Abstract
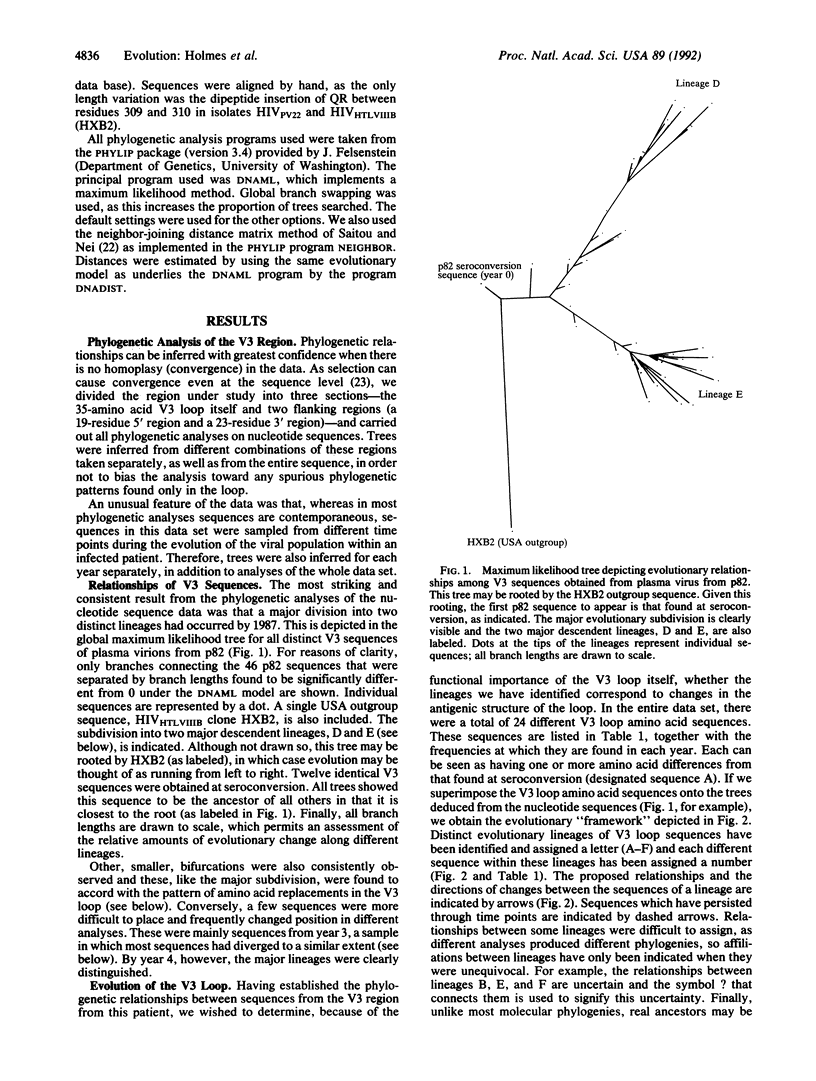
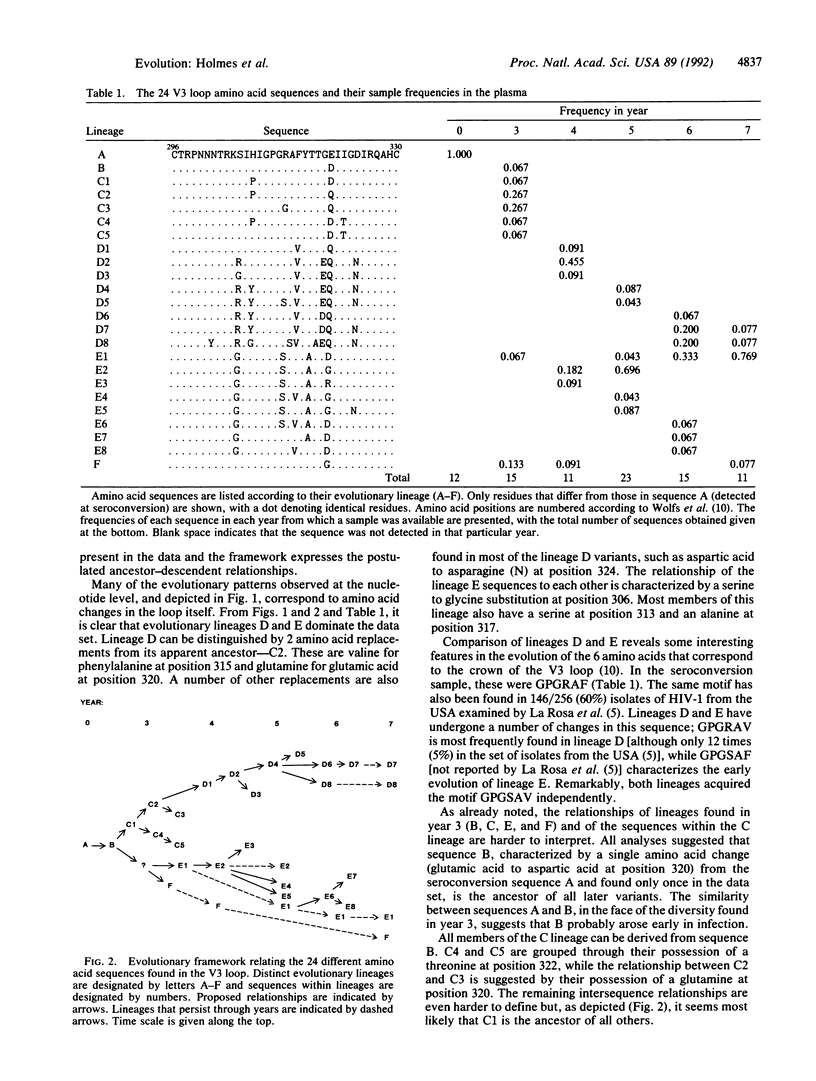
In an investigation of the evolution of the third hypervariable loop of gp120 (V3), the principal neutralization determinant of human immunodeficiency virus type 1, we have analyzed 89 V3 sequences of plasma viral RNA purified from peripheral blood samples donated over 7 years by an infected hemophiliac. Considerable sequence diversity in the V3 region was found at all time points after seroconversion. Phylogenetic analysis revealed that an important diversification had occurred by 3 years postinfection and that, subsequently, most sequences could be allocated to either one of two major lineages that persisted throughout the remainder of the infection. Rapid changes in frequency of the most common sequences and the observation that the same hexapeptide motif (GPGSAV) at the crown of the V3 loop has evolved convergently provide strong evidence that selective processes determine the evolutionary fate of sequence variants in this region.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. M., May R. M. Coevolution of hosts and parasites. Parasitology. 1982 Oct;85(Pt 2):411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Balfe P., Simmonds P., Ludlam C. A., Bishop J. O., Brown A. J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990 Dec;64(12):6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. L., Monaghan P. Evolution of the structural proteins of human immunodeficiency virus: selective constraints on nucleotide substitution. AIDS Res Hum Retroviruses. 1988 Dec;4(6):399–407. doi: 10.1089/aid.1988.4.399. [DOI] [PubMed] [Google Scholar]
- Clements J. E., Gdovin S. L., Montelaro R. C., Narayan O. Antigenic variation in lentiviral diseases. Annu Rev Immunol. 1988;6:139–159. doi: 10.1146/annurev.iy.06.040188.001035. [DOI] [PubMed] [Google Scholar]
- Delassus S., Cheynier R., Wain-Hobson S. Evolution of human immunodeficiency virus type 1 nef and long terminal repeat sequences over 4 years in vivo and in vitro. J Virol. 1991 Jan;65(1):225–231. doi: 10.1128/jvi.65.1.225-231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. G., Kuiken C., Blumberg B. M., Hartman S., Sharer L. R., Clement M., Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991 Feb;180(2):583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- Hahn B. H., Shaw G. M., Taylor M. E., Redfield R. R., Markham P. D., Salahuddin S. Z., Wong-Staal F., Gallo R. C., Parks E. S., Parks W. P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986 Jun 20;232(4757):1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Langlois A. J., McDanal C., Ross K. L., Eckler L. I., Jellis C. L., Profy A. T., Rusche J. R., Bolognesi D. P., Putney S. D. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa G. J., Davide J. P., Weinhold K., Waterbury J. A., Profy A. T., Lewis J. A., Langlois A. J., Dreesman G. R., Boswell R. N., Shadduck P. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990 Aug 24;249(4971):932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990 Jun 25;265(18):10373–10382. [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol. 1988 Jul;5(4):313–330. doi: 10.1093/oxfordjournals.molbev.a040503. [DOI] [PubMed] [Google Scholar]
- McKeating J. A., Gow J., Goudsmit J., Pearl L. H., Mulder C., Weiss R. A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989 Dec;3(12):777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- Meyerhans A., Cheynier R., Albert J., Seth M., Kwok S., Sninsky J., Morfeldt-Månson L., Asjö B., Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989 Sep 8;58(5):901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- Modrow S., Hahn B. H., Shaw G. M., Gallo R. C., Wong-Staal F., Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987 Feb;61(2):570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. A., Anderson R. M., McLean A. R., Wolfs T. F., Goudsmit J., May R. M. Antigenic diversity thresholds and the development of AIDS. Science. 1991 Nov 15;254(5034):963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- Nowak M. A., May R. M., Anderson R. M. The evolutionary dynamics of HIV-1 quasispecies and the development of immunodeficiency disease. AIDS. 1990 Nov;4(11):1095–1103. doi: 10.1097/00002030-199011000-00007. [DOI] [PubMed] [Google Scholar]
- Saag M. S., Hahn B. H., Gibbons J., Li Y., Parks E. S., Parks W. P., Shaw G. M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature. 1988 Aug 4;334(6181):440–444. doi: 10.1038/334440a0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salinovich O., Payne S. L., Montelaro R. C., Hussain K. A., Issel C. J., Schnorr K. L. Rapid emergence of novel antigenic and genetic variants of equine infectious anemia virus during persistent infection. J Virol. 1986 Jan;57(1):71–80. doi: 10.1128/jvi.57.1.71-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. V., Stowring L., Haase A. T., Narayan O., Vigne R. Antigenic variation in visna virus. Cell. 1979 Oct;18(2):321–327. doi: 10.1016/0092-8674(79)90051-5. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Balfe P., Ludlam C. A., Bishop J. O., Brown A. J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990 Dec;64(12):5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Balfe P., Peutherer J. F., Ludlam C. A., Bishop J. O., Brown A. J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990 Feb;64(2):864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Zhang L. Q., McOmish F., Balfe P., Ludlam C. A., Brown A. J. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J Virol. 1991 Nov;65(11):6266–6276. doi: 10.1128/jvi.65.11.6266-6276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. B., Schilling J. W., Wilson A. C. Adaptive evolution in the stomach lysozymes of foregut fermenters. 1987 Nov 26-Dec 2Nature. 330(6146):401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- Wolfs T. F., Zwart G., Bakker M., Valk M., Kuiken C. L., Goudsmit J. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology. 1991 Nov;185(1):195–205. doi: 10.1016/0042-6822(91)90767-6. [DOI] [PubMed] [Google Scholar]
- Wolfs T. F., de Jong J. J., Van den Berg H., Tijnagel J. M., Krone W. J., Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Gojobori T. Molecular evolution and phylogeny of the human AIDS viruses LAV, HTLV-III, and ARV. J Mol Evol. 1987;24(4):330–336. doi: 10.1007/BF02134131. [DOI] [PubMed] [Google Scholar]
- Zhang L. Q., Simmonds P., Ludlam C. A., Brown A. J. Detection, quantification and sequencing of HIV-1 from the plasma of seropositive individuals and from factor VIII concentrates. AIDS. 1991 Jun;5(6):675–681. doi: 10.1097/00002030-199106000-00006. [DOI] [PubMed] [Google Scholar]


