Summary
Soil microbial communities are essential for ecosystem function, but linking community composition to biogeochemical processes is challenging because of high microbial diversity and large spatial variability of most soil characteristics. We investigated soil bacterial community structure under switchgrass planted on soil historically supporting grassland vegetation at high spatial resolution to determine whether biogeographic trends occurred at the centimeter scale. Moreover, we tested whether such heterogeneity, if present, influenced community structure between treatments within our field site or among ecosystems at a global scale. Pronounced heterogeneity was observed at centimeter scales, with abrupt changes in relative abundance of phyla from sample to sample. At the ecosystem scale (>10 m), however, bacterial community composition and structure were subtly, but significantly, altered by fertilization, with higher alpha diversity in fertilized plots. Moreover, by comparing these data with data from 1,772 soils from the Earth Microbiome Project, we found that 20% of bacterial taxa were shared between our site and diverse globally sourced soil samples, while grassland soils shared ∼40% of their OTUs with the current study. By spanning several orders of magnitude, our analysis suggests that extreme patchiness characterizes community structure at smaller scales but that coherent patterns emerge at longer length scales.
Keywords: biogeography, microbial community, 16S rRNA, Earth Microbiome Project, soil, bacteria, Panicum virgatum
Introduction
Soil microbial communities are essential to the character of terrestrial ecosystems because they are largely responsible for decomposition of organic residues, mineralization of nutrients, and formation of stable soil organic matter (Cotrufo et al., 2013). The structure of microbial communities (presence and relative abundances of various organisms) can have important ramifications for these processes. For instance, microbial diversity promotes plant coexistence and ecosystem functions such as productivity and N cycling (Miki et al., 2010; Philippot et al., 2013), and shifts in microbial community structure can affect important processes such as plant litter decomposition (Strickland et al., 2009; McGuire and Treseder, 2010) and denitrification rates (Philippot et al., 2009; Jones et al., 2014). However, quantifying the effects of microbial community structure on such functions has been challenging because soil microbial communities are highly diverse (Fierer et al., 2007) and microbial communities, like other soil characteristics, are extremely variable across spatial scales (Ettema and Wardle, 2002).
Microbial biogeography has been examined over regional to continental gradients (Fierer et al., 2009; Bru et al., 2011; Barberán et al., 2012; Fierer, et al., 2012; Fierer et al., 2013; Liu et al., 2014; Whitaker et al., 2014), and at ranges of 1 m to a few 100 m within a plot (Klironomos et al., 1999; Franklin & Mills, 2003; Noguez et al. 2005; Zhou et al., 2008; Philippot et al., 2009; Ushio et al., 2010; Correa-Galeote et al., 2013; Barberán et al., 2014), but the centimeter scale has been explored in only a few cases (Morris, 1999; Grundmann and Debouzie, 2000; Franklin and Mills, 2003; Oline et al., 2006; Keil et al., 2011). Communities are known to differ among widely different biomes (e.g., Fierer, et al., 2012), or across regional gradients (e.g., Griffiths et al., 2011; Ranjard et al., 2013). Evidence for biogeographical patterns among microbes is also available at smaller scales (e.g. Noguez et al., 2005). Several studies have revealed striking heterogeneity in community fingerprints (Franklin and Mills, 2003) and N-cycle genes (Keil et al., 2011) over distances shorter than 1 m and in microbial biomass (Morris, 1999) and nitrifiers (Grundmann and Debouzie, 2000) across distances of only a few centimeters. Still, the very large sample volumes typically collected in a soil core are so large compared to microbial cell size that bacterial biogeographical patterns at scales shorter than 1 m are virtually unknown (Vos et al., 2013). Thus, it is still unclear how small-scale spatial heterogeneity influences the relative abundance of the myriad taxa that constitute soil communities, or how this influences spatial patterns at ecosystem or continental scales.
Understanding the small-scale distributions of soil microbial communities has several benefits. For example, it could narrow down the factors driving community structure patterns and link that structure to observed heterogeneity in biogeochemical processes. Such mechanisms could then be included in Earth system models, which currently treat microbial communities as a black box represented by a fixed or variable parameter (Todd-Brown et al., 2011) despite growing evidence that explicitly including microbial processes in global soil carbon models can improve spatial representation of soil C by as much as 50% (Wieder et al., 2013). Characterizing spatial variability of microbial communities at short length scales has practical implications as well. First, information on the natural variability of a parameter is required for designing experiments with adequate sampling replication and for making inferences based on the results (Klironomos et al., 1999; Peigné et al., 2009). Second, spatial variability could hamper detection of the many microbial and ecosystem processes that are dynamic over short time scales (i.e., minutes to days). Given the paucity of sensors to conduct real-time, nondestructive measurements of microbial activity (metabolism, biomass, physiology) and the rates of resultant biogeochemical processes, destructive serial sampling is necessary in order to construct time series datasets. However, little evidence exists to support the somewhat optimistic assumption that two points close in space are similar enough that they can reliably serve as temporal replicates.
Here we applied an intensive sampling scheme to determine whether the fine-scale (cm) geographic distribution of bacterial communities leads to differences in community structure at larger scales, i.e. within an experimentally altered ecosystem (m) or among ecosystems (km). We used a high-resolution sampling approach to characterize bacterial communities in a grassland soil and a replicated fertilization experiment with switchgrass to test if nutrient availability impacts community structure or its spatial structure. We analyzed these soil community profiles using the 16S rRNA amplicon sequencing protocols outlined by the Earth Microbiome Project (EMP; www.earthmicrobiome.org (Gilbert et al., 2014)) and then compared them to communities found in other studies sequenced by the EMP. We hypothesized that (1) communities would exhibit significant spatial autocorrelation at the centimeter scale, given that the soil type and plant community were consistent among samples; (2) fertilizer application would increase the range of spatial autocorrelation by smoothing uneven resource availability; and (3) few OTUs would be shared between the switchgrass experimental site and disparate soil ecosystem types (e.g., desert, forest) in the EMP database, demonstrating that bacterial populations are more geographically restricted in soils than in other well-mixed ecosystems such a the marine environment (e.g., Gibbons et al., 2013).
Results
Relationships between edaphic variables and bacterial community composition
Twenty-eight of the 750 samples collected were excluded from the final analysis because of inadequate soil volume, sequence depth below the rarefacation threshold, or poor sequence quality. Gravimetric soil moisture was 36%, on average (SE = 0.26), and was unaffected by fertilizer or block (treatment P = 0.19, block P = 0.08, n = 6). Moisture did not predict the Shannon index (P= 0.82, n=30), total species richness (P = 0.17, n = 30), or proportion of the dominant group, Verrucomicrobia (P = 0.95, n = 30). Soil pH (measured on dry material composited by grid after gravimetric moisture quantification), did not differ between fertilization treatments (fertilized = 5.58, unfertilized = 5.60; P = 0.616, n = 6). Relative abundance of Verrucomicrobia, richness, and moisture were not good predictors of CO2 efflux (n = 24; P = 0.612, 0.182, and 0.100, respectively). Total carbon (39.9 ± 0.5 mg g-1) and total N (3.46 ± 0.04 mg g-1) did not differ between treatments (C: P = 0.64 N: P = 0.71) and did not explain variation in bacterial diversity (Shannon index; C: P = 0.891 N: P = 0.618, n= 48; Fig. S2) or proportion of Verrucomicrobia (C: P = 0.9650 N: P = 0.750, n = 48).
Centimeter-scale spatial structure of bacterial communities
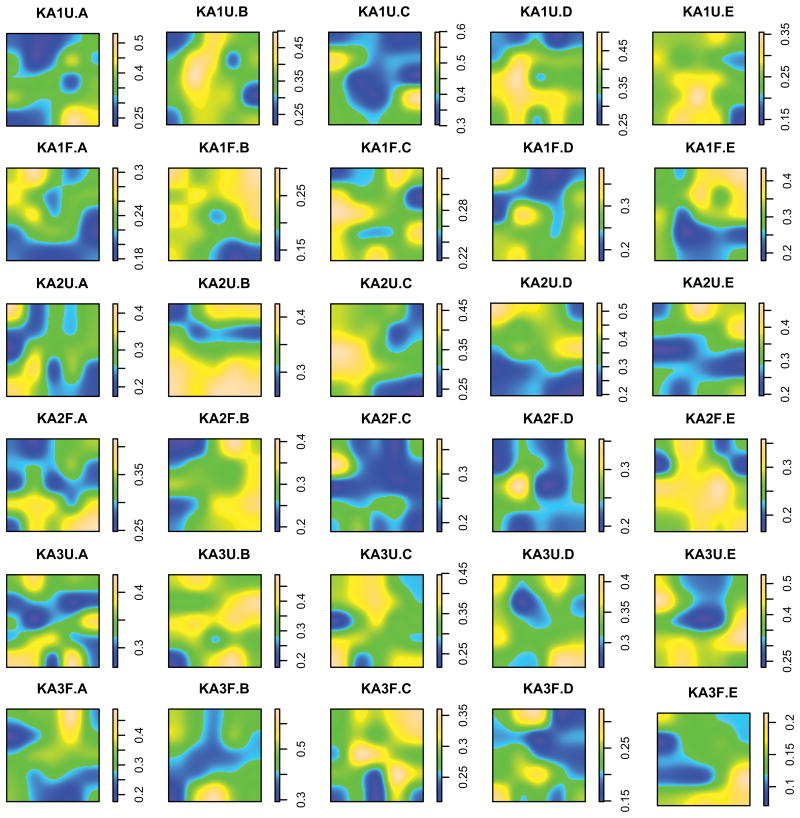
Bacterial community structure was remarkably patchy at centimeter distance scales. For instance, relative abundances of Verrucomicrobia (the dominant phylum) ranged roughly 2.5-fold within any single 10 cm × 10 cm grid (average min = 0.18 ± 0.01, average max = 0.46 ± 0.02; Table 1; Figs. 1, 2a). Although other phyla were less abundant, the relative magnitude of core-to-core variability was similar (Table 1). No significant co-occurrence structure was observed between samples in the full dataset (C-score = 0.46; P = 0.61; 20,000 iterations) or within treatments (fertilized C-score = 0.44, P = 0.52; unfertilized C-score = 0.49, P = 0.76) and few significant correlations were observed among taxa (Fig S3). Furthermore, Moran's I, a metric of spatial autocorrelation, was significant for only 15% of instances tested (six dominant taxonomic groups [phyla or, in the case of Proteobacteria, classes] in thirty grids yielded twenty-seven significant cases), and the magnitude of the effect was nearly zero (-0.024 ± 0.003), indicating a lack of coherent spatial structure at centimeter length scales (Table S2). Similarly, Mantel tests of the correlation between pairwise UniFrac distances and spatial distances in each grid yielded no significant results after Bonferroni correction for multiple comparisons (P>0.002), and all were nearly zero (0.039 ± 0.015; Table S3). Despite a low degree of spatial autocorrelation, the degree of spatial patchiness itself was nonrandom. In all thirty grids, the mean pairwise beta diversity distance (weighted UniFrac) between samples within a grid was significantly greater than expected by chance (Table S4).
Table 1. Variation in relative abundance and of ten most abundant phyla (or classes, for Proteobacteria).
| Phylum | Mean | Range | CV |
|---|---|---|---|
| Verrucomicrobia | 0.316 | 0.053–0.701 | 30.9 |
| Acidobacteria | 0.153 | 0.055–0.418 | 23.2 |
| Deltaproteobacteria | 0.088 | 0.029–0.179 | 26.3 |
| Bacteroidetes | 0.077 | 0.017–0.433 | 40.1 |
| Alphaproteobacteria | 0.067 | 0.019–0.157 | 34.1 |
| Betaproteobacteria | 0.057 | 0.012–0.122 | 35.1 |
| Gammaproteobacteria | 0.040 | 0.008–0.361 | 52.9 |
| Actinobacteria | 0.038 | 0.007–0.141 | 50.9 |
| Planctomycetes | 0.031 | 0.007–0.058 | 25.4 |
| Chloroflexi | 0.012 | 0.001–0.031 | 44.2 |
N=722
CV=coefficient of variation
Figure 1.
Spatially explicit, heatmaps representing relative abundance of Verrucomicrobia in thirty 10 cm × 10 cm sampling grids. Values between sampling points were interpolated based on ordinary kriging. Blue represents areas of relatively low abundance, and yellow represents areas of relatively high abundance.
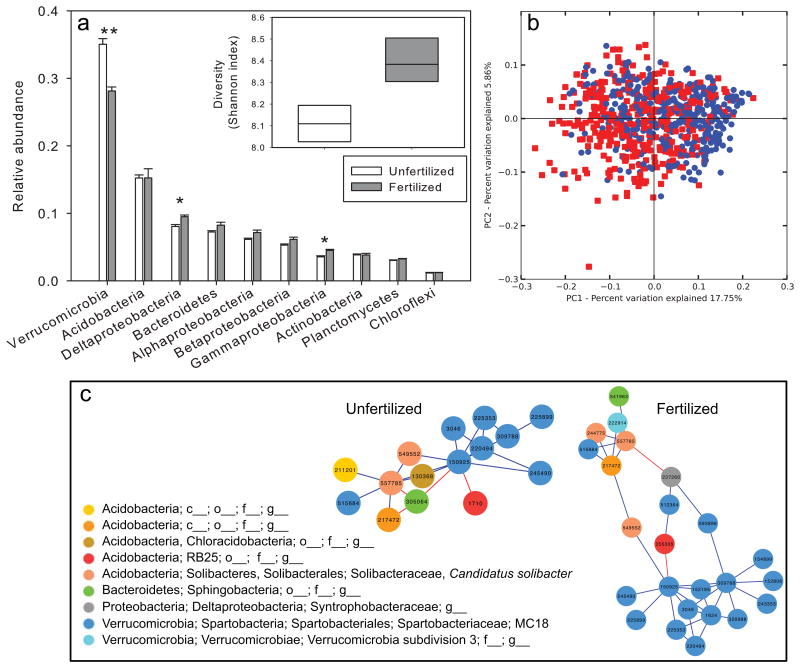
Figure 2.
Comparison of bacterial community structure in fertilized vs. unfertilized soil. (a)Relative abundance of the ten most abundant phyla (or classes, for Proteobacteria). Bars represent means and error bars are se (n=3). Symbols denote significant difference between treatments for that phylum based on Tukey's HSD. On average, 529 ± 7 OTUs were present per sample. * <0.05, **<0.01. (b) Principal coordinates analysis of all 722 individual soil cores based on weighted unifrac distances. Red squares represent samples collected from fertilized plots, and blue circles represent samples from unfertilized plots. (c) Network diagrams of significant SparCC correlations among taxa in unfertilized and fertilized soil. Blue edges indicate positive correlations among taxa, and red edges indicate negative correlations. Nodes are colored by phylogeny (see legend).
Ecosystem-scale bacterial response to fertilizer application
Fertilizer treatment induced a significant or marginally significant increase in most measures of diversity (n = 6; richness: F = 548 ± 14, U = 511 ± 10 P = 0.106; Shannon index: F = 8.40 ± 0.04, U = 8.11 ± 0.03, P = 0.041; Simpson index: F = 0.990 ± 0.003, U = 0.986 ± 0.002, P = 0.045; Chao1: F = 1123 ± 52 U = 1014 ± 14 P = 0.163; Phylogenetic branch length: F = 79.1 ± 0.8, U = 74.8 ± 0.7 P = 0.09; Phylogenetic distance per OTU: F = 0.1451 ± 0.0006, U= 0.1471 ± 0.0004, P = 0.121; Fig 2a) which was attributable to changes in community structure. Verrucomicrobia, the most abundant phylum with 31.6 ± 1.6 % of reads on average, significantly decreased in relative abundance in response to fertilizer treatment (Fig. 2a; Table 1). Of the 248 OTUs whose abundance was significantly different between the fertilized and unfertilized soils (P<0.05 after false discovery rate correction; Table S5), roughly 40% were related to Verrucomicrobia (18.5%) or Acidobacteria (19.4%) (Table S5). In addition, subtler but still significant increases were also observed in Deltaproteobacteria and Gammaproteobacteria (Fig. 2a). A small but significant difference in beta diversity (weighted UniFrac, ANOSIM P-value < 0.05) was observed, although principal coordinates analysis showed no clear differences between treatments (Fig. 2b). Fertilizer treatment resulted in a larger OTU-OTU correlation network, with 24 OTUs compared to 14 OTUs in the network from unfertilized plots (Fig 2c). The most-connected OTUs (highest degree nodes) were Candidatus Solibacter in the Acidobacteria phylum and three members of the MC18 genus in the Spartobacteriaceae family of the Verrrucomicrobia phylum. No differences in spatial autocorrelation were detected, with no significant differences in mean Moran's I between fertilized and unfertilized treatments (Acidobacteria P = 0.40, Alphaproteobacteria P = 0.53, Betaproteobacteria P = 0.818, Deltaproteobacteria P = 0.753, Gammaproteobacteria P = 0.159, Verrucomicrobia P = 0.975; Table S2).
Regional to global-scale community overlap
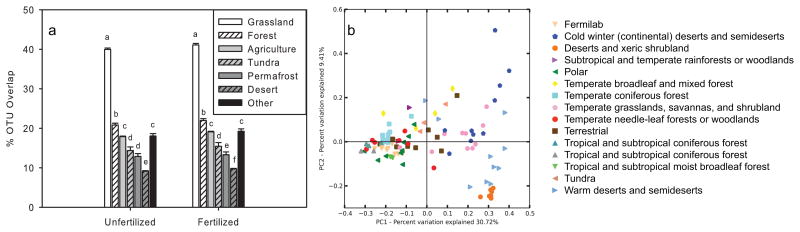
We compared the diversity present in our dataset against that in other soils that were similarly processed to test whether observed diversity scales with sequencing effort or if substantially more data would reveal a “core” microbiome for soil (similar to methods in Gibbons et al., 2013). The 1,772 EMP soils samples had 2,041 ± 17 OTUs on average, whereas the “deeply sequenced” fertilized and unfertilized Fermilab samples had 86,023 and 76,851 OTUs, respectively. On average 22% of the OTUs (97% clusters) found in individual EMP soil samples were also observed in soil from our experimental site (Fig. 3a). Little clustering was found among EMP sites, although separations along the first axis appear to be related to moisture availability (Fig. 3b). OTUs in the Verrucomicrobia phylum were the most commonly shared organisms between our study and at least 69% of the EMP soil datasets (Fig. S4), with a member of the DA101 genus of the Chthoniobacteraceae family being highly abundant in our soils and present in 70% of the EMP samples. When the “core” community (i.e., those organisms found in both our data and in EMP soil data) was defined by a lower threshold of shared sites (e.g., shared with 15% of EMP sites), its membership was more diverse. The OTU overlap was even higher when EMP grassland soils were considered separately, with an average of 40% of the taxa in individual EMP grassland samples appearing in the “deeply sequenced” Fermilab soil (Fig. 3a).
Figure 3.
Comparison of bacterial community structure in Fermilab soils vs. other soils in the Earth Microbiome Project database. (a) Proportion of operational taxonomic units (OTUs) present in both composite Fermilab soils (unfertilized and fertilized treatments) and soils in the EMP10k database from various biomes. Letters indicate significantly different overlap for that treatment among biomes (P<0.05 based on one-way ANOVA followed by means separation by least significant difference). Sample sizes: Grassland = 337, Forest = 261, Agriculture = 696, Tundra = 136, Permafrost = 66, Desert = 104, Other = 172. (b) Principal coordinates analysis with 10 randomly selected representatives from Fermilab and soils from terrestrial biomes in the EMP database. Biomes were identified from user-generated metadata.
Discussion
Centimeter-scale spatial structure of bacterial communities
Our sampling scheme revealed unexpected heterogeneity in soil community structure at centimeter-distance scales, with the relative abundance of bacterial taxa varying considerably between soil cores collected only centimeters apart (Fig 1; Table 1; Table S2; Table S3). Such heterogeneity could arise from some unmeasured environmental parameter that varies stochastically across the spatial scales sampled in this study (Landesman et al., 2014). Indeed, turnover of bacterial diversity has been linked to shifts in edaphic properties over regional gradients (e.g., Ranjard et al., 2013). At the cm-scale, our samples included rhizosphere soil, and so the uneven influence of roots could have contributed to patchiness in bacterial community structure. Because we aimed to characterize bacterial community distribution patterns at small spatial scales, our samples were necessarily too small to measure many other edaphic properties that might explain variability in community structure. However, we also took care to reduce edaphic influences by restricting our sampling to a single soil series (Grays silt loam). In addition, heterogeneity could arise from fine-scale historical environmental differences (Andersson et al., 2014), or as a result of variation in the size and versatility in bacterial genomes, which are hypothesized to underlie habitat breadth (Barberán et al., 2014). However, bacterial patchiness could also be due to dispersal limitation, which might allow less-adaptable taxa to inhabit niches where they would otherwise be excluded by competition.
In our study, taxa appeared to co-occur randomly rather than in a structured way (Fig. S3; Table S2). Significant correlations between the presence of taxa have been found in other soils. These correlations have been used to identify keystone taxa within land use types (Lupatini et al., 2014) and to assign ecological roles to taxonomic groups across sites (Barberán et al., 2012). Significant spatial autocorrelation at centimeter or meter scales has also been reported (Morris, 1999; Grundmann & Debouzie, 2000; Franklin & Mills, 2003; Noguez et al. 2005; Keil et al., 2011). However, these studies all characterized microbial communities at a much coarser taxonomic resolution than we report here. Our data indicate a lack of co-occurrence structure and a lack of strong spatial autocorrelation, which suggests that bacterial populations are subject to small-scale dispersal limitation. If, as is likely, they respond to variation in environmental factors at scales shorter than 1 cm, it would indicate that the environmental conditions driving niche competition at this site occur at length scales shorter than 1 cm. This might explain why we observed beta diversity within each grid that was greater than expected by chance (Table S4). It is possible that recent disturbances associated with establishing the switchgrass stands disrupted larger-scale patterns, and if so, such patterns could develop again over time.
Heterogeneity in microbial habitats at even smaller scales than we measured—within the inter- and intra-aggregate pore network—undoubtedly drives spatial structure in bacterial communities at scales more relevant to cellular processes. Cells likely inhabit distinct microenvironments that promote diversity by encouraging trade-offs in resource utilization and relieving competition (Vos et al., 2013) whereby isolation derived from low pore connectivity increases diversity (Carson et al., 2010). Community structure has been shown to differ among different size classes of soil aggregates (Davinic et al., 2012), between individual aggregates (Bailey et al., 2013), from aggregate interiors to their surfaces (Chenu et al., 2001; Mummey et al., 2006), among particle size fractions (Neumann et al., 2013), and even on surfaces of clays with differing mineralogy (Carson et al., 2009). Indeed, spatial autocorrelation at sub-millimeter scales has been observed by using direct counts of stained cells from thin sections (Nunan et al., 2002). Microscale habitat diversity (e.g., heterogeneous inter- and intra-aggregate pore networks) would explain why we observed significant within-grid beta diversity values (Table S4) and little correlation between community structure and factors such as soil N and CO2 flux (Fig S2). If the cellular-scale environmental profile varies stochastically with distance, then presence of an organism in one location would have little bearing on the presence of another species in a nearby spot, even within the same small soil core. Indeed, our sampling design captured heterogeneity not observed before, but our samples were still was quite large compared to the scale of bacteria.
Several unmeasured biotic factors could also play a role in the remarkable small-scale heterogeneity we measured. For instance, roots can shape microbial communities by altering the physical environment (e.g. soil water relations, gas exchange; Young et al., 2009), affecting resource availability (e.g., exudation; (Paterson et al., 2007), or by associations with fungi. Fungi might also influence bacterial community structure and contribute to soil biodiversity. Fungi compete with bacteria for resources, consume bacterial biomass or are consumed by mycolytic bacteria, and create unique microhabitats that could drive bacterial niche differentiation (De Boer et al., 2005). Mycorrhizal fungi also exhibit significant spatial heterogeneity (Bahram et al., 2015) that could impact bacterial diversity patterns. This effect might become more important over time in our experiment, since fungal-to-bacterial ratios tend to increase with time since disturbance at this and other sites (Allison et al., 2005; Bach et al., 2010).
If the total bacterial community we measured does not reflect functional heterogeneity of the community, either because of dormancy or high functional redundancy (Lennon and Jones, 2011), then the heterogeneity we observed might not manifest in differential ecosystem function. However, community structure has been shown to affect function (Strickland et al., 2009; Fierer et al., 2011; Philippot et al., 2013; Jones et al., 2014), and community activity has been shown to vary dramatically at centimeter distances (Becker et al., 2006). Further, high fidelity between 16S rRNA profiles and protein-encoding genes from shotgun metagenomic data (Fierer, et al., 2012) suggest that the heterogeneity in community profiles we found is likely meaningful for soil function.
Our findings also have critical implications for design of future soil microbiology studies. Morris (1999) found hotspots and cold spots of microbial biomass that averaged approximately 2 cm, roughly the diameter of commonly used soil cores. Our community composition results are consistent with Morris's biomass findings, implying that multiple cores should be composited or that substantial replication is necessary. Given the variability we observed, we estimate that an ANOVA would require roughly 235 samples to detect significant difference (95% confidence interval) in the relative abundance of the most dominant group (Verrucomicrobia, power = 0.9). These results are particularly important for experiments where samples will be collected at high temporal frequency or where sampling is restricted to small volumes in order to limit site disturbance.
Ecosystem-scale bacterial response to nitrogen addition
Fertilizer induced a significant increase in diversity (Fig 2a), which is consistent with other observations (Turlapati et al., 2013). In contrast, long-term fertilizer suppressed diversity in the organic layer of moist acidic tundra (Campbell et al., 2010) and had no effect on diversity in grassland or agricultural sites (Fierer, et al., 2011). Community composition can affect microbial activity responsible for ecosystem processes such as productivity, litter decomposition, and N cycling, and carbon storage (Carney et al. 2007; Philippot et al., 2009; Strickland et al., 2009; McGuire & Treseder, 2010; Miki et al., 2010; Philippot et al., 2013; Jones et al., 2014). Although fertilizer increased diversity at our site, in contrast to our expectations, the spatial heterogeneity of bacterial communities was similar regardless of N addition (Fig. 1; Table S2), suggesting that fertilizer did not promote resource uniformity at a scale that affected bacterial biogeography. The connections between bacterial diversity, activity, and ecosystem processes such as decomposition have been shown to be more responsive to N than other factors (Matulich and Martiny, 2015), but the mechanisms underlying these observations require further study.
The increase in diversity we detected was attributable to changes in community structure, which is commonly observed with fertilizer addition (Bradley et al., 2006; Turlapati et al., 2013; Pan et al., 2014). This change in structure was most attributable to the Verrucomicrobia (Fig. 2a; Table 1; Table S5). N-induced suppression of Verrucomicrobia could indicate a shift toward a community favoring copiotrophs over oligotrophs, if easing N limitation gives a competitive advantage to more copiotrophic taxa that have higher N demands and specialize on more labile C pools such as root exudates (Bergmann et al., 2011; Ramirez et al., 2012; Fierer et al., 2013). The correlation network was larger and more connected for the fertilized soil (Fig 2c), and together with the increase in Shannon index, suggests perhaps that more taxa were able to thrive via competitive release when N limitation was eased by fertilization. Such changes have been reported before, although there is substantial variation among sites (Ramirez et al., 2012; Pan et al., 2014), which could result from inconsistencies among experimental parameters, the quantity of N added (Fierer, et al., 2011), the length of exposure (Bradley et al., 2006), or differences in the structure of communities before fertilization. Our N addition rate of 67 kg N ha-1 is relatively low, and our experimental period (two years) is relatively short compared with that of other experiments, so we expected the effects to be subtle. Furthermore, the weak relationship between community structure and total soil N (Fig S2) suggests that bacteria were utilizing only a portion of the total N pool or that fertilization effects were indirect (but not a result of fertilizer-induced change in pH, which did not differ between treatments) which is consistent with fertilization experiments in other grasslands (Bradley et al., 2006).
Regional to global-scale community overlap
Despite the centimeter-scale patchiness, we found that on average 22% of the OTUs found in EMP soil samples could be mapped back to our experimental site, suggesting that, as with marine bacteria (Gibbons et al., 2013), soil bacteria demonstrate a more cosmopolitan distribution than previously assumed. Recently, global soil microbial diversity was reported to be well represented within one sampling site, where the site comprised a wide range of soil characteristics (Ramirez et al., 2014). However, our study is the first time such a substantial core community has been identified between samples from a single soil type under a single plant species and sites from around the world. Little clustering was found among EMP sites, although separations along the first axis appear to be related to moisture availability, with deserts tending to be on the right and moister ecosystems tending to be on the left (Fig. 3b). Verrucomicrobia dominated the OTUs shared between our study and at least 69% of the EMP soil datasets (Fig. S4). The most representative OTU, present in 70% of the EMP samples and highly abundant in our data, was a member of the Verrucomicrobia in the DA101 genus of the Chthoniobacteraceae family. Despite its highly cosmopolitan distribution, few cultivated representatives are available. However, organisms in this family are known to utilize saccharides from plant biomass or engage in symbiosis with soil nematodes (Sangwan et al., 2004).
The overlap was even higher when EMP grassland soils were considered separately, with 40% of the taxa in globally distributed EMP grassland soils appearing in the “deeply sequenced” Fermilab samples (Fig. 3a). This degree of overlap in soil, which is similar to the overlap found in marine systems (at an equivalent sampling depth; Gibbons et al., 2013), is somewhat surprising because we would assume that slow mixing of the solid soil matrix generates wide variability in soil microsites. There is some evidence for long distance distribution of bacteria associated with dust (Kellogg and Griffin, 2006), but this regional- to global-scale mechanism for terrestrial redistribution of bacteria is likely much less efficient than the physical mixing of ocean waters. This sharing of taxa is likely indicative of a ubiquitous “seed bank” of soil microorganisms that is selected for and structured (abundance of each taxon) by local edaphic and plant distribution characteristics. Evidence for microbial cosmopolitanism is mixed, however, which could be due to undersampling and the relatively coarse taxonomic resolution of previous analyses (Green and Bohannan, 2006). Moreover, the 97% sequence similarity definition of OTUs (16S rRNA gene) is often inadequate to resolve pertinent functional differences among sites given the ecological distinctions that can be found among closely related bacteria (Eren et al., 2013).
Conclusions
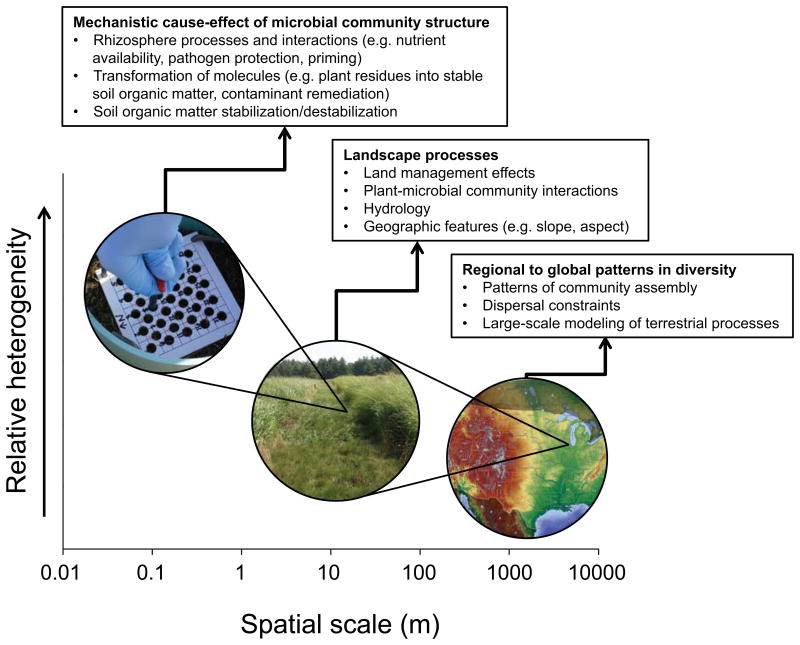
By spanning length scales across several orders of magnitude, our dataset captured a discontinuity in the spatial trends in soil bacterial community structure. This scale-dependent variation in heterogeneity can substantially influence how researchers approach experimental design and analysis (Fig. 4). We liken this to a photomosaic, where a large picture is composed of many smaller photographs; fine details can be observed in any of the small contributing photographs when viewed up close, but they blur together to give way to the larger composite image when viewed from farther away. Similarly, communities in our 100-cm2 grids were enormously patchy, but together they represented a large proportion of the diversity found in grasslands worldwide. Yet, at scales somewhere in between, measurable differences in community structure (e.g., in response to resource manipulation) could be detected. While the implications of spatially nested patterns in soil ecology are manifold, further investigation is essential to determine how hierarchical heterogeneity in soil bacterial communities influences emergent biogeochemical properties at the local and landscape scales.
Figure 4. Conceptual figure highlighting ecological issues that could be impacted by heterogeneity occurring at each of the scales observed in this study.
Experimental Procedures
Site description and field sampling
The study was carried out in the Sustainable Bioenergy Crop Production Research Facility at Fermi National Accelerator Laboratory (Fermilab) in Batavia, IL, USA (88°13′47′W, 41°50′29′N). In the fall of 2007, standing vegetation (a mixture of the perennial, cool-season C3 grasses) was treated with herbicide and removed by burning. The following spring, vegetative regrowth was suppressed with another herbicide treatment. In June 2008, 42 plots (36 × 20 m) were established by no-till drill seeding with seven vegetative treatments replicated six times each over three blocks. Nitrogen fertilizer was applied to half the plots at a rate of 67 kg N ha-1 as urea in early June 2009 and 2010. The soil series was Grays silt loam (2–4% slopes; fine-silty, mixed, superactive, mesic Mollic Oxyaquic Hapludalf; https://soilseries.sc.egov.usda.gov/OSD_Docs/G/GRAYS.html; United States Department of Agriculture, Natural Resources Conservation Service Web Soil Survey). Edaphic variables measured at the time of plot establishment in the areas of sampling are given in Table S1. Average air temperature was 9.7 °C, and average annual precipitation was 797 mm for the period from 2001 to 2010 (www.wunderground.com/history for Batavia, IL).
In this study, we focused on the 6 plots (3 fertilized and 3 unfertilized) that were planted with Kanlow, a lowland cultivar of switchgrass (Panicum virgatum L.), originating from central Oklahoma. By selecting a plant community dominated by a single cultivar of a single species (notwithstanding the presence of non-planted, mostly low-stature cool-season grasses), we minimized spatial variability arising from plant influences (Millard and Singh, 2009; Parker et al., 2012). In late November 2010, we collected soil from five sampling stations placed roughly 6 m apart along a 30 m transect centered on the short edge of each of the six Kanlow plots (Fig. S1). Sampling points were placed 1 m away from the transect in a random compass direction. Four of the sampling stations in each plot were randomly selected for soil CO2 efflux measurement using a LI-COR 8100 in survey mode with a portable chamber (LI-COR Environmental, Lincoln, NE). Collars (20 cm diameter × 10 cm tall) made from PVC pipe were inserted 4 cm into the soil next to a switchgrass crown at each location approximately 24 h before flux measurements were made. Within an hour of each respiration measurement, sterile cork borers (7 mm dia × 50 mm deep, yielding 1.9 g dry soil, on average) were used to collect twenty-five evenly spaced soil samples from a 10 cm × 10 cm grid placed within the collar (stations where respiration measurements were not made were sampled similarly). The samples, still in the borers, were placed individually in sterile plastic collection bags (Whirl-Pak; Nasco, Fort Atkinson, WI) and immediately frozen on dry ice and stored at -80°C at the end of each day. An additional sixteen evenly spaced cores were collected from each grid for gravimetric moisture quantification with the same type of corers used to collect samples destined for DNA extraction. Total carbon and nitrogen (N) were measured on a subset of 50 samples (those from one fertilized and one unfertilized grid) by dry combustion with a Carlo Erba NC2500 elemental analyzer (Milan, Italy).
DNA extraction, amplicon library preparation, and sequencing
Frozen soil was removed from the core tubes and briefly homogenized by manual manipulation through the collection bag, and then subsamples of each soil core were manually loaded into deep 96-well plates included with the PowerSoil®-htp 96 Well Soil DNA Isolation Kit (MO BIO Laboratories, Inc. Carlsbad, CA). DNA was extracted according to the manufacturer's protocol with an additional 20 minute 65°C heating step before being placed on the plate shaker. Fluorometric-based determination of DNA concentration was performed using PicoGreen (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Genomic DNA was amplified by using the Earth Microbiome Project barcoded primer set (http://www.earthmicrobiome.org/emp-standard-protocols/16s/), adapted for the Illumina MiSeq by adding nine extra bases in the adapter region of the forward amplification primer that support paired-end sequencing. The V4 region of the 16S rRNA gene (515F-806R) was amplified with region-specific primers that included the Illumina flowcell adapter sequences. The reverse amplification primer contained a twelve-base barcode sequence that supports pooling of up to 2,167 different samples in each lane(Caporaso, et al., 2010; Caporaso et al., 2012). Each 25 μl PCR reaction contained 12 μl of MO BIO PCR Water (Certified DNA-Free), 10 μl of 5 Prime HotMasterMix (1x), 1 μl of Forward Primer (5μM concentration, 200 pM final), 1 μl Golay Barcode Tagged Reverse Primer (5 μM concentration, 200 pM final), and 1μl of template DNA. The conditions for PCR were as follows: 94 °C for 3 minutes to denature the DNA, with 35 cycles at 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s with a final extension of 10 min at 72 °C to ensure complete amplification. Triplicate PCR amplicon libraries were pooled and then quantified fluorometrically (PicoGreen, Invitrogen), and then pooled again into a single tube so that each amplicon library was represented equally. This pool was cleaned up using UltraClean® PCR Clean-Up Kit (MO BIO), and then quantified using the Qubit (Invitrogen). After quantification, the pool was diluted to 2nM, denatured, and then diluted to a final concentration of 6.1 pM with a 30% PhiX spike for paired-end sequencing (151 bp × 12 bp × 151 bp) on the Illumina MiSeq. Sequence data are available in MG-RAST (metagenomics.anl.gov) under Project ID 342.
Data analysis
Sequence data processing was carried out using the Quantitative Insights into Microbial Ecology pipeline (QIIME; Caporaso, et al., 2010). Data were demultiplexed, the paired reads were joined, and sequences that had any ambiguous bases or a phred score below 20 over the entire read length were discarded. Operational taxonomic units (OTUs) were identified through open-reference OTU picking (97% similarity) against the Greengenes database (2012 release; McDonald et al., 2012). Singleton OTUs were removed and the data were rarefied to 1,000 sequences per sample. Representative sequences for each OTU centroid were aligned with PyNAST (Caporaso, et al., 2010), and phylogenetic trees were constructed using FastTree 2.0 (Price et al., 2010). Alpha diversity metrics (species richness, Shannon diversity, and Simpson diversity) were calculated for all read-depth-normalized samples by using the alpha_diversity.py script in QIIME. For beta-diversity analysis, distance matrices were constructed by using weighted and unweighted UniFrac scores (Lozupone and Knight, 2005). Plotting and statistical analyses were carried out using R v.2.15.2 (R Development Core Team, 2013) and python (Ascher et al., 2001; Rossum and Drake, 2001; Thiruvathukal and Hunter, 2007).
Effects of N fertilization on alpha diversity and phylum relative abundances were tested by using one-way ANOVA with plot as the sampling unit (n = 6). One-way ANOVA was used to compare observed beta diversity (weighted UniFrac distances) with simulated beta diversity in order to test whether the within-grid patchiness was greater than expected by chance (n = 22–25). Beta diversity was simulated by randomly resampling from pooled data for each grid. Spatially explicit heatmaps were generated to visualize the structure of dominant members of the community in each of the thirty 10 cm × 10 cm sampling grids. Values between sampling points were interpolated based on ordinary kriging by using the R package SpatStat (Baddeley and Turner, 2005). Values for missing relative abundance data (see Results) were estimated by interpolation using the R package Zoo before spatial analyses (which prohibit missing data) but were omitted from all other statistical tests. Moran's index was used to test for spatial autocorrelation in relative abundances of the five most abundant phyla (divided by class for Proteobacteria) within each grid using the R package Ape (Paradis et al., 2004). Mantel tests were used to test for correlations between similarity in overall community structure (weighted UniFrac distance) and spatial distance within each grid using the compare_distance_matrices.py script in QIIME. Checkerboard scores were calculated to test for significant co-occurrence in the entire dataset (Stone and Roberts, 1990) by using the R package Vegan (Oksanen et al., 2014), and Spearman's correlation was used to test for rank-order relationships among the presence of individual taxonomic groups (phylum, class, and family levels). Additional OTU-OTU correlations were conducted using SparCC, which appropriately handles compositional data, and significant correlative relationships were visualized using network analysis (Friedman and Alm, 2012). Data were grouped by treatment, and rare taxa (<0.1% of the community) were pruned, leaving 150–170 OTUs. Resulting OTU tables were normalized (500 sequences per sample) and then tested for SparCC correlations (pairwise OTU-OTU correlations, with 5 iterations). Bootstrapping (500 times) on randomized OTU tables was used to generate P-values for each SparCC correlation coefficient. Correlations with coefficients smaller than 0.3 (absolute value) or a bootstrap P-value larger than 0.05 were discarded.
We compared the diversity present in our dataset against that in other soils that were similarly processed to test whether observed diversity scales with sequencing effort, or if substantially more data would reveal a “core” microbiome for soil (similar to methods in Gibbons et al., 2013). Combining EMP soil 16S rRNA data with the data from this study, OTUs were repicked by using open reference workflow (Gibbons et al., 2013). EMP samples were rarified to 5,000 sequences per sample, whereas our data were pooled into two samples representing “deeply sequenced” soil (one fertilized and one unfertilized). These two “samples” were rarified to 1,000,000 sequences each and then compared with EMP soil data (∼1,700 samples) by dividing the number of co-occurring OTUs by the total OTU richness for each sample to yield a percentage of shared community.
Results are presented as means ± standard error. Heatmap, bar, and box-plots were generated using R v.2.15.2. Principal coordinate plots were constructed using the beta_diversity_through_plots.py script in QIIME.
Supplementary Material
Supplementary Figure 1. The experiment consisted of 36 m × 20 m plots in a block design with fertilizer and control treatments replicated three times (six plots). Five sampling stations were located along a transect centered on the short edge of each plot. Twenty-five soil samples were collected from a 10 cm × 10 cm grid at each station, for a total of 750 soil samples.
Supplementary Figure 2. Relationship between Shannon index and (a) total soil C and (b) total soil N. n = 50.
Supplementary Figure 3. Heatmap representing Spearman's correlation coefficients between phyla (or classes, for Proteobacteria) in Fermilab soil (n = 722). Blue indicates a strong positive correlation, red indicates a strong negative correlation, and white indicates no correlation. Results were similar with few strong correlations at the class or family levels (not shown).
Supplementary Figure 4. OTU membership of the “core” microbial community between EMP and Fermilab soils. Bars represent specific thresholds for membership in the core community, where a taxon from Fermilab must be found in a defined fraction of EMP soil samples (e.g., 15%, 21%). Stacks within bars indicate the proportion of the shared OTUs categorized by phylum, and numbers at the top of each bar indicate how many OTUs were shared.
Supplementary Table 1. Edaphic properties of study soil. Values are mean ± standard error of 5 replicates collected to 15 cm depth in early spring 2007 (as part of the site selection process for establishing the experimental facility).
Supplementary Table 2. Magnitude and significance of Moran's I results for relative abundance of dominant taxa in each of thirty 10cm × 10cm sampling grids (n = 22–25). Values can range from -1 (complete dispersal) to 1 (perfect spatial autocorrelation). Boldface indicates a significant result.
Supplementary Table 3. Magnitude and significance of Mantel test results of correlations between weighted UniFrac distances and geographic distances in each of thirty 10cm × 10cm sampling grids (n = 22–25). Values can range from -1 (perfect negative correlation) to 1 (perfect positive correlation).
Supplementary Table 4. ANOVA results comparing observed beta diversity to beta diversity simulated by randomly resampling pooled data from each grid. n = 22–25 for each.
Supplementary Table 5. Indicator organisms demonstrating differences in abundance in response to fertilizer application (Microsoft Excel Workbook).
Originality-Significance Statement.
We conducted a novel set of analyses examining soil bacterial communities at three scales: centimeter, ecosystem, and regional-to-global. We found that (1) profound heterogeneity in bacterial communities is present at centimeter scales, despite samples being collected from a single soil series under a single plant cultivar; (2) fertilizer addition increased bacterial diversity and connectedness of OTUs, likely by relieving competition for soil nitrogen, but did not change spatial structure of bacterial communities; and (3) bacteria display unexpected cosmopolitanism in soil, where, unlike ocean waters, physical mixing is very slow and so mechanisms of dispersal are unclear. This is the first time centimeter-scale heterogeneity of this magnitude has been observed in soil microbial community structure, while also being the first example of such a large number of taxa shared between global soils and just one site.
Acknowledgments
T. Flynn and R. Matamala engaged in helpful discussion, T. Vugetveen, and J. Droge provided valuable field sampling assistance, and E. Rakowski patiently loaded sample extraction plates. We thank D. Braithwaite, N. Scott, and D. Smith for help with statistical software. We appreciate the constructive comments of five anonymous reviewers. This work was completed in part with resources provided by the University of Chicago Research Computing Center. SMG was supported by an EPA STAR Fellowship and the National Institutes of Health Training Grant 5T-32EB-009412. This material was based upon research supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
Supplementary information is available at Environmental Microbiology's website.
The authors declare no conflict of interest.
References
- Allison VJ, Miller RM, Jastrow JD, Matamala R, Zak DR. Changes in Soil Microbial Community Structure in a Tallgrass Prairie Chronosequence. Soil Sci Soc Am J. 2005;69:1412–1421. [Google Scholar]
- Andersson MG, Berga M, Lindström ES, Langenheder S. The spatial structure of bacterial communities is influenced by historical environmental conditions. Ecology. 2014;95:1134–1140. doi: 10.1890/13-1300.1. [DOI] [PubMed] [Google Scholar]
- Ascher D, Dubois PF, Hinsen K, Hugunin J, T O. Numerical Python, Lawrence Livermore National Laboratory, Livermore, California, USA. 2001 Available at http://www.pfdubois.com/numpy.
- Bach EM, Baer SG, Meyer CK, Six J. Soil texture affects soil microbial and structural recovery during grassland restoration. Soil Biol Biochem. 2010;42:2182–2191. [Google Scholar]
- Baddeley A, Turner R. Spatstat: an R package for analyzing spatial point patterns. J Stat Softw. 2005;12:1–42. [Google Scholar]
- Bahram M, Peay KG, Tedersoo L. Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol. 2015;205:1454–1463. doi: 10.1111/nph.13206. [DOI] [PubMed] [Google Scholar]
- Bailey VL, McCue LA, Fansler SJ, Boyanov MI, DeCarlo F, Kemner KM, Konopka A. Micrometer-scale physical structure and microbial composition of soil macroaggregates. Soil Biol Biochem. 2013;65:60–68. [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ramirez KS, Leff JW, Bradford Ma, Wall DH, Fierer N. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria Ecol Lett. 2014;17:794–802. doi: 10.1111/ele.12282. [DOI] [PubMed] [Google Scholar]
- Becker JM, Parkin T, Nakatsu CH, Wilbur JD, Konopka A. Bacterial activity, community structure, and centimeter-scale spatial heterogeneity in contaminated soil. Microb Ecol. 2006;51:220–231. doi: 10.1007/s00248-005-0002-9. [DOI] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Bradley K, Drijber RA, Knops J. Increased N availability in grassland soils modifies their microbial communities and decreases the abundance of arbuscular mycorrhizal fungi. Soil Biol Biochem. 2006;38:1583–1595. [Google Scholar]
- Bru D, Ramette A, Saby NPA, Dequiedt S, Ranjard L, Jolivet C, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2011;5:532–42. doi: 10.1038/ismej.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EaG. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol. 2010;12:1842–54. doi: 10.1111/j.1462-2920.2010.02189.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high- throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters Wa, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2010;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JK, Campbell L, Rooney D, Clipson N, Gleeson DB. Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol Ecol. 2009;67:381–8. doi: 10.1111/j.1574-6941.2008.00645.x. [DOI] [PubMed] [Google Scholar]
- Carson JK, Gonzalez-Quiñones V, Murphy DV, Hinz C, Shaw Ja, Gleeson DB. Low pore connectivity increases bacterial diversity in soil. Appl Environ Microbiol. 2010;76:3936–42. doi: 10.1128/AEM.03085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenu C, Hassink J, Bloem J. Short-term changes in the spatial distribution of microorganisms in soil aggregates as affected by glucose addition. Biol Fertil Soils. 2001;34:349–356. [Google Scholar]
- Correa-Galeote D, Marco DE, Tortosa G, Bru D, Philippot L, Bedmar EJ. Spatial distribution of N-cycling microbial communities showed complex patterns in constructed wetland sediments. FEMS Microbiol Ecol. 2013;83:340–51. doi: 10.1111/j.1574-6941.2012.01479.x. [DOI] [PubMed] [Google Scholar]
- Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob Chang Biol. 2013;19:988–95. doi: 10.1111/gcb.12113. [DOI] [PubMed] [Google Scholar]
- Davinic M, Fultz LM, Acosta-Martinez V, Calderón FJ, Cox SB, Dowd SE, et al. Pyrosequencing and mid-infrared spectroscopy reveal distinct aggregate stratification of soil bacterial communities and organic matter composition. Soil Biol Biochem. 2012;46:63–72. [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema CH, Wardle DA. Spatial soil ecology. Trends Ecol Evol. 2002;17:177–183. [Google Scholar]
- Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73:7059–66. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Carney KM, Horner-Devine MC, Megonigal JP. The biogeography of ammonia-oxidizing bacterial communities in soil. Microb Ecol. 2009;58:435–45. doi: 10.1007/s00248-009-9517-9. [DOI] [PubMed] [Google Scholar]
- Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, et al. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science. 2013;342:621–4. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2011;6:1007–17. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A. 2012;109:21390–5. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RB, Mills AL. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol. 2003;44:335–46. doi: 10.1016/S0168-6496(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput Biol. 2012;8:1–11. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci U S A. 2013;110:4651–5. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–7. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- Grundmann GL, Debouzie D. Geostatistical analysis of the distribution of NH4+ and NO2- -oxidizing bacteria and serotypes at the millimeter scale along a soil transect. FEMS Microbiol Ecol. 2000;34:57–62. doi: 10.1111/j.1574-6941.2000.tb00754.x. [DOI] [PubMed] [Google Scholar]
- Jones CM, Spor A, Brennan FP, Breuil MC, Bru D, Lemanceau P, et al. Recently identified microbial guild mediates soil N2O sink capacity. Nat Clim Chang. 2014;4:801–805. [Google Scholar]
- Keil D, Meyer A, Berner D, Poll C, Schützenmeister A, Piepho HP, et al. Influence of land-use intensity on the spatial distribution of N-cycling microorganisms in grassland soils. FEMS Microbiol Ecol. 2011;77:95–106. doi: 10.1111/j.1574-6941.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- Kellogg CA, Griffin DW. Aerobiology and the global transport of desert dust. Trends Ecol Evol. 2006;21:638–644. doi: 10.1016/j.tree.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Klironomos JN, Rillig MC, Allen MF. Designing belowground field experiments with the help of semi-variance and power analyses. Appl Soil Ecol. 1999;12:227–238. [Google Scholar]
- Landesman WJ, Nelson DM, Fitzpatrick MC. Soil properties and tree species drive β-diversity of soil bacterial communities. Soil Biol Biochem. 2014;76:201–209. [Google Scholar]
- Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- Liu J, Sui Y, Yu Z, Shi Y, Chu H, Jin J, et al. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol Biochem. 2014;70:113–122. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupatini M, Suleiman AKA, Jacques RJS, Antoniolli ZI, de Siqueira Ferreira A, Kuramae EE, Roesch LFW. Network topology reveals high connectance levels and few key microbial genera within soils. Front Environ Sci. 2014;2 doi: 10.3389/fenvs.2014.00010. [DOI] [Google Scholar]
- Matulich KL, Martiny JBH. Microbial composition alters the response of litter decomposition to environmental change. Ecology. 2015;96:154–163. doi: 10.1890/14-0357.1. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire KL, Treseder KK. Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol Biochem. 2010;42:529–535. [Google Scholar]
- Miki T, Ushio M, Fukui S, Kondoh M. Functional diversity of microbial decomposers facilitates plant coexistence in a plant-microbe-soil feedback model. Proc Natl Acad Sci U S A. 2010;107:14251–6. doi: 10.1073/pnas.0914281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P, Singh BK. Does grassland vegetation drive soil microbial diversity? Nutr Cycl Agroecosystems. 2009;88:147–158. [Google Scholar]
- Morris SJ. Spatial distribution of fungal and bacterial biomass in southern Ohio hardwood forest soils: fine scale variability and microscale patterns. Soil Biol Biochem. 1999;31:1375–1386. [Google Scholar]
- Mummey D, Holben W, Six J, Stahl P. Spatial stratification of soil bacterial populations in aggregates of diverse soils. Microb Ecol. 2006;51:404–11. doi: 10.1007/s00248-006-9020-5. [DOI] [PubMed] [Google Scholar]
- Neumann D, Heuer A, Hemkemeyer M, Martens R, Tebbe CC. Response of microbial communities to long-term fertilization depends on their microhabitat. FEMS Microbiol Ecol. 2013;86:71–84. doi: 10.1111/1574-6941.12092. [DOI] [PubMed] [Google Scholar]
- Noguez AM, Arita HT, Escalante AE, Forney LJ, García-Oliva F, Souza V. Microbial macroecology: Highly structured prokaryotic soil assemblages in a tropical deciduous forest. Glob Ecol Biogeogr. 2005;14:241–248. [Google Scholar]
- Nunan N, Wu K, Young IM, Crawford JW, Ritz K. In situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil. Microb Ecol. 2002;44:296–305. doi: 10.1007/s00248-002-2021-0. [DOI] [PubMed] [Google Scholar]
- Oksanen AJ, Blanchet FG, Kindt R, Legen- P, Minchin PR, Hara RBO, et al. vegan: Community ecology package. R package version 2.0-7. 2014 http://CRAN.R-project.org/package=vegan.
- Oline DK, Schmidt SK, Grant MC. Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microb Ecol. 2006;52:480–490. doi: 10.1007/s00248-006-9101-5. [DOI] [PubMed] [Google Scholar]
- Pan Y, Cassman N, de Hollander M, Mendes LW, Korevaar H, Geerts RHEM, et al. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol Ecol. 2014;90:195–205. doi: 10.1111/1574-6941.12384. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parker SS, Seabloom EW, Schimel JP. Grassland community composition drives small-scale spatial patterns in soil properties and processes. Geoderma. 2012;170:269–279. [Google Scholar]
- Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Peigné J, Vian JF, Cannavacciuolo M, Bottollier B, Chaussod R. Soil sampling based on field spatial variability of soil microbial indicators. Eur J Soil Biol. 2009;45:488–495. [Google Scholar]
- Philippot L, Cuhel J, Saby NPa, Chèneby D, Chronáková A, Bru D, et al. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol. 2009;11:1518–26. doi: 10.1111/j.1462-2920.2009.01879.x. [DOI] [PubMed] [Google Scholar]
- Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, et al. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013;7:1609–19. doi: 10.1038/ismej.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol. 2012;18:1918–1927. [Google Scholar]
- Ramirez KS, Leff JW, Barberan A, Bates ST, Betley J, Crowther TW, et al. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc R Soc B. 2014;20141988 doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjard L, Dequiedt S, Chemidlin Prévost-Bouré N, Thioulouse J, Saby NPA, Lelievre M, et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat Commun. 2013;4:1434. doi: 10.1038/ncomms2431. [DOI] [PubMed] [Google Scholar]
- Van Rossum G, Drake FL. Python Reference Manual, PhythonLabs. Virginia, USA: 2001. Available at http://www.python.org. [Google Scholar]
- Sangwan P, Chen X, Hugenholtz P, Janssen PH. Chthoniobacter flavus gen. nov., sp nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl Environ Microbiol. 2004;70:5875–81. doi: 10.1128/AEM.70.10.5875-5881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone L, Roberts A. The checkerboard score and species distributions. Oecologia. 1990;85:74–79. doi: 10.1007/BF00317345. [DOI] [PubMed] [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- Thiruvathukal EGK, Hunter BJD. MATPLOTLIB: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- Todd-Brown KEO, Hopkins FM, Kivlin SN, Talbot JM, Allison SD. A framework for representing microbial decomposition in coupled climate models. Biogeochemistry. 2011;109:19–33. [Google Scholar]
- Turlapati SA, Minocha R, Bhiravarasa PS, Tisa LS, Thomas WK, Minocha SC. Chronic N-amended soils exhibit an altered bacterial community structure in Harvard Forest, MA, USA. FEMS Microbiol Ecol. 2013;83:478–93. doi: 10.1111/1574-6941.12009. [DOI] [PubMed] [Google Scholar]
- Ushio M, Kitayama K, Balser TC. Tree species-mediated spatial patchiness of the composition of microbial community and physicochemical properties in the topsoils of a tropical montane forest. Soil Biol Biochem. 2010;42:1588–1595. [Google Scholar]
- Vos M, Wolf AB, Jennings SJ, Kowalchuk Ga. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev. 2013;37:936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]
- Whitaker J, Ostle N, Nottingham AT, Ccahuana A, Salinas N, Bardgett RD, et al. Microbial community composition explains soil respiration responses to changing carbon inputs along an Andes-to-Amazon elevation gradient. J Ecol. 2014;102:1058–1071. doi: 10.1111/1365-2745.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieder WR, Bonan GB, Allison SD. Global soil carbon projections are improved by modelling microbial processes. Nat Clim Chang. 2013;3:909–912. [Google Scholar]
- Young IM, Crawford JW, Nunan N, Otten W, Spiers a. Chapter 4 Microbial Distribution in Soils Physics and Scaling. 1st. Elsevier Inc; 2009. [Google Scholar]
- Zhou J, Kang S, Schadt CW, Garten CT. Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci U S A. 2008;105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The experiment consisted of 36 m × 20 m plots in a block design with fertilizer and control treatments replicated three times (six plots). Five sampling stations were located along a transect centered on the short edge of each plot. Twenty-five soil samples were collected from a 10 cm × 10 cm grid at each station, for a total of 750 soil samples.
Supplementary Figure 2. Relationship between Shannon index and (a) total soil C and (b) total soil N. n = 50.
Supplementary Figure 3. Heatmap representing Spearman's correlation coefficients between phyla (or classes, for Proteobacteria) in Fermilab soil (n = 722). Blue indicates a strong positive correlation, red indicates a strong negative correlation, and white indicates no correlation. Results were similar with few strong correlations at the class or family levels (not shown).
Supplementary Figure 4. OTU membership of the “core” microbial community between EMP and Fermilab soils. Bars represent specific thresholds for membership in the core community, where a taxon from Fermilab must be found in a defined fraction of EMP soil samples (e.g., 15%, 21%). Stacks within bars indicate the proportion of the shared OTUs categorized by phylum, and numbers at the top of each bar indicate how many OTUs were shared.
Supplementary Table 1. Edaphic properties of study soil. Values are mean ± standard error of 5 replicates collected to 15 cm depth in early spring 2007 (as part of the site selection process for establishing the experimental facility).
Supplementary Table 2. Magnitude and significance of Moran's I results for relative abundance of dominant taxa in each of thirty 10cm × 10cm sampling grids (n = 22–25). Values can range from -1 (complete dispersal) to 1 (perfect spatial autocorrelation). Boldface indicates a significant result.
Supplementary Table 3. Magnitude and significance of Mantel test results of correlations between weighted UniFrac distances and geographic distances in each of thirty 10cm × 10cm sampling grids (n = 22–25). Values can range from -1 (perfect negative correlation) to 1 (perfect positive correlation).
Supplementary Table 4. ANOVA results comparing observed beta diversity to beta diversity simulated by randomly resampling pooled data from each grid. n = 22–25 for each.
Supplementary Table 5. Indicator organisms demonstrating differences in abundance in response to fertilizer application (Microsoft Excel Workbook).