Abstract
Rhodopsin is the visual pigment responsible for initiating the phototransduction cascade in vertebrate rod photoreceptors. Although well‐characterized in a few model systems, comparative studies of rhodopsin function, particularly for nonmammalian vertebrates are comparatively lacking. Bowerbirds are rare among passerines in possessing a key substitution, D83N, at a site that is otherwise highly conserved among G protein‐coupled receptors. While this substitution is present in some dim‐light adapted vertebrates, often accompanying another unusual substitution, A292S, its functional relevance in birds is uncertain. To investigate functional effects associated with these two substitutions, we use the rhodopsin gene from the great bowerbird (Ptilonorhynchus nuchalis) as a background for site‐directed mutagenesis, in vitro expression and functional characterization. We also mutated these sites in two additional rhodopsins that do not naturally possess N83, chicken and bovine, for comparison. Both sites were found to contribute to spectral blue‐shifts, but had opposing effects on kinetic rates. Substitutions at site 83 were found to primarily affect the kinetics of light‐activated rhodopsin, while substitutions at site 292 had a larger impact on spectral tuning. The contribution of substitutions at site 83 to spectral tuning in particular depended on genetic background, but overall, the effects of substitutions were otherwise surprisingly additive, and the magnitudes of functional shifts were roughly similar across all three genetic backgrounds. By employing a comparative approach with multiple species, our study provides new insight into the joint impact of sites 83 and 292 on rhodopsin structure‐function as well as their evolutionary significance for dim‐light vision across vertebrates.
Keywords: passerine birds, evolution of vision, visual pigment, evolution of protein structure and function, comparative biochemistry
Introduction
Rhodopsin is one of the best‐characterized members of the G protein‐coupled receptor (GPCR) superfamily, and is responsible for initiating the phototransduction cascade in vertebrate rod photoreceptors.1 It has been the subject of numerous evolutionary and biochemical studies across vertebrates due to its instrumental role in dim‐light vision, inherently high stability, and high levels of expression in the retina.2, 3 Moreover, rhodopsin is also a model in GPCR research as well as retinal disease research, as many mutations impacting its structure and function have known links to retinal degeneration.4, 5, 6
Examining visual pigment function in different vertebrate groups can be a fruitful approach for studying the relationship between natural variation in protein sequences and structure−function mechanisms, because vision plays a major role in the adaptation of organisms to a variety of environments.7, 8, 9, 10 Curiously, although birds have been the focus of much research on cone visual pigments,11, 12, 13, 14, 15, 16 avian rhodopsin is comparatively understudied in vitro relative to other vertebrate taxa, with the notable exception of the chicken.17 This is surprising, considering birds comprise one of the most diverse groups of tetrapods, and live in a variety of photic environments with a wide range of life histories.18 Moreover, they have numerous behavioural, morphological, and physiological adaptations that are of relevance from a visual perspective.8, 19
Bowerbirds (Ptilonorynchidae) are a charismatic family of passerines that dwell in dense vegetative jungle forest cover, and perform courtship rituals that involve the construction of “bowers,” large structures designed to house colourful arrangements of found ornaments to attract mates.20, 21, 22 Males of the great bowerbird, Ptilonorhynchus (=Chlamydera) nuchalis, also possess colorful patches at the nape of their neck, which are thought to be involved in mate attraction.8, 20 Bowerbirds generally prefer a frugivorous diet and primarily forage in daytime, and the ornaments used by males to attract mates may exploit biases in female visual perception associated with foraging.8 Evidently, bowerbirds are highly visual organisms, but while their cone pigments have been studied in detail,14, 23 their rod pigment has received little attention.
Rhodopsin is a molecular complex, consisting of a vitamin A‐derived retinal chromophore, 11‐cis‐retinal, covalently bound to a seven transmembrane domain protein moiety. Isomerization of the chromophore to all‐trans‐retinal is triggered by absorption of a photon, thereby activating the pigment.24 Upon activation, a series of intermediates are formed. The biologically active intermediate, metarhodopsin II (meta II) is responsible for catalyzing the GDP‐for‐GTP exchange in the G protein transducin, thereby activating transducin and generating an electrical response in the photoreceptor cell.2 Much like the dark state of rhodopsin, meta II has been functionally studied and a crystal structure has been resolved.25 After activation, all‐trans‐retinal exits the protein, and a new 11‐cis‐retinal molecule enters, serving as an inverse agonist for the pigment and stabilizing its inactive state.26
The most prominent aspect of rhodopsin function studied to date is its wavelength of maximum absorbance (λ max), which is typically ∼500 nm.27 Numerous comparative studies of rhodopsin have focused on spectral tuning, wherein certain substitutions in rhodopsin, typically found within the chromophore binding pocket, shift λ max.28, 29, 30, 31, 32 Some substitutions produce large effects, while several substitutions working in concert may provide only minimal shifts. The majority of spectral tuning studies to date have investigated these substitutions through site‐directed mutagenesis experiments in a bovine rhodopsin background.27, 33, 34, 35 However, more recent works have characterized spectral shifts in different vertebrate rhodopsin backgrounds, revealing previously overlooked insights into both structure−function and evolutionary significance.36, 37, 38
In addition, although there are increasing numbers of spectral tuning studies across vertebrates, investigations of other aspects of visual pigment function have only recently been given more attention in a comparative context. There is evidence that sites known to mediate spectral tuning may also have additional effects on rhodopsin kinetics. A key study by Sugawara and colleagues36 suggested that an asparagine (N) at an otherwise highly conserved site across Family A GPCRs, D83 (aspartic acid), may confer an advantage under dim‐light conditions by increasing the rate of rhodopsin kinetics such that the active Meta‐II state is favoured, therefore increasing photosensitivity. While Sugawara et al.36 measured the rate of Meta‐II formation, Meta‐II stability can also be inferred through measurements of the rate of all‐trans retinal release from light‐activated rhodopsin.37 In fact, N83 has been found in a variety of dim‐light adapted organisms, including deep‐sea fishes, marine mammals, and bats, and is often found in concert with a substitution at another otherwise highly conserved site, A292S.28, 31, 36, 39, 40, 41 In previous mutagenesis studies, this substitution at 292 has resulted in shifts in spectral sensitivity towards shorter wavelengths,39, 41 but other aspects of rhodopsin function that may be affected by this site, such as retinal release rates, have yet to be investigated.
Previous studies have found, somewhat surprisingly given that the substitution is typically associated with nocturnal or aquatic species, that the rhodopsin of all bowerbird species examined to date contains N83, but its functional relevance in birds remains unknown.14, 42 Here, we use site‐directed mutagenesis and in vitro opsin expression in the great bowerbird rhodopsin (RH1) as a model system to investigate the effects of substitutions at two sites that have been suggested as dim‐light adaptations, D83N and A292S. We compare the effects of mutations at these sites in the bowerbird to similar mutations in another bird, the chicken, which does not possess N83, as well as to bovine rhodopsin. We find that substitutions at site 83 have a large impact on the kinetics of light‐activated rhodopsin, whereas spectral tuning effects are relatively minor. The opposite is true of site 292, where large effects on spectral tuning are seen, with much smaller effects on light‐activated rhodopsin kinetics. The functional effects of the mutations at these sites were also found to be similar in magnitude across the different genetic backgrounds. These results are discussed in light of other comparative studies of visual pigment structure and function, as well as implications for dim‐light vision, and the evolution of bowerbird behaviour and ecology.
Results
Sequence analysis of great bowerbird RH1
The translated amino acid sequence of great bowerbird RH1 contained conserved residues and motifs known to be important for visual pigment function (reviewed in Ref. 43). For example, both K296, the residue that forms a Schiff base linkage with the chromophore44 and E113, the primary counterion in the dark state27 are present. Unlike bovine RH1, bowerbird RH1 contains additional serine and threonine residues at the C terminus, typical of birds and other vertebrates but not of eutherian mammals; these are likely additional sites of phosphorylation important for rhodopsin deactivation.45 Aside from the highly variable C terminus, the chicken and bowerbird sequences differ at only 9 sites (∼3%), including site 83, while the bovine sequence differs from both birds at 31 sites (∼9%).
The majority of bird and vertebrate rhodopsins possess D83, which is conserved among GPCRs in general.46 However, a variety of vertebrate lineages have N83 at this site, which has been associated with shifts in rhodopsin spectral tuning and activation kinetics.36, 37, 41 Though RH1 genes from several disparate avian families have N83, among passerines the substitution appears to be rare, possessed only by the bowerbirds and at least one species of Acanthisittidae (New Zealand wrens). Site 292 varies between A and S among vertebrate rhodopsins, with the less common identity, S292, causing large blue‐shifts in teleost fish and mammals where it often co‐occurs with N83.28, 31, 36, 39, 40, 41 Among birds, however, all currently available RH1 sequences, including bowerbirds, have A292.
Characterizing wild‐type P. nuchalis rhodopsin
Wild‐type bowerbird, bovine, and chicken rhodopsin were expressed in vitro, regenerated with 11‐cis‐retinal, and purified, producing stable visual pigments with λ max values of 500 nm, 499 nm, and 503 nm, respectively (Fig. 1, Table 1). When bleached with light, the absorption peak of each rhodopsin shifted to ∼380 nm, characteristic of the biologically active metarhodopsin II intermediate; difference spectra were calculated by subtracting light‐bleached spectra from respective dark spectra (Fig. 1). When denatured with acid (100 mM HCl), the λ max of all three rhodopsins shifted to 440 nm (Fig. 1), characteristic of 11‐cis‐retinal covalently bound to a denatured opsin,47 and indicative of a proper Schiff base linkage. Bowerbird rhodopsin was also not initially susceptible to hydroxylamine, similar to bovine rhodopsin, indicating an inaccessible binding pocket typical of mammalian rhodopsins27, 37 (Fig. 2). The rate of all‐trans‐retinal release from opsin following light‐activation was monitored using a fluorescence assay. The half‐life of retinal release for bowerbird rhodopsin was 29.4 minutes, nearly twice as slow as half‐life values calculated for bovine and chicken rhodopsins (15.6 and 17.9 min, respectively) (Fig. 3; Table 1), which are comparable to previously reported values.48, 49, 50, 51
Figure 1.
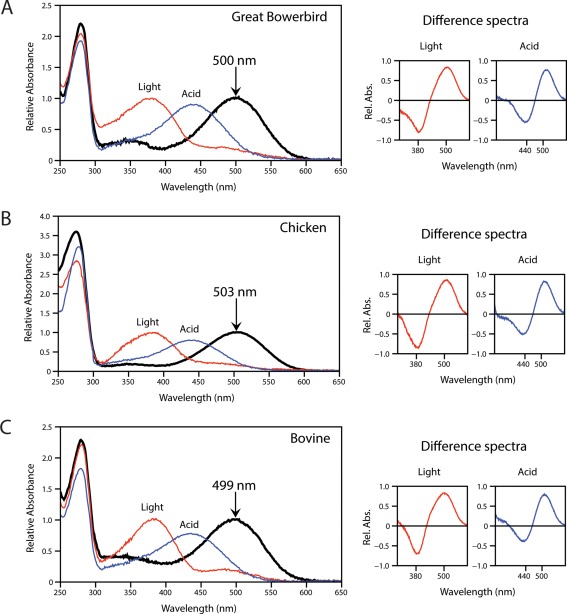
Characterization of great bowerbird rhodopsin (A) as compared with chicken (B) and bovine (C) rhodopsin controls. Absorption spectra show bleaching by light (red) and acid (blue) with the shifted peaks that result after sample exposure to white light (λ max = 380 nm), or to 100 mM hydrochloric acid (λ max = 440 nm), indicating a functional active state and Schiff base respectively. Dark‐light and dark‐acid difference spectra are shown to the right of each panel. The indicated λ max values were estimated according to the curve‐fitting methodology of Govardovskii.71
Table 1.
Spectral Tuning and Retinal Release Half‐Lives of Great Bowerbird, Chicken, and Bovine Rhodopsins Measured in vitro
| Species | Mutant | 83, 292a | λ max (nm) | t 1/2 (min) |
|---|---|---|---|---|
| P. nuchalis | Wild‐type | N A | 500 | 29.4 |
| P. nuchalis | N83D | D A | 502 (+2) | 18.5 (−10.9) |
| P. nuchalis | A292S | N S | 490 (−10) | 24.4 (−5) |
| G. gallus | Wild‐type | D A | 503 | 17.9 |
| G. gallus | D83N | N A | 501 (−2) | 27.3 (+9.4) |
| B. taurus | Wild‐type | D A | 499 | 15.6 |
| B. taurus | D83N | N A | 495 (−4) | 28.3 (+12.7) |
| B. taurus | A292S | D S | 489 (−10) | 11.4 (−4.2) |
| B. taurus | D83N+A292S | N S | 485 (−14) | 22.2 (+6.6) |
83 and 292 refer to the amino acid positions of the rhodopsin protein sequence. The amino acid identities are indicated with standard abbreviations (A, alanine; D, aspartic acid; N, asparagine; S, serine), with colours indicating whether the residue is associated with either a spectral red‐ or blue‐shift.
Numbers in parentheses indicate the functional shift relative to each species' wild‐type condition.
Figure 2.
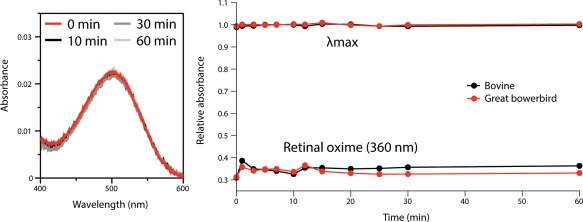
Hydroxylamine (50 mM) exposure for great bowerbird rhodopsin shows dark state peak (500 nm) does not decay over time (left panel), and shows comparable stability with bovine rhodopsin at λ max (499 nm) and at the retinal oxime peak (360 nm) (right panel), indicating the chromophore is protected from hydrolyzing solvent.
Figure 3.
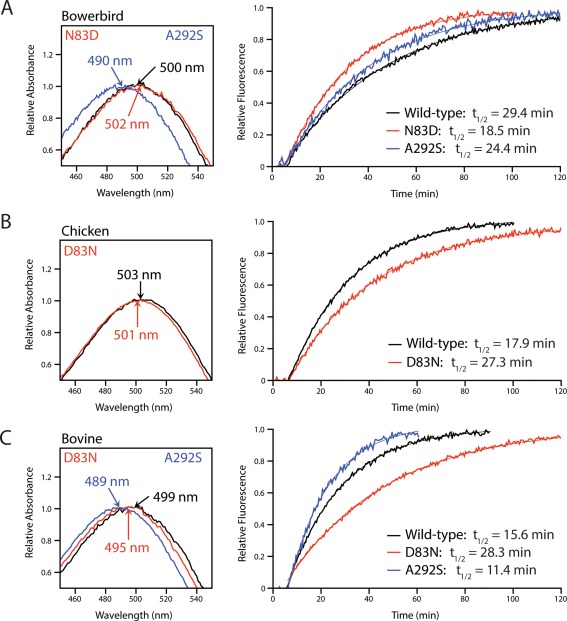
The effect of substitutions at sites 83 and 292 on spectral tuning and rates of retinal release in great bowerbird (A), chicken (B), and bovine (C) rhodopsin. N83 slows down retinal release and blue‐shifts λ max while D83 accelerates retinal release and red‐shifts λ max relative to all three wild‐type conditions. S292 accelerates retinal release and greatly blue‐shifts λ max relative to wild‐type conditions in both great bowerbird and bovine rhodopsin. The indicated λ max values were estimated according to the curve‐fitting methodology of Govardovskii,72 and half‐lives were estimated by fitting time courses to first‐order exponential curves.
Functional characteristics of site 83 and 292 mutants
To investigate the functional consequences of substitutions at sites 83 and 292, we used site‐directed mutagenesis to introduce several mutations at these sites in bowerbird (N83D, A292S), bovine (D83N, A292S), and chicken (D83N) rhodopsin, then assessed whether these mutations altered λ max and/or light‐activated retinal release rates.
Mutations at site 83 caused smaller changes to spectral tuning in the avian rhodopsins (bowerbird, chicken) compared to bovine rhodopsin. The bowerbird N83D mutant had a 2 nm red‐shift in λ max compared to wild‐type, which was consistent with the reverse D83N mutation in chicken rhodopsin that resulted in a 2 nm blue‐shift [Fig. 3(A,B), Table 1]. However, the D83N mutation in bovine rhodopsin caused a 4 nm blue‐shift, twice as big as the shift seen in either avian rhodopsin [Fig. 3(C), Table 1], and similar to previous studies of bovine rhodopsin mutants.42, 52, 53
The bowerbird N83D mutant also led to a 10.9 minute decrease in retinal release half‐life compared to wild‐type, which was consistent with the reverse D83N mutation in chicken rhodopsin that resulted in a 9.4 minute increase to the half‐life [Fig. 3(A,B), Table 1]. The half‐life value for the bovine D83N mutant was increased by 12.7 minutes relative to wild‐type, once again showing a slightly larger shift than in the avian rhodopsins [Fig. 3(C), Table 1].
Unlike mutations at site 83, the A292S mutation had the same effect on spectral tuning and retinal release in both bowerbird and bovine rhodopsin, causing a 10 nm blue‐shift to λ max and a decrease of ∼5 min in retinal release half‐life relative to the wild‐types (Fig. 4, Table 1).
Figure 4.
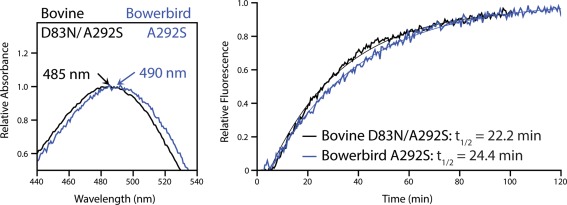
The effect of N83 and S292 on spectral tuning and rates of retinal release in different species backgrounds (bovine D83N/A292S and bowerbird A292S). While rates of retinal release are comparable, spectral tuning differs by ∼5 nm. This suggests that function cannot be predicted in different species' rhodopsins based on the residues at these sites alone.
Because the natural occurrence of S292 often coincides with N83,28, 31, 39, 40, 41 we also created a bovine D83N/A292S double mutant. The effect on both spectral tuning and retinal release was essentially additive, resulting in a 14 nm blue‐shift to λ max and a 6.6‐min increase in retinal release half‐life relative to wild‐type (Fig. 4, Table 1). The less consistent influence of site 83 on rhodopsin function between avian and bovine rhodopsin was evident when this double mutant was compared with the bowerbird A292S mutant. Both these pigments had the same residues at sites 83 and 292 (N, S), yet retinal release rates, and spectral tuning especially, were different (Fig. 4, Table 1).
Discussion
By employing a comparative approach involving multiple species and aspects of rhodopsin function, our study offers new insights into the mechanistic and molecular evolutionary significance of amino acid substitutions at two key sites in rhodopsin that have been previously suggested to be important for dim‐light vision. In our study, two naturally occurring mutations, at sites 83 and 292, had opposing effects on rhodopsin spectral vs. kinetic functions that were relatively consistent in both avian and mammal backgrounds. The substitution, D83N, greatly reduced rates of light‐activated retinal release while causing only minor blue‐shifts in spectral tuning. In contrast, the A292S substitution slightly increased retinal release rates while causing large blue‐shifts in spectral tuning. Moreover, the functional patterns we observed for both sites were congruent with their structural locations in the rhodopsin protein. Though prior in vitro investigations of rhodopsin function in nonmodel organisms have overwhelmingly focused on spectral tuning, our results demonstrate previously underappreciated shifts in kinetic function as well, implying that functional trade‐offs may influence vertebrate rhodopsin evolution.
The role of sites 83 and 292 in rhodopsin activation
Our results suggest that mutations at sites 83 and 292 influence rhodopsin function through different molecular mechanisms, as mutations at site 83 were found to have a larger influence on retinal release, while those at site 292 caused greater shifts in λ max. In rhodopsin, D83 is known to be involved in an important hydrogen bond network in the dark state. This includes both a direct hydrogen bond with the side chain of N55, as well as an indirect hydrogen bond with N302 of the “NPxxY” motif on helix 7, mediated by a water molecule.54 This hydrogen‐bonded chain of N55‐D83‐N302 (Fig. 5) is thought to result in inter‐helical constraints that stabilize the dark state of rhodopsin,55 with changes in these hydrogen bonds occurring during the transition from meta I to meta II.55 The D83N substitution, identified in several groups of vertebrates including fish,28, 31 mammals,37, 39, 40 and reptiles,56 favors meta II in the metarhodopsin equilibrium,36, 54, 57 likely by weakening interactions with N55 and N302 that stabilize the dark state and the meta I intermediate. By favoring meta II, N83 prioritizes the stability of activated rhodopsin, which likely explains why rhodopsins with N83 show large increases (9.4 to 12.7 min) in retinal release half‐life values. D83 is also ∼12 Å from the Schiff base linkage in dark state rhodopsin,58 which is fairly distant for a spectral tuning site (Fig. 5). This distance likely explains the relatively minor shifts in λ max (2 − 4 nm) caused by substitutions between D and N at site 83, which are consistent with previous mutagenesis studies.39, 59
Figure 5.
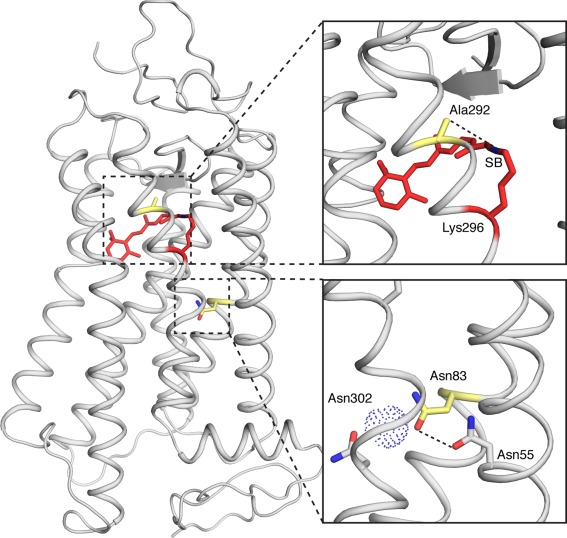
Homology model of bowerbird rhodopsin, illustrating the relative difference between A292 and N83 in distance from the chromophore. The A292 side chain is 4.5 Å from the Schiff Base of the chromophore (SB). N83 is more distant from the chromophore; however, it interacts with N55 (3.4 Å), and indirectly with N302 via a water molecule. Model was generated from bovine rhodopsin.48
Meanwhile, A292 is much more proximal (∼4 Å) to the Schiff base compared to D83 in dark state rhodopsin (Fig. 5), only a single α‐helical turn away from the Schiff base linkage.60 This likely explains why A292S causes much larger shifts in λ max (10 nm), also consistent with previous spectral tuning studies,39, 41 through alterations to the electrostatic environment of the chromophore binding pocket. The close proximity of A292 to the Schiff base may also be why the spectral tuning effect of A292S is more consistent in both bovine and bowerbird rhodopsin, compared to the differences seen between bovine and avian opsins for substitutions at site 83. Site 292 is likely to interact directly with the Schiff base end of the chromophore, whereas 83 may affect spectral tuning more indirectly via an H‐bond network, likely influenced by neighboring residues that may vary in different rhodopsin backgrounds. As A292S also shows moderately decreased (∼5 min) retinal release half‐life values, S292 may be involved in stabilizing dark state rhodopsin via H‐bond interactions in the vicinity of the chromophore.61 Therefore, while D83N and A292S are both spectrally blue‐shifting substitutions (2 − 4 nm and 10 nm, respectively), they seem to have opposite effects on the kinetic properties of rhodopsin, such as retinal release and meta II formation.36
Potential influences of avian ecology and evolution on rhodopsin function
In other vertebrates, teleost fish and mammals in particular, substitutions N83 and S292 have both been implicated in visual adaptation to dim‐light environments through blue‐shifts in spectral sensitivity,28, 31, 39, 41, 62 and additionally for N83 through increased stability of the active meta II state.36, 37 Nevertheless, the relative functional importance of these substitutions in different ecological and evolutionary contexts has not yet been well explored. Following significant expansions in genome resources over recent years, the N83 substitution appears to have convergently evolved in an ever more scattered collection of vertebrate rhodopsins, with no obvious correlations to life history traits.13 Furthermore, studies that emphasize the multi‐faceted structural roles of this site48, 63 make it clear that resolving the functional mechanisms of the N83 substitution, and its relationship to organism lifestyle, will depend on better understanding its interactions with other sites.
The spectral tuning of bowerbird rhodopsin was highly similar to the chicken and bovine rhodopsin controls (1 − 2 nm) despite the presence of the D83N substitution. Nevertheless, the major increase in retinal release half‐life (11 min) that results from the N83 substitution in bowerbirds may contribute to an increase in photosensitivity that is important for the family's unique reliance on vision for bower‐building. Anecdotal evidence from field observations has suggested that male bowerbirds will conduct intricate repair work during the night on bowers that have been damaged by competing males (Endler, pers. obs). The birds must be able to see well enough for very high quality thatching under dim‐light conditions. In contrast to the minor shift in spectral sensitivity resulting from the D83N substitution, the large increase in retinal release half‐life suggests this substitution may be adaptive due to increased stability of the Meta‐II state, allowing for increased photosensitivity as has been suggested for similar substitutions in bats and deep water cichlids.36 Aside from bowerbirds, the N83 substitution seems to be present in few other bird families, appearing in only one or two species each of eagles, cormorants, loons, tropicbirds, and hummingbirds. This is a diverse and scattered spread of species in the Aves phylogeny, and similarly to mammals, any patterns between the substitution and specific aspects of lifestyle are not readily discernible. To further investigate the role of rhodopsin in bowerbird nocturnal vision, future work is needed directly relating ambient light intensity and rhodopsin sensitivity with bower construction.
Alternatively, the slower kinetic rates of bowerbird light‐activated rhodopsin may be related to thermoregulation. Recent studies of rhodopsins from poikilothermic vertebrates suggest they have generally increased kinetic rates relative to mammalian rhodopsins, possibly because at higher body temperatures, slower kinetics may result from increased thermal stability.38 Birds are known for generally having higher metabolic rates (and body temperatures) than mammals,64 which may in part explain the occurrence of N83 in some avian rhodopsins. Future in vitro investigations of rhodopsins from other bird species, as well as a more thorough investigation of the substitution's phylogenetic distribution will be necessary to refine hypotheses about the functional relevance of N83 in birds.
The co‐occurrence of N83 with S292 appears to be more strictly tied to ecology, present only in organisms that live or are active in extreme dimly lit environments such as the deep sea or darkly stained rivers.30, 31, 41, 62, 65 As of yet, the S292 substitution has not been found in any avian species. Given the prior work on the distribution and function of N83 and S292 among vertebrates, our results support the notion that while S292 is very likely a spectral tuning adaptation, the advantages conferred by the N83 substitution may be primarily kinetic.36
Extensive comparative studies of biochemical function are lacking outside of a few models systems, such as hemoglobins66, 67 and opsins. Even within the opsins most comparative studies of function have centered on spectral tuning, neglecting other important aspects of rhodopsin function. Ours is one of few comparative studies of opsin kinetics, which likely play a critical role in important aspects of vision such as photosensitivity and rates of dark adaptation.52, 68 The contrasting functional effects between 83 and 292 that we found highlight the importance of considering functional trade‐offs in comparative studies of protein structure and function. Potentially overlooked interactions among different functions that arise from highly convergent substitutions, such as N83, may be key to characterizing their broader evolutionary relevance. Our in vitro investigation of bowerbird rhodopsin is a case example illustrating how comparative research involving nonmodel organisms can highlight protein structural‐functional mechanisms of evolutionary relevance that may otherwise go unnoticed in more traditional protein biochemistry study settings.
Methods
Great bowerbird rhodopsin isolation, sequencing, and site‐directed mutagenesis
For details regarding the collection of great bowerbirds and retinal RNA extraction see van Hazel et al.14 Briefly, degenerate primers and RACE (Rapid Amplification of cDNA Ends) PCR were used to amplify the rhodopsin coding sequence from cDNA libraries constructed with the extracted RNA. The complete RH1 coding sequence was then inserted into the p1D4‐hrGFP II expression vector.69 Site‐directed mutagenesis, preformed using the QuickChange mutagenesis kit (Stratagene), was used to generate the mutants N83D and A292S in the bowerbird RH1, D83N in chicken RH1, and D83N, A292S, and D83N + A292S in bovine RH1. All mutant sequences were confirmed using a 3730 DNA Analyzer (Applied Biosystems).
Great bowerbird rhodopsin expression and functional assays
Bowerbird, chicken, and bovine rhodopsins were transfected, harvested, and purified as reported previously.69 In summary, rhodopsin was inserted into the p1D4‐hrGFP II expression vector, which was used to transiently transfect HEK293T cells using Lipofectamine 2000 (Invitrogen). Rhodopsin samples were harvested 48 h post‐transfection, regenerated with 11‐cis‐retinal, solubilized in 1% N‐dodecyl‐D‐maltoside, and immunoaffinity purified using the 1D4 monoclonal antibody.70 The UV‐visible absorption spectra of purified rhodopsin samples were measured using a Cary 4000 double‐beam spectrophotometer (Varian) at 20°C in the dark, and after 60 s of white light bleaching. Difference spectra were calculated by subtracting light spectra from dark spectra. To estimate λ max, the dark absorbance spectra were fit to a standard template for A1 visual pigments.71 The wild‐type bowerbird rhodopsin was also exposed to hydrochloric acid (HCl; 100 mM) to ensure the presence of a proper Schiff base linkage with the chromophore, and to hydroxylamine (NH2OH; 50 mM).
The rate of all‐trans‐retinal release from photoactivated rhodopsins was measured as the increase in intrinsic tryptophan fluorescence following retinal release from the chromophore binding pocket.48, 72 The intrinsic fluorescence signal was measured using a Cary Eclipse fluorescence spectrophotometer (Varian) at 20°C as previously described.38 Fluorescence time courses were fit to a first‐order exponential curve (y = y 0 + a(1‐e‐kx)), with half‐time values calculated from the rate constant “k” (t 1/2 = ln(2)/k).
Homology modeling of bowerbird rhodopsin
The 3D structure of the bowerbird rhodopsin was inferred with homology modeling, using dark state bovine rhodopsin (PDB code: 1U1960) as a template. The Modeller package58 was used to generate 100 models that were then ranked according to DOPE score,73 the model with the lowest value being selected for visualization in MacPyMOL (The PyMOL Molecular Graphics System, Version 1.7.4.4 Schrödinger, LLC). The generated bowerbird rhodopsin structure had a total energy that was comparable with bovine rhodopsin (comparable z‐scores in Pro‐SA‐web74), and amino acid bond conformations with high probabilities (assessed with ProCheck75).
Acknowledgment
11‐cis‐retinal was generously provided by Rosalie Crouch (Medical University of South Carolina).
References
- 1. Burns ME, Baylor DA (2001) Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Ann Rev Neurosci 24:779–805. [DOI] [PubMed] [Google Scholar]
- 2. Ridge KD, Palczewski K (2007) Visual rhodopsin sees the light: structure and mechanism of G protein signaling. J Biol Chem 282:9297–9301. [DOI] [PubMed] [Google Scholar]
- 3. Yanagawa M, Kojima K, Yamashita T, Imamoto Y, Matsuyama T, Nakanishi K, Yamano Y, Wada A, Sako Y, Shichida Y (2015) Origin of the low thermal isomerization rate of rhodopsin chromophore. Sci Rep 5:11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL (1990) A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature 343:364–366. [DOI] [PubMed] [Google Scholar]
- 5. Sung CH, Davenport CM, Nathans J (1993) Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J Biol Chem 268:26645–26649. [PubMed] [Google Scholar]
- 6. Kaushal S, Khorana HG (1994) Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 33:6121–6128. [DOI] [PubMed] [Google Scholar]
- 7. Endler JA (1991) Variation in the appearance of guppy color patterns to guppies and their predators under different visual conditions. Vision Res 31:587–608. [DOI] [PubMed] [Google Scholar]
- 8. Endler JA, Westcott DA, Madden JR, Robson T (2005) Animal visual systems and the evolution of color patterns: sensory processing illuminates signal evolution. Evolution 59:1795–1818. [DOI] [PubMed] [Google Scholar]
- 9. Bowmaker JK, Hunt DM (2006) Evolution of vertebrate visual pigments. Curr Biol 16:R484–R489. [DOI] [PubMed] [Google Scholar]
- 10. Bowmaker JK (2008) Evolution of vertebrate visual pigments. Vision Res 48:2022–2041. [DOI] [PubMed] [Google Scholar]
- 11. Wilkie SE, Robinson PR, Cronin TW (2000) Spectral tuning of avian violet‐and ultraviolet‐sensitive visual pigments. Biochemistry 39:7895–7901. [DOI] [PubMed] [Google Scholar]
- 12. Hart NS, Hunt DM (2007) Avian visual pigments: characteristics, spectral tuning, and evolution. Am Nat 169 (Suppl 1):S7–S26. [DOI] [PubMed] [Google Scholar]
- 13. Hunt DM, Carvalho LS, Cowing JA, Davies WL (2009) Evolution and spectral tuning of visual pigments in birds and mammals. Phil Trans Roy Soc B 364:2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Hazel I, Sabouhanian A, Day L, Endler JA, Chang BSW (2013) Functional characterization of spectral tuning mechanisms in the great bowerbird short‐wavelength sensitive visual pigment (SWS1), and the origins of UV/violet vision in passerines and parrots. BMC Evol Biol 13:250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bloch NI, Price TD, Chang BSW (2015) Evolutionary dynamics of Rh2 opsins in birds demonstrate an episode of accelerated evolution in the New World warblers (Setophaga). Mol Ecol 24:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bloch NI, Morrow JM, Chang BSW, Price TD (2015) SWS2 visual pigment evolution as a test of historically contingent patterns of plumage color evolution in warblers. Evolution 69:341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imai H, Mizukami T, Imamoto Y, Shichida Y (1994) Direct observation of the thermal equilibria among lumirhodopsin, metarhodopsin I, and metarhodopsin II in chicken rhodopsin. Biochemistry 33:14351–14358. [DOI] [PubMed] [Google Scholar]
- 18. Hart NS (2001) The visual ecology of avian photoreceptors. Prog Ret Eye Res 20:675–703. [DOI] [PubMed] [Google Scholar]
- 19. Badyaev AV, Hill GE (2003) Avian sexual dichromatism in relation to phylogeny and ecology. Ann Rev Ecol 24:27–49. [Google Scholar]
- 20. Marshall AJ (1954) Bower birds. Biol Rev 29:1–45. [Google Scholar]
- 21. Borgia G (1995) Why do bowerbirds build bowers? Am Sci 83:542–547. [Google Scholar]
- 22. Endler JA, Day LB (2006) Ornament colour selection, visual contrast and the shape of colour preference functions in great bowerbirds, Chlamydera nuchalis. Animal Behav 72:1405–1416. [Google Scholar]
- 23. Coyle BJ, Hart NS, Carleton KL, Borgia G (2012) Limited variation in visual sensitivity among bowerbird species suggests that there is no link between spectral tuning and variation in display colouration. J Exp Biol 215:1090–1105. [DOI] [PubMed] [Google Scholar]
- 24. Pugh EN, Lamb TD (1993) Amplification and kinetics of the activation steps in phototransduction. Biochim Biophys Acta 1141:111–149. [DOI] [PubMed] [Google Scholar]
- 25. Choe H‐W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauß N, Hofmann KP, Scheerer P, Ernst OP (2011) Crystal structure of metarhodopsin II. Nature 471:651–655. [DOI] [PubMed] [Google Scholar]
- 26. Corson DW, Cornwall MC, MacNichol EF, Jin J, Johnson R, Derguini F, Crouch RK, Nakanishi K (1990) Sensitization of bleached rod photoreceptors by 11‐cis‐locked analogues of retinal. Proc Natl Acad Sci USA 87:6823–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakmar TP, Franke RR, Khorana HG (1989) Glutamic acid‐113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci USA 86:8309–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunt DM, Fitzgibbon J, Slobodyanyuk SJ, Bowmaker JK (1996) Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Res 36:1217–1224. [DOI] [PubMed] [Google Scholar]
- 29. Sun H, Macke JP, Nathans J (1997) Mechanisms of spectral tuning in the mouse green cone pigment. Proc Natl Acad Sci USA 94:8860–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fasick JI, Robinson PR (2000) Spectral‐tuning mechanisms of marine mammal rhodopsins and correlations with foraging depth. Vis Neurosci 17:781–788. [DOI] [PubMed] [Google Scholar]
- 31. Sugawara T, Terai Y, Imai H, Turner GF, Koblmüller S, Sturmbauer C, Shichida Y, Okada N (2005) Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep‐water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad Sci USA 102:5448–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies WL, Cowing JA, Carvalho LS, Potter IC, Trezise AEO, Hunt DM, Collin SP (2007) Functional characterization, tuning, and regulation of visual pigment gene expression in an anadromous lamprey. Faseb J 21:2713–2724. [DOI] [PubMed] [Google Scholar]
- 33. Karnik SS, Sakmar TP, Chen HB, Khorana HG (1988) Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci USA 85:8459–8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamashita T, Terakita A, Shichida Y (2000) Distinct roles of the second and third cytoplasmic loops of bovine Rhodopsin in G protein activation. J Biol Chem 275:34272–34279. [DOI] [PubMed] [Google Scholar]
- 35. Altun A, Yokoyama S, Morokuma K (2008) Mechanism of spectral tuning going from retinal in vacuo to bovine Rhodopsin and its mutants: Multireference ab initio quantum mechanics/molecular mechanics studies. J Phys Chem B 112:16883–16890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugawara T, Imai H, Nikaido M, Imamoto Y, Okada N (2010) Vertebrate rhodopsin adaptation to dim light via rapid meta‐II intermediate formation. Mol Biol E 27:506–519. [DOI] [PubMed] [Google Scholar]
- 37. Bickelmann C, Morrow JM, Müller J, Chang BSW (2012) Functional characterization of the rod visual pigment of the echidna (Tachyglossus aculeatus), a basal mammal. Vis Neurosci 29:211–217. [DOI] [PubMed] [Google Scholar]
- 38. Morrow JM, Chang BSW (2015) Comparative mutagenesis studies of retinal release in light‐activated zebrafish Rhodopsin using fluorescence spectroscopy. Biochemistry 54:4507–4518. [DOI] [PubMed] [Google Scholar]
- 39. Fasick JI, Robsinson PR (1998) Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry 37:433–438. [DOI] [PubMed] [Google Scholar]
- 40. Zhao H, Ru B, Teeling EC, Faulkes CG, Zhang S, Rossiter SJ (2009) Rhodopsin molecular evolution in mammals inhabiting low light environments. PLoS One 4:e8326–e8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dungan SZ, Kosyakov A, Chang BSW (2016) Spectral tuning of killer whale (Orcinus orca) Rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol Biol E 33:323–336. [DOI] [PubMed] [Google Scholar]
- 42. van Hazel I (2012) Molecular evolution and functional characterization of the visual pigment proteins of the Great Bowerbird (Chlamydera nuchalis) and other vertebrates (Doctoral dissertation).
- 43. Smith SO (2010) Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys 39:309–328. [DOI] [PubMed] [Google Scholar]
- 44. Zhukovsky E, Robinson P, Oprian D (1991) Transducin activation by rhodopsin without a covalent bond to the 11‐cis‐retinal chromophore. Science 251:558–560. [DOI] [PubMed] [Google Scholar]
- 45. Greene NM, Williams DS, Newton AC (1997) Identification of protein kinase C phosphorylation sites on bovine rhodopsin. J Biol Chem 272:10341–10344. [DOI] [PubMed] [Google Scholar]
- 46. Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y (2002) Functional role of internal water molecules in rhodopsin revealed by X‐ray crystallography. Proc Natl Acad Sci USA 99:5982–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kito Y, Suzuki T, Azuma M, Sekoguti Y (1968) Absorption spectrum of rhodopsin denatured with acid. Nature 218:955–957. [DOI] [PubMed] [Google Scholar]
- 48. Chen M‐H, Kuemmel C, Birge RR, Knox BE (2012) Rapid release of retinal from a cone visual pigment following photoactivation. Biochemistry 51:4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Janz JM, Farrens DL (2001) Engineering a functional blue‐wavelength‐shifted Rhodopsin mutant. Biochemistry 40:7219–7227. [DOI] [PubMed] [Google Scholar]
- 50. Janz JM, Fay JF, Farrens DL (2003) Stability of dark state rhodopsin is mediated by a conserved ion pair in intradiscal loop E‐2. J Biol Chem 278:16982–16991. [DOI] [PubMed] [Google Scholar]
- 51. Sommer ME, Farrens DL (2006) Arrestin can act as a regulator of rhodopsin photochemistry. Vis Res 46:4532–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nathans J (1990) Determinants of visual pigment absorbance: role of charged amino acids in the putative transmembrane segments. Biochemistry 29:937–942. [DOI] [PubMed] [Google Scholar]
- 53. Breikers G, Bovee‐Geurts PHM, DeCaluwé GLJ, DeGrip WJ (2001) A structural role for Asp83 in the photoactivation of rhodopsin. Biol Chem 382:1–8. [DOI] [PubMed] [Google Scholar]
- 54. Palczewski K (2000) Crystal structure of rhodopsin: A G protein‐coupled receptor. Science 289:739–745. [DOI] [PubMed] [Google Scholar]
- 55. Rath P, DeCaluwé LL, Bovee‐Geurts PH, DeGrip WJ, Rothschild KJ (1993) Fourier transform infrared difference spectroscopy of rhodopsin mutants: light activation of rhodopsin causes hydrogen‐bonding change in residue aspartic acid‐83 during meta II formation. Biochemistry 32:10277–10282. [DOI] [PubMed] [Google Scholar]
- 56. Kawamura S, Yokoyama S (1998) Functional characterization of visual and nonvisual pigments of American chameleon (Anolis carolinensis). Vis Res 38:37–44. [DOI] [PubMed] [Google Scholar]
- 57. Weitz CJ, Nathans J (1993) Rhodopsin activation: effects of the metarhodopsin I‐metarhodopsin II equilibrium of neutralization or introduction of charged amino acids within putative transmembrane segments. Biochemistry 32:14176–14182. [DOI] [PubMed] [Google Scholar]
- 58. Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. [DOI] [PubMed] [Google Scholar]
- 59. Janssen J, De Caluwé G, de Grip WJ (1990) Asp 83, Glu 113 and Glu 134 are not specifically involved in Schiff base protonation or wavelength regulation in bovine rhodopsin. FEBS Lett 260:113–118. [DOI] [PubMed] [Google Scholar]
- 60. Okada T, Sugihara M, Bondar A‐N, Elstner M, Entel P, Buss V (2004) The retinal conformation and its environment in Rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol 342:571–583. [DOI] [PubMed] [Google Scholar]
- 61. Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP (1998) Mechanisms of spectral tuning in blue cone visual pigments: visible and raman spectroscopy of blue‐shifted rhodopsin mutants. J Biol Chem 273:24583–24591. [DOI] [PubMed] [Google Scholar]
- 62. Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK (2001) The molecular basis for spectral tuning of rod visual pigments in deep‐sea fish. J Exp Biol 204:3333–3344. [DOI] [PubMed] [Google Scholar]
- 63. Pope A, Eilers M, Reeves PJ, Smith SO (2014) Amino acid conservation and interactions in rhodopsin: Probing receptor activation by NMR spectroscopy. Biochim Biophys Acta Bioenergetics 1837:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heinrich B (1977) Why have some animals evolved to regulate a high body temperature? Am Nat 111:623–640. [Google Scholar]
- 65. Van Nynatten A, Bloom D, Chang BSW, Lovejoy NR (2015) Out of the blue: adaptive visual pigment evolution accompanies Amazon invasion. Biol Lett 11:20150349–20150345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A (2009) Evolutionary and functional insights into the mechanism underlying high‐altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci USA 106:14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tufts DM, Natarajan C, Revsbech IG, Projecto‐Garcia J, Hoffmann FG, Weber RE, Fago A, Moriyama H, Storz JF (2015) Epistasis constrains mutational pathways of hemoglobin adaptation in high‐altitude pikas. Mol Biol E 32:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lamb TD, Pugh EN (2004) Dark adaptation and the retinoid cycle of vision. Prog Ret Eye Res 23:307–380. [DOI] [PubMed] [Google Scholar]
- 69. Morrow JM, Chang BSW (2010) The p1D4‐hrGFP II expression vector: a tool for expressing and purifying visual pigments and other G protein‐coupled receptors. Plasmid 64:162–169. [DOI] [PubMed] [Google Scholar]
- 70. Molday RS, MacKenzie D (1983) Monoclonal antibodies to rhodopsin: characterization, cross‐reactivity, and application as structural probes. Biochemistry 22:653–660. [DOI] [PubMed] [Google Scholar]
- 71. Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K (2000) In search of the visual pigment template. Vis Neurosci 17:509–528. [DOI] [PubMed] [Google Scholar]
- 72. Farrens DL, Khorana HG (1995) Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J Biol Chem 270:5073–5076. [DOI] [PubMed] [Google Scholar]
- 73. Shen M‐Y, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wiederstein M, Sippl MJ (2007) ProSA‐web: interactive web service for the recognition of errors in three‐dimensional structures of proteins. Nucl Acid Res 35:W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laskowski RA, MacArthur MW, Moss DS Thornton JM, IUCr (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]


