Dynamic regulation of covalent histone modifications at enhancers and promoters has a key role in the modulation of gene expression and consequently fate specification 1–6. Recent findings emerging from human cancer genome sequencing efforts have revealed that many genes that encode chromatin regulators that modify histones are frequently mutated across a wide variety of cancers7–11. Converging lines of investigation have particularly highlighted links between enhancer of zeste homologue 2 (EZH2) and cancer.
EZH2 is the enzymatic subunit of Polycomb repressive complex 2 (PRC2), a complex that methylates lysine 27 of histone H3 to promote transcriptional silencing 12,13. Distinct cancer-associated perturbations of this regulatory axis have emerged and include both gain-of-function and loss-of-function mutations in EZH2, overexpression of EZH2, mutations in the H3K27 demethylase UTX, and frequent mutations in the SWI/SNF chromatin remodeling complex that partially antagonizes Polycomb function. Recurrent missense mutations of the H3K27 site itself have also been identified. Interest in elucidating the roles of EZH2 in cancer has been further enhanced by the development of small molecules that effectively inhibit the enzymatic activity of EZH2 and the translation of these molecules into early phase trials with preliminary evidence of clinical responses.
Here we begin by reviewing the spectrum of EZH2 and H3K27 perturbations found in cancer and synthesize a perspective that unites the function of EZH2 as a master regulator of transcription. We then review the development, translation, and early clinical findings from therapeutic targeting of EZH2.
Mutations Resulting in Gain of Function of EZH2 in Cancer
EZH2 is the enzymatically active core subunit of the PRC2 complex, which also includes EED, SUZ12, and RbAp46/48. PRC2 methylates of the lysine residue at position 27 of histone 3 (H3K27) 14,15, which facilitates chromatin compaction and gene silencing (Figure 1). Several lines of evidence have implicated EZH2 in the development and progression of a variety of cancers. An early indication came from the observation that EZH2 overexpression is associated with worse progression of prostate cancer 16. Similar findings have emerged in other human cancers including breast cancer, bladder cancer, endometrial cancer, and melanoma as high levels of EZH2 were shown to correlate with aggressiveness and advanced disease in each of these cancer types 16–19 (Table 1). EZH2 has been shown to be essential for proliferation of cancer cell lines and independently ectopic EZH2 expression to confer a proliferative advantage upon non-cancerous cells 17. Forced expression of EZH2 leads to the development of myeloproliferative disorder in mice 20. In an immortalized human epithelial cell line, expression of causes neoplastic transformation of breast epithelial cells, a phenotype that is dependent upon its methyltransferase domain 21.
Figure 1. The PRC2 complex and its function in transcriptional regulation.
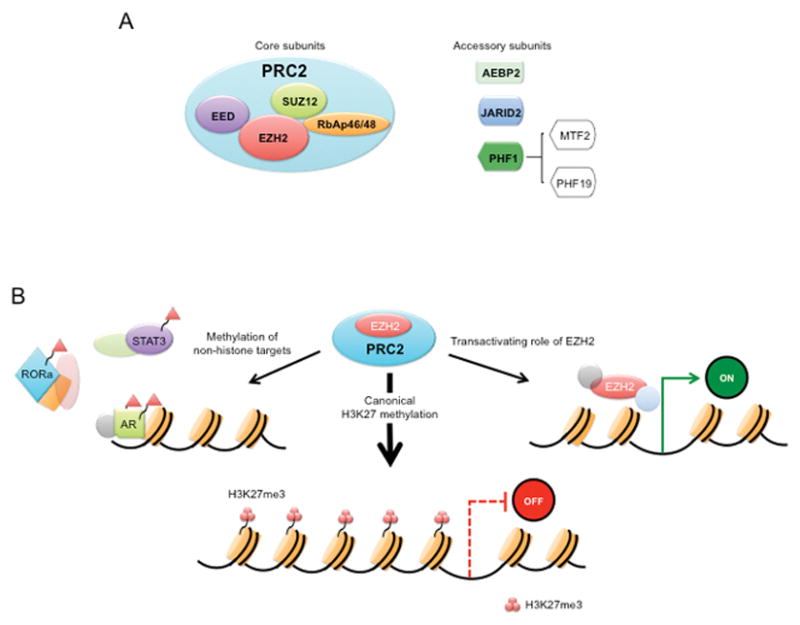
a. The mammalian PRC2 complex consists of four core subunits: the catalytic subunit enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED), suppressor of zeste 12 (SUZ12), and retinoblastoma (Rb)-associated protein 46/48 (RbAp46/48) and additional proteins, including AEBP2, PCL, and JARID2 have also at times been found to be associating with PRC2 complex to modulate the activity of PRC2 in different context. b. EZH2 regulates transcriptional activity: 1) EZH2 is also capable of methylating a number of non-histone protein substrates, 2) PRC2 methylates Histone 3 on lysine 27, which contributes to transcriptional silencing, 3) EZH2 also has a PRC2-independent role in transcriptional activation.
Table 1.
EZH2 overexpression and gain-of-function mutations identified in different cancer types and affected target genes
| EZH2 status | Associated cancer type | Target genes | Reference |
|---|---|---|---|
| EZH2 overexpression | Breast | CK5/6. P-cadherin, RAD51 paralogs, RUNX3, CDKN1C, FOXC1, RAF1-B-catenin signaling, DNA damage repair RKIP, CIITA, KLF2, | 16–18,20,48,52, 96–100 |
| Prostate | DAB2IP, p16, CDK4, MSMB, ADRB2, E-Cadherin, Ras, NF-kappaB, SLIT2, TIMP-2/3, EMT regulator, | 16,18,53,59, 101–105 | |
| Endometrial | p16, E-cadherin, sFRP1, DKK3, B-Catenin | 18,107,108 | |
| Melanoma | p21/CDKN1A, DCK, AMD1, WDR19, | 17,18,109,110 | |
| Bladder | unknown | 17,111,112 | |
| Glioblastoma | BMPR1B | 17,113 | |
| Liver | unknown | 114 | |
| Lung | Dkk-1 | 115 | |
| Natural killer/T-cell lymphoma | Cyclin D1 | 116 | |
| Ovarian | VASH1 | 117 | |
| EZH2 mutation (gain-of-function) | Non-Hodgkin’s Lymphoma (FL, GCB-subtype DLBCL), melanoma | Cdkn2a, Cdkn1a/p21, BLIMP1/PRDM1 | 22,24–26,118 |
Additional findings to support an oncogenic role for EZH2 have more recently emerged. Studies have shown that recurrent heterozygous point mutations at tyrosine 641 (Y641) within the C-terminal catalytic SET domain of EZH2 occur in 22% of germinal center B-cell (GCB) diffuse large cell B-cell lymphomas (DLBCL) and in 7% to 12% of follicular lymphomas (FL) 22,23 (Table 1). The Y641 mutation was initially thought to be a loss-of-function mutation. However, via in vitro biochemical enzymatic assays this mutation was subsequently shown to confer gain-of-function of enzyme activity resulting in augmented conversion of H3K27 to the trimethylated form 24. Y641 mutants (Y641F, Y641N, Y641S, Y641C, and Y641H) have reduced methylation activity of unmethylated H3K27 but enhanced activity for the dimethylated version of H3k27. The mutant thus cooperates together with wild-type EZH2 to shift the steady state of H3K27 to favor trimethylation and thus represses expression of Polycomb targets 25. EZH2 point mutations at the A677 and A687 residues have also been identified in non-Hodgkin lymphomas (NHL), where they similarly result in hypertrimethylation of H3K27 26,27.
Additional support for a gain-of-function role for mutant EZH2 in cancer comes from the identification of cancer-associated loss-of-function mutations in other chromatin regulators that normally antagonize EZH2 activity. UTX (ubiquitously transcribed tetratricopeptide repeat gene on X chromosome) is a histone demethylase that functions in part by antagonizing EZH2 activity by removing methyl groups from di- and trimethylated H3K27. Inactivating mutations affecting UTX occur in several types of human cancer, including multiple myeloma, medulloblastoma, esophageal cancer, bladder cancer, pancreatic cancer, and renal cancers 8,28–30. These mutations include homozygous (in females) or hemizygous (in males) large deletions, nonsense mutations, small frame-shifting insertion/deletions, and consensus splice site mutations that lead to a premature termination codon 31. Almost all mutations are predicted to result in loss of the JmjC domain of UTX, which is essential for its demethylase activity, and have been shown to cause increased levels of H3K27 trimethylation 31,32. Consequently, the loss-of-function mutations in UTX may be analogous to those caused by gain-of-function mutations in EZH2.
Genetic studies in many different organisms have also revealed an evolutionarily conserved antagonistic relationship between Polycomb proteins and SWI/SNF complexes, which utilizes the energy of ATP hydrolysis to remodel chromatin 33–36. SWI/SNF complexes are comprised of 12 to 15 subunits which have collectively been found to be mutated in 20% of all human cancers 37,38. Unopposed EZH2 activity is also a driver of cancers driven by loss of the SWI/SNF core subunit SNF5/SMARCB1 as originally shown in malignant rhabdoid tumor, a highly aggressive type of pediatric cancer 39,40. Recent studies have extended this antagonistic relationship to cancers linked to inactivation of other SWI/SNF subunits as EZH2 inhibition is synthetic lethal in ovarian cancer xenografts mutant for the SWI/SNF subunit ARID1A and sensitizes lung cancer xenografts mutant for the SWI/SNF ATPase core subunit BRG1/SMARCA4-to chemotherapy in mice 41,42. Most recently, a broad role for EZH2 in progression of cancers that have mutations in the SWI/SNF tumor suppressor subunits ARID1A, PBRM1, and SMARCA4 has been demonstrated in both cell lines and in vivo models 43. Notably, a non-catalytic role for EZH2 was identified in this context, indicating that dependency upon EZH2 for cancer progression can be derived from both catalytic and non-catalytic functions of EZH2.
Mechanism of EZH2 Oncogenic Activity
The extensive evidence linking EZH2 activity to cancer has prompted interest in the underlying mechanism. EZH2/PRC2 is known to be recruited to specific loci during development to silence genes associated with alternative fates 44–46. Analogous to its role in normal stem cells where EZH2 suppresses differentiation by repressing lineage-specifying factors 45–47, it is expressed at higher levels in cancer stem cell (CSC) populations isolated from human breast cancer xenografts and primary breast tumor cells compared to noncancer cell lines and is essential for the maintenance of these populations 48. In at least some CSC models, EZH2 suppresses expression of genes associated with lineage specification leading to the hypothesis that EZH2 facilitates transformation by blocking differentiation 47,49. However, it is important to note that EZH2 has also been shown to be essential to facilitate some differentiation programs of several tissue types 50,51. Ultimately, the central function of EZH2/PRC2 during development may not be to either promote stemness or differentiation per se, but rather to suppress transcriptional programs that underlie alternate fates. Thus, the consequences of perturbing EZH2 on fate control are likely to be highly cell-type specific.
Given its role as a transcriptional regulator, substantial efforts have been dedicated to the identification of downstream targets or pathways that contribute to transformation driven by EZH2. EZH2 has been shown to be downstream of the cell cycle regulatory retinoblastoma-E2F pathway and was in turn required for the expression of proliferative genes and for E2F-driven proliferation 17. The Ink4a/Arf tumor suppressor locus is thought to be another key target that can be silenced by Polycomb activity, and EZH2-mediated silencing of E-cadherin and FOXC1 and DNA damage repair pathways have also been shown to contribute to oncogenesis 48,52–54. An unresolved question that carries potential therapeutic relevance is whether any EZH2 targets are universally essential for EZH2-mediated transformation independent of cell lineage or whether EZH2-induced oncogenic transcriptional changes are context-specific depending upon cancer type and cell-of-origin.
Several studies have also identified a PRC2-independent function of EZH2 in transcriptional activation rather than repression 55–58 (Figure 1). In a model of castration-resistant prostate cancer, the oncogenic function of EZH2 was shown to be independent of its role as a Polycomb transcriptional repressor, but instead due to a PRC2-independent coactivator role of EZH2 for transcription factors including androgen receptor 59. An activating role of EZH2 was also demonstrated in breast cancer cells via activation of NF-κB targets and NOTCH1 57,58. EZH2 has also been implicated in transcriptional activation of gene expression in breast cancer, where it has been shown to induce the expression of genes that are regulated by estrogen receptor (ER) and Wnt signaling transcription factors by physically bridging between the ER and components of Wnt signaling (β–catenin) 55. In addition to its known roles in histone modification and transcriptional regulation, it has been shown that EZH2 can methylate non-histone substrates. EZH2 binds and methylates STAT3, which promotes tumorigenicity of CSC in glioblastoma and in a prostate cancer model, EZH2 has been shown methylate the androgen receptor (AR), modulating AR recruitment to the sites bound by both AR and EZH2 59,60. EZH2 has also been implicated in the methylation of non-histone substrates, this mechanism may facilitate recognition by the ubiquitination machinery driving degradation of the methylated proteins 61,62. Collectively, these results suggest that in addition to its known role as an H3K27 histone methyltransferase and transcriptional suppressor, EZH2 may also have PRC2-independent roles as a transcriptional coactivator and directly modulate the activity of transcription factors and other proteins. However, the contribution of these noncanonical functions to the overall cellular role of EZH2 and their link to any role of EZH2 in oncogenic transformation remain unclear.
EZH2 as a therapeutic target
Given the evidence for EZH2 enzymatic gain-of-function being a cancer driver, development of EZH2-specific inhibitors has been an active area of investigation and multiple biotech and pharmaceutical companies have been developing such compounds. Promising preclinical results have been obtained and human phase I trials are now underway, with early results suggesting potential clinical activity (Table 2).
Table 2.
EZH2 inhibitors and their status in clinical development
| Compound | Mechanism | Potency (Ki, nM) | Selectivity | Status | Reference |
|---|---|---|---|---|---|
| DZNep | SAH hydrolase inhibitor of methyltransferases | Unknown | Unknown | Preclinical | 119–122 |
| EI1 | SAM-competitive inhibitor of PRC2 | 13−/+3 nM | >10,000-fold over other HMTs, ~90-fold over EZH1 | Preclinical | 69 |
| EPZ005687 | SAM-competitive inhibitor of PRC2 | 24−/+7 nM | >500-fold over other HMTs, ~50-fold over EZH1 | Preclinical | 66 |
| GSK343 | SAM-competitive inhibitor of PRC2 | 1.2−/+0.2 nM | 1,000-fold over other HMTs, 60-fold over EZH1 | Preclinical | 67 |
| GSK126 | SAM-competitive inhibitor of PRC2 | 0.5−/+3 nM | >1,000-fold over 20 other HMTs, 150-fold over EZH1 | Phase I | 68 |
| UNC1999 | SAM-competitive inhibitor of PRC2 | 4.6−/+0.8 nM | 10,000-fold over other HMTs, 10-fold over EZH1 | Preclinical | 70 |
| EPZ-6438 | SAM-competitive inhibitor of PRC2 | 2.5−/+0.5 nM | >4,500-fold over 14 other HMTs, 35-fold over EZH1 | Phase I/II | 71,72 |
| Stabilized a-helix of EZH2 peptide (SAH-EZH2) | Hydrocarbon-stapled peptides that mimics the a-helical EED binding domain of EZH2 (aa 40–68) to disrupt the protein interation between EZH2 and EED | Unknown | More selective for EZH2 over EZH1 | Preclinical | 73 |
Enzymatic inhibition of EZH2
The first EZH2 inhibitor that was widely used for experimental work was 3-deazaneplanocin A (DZNep). It is a cyclopentanyl analog of 3-deazaadenosine that potently interferes with S-adenosyl-L-homocysteine hydrolase (SAH), a component of the methionine cycle, which causes cellular SAH levels to increase, repressing the activity of S-adenosyl-L-methionine-dependent histone lysine methyltransferase activity 63. Therefore, the effect of DZNep on inhibition of histone methylation is not specific to EZH2. Treatment with DZnep induces significant antitumor activity in various cancer types corresponding with inhibition of PRC2, and removal of H3K27me3 marks 64. In spite of potentially promising results in vitro and in vivo, DZNep has a very short plasma half-life, confers non-specific inhibition of histone methylation, and is toxic in animal models 65.
In order to improve anti-tumor activity and reduce toxicity, significant efforts have been directed toward developing compounds that are potent and selective inhibitors of EZH2. High-throughput biochemical screens have yielded several potent inhibitors based on the conserved SET domain architecture, which enabled prediction of two essential binding pockets for the S-adenosyl-L-methionine (SAM) methyl donor and the H3K27 substrate. In 2012, several groups announced independent development of SAM-competitive inhibiting compounds derived from high throughput screens. EPZ005687 binds to wild-type and Y641-mutant EZH2 and displays greater than 500-fold selectivity for EZH2 compared with 15 other human protein methyltransferases and 50-fold selectivity over EZH1 66. EPZ005687 shows dose-dependent inhibition of H3K27me3 in EZH2–wild-type and, Y641- and A677-mutant lymphoma cells as well as in cell lines of other cancer types, including breast and prostate cancer. The simultaneously developed small-molecule EZH2 inhibitor, GSK126, inhibits both wild-type and mutant EZH2 and has greater than 1,000-fold selectivity for EZH2 compared with other methyltransferases and 150-fold compared to EZH1 67,68. GSK126 markedly inhibits the growth of lymphomas carrying activating EZH2 mutations in vivo 68. A third independent SAM-competitive inhibitor, EI1, inhibits both wild-type and mutant EZH2 with an IC50 15 nM, shows >10,000 fold selectivity for EZH2 over other methyltransferases and 90-fold selectivity over EZH1 69. EI1 inhibits H3K27me2/3 levels without affecting EZH2 protein levels in EZH2-mutant DLBCL cells and a SMARCB1-mutant rhabdoid tumor cell line and inhibits cell growth and causes cell cycle arrest and apoptosis in cells carrying EZH2 mutations. These effects were accompanied by downregulation of a proliferation gene expression signature and increased expression of PRC2 targets. In pursuit of compounds more suitable for long-term animal studies by not requiring frequent injection, UNC1999 was synthesized as the first orally bioavailable inhibitor that was highly selective for wild-type EZH2 and the EZH2 Y641 mutant. UNC1999 is also relatively active against EZH1 with only 10-fold less potency than for EZH2, therefore offering the potential to target both EZH2 and EZH1 70. EPZ-6438 was subsequently developed and is more potent compared to EPZ005687 with improved pharmacokinetic properties including good oral bioavailability 71. Treatment of mice carrying SMARCB1-mutant malignant rhabdoid tumors with EPZ-6438 resulted in decreased levels of H3K27me3 and dose-dependent tumor regression 72. In June 2013, a phase I/II clinical trial of EPZ-6438 in patients with advanced solid tumors or with B-cell lymphomas was launched (NCT01897571). Preliminary reports from the study have been presented at scientific meetings (http://www.epizyme.com/wp-content/uploads/2014/11/Ribrag-ENA-FINAL.pdf and http://www.epizyme.com/wp-content/uploads/2015/07/ICML-Slides-Presented-062015-v2.pdf). The data show potentially encouraging activity of the drug: partial or complete responses in nine of fifteen NHL patients, including a partial response in the one patient who had an EZH2 mutation (EZH2 Y646H), and one complete response and partial responses in malignant rhabdoid tumor patients were observed. Two other clinical trials (NCT02395601 and NCT02082977) are actively enrolling patients and further investigation in EZH2-mutant B-cell NHL and SMARCB1-deficient tumors is currently being pursued.
Inhibitors that Disrupt PRC2 Stability
The discovery of non-enzymatic functions for EZH2 and the implication of these in SWI/SNF-mutant cancers raises the possibility that the enzymatic inhibitors currently in clinical trials may not fully suppress the transformation promoting activity of EZH2. EZH2 can also be inhibited by disrupting its interaction with other PRC2 subunits with a peptide known as stabilized alpha-helix of EZH2 (SAH-EZH2) that is derived from the domain of EZH2 that interacts with EED 73. SAH-EZH2 disrupts the EZH2–EED complex, reduces EZH2 protein levels, and selectively inhibits H3K27 trimethylation in a dose-dependent manner. This peptide is efficacious against EZH2-dependent MLL–AF9 leukemia and EZH2-mutant lymphoma cells but has no effect on non-transformed and EZH2–wild-type controls. Notably, whereas the anti-proliferative effect of the SAH-EZH2 correlated with the reduction in H3K27me3 levels, the effect seemed to correlate even more strongly with reduction of EZH2 protein levels, consistent with the findings of dependence upon non-enzymatic roles of EZH2 in SWI/SNF-mutant cancers 43.
Therapy Resistance and Combination Therapies
Therapy resistance typically enables cancer cells to escape the effects of any single agent and mechanisms of resistance to EZH2 inhibitors are beginning to emerge. In a cell line model, two novel secondary mutations of EZH2 (Y111L and Y661D) were identified in resistant cells following prolonged exposure to EZH2 inhibitors and were found to cooperate to confer resistance 74. Separately, it had earlier been reported that loss of PRC2 subunits can amplify Ras-driven transcription in PNS tumors, high-grade gliomas, and melanomas and co-occurrence of a Ras pathway mutation with mutations in SWI/SNF correlated with resistance to EZH2 inhibition 75,76 (Figure 2). Consequently, there is interest in identifying therapies that have the potential to cooperate with EZH2 inhibitors. In preclinical models of EZH2-mutant NHL, combining EPZ-6438 with conventional NHL-directed chemotherapy was synergistic in preventing tumor growth 77. The combination of EPZ-6438 and a glucocorticoid receptor agonist (GRag) also enhanced inhibition of proliferation both in cells harboring EZH2 mutations and in germinal center NHL. In prostate cancer models, combination of the chemotherapeutic agent etoposide with GSK126 significantly increased death of murine and human prostate cancer cell lines 78. Finally, in the preclinical study of non-small cell lung cancers, EZH2 inhibition was found to have differing effects in subsets of these cancers as defined by their differing mutations. In tumors that carried either BRG1/SMARCA4 loss-of-function mutations or EGFR gain-of-function mutations, EZH2 inhibition sensitized the malignancies to TopoII inhibitors. In contrast, in tumors that lacked these mutations, EZH2 inhibition conversely promoted resistance to TopoII inhibitors 42. Collectively, while development of EZH2 inhibition as a therapeutic is only in early stages, evidence of resistance mechanisms is beginning to emerge as are potential approaches for combination therapy, both areas of ongoing study.
Figure 2. EZH2 as a therapeutic target in cancer.
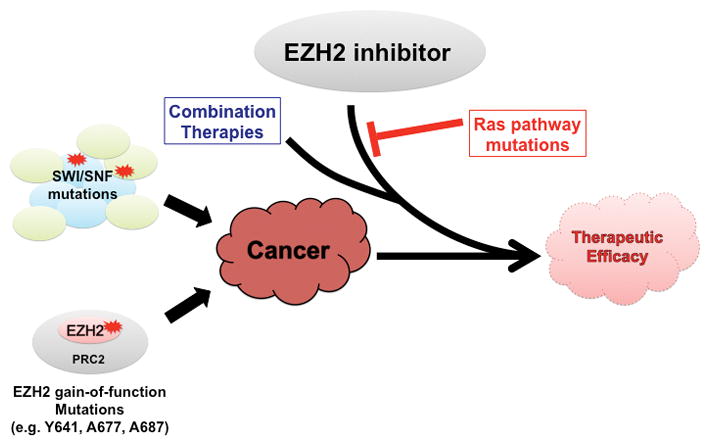
a. The roles of EZH2 mediated transcriptional silencing are context specific. For cell types in which its hyperactivity drives oncogenesis, contributions from silencing of lineage specification genes, the tumor suppressor Rb and DNA repair genes have been identified. b. Cancers harboring SWI/SNF mutations and gain-of-function EZH2 mutations confer dependency on EZH2 inhibition. Early pre-clinical evidence suggests potential benefit of combination therapy with an EZH2 inhibitor. At least in the case of cancers driven by SWI/SNF mutation, Ras pathway mutations can confer resistance to EZH2 inhibition.
Loss-of-function EZH2 mutations in cancer
In the case of EZH2, although overexpression and gain-of-function mutations suggest oncogenic activity, there is also evidence suggesting that EZH2 acts as a tumor suppressor in some cancer types 79–81 (Table 3). Recurrent inactivating deletion, frameshift, nonsense, and missense mutations in EZH2 occur in a subset of myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPN), and MDS/MPN overlap disorders 80,81. Whereas the truncating mutations are dispersed throughout the gene, the missense mutations preferentially occur in highly conserved residues in the domain required for interaction with SUZ12 of PRC2 and the CXC-SET domain required for EZH2 catalytic activity. These mutations can be found in both monoallelic and biallelic states and individuals whose myeloid disorders had homozygous mutations were shown to have reduced survival compared to those with heterozygous mutations 80. Mice lacking Ezh2 had promoted initiation and propagation of Runx1-mutant MDS, consistent with a tumor suppressive role of EZH2 in MDS 79. Loss-of-function mutations and deletions of EZH2 also occur in human T cell acute lymphoblastic leukemia (T-ALL) 82,83 (Table 3). Notably, EZH2 was not the only PRC2 subunit affected, as missense mutations were identified in both EZH2 (11 of 68 cases) and SUZ12 (3 of 68 cases) 82. The frequency of PRC2 mutations was even higher in a pediatric subtype of T-ALL, early T cell precursor (ETP) ALL where deletions and sequence mutations of EED, EZH2, and SUZ12 are found in 42% of ETP ALL, and are also found in 12% of non-ETP pediatric T-ALL 84. Mutation of genes encoding PRC2 subunits other than EZH2 also occurs in other cancers 85–87 (Table 3). In endometrial stromal tumors, fusion of SUZ12 (also known as JJAZ1) with JAZF1 was identified 86,87. Loss-of-function somatic alterations of EED or SUZ12 occur in 70–90% of malignant peripheral nerve sheath tumors (MPNSTs) where they correspond with complete loss of H3K27me3 and activation of multiple developmentally suppressed pathways 85,88. EED mutations that affect EED protein stability, its interaction with EZH2, or its binding to H3K27me3 also occur in a subset of patients with MDS and related diseases and haploinsufficiency of EED leads to a myeloproliferative disorder in mice 89–91.
Table 3.
EZH2/PRC2 loss-of-function mutations identified in different cancer types and affected target genes
| EZH2/PRC2 status | Associate cancer type | Target genes | Reference |
|---|---|---|---|
| EZH2 mutation (loss-of-function) | MDS, MPN, T-ALL | NOTCH/JAK-STAT, NOTCH1 | 79–83 |
| SUZ12 fusion | Endometrial stromal tumors | unknown | 86,87 |
| SUZ12/EED mutation | T-ALL, ETP ALL, MDS/MPN/MPD, MPNSTs | NOTCH1, JAK-STAT, FOXN4, IGF2, PAX2, TLX1 | 82–85,89,90 |
The canonical substrate of PRC2, lysine residue 27 of histone H3 and its variants, has itself been found to harbor specific recurrent missense mutations in highly restricted cancer types, including 31% of pediatric glioblastoma multiforme (GBM), 78% of diffuse intrinsic pontine gliomas (DIPG), and 50% of pediatric high-grade gliomas (pHGG) 92–94. In vitro, the H3K27M mutation has been shown to block PRC2 activity as it has an increased affinity for the protein, causing it to act as a sink for EZH2 binding and thus inhibit methylation at other H3 sites 94. Expression of H3.3 K27M also increases H3K27ac, a modification mutually exclusive to H3K27me3 that correlates with transcriptional activation rather than repression 95. Therefore, histone H3 mutations may contribute to tumorigenesis through aberrant activation of key regulatory loci by reducing H3K27 trimethylation and facilitating acetylation. Correspondingly, the high frequency of H3K27 mutations in these pediatric brain cancers but their rarity in other cancers suggests a highly context-dependent cancer-promoting role, perhaps reflecting perturbation of a role for EZH2/PRC2 in regulation of neural stem cell fate. However, PRC2 subunit gene mutations have not been detected in pediatric GBM or DIPG, so it remains to be determined whether the H3K27M mutations are similar in mechanism to EZH2/PRC2 loss-of-function mutations or whether the these mutations exert effects via additional non-Polycomb mechanisms. Collectively, the discoveries that EZH2/PRC2 loss-of-function can drive oncogensis in certain contexts suggest that some caution is warranted in the clinical application of EZH2 inhibitors.
Conclusions, questions, and future directions
The parallel development and testing of independent EZH2 inhibitors, three of which have now moved to clinical trials, should soon yield a trove of data on the toxicity as well as potential efficacy of this approach to enzymatic inhibition in cancers that carry a variety of EZH2 activation mechanisms. As only data from patients with EZH2–wild-type tumors has been reported thus far, it will be of great interest to determine efficacy of these inhibitors in cancers that carry activating EZH2 mutations. Given the emerging non-PRC2–based functions of EZH2, it will also be of interest to understand the extent to which EZH2 mutations promote cancer via PRC2-dependent versus independent effects. Similarly, it will also be important to understand the extent to which the cancer-promoting effects of EZH2 depend upon its enzymatic activity compared to non-enzymatic structural contributions to PRC2 integrity as this carries substantial implications for drug development.
Given its clear gain-of-function contributions to cancer and the capability of directly inhibiting its enzymatic function, EZH2 constitutes a compelling target for anti-cancer therapy, albeit with potential caveats from its developmental and tumor suppressor roles. The story of EZH2, from discovery as a regulator of body patterning in fruit flies to transcriptional regulator of chromatin structure to driver of cancer should soon have a new chapter when clinical trials reveal whether targeting EZH2 can bring a substantial therapeutic advance.
Acknowledgments
K.H.K. was supported by an award from National Cancer Center. C.W.M.R. was supported by R01CA172152 and R01CA113794. The Garrett B. Smith Foundation, the Cure AT/RT Now foundation, The Avalanna Fund, and Miles for Mary provided additional support.
Footnotes
Conflict of Interest Statement: As a recipient of a Dana-Farber Cancer Institute-Novartis Institutes for Biomedical Research Drug Discovery Program research grant, C.W.M.R. received research funding and consulting fees from the Novartis Institutes for Biomedical Research.
References
- 1.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell stem cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature genetics. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AS, et al. The mutational landscape of adenoid cystic carcinoma. Nature genetics. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nature communications. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annual review of pathology. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 12.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 14.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 15.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 16.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 17.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 19.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell stem cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera-Merchan A, et al. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nature communications. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 21.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodor C, et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011;25:726–729. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 24.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majer CR, et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS letters. 2012;586:3448–3451. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 28.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 34.Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- 35.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 36.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Molecular cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 37.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 38.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Molecular and cellular biology. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nature medicine. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fillmore CM, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nature medicine. 2015 doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornton SR, Butty VL, Levine SS, Boyer LA. Polycomb Repressive Complex 2 regulates lineage fidelity during embryonic stem cell differentiation. PloS one. 2014;9:e110498. doi: 10.1371/journal.pone.0110498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 47.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CJ, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz D, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141:867–877. doi: 10.1242/dev.094342. [DOI] [PubMed] [Google Scholar]
- 52.Du J, et al. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast cancer research and treatment. 2012;131:65–73. doi: 10.1007/s10549-011-1396-3. [DOI] [PubMed] [Google Scholar]
- 53.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruggeman SW, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung HY, et al. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Molecular cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee ST, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Molecular cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 58.Gonzalez ME, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higa LA, et al. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 62.Lee JM, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Molecular cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Glazer RI, et al. 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem Biophys Res Commun. 1986;135:688–694. doi: 10.1016/0006-291x(86)90048-3. [DOI] [PubMed] [Google Scholar]
- 64.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miranda TB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 67.Verma SK, et al. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 69.Qi W, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konze KD, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knutson SK, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 73.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibaja V, et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 2015 doi: 10.1038/onc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baude A, Lindroth AM, Plass C. PRC2 loss amplifies Ras signaling in cancer. Nat Genet. 2014;46:1154–1155. doi: 10.1038/ng.3124. [DOI] [PubMed] [Google Scholar]
- 76.De Raedt T, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer cell. 2011;20:400–413. doi: 10.1016/j.ccr.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Knutson SK, et al. Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas. PloS one. 2014;9:e111840. doi: 10.1371/journal.pone.0111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirk JS, et al. Top2a identifies and provides epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer. Oncotarget. 2015;6:3136–3146. doi: 10.18632/oncotarget.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sashida G, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nature communications. 2014;5:4177. doi: 10.1038/ncomms5177. [DOI] [PubMed] [Google Scholar]
- 80.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 81.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 82.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simon C, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee W, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koontz JI, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, et al. Effects of rearrangement and allelic exclusion of JJAZ1/SUZ12 on cell proliferation and survival. Proc Natl Acad Sci U S A. 2007;104:20001–20006. doi: 10.1073/pnas.0709986104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang M, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ueda T, et al. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 2012;26:2557–2560. doi: 10.1038/leu.2012.146. [DOI] [PubMed] [Google Scholar]
- 90.Lessard J, et al. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 1999;13:2691–2703. doi: 10.1101/gad.13.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Score J, et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood. 2012;119:1208–1213. doi: 10.1182/blood-2011-07-367243. [DOI] [PubMed] [Google Scholar]
- 92.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 93.Chen HW, et al. A novel infusible botanically-derived drug, PG2, for cancer-related fatigue: a phase II double-blind, randomized placebo-controlled study. Clin Invest Med. 2012;35:E1–11. doi: 10.25011/cim.v35i1.16100. [DOI] [PubMed] [Google Scholar]
- 94.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 95.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. The Journal of biological chemistry. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang X, et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PloS one. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ren G, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer research. 2012;72:3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 99.Truax AD, Thakkar M, Greer SF. Dysregulated recruitment of the histone methyltransferase EZH2 to the class II transactivator (CIITA) promoter IV in breast cancer cells. PloS one. 2012;7:e36013. doi: 10.1371/journal.pone.0036013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taniguchi H, et al. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene. 2012;31:1988–1994. doi: 10.1038/onc.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. The Journal of biological chemistry. 2005;280:22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 102.Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;26:4590–4595. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- 103.Chen Y, et al. Proteomic analysis of EZH2 downstream target proteins in hepatocellular carcinoma. Proteomics. 2007;7:3097–3104. doi: 10.1002/pmic.200700019. [DOI] [PubMed] [Google Scholar]
- 104.Yu J, et al. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene. 2010;29:5370–5380. doi: 10.1038/onc.2010.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Min J, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nature medicine. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shin YJ, Kim JH. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PloS one. 2012;7:e30393. doi: 10.1371/journal.pone.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia N, et al. Enhancer of zeste homolog 2 is involved in the proliferation of endometrial carcinoma. Oncology letters. 2014;8:2049–2054. doi: 10.3892/ol.2014.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eskander RN, et al. Inhibition of enhancer of zeste homolog 2 (EZH2) expression is associated with decreased tumor cell proliferation, migration, and invasion in endometrial cancer cell lines. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2013;23:997–1005. doi: 10.1097/IGC.0b013e318296a265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan T, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Molecular cancer research : MCR. 2011;9:418–429. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nature communications. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 111.Weikert S, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. International journal of molecular medicine. 2005;16:349–353. [PubMed] [Google Scholar]
- 112.Raman JD, et al. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 113.Lee J, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sudo T, et al. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. British journal of cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hussain M, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer research. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan J, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121:4512–4520. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 117.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Caganova M, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. The Journal of clinical investigation. 2013;123:5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hayden A, Johnson PW, Packham G, Crabb SJ. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast cancer research and treatment. 2011;127:109–119. doi: 10.1007/s10549-010-0982-0. [DOI] [PubMed] [Google Scholar]
- 120.Kemp CD, et al. Polycomb repressor complex-2 is a novel target for mesothelioma therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:77–90. doi: 10.1158/1078-0432.CCR-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Suva ML, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer research. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 122.Smits M, et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PloS one. 2011;6:e16282. doi: 10.1371/journal.pone.0016282. [DOI] [PMC free article] [PubMed] [Google Scholar]


