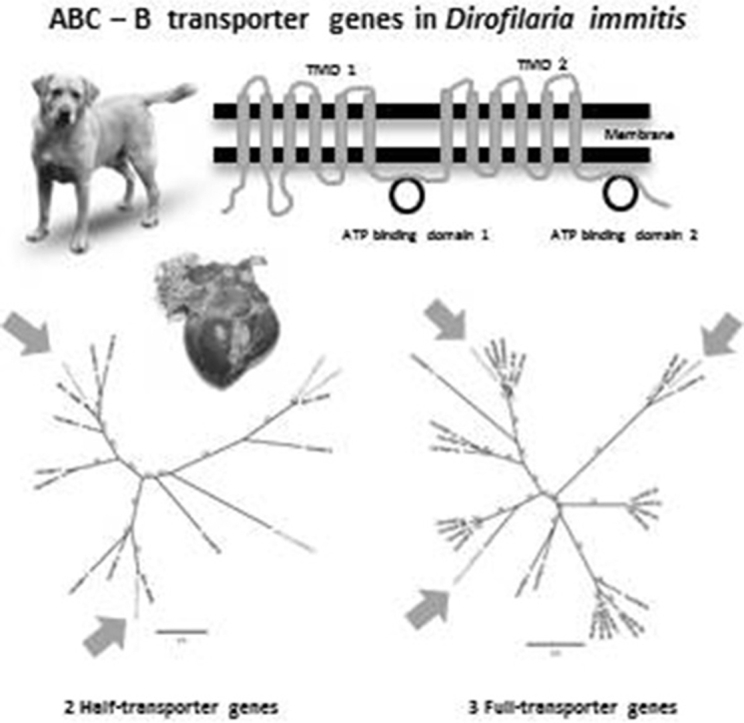
Abstract
Dirofilaria immitis is a filarial nematode causing infection and heartworm disease in dogs and other canids, cats, and occasionally in humans. Prevention with macrocyclic lactones (ML) is recommended during the mosquito transmission season. Recently, ML resistance has been reported. ABC-B transporter genes are thought to be involved in the mechanism of ML resistance in other nematodes. This study aimed to identify all the ABC-B transporter genes in D. immitis using as a reference the nDi.2.2 D. immitis whole genome, which is not completely annotated. Using bioinformatic tools and PCR amplification on pooled D. immitis genomic DNA and on pooled cDNA, nine ABC transporter genes including one pseudogene were characterized. Bioinformatic and phylogenetic analyses allowed identification of three P-glycoproteins (Pgps) (Dim-pgp-3 Dim-pgp-10, Dim-pgp-11), of two ABC-B half transporter genes (one ortholog of Cel-haf-4 and Cel-haf-9; and one ortholog of Cel-haf-1 and Cel-haf-3), of one ABC half transporter gene (ortholog of Cel-haf-5) that contained an ABC-C motif, and of one additional half transporter that would require functional study for characterization. The number of ABC-B transporter genes identified was lower than in Caenorhabditis elegans and Haemonchuscontortus. Further studies are needed to understand their possible role in ML resistance in D. immitis. These ABC transporters constitute a base for ML resistance investigation in D. immitis and advance our understanding of the molecular biology of this parasite.
Keywords: Dirofilaria immitis, P-glycoprotein, Genetic identification
Abbreviations: ML, macrocyclic lactones; LOE, loss of efficacy; Kb, kilobase; Tm, melting temperature; TM, transmembrane domain; Pgp, P-glycoprotein; ATP, adenosine triphosphate; IVM, ivermectin
Graphical abstract
Highlights
-
•
Identification of ABC-B full and half transporter genes in Dirofilaria immitis.
-
•
Phylogenetic analysis of the D. immitis ABC-B transporter genes.
-
•
Lower number of ABC-B transporter genes in D. immitis compared with Clade V nematodes.
1. Introduction
Dirofilaria immitis is a filarial nematode infecting and sometimes causing heartworm disease in dogs and other canids, cats, and occasionally in humans (Lee et al., 2010, Damle et al., 2014). The parasite is transmitted to the host through infected mosquitoes. Chronic heartworm disease includes symptoms such as lethargy, exercise intolerance, loss of appetite, weight loss, coughing, cardiac insufficiency and difficulty breathing. In the host, D. immitis juvenile worms migrate to pulmonary arteries and mature, sometimes leading to the arteries becoming blocked. In heavy infections, adult worms may be present in the heart chambers. These conditions can lead to death of the animal in the most severe cases. Heartworm is distributed worldwide (Morchon et al., 2012, Simon et al., 2012).
Melarsomine, an arsenic-based drug, is used to kill adult worms and worms as young as 4 months old (Raynaud, 1992). Prevention with macrocyclic lactones (ML) is recommended all year-round in USA, or during the mosquito transmission season in other regions. In 2005, a first report on ML loss of efficacy (LOE) in the USA, was published (Hampshire, 2005). Then, other studies on ML LOE and ML resistance in D. immitis have been reported (Bourguinat et al., 2011a, Bourguinat et al., 2011b, Bourguinat et al., 2015, Pulaski et al., 2014).
The mechanism of ML resistance in nematodes is still not well understood. However, several studies reported that ML resistance may be polygenic (Prichard, 2001, Vercruysse and Rew, 2002, McCavera et al., 2007, Sutherland and Scott, 2009). P-glycoproteins (Pgp) have been shown to be implicated in several drug resistance processes such as chemotherapy resistance in tumour cells in humans (Lespine et al., 2012) and also to be associated with ML resistance in C. elegans and in parasitic nematodes (Blackhall et al., 1998, Xu et al., 1998, Ardelli et al., 2005, Ardelli et al., 2006, Ardelli and Prichard, 2007, Prichard and Roulet, 2007, James and Davey, 2009, Lespine et al., 2012, Ardelli, 2013, Janssen et al., 2013a). A correlation between loss of efficacy of ML heartworm preventives and Pgp genotype was reported in D. immitis (Bourguinat et al., 2011b). Pgps are members of the ATP binding cassette family of proteins also called ABC transporters (from A to H) (Ambudkar et al., 2003). Pgps belong to the ABC-B group of transporters. Pgps are full size ABC-B proteins composed of two transmembrane (TM) domains that each contains six TM helices. Each TM domain is followed by a nucleotide binding domain (Ambudkar et al., 2003). The two sections of the protein are connected by a linker region. However, half size ABC-B transporter proteins are composed of one TM, including six TM fragments, and one nucleotide binding domain (Sheps et al., 2004). Pgps play the role of pumps that can enable a substrate to be transported outside the cell using ATP as energy. Substrates of Pgp are neutral and cationic hydrophobic compounds. Ivermectin (IVM) (Lespine et al., 2007, Kerboeuf and Guegnard, 2011) and selamectin (Griffin et al., 2005) are reported to be good substrates for Pgps, with moxidectin being less transported by Pgps (Cobb and Boeckh, 2009, Godoy et al., 2015). So far, 15 Pgps have been reported in C. elegans (Sheps et al., 2004) including a pseudogene (Lespine et al., 2008) and 10 in H. contortus (Laing et al., 2013). Based on their respective genomes, the filarial worms, Brugia malayi, Onchocerca volvulus and Loa loa appear to have fewer Pgps than C. elegans or H. contortus. It is not clear how many Pgps are presents in D. immitis. The purpose of this study was to identify all the ABC-B transporters in heartworm, using the D. immitis whole genome that is available (Godel et al., 2012) but not yet completely annotated. The corresponding information obtained is valuable to improve knowledge of the structure of ABC-B transporters genes in D. immitis which may be useful for further investigation of ML resistance mechanisms in D. immitis. Also this information could be used to identify potential additional genetic marker that could predict ML resistance in D. immitis, and adds to our knowledge of the molecular biology of this parasite.
2. Material and methods
2.1. Bioinformatic identification of D. immitis ABC-B transporter genes
All ABC-B transporter genes reported (Sheps et al., 2004, Lespine et al., 2008) in the nematode model organism C. elegans (25 genes) were collected from WormBase (http://www.wormbase.org) and from GenBank NCBI (http://www.ncbi.nlm.nih.gov/). Also, based on the analysis of 15 genomic sequences that could predict ATP-binding cassette (ABC) systems in B. malayi (Ardelli et al., 2010), Pgp genes were identified using NCBI Blast tool, BLASTN 2.3.1+ (Zhang et al., 2000) and BLASTX 2.3.1+ (Altschul et al., 1997) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Additional Blast analysis on these 15 sequences was performed with the Broad Institute Blast tools (http://www.broadinstitute.org/), to identify additional sequences in filarial nematodes. In total, 69 sequences were collected from different organisms (Supplementary data S1): 25 in C. elegans, 9 in Caenorhabditis remanei, 9 in Loa. loa, 15 in B. malayi, 1 in D. immitis, 5 in Wuchereria bancrofti and 5 in O. volvulus. These 69 sequences were used to interrogate the D. immitis genome nDi.2.2 (http://xyala.cap.ed.ac.uk/downloads/959nematodegenomes/blast/filareu.php) with BLASTN 2.2.25 and TBLASTN 2.2.25 (Altschul et al., 1997). This allowed location of ABC-B transporter genes on D. immitis scaffolds. Additionally, sequences annotated as nematode orthologs of Pgps in the nDi.2.2 genome browser (http://salmo.bio.ed.ac.uk/cgi-bin/gbrowse/gbrowse/nDi.2.2.2/) were collected.
2.2. Confirmation sequencing of ABC-B transporter genes
Confirmation of sequences was performed from pooled D. immitis samples shipped from TRS Labs (TRS Labs Inc., Athens, GA, USA) on dry ice. Worms were thawed in petri dishes containing RNAlater™ RNA Stabilization Reagent (Qiagen Inc., Toronto, ON, Canada). The worms were sectioned: one third of each worm was used for gDNA isolation using DNeasy®Blood and Tissue kit (Qiagen). Two third of each worm was used for RNA isolation, using TRIzol®Reagent (Ambion®, Life technologies™ Inc., Burlington, ON, Canada). QuantiTect Reverse Transcription kit (Qiagen) was used to prepare cDNA.
Several specific primers per gene (Supplementary data S2) were designed, based on the genomic DNA of the nDi.2.2 D. immitis genome, from the 7 scaffolds identified from Blast analysis and from 1 scaffold identified from the genome browser, to amplify full length genomic DNA sequences of full and half ABC-B transporter genes. Simultaneously, open reading frame (ORF) finder from Geneious Pro.5.6.3 software (http://www.geneious.com/; Kearse et al., 2012) was used to predict cDNA sequences. Forward splice leader SL1 (Blaxter and Liu, 1996) primer and reverse specific primers (Supplementary data S2) in the 3′UTR region were used to amplify full cDNA sequences of ABC-B transporter genes.
One microliter of 10 μM forward and reverse primers, 2 μl of 50 mM MgSO4 and 1 μl of 10 mM dNTPs were used with 0.3 μl High Fidelity Platinium® Taq DNA polymerase (Invitrogen Inc., Burlington, ON, Canada) in 50 μl reaction for all PCR amplifications. DNA template was adjusted to 80 ng. PCR amplifications were performed on a MJ Research PTC-200 thermal cycler following the cycling parameters: an initial denaturation step at 94 °C for 2 min followed by 36 amplification cycles, with each cycle including a denaturation step at 94 °C for 30 s, an annealing step at Tm°C, depending on the primer set (Tm included in Supplementary data S2) for 30 s and an extension step at 68 °C for 1 min/kb. A final extension was performed at 68 °C for 10 min.
PCR products were examined by agarose gel electrophoresis. When multiple amplicons were observed on agarose gels, bands were cut, DNA purified using Zymoclean™ Gel DNA Recovery Kit (Zymo Research Corporation, CA, USA) and Sanger sequenced (Sanger and Coulson, 1975, Sanger et al., 1977) using a 3730XL DNA Analyzer system at McGill University/Génome Québec Innovation Centre. When only one band was observed on agarose gels, PCR products were directly sent for Sanger sequencing at McGill University/Génome Québec Innovation Centre. All the sequences obtained from the PCR amplicons were assembled and analysed using Geneious Pro.5.6.3, Sequencher® version 4.10 software (Gene Codes Corporation, Ann Arbor, MI, USA) and the nuclear genome assembly nDi.2.2 as D. immitis reference genome (http://nematodes.org/genomes/dirofilaria_immitis/).
2.3. Identification of ABC transporter protein features
Amino acids were predicted from cDNA sequences using Geneious Pro.5.6.3 software translation tool. Bioinformatics Resource Portal called ScanProsite tool (released 20.123; http://prosite.expasy.org/scanprosite/) was used to identify the number and positions of transmembrane domains, ATP-binding cassette domains and ABC signature motifs of each predicted protein sequence from cDNA sequences. Based on the location of the predicted TM, the predicted number of transmembrane helices within the TMs were estimated using Transmembrane Hidden Markov models (TMHMM) (v2.0) (Sonnhammer et al., 1998) in Geneious Pro.5.6.3 software. Conserved Walker A and Walker B sites were located with NCBI server using BLASTP 2.3.1+ (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1997).
2.4. Phylogenetic analysis
ABC-B transporter amino acid sequences from C. elegans, B. malayi, O. volvulus and L. loa, listed in Table 1, were included in the phylogenetic analysis with the different predicted protein sequences from D. immitis. Only complete amino acid sequences were included in the analyses. The different isoforms of Cel-PGP-2, Cel-PGP-5 and Cel-PGP-6 were also included in this analysis. The two ClustalW alignments of full length amino acid sequences for ABC–B full and half transporters were performed with Geneious Pro.5.6.3 software using Matrix Blosum. The two alignments available in FASTA format were trimmed as indicated in the Supplementary data S3 for the full transporters and in Supplementary data S4 for the half transporters. The criterion for trimming was to work on a common region, for each of the amino acid sequences, that started and finished with a conserved region. An exception was made for Cel-PGP-5b and Cel-PGP-6b as they had shorter complete amino acid sequences compared to the others. Subsequently, with Geneious Pro version 5.6.3, the corresponding two un-rooted phylogenetic trees were built from the trimmed alignments using PHYML, an appropriate and accurate maximum likelihood method (Guindon and Gascuel, 2003) with Blosum 62, and using Bootstrapping. Bootstrap for higher confidence was performed at the level of 1000. Bootstrap proportions are indicated on the tree branches.
Table 1.
Accession numbers of amino acid sequences used for the phylogenetic analysis. The sequences were either collected on WormBase (http://www.wormbase.org/#01-23-6) or GenBank NCBI (http://www.ncbi.nlm.nih.gov/genbank/).
| Genes | Accession numbers | Genes | Accession numbers | |
|---|---|---|---|---|
| Full ABC-B transporter | C. elegans | B. malayi | ||
| Cel-PGP-1 | WP:CE11932 | Bm1_43165 | XP_001900095.1 | |
| Cel-PGP-2a | WP:CE41207 | |||
| Cel-PGP-2b | WP:CE50744 | O. volvulus | ||
| Cel-PGP-2c | WP:CE08576 | Ovo-PGP-10 | WP:OVP13603 | |
| Cel-PGP-2d | WP:CE29212 | Ovo-PGP-11 | WP:OVP14503 | |
| Cel-PGP-3 | WP:CE03818 | AAX82635.1 | AAX82635.1 | |
| Cel-PGP-C | CAA46191.1 | AAD49436.1 | AAD49436.1 | |
| Cel-PGP-4a | WP:CE44238 | |||
| Cel-PGP-4b | WP:CE03308 | L. loa | ||
| Cel-PGP-5a | WP:CE43003 | LOAG_00381 | EFO28095.2 | |
| Cel-PGP-5b | WP:CE43182 | LOAG_03722 | XP_003139307 | |
| Cel-PGP-6a | WP:CE40818 | |||
| Cel-PGP-6b | WP:CE40819 | D. immitis | ||
| Cel-PGP-6c | WP:CE46295 | “nDi2.2.scaf00004″ | Genbank: KP296255 | |
| Cel-PGP-7 | WP:CE36668 | “nDi2.2.scaf00046″ | Genbank: KP296249 | |
| Cel-PGP-8 | WP:CE31624 | “nDi2.2.scaf00048″ | Genbank: KP296245 | |
| Cel-PGP-9 | WP:CE15714 | |||
| Cel-PGP-10 | WP:CE40807 | |||
| Cel-PGP-11 | WP:CE34788 | |||
| Cel-PGP-12 | WP:CE03260 | |||
| Cel-PGP-13 | WP:CE40253 | |||
| Cel-PGP-14 | WP:CE03262 | |||
| Half ABC transporter | C. elegans | B. malayi | ||
| Cel-HAF-1 | WP:CE39331 | Bm1_15490 | XP_001894558.1 | |
| Cel-HAF-2 | WP:CE07240 | Bm1_30435 | XP_001897525.1 | |
| Cel-HAF-3 | WP:CE41666 | Bm1_50255 | XP_001901519.1 | |
| Cel-HAF-4 | WP:CE28355 | |||
| Cel-HAF-5/HMT-1 | WP:CE31731 | D. immitis | ||
| Cel-HAF-6 | WP:CE39850 | “nDi2.2.scaf00023″ | Genbank: KP296251 | |
| Cel-HAF-7 | WP:CE24404 | “nDi2.2.scaf00101″ | Genbank: KP296253 | |
| Cel-HAF-8 | WP:CE14926 | “nDi2.2.scaf00496.1″ | Genbank: KP296257 | |
| Cel-HAF-9 | WP:CE27353 | “nDi2.2.scaf00496.2″ | Genbank: KP296258 | |
3. Results and discussion
3.1. Bioinformatic identification and confirmation of sequences of D. immitis ABC-B transporter genes
All the cDNA sequences discussed contained the SL1 sequence (Blaxter and Liu, 1996) and a stop codon. The gene, annotated as being an ortholog of Cre-pgp-13 in nDi.2.2.scaf00049:93378..99486, was amplified by PCR. From the two amplicons identified, one sequence had a repeat region of 49bp similar to the sequence from nDi.2.2.scaf00049 and one sequence did not carry the extra repeated 49bp. Each amplicon was approximately 5 kb. Additional Blast analysis did not allow identification of the ABC signature. In nDi.2.2.scaf00049, the region from 93378 to 99486 hit protein Bm360 from B. malayi. Additional investigations and analysis in this scaffold and the amplified sequences did not reveal any additional evidence of a full-length ABC transporter gene for this previously annotated sequence and raises a question about this annotation. Thus, it was not included in the subsequent work.
Blast analysis in D. immitis nDi.2.2 with Pgp sequences from C. elegans, C. remanei, B. malayi, W. bancrofti, D. immitis, O. volvulus, and L. loa allowed 7 genes, that contained ABC-B motifs, to be identified in the following scaffolds (nDi.2.2.scaf00004, nDi.2.2.scaf00023, nDi.2.2.scaf00041, nDi.2.2.scaf00046, nDi.2.2.scaf00048, nDi.2.2.scaf00101 and nDi.2.2.scaf00496) (Supplementary data S5). In total, 7 complete gDNA sequences and 12 cDNA sequences (all including SL1 sequence (Blaxter and Liu, 1996) were successfully amplified, sequenced and confirmed. GenBank accession numbers of these sequences are available in Table 2. NCBI BLASTX 2.3.1+ results of the ABC-B transporter gene located in nDi.2.2.scaf00041 identified three fragments, nDi.2.2.scaf00041:17947..19267 (1321bp), corresponding to hypothetical protein LOAG_07988, nDi.2.2.scaf00041:19268..20612 corresponding to hypothetical protein Bm1_15040, and nDi.2.2.scaf00041:20612..27954 (7343bp) corresponding to hypothetical protein LOAG_07989 and Cbr-PGP-4 protein. Predicted cDNA sequences showed more similarity to pgp-16 from Haemonchus contortus (Supplementary data S5). Subsequent analysis on the consensus sequence generated from nDi.2.2.scaf00041, revealed that the gDNA sequence contained 7 stop codons located in predicted exon regions (Supplementary data S6). Two stop codons were located in exon 25, which carries the ABC signature motif. The sequence could be considered as a pseudogene (Vanin, 1985) as the gene contains multiple genetic lesions that would prevent the translation of a functional protein. In this regard, cDNA could not be translated to a predicted amino acid sequence and thus could not be included in the phylogenetic analysis. No specific name was attributed to this pseudogene. Interestingly, in C. elegans, Cel-pgp-15 was reported to be a pseudogene (Lespine et al., 2008). In Entamoeba histolytica, two Pgps have been reported with a frame shift and stop codons within their ATP binding domain (Descoteaux et al., 1992). However, even though the corresponding protein will not be functional, polymorphism in pseudogenes may lead to important physiological changes (Macphee et al., 2002). Alignments between gDNA sequences and cDNA allowed the observation that the ABC-B transporter transcript frame, from nDi.2.2.scaf00004, nDi.2.2.scaf00023, nDi.2.2.scaf00046, nDi.2.2.scaf00048, nDi.2.2.scaf00101 and nDi.2.2.scaf00496, contained 24, 14, 26, 29, 10, 13 introns and 25, 15, 27, 30, 11, 14 exons, respectively (Supplementary data S7, one worksheet for each scaffold).
Table 2.
GenBank accession numbers of the D. immitis ABC-B transporters genes and cDNA newly identified.
| Name | Gene/cDNA | GenBank Accession number |
|---|---|---|
| Pseudogene | gene | KP296247 |
| Dim-pgp-3 | gene | KP296248 |
| Dim-pgp-3 | cDNA | KP296249 |
| Dim-pgp-10 | cDNA | KP296245 |
| Dim-pgp-10 | gene | KP296246 |
| Dim-pgp-11 | gene | KP296254 |
| Dim-pgp-11 | cDNA | KP296255 |
| Dim-haf-1 | gene | KP296250 |
| Dim-haf-1 | cDNA | KP296251 |
| Dim-haf-4 | gene | KP296252 |
| Dim-haf-4 | cDNA | KP296253 |
| Dim-haf-5 | gene | KP296256 |
| Dim-haf-5.1 | cDNA isoform 1 | KP296257 |
| Dim-haf-5.2 | cDNA isoform 2 | KP296258 |
In this study, cDNA translation to amino acid sequences allowed identification of the structure of ABC transporter proteins (Table 3). From the cDNA amplification related to nDi.2.2.scaf00048, five amplicons with different sizes were obtained (Table 3). Based on TM prediction and subsequent transmembrane helix prediction in TM domains from the translated nucleotides, the amino acid sequence SEQ-1 did not carry the first TM, and SEQ-2 contained, from the first methionine, a total of 2 transmembrane helices in the first TM domain. The rest of the sequences were both followed by an ATP binding cassette domain, a complete TM containing 6 transmembrane helices, and a second ATP binding cassette domain. SEQ-1 and SEQ-2 from nDi.2.2.scaf00048 were not considered complete and were not investigated further. The amino acid sequence from the translated nucleotides of SEQ-3 contained 2 TM domains including 6 transmembrane helices and 2 ATP binding cassette domains. SEQ-3 from nDi.2.2.scaf00048 was included in the phylogenetic analysis as a full ABC-B transporter. Additionally, the amino acid sequences from the translated nucleotides of SEQ-4 and SEQ-5 contained, from their first methionine, respectively 2 and 6 transmembrane helices in their TM and one ATP binding cassette domain. Only SEQ-5 from nDi.2.2.scaf00048 was included in the phylogenetic analysis and called in the tree “nDi.2.2.scaf00048.5”. As a general observation for SEQ-1 to SEQ-5 from nDi.2.2.scaf00048, the difference between the sequences was only the length. Beside this, the sequences were identical. In this regard, the first methionine of SEQ-5 was located at position 708 of the amino acid sequence SEQ-3 related to nDi.2.2.scaf00048 or at position 2122 in nucleotide cDNA sequence of SEQ-3.
Table 3.
Analysis of potential ABC-B transporter cDNA amplicons and predicted structural organization of the amino acid sequences D. immitis.* indicates new sequences submitted to GenBank. TM and ABC refer to transmembrane domain and ATP-binding cassette domain, respectively. Nucleotide translation was performed with the translation tool of Geneious Pro.5.6.3 software. Predictions of transmembrane domain, ATP-binding cassette, and of ABC transporter family signature, were performed using ScanProsite (released 20.123; http://prosite.expasy.org/scanprosite/). Prediction of transmembrane helices was done with TMHMM (v.2.0) in Geneious Pro.5.6.3 software.
| Location on nDi.2.2 genome | Variants | CDNA length (bp) | Predicted Amino Acid (AA) sequences | Positions based on the AA sequence |
Predicted motif in AA sequences | ||
|---|---|---|---|---|---|---|---|
| Predicted TM | Predicted ABC domain | ABC transporters family Signature | |||||
| scaf00048: 79265..63901 | SEQ-1 | 3111 | 1036 | 476–761 | 1: 108–344 | 247–261: LSGGQKQRVAIARAV |  |
| 2: 796–1034 | 937–951: LSGGQKQRIAIARAI | ||||||
| SEQ-2 | 3336 | 1111 | 1: 10–129 | 1: 183–419 | 322–336: LSGGQKQRVAIARAV | 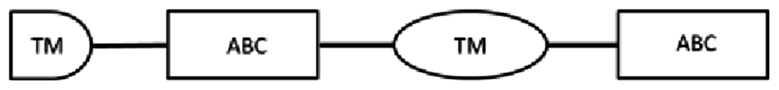 |
|
| 2: 551–836 | 2: 871–1109 | 1012–1026: LSGGQKQRIAIARAI | |||||
| SEQ-3 | 3978* | 1325 | 1: 29–343 | 1: 397–633 | 536–550: LSGGQKQRVAIARAV | 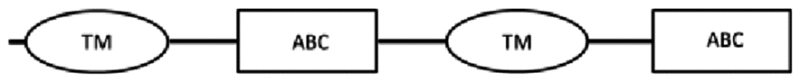 |
|
| 2: 765–1050 | 2:1085–1323 | 1226–1240: LSGGQKQRIAIARAI | |||||
| SEQ-4 | 1173 | 390 | 1–115 | 150–388 | 291–305: LSGGQKQRIAIARAI | 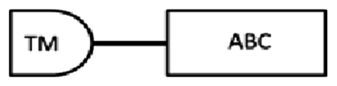 |
|
| SEQ-5 | 1857 | 618 | 58–343 | 378–616 | 519–533: LSGGQKQRIAIARAI | 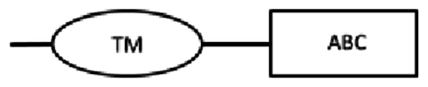 |
|
| scaf00004: 88673..79181 | 3861* | 1286 | 1: 64–354 | 1: 389–625 | 528–542: LSGGQKQRIAIARTI | 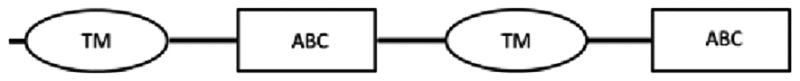 |
|
| 2: 726–1012 | 2:1045–1281 | 1184–1198: LSGGQKQRIAIARAL | |||||
| scaf00046: 226523..237100 | SEQ-1 | 1317 | 438 | 1–164 | 198–434 | 337–351: LSGGQKQRIAIARAI | 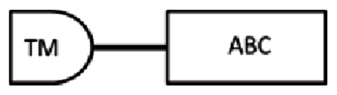 |
| SEQ-2 | 3759* | 1252 | 1: 41–333 | 1: 368–604 | 507–521: LSGGQKQRIAIARIL | 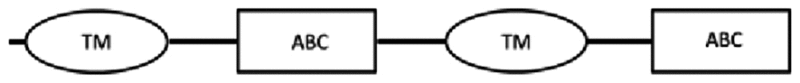 |
|
| 2: 691–978 | 2:1012–1248 | 1151–1165: LSGGQKQRIAIARAI | |||||
| scaf00023: 388438..395064 | 2040* | 679 | 100–399 | 432–672 | 573–587: LSGGQRQRIAIARAL | 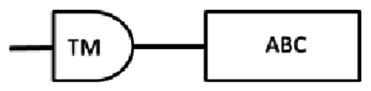 |
|
| scaf00101: 45240..51224 | 2277* | 758 | 199–479 | 511–747 | 650–664: MSGGQKQRIAIARAL |  |
|
| scaf00496: 20854..25667 | SEQ-1 | 2142* | 713 | 102–397 | 433–667 | 570–584: LSGGEKQRVAIARAL | 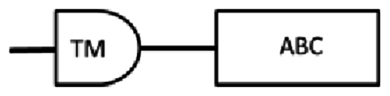 |
| SEQ-2 | 1896* | 631 | 20–315 | 351–585 | 488–502: LSGGEKQRVAIARAL | 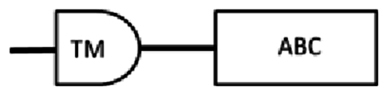 |
|
For cDNA amplification related to nDi.2.2.scaf00004, one amplicon was observed and sequenced. Based on TM prediction and subsequent transmembrane helix predictions in the TM, from the translated nucleotides, the amino acid sequence contained 2 complete sets of 6 transmembrane helices each in a TM domain, and two nucleotide binding domains as illustrated in Table 3. The amino acid sequence was inserted in the phylogenetic analysis of the full ABC-B transporters.
From the cDNA amplification related to nDi.2.2.scaf00046, two amplicons with different sizes were obtained (Table 3). Based on TM domain prediction and subsequent transmembrane helix predictions in the TM, from the translated nucleotides, the amino acid sequence SEQ-1 from nDi.2.2.scaf00046 contained, from the first methionine, an incomplete TM carrying 3 predicted transmembrane helices followed by one ATP binding cassette domain. The amino acid sequence SEQ-2 from nDi.2.2.scaf00046 contained 2 complete TM domains, each of which included 6 predicted transmembrane helices, and two ATP binding cassette domains as illustrated in Table 3. Only the amino acid sequence SEQ-2 from nDi.2.2.scaf00046 was inserted in the phylogenetic analysis of the full ABC-B transporters. As a general observation for SEQ-1 to SEQ-2 from nDi.2.2.scaf00046, the difference between the sequences was only the length. Beside this, the sequences were identical. Interestingly, in the D. immitis genome nuclear browser nDi.2.2.2 (http://salmo.bio.ed.ac.uk/cgi-bin/gbrowse/gbrowse/nDi.2.2.2/), a transcript was annotated as being an ortholog of Cre-PGP-4 in nDi.2.2.scaf00046 at position 226523..238764. The SEQ-2 cDNA sequence that we amplified from this scaffold started at position 226523 also in nDi.2.2.scaf00046. However, a 1664 bp difference was observed at the end of the amplicons between the annotated sequence from nDi.2.2 genome and SEQ-2 amplified in this study.
From the cDNA amplification related to nDi.2.2.scaf00023, nDi.2.2.scaf00101, nDi.2.2.scaf00496, one, one and two amplicons were observed, respectively and each sequenced. Based on TM predictions and subsequent transmembrane helix predictions in the TM domain from the translated nucleotides, the amino acid sequences contained one TM domain including 4 transmembrane helices followed by 1 ATP binding cassette domain.
All 4 amino acid sequences related to nDi.2.2.scaf00023, nDi.2.2.scaf00101, and nDi.2.2.scaf00496 were included in the phylogenetic analysis of the half transporters. The two sequences from nDi.2.2.scaf00496 were called “nDi.2.2.scaf00496.1” and “nDi.2.2.scaf00496.2”. Between these last 2 sequences, only the beginning of the sequences differed.
As a summary, one pseudogene, three potential full, and five potential half ABC-B transporters were identified.
3.2. Phylogenetic analysis
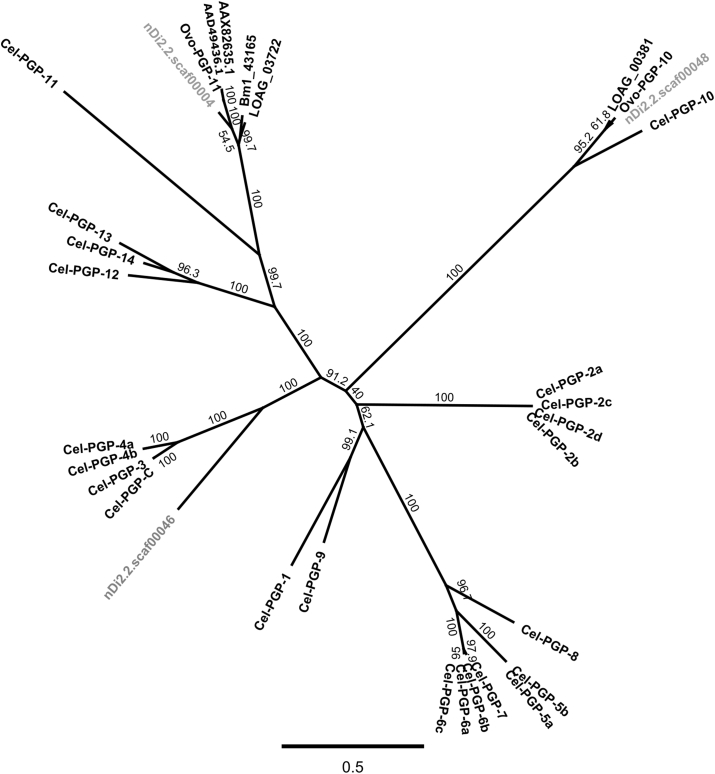
Two phylogenetic trees have been generated allowing orthology identification of 8 out of the 9 amino acid sequences (pseudogene not included). The phylogenetic tree of the full ABC–B transporters is presented in Fig. 1. Even though the amino acid sequences from Cel-PGP-5b and Cel-PGP-6b were shorter than the others in the ClustalW alignment, in the phylogenetic tree, Cel-PGP-5b and Cel-PGP-6b grouped respectively with their isoforms Cel-PGP-5a and Cel-PGP-6a. The nucleotide translated sequence related to nDi.2.2.scaf00004 was an ortholog of Cel-PGP-11. Based on Beech et al. (Beech et al., 2010), the gene should be named Dim-pgp-11.The nucleotide translated sequence related to nDi.2.2.scaf00046 was an ortholog of Cel-PGP-3, of its spliced variant Cel-PGP-C, of Cel-PGP-4a and of Cel-PGP-4b. The corresponding gene is called Dim-pgp-3. The nucleotide translated sequence related to nDi.2.2.scaf00048 was an ortholog of Cel-PGP-10. The corresponding gene is called Dim-pgp-10.
Fig. 1.
Phylogenetic tree of full ABC-B transporter genes constructed from PHYML (Maximum likelihood) and Bootstrap 1000 with Geneious Pro.5.6.3 software. Numbers over the branches correspond to the percentage of bootstrap values (calculation based on 1000 pseudoreplicates). The three letter prefixes in Pgp gene names: Cel, Dim and Ovo refer to C. elegans, D. immitis, and O. volvulus. LOAG and Bm1 prefixes refer to L. loa and B. malayi sequences. AAX82635.1 and AAD49436.1 are GenBank accession numbers referring to O. volvulus Pgps. All the GenBank accession numbers for amino acid sequences are provided in Table 1 nDi.2.2.scaf00004, nDi.2.2.scaf00046 and nDi.2.2.scaf00048 labelling correspond to D. immitis amino acid sequences that were identified based on work on these scaffolds.
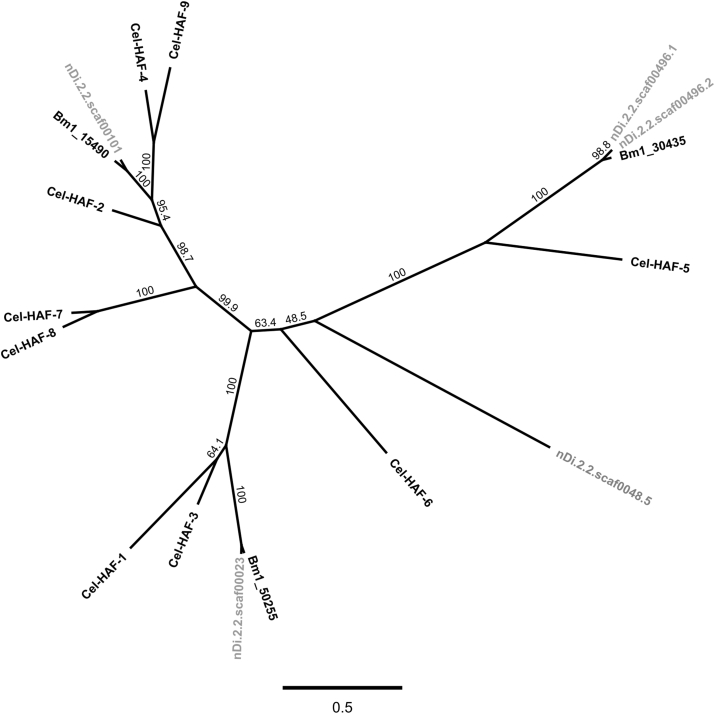
The phylogenetic tree of the half ABC–B transporters is presented in Fig. 2. The amino acid sequence corresponding to SEQ-5 related to nDi.2.2.scaf00048 and called “nDi.2.2.scaf00048.5” in Fig. 2 does not show clear orthology with other C. elegans half transporter. An additional phylogenetic tree (Supplementary data S8) including all the full ABC-B transporters and half-ABC-B transporters was constructed with the same trimmed sequences used in Fig. 1 and Fig. 2. In Supplementary data S8, “nDi.2.2.scaf00048.5” is localized between the branches of the full ABC-B transporter and the branches of the half transporter. We did not have enough data on which to name this sequence. Functional analysis may be required to confirm if this potential half transporter is functional. However, as explained previously, this sequence shares the same characteristics as Dim-pgp-10, as the second part of Dim-pgp-10 is the same as SEQ-5 (“nDi.2.2.scaf00048.5”) and could represent a splice variant. The nucleotide translated sequence related to nDi.2.2.scaf00023 was an ortholog of Cel-HAF-1 and of Cel-HAF-3. The corresponding gene is called Dim-haf-1. The nucleotide translated sequence related to nDi.2.2.scaf00101 was an ortholog of Cel-HAF-4 and Cel-HAF-9. The gene is called Dim-haf-4. The nucleotide translated sequences related to nDi.2.2.scaf00496.1 and nDi.2.2.scaf00496.2 were orthologs of Cel-HAF-5 also called HMT-1. The corresponding genes in D. immitis are called Dim-haf-5.1 and Dim-haf-5.2. With additional Blast analysis using BLASTP 2.3.1+ (NCBI), Dim-HAF-5.1 and Dim-HAF-5.2 carried ABC-C motifs, as does Cel-HAF-5. As it was predicted in the first part of the study, the TM related to Dim-HAF-1, Dim-HAF-4 and Dim-HAF-5.1, and Dim-HAF-5.2 carried only 4 transmembrane helices. We additionally analysed all the amino acid sequences of the C. elegans half transporter orthologs using the same procedure as for D. immitis with Geneious Pro.5.6.3 software. Interestingly, from the analysis of Cel-HAF-1, Cel-HAF-3, Cel-HAF-4, Cel-HAF-5 and Cel-HAF-9, all carried 4 predicted transmembrane helices in their TM as did the D. immitis half transporters. Consequently, the structure of the TM presented in Table 3 for Dim-HAF-1 (nDi.2.2.scaf00023), Dim-HAF-4 (nDi.2.2.scaf00101) and Dim-HAF-5.1, and Dim-HAF-5.2 (nDi.2.2.scaf00496) should be considered as complete. In both Fig. 1, Fig. 2, Clade III (Filariae) and Clade V (C. elegans) (Blaxter et al., 1998) clearly separated with the branches. The sequences from B. malayi, L. loa, O. volvulus and D. immitis always grouped together. With 1 pseudogene, 3 full, and 2 half ABC-B transporters genes identified from the genome of D. immitis, this organism carries only 20% of the number of ABC-B transporters as C. elegans (Sheps et al., 2004) and one third the number found in H. contortus (Laing et al., 2013). C. elegans is a free living nematode, while H. contortus (also from Clade V (Blaxter et al., 1998)) and the different filarial nematodes are parasitic. The difference in number of Pgps between H. contortus and D. immitis remains interesting. They have distinct life cycles with D. immitis being an obligate parasite at all stages of its life cycle, and requiring a mosquito vector (Grieve et al., 1983). Similar to C. elegans, H. contortus has several free living stages in its life cycle that may expose the parasite to considerable levels of environmental toxins, compared with D. immitis which only lives in mosquito or mammalian hosts. Such observation may explain the higher number of ABC-B transporter genes in Trichostrongyles and other Rhabditia, as they may need a high level of protection against natural toxins in the terrestrial environment (Broeks et al., 1995).
Fig. 2.
Phylogenetic tree of half ABC transporter genes constructed from PHYML (Maximum likelihood) and Bootstrap 1000 with Geneious Pro.5.6.3 software. Numbers over the branches correspond to the percentage of bootstrap values (calculation based on 1000 pseudoreplicates). Cel, Dim, and Bm1 refer to C. elegans, D. immitis, and B. malayi sequences. All the GenBank accession numbers for amino acid sequences are provided in Table 1 nDi.2.2.scaf00023, nDi.2.2.scaf00101, nDi.2.2.scaf00496.1, nDi.2.2.scaf00496.2 and “nDi.2.2.scaf00048.5” (SEQ-5 from nDi.2.2.scaf00048) labelling correspond to D. immitis amino acid sequences that were identified, based on work with these scaffolds.
Besides differences observed between C. elegans, H. contortus and D. immitis, studies on different nematodes could be predictive in terms of the likely function and/or properties of ABC-B transporters in D. immitis. In C. elegans, Cel-HAF-1 was localized on the inner mitochondrial membrane and was reported to be necessary for peptide transport and for the mitochondrial unfolded protein response (UPRmt) signaling pathway (Haynes et al., 2010). Cel-HAF-4 and Cel-HAF-9 were localized on the membranes of intestinal granular organelles (Kawai et al., 2009). Cel-HAF-4 and Cel-HAF-9 were reported to form a heterodimer that was required for their own stabilization (Tanji et al., 2013). They both were reported to be necessary for a normal defecation cycle, for proper growth and for normal brood size (Kawai et al., 2009). Cel-pgp-3 and Cel-pgp-4 were located on the same chromosome X (Lincke et al., 1992). The expression of Cel-PGP-3 was identified in the apical membrane of the excretory and intestinal cells (Broeks et al., 1995, Ardelli and Prichard, 2013) while expression of Cel-PGP-4 was reported in the larval excretory cell (Zhao et al., 2004). Cel-PGP-3 expression was reported in developmental stages (Broeks et al., 1995). The deletion of Cel-pgp-3 was not associated with an increased sensitivity to IVM. However, it was associated with increased sensitivity to colchicine and chloroquine (Broeks et al., 1995). In C. elegans, mRNA levels of Cel-pgp-4, Cel-haf-1 and Cel-haf-3 significantly increased in IVR10 (C. elegans able to survive at 10 ng/ml IVM) compared with the N2 wild-type strain (Yan et al., 2012). Also transcription down regulation of Cel-PGP-4, and Cel-HAF-9, induced by 15 or 20 ng/ml IVM, significantly increased the effect of IVM to reduce egg production (Yan et al., 2012). Additionally, down regulation of transcription of Cel-PGP-3, Cel-PGP-4 and Cel-HAF-9 increased motility at high IVM concentrations (Yan et al., 2012). Cel-PGP-10 was expressed in the worm's intestine as is Cel-PGP-3. Cel-PGP-3 showed a higher expression level after a short exposure of IVM in N2 wild-type strain (Ardelli and Prichard, 2013).
In C. elegans, an increase of IVM susceptibility was reported after loss-of-function of several Pgps including Cel-PGP-3 and Cel-PGP-11, while loss of function of Cel-PGP-10 had a limited effect (Janssen et al., 2013b). Interestingly, Cel-PGP-11 plays an important role in IVM detoxification (Janssen et al., 2013b). In ML resistant Cooperia oncophora adult worms and L3, an upregulation of the transcript level of Con-pgp-11 was reported while it was not observed in ML susceptible parasites (De Graef et al., 2013). In Parascaris equorum, IVM resistance was correlated with an increase in the level of Peq-pgp-11 mRNA and with the presence of 3 individual SNPs (Janssen et al., 2013a). In a motility assay, the expression of Peq-PGP-11 in C. elegans deficient with Cel-PGP-11 significantly reduced IVM susceptibility (Janssen et al., 2015). Interestingly, the “GG-GG” genotype which was associated with IVM response phenotype in D. immitis (Bourguinat et al., 2011b) is located in Dim-pgp-11.
In H. contortus, Hco-pgp-3, Hco-pgp-4, Hco-pgp-10, and Hco-pgp-11 partial sequences had no significant change in their mRNA expression levels when pools of L3 larvae were compared between a rapidly selected IVM-resistant parasite isolate and its drug-sensitive parent (Williamson and Wolstenholme, 2012). However, there was great variability in these results which may have been due to the presence of larvae in different states of viability and development in the pools. Nine different H. contortus Pgp genes, including Hco-pgp-3 and Hco-pgp-11, were specifically up-regulated in parasitic life stages, which suggested a potential involvement of these Pgps in the detoxification of eosinophil granule products (Issouf et al., 2014). Also eosinophil granules had induced a dose dependent overexpression of Hco-pgp-3. It was hypothesized that some helminth Pgps could be involved in the detoxification of host products, and thus may help the worms to escape the host immune response (Issouf et al., 2014). In three subpopulations of H. contortus larvae, that were able to survive increasing concentrations of levamisole, Pgp gene expression levels were measured. Expression of Hco-pgp-3, Hco-pgp-4 and Hco-pgp-10 genes increased by 1.5–3 fold in the subpopulation surviving the lowest levamisole concentrations (Sarai et al., 2014).
4. Conclusions
This study identified, for the first time, 9 complete ABC transporter genes in D. immitis, including three full size Pgp-type sequences (Dim-pgp-3, Dim-pgp-10, Dim-pgp-11), and another pseudogene, two ABC-B half transporter genes (Dim-haf-1 and Dim-haf-4), two ABC-C transporter genes (Dim-haf-5.1 and Dim-haf-5.2) and one additional half transporter that may require additional characterization. Filarial nematodes seem to carry fewer ABC-B transporter genes than C. elegans and H. contortus. Thus, different ABC transporters may be involved in the development of ML resistance in the two clades. ABC-B transporters are considered important in ML resistance mechanisms. The ABC transporters identified may be used as tools to identify genetic markers for ML resistance prediction in D. immitis. Monitoring tools are urgently needed, particularly in USA where D. immitis resistance has developed in the Mississippi Delta region. Furthermore, this characterization of the ABC-B transporter genes in D. immitis should be a precursor for studies on the mechanism of ML resistance in this parasite.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Bayer Animal Health, the Natural Sciences and Engineering Research Council, Canada (grant number: RGPIN/2777), and the FRQNT Centre for Host-Parasite Interactions, Quebec for funding support. We thank Dr Pascal Maeser, Dr Christelle Godel and Dr Mark Blaxter for letting us use the nDi.2.2 Dirofilaria immitis genome and Dr Robin Beech for bioinformatic advice. We also acknowledge the help of the McGill University and Génome Québec Innovation Centre.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijpddr.2016.04.001.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar S.V., Kimchi-Sarfaty C., Sauna Z.E., Gottesman M.M. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F. Transport proteins of the ABC systems superfamily and their role in drug action and resistance in nematodes. Parasitol. Int. 2013;62:639–646. doi: 10.1016/j.parint.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Guerriero S.B., Prichard R.K. Genomic organization and effects of ivermectin selection on Onchocerca volvulus P-glycoprotein. Mol. Biochem. Parasitol. 2005;143:58–66. doi: 10.1016/j.molbiopara.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Guerriero S.B., Prichard R.K. Ivermectin imposes selection pressure on P-glycoprotein from Onchocerca volvulus: linkage disequilibrium and genotype diversity. Parasitology. 2006;132:375–386. doi: 10.1017/S0031182005008991. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Prichard R.K. Reduced genetic variation of an Onchocerca volvulus ABC transporter gene following treatment with ivermectin. Trans. R. Soc. Trop. Med. Hyg. 2007;101:1223–1232. doi: 10.1016/j.trstmh.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Prichard R.K. Inhibition of P-glycoprotein enhances sensitivity of Caenorhabditis elegans to ivermectin. Veterinary Parasitol. 2013;191:264–275. doi: 10.1016/j.vetpar.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Stitt L.E., Tompkins J.B. Inventory and analysis of ATP-binding cassette (ABC) systems in Brugia malayi. Parasitology. 2010;137:1195–1212. doi: 10.1017/S0031182010000120. [DOI] [PubMed] [Google Scholar]
- Beech R.N., Wolstenholme A.J., Neveu C., Dent J.A. Nematode parasite genes: what's in a name? Trends Parasitol. 2010;26:334–340. doi: 10.1016/j.pt.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Blackhall W.J., Liu H.Y., Xu M., Prichard R.K., Beech R.N. Selection at a P-glycoprotein gene in ivermectin- and moxidectin-selected strains of Haemonchus contortus. Mol. Biochem. Parasitol. 1998;95:193–201. doi: 10.1016/s0166-6851(98)00087-5. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Liu L. Nematode spliced leaders–ubiquity, evolution and utility. Int. J. Parasitol. 1996;26:1025–1033. [PubMed] [Google Scholar]
- Blaxter M.L., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vida J.T., Thomas W.K. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Bourguinat C., Keller K., Bhan A., Peregrine A., Geary T., Prichard R. Macrocyclic lactone resistance in Dirofilaria immitis. Veterinary Parasitol. 2011;181:388–392. doi: 10.1016/j.vetpar.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Bourguinat C., Keller K., Blagburn B., Schenker R., Geary T.G., Prichard R.K. Correlation between loss of efficacy of macrocyclic lactone heartworm anthelmintics and P-glycoprotein genotype. Veterinary Parasitol. 2011;176:374–381. doi: 10.1016/j.vetpar.2011.01.024. [DOI] [PubMed] [Google Scholar]
- Bourguinat C., Lee A.C., Lizundia R., Blagburn B.L., Liotta J.L., Kraus M.S., Keller K., Epe C., Letourneau L., Kleinman C.L., Paterson T., Gomez E.C., Montoya-Alonso J.A., Smith H., Bhan A., Peregrine A.S., Carmichael J., Drake J., Schenker R., Kaminsky R., Bowman D.D., Geary T.G., Prichard R.K. Macrocyclic lactone resistance in Dirofilaria immitis: failure of heartworm preventives and investigation of genetic markers for resistance. Veterinary Parasitol. 2015;210:167–178. doi: 10.1016/j.vetpar.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Broeks A., Janssen H.W., Calafat J., Plasterk R.H. A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J. 1995;14:1858–1866. doi: 10.1002/j.1460-2075.1995.tb07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R., Boeckh A. Moxidectin: a review of chemistry, pharmacokinetics and use in horses. Parasites Vectors. 2009;2(Suppl. 2):S5. doi: 10.1186/1756-3305-2-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle A.S., Iravane Bajaj J.A., Khaparkhuntikar M.N., Maher G.T., Patil R.V. Microfilaria in human subcutaneous dirofilariasis: a case report. J. Clin. Diagn. Res. JCDR. 2014;8:113–114. doi: 10.7860/JCDR/2013/6886.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graef J., Demeler J., Skuce P., Mitreva M., Von Samson-Himmelstjerna G., Vercruysse J., Claerebout E., Geldhof P. Gene expression analysis of ABC transporters in a resistant Cooperia oncophora isolate following in vivo and in vitro exposure to macrocyclic lactones. Parasitology. 2013;140:499–508. doi: 10.1017/S0031182012001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux S., Shen P.S., Ayala P., Orozco E., Samuelson J. P-glycoprotein genes of Entamoeba histolytica. Arch. Med. Res. 1992;23:23–25. [PubMed] [Google Scholar]
- Godel C., Kumar S., Koutsovoulos G., Ludin P., Nilsson D., Comandatore F., Wrobel N., Thompson M., Schmid C.D., Goto S., Bringaud F., Wolstenholme A., Bandi C., Epe C., Kaminsky R., Blaxter M., Maser P. The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012;26:4650–4661. doi: 10.1096/fj.12-205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy P., Lian J., Beech R.N., Prichard R.K. Haemonchus contortus P-glycoprotein-2: in situ localisation and characterisation of macrocyclic lactone transport. Int. J. Parasitol. 2015;45:85–93. doi: 10.1016/j.ijpara.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Grieve R.B., Lok J.B., Glickman L.T. Epidemiology of canine heartworm infection. Epidemiol. Rev. 1983;5:220–246. doi: 10.1093/oxfordjournals.epirev.a036260. [DOI] [PubMed] [Google Scholar]
- Griffin J., Fletcher N., Clemence R., Blanchflower S., Brayden D.J. Selamectin is a potent substrate and inhibitor of human and canine P-glycoprotein. J. Vet. Pharmacol. Ther. 2005;28:257–265. doi: 10.1111/j.1365-2885.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hampshire V.A. Evaluation of efficacy of heartworm preventive products at the FDA. Vet. Parasitol. 2005;133:191–195. doi: 10.1016/j.vetpar.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Haynes C.M., Yang Y., Blais S.P., Neubert T.A., Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issouf M., Guegnard F., Koch C., Le Vern Y., Blanchard-Letort A., Che H., Beech R.N., Kerboeuf D., Neveu C. Haemonchus contortus P-glycoproteins interact with host eosinophil granules: a novel insight into the role of ABC transporters in host-parasite interaction. PloS one. 2014;9:e87802. doi: 10.1371/journal.pone.0087802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Janssen I.J., Krucken J., Demeler J., Basiaga M., Kornas S., von Samson-Himmelstjerna G. Genetic variants and increased expression of Parascaris equorum P-glycoprotein-11 in populations with decreased ivermectin susceptibility. PLoS One. 2013;8:e61635. doi: 10.1371/journal.pone.0061635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen I.J., Krucken J., Demeler J., von Samson-Himmelstjerna G. Caenorhabditis elegans: modest increase of susceptibility to ivermectin in individual P-glycoprotein loss-of-function strains. Exp. Parasitol. 2013;134:171–177. doi: 10.1016/j.exppara.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Janssen I.J., Krucken J., Demeler J., von Samson-Himmelstjerna G. Transgenically expressed Parascaris P-glycoprotein-11 can modulate ivermectin susceptibility in Caenorhabditis elegans. Int. J. Parasitol. Drugs Drug Resist. 2015;5:44–47. doi: 10.1016/j.ijpddr.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H., Tanji T., Shiraishi H., Yamada M., Iijima R., Inoue T., Kezuka Y., Ohashi K., Yoshida Y., Tohyama K., Gengyo-Ando K., Mitani S., Arai H., Ohashi-Kobayashi A., Maeda M. Normal formation of a subset of intestinal granules in Caenorhabditis elegans requires ATP-binding cassette transporters HAF-4 and HAF-9, which are highly homologous to human lysosomal peptide transporter TAP-like. Mol. Biol. Cell. 2009;20:2979–2990. doi: 10.1091/mbc.E08-09-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf D., Guegnard F. Anthelmintics are substrates and activators of nematode P glycoprotein. Antimicrob. Agents Chemother. 2011;55:2224–2232. doi: 10.1128/AAC.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S.L., Kryukov I., Li K., Morrison A.A., Reid A.J., Sargison N., Saunders G.I., Wasmuth J.D., Wolstenholme A., Berriman M., Gilleard J.S., Cotton J.A. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.C., Montgomery S.P., Theis J.H., Blagburn B.L., Eberhard M.L. Public health issues concerning the widespread distribution of canine heartworm disease. Trends Parasitol. 2010;26:168–173. doi: 10.1016/j.pt.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Lespine A., Alvinerie M., Vercruysse J., Prichard R.K., Geldhof P. ABC transporter modulation: a strategy to enhance the activity of macrocyclic lactone anthelmintics. Trends Parasitol. 2008;24:293–298. doi: 10.1016/j.pt.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Lespine A., Martin S., Dupuy J., Roulet A., Pineau T., Orlowski S., Alvinerie M. Interaction of macrocyclic lactones with P-glycoprotein: structure-affinity relationship. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2007;30:84–94. doi: 10.1016/j.ejps.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Lespine A., Menez C., Bourguinat C., Prichard R.K. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: prospects for reversing transport-dependent anthelmintic resistance. Int. J. Parasitol. Drugs Drug Resist. 2012;2:58–75. doi: 10.1016/j.ijpddr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincke C.R., The I., van Groenigen M., Borst P. The P-glycoprotein gene family of Caenorhabditis elegans. Cloning and characterization of genomic and complementary DNA sequences. J. Mol. Biol. 1992;228:701–711. doi: 10.1016/0022-2836(92)90855-e. [DOI] [PubMed] [Google Scholar]
- Macphee I.A., Fredericks S., Tai T., Syrris P., Carter N.D., Johnston A., Goldberg L., Holt D.W. Tacrolimus pharmacogenetics: polymorphisms associated with expression of cytochrome p4503A5 and P-glycoprotein correlate with dose requirement. Transplantation. 2002;74:1486–1489. doi: 10.1097/00007890-200212150-00002. [DOI] [PubMed] [Google Scholar]
- McCavera S., Walsh T.K., Wolstenholme A.J. Nematode ligand-gated chloride channels: an appraisal of their involvement in macrocyclic lactone resistance and prospects for developing molecular markers. Parasitology. 2007;134:1111–1121. doi: 10.1017/S0031182007000042. [DOI] [PubMed] [Google Scholar]
- Morchon R., Carreton E., Gonzalez-Miguel J., Mellado-Hernandez I. Heartworm disease (Dirofilaria immitis) and their vectors in Europe – new distribution trends. Front. Physiol. 2012;3:196. doi: 10.3389/fphys.2012.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard R. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17:445–453. doi: 10.1016/s1471-4922(01)01983-3. [DOI] [PubMed] [Google Scholar]
- Prichard R.K., Roulet A. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134:1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Pulaski C.N., Malone J.B., Bourguinat C., Prichard R., Geary T., Ward D., Klei T.R., Guidry T., Smith G., Delcambre B., Bova J., Pepping J., Carmichael J., Schenker R., Pariaut R. Establishment of macrocyclic lactone resistant Dirofilaria immitis isolates in experimentally infected laboratory dogs. Parasites Vectors. 2014;7:494. doi: 10.1186/s13071-014-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud J.P. Thiacetarsamide (adulticide) versus melarsomine (RM 340) developed as macrofilaricide (adulticide and larvicide) to cure canine heartworm infection in dogs. Annales de recherches veterinaires. Ann. Veterinary Res. 1992;23:1–25. [PubMed] [Google Scholar]
- Sanger F., Coulson A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai R.S., Kopp S.R., Coleman G.T., Kotze A.C. Drug-efflux and target-site gene expression patterns in Haemonchus contortus larvae able to survive increasing concentrations of levamisole in vitro. Int. J. Parasitol. Drugs Drug Resist. 2014;4:77–84. doi: 10.1016/j.ijpddr.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheps J.A., Ralph S., Zhao Z., Baillie D.L., Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon F., Siles-Lucas M., Morchon R., Gonzalez-Miguel J., Mellado I., Carreton E., Montoya-Alonso J.A. Human and animal dirofilariasis: the emergence of a zoonotic mosaic. Clin. Microbiol. Rev. 2012;25:507–544. doi: 10.1128/CMR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sutherland I., Scott I. Wiley; 2009. Gastrointestinal Nematodes of Sheep and Cattle: Biology and Control. [Google Scholar]
- Tanji T., Nishikori K., Shiraishi H., Maeda M., Ohashi-Kobayashi A. Co-operative function and mutual stabilization of the half ATP-binding cassette transporters HAF-4 and HAF-9 in Caenorhabditis elegans. Biochem. J. 2013;452:467–475. doi: 10.1042/BJ20130115. [DOI] [PubMed] [Google Scholar]
- Vanin E.F. Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Vercruysse J., Rew R.S. CABI; 2002. Macrocyclic Lactones in Antiparasitic Therapy. [Google Scholar]
- Williamson S.M., Wolstenholme A.J. P-glycoproteins of Haemonchus contortus: development of real-time PCR assays for gene expression studies. J. Helminthol. 2012;86:202–208. doi: 10.1017/S0022149X11000216. [DOI] [PubMed] [Google Scholar]
- Xu M., Molento M., Blackhall W., Ribeiro P., Beech R., Prichard R. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol. Biochem. Parasitol. 1998;91:327–335. doi: 10.1016/s0166-6851(97)00215-6. [DOI] [PubMed] [Google Scholar]
- Yan R., Urdaneta-Marquez L., Keller K., James C.E., Davey M.W., Prichard R.K. The role of several ABC transporter genes in ivermectin resistance in Caenorhabditis elegans. Veterinary Parasitol. 2012;190:519–529. doi: 10.1016/j.vetpar.2012.06.038. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Sheps J.A., Ling V., Fang L.L., Baillie D.L. Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J. Mol. Biol. 2004;344:409–417. doi: 10.1016/j.jmb.2004.09.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.