reduced IL-17+ cells in the central nervous system (CNS) of OX86 treated mice (by >50% reduction) as compared to the controls (Figures 7C and 7D).
Figure 7. Critical role of the OX40-RelB pathway in suppression of EAE in vivo.
(A) Incidence and clinical scores of EAE in Ifng−/− mice treated with OX86 or IgG. Data are presented as mean ± SD (n=7). ** p <0.01.
(B) Tissue pathology of spinal cord sections in Ifng−/− mice. Arrowhead indicates inflammatory cell infiltration (H&E) and area of severe demyelination. Bar scale: 100μm.
(C and D) FACS plots showing IL-17+CD4+ T cells obtained from the central nervous system (CNS) (C). Summaries of relative percentage (left) and absolute numbers (right) of IL-17-producing CD4+ cells in the CNS (D). Data are presented as mean ± SD (n=3). ** p <0.01.
(E) Incidence and clinical scores of EAE in Rag1−/− mice adoptively transferred with WT or Relb−/− CD4+ T cells. Data are presented as mean ± SD (n=5 and 7). ** p <0.01.
(F and G) Tissue histology of spinal cord sections. Arrowheads indicate inflammatory cell infiltration (H&E) (F), and areas of severe demyelination (Luxol fast blue, LFB) (G). Bar scale: 100μm.
(H) FACS plots showing IL-17-producing CD4+ T cells obtained from CNS of host mice. (I) The graph shows relative percentage and absolute numbers of IL-17-producing CD4+ cells in the CNS. Data are presented as mean ± SD of 3 experiments (n=3). ** p <0.01.
We also examined the role of RelB in autoimmune EAE. As Relb−/− mice are autoimmune and are not ideal as hosts (Millet et al., 2013), we used a passive transfer model in which Rag1−/− hosts were transferred with either WT B6 CD4+ T cells or Relb−/− CD4+ T cells to examine the role of RelB in EAE. The host mice were then immunized with MOG, and induction of EAE was monitored. As shown in Figure 7E, Rag1−/− mice transferred with Relb−/− CD4+ T cells developed far more severe EAE scores than those with WT B6 CD4+ T cells, and 28 days after MOG immunization, all Rag1−/− hosts with Relb−/− CD4+ T cells required euthanasia, whereas those with WT CD4+ T cells showed no or minimal signs of EAE (Figure 7E). Histologically, mice with Relb−/− CD4+ T cells showed intense cellular infiltration and prominent tissue destruction in the spinal cord (Figure 7F). Furthermore, Luxol fast blue staining revealed extensive demyelination induced by Relb−/− CD4+ T cells as compared to that induced by WT CD4+ T cells (Figure 7G). Analysis of CD4+ T cells infiltrating the CNS in mice transferred with Relb−/− CD4+ T cells showed markedly increased Th17 cells (Figures 7H and 7I), demonstrating that in the absence of RelB, more CD4+ T cells are converted to Th17 cells in vivo.
Discussion
In the present study, we demonstrate that OX40 inhibits IL-17 and Th17 cells through activation of histone methyltransferases G9a and SETDB1; they deposit repressive chromatin marks at the IL-17 locus by catalyzing H3K9 trimethylation, resulting in the closure of the IL-17 locus and suppression of Th17 cell function. Mechanistically, we provide evidence that OX40 activates the NF-κB family member RelB, and RelB binds G9a and SETDB1, and positions them at the kB sites in the IL-17 locus. Importantly, RelB acts independently of both p50 and p52 in activating G9a and SETDB1, as well as in mediating repressive chromatin remodeling. Thus, we identify RelB as an important molecular switch, and in addition to dimerizing with p52 or p50 and acting as a transcription factor (Vallabhapurapu and Karin, 2009), RelB can also activate repressive chromatin remodeling pathways (through G9a and SETDB1) and mediate gene repression. This repressive chromatin remodeling links OX40 costimulatory signals, which traditionally support cell survival and proliferation (Croft, 2003), to suppression of IL-17 and Th17 cell function. Our findings suggest that approaches that target such chromatin remodeling pathways may provide additional means of modulating Th17-induced autoimmune diseases.
Our data uncover a role for OX40 in the control of Th cell fates beyond its effects in cell survival and proliferation (Croft, 2003). For decades, OX40 has been extensively studied as a positive costimulatory molecule, favoring cell survival as well as the generation of Th1, Th2, Th9, and memory CD4+ T cells (Croft, 2010). The suppression of Th17 cells, in addition to Foxp3+ Treg cells (Vu et al., 2007), is rather unexpected and highlights a unique role for OX40 in T cell responses. The discovery that OX40 triggers a robust repressive chromatin remodeling response is particularly important, as it explains mechanistically how OX40 exerts its diverse effects, and will broaden our understanding as well as therapeutic interventions of OX40 in multiple disease settings. We provide evidence that OX40 controls the activity of G9a and SETDB1, in which RelB serves as a critical intermediary. Specifically, OX40 costimulation activates RelB, and RelB then binds G9a and SETDB1, and positions them in a site-specific manner to mediate chromatin remodeling. This notion is supported by our finding that in CD4+ T cells activated under Th17 conditions, OX40 signaling results in the deposition of repressive chromatin marks (H3K9me2, H3K9me3) at the promoter and CNS regions of Il17a gene, which prevents binding of Th17-associated transcription factors to mediate IL-17 expression. Furthermore, in activated CD4+ T cells stimulated by OX40, forced expression of RORγt failed to induce IL-17 expression because of a closed IL-17 locus. It has been shown that conditional deletion of G9a in CD4+ T cells also resulted in an enhanced induction of Th17 cells, while induction of Th2 cells was impaired, and this effect was associated with the loss of H3K9 dimethylation (Lehnertz et al., 2010), a finding that is consistent with our current study. In other models, G9a deletion led to enhanced induction of Foxp3+ Tregs, which was due to an increased sensitivity of G9a-deficient T cells to TGF-β (Antignano et al., 2014). Thus, under different conditions, G9a may mediate diverse effects that can determine the functional attributes of CD4+ T cells.
Another important finding of our study is that RelB, which is a classical transcription factor in the NF-κB family, is able to branch out to activate chromatin-remodeling pathways to repress gene transcription. This finding places RelB as an important molecular switch, capable of mediating both feed forward and feedback regulation of gene transcription. Like other Rel family members, RelB contains a TAD domain, and upon dimerizing with p52 or p50 through the RHD domain, it drives robust expression of NF-κB responsive genes (Vallabhapurapu and Karin, 2009). In our study, RelB is independent of both p50 and p52 in driving chromatin remodeling responses in suppression of Th17 cells. This is supported by out finding that in p50 and p52 double deficient T cells, RelB remains suppressive in IL-17 expression. Also, a point mutation that destroys RelB from dimerizing with p50 or p52 fails to alter the role of RelB in suppression of Th17 cells. Furthermore, deletion of the TAD domain in RelB also fails to affect RelB in suppression of Th17 cells. These data clearly show that the transcriptional activity of RelB is different from its chromatin remodeling activities. However, the selectivity of RelB in engaging G9a and SETDB1, amongst other histone methyltransferases and demethylases, is a question that deserves further investigation. G9a and SETDB1 are known to possess multiple domains that can interact with other proteins (Shinkai and Tachibana, 2011), but what determines their preference to RelB is unclear. Unlike other Rel family members, RelB contains a LZ domain (Vallabhapurapu and Karin, 2009), and the LZ domain is clearly required in suppression of Th17 cells. Whether the LZ domain explains the selectivity of RelB to G9a and SETDB1 remains to be clarified in future studies.
It is remarkable that H3K9 is the focal point where IL-17 expression is controlled, considering the complexity of chromatin modifications in T cells. It has been suggested that Il17 gene is devoid of CpG islands (Yang et al., 2013), places where DNA modifications take place (Lal and Bromberg, 2009). But the potency of H3K9 methylation, as opposed to other lysine residues in the control of Th17 cells, is intriguing. H3K9me2 are known to be catalyzed by G9a and GLP, and under certain conditions, G9a forms complexes with GLP to achieve optimal function (Shinkai and Tachibana, 2011). H3K9me3 is mediated by SETDB1 and SUV39H1 (Tachibana et al., 2008). In our studies, we observed that G9a and SETDB1 play a dominant role in H3K9 trimethylation at IL-17 locus in response to OX40. However, GLP and SUV39H1 are clearly induced by OX40 in activated CD4+ T cells, and their exact contributions to Th17 cells as well as other cellular responses require further clarification in future studies. After all, activation of different chromatin modifiers likely controls the specificity of chromatin remodeling process in the generation of different Th cell subsets. Thus, a detailed appreciation of chromatin modifiers and the context in which they regulate gene expression versus repression is an important area of future studies.
In conclusion, OX40 links a costimulatory receptor to a repressive chromatin remodeling pathway, and this response provides a powerful inhibitory mechanism in Th17 cell functions. Our finding suggests that costimulatory molecules can act far beyond their initial roles in cell survival to become regulators of Th cell fate decisions. Furthermore, our study highlights the importance of epigenetic mechanisms in the control of Th17 cells, in addition to the induction of lineage-specific transcription factors (Korn et al., 2009). This line of inquiry may lead to the design of new therapeutics in treatment of autoimmune diseases.
Experimental procedures
Animals
Relb−/−, Ifng−/−, OT-II, and Rag1−/− mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Tnfrsf4−/−, Nfkb1−/−, Nfkb2−/−, OX40L-Tg, and Foxp3gfp reporter mice have been described before (Xiao et al., 2012b). Generation of Nfkb1−/−Nfkb2−/−double knockout mice was accomplished by crossing Nfkb1−/− and Nfkb2−/− mice, and mice deficient for both Nfkb1−/− and Nfkb2−/−, were selected by PCR assisted genotyping and used for the studies. Some strains were crossed with Foxp3gfp mice to genetically mark Foxp3+ Treg cells for sorting purposes. All animals were maintained in a specific pathogen free facility at Houston Methodist Research Institute in Houston, Texas. Animal use and care were approved by the Houston Methodist Animal Care Committee in accordance with institutional animal care and use guidelines.
Co-immunoprecipitation assay
Naive CD4+Foxp3− T cells were transduced with retroviral vector expressing FLAG-RelB, and polarized under Th17 condition for 48 hrs. Cells (15 × 106 cells) were collected in IP lysis buffer (87788; Thermo Scientific). Subsequently, cell lysate was incubated with 10ug anti-FLAG (F1804; Sigma-Aldrich) or control antibody (ab18414; Abcam) at 4° C overnight, followed by incubation with Protein A agarose beads (9863; Cell Signaling Technology) for additional 4hr. Beads were collected by centrifugation and washed 4 times with IP lysis buffer. Proteins were eluted in reducing sample buffer (39000; Thermo Scientific) and analyzed by immunoblot.
Chromatin immunoprecipitation (ChIP) assay
Chromatin was extracted from naive CD4+Foxp3− T cells polarized for 48–72 h under various conditions (1×106 cells) after fixation with formaldehyde. Anti-H3K9me3 (61013; Active Motif), anti-dimethyl-H3 (diMe-Lys9) (D5567; Sigma-Aldrich), anti-H3K27me2 (ab24684; Abcam), anti-H3K4me3 (17-678; Millipore), anti-H3K36me3 (17-10493; Millipore), anti-H3K79me3 (17-10130; Millipore), anti-RORγt (sc-28559; Santa Cruz) and isotype-matched control antibody (sc-2027; Santa Cruz) were used for the immunoprecipitation of chromatin with an EZ ChIP kit according to the manufacturer’s instructions (Millipore). For cells transduced with retrovirus expressing FLAG-RelB, anti-FLAG (F1804; Sigma-Aldrich) was used. The precipitated DNA was then analyzed by real-time PCR. Data are presented as relative binding based on normalization to input DNA (Xiao et al., 2015). The sequences of all primers used in this study are given in Table S2.
Gene knockdown assay using short hairpin RNA (shRNA)
Targeting sequences for Emht1(Glp), Emht2 (G9a), Setdb1, and Suv39h1 were designed using online software BLOCK-iT™ RNAi Designer at https://rnaidesigner.thermofisher.com/rnaiexpress. Ten oligos for each gene were selected, and synthesized oligonucleotides were annealed and ligated into RNAi-Ready pSIREN-RetroQ-ZsGreen1 vector (#632455, Clontech Laboratories, Inc) by restriction enzymes BamHI and EcoRI. The inserted shRNA sequences were validated by direct sequencing. The shRNA oligos for each target gene were listed in Table S3. To generate retrovirus particles, retroviral vectors were transfected into retrovirus packaging Platinum E cells (Cell Biolabs) and the supernatants from transfected cells were used to directly infect T cells. Infected T cells were sorted by flow cytometry based on GFP expression, and the sorted GFP+ T cells were used to confirm the knockdown efficacy (Figure S7A), and used for further studies.
RNA-Sequencing (RNA-seq)
RNA-Seq was performed as previously reported (Huang et al., 2015). For RNA preparation, 5 million CD4+ T cells cultured under Th17-polarizing conditions were used for RNA extraction using the miRNAeasy Mini Kit (Qiagen). Complementary DNAs (cDNAs) were then synthesized using Superscript III kits (Invitrogen), and RNA-seq libraries were prepared and sequenced at BGI Genomics Cooperation (Hong Kong, China). Sequencing reads were mapped by Tophat and transcripts called by Cufflinks (Felzmann et al.). For RNA-seq analysis, volcano scores were calculated for each transcript as a function of its P value and fold changes between OX40 stimulated and control Th17 cells. The data in BAM files were converted to .tdf format by cluster software for viewing with the java TreeView (Huang et al., 2015).
Induction of EAE
EAE was induced in host mice using active EAE induction kit from Hooke Labs (EK-0113, Hooke Labs, MA) according to manufacturer’s instructions. For EAE induction in Rag1−/− mice, the mice were transferred with CD4+ T cells isolated from WT B6 and Relb−/− mice using the CD4+ T Cell Isolation Kit (Miltenyi Biotec) (7 × 106 per mouse), and immunized one day later. In some experiments, female Ifng−/− mice were used as hosts for EAE induction. Ifng−/− mice were treated i.p. with agonist anti-OX40 mAb or control IgG at 0.25mg (BioXcell, Lebanon, NH) on days 0, 1, 3 and 6 following immunization. Mice were monitored daily for signs of EAE according to the previously reported criteria (Lee et al., 2012). At the peak of disease, mice were killed for analysis of CNS-infiltrating cells as previous reported (Lee et al., 2012). Briefly, CNS tissue was minced with a sharp razor bla de and digested for 20 min at 37 °C with collagenase D (2.5 mg/ml; Roche Diagnostics) and DNaseI (1 mg/ml; Sigma). Mononuclear cells were isolated by passage of the tissue through a cell strainer (70 μm), followed by centrifugation through a Percoll gradient (37% and 70%). Mononuclear cells in the interphase were removed, washed and resuspended in culture medium, stimulated with PMA, ionomycin in the presence of GolgiStop for 4 h, and then were used for analysis by intracellular staining.
For histopathological analysis, spinal cords from host mice were fixed in 10% formaldehyde. Fixed spinal cords were embedded in paraffin and sectioned into 5-um slices. Sections were either stained with Luxol fast blue or H&E. All sections were viewed using a Nikon Eclipse E600 microscope, and images were processed in Image J (NIH, Bethesda, MD).
Statistical analysis
Data were represented as mean ± SD and analyzed with Prism version 6.04 for Windows (GraphPad Software). The Mann-Whitney test was used for EAE clinical score analysis. Other measurements were performed using unpaired two-tailed Student’s t test. A p value < 0.05 was considered significant. P values are denoted in figures as follows: * p < 0.05; ** p < 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Yang-Xin Fu at University of Chicago for providing Nfkb1−/− and Nfkb2−/−mice, Dr. Naoto Ishii at Tohoku University for the OX40Ltg mice. We also thank the flow cytometry and the pathology core facilities at Houston Methodist for excellent services and Dr. Roger Sciammas for discussions. This study was supported by grants from the National Institutes of Health (R01106200, R01070315, and R01080779).
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq data reported in this paper is GSE80010.
Author contributions
XX, XS, YF, CW, and XZ designed and performed experiments, LM contributed to operation support and animal handling, PL and WL provided technical supports in key experiments, RMG provided helpful discussions. XX and XCL supervised the studies and wrote the paper. XX, XS, YF contributed equally.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antignano F, Burrows K, Hughes MR, Han JM, Kron KJ, Penrod NM, Oudhoff MJ, Wang SK, Min PH, Gold MJ, et al. Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation. J Clin Invest. 2014;124:1945–1955. doi: 10.1172/JCI69592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittencourt D, Wu DY, Jeong KW, Gerke DS, Herviou L, Ianculescu I, Chodankar R, Siegmund KD, Stallcup MR. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursey TG, Chen PW, Niederkorn JY. IL-17-dependent, IFN-gamma-independent tumor rejection is mediated by cytotoxic T lymphocytes and occurs at extraocular sites, but is excluded from the eye. J Immunol. 2011;187:4219–4228. doi: 10.4049/jimmunol.1100826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M. Co-stimulatory members of the TNFR family: keys to effective T cell immunity? Nat Rev Immunol. 2003;3:609–620. doi: 10.1038/nri1148. [DOI] [PubMed] [Google Scholar]
- Croft M. Control of immunity by the TNFR-related molecule OX40 (CD134) Annu Rev Immunol. 2010;28:57–78. doi: 10.1146/annurev-immunol-030409-101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci G, Amanullah F, Kewalaramani R, Yagita H, Strom TB, Sayegh MH, Li XC. Critical role of OX40 in CD28 and CD154 independent rejection. Journal of Immunology. 2004;172:1691–1698. doi: 10.4049/jimmunol.172.3.1691. [DOI] [PubMed] [Google Scholar]
- Dong C. Diversification of T helper cell lineages: finding the family root of IL-17-producing cells. Nat Rev Immunol. 2006;6:329–334. doi: 10.1038/nri1807. [DOI] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Falvo JV, Jasenosky LD, Kruidenier L, Goldfeld AE. Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv Immunol. 2013;118:37–128. doi: 10.1016/B978-0-12-407708-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini MC, Rizzo A, Fina D, Caruso R, Becker C, Neurath MF, Macdonald TT, Pallone F, Monteleone G. IL-21 regulates experimental colitis by modulating the balance between Treg and Th17 cells. Eur J Immunol. 2007;37:3155–3163. doi: 10.1002/eji.200737766. [DOI] [PubMed] [Google Scholar]
- Felzmann T, Gadd S, Majdic O, Maurer D, X***X, Smolen J, Knapp W. Analysis of function-associated receptor molecules on peripheral blood and synovial fluid granulocytes from patients with rheumatoid and reactive arthritis. Journal of Clinical Immunology. 1991;11:205–212. doi: 10.1007/BF00917426. [DOI] [PubMed] [Google Scholar]
- Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo FW, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ito T, Wang Y-H, Duramad O, Hori T, Delespesse GJ, Watanabe N, Qin FX-F, Yao Z, Cao W, Liu Y-J. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J Exp Med. 2005;202:1213–1223. doi: 10.1084/jem.20051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kebir H, Kreymborg K, Ifergan I, Dodelet-Devillers A, Cayrol R, Bernard M, Giuliani F, Arbour N, Becher B, Prat A. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med. 2007;13:1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. Journal of Experimental Medicine. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Northrop JP, Antignano F, Burrows K, Hadidi S, Mullaly SC, Rossi FM, Zaph C. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J Exp Med. 2010;207:915–922. doi: 10.1084/jem.20100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li L, Shang X, Benson J, Merle Elloso M, Schantz A, Bracht M, Orlovsky Y, Sweet R. Negative regulation of IL-17 production by OX40/OX40L interaction. Cell Immunol. 2008;253:31–37. doi: 10.1016/j.cellimm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol Rev. 2014;261:141–156. doi: 10.1111/imr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HL, Liang S, Li J, Napierata L, Brown T, Benoit S, Senices M, Gill D, Dunussi-Joannopoulos K, Collins M, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest. 2008;118:597–607. doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Millet P, McCall C, Yoza B. RelB: an outlier in leukocyte biology. J Leukoc Biol. 2013;94:941–951. doi: 10.1189/jlb.0513305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DD, Kuchroo VK. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Piconese S, Valzasina B, Colombo MP. OX40 triggering blocks suppression by regulatory T cells and facilitates tumor rejection. J Exp Med. 2008;205:825–839. doi: 10.1084/jem.20071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PR, Song J, Gramaglia I, Killeen N, Croft M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity. 2001;15:445–455. doi: 10.1016/s1074-7613(01)00191-1. [DOI] [PubMed] [Google Scholar]
- Sato T, Ishii N, Murata K, Kikuchi K, Nakagawa S, Ndhlovu LC, Sugamura K. Consequences of OX40-OX40 ligand interactions in langerhans cell function: enhanced contact hypersensitivity responses in OX40L-transgenic mice. Eur J Immunol. 2002;32:3326–3335. doi: 10.1002/1521-4141(200211)32:11<3326::AID-IMMU3326>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Sharpe AH. Mechanisms of costimulation. Immunol Rev. 2009;229:5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes & development. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So T, Choi H, Croft M. OX40 Complexes with Phosphoinositide 3-Kinase and Protein Kinase B (PKB) To Augment TCR-Dependent PKB Signaling. J Immunol. 2011a;186:3547–3555. doi: 10.4049/jimmunol.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So T, Soroosh P, Eun SY, Altman A, Croft M. Antigen-independent signalosome of CARMA1, PKC{theta}, and TNF receptor-associated factor 2 (TRAF2) determines NF-{kappa}B signaling in T cells. Proc Natl Acad Sci U S A. 2011b;108:2903–2908. doi: 10.1073/pnas.1008765108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugamura K, Ishii N, Weinberg AD. Therapeutic targeting of the effector T cell costimulatory molecule OX40. Nature Rev Immunol. 2004;4:420–431. doi: 10.1038/nri1371. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. The EMBO journal. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. The EMBO journal. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kB transcription factors in the immune system. Ann Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Vu MD, Xiao X, Gao W, Degauque N, Chen M, Kroemer A, Killeen N, Ishii N, Li XC. OX40 costimulation turns off Foxp3+ Tregs. Blood. 2007;110:2501–2510. doi: 10.1182/blood-2007-01-070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- Xiao X, Balasubramanian S, Liu W, Chu X, Wang H, Taparowsky EJ, Fu YX, Choi Y, Walsh MC, Li XC. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. 2012a;13:981–990. doi: 10.1038/ni.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Gong W, Demirci G, Liu W, Spoerl S, Chu X, Bishop DK, Turka LA, Li XC. New insights on OX40 in the control of T cell immunity and immune tolerance in vivo. J Immunol. 2012b;188:892–901. doi: 10.4049/jimmunol.1101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Shi X, Fan Y, Zhang X, Wu M, Lan P, Minze L, Fu YX, Ghobrial RM, Liu W, Li XC. GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nature communications. 2015;6:8266. doi: 10.1038/ncomms9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Jiang TT, Chaturvedi V, Kinder JM, Ertelt JM, Rowe JH, Steinbrecher KA, Way SS. Commensal microbes drive intestinal inflammation by IL-17-producing CD4+ T cells through ICOSL and OX40L costimulation in the absence of B7-1 and B7-2. Proc Natl Acad Sci U S A. 2014;111:10672–10677. doi: 10.1073/pnas.1402336111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, Kerketta R, Lee YH, Chang SH, Corry DB, et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol. 2013;14:732–740. doi: 10.1038/ni.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Paez-Cortez J, Schmitt-Knosalla I, D’Addio F, Mfarrej B, Donnarumma M, Habicht A, Clarkson MR, Iacomini J, Glimcher LH, et al. A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J Exp Med. 2008;205:3133–3144. doi: 10.1084/jem.20081937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhong W, Hinrichs D, Wu X, Weinberg A, Hall M, Spencer D, Wegmann K, Rosenbaum JT. Activation of OX40 augments Th17 cytokine expression and antigen-specific uveitis. Am J Pathol. 2010;177:2912–2920. doi: 10.2353/ajpath.2010.100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.