Abstract
Streptomyces species produce many important antibiotics and have a crucial role in soil nutrient cycling. However, their evolutionary history remains poorly characterized. We have evaluated the impact of homologous recombination on the evolution of Streptomyces using multi-locus sequence analysis of 234 strains that represent at least 11 species clusters. Evidence of inter-species recombination is widespread but not uniform within the genus and levels of mosaicism vary between species clusters. Most phylogenetically incongruent loci are monophyletic at the scale of species clusters and their subclades, suggesting that these recombination events occurred in shared ancestral lineages. Further investigation of two mosaic species clusters suggests that genes acquired by inter-species recombination may have become fixed in these lineages during periods of demographic expansion; implicating a role for phylogeography in determining contemporary patterns of genetic diversity. Only by examining the phylogeny at the scale of the genus is apparent that widespread phylogenetically incongruent loci in Streptomyces are derived from a far smaller number of ancestral inter-species recombination events.
Introduction
Homologous recombination (HR), the acquisition of genetic material through genetic exchange of similar DNA sequences, is an important mechanism in the evolution of many microbial lineages. As a consequence of HR, microbial genomes contain genes having diverse evolutionary histories (Boussau et al., 2008; Boyer et al., 2009; Zhaxybayeva et al., 2009; Caro-Quintero et al., 2012), which may be different from the history of the organism itself. HR has significant impacts on the evolution and diversification of diverse taxa of Bacteria and Archaea (Hernández-López et al., 2013; Paul et al., 2013; Darch et al., 2015; Huang et al., 2015). Extensive HR also coincides with indistinct or ‘fuzzy' boundaries between genetic clusters (Hanage et al., 2005; Papke et al., 2007). These observations have generated lively debates about mechanisms of microbial evolution and the concept of species in the microbial world (Bapteste and Boucher, 2009; Fraser et al., 2009; Riley and Lizotte-Waniewski, 2009; Caro-Quintero and Konstantinidis, 2012; Segerman, 2012).
Discussions of microbial evolution often focus on adaptive traits as they relate to ecological niche (Koeppel et al., 2008). However, neutral processes such as genetic drift comprise a cornerstone of evolutionary theory, and genetic drift can promote allopatric speciation in microorganisms (Whitaker et al., 2003) independent of niche dynamics. In addition, neutral changes resulting from HR can have major impacts on the evolution of microbial genomes (Vetsigian and Goldenfeld, 2005; Fraser et al., 2007; Sheppard et al., 2008; Doroghazi and Buckley, 2011). Multi-locus sequence analysis (MLSA) can be used to characterize extant patterns of genetic diversity in microbial lineages and the degree to which these patterns result from HR within and between species. Knowledge of these patterns is required to infer the evolutionary processes that govern microbial diversification.
There is evidence for widespread HR both within and between Streptomyces species (Doroghazi and Buckley, 2010, 2014; Cheng et al., 2015). Streptomyces use an unusual method of gene exchange in which conjugation mobilizes double-stranded DNA (Kataoka et al., 1991). In the model organism, Streptomyces coelicolor A3(2), long considered as the genetic workhorse in Streptomyces biology, chromosomal markers are mobilized through this mechanism at a frequency of 0.1–1% (Hopwood et al., 1985), with plasmid integration causing mobilization of chromosomal genes at an efficiency that approaches 100% (Chater et al., 1982). However, whether this is true for other members of this genus remains unclear, as studies comparing DNA mobilization and conjugation rates in other species of Streptomyces are lacking. Nevertheless, this double-stranded DNA conjugation system permits inter-species recombination between isolates in the laboratory (Alacevic, 1963) and can generate hybrid strains with genomes having nearly equal genetic contributions from each parent (Wang et al., 1999). Such laboratory-generated hybrids are described to display new combinations of parent phenotypes including changes in phage sensitivity (Lomovskaya et al., 1977) and antibiotic production. Furthermore, acquisition of antibiotic biosynthesis and resistance genes has been documented in wild strains of Streptomyces (Deng et al., 2011; Kinashi, 2011), suggesting that the horizontal gene exchange may have contributed to the tremendous biosynthetic diversity within the genus Streptomyces.
There have been few efforts to characterize the impact of recombination on the evolutionary history of Streptomyces. MLSA has revealed that many species of Streptomyces have mosaic ancestry because of widespread gene exchange (Doroghazi and Buckley, 2010). Not surprisingly, recombination rates are much higher within species than between species (Doroghazi and Buckley, 2010). High rates of gene exchange between closely related strains may act as a cohesive force that reinforces the genetic similarity of species and species clusters (Doroghazi and Buckley, 2011). For example, strains of Streptomyces pratensis are in linkage equilibrium over much of their genome despite having a geographic range that spans >1000 km (Doroghazi and Buckley, 2014). This pattern of ancestry can result only if the S. pratensis genomes are recombining freely across a large geographic range, or if S. pratensis has experienced evolutionarily recent demographic expansion from an ancestral population that was in linkage equilibrium. Ecological niche differences (marine, soil, insect host and plant host) can pose barriers to gene exchange and these factors may influence lineage diversification (Cheng et al., 2015), but geographic and phylogenetic factors that influence gene exchange in Streptomyces remain poorly described.
We use MLSA to investigate patterns of inter-species HR among diverse Streptomyces species from 11 species clusters. In particular, we seek to determine if patterns of inter-species HR are random or if they are associated with phylogeny and/or geography. We select five species clusters for further analysis, including three that demonstrate mosaic ancestry and two that have contributed ancestry to the mosaic clusters. We screened Streptomyces isolates from diverse geographic sites to identify wild-type strains representing these five species clusters.
Materials and methods
Strain isolation and sequence data
Analysis of Streptomyces genetic diversity and phylogeny used the MLSA scheme of Guo et al. (2008). Nucleotide sequences of six loci from 150 strains were obtained from the Streptomyces database of the pubMLST website (http://pubmlst.org/streptomyces/ as of December 2012). These include partial sequences of six loci—16S ribosomal RNA (rRNA) gene (432 bp), trpB (571 bp), gyrB (423 bp), rpoB (540 bp), atpD (496 bp) and recA (504 bp). Three isolates (S. cremeus strain AS 4.1625 ID# 23, S. flavidofuscus strain AS 4.1617 ID# 21 and S. kasugaensis strain DSM 40819 ID# 99) were excluded from our analyses on account of missing sequence information for at least one locus. For strains with identical sequence types, only one representative was selected. We also excluded three strains (S. atratus strain AS 4.1632 ID# 124, S. gelaticus strain AS 4.1444 ID# 132 and S. sanglieri strain AS 4.1146 ID# 144) that have divergent rpoB and trpB genes and are more similar to other genera of Actinobacteria than to Streptomyces when compared with the non-redundant database available from the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990). S. griseoplanus was renamed S. pratensis following the recent report on its taxonomical characterization (Rong et al., 2013). In total, 139 strains from pubMLST representing 128 species were used in the analysis (Supplementary Table S1).
Additional strains for MLSA were derived from a geographically explicit Streptomyces culture collection. The culture collection consists of >1000 isolates obtained from 15 temperate grassland sites across the continental United States (Supplementary Table S2). Sites were selected to represent a narrow range of ecological characteristics including meadow, pasture or native grasslands dominated by perennial grasses and having moderately acidic soil (pH: 6.0±1.0; ave.±s.d., Supplementary Table S2). Strains were isolated using uniform conditions and this will select for strains having similar physiological characteristics. Briefly, soil was sampled from 0 to 5 cm, air dried and sieved to 4 mm. Streptomyces strains were isolated at room temperature on glycerol-arginine agar, pH 8.7, containing 300 mg l–1 cycloheximide and 30 mg l–1 Rose Bengal as previously described (Doroghazi and Buckley, 2010). The analysis of physiologically similar strains from ecologically similar sites improves our ability to detect biogeographical patterns that result from neutral processes such as genetic drift (as reviewed in Hanson et al., 2012). We screened these isolates by sequencing rpoB to identify strains (n=95; Supplementary Table S2) that belonged to species clusters 4, 5, 6, 7 and 10 (as identified in Figure 1, see Results section). These species clusters represent three mosaic species clusters (4, 6 and 7) and two ancestral species clusters (5 and 10) identified in the analysis of pubMLST strains. We focused on these species clusters to evaluate in greater detail the phylogeny and phylogeography of strains in admixed lineages. For each isolate, we then sequenced the genes atpD, gyrB, recA and trpB using the method described in Doroghazi and Buckley (2010). We did not include the 16S rRNA gene because of its low genetic variability among taxa in our selected clusters.
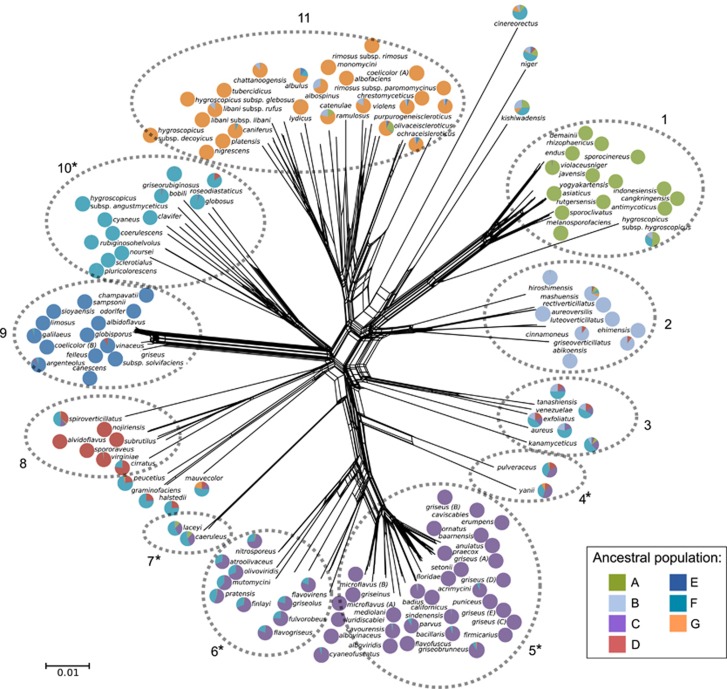
Figure 1.
A phylogenetic network for Streptomyces generated using SplitsTree with default settings. The network was built using concatenated sequences of 16S rRNA, trpB, atpD, gyrB, rpoB and recA loci. The pie charts associated with each species show the ancestry of each species, with each color representing the seven ancestral lineages inferred by Structure. The dotted circles highlight distinct clusters. Asterisks indicate clusters targeted for further analysis including those with high (4 and 6) and low (5 and 10) admixture. Admixed clusters 4, 6 and 7 have genetic contributions from clusters 5 and 10. Scale bar indicates 0.01 substitutions per site.
Phylogenetic analyses
Multiple sequence alignments were performed using MUSCLE v3.5 (Edgar, 2004) with default parameters. A maximum likelihood phylogenetic tree using PhyML v.3.0 (Guindon et al., 2010) was built for each gene under the Generalized Time Reversible (Tavaré, 1986) substitution model, estimated gamma parameter, estimated proportion of invariable sites, four substitution rate categories and 100 bootstrapped replicates. The six genes were then concatenated and used as input for the NeighborNet program implemented in SplitsTree v4.13.1 (Huson, 1998) to further analyze species relationships. ClonalFrame was run with default settings to generate a 50% consensus tree (Didelot and Falush, 2007). We used LDhat (McVean et al., 2002) to estimate the number of segregating sites and average pairwise differences for each of the five clusters in the second data set, using the concatenated alignment as input. Nucleotide diversity (π), number of synonymous substitutions per non-synonymous site (dN) and number of synonymous substitutions per synonymous site (dS) were calculated using DnaSP v5.10.01 (Librado and Rozas, 2009).
Clustering analysis
To examine the population structure and estimate the extent of genetic mixing, we utilized the model-based clustering algorithm implemented in Structure (Falush et al., 2003). First, we performed clustering analysis for all 139 strains from pubMLST. We tested values of K from 2 to 10, where K is the number of inferred populations. For each specific K value, we performed 10 replicates, with each run lasting for 300 000 iterations and the initial 50 000 iterations discarded as burnin. Based on the criterion recommended by Evanno et al. (2005) and Prunier and Holsinger (2010), we selected the K value of 7 that best represents our data. Structure allowed us to identify species that belong to one ancestral lineage (un-admixed) and to estimate the proportion of ancestry for species that originate from more than one ancestral lineage (admixed or mosaic). The individual proportion of membership (qi) determines the proportion of genetic ancestry that each individual derives from a given ancestral lineage. Empirical and simulation results (Barilani et al., 2007) suggest qi⩾0.90 for un-admixed individuals. Using this threshold, we assigned each individual to one lineage if qi⩾0.90 or as admixed if the proportion of membership from each ancestral lineage was qi<0.90.
Tree topology tests
The program IQ-TREE v.0.9.5 (Nguyen et al., 2015) was used to perform tests of phylogenetic incongruence including: the RELL method (Kishino et al., 1990), the one-sided Kishino–Hasegawa test (Kishino and Hasegawa, 1989), the Shimodaira–Hasegawa test (Shimodaira and Hasegawa, 1999) and the Expected Likelihood Weight test (Strimmer and Rambaut, 2002). Significance was determined by permutation with 1000 re-samplings. The output shows the log likelihood score of all trees, the difference in log likelihood between each tree and the best tree, and the associated P-value for the probability that a given tree can explain the sequence data (Supplementary Table S3).
Recombination analyses
We used two programs in examining HR in Streptomyces. The Recombination Detection Program (RDP v3.44), which implements nine different detection methods, was used to analyze both data sets (Martin et al., 2010). Detection was performed using default settings. We included only those recombination events that are supported by at least three out of the nine methods in RDP to avoid dependence on a single methodology (RDP, BOOTSCAN, GENECONV, MAXCHI, CHIMAERA, SISCAN, PhylPro, LARD and 3Seq). Previous recombination studies using RDP have used the 3/9 criterion, with the expectation that true recombination events will likely be detected using different approaches (Bilhère et al., 2009; Cadillo-Quiroz et al., 2012; Alvarez-Pérez et al., 2013). The strength of RDP is its implementation of a variety of detection approaches to both identify and characterize recombination events, such as phylogenetic conflicts clustering of nucleotide substitutions and fitting of substitutions to an expected statistical distribution. Hence, using a 3/9 criterion will give us a stronger support for a recombination event, and a higher threshold will only slightly improve the results. Reported P-values were calculated using the Bonferroni correction within the program and statistical significance was set at P<0.05 level. Using the alignment of the concatenated genes of each of the five clusters in the second data set as input, we also ran the Pairwise Homoplasy Index (PHI) test for recombination implemented in SplitsTree v4.13.1 (Huson, 1998).
Results and Discussion
Contemporary and ancestral inter-species recombination
We evaluated patterns of HR among Streptomyces species by analyzing sequences of 16S rRNA, trpB, atpD, gyrB, rpoB and recA loci for 139 Streptomyces species available from the pubMLST database (www.pubmlst.org). Seven ancestral populations were chosen to represent the collective ancestry of these species. The criteria for identifying the most likely number of ancestral populations has been described previously (Evanno et al., 2005; Prunier and Holsinger, 2010); briefly we selected the smallest number of populations that explained the greatest probability of the data whereby successive increases in population number had only a marginal impact on the probability of the data. The species were then grouped into 11 species clusters on the basis of phylogenetic affinity (Figure 1). These 11 clusters roughly correspond to 16S rRNA gene clusters identified in a recent study of 615 Streptomyces species (Labeda et al., 2012). Phenotypic conservation by species in these gene clusters is generally unremarkable (as assessed from Labeda et al., 2012), although species clusters 1 and 11 contain strains that primarily have rugose spores (corresponding to clusters 75–80 in Labeda et al., 2012) and species cluster 9 contains species that produce smooth-surfaced, olive-buff colored spores carried in flexuous chains (corresponding to cluster 112 in Labeda et al., 2012). Biosynthetic gene clusters generally differ between species of Actinobacteria (Doroghazi and Metcalf, 2013; Ziemert et al., 2014), and such gene clusters are often exchanged horizontally within and between species (Gontang et al., 2010; Ziemert et al., 2014), although conservation of these gene clusters within species clusters remains uncharacterized.
Individual gene trees for the six loci all exhibit conflicting topologies (Supplementary Figure S1,Supplementary Table S3) supporting the finding of widespread inter-species HR. Similarly, analysis of genetic ancestry using Structure (Falush et al., 2003) suggests widespread admixture between species clusters (Figure 1). Using the recombination detection software RDP, we detected 32 significantly supported HR events in the concatenated sequence alignment (Figure 2). A total of 98 of the strains (71%), and 122 of the 834 total genes (15%), were found by RDP to have evidence of inter-species HR, and these results are broadly consistent with prior observations (Doroghazi and Buckley, 2010). However, the statistical criteria used by RDP to detect these HR events are insensitive to exchange of short similar sequences, and hence should be viewed as a conservative lower bound on the actual number of events that have occurred.
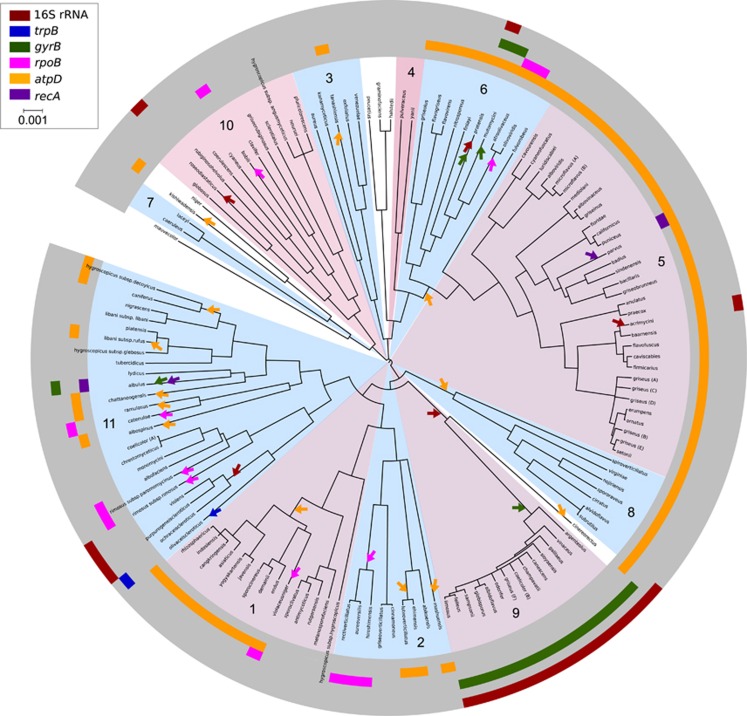
Figure 2.
Clonal phylogeny for Streptomyces generated using ClonalFrame (50% consensus tree). Numbers and alternating blue and pink colors in the middle of the tree correspond to species clusters from Figure 1. Arrows indicate HR events at nodes of the tree as inferred by RDP and their position relative to the length of each branch is arbitrary. Colored bars in the outer rings indicate genes that are affected by HR as determined by RDP analysis. The loci inferred to be acquired by HR are identified by color as indicated in the scale. The scale bar in the ClonalFrame genealogy represents time in coalescent units.
Genes observed to have evidence of HR can be derived both from contemporary and ancestral HR events. The fixation of ancestral HR events in ancestral lineages will result in an indefinitely large number of contemporary lineages that have inherited ‘horizontally acquired' genes through vertical descent. Thus, widespread evidence of HR within a lineage can result from both prolific contemporaneous gene exchange and/or accumulation of ancestral HR events over evolutionary time. We mapped the evolutionary history of HR events by examining patterns of shared ancestry (that is, recombination donors and recipients inferred by RDP) among horizontally acquired genes (Figure 2). Of the 32 HR events detected, 9 are inferred to have taken place in ancestral lineages. These ancestral HR events included four that occurred in the last common ancestors of species clusters (subsequently inherited by 63 strains) and five that occurred in the last common ancestors of various subclades (subsequently inherited by 23 species). Hence, ancestral HR accounts for 86 of the 98 strains (88% of strains) in which HR was detected by RDP analysis. The remaining 23 HR events were unique to a single strain (note that sums are not additive because several strains have multiple HR events). Furthermore, it is likely that many of the HR events detected in a single lineage may have also occurred ancestrally, but are observed at the tips of the tree because related strains that share these HR events remain unsampled. Hence, to more rigorously evaluate the role of ancestral HR on the genetic diversity of Streptomyces, it is necessary to more rigorously sample from species clusters affected by ancestral HR.
Phylogenetic relationships of targeted species clusters
To further evaluate the role of ancestral HR in determining the extant genetic diversity of Streptomyces, we focused upon three admixed species clusters (4, 6 and 7) and two additional species clusters (5 and 10) that contain extant representatives of the ancestral populations that contributed genetic ancestry to these admixed clusters (Figure 1). Specifically, we sought to determine whether the HR events that contribute to phylogenetic incongruence were due to contemporary or ancestral HR events. The pubMLST database did not contain sufficient numbers of strains to permit robust analysis of these five species clusters; hence, additional strains that belonged to these clusters were identified from a collection of wild-type strains that we isolated from a geographically diverse set of soil samples from temperate grassland sites across the United States (as described in Materials and methods section). We identified and sequenced loci from 95 new strains bringing the total number of strains representing the five clusters to 152. The concatenated sequence alignment for the five genes is 2534 bp. Pairwise nucleotide identity of MLSA loci within species clusters ranged from 91.9% to 100%, whereas pairwise nucleotide identity between species clusters ranged from 87.7% to 95.5% (Supplementary Figure S2).
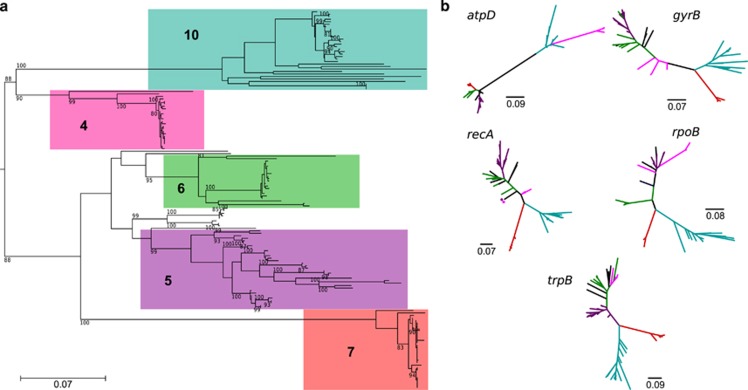
The individual gene phylogenies show significant incongruence across the five species clusters (Figure 3 and Supplementary Figure S4,Supplementary Table S4). As expected, and as described below, many of these inter-species HR events took place in a common ancestor of a species cluster or subclade and were inherited vertically by extant strains. For example, strains from cluster 7 are most similar to cluster 10 strains at the loci trpB, gyrB, rpoB and recA (Supplementary Figure S4), and across these four loci they differ considerably from cluster 6 strains (>10% substitutions per site). However, an incongruent result is obtained with atpD, as cluster 7 strains are instead more similar to cluster 6 strains (<5% substitutions per site; Supplementary Figure S4) than to cluster 10 strains. This result suggests that a recent common ancestor of cluster 7 exchanged either part or all of its atpD gene with an ancestor of cluster 6. Similarly, strains in cluster 4 share affinity with those from clusters 5 and 6 for trpB, gyrB, rpoB and recA but affiliate with cluster 10 at atpD (Supplementary Figure S4). In addition, S. olivoviridis and S. atroolivaceus, each have four genes that associate with cluster 6, but have gyrB genes that affiliate with cluster 5 (Supplementary Figure S4).
Figure 3.
Maximum likelihood phylogeny of species clusters 4, 5, 6, 7 and 10, with the addition of isolates collected from diverse geographic sites (n=152). (a) Phylogeny calculated from the concatenated alignment of the five housekeeping genes trpB, gyrB, rpoB, atpD and recA (see Supplementary Figure S3 for names of taxa). Only bootstrap values ⩾50% are shown. (b) Individual gene trees for each of the five genes (see Supplementary Figure S3 for names of taxa). Taxa labeled in black indicate ambiguous cluster membership. Scale bars indicate substitutions per site.
We also observed 11 species and strains with ambiguous cluster affiliation intermediate between clusters 4, 5 and 6, including: S. finlayi, S. nitrosporeus, S. fulvorobeus, S. flavogriseus, S. flavovirens, S. griseolus, or75, or89, wa23, wa1022 and wa1068 (taxa labeled in black; Figures 3 and 4, Supplementary Figures S3 and S4). The latter eight of these form a cohesive group (99% bootstrap value) in the concatenated phylogeny and in all gene trees (Figure 3, Supplementary Figures S3 and S4); however, the placement of this group varies considerably in the different gene trees (Supplementary Figure S4). For example, this group of eight is most closely related to cluster 6 in the gyrB and recA gene trees, to cluster 5 in the atpD and rpoB gene trees, and to cluster 4 in the trpB tree (Supplementary Figure S4). Again, these results indicate that the most recent common ancestor of this group acquired multiple genes by HR and extant species subsequently inherited these genes from this common ancestor. The placement of the remaining three species, S. finlayi, nitrosporeus, and S. fulvorobeus varies considerably between clusters 4, 5 and 6 for each of the different genes (Supplementary Figure S4) suggesting that these lineages may have been recipients of a number of HR events from species in these different clusters.
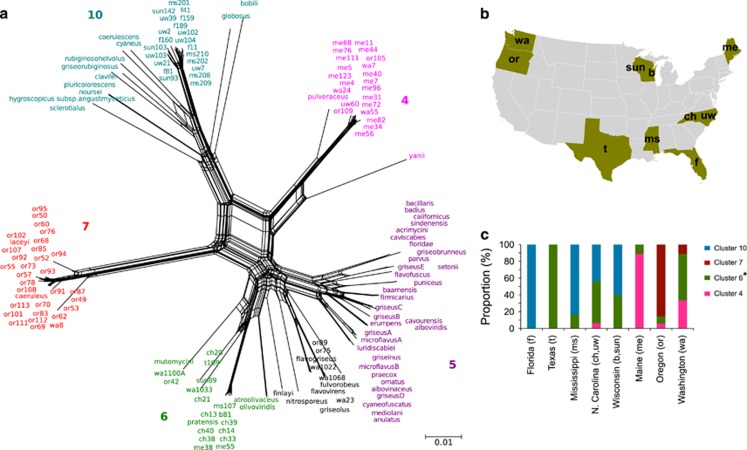
Figure 4.
Evolutionary relationships and geographic sources of isolates from the second data set, consisting of species clusters 4, 5, 6, 7 and 10. (a) SplitsTree network of the five selected clusters, genetic relationships were calculated from the concatenated alignment of the five housekeeping genes trpB, gyrB, rpoB, atpD and recA. Colors correspond with Figure 3. Taxa labeled in black indicate ambiguous cluster membership from Figure 3. Scale bar indicates 0.01 substitutions per site. (b) Location of sites from which strains were isolated (see also Supplementary Table S2). The labels for each site correspond to the sampling names used for the isolates. (c) Distribution of the five clusters in each state in the United States. Colors of the bars correspond to those found in panel a. Cluster 6 consists of isolates labeled in green and black in the SplitsTree.
Contributions of geography to recombination
Geographical and ecological factors have been previously shown to influence patterns of gene flow in diverse microorganisms (Reno et al., 2009; Cadillo-Quiroz et al., 2012; Shapiro et al., 2012). It is possible to evaluate whether patterns of admixture exhibit geographical partitioning in Streptomyces, as the geographic origin of our strains is known. Analysis of strains with similar characteristics (all strains were isolated under the same conditions on the same medium, see Materials and methods section) from similar habitats maximizes our ability to observe neutral genetic variation associated with biogeography (as discussed in Hanson et al., 2012). HR in housekeeping genes would generally be expected to result in neutral variation, which can readily be detected through MLSA (Falush et al., 2003).
We observe that species clusters 4 and 7 are found almost exclusively (98% of strains, Figure 4) in higher latitude sites (WA, ME and OR), whereas species clusters 6 and 10 occurred almost exclusively (95% of strains, Figure 4) in lower latitude sites (TX, FL, MS, NC and WI). This pattern corresponds to historical patterns of glaciation as all higher latitude sites were subject to late Pleistocene glaciation (Peltier, 1994). We note that much of Wisconsin (WI) was within the driftless zone and did not experience glacial cover during the last glacial maximum (Holliday et al., 2002). Species clusters 4 and 7 have mosaic ancestry with asymmetric contributions from ancestral populations represented by strains in clusters 5, 6 and 10 (Figures 1 and 3b, Supplementary Figure S4). Species clusters 4 and 7 also have lower nucleotide diversity and higher dN/dS than those of clusters 6 and 10 (Table 1). In addition, significant intra-cluster recombination is observed within clusters 6 and 10 but not within clusters 4 and 7 (PHI test, Table 1). These results suggest that the fixation of ancestral HR events in species clusters 4 and 7 may have occurred as the result of rapid demographic expansion into virgin habitat exposed by glacial retreat, as discussed below.
Table 1. Estimated population and evolutionary parameters.
| Cluster | Number of | Length | No. segregating | Average pairwise | Nucleotide | dN/dS | PHI test |
|---|---|---|---|---|---|---|---|
| sequences | sites | differences | diversity π | P-value | |||
| 4 | 22 | 2528 | 34 | 5.874 | 0.00232 | 0.6637 | 0.1293 |
| 6* | 22 | 2528 | 292 | 78.515 | 0.03108 | 0.6250 | 0.0 |
| 7 | 30 | 2534 | 98 | 12.577 | 0.00496 | 0.6643 | 0.6412 |
| 10 | 21 | 2519 | 91 | 21.238 | 0.00844 | 0.583 | 4.65E–03 |
Abbreviation: PHI, Pairwise Homoplasy Index. Values for number of segregating sites and average pairwise differences were calculated by LDhat. The P-value for the PHI test was calculated under the null hypothesis of no recombination. Nucleotide diversity (π), defined as the average number of nucleotide differences per site, as well as dN and dS values were calculated using DnaSP.
Glaciation has had strong impacts on the biogeography and genetic diversity of a range of terrestrial species (Conroy and Cook, 2000; Milá et al., 2006; Wilson and Eigenmann Veraguth, 2010; Pointing et al., 2014; Newman and Austin, 2015) and has been shown to impact microbial diversity in soil (Vercken et al., 2010; Boyd et al., 2011; Park et al., 2011; Eisenlord et al., 2012; Stibal et al., 2012; Hamilton et al., 2013). Rapid demographic expansion resulting from glacial retreat has had profound impacts on the genetics of species (Hewitt, 1996; Conroy and Cook, 2000; Hewitt, 2000, 2004; Wilson and Eigenmann Veraguth, 2010). In particular, demographic expansion should manifest in lower nucleotide diversity (Hewitt, 1996), the potential for relaxed selection because of founder effects (Templeton, 2008) and less recombination because of epidemic population growth (Smith et al., 2003). These predictions are broadly consistent with our observations for species clusters 4 and 7 (as described in the paragraph above).
We hypothesize that inter-species HR has occurred at relatively low frequency between species clusters 5 and 10 (relative to intra-cluster HR), but demographic expansion caused genes acquired by inter-species HR to reach fixation in the populations that ultimately founded species clusters 4 and 7. Furthermore, even if mosaic (or hybrid) strains resulting from inter-species HR are maladapted because of genetic incompatibility (Berg and Kurland, 2002), demographic expansion would create an opportunity for maladapted stains to increase in frequency in the absence of competition from ancestral strains (as described in Templeton, 2008). Over time, compensatory mutations would be expected to accumulate, eliminating fitness drag from maladapted alleles (Bouma and Lenski, 1988).
The Pleistocene was characterized by repeated glacial–interglacial cycles (Cox and Moore, 2000), generating many opportunities for lineage divergence because of demographic expansion and contraction (Hewitt, 1996). Indeed, glacial–interglacial cycles of population expansion and contraction have been widely recognized for their impacts on hybridization and lineage divergence of terrestrial species (Swenson and Howard, 2005). It may be possible that species cluster 6, which has mosaic ancestry (Figure 1) but is found in southern sites and has higher nucleotide diversity than species clusters 4 and 7, was generated in a prior glacial–interglacial cycle. Species cluster 6 also exhibits multiple patterns of incongruence across strains, whereas clusters 4 and 7 have each inherited a single incongruent locus (Supplementary Figure S4).
There is widespread evidence of mosaicism across Streptomyces species (Doroghazi and Buckley, 2010), but most of the incongruence in gene phylogenies is due to relatively few ancestral recombination events that have been inherited by many extant strains (Figure 2). Looking only at six loci, it appears that even if inter-species and inter-cluster recombination was relatively rare on a per generation basis, such events could accumulate over evolutionary time leading to complex patterns of genetic mosaicism. Furthermore, we hypothesize that the fixation of incongruent loci is a consequence of evolutionary dynamics associated with historical biogeography and that such dynamics may explain the ancestral formation of mosaic lineages. If the fixation of inter-species recombination events is associated with historical demographic events, then it means that the frequency of genetic mosaicism in Streptomyces may be less a function of inter-species recombination frequency and more a function of historical biogeography. Future tests of this hypothesis will require analysis of additional strains and sites to better establish whether species and species clusters have distinct geographical range, to establish range boundaries, and to perform comparative population genomic analysis to evaluate the frequency of ancestral HR and the extent of genetic mosaicism in relation to phylogeography.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant no. DEB-1050475. We thank the Cornell Life Sciences Core Laboratories Center for technical assistance with DNA sequencing.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alacevic M. (1963). Interspecific recombination in Streptomyces. Nature 197: 1323. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Pérez S, de Vega C, Herrera CM. (2013). Multilocus sequence analysis of nectar pseudomonads reveals high genetic diversity and contrasting recombination patterns. PloS One 8: e75797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, Boucher Y. (2009). Epistemological impacts of horizontal gene transfer on classification in microbiology. Methods Mol Biol Clifton NJ 532: 55–72. [DOI] [PubMed] [Google Scholar]
- Barilani M, Bernard-Laurent A, Mucci N, Tabarroni C, Kark S, Perez Garrido JA et al. (2007). Hybridisation with introduced chukars (Alectoris chukar threatens the gene pool integrity of native rock (A. graeca and red-legged (A. rufa partridge populations. Biol Cons 137: 57–69. [Google Scholar]
- Berg OG, Kurland CG. (2002). Evolution of microbial genomes: sequence acquisition and loss. Mol Biol Evol 19: 2265–2276. [DOI] [PubMed] [Google Scholar]
- Bilhère E, Lucas PM, Claisse O, Lonvaud-Funel A. (2009). Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl Environ Microbiol 75: 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma JE, Lenski RE. (1988). Evolution of a bacteria/plasmid association. Nature 335: 351–352. [DOI] [PubMed] [Google Scholar]
- Boussau B, Guéguen L, Gouy M. (2008). Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evol Biol 8: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Lange RK, Mitchell AC, Havig JR, Hamilton TL, Lafrenière MJ et al. (2011). Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem. Appl Environ Microbiol 77: 4778–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L et al. (2009). Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA 106: 21848–21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML et al. (2012). Patterns of gene flow define species of thermophilic Archaea. PLoS Biol 10: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, Konstantinidis KT. (2012). Bacterial species may exist, metagenomics reveal. Environ Microbiol 14: 347–355. [DOI] [PubMed] [Google Scholar]
- Caro-Quintero A, Ritalahti KM, Cusick KD, Löffler FE, Konstantinidis KT. (2012). The chimeric genome of Sphaerochaeta: nonspiral spirochetes that break with the prevalent dogma in spirochete biology. mBio 3: : e00025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF, Hopwood DA, Kieser T, Thompson CJ. (1982). Gene cloning in Streptomyces. Curr Top Microbiol Immunol 96: 69–95. [DOI] [PubMed] [Google Scholar]
- Cheng K, Rong X, Pinto-Tomás AA, Fernández-Villalobos M, Murillo-Cruz C, Huang Y. (2015). Population genetic analysis of Streptomyces albidoflavus reveals habitat barriers to homologous recombination in the diversification of streptomycetes. Appl Environ Microbiol 81: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy CJ, Cook JA. (2000). Phylogeography of a post-glacial colonizer: Microtus longicaudus (Rodentia: Muridae). Mol Ecol 9: 165–175. [DOI] [PubMed] [Google Scholar]
- Cox C, Moore P. (2000) Biogeography: An Ecological and Evolutionary Approach. Blackwell Science: London, UK. [Google Scholar]
- Darch SE, McNally A, Harrison F, Corander J, Barr HL, Paszkiewicz K et al. (2015). Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep 5: 7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M-R, Guo J, Li X, Zhu C-H, Zhu H-H. (2011). Granaticins and their biosynthetic gene cluster from StreptomycesVietnamensis: evidence of horizontal gene transfer. Antonie Van Leeuwenhoek 100: 607–617. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. (2007). Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Buckley DH. (2011). A model for the effect of homologous recombination on microbial diversification. Genome Biol Evol 3: 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Buckley DH. (2014). Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genomics 15: 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Buckley DH. (2010). Widespread homologous recombination within and between Streptomyces species. ISME J 4: 1136–1143. [DOI] [PubMed] [Google Scholar]
- Doroghazi JR, Metcalf WW. (2013). Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenlord SD, Zak DR, Upchurch RA. (2012). Dispersal limitation and the assembly of soil Actinobacteria communities in a long-term chronosequence. Ecol Evol 2: 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. (2009). The bacterial species challenge: making sense of genetic and ecological diversity. Science 323: 741–746. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. (2007). Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Gaudêncio SP, Fenical W, Jensen PR. (2010). Sequence-based analysis of secondary-metabolite biosynthesis in marine Actinobacteria. Appl Environ Microbiol 76: 2487–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zheng W, Rong X, Huang Y. (2008). A multilocus phylogeny of the Streptomyces griseus 16 S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58: 149–159. [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Peters JW, Skidmore ML, Boyd ES. (2013). Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J 7: 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Spratt BG. (2005). Fuzzy species among recombinogenic bacteria. BMC Biol 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 497–506. [DOI] [PubMed] [Google Scholar]
- Hernández-López A, Chabrol O, Royer-Carenzi M, Merhej V, Pontarotti P, Raoult D. (2013). To tree or not to tree? Genome-wide quantification of recombination and reticulate evolution during the diversification of strict intracellular bacteria. Genome Biol Evol 5: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. (2000). The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci 359: 183–195; discussion 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58: 247–276. [Google Scholar]
- Holliday VT, Knox JC, Running IV GL, mandel RD, Ferring CR. (2002) The central lowlands and great plains. In: The Physical Geography of North America Orme AR (ed). Oxford University Press: New York, NY, USA, pp 335–362. [Google Scholar]
- Hopwood DA, Lydiate DJ, Malpartida F, Wright HM. (1985). Conjugative sex plasmids of Streptomyces. Basic Life Sci 30: 615–634. [DOI] [PubMed] [Google Scholar]
- Huang C-L, Pu P-H, Huang H-J, Sung H-M, Liaw H-J, Chen Y-M et al. (2015). Ecological genomics in Xanthomonas: the nature of genetic adaptation with homologous recombination and host shifts. BMC Genomics 16: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinforma Oxf Engl 14: 68–73. [DOI] [PubMed] [Google Scholar]
- Kataoka M, Seki T, Yoshida T. (1991). Regulation and function of the Streptomyces plasmid pSN22 genes involved in pock formation and inviability. J Bacteriol 173: 7975–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi H. (2011). Giant linear plasmids in Streptomyces: a treasure trove of antibiotic biosynthetic clusters. J Antibiot (Tokyo) 64: 19–25. [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. (1989). Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol 29: 170–179. [DOI] [PubMed] [Google Scholar]
- Kishino H, Miyata T, Hasegawa M. (1990). Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol 31: 151–160. [Google Scholar]
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM et al. (2008). Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci USA 105: 2504–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M et al. (2012). Phylogenetic study of the species within the family. Streptomycetaceae. Antonie Van Leeuwenhoek 101: 73–104. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma Oxf Engl 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lomovskaya ND, Voeykova TA, Mkrtumian NM. (1977). Construction and properties of hybrids obtained in interspecific crosses between Streptomyces coelicolor A3(2) and Streptomycesgriseus Kr-15. J Gen Microbiol 98: 187–198. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. (2010). RDP3: a flexible and fast computer program for analyzing recombination. Bioinforma Oxf Engl 26: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. (2002). A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milá B, Smith TB, Wayne RK. (2006). Postglacial population expansion drives the evolution of long-distance migration in a songbird. Evol Int J Org Evol 60: 2403–2409. [PubMed] [Google Scholar]
- Newman CE, Austin CC. (2015). Thriving in the cold: glacial expansion and post-glacial contraction of a temperate terrestrial salamander (Plethodon serratus. PloS One 10: e0130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke RT, Zhaxybayeva O, Feil EJ, Sommerfeld K, Muise D, Doolittle WF. (2007). Searching for species in haloarchaea. Proc Natl Acad Sci USA 104: 14092–14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Park B-J, Jung M-Y, Kim S-J, Chae J-C, Roh Y et al. (2011). Influence of deglaciation on microbial communities in marine sediments off the coast of Svalbard, Arctic Circle. Microb Ecol 62: 537–548. [DOI] [PubMed] [Google Scholar]
- Paul S, Linardopoulou EV, Billig M, Tchesnokova V, Price LB, Johnson JR et al. (2013). Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J Bacteriol 195: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier WR. (1994). Ice age paleotopography. Science 265: 195–201. [DOI] [PubMed] [Google Scholar]
- Pointing SB, Bollard-Breen B, Gillman LN. (2014). Diverse cryptic refuges for life during glaciation. Proc Natl Acad Sci USA 111: 5452–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier R, Holsinger KE. (2010). Was it an explosion? Using population genetics to explore the dynamics of a recent radiation within Protea (Proteaceae L.). Mol Ecol 19: 3968–3980. [DOI] [PubMed] [Google Scholar]
- Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. (2009). Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci USA 106: 8605–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Lizotte-Waniewski M. (2009). Population genomics and the bacterial species concept. Methods Mol Biol Clifton NJ 532: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X, Doroghazi JR, Cheng K, Zhang L, Buckley DH, Huang Y. (2013). Classification of Streptomyces phylogroup pratensis (Doroghazi and Buckley, 2010) based on genetic and phenotypic evidence, and proposal of Streptomycespratensis sp. nov. Syst Appl Microbiol 36: 401–407. [DOI] [PubMed] [Google Scholar]
- Segerman B. (2012). The genetic integrity of bacterial species: the core genome and the accessory genome, two different stories. Front Cell Infect Microbiol 2: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G et al. (2012). Population genomics of early events in the ecological differentiation of bacteria. Science 336: 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MCJ. (2008). Convergence of Campylobacter species: implications for bacterial evolution. Science 320: 237–239. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. (1999). Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16: 1114. [Google Scholar]
- Smith NH, Dale J, Inwald J, Palmer S, Gordon SV, Hewinson RG et al. (2003). The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc Natl Acad Sci USA 100: 15271–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibal M, Hasan F, Wadham JL, Sharp MJ, Anesio AM. (2012). Prokaryotic diversity in sediments beneath two polar glaciers with contrasting organic carbon substrates. Extrem Life Extreme Cond 16: 255–265. [DOI] [PubMed] [Google Scholar]
- Strimmer K, Rambaut A. (2002). Inferring confidence sets of possibly misspecified gene trees. Proc Biol Sci 269: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson NG, Howard DJ. (2005). Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat 166: 581–591. [DOI] [PubMed] [Google Scholar]
- Tavaré S. (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. In: American Mathematical Society: Lectures on Mathematics in the Life Sciences Vol. 17. American Mathematical Society: Providence, RI, USA, pp 57–86. [Google Scholar]
- Templeton AR. (2008). The reality and importance of founder speciation in evolution. BioEssays News Rev Mol Cell Dev Biol 30: 470–479. [DOI] [PubMed] [Google Scholar]
- Vercken E, Fontaine MC, Gladieux P, Hood ME, Jonot O, Giraud T. (2010). Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLoS Pathog 6: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsigian K, Goldenfeld N. (2005). Global divergence of microbial genome sequences mediated by propagating fronts. Proc Natl Acad Sci USA 102: 7332–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Chang HM, Lin YS, Huang CH, Chen CW. (1999). Streptomyces genomes: circular genetic maps from the linear chromosomes. Microbiol Read Engl 145(Pt 9): 2209–2220. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301: 976–978. [DOI] [PubMed] [Google Scholar]
- Wilson AB, Eigenmann Veraguth I. (2010). The impact of Pleistocene glaciation across the range of a widespread European coastal species. Mol Ecol 19: 4535–4553. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT et al. (2009). On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci USA 106: 5865–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci USA 111: E1130–E1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.