Abstract
We present a hybrid computational methodology to predict multiple energetically accessible conformations for G protein-coupled receptors (GPCRs) that might play a role in binding to ligands and different signaling partners. To our knowledge, this method, termed ActiveGEnSeMBLE, enables the first quantitative energy profile for GPCR activation that is consistent with the qualitative profile deduced from experiments. ActiveGEnSeMBLE starts with a systematic coarse grid sampling of helix tilts/rotations (∼13 trillion transmembrane-domain conformations) and selects the conformational landscape based on energy. This profile identifies multiple potential active-state energy wells, with the TM3–TM6 intracellular distance as an approximate activation coordinate. These energy wells are then sampled locally using a finer grid to find locally minimized conformation in each energy well. We validate this strategy using the inactive and active experimental structures of β2 adrenergic receptor (hβ2AR) and M2 muscarinic acetylcholine receptor. Structures of membrane-embedded hβ2AR along its activation coordinate are subjected to molecular-dynamics simulations for relaxation and interaction energy analysis to generate a quantitative energy landscape for hβ2AR activation. This landscape reveals several metastable states along this coordinate, indicating that for hβ2AR, the agonist alone is not enough to stabilize the active state and that the G protein is necessary, consistent with experimental observations. The method’s application to somatostatin receptor SSTR5 (no experimental structure available) shows that to predict an active conformation it is better to start from an inactive structure template based on a close homolog than to start from an active template based on a distant homolog. The energy landscape for hSSTR5 activation is consistent with hβ2AR in the role of the G protein. These results demonstrate the utility of the ActiveGEnSeMBLE method for predicting multiple conformations along the pathways for activating GPCRs and the corresponding energy landscapes, thereby providing detailed structural insights into the initial molecular events of GPCR function that are not easily accessible by experiments.
Introduction
G protein-coupled receptors (GPCRs) serve critical signaling functions in numerous cellular processes and thus are important targets for therapeutics. The development of such therapeutics is complicated because the activation of GPCRs, which is integral to their function, involves multiple distinct conformations along the pathway for activation. Moreover, since some GPCRs are capable of activating more than one intracellular (IC) signaling pathway (1), it is essential to identify multiple active conformations that may be involved and have different functions. To understand GPCR activation mechanisms and carry out structure-based drug design, it is necessary to obtain accurate three-dimensional (3D) structures for each of these important conformations. This creates a huge problem for structure-determination experiments, since each one of these structures must be sufficiently stabilized to obtain an ordered crystal. Indeed, despite huge efforts and remarkable experimental breakthroughs, experimental structures are available for only 3% of the ∼800 human GPCRs. Most of these structures are for an inactive conformation and only four are for active-like conformations. In addition, of the four GPCRs with both the active and inactive structures crystallized, only one (rhodopsin) has more than one active experimental structure.
There is a huge potential role for theory and simulation to fill in this crystal structure gap for GPCRs. However, there are major problems with using theory to predict the activation of such complex membrane-bound proteins. Millisecond-long molecular dynamics (MD) simulations have not been successful in following activation from an inactive state to an active-like state along an activation pathway. In addition, the sequence homologies between most GPCRs and available active-state GPCR templates are too low to get active structures for the remaining GPCRs. To begin to address this problem, we previously developed the GEnSeMBLE (2) method to predict the ensemble of low-energy (stable) 3D structures of GPCRs. This method has successfully predicted the structures for several class A and class B GPCRs: C-C chemokine receptor type 5 (CCR5) (3), adenosine A3 receptor (AA3R) (4), cannabinoid receptor type 1 (CB1) (5), taste receptor type 2 member 38 (Tas2R38) (6), olfactory receptor 1G1 (OR1G1) (7), glucagon-like peptide 1 receptor (GLP1R) (8), and prostaglandin D2 receptor (DP) (9). Most of the predictions are for inactive-state structures, but we were able to predict and validate active-state structures of AA3R (4) and CB1 (5).
The GEnSeMBLE methodology starts with several template configurations that specify the initial helix packing (helix locations, tilts, and rotations) for the seven helical transmembrane domains (TMDs) based either on an experimental structure of a similar GPCR or on a previously predicted structure. Then, we consider all possible simultaneous rotations by multiples of 30° about the helical axes for all seven TMDs. We estimate the energies of all 127≈35 million conformations sampling helix rotations by calculating the energies for all combinations of the 12 interacting pairs of helices (BiHelix), including optimized side chains. We build the best 1000 of these conformations by energy into seven-helix bundles with optimized side chains and then select ∼20 of the lowest conformations by energy for further consideration. This is done for all plausible templates to identify which templates are the best and which rotations best accommodate the target sequence. This is followed by an exhaustive sampling of simultaneous tilts and rotations of the seven helices (SuperBiHelix), leading to ∼13 trillion helix tilts/rotation combinations. The energies for all 13 trillion conformations are again estimated by combining the BiHelix energies to identify the best 2000 conformations, which are then built into seven-helix bundles for final side-chain optimization. From this list, the ∼20 lowest-energy conformations are selected for binding to different ligands and for further studies (2).
Although such conformational sampling is exhaustive, it tends to be biased toward inactive conformations because 1) the available templates are mainly inactive, 2) inactive conformations usually have lower energy than active conformations and the procedure seeks lower-energy structures, and 3) the agonists that might stabilize the active configurations are not present during the reduction from 13 trillion to 20 structures. Even so, GEnSeMBLE has successfully modeled some active GPCRs. In this work, we propose a hybrid method, denoted ActiveGEnSeMBLE, that builds upon the original GEnSeMBLE method to systematically predict multiple potentially active conformations of GPCRs. It utilizes a hierarchical sampling scheme that first samples conformations on a coarse grid, followed by another conformational sampling with a finer grid. In addition, rather than using only energy-based scoring, the ActiveGEnSeMBLE method uses both structural and energy information to identify the higher-energy candidates for active conformations that may reside in local energy wells stabilized by appropriate agonists or other cellular effectors (e.g., G protein or β-arrestin).
In this work, we first validated the ActiveGEnSeMBLE method against experimental structures of GPCRs that were previously obtained in active conformations with noncovalently bound ligands: human β2 adrenergic receptor (hβ2AR) (10, 11) and human M2 muscarinic acetylcholine receptor (hM2) (12, 13). (The x-ray structure of the agonist-bound mouse μ-opioid receptor (mOPRM or μOR) stabilized with a nanobody (14) was not available at the time of this study, so we did not include it as a test case.) We then applied ActiveGEnSeMBLE to predict multiple active and inactive forms of human somatostatin receptor subtype 5 (hSSTR5). We selected hSSTR5 because 1) it plays an important role in antiproliferation, hormone secretion, and human diseases such as pancreas cancer (15); 2) no experimental structure for hSSTR5 is available for use in drug development; and 3) a recent study identified hSSTR5 as the most valuable template for homology modeling of the nonorphan and nonolfactory class A GPCRs that constitute the majority of this class of GPCRs (16).
Materials and Methods
Protocol
Our original GEnSeMBLE method involves the following steps, as described in detail elsewhere (2):
Step 1. Align the target sequence to the other GPCR sequences homologous up to an Expect (E) value of 0.1, and use the PredicTM method to determine the lengths and ranges of the helical hydrophobic core regions. Then use secondary-structure prediction servers to predict helical regions that might extend beyond the hydrophobic core outside the membrane.
Step 2. Generate the structures for the helical regions of the target receptor with a variety of helical shapes using either OptHelix, which generates a helical shape using minimization and MD, or homology modeling based on TMD from known GPCR crystal structures with high sequence identity to the target protein sequence.
Step 3. For each template-based structure (using OptHelix or homology helix shapes), sample 127≈35 million combinations of simultaneous rotations (η) of all seven helices (using BiHelix), and then select the best 1000 conformations (based on energy) to build full seven-helix bundles (CombiHelix) with optimized side chains. From these conformations, select a few diverse structures with the lowest predicted energies as the starting points for subsequent simultaneous optimization of tilt angles (θ, φ) and helix rotation angles (η). This SuperBiHelix sampling generally involves (5 × 5 × 3)7 ≈ 13 trillion combinations. The best 2000 of these are selected and then built into seven-helix bundles (SuperComBiHelix), from which the best ∼20 conformations are selected (by energy) as the conformational ensemble that might play a role in GPCR function. These ∼20 conformations are then analyzed in terms of interhelical hydrogen bonds, particularly with regard to whether there are salt bridges between different TMDs, including the salt-bridge interaction between domains TM3 and TM6, which is associated with inactive conformations for many class A GPCRs (17). Typically, helix shapes based on homology lead to more stable conformations, except for CB1 (5) and DP (9), where helix shape based on OptHelix was best.
Step 4. Use the DarwinDock/GenDock (18, 19) method to validate the predicted GPCR structures by exhaustive sampling of poses of known agonists and antagonists (including numerous torsional conformations) over possible binding regions. This involves assessing the contributions of binding from various protein residues in the ligand-binding cavity (cavity analysis) and often comparisons of binding for a range of ligands (structure-activity relationship (SAR) analysis).
Step 5. Use homology or Monte Carlo procedures to add the loops and the amino (N-) and carboxyl (C-) terminal segments to the TMD of the predicted ligand-GPCR structures and optimize the loops by annealing. Build these complete predicted structures into the lipid membrane surrounded by explicit water and salt (∼60,000 atoms per cell) and carry out a modest MD simulation (10–50 ns) to validate the stability of the predicted structures. The goal of the MD simulations is to allow water and ions to interact with the ligand-protein complex to relax the predicted structures. Here, we analyze the changes in the strong interhelical interactions within the GPCR and strong ligand-protein interactions. For a valid structure, we expect to gain new interactions (sometimes due to water molecules) while not losing the original strong couplings. Such a short MD simulation would not allow an inactive structure to become activated.
Two GPCRs (rhodopsin and hβ2AR) have reported crystal structures in which the full or partial Gα subunit C-terminus of the G protein is bound to the IC side of the GPCR. It is believed that these structures capture a stable GPCR active state. A comparison of these active structures with their inactive-state counterparts shows that the active states have a different packing of the TM helices and that TM6 changes shape during activation. In hβ2AR, bovine rhodopsin (bRho), hM2, and mOPRM, the TM6 IC end moves horizontally away from its position relative to TM3 in the inactive state with a residue near the hydrophobic plane as the pivot point, resulting in the TM3–TM6 space on the IC side opening up by ∼3–5 Å (Table S1 in the Supporting Material). Both experiment and previous predictions provide strong evidence that each GPCR sequence may have multiple active states, with the active states generally being higher in energy (less stable) than the inactive state (20).
These insights inspired us to modify steps 2 and 3 of GEnSeMBLE to develop the ActiveGEnSeMBLE method. In ActiveGEnSeMBLE, for step 2, the template for the homology model is based (in addition to an inactive-state crystal structure) either on an available active-state crystal structure (for validation purposes only) or on another model based on a hybrid template in which only TM6 comes from an active-state crystal structure, whereas the other six TMs (TM1–5 and TM7) come from an inactive-state crystal structure. In ActiveGEnSeMBLE, step 3 of GEnSeMBLE is replaced by a two-step conformational sampling scheme for SuperBiHelix that includes a coarse conformational sampling aimed at locating structures in the active-state potential energy wells, followed by a finer conformational sampling starting from specific potential functionally diverse conformations identified by the coarse sampling. Coarse conformational sampling casts a wide net to catch conformations that are potentially active. Specific conformations from coarse sampling are identified along an activation coordinate by using a geometric criterion before the energetic criterion as described below. Fine conformational sampling starting from these conformations relaxes them in their local potential energy wells. We contend that this procedure is much faster and efficient than using a standard MD simulation to identify and relax active-like conformations.
In summary, the ActiveGEnSeMBLE method (Fig. S1) is performed as follows:
Step 1. Same as step 1 of GEnSeMBLE.
Step 2. Same as step 2 of GEnSeMBLE, except that we include a template based on an active-state crystal structure (for validation purposes only) and a hybrid template based on the inactive-state template with an active-state TM6 for the active conformation prediction.
-
Step 3.1. We sample the orientations of the helices using BiHelix/CombiHelix as in GEnSeMBLE, followed by a coarse SuperBiHelix/SuperComBiHelix (Δφ from −90° to 90° in 45° increments; Δθ = 0, ±15° and Δη = 0, ±30° as in GEnSeMBLE). In contrast to GEnSeMBLE, in which we select the conformations corresponding to the 20 lowest-energy states for further analysis, in ActiveGEnSeMBLE we measure the distance between the TM3 and TM6 IC ends (R36) of the 1000 lowest-energy structures generated during coarse conformational sampling. We also measure R36 for the inactive-state template, and denote it as R36(it).
To select potential active-state structures from the coarse conformational sampling, we search for the lowest-energy structure with R36 − R36(it) > 4 Å. We denote the selected structures S2.1 (for the case with hybrid or active-state initial template) and S3.1 (for the case with inactive-state initial template). We select potential inactive-state structures from the coarse conformational sampling (S4.1) using the same criteria described for GEnSeMBLE.
Step 3.2. All the structures identified in Step 3.1 go through Step 3.2a. In addition, for structure S3.1, there is an alternative treatment Step 3.2b:
Step 3.2a. Structure S2.1, S3.1, or S4.1 is used directly as the starting structure of a finer SuperBiHelix/SuperComBiHelix (φ from −30° to 30° in 15° increments; θ and η similar to GEnSeMBLE).
Step 3.2b. TM6 in structure S3.1 is replaced by a TM6 shape from an existing active-state crystal structure as described in Supporting Materials and Methods. The resulting structure, S3.1b, is then used as the starting structure for fine conformational sampling (Δφ from −30° to 30° in 15° increments; Δθ and Δη similar to GEnSeMBLE).
Step 3.3. To select final active-state candidates, check that the lowest-energy structure from step 3.2a or step 3.2b at least satisfies the criterion R36 − R36(it) > 3 Å. If it does, select this structure as a potential active-state conformation for the target protein. If it does not, check the second lowest-energy structure and so on. This step is carried out separately for different initial templates to have a set of candidate structures diverse in TMD shapes. The final inactive-state candidates are again selected using the same criteria as employed in GEnSeMBLE.
Step 4. Same as step 4 of GEnSeMBLE.
Step 5. Same as step 5 of GEnSeMBLE, except that an MD simulation of the docked active-state candidates with the agonist bound and the G protein bound is performed.
Defining the distance between the TM3 and TM6 IC ends, R36
We define R36 as the minimal approach distance between the IC ends of the TM3 and TM6 backbone atoms. We do not define it as the distance between residues 3.50 and 6.30 (denoted in the Ballesteros-Weinstein numbering scheme) (21), which usually form a salt bridge in the inactive-state crystal structure, because pure rotations of TM3 and TM6 can increase the distance between these two (or any two) residues without opening any space between the two TMs for Gα to couple to the receptor. To calculate R36, we use the following algorithm:
Step 1. Orient the GPCR such that its hydrophobic plane is in the x-y plane (z = 0), the extracellular (EC) end has positive z-coordinates, and the IC end has negative z-coordinates. For the IC ends of domains TM3 and TM6, the one with the less negative z-coordinate value is termed shortTM and the other one is termed longTM.
Step 2. Select a range of neighboring residues r1,r2,…,rn starting from the most IC residue of shortTM. In the example presented here, we used n = 4 because there are usually four residues per turn on a peptide α-helix.
Step 3. For each given residue, rm, selected in step 2, determine the z-coordinate zm for each of its backbone atoms. Calculate all distances between the shortTM backbone atoms in rm and the longTM backbone atoms with z-coordinates in the range of (zm − ΔzIC, zm + ΔzEC). In general, the value of ΔzIC is chosen to be 5.4 Å because this is the height of one turn of the α-helix. For structure-prediction steps, ΔzEC is chosen to be 5.4 Å. To analyze the trajectory from the MD simulation step discussed below, the value of ΔzEC is chosen such that zm + ΔzEC is about the same as the least-negative z-coordinate of the Gα subunit C-terminus in the Gα-coupled case. For the structure-prediction cases discussed in this work, the latter choice of ΔzEC gives the same R36 value as choosing ΔzEC = 5.4 Å.
Step 4. The smallest distance among all distances between TM3 and TM6 calculated in step 3 is R36.
This definition provides a robust geometric and steric measure of the IC distance between TM3 and TM6, which correlates with the potential of G protein coupling to the active conformations.
Using R36 values to facilitate selection of the active-state candidates
Let R36 for the active-state structure be R36(a) and that for the inactive-state structure be R36(i). Define ΔR36 = R36(a) − R36(i). Class A GPCRs hβ2AR and bRho are crystallized with the G protein or the C-terminus of the Gα subunit in complex with the GPCR, and they both have ΔR36 ≈ 4 Å (Table S1). For hM2 and mOPRM, their active states are crystallized with a G protein mimetic camelid antibody fragment and their ΔR36 values are ∼3 Å and 5 Å, respectively. Since the G protein couples to the receptor with the C-terminus of the Gα subunit inserted into the IC side of the GPCR in between TM3 and TM6, it is reasonable to estimate ΔR36 by adding the diameter of a peptide α-helix (2.3 Å) to a C-C single bond length (1.5 Å), which leads to 3.8 Å. Thus, we find ΔR36 > 3 Å as a reasonable target separation to locate active-like conformations. Below, we show that our final predicted active hSSTR5 structures result in ΔR36 ≈ 3.5 Å.
Details
Validation
Structure prediction
Starting from the TMD of the crystal structures of each validation case, hβ2AR (PDB ID: 3SN6 and 2RH1) and hM2 (PDB ID: 4MQS and 3UON), we performed step 3 according to the ActiveGEnSeMBLE protocol as described above. The energy ECNti was used in energy ranking and is defined in Supporting Materials and Methods. The resulting active and inactive conformations were compared with those observed in experimental structures.
MD of hβ2AR crystal structures
We performed an MD simulation of hβ2AR starting from its active-state crystal structure (PDB ID: 3SN6) and inactive-state crystal structure (PDB ID: 2RH1). The MD simulation was carried out for the following cases with an explicit lipid and water environment: agonist+active GPCR+Gαs, agonist+active GPCR, active GPCR (apo)+Gαs, active GPCR (apo), agonist+inactive GPCR, inactive GPCR (apo), Gαs, and agonist. In the absence of experimental structures, this can be done with the receptor conformations from ActiveGEnSeMBLE.
We chose the agonist to be BI-167107, which is in the binding site of the active-state crystal structure for hβ2AR. Only the Gα subunit of the G protein was included in the simulation because our main focus was on the binding interface between the GPCR and the G protein, which only involves the Gα subunit and the GPCR. The active GPCR (apo)+Gαs case starts from the equilibrated agonist+active GPCR (apo)+Gαs with the agonist removed.
MD simulation
For the MD simulation, we used the AMBER force-field engine implemented in NAMD 2.9 (22). The conjugate-gradient method was used in minimization. The Nosé-Hoover Langevin piston pressure control was used in the NPT dynamics. The 51 ns MD simulation was performed as detailed in Supporting Materials and Methods.
Energy analysis of the MD trajectories
We used the self-interaction energy function of NAMD 2.9 to perform a single-point energy calculation of each component in the complexes along the trajectories obtained from the MD simulation above. We carried out a minimization of 5000 steps on each frame before calculating the single-point energy. We then computed the energies for the receptor, the Gα protein, the ligand, and the interactions of Gα/ligand with the receptor, for whichever nonsolvent molecules were present in the complex to be studied. We then clustered the complexes along each trajectory that were saved every 100 ps, such that complexes within a root mean-square deviation (RMSD) of 2 Å of each other were grouped into one family. The families were then classified as inactive, intermediate, or active states, defined as R36 < 10 Å, 10 Å < R36 < 13 Å, and R36 > 13 Å, respectively. For each of these activation states of a trajectory, the mean value of the energy of each component was calculated and labeled as ER, EG, EL, ELR, ERG, or ELRG, with the component under consideration subscripted (R denotes the receptor, G denotes the Gα protein, L denotes the ligand, LRG denotes the ligand+receptor+Gα complex, LR denotes the ligand+receptor complex, and RG denotes the receptor+Gα complex). We also calculated the corresponding standard deviations. We were then able to calculate the total energy of the receptor plus the stabilization from the interaction between the receptor and the ligand and/or the Gα protein as follows:
The total energy of the system, with the internal energy of the ligand and the Gα protein also considered, was calculated as follows:
The superscript letters indicate the MD simulation case from which the energy was obtained, and the subscript letters indicate which components of the case were grouped to obtain the energy.
Application
Structure prediction and ligand docking
We applied the ActiveGEnSeMBLE method to the hSSTR5 receptor, for which no experimental structures are available. The exact procedures of steps 1–3 follow the ActiveGEnSeMBLE protocol mentioned above and are described in detail in our previous SSTR5 publication (23). The ligands (L-817,818 and F21) were docked to each of the five predicted hSSTR5 structures (InactiveConf1,2,3 and ActiveConf1,2) as described in Supporting Materials and Methods.
MD simulations and analysis
We carried out MD simulations of the following complexes: agonist+ActiveConf2+Gαi, agonist+ActiveConf2, apo-ActiveConf2+Gαi, apo-ActiveConf2, agonist+InactiveConf2, apo-InactiveConf2, Gαi alone, and agonist alone.
As with the MD simulation of hβ2AR, we chose to use these systems so that we could perform an interaction energy analysis of the MD trajectories to extract meaningful information regarding the GPCR activation mechanism. The starting agonist-GPCR complexes were the lowest-energy L-817,818-bound inactive- and active-state structures obtained from docking. We obtained the starting structure for apo-ActiveConf2+Gαi by removing the agonist from the last frame of a 51 ns MD simulation of agonist+ActiveConf2+Gαi.
The procedure used to build the starting system for the MD simulation is described in detail in Supporting Materials and Methods. The MD procedure and energy analysis were performed as described above. The inactive, intermediate, and active states in the energy analysis are defined as R36 < 8 Å, 8 Å < R36 < 11 Å, and R36 > 11 Å, respectively.
Results and Discussion
Method validation
Structure prediction
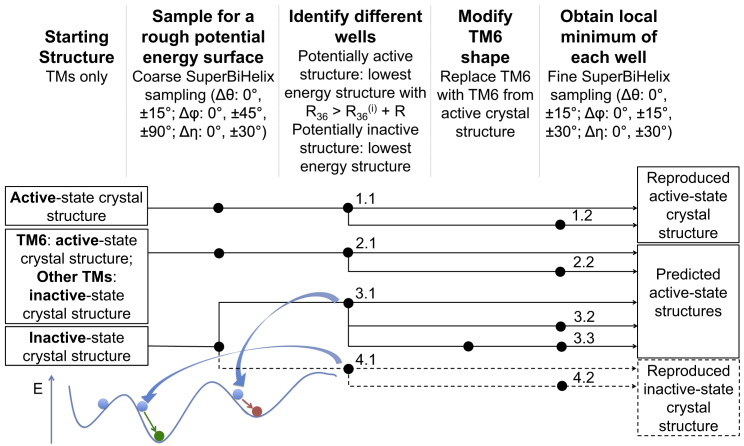
We validated the ActiveGEnSeMBLE method using the hβ2AR and hM2 receptor systems, for which both active-state and inactive-state structures have been crystallized. The ligand of bRho, retinal, is covalently bound to the GPCR. Since this does not represent the majority of class A GPCRs, which do not have covalently bound ligands, we did not consider bRho as a validation case. In addition, it is nontrivial to quantify the energies of the receptor with and without the ligand for a covalently bound ligand to account for the effect of the receptor-ligand interaction energy on the thermodynamic state of the receptor system. Fig. 1 summarizes the methods tested, with each final structure sharing the same numbering as the method that generated the structure. Starting from an active-state structure, an inactive-state structure, and a hybrid structure mixing active-state (TM6) and inactive-state (TM1–5 and TM7) helices, we compared the best final structures obtained by different methods with the active-state crystal structure. The structural features and energy value of the last node in every pipeline in Fig. 1 (i.e., the final structure from each method) are summarized in Table S2, with further details in Table S3. We assumed that the energy values and R36 values would be the only information that would be available for selecting candidate structures to predict an unknown structure. For hβ2AR and hM2, using the active-state crystal structure as a starting structure to predict active-state structures (method 1.x), the procedure is able to reproduce the TM orientations in the active-state crystal structure (Tables S2 and S3). This is a good (and necessary) test of the overall methodology and the force field, as they are able to identify a conformation close to the experimentally observed conformation out of (5 × 3 × 3)7 ≈ 374 billion conformations sampled. Similarly, using the inactive-state crystal structure as a starting structure to predict inactive-state structures (method 4.x), the procedure is able to reproduce the TM orientations in the inactive-state crystal structure (Tables S2 and S3).
Figure 1.
Methods for validating inactive- and active-state predictions using hβ2AR and hM2 as test cases. The black node indicates that the corresponding step is applied to the structure from the previous black node on the same line on the left. Each number beside a black node denotes the optimal structure obtained from the method that is denoted by the same number. To see this figure in color, go online.
Next, as a test of ActiveGEnSeMBLE on more practical cases, we find that both active-state prediction methods starting from either the inactive-state crystal structure or the hybrid structure (methods 2.x and 3.x; see rows with the first column 2.x and 3.x in Table S2) can reduce the RMSD of the predicted active-state candidate to the active-state crystal structure by 1.0 Å for hβ2AR and 0.8 Å for hM2. These numbers are significant because the RMSDs between the inactive-state and active-state crystal structures are 2.48 Å and 2.30 Å for hβ2AR and hM2, respectively. A comparison of different methods that start from the inactive-state crystal structure or the hybrid structure (method 2.1 versus 3.1, and method 3.3 versus 3.2; Table S2) shows that replacing the inactive-state TM6 shape with the active-state TM6 shape further lowers the RMSD between the candidate structure from sampling and the actual active-state crystal structure by ∼0.4 Å. This implies that the active-like TM6 shape plays an important role and may be necessary for a high-accuracy computational prediction of active-state GPCR structures.
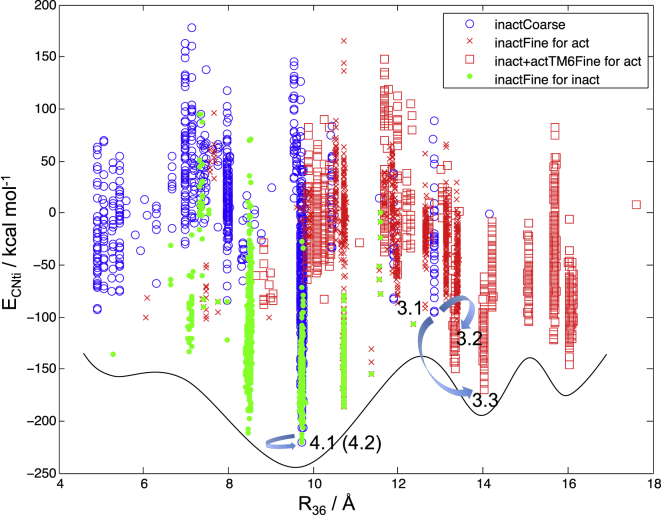
From the coarse sampling results, two structures are picked. One is for the inactive-state prediction (structure 4.1 of Fig. 1; Tables S2 and S3) and the other is for the active-state prediction (structure 3.1 of Fig. 1; Tables S2 and S3). From the energy profiles in Fig. 2 (hβ2AR) and Fig. S3 (hM2), it can be seen that the fine samplings can effectively achieve lower energies while keeping the R36 values of the lowest-energy final structures in the active-like energy well similar (within 1 Å) to those of the starting structures of the corresponding fine sampling. In other words, the fine sampling helps to locate the local minimum of a potential energy well (defined by R36) even when the activation coordinate R36 is not sampled explicitly. Although these potential energy profiles are crude, they are consistent with the hypothesis that structures with R36 values that deviate by >3–4 Å from R36(i) correspond to potential energy wells with higher energy local minima. In addition, they show that there are multiple higher-energy potential energy wells, which is consistent with biophysical evidence of multiple active states for a given GPCR. Furthermore, the recently published arrestin-bound receptor structure (24) was used to match arrestin to all the minimum energy structures of these energy wells for hβ2AR. We found conformations for hβ2AR that could accommodate arrestin, but not the Gs protein (Fig. S2). This is also consistent with the hypothesis that some of these different active states might be capable of activating different signaling pathways by coupling to different regulators. These results demonstrate that the ActiveGEnSeMBLE method is able to predict functionally distinct active-like conformations of GPCRs that might be responsible for coupling to different signaling pathways. The method has the potential to map out the activation landscape of a GPCR for multiple signaling pathways efficiently.
Figure 2.
Potential energy profile from sampling hβ2AR conformations. The black curve illustrates how our sampling results can be qualitatively translated into a potential energy curve using R36 as the x axis and does not quantitatively represent any real data. Results of the coarse sampling starting from the inactive-state crystal structure are in blue circles. Starting from structure 3.1, results of methods that generated structures 3.2 and 3.3 are shown in red crosses and red squares, respectively. Starting from structure 4.1, results of the fine sampling that generated structure 4.2 are in green dots. Every blue arrow points from a starting structure toward the optimal structure from the fine sampling of the corresponding method.
MD simulation and analysis
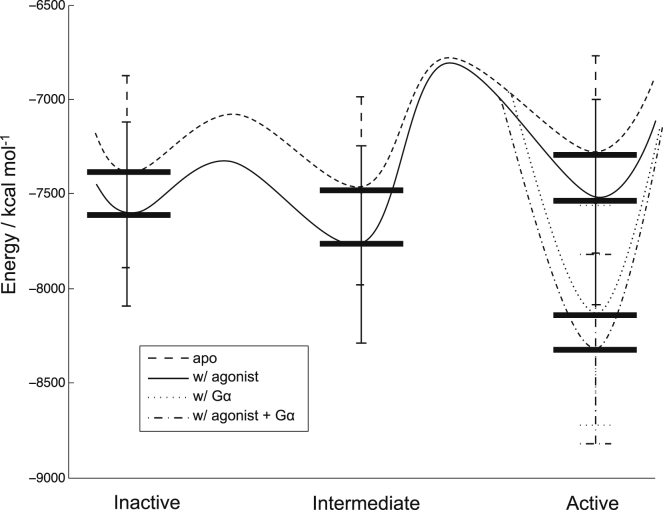
So far, only qualitative energy landscapes have been generated for GPCR activation, and the best-studied case is hβ2AR (25). To obtain a quantitative energy landscape that might provide more insights into GPCR activation to develop a strategy to elucidate the activation of GPCRs for which only predicted structures are available, we carried out MD simulations of hβ2AR starting from its crystal structures as described in Materials and Methods. Analysis of R36 during the MD simulations showed that the inactive conformation with and without the agonist remained inactive (Fig. S17), the Gαs-bound active conformation remained stable during MD, and the active conformation that was not bound to Gαs slowly lost its activity (i.e., a decrease in R36) during the 51 ns MD simulation as expected (Fig. S18). In addition, the apo-GPCR on average always had a slightly smaller R36 value than the agonist-bound GPCR toward the end of the 51 ns, which is consistent with the notion that the agonist shifts the equilibrium toward more activated states. We then grouped the conformations in corresponding trajectories by R36, with inactive, intermediate, and active states defined as R36 < 10 Å, 10 Å < R36 < 13 Å, and R36 > 13 Å, respectively, and calculated ER+interaction for each group. Note that each inactive, intermediate, and active state here contains multiple 3D conformational states that satisfy the respective R36 criterion. The resulting energy landscape (Fig. 3) is overall consistent with the qualitative picture obtained from experiments (Fig. S21, adapted from Ref. (25)).
Figure 3.
Energy landscape of hβ2AR activation from MD simulation. The horizontal bars are ER+interaction calculated as described in the Materials and Methods section. The curved lines are fictitious energy surfaces, with the barriers being qualitative and the minima defined by the corresponding ER+interaction values. Inactive, intermediate, and active states in the figure are defined as R36 < 10 Å, 10 Å < R36 < 13 Å, and R36 > 13 Å, respectively.
For agonist-bound (with BI-167107 being the agonist) and apo-GPCR systems, the energy is lower in the inactive state than in the active state, but higher than in the intermediate state (Fig. 3). This is a signature of BI-167107-bound hβ2AR as opposed to isoproterenol-bound hβ2AR, and was found in previous experiments (25).
When the Gα protein was present, both the agonist-bound and apo-GPCR energies were significantly lowered for the active state, which comprises the only group of states that can accommodate the G protein. This finding of a lower energy for agonist-bound GPCR upon coupling with Gα is consistent with experiments. Apo-GPCR coupled with the G protein is not in the picture of the experimental energy landscape, but we were able to do an MD simulation and analysis of this system, and rationalize our finding regarding its relative energy. To be specific, our results suggest that if an apo-GPCR can indeed couple with the G protein, its energy will be lower upon coupling, but not as low as that of the agonist-bound GPCR. This explains the basal activity of hβ2AR and suggests that an agonist is able to increase the activity of a GPCR by shifting the equilibrium from the apo-GPCR+Gα complex toward the more stable agonist+GPCR+Gα complex.
Furthermore, the agonist-bound GPCR had lower energy than the apo-GPCR for all other states as well, including the inactive states. This was also found in previous experiments (25). As the agonist-bound intermediate state remains the lowest-energy state among the three states, and the energy is lowered to a greater extent than in the other states, this supports the notion of a second route for GPCR activation that starts from an increased equilibrium population of the intermediate state upon binding with the agonist. Since the agonist-bound receptor is more dynamic in conformation than the apo-GPCR (25), the agonist-bound receptor can then be stabilized by the G protein as it transits from the intermediate state to the active state.
The above analysis is based on the quantitative energy landscape plotted with ER+interaction, which we chose because we wanted to eliminate any possible internal energy changes in the Gα and the ligand caused by Gα not coupling with the other subunits of the G protein and by the ligand strain, which may not be captured accurately by computational methods. Nevertheless, the energy profiles of ER and ETotal are shown in Figs. S19 and S20, respectively. Although ETotal is qualitatively the same as ER+interaction for hβ2AR, a subsequent analysis showed that they are different for hSSTR5 (Fig. S13). The energy profile of the receptor by itself (ER) shows that the receptor is actually destabilized by the G protein if the agonist is not present. Based on these results and our previous analysis, we conclude that upon GPCR coupling to the G protein, it is the interaction between the G protein and the GPCR that stabilizes the system, rather than the G protein directly lowering the energy of the GPCR by itself. This insight was lacking from experimental profiles, but we are able to deduce it from our analysis.
Application to hSSTR5
Structure prediction
We applied ActiveGEnSeMBLE to a GPCR without a known experimental structure, hSSTR5. The workflow is shown in Fig. S1, with mOPRM as the starting crystal structure template. When generating active conformations from hybrid templates, we carried out the optional BiHelix step in the flow chart in Fig. S1. The methods used for the inactive-state structural prediction were discussed in detail in our previous publication (23). When selecting the potentially active structure from the coarse sampling, we used 10.04 Å as a criterion because R36(it) is 6.04 Å. Similar to the cases with hβ2AR and hM2, Table S4 shows that fine conformational sampling is able to lower the energies starting from the results of coarse conformational sampling. In addition, all structures 3.x have R36 within 1.14 Å of each other, and all structures 4.x have R36 within 1.58 Å of each other. In other words, the R36 values of the lowest-energy structures from fine samplings are similar to those of the structures the fine samplings started with. This means that we successfully explored lower-energy structures for the inactive- and active-like states in their respective energy wells.
In addition to the methods described above, we also tested ActiveGEnSeMBLE using the hβ2AR active-state x-ray structure as the template for all of the hSSTR5 TMs. The lowest-energy structure has ECNti of −224.7 kcal mol−1, which is less stable than any of structures 3.x (−265.8 to −306.6 kcal mol−1), for which we used either the mOPRM inactive-state template or the hybrid template. As a result, we did not use this structure for the later steps. This suggests that the crystal structure of the hβ2AR active-state conformation may not be the best template for building an active-state homology model for many GPCRs. Since there are more inactive-state templates than active ones, if a target receptor has the best sequence homology to a receptor with only an inactive-state template, one should build the starting structure using that template. Then, one can use ActiveGEnSeMBLE on that structure to predict the active-state and inactive-state conformations of the target receptor.
The energy profile of hSSTR5 plotted against R36 is illustrated in Fig. S4. Compared with hβ2AR and hM2, hSSTR5 seems to have a flatter energy surface. This may be a feature of hSSTR5, but it could also be affected by the homology template used. As the energies of ActiveConf1 and ActiveConf2 are comparable, and we wanted to have a diverse set of candidate structures, we chose both of these structures as active-state candidates. To determine whether our predicted structures were reasonable, we carried out ligand-binding studies and MD simulations.
Ligand binding studies
To verify the predicted hSSTR5 structures, we docked five antagonists (26) (Fig. S5) and two agonists (27) (Fig. S6) to all five candidate structures (InactiveConf1, InactiveConf2, InactiveConf3, ActiveConf1, and ActiveConf2). Binding studies of the antagonists docking are discussed in detail in Dong et al. (23), in which the predicted binding energy of the antagonist series is consistent with experimental binding constants.
The agonists L-817,818 and F21 were selected because they have very high affinities (subnanomolar binding constants) for hSSTR5 and display both similar and different structural features. We did not select a series of agonists with the same scaffold for SAR analysis due to the lack of published experimental SAR data. The selected agonists’ structures are shown in Fig. S6. They are both peptide mimics derived from the endogenous ligand, somatostatin. They both have polycyclic aromatic groups and have a group mimicking lysine. However, L-817,818 has one more positively charged amine group than F21.
For each agonist, docking to the five predicted structures shows that the lowest-energy poses with the inactive state and the active state are only slightly different (Table S5; Figs. S7 and S8). This suggests an easy pathway for the agonist-bound receptor to interconvert between the inactive and active states, consistent with experiments on hβ2AR (25). Binding energies from docking have also shown that, in the absence of a G protein, the agonist stabilizes its inactive conformation. This again is consistent with experiments on hβ2AR (25) and is further supported by an interaction energy analysis of MD trajectories described in the next section.
We also found that for both agonists (L-817,818 and F21), the best active-state pose was achieved with ActiveConf2 (Table S5). Since ActiveConf2 was generated using structure-prediction method 3.3, this again suggests that the TM6 shape is an important factor in GPCR activation, and a more active-like TM6 shape makes computational prediction of active-state conformations more likely to succeed. Contrary to the antagonist M59 (the highest-binding-affinity antagonist we docked), which favored InactiveConf1, the best inactive-state poses for both agonists were observed with InactiveConf2, which had a slightly higher energy (by 5 kcal mol−1) and greater R36 (by 0.4 Å) than InactiveConf1. This implies that agonists may stabilize a slightly more active inactive state than antagonists, and is a direct demonstration of the ability of ActiveGEnSeMBLE to predict multiple states, which is crucial for elucidating GPCR activation mechanisms.
To date, no published mutation studies have probed the interaction between hSSTR5 and small-molecule ligands. In addition, only limited mutation data are available regarding the somatostatin receptors (SSTRs) binding with the endogenous somatostatin-14 (S-14) and somatostatin-28 (S-28), especially in the TM regions. Here, we will attempt to compare our binding analysis with the available experimental mutation data, and suggest mutations that can be experimentally tested to probe the binding of agonists L-817,818 and F21.
As shown in Fig. S7, the positively charged amine group in the ligand F21 forms a salt bridge with D1193.32 on TM3, which is common among the closely related opioid receptors. We can also see that ActiveConf2 forms a hydrophobic pocket that is in contact with the majority of nonpolar groups in F21. Note that the hydrophobic pocket includes F2646.51 on TM6. It is known that the mutation F6.52Y in rat SSTR5 (rSSTR5) can increase the binding affinity of S-14 to rSSTR5 by 20-fold (28), and this means that F2656.52 is involved in SSTR5-agonist binding. Since F2656.52 and F2646.51 are neighboring residues and the ligand F21 is much smaller than S-14, we can hypothesize that F2646.51 is important for F21 binding with the activated hSSTR5. Notice that residue 6.52 is tyrosine for all other SSTRs, but F6.51 is conserved in all SSTRs. As a result, we suggest that F6.51 is responsible for the affinity of SSTRs to the ligand F21, but not for the selectivity.
L-817,818 has different binding modes because of its two positively charged amine groups. The 3D visualization and ligand interaction diagrams (LIDs) of the best pose with the inactive state (L_i2) and the active state (L_a2) are shown in Fig. S8. Similar to what was observed for ligand F21, the salt bridge between one amine group with D1193.32 is present, and there is a π-π interaction between the aromatic groups in L-817,818 and F2646.51. The other lysine-like amine group in L-817,818 forms a hydrogen bond with Q1233.36 and has the potential to have an electrostatic interaction with D862.50. In the MD simulation discussed in more detail in the next section, we found that this amine group was indeed forming a salt bridge with D862.50 in the active-state simulation. Thus, we suggest that D862.50 is involved in hSSTR5 activation by L-817,818. Also, the hSSTR5 mutant F264Y is predicted to have a higher affinity for L-817,818 due to the potential of an additional hydrogen bond between the ester oxygen atom in L-817,818 and Y264.
MD simulation and analysis
To further investigate the activation mechanism of hSSTR5, we carried out MD simulations on the L-817,818-bound and apo predicted structures. We considered six cases: 1) agonist+ActiveConf2+Gαi, 2) agonist+ActiveConf2, 3) apo-ActiveConf2+Gαi, 4) apo-ActiveConf2, 5) agonist+InactiveConf2, and 6) apo-InactiveConf2. The binding site of agonist+ActiveConf2 is from L_a2, and that of agonist+InactiveConf2 is from L_i2. As expected, similar to the case with hβ2AR, the inactive state remained inactive during the MD simulation (Fig. S9), and the coupling of Gαi was able to keep both the agonist-bound GPCR and apo-GPCR active during the MD simulation (Fig. S11). Interestingly, contrary to the case with hβ2AR, where both the agonist-bound GPCR and apo-GPCR lost their activity during the MD simulation, for hSSTR5, starting from active-state conformations without Gαi, the apo-GPCR was able to keep hSSTR5 with a large R36 capable of coupling to Gαi, whereas the agonist-bound GPCR returned to its intermediate state within 5 ns and to its inactive state in 28 ns (Fig. S10). This suggests that the constitutive activity of hSSTR5 plays an important role in its activation mechanism. The G protein is more likely to couple to hSSTR5 before the agonist binds.
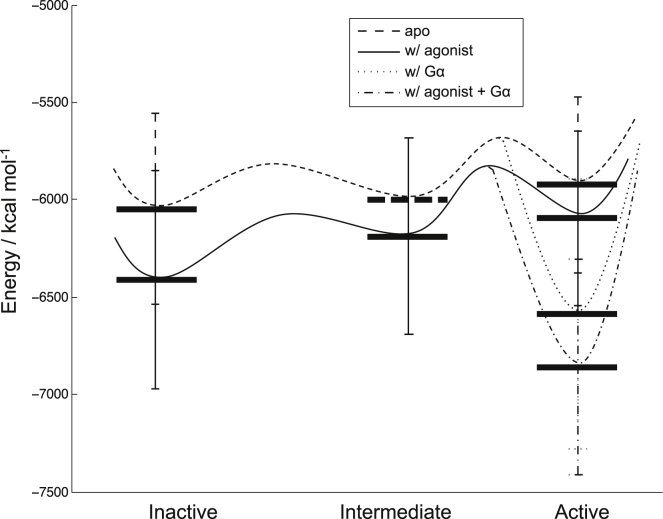
For each case, we analyzed the potential energy of the agonist-bound GPCR and apo-GPCR along the MD trajectory, and the resulting energy landscape (Fig. 4) is consistent with the picture derived from experimental findings for hβ2AR (20, 25). To be more specific, although the active state of a GPCR has higher energy than its inactive state, binding of the G protein lowers the energy of the active state of the GPCR, and ER+interaction of the apo-GPCR+Gαi complex is not as low as the agonist-bound complex. This activation picture is another example (besides hβ2AR) that quantitatively confirms the notion that the G protein facilitates GPCR activation by stabilizing both the agonist-bound GPCR and apo-GPCR. The former leads to agonist-induced activity and the latter leads to constitutive activity.
Figure 4.
Energy landscape of hSSTR5 activation from MD simulation. The solid horizontal bars are ER+interaction calculated as described in the Materials and Methods section. The dashed horizontal bar is fictitious. The curved lines are fictitious energy surface, with the barriers being qualitative and minima defined by the corresponding ER+interaction values. Inactive, intermediate, and active states in the figure are defined as R36 < 8 Å, 8 Å < R36 < 11 Å, and R36 > 11 Å, respectively.
When we compare the energy landscape of hβ2AR in Fig. 3 with that of hSSTR5 in Fig. 4, we find that their intermediate states have different features. The intermediate states of the BI-167107-bound hβ2AR have lower energy than the inactive states, whereas those of the L-817,818-bound hSSTR5 have higher energy than the inactive states. This can be reasonable because the energy landscape of even the same receptor can be altered by different agonists. For example, another hβ2AR agonist, isoproterenol, makes hβ2AR have higher-energy intermediate states than inactive states (Fig. S21) (25). In addition, the definition of intermediate state here includes many different states that have R36 between the inactive state and the states that can couple to the G protein, and the average energy of these intermediate states depends on the distribution of these states in the MD trajectory.
Furthermore, experimental structures of several agonist-bound GPCRs have exhibited features ranging from inactive to various degrees of partially active without the presence of the G protein, suggesting that the agonist plays a role in the initial steps of GPCR activation that varies with different GPCRs. For example, for the turkey β1 adrenergic receptor, the agonist-bound structure is nearly identical to the antagonist-bound inactive-state structure except for a 1 Å contraction of the binding pocket (29), whereas for the human adenosine A2A receptor, the agonist-bound structure has all active-like features except that the IC end of TM6 is only partially opened for coupling to the G protein (Table S1) (30).
The agonist-bound human serotonin 1B (5-HT1B) receptor, human serotonin 2B (5-HT2B) receptor, and rat neurotensin 1 receptor (NTSR1) do not have an antagonist-bound inactive-state structure of the same receptor for comparison, but they show partially active features. Bound with the same agonist, the 5-HT2B receptor has a less active TM6 and a more active TM7 than the 5-HT1B receptor, and they both exhibit an outward shift of the TM6 IC end compared with the inactive-state structures of other aminergic receptors (31, 32). NTSR1 also shows active-like features found in bRho and hβ2AR, with a TM6 position similar to that of the active-state bRho, but not to the extent of active-state hβ2AR (33).
Therefore, we conclude that different GPCRs may adopt different activation pathways with different agonists in terms of the energy ordering of different states dictated by the respective energy landscapes of activation. It is reasonable that our agonist-bound inactive states of hSSTR5 have the lowest energy among its various states that display different degrees of activation. Thus, we consider that the computation of the energy landscape of GPCRs using the method detailed in this work is valuable because it may allow for the activation mechanisms of a broader variety of GPCRs to be mapped out efficiently.
Similar to the case with hβ2AR, the hSSTR5 energy profile plotted with ER (Fig. S12) also shows that the G protein destabilizes the receptor, except that in the case of hSSTR5 this occurs regardless of whether the agonist is bound. This may be reasonable because the hSSTR5 agonist L-817,818 destabilizes hSSTR5, whereas the hβ2AR agonist BI-167107 stabilizes hβ2AR. Since ER+interaction shows a lower energy for the agonist-bound hSSTR5, we can conclude that the stabilization of hSSTR5 comes from the interaction between the agonist and the receptor, in addition to the interaction between Gαi and the receptor.
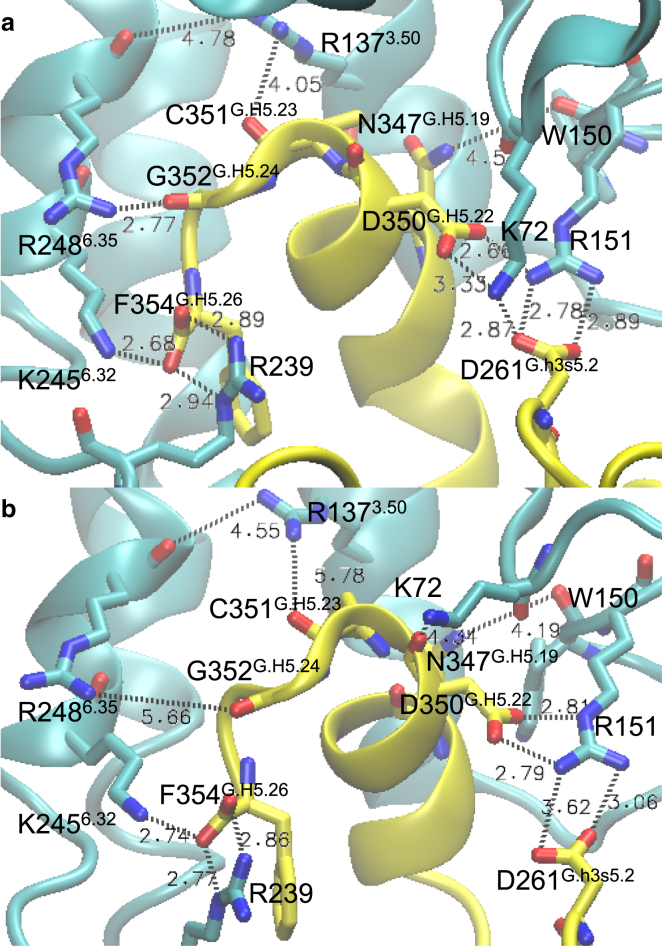
The stabilizing effect of the G protein on the active state of the GPCR can indeed be explained by the specific interactions between the Gα subunit and the GPCR. During the MD simulation of agonist+ActiveConf2+Gαi, salt-bridge and hydrogen-bond networks were able to form between the C-terminal helix of Gαi and ActiveConf2, as shown in Fig. 5. In particular, the formation of a salt-bridge network involving Gαi’s D261G.h3s5.2 and D350G.H5.22, and hSSTR5’s K72 on IC loop 1 (ICL1) and R151 on ICL2, and the formation of a hydrogen bond between R1373.50 and Gαi’s C351G.H5.23 replace the inactive state’s R151-D1363.49-R1373.50-T2476.34 network. An additional salt-bridge network is formed between the carboxylate group on the Gαi C-terminal residue F354G.H5.26 and hSSTR5’s K2456.32 on TM6 and R239 on ICL3. In addition, a hydrogen bond is formed between R2486.35 and G352G.H5.24, and a weaker hydrogen bond is formed between W150 on ICL2 and N347G.H5.19. Furthermore, the highly conserved L348G.H5.20 and L353G.H5.25 are in a hydrophobic pocket that consists of V1413.54, I2245.61, V2466.33, M2496.36, and V228 on ICL3. Since an experimental mutagenesis study (34) showed that the mutations L348AG.H5.20, L353AG.H5.25, and G352AG.H5.24 severely hindered coupling between Gαi and light-activated bRho (bRho∗), which has the conserved residues V1393.54, L2265.61, V2506.33, M2536.36, and R2526.35, we may conclude that the corresponding interactions we found between hSSTR5 and Gαi are consistent with experiments. The same experimental study showed that N347AG.H5.19 did not have a significant effect on coupling between Gαi and bRho∗, which is consistent with the weak interaction we found between W150 and N347G.H5.19. Interestingly, the mutation D350AG.H5.22 seems to stabilize the bRho∗-Gαi complex. This may be a property specific to bRho∗-Gαi arising from the differences in helix packing and ICL sequences between bRho and hSSTR5.
Figure 5.
(a and b) Interactions between the Gαi C-terminus and hSSTR5 after a 51 ns MD simulation of (a) agonist+ActiveConf2+Gαi and (b) apo-ActiveConf2+Gαi. Gαi is shown in yellow and hSSTR5 is shown in cyan. The superscript of Gα residue numbers follows the common Gα numbering system (53). The coordinates of the agonist+ActiveConf2+Gαi complex shown are included in the Supporting Material as a separate file in PDB format.
After the agonist is removed, K72 on ICL1 breaks away from D350G.H5.22 and D261G.h3s5.2, and forms a hydrogen bond with the backbone oxygen atom of D350G.H5.22. In addition, the hydrogen bond between R1373.50 and the C351G.H5.23 backbone oxygen atom, and the hydrogen bond between R2486.35 and the G352G.H5.24 backbone oxygen atom become water mediated. The weakening of the interaction between the apo-GPCR and Gα is consistent with the notion that the agonist stabilizes the binding of the G protein with GPCR.
When we examined the protein-ligand interactions in more detail, we found a characteristic interaction that formed at ∼25 ns of the agonist+ActiveConf2+Gαi MD simulation but was absent in the entire agonist+InactiveConf2 MD trajectory: a salt bridge between the lysine-like positively charged amine of L-817,818 and D862.50 on hSSTR5 (Figs. S14–S16). The highly conservative residue 2.50 has been studied in several GPCRs, but its role is not well understood and varies across different systems (35, 36). In particular, the mutation D2.50N has different effects in SSTR1 and SSTR2. Although D2.50 is widely viewed as an allosteric site, our result suggests that the orthosteric site of L-817,818 in hSSTR5 may extend to D2.50. Therefore, our result raises the possibility that D862.50 plays a crucial role in hSSTR5 activation by engaging in the interaction with the agonist, and further experimental investigations of this residue would be worthwhile. If the significance of D862.50 is verified, designing agonists that are able to form salt bridges with both D1193.32 and D862.50 may be a desirable path toward developing drugs that target hSSTR5.
Conclusions
We have presented a new, to our knowledge, method for GPCR structure prediction, termed ActiveGEnSeMBLE, that overcomes the conformational sampling limits of MD simulations. This method can be used to identify multiple energetically accessible conformations for a GPCR that might play a role in its activation in addition to multiple lower-energy structures that might correspond to in inactive states. We validated ActiveGEnSeMBLE by predicting active hβ2AR and hM2 crystal structures. We found that ActiveGEnSeMBLE sampled the orientations of the TM helices and located structures in various energy wells spanning the range of TM3–TM6 distances (R36) traversed in the process of activation. Subsequent analysis revealed a local minimum in each of these energy wells that was close or identical to a crystal-structure conformation with a similar R36 value. MD simulations of the crystal structures of hβ2AR with and without the G protein and the agonist generated energy profiles that are consistent with the qualitative energy landscape of hβ2AR obtained from experiments, providing information about how the ligand and G protein may play a role in activation. These results indicate that the agonist alone is not enough to stabilize the active state, and that the Gα C-terminal chain needs to be bound to the GPCR to promote activation, in agreement with conclusions from experiments.
We then applied the validated ActiveGEnSeMBLE method to the hSSTR5 receptor, for which there is no available experimental structure. Importantly, we found that a hybrid template consisting of the TM6 from the available active-state crystal structure combined with TM1–5 and TM7 of inactive-state crystal structures from GPCRs with high sequence identity generated even lower-energy active-like structures than a template based purely on the available active-state crystal structures. Thus, it is not necessary to have the full structure for an active GPCR to apply ActiveGEnSeMBLE. Docking of agonists and subsequent MD simulations identified important residues involved in hSSTR5 activation by the respective agonists. MD simulations of the predicted structures of hSSTR5 with and without the G protein and the agonist generated energy profiles that are consistent with the qualitative energy landscape of hβ2AR obtained from experiments and also with the quantitative energy landscape of hβ2AR presented in this study. The differences are compatible with previous findings from agonist-bound experimental structures for various GPCRs in that the agonist promoted the initial steps of GPCR activation to degrees that varied among different GPCRs. These energy profiles indicate that the G protein helps to stabilize the agonist-bound GPCR. These results confirm that ActiveGEnSeMBLE is effective in predicting the active-state conformations of at least class A GPCRs, and provides a powerful new tool for elucidating the activation mechanisms of GPCRs by identifying the sequence of conformations along the pathway for activation. We hope that this will accelerate the rational design of new, more potent and selective agonists.
Author Contributions
S.S.D. designed and performed research, and wrote the manuscript. W.A.G. and R.A. designed research and wrote the manuscript.
Acknowledgments
We thank Dr. Soo-Kyung Kim, Adam Griffith, and Dr. Vaclav Cvicek for helpful discussions.
Partial support was provided by the NSF (EFRI-1332411), the GIST-Caltech Project, the NIH (R01AI040567), and donors to the Materials and Process Simulation Center at California Institute of Technology. Partial support for R.A. was provided from startup funds from Cedars-Sinai Medical Center.
Editor: Bert de Groot.
Footnotes
Supporting Materials and Methods, twenty-one figures, five tables, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30231-4.
Contributor Information
William A. Goddard, III, Email: wag@wag.caltech.edu.
Ravinder Abrol, Email: abrolr@csmc.edu.
Supporting Citations
References (37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52) appear in the Supporting Material.
Supporting Material
References
- 1.Kenakin T., Miller L.J. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrol R., Griffith A.R., Goddard W.A., 3rd Structure prediction of G protein-coupled receptors and their ensemble of functionally important conformations. Methods Mol. Biol. 2012;914:237–254. doi: 10.1007/978-1-62703-023-6_14. [DOI] [PubMed] [Google Scholar]
- 3.Abrol R., Trzaskowski B., Irons C. Ligand- and mutation-induced conformational selection in the CCR5 chemokine G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2014;111:13040–13045. doi: 10.1073/pnas.1413216111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S.K., Riley L., Goddard W.A., 3rd Predicted structures of agonist and antagonist bound complexes of adenosine A3 receptor. Proteins. 2011;79:1878–1897. doi: 10.1002/prot.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott C.E., Abrol R., Goddard W.A., 3rd Molecular basis for dramatic changes in cannabinoid CB1 G protein-coupled receptor activation upon single and double point mutations. Protein Sci. 2013;22:101–113. doi: 10.1002/pro.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan J., Abrol R., Goddard W.A., 3rd 3D structure prediction of TAS2R38 bitter receptors bound to agonists phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP) J. Chem. Inf. Model. 2012;52:1875–1885. doi: 10.1021/ci300133a. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.-K., Goddard W.A., 3rd Predicted 3D structures of olfactory receptors with details of odorant binding to OR1G1. J. Comput. Aided Mol. Des. 2014;28:1175–1190. doi: 10.1007/s10822-014-9793-4. [DOI] [PubMed] [Google Scholar]
- 8.Kirkpatrick A., Heo J., Goddard W.A., 3rd Predicted structure of agonist-bound glucagon-like peptide 1 receptor, a class B G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2012;109:19988–19993. doi: 10.1073/pnas.1218051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., Zhu F., Laoui A. Prediction of the 3D structure and dynamics of human DP G-protein coupled receptor bound to an agonist and an antagonist. J. Am. Chem. Soc. 2007;129:10720–10731. doi: 10.1021/ja070865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherezov V., Rosenbaum D.M., Stevens R.C. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen S.G.F., DeVree B.T., Kobilka B.K. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haga K., Kruse A.C., Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruse A.C., Ring A.M., Kobilka B.K. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W., Manglik A., Kobilka B.K. Structural insights into μ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Tanaka M., Gingras M.C. Association between somatostatin receptor 5 gene polymorphisms and pancreatic cancer risk and survival. Cancer. 2011;117:2863–2872. doi: 10.1002/cncr.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobarec J.C., Sanchez R., Filizola M. Modern homology modeling of G-protein coupled receptors: which structural template to use? J. Med. Chem. 2009;52:5207–5216. doi: 10.1021/jm9005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trzaskowski B., Latek D., Filipek S. Action of molecular switches in GPCRs—theoretical and experimental studies. Curr. Med. Chem. 2012;19:1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floriano W.B., Vaidehi N., Goddard W.A., 3rd HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J. Med. Chem. 2004;47:56–71. doi: 10.1021/jm030271v. [DOI] [PubMed] [Google Scholar]
- 19.Goddard W.A., 3rd, Kim S.-K., Abrol R. Predicted 3D structures for adenosine receptors bound to ligands: comparison to the crystal structure. J. Struct. Biol. 2010;170:10–20. doi: 10.1016/j.jsb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Manglik A., Kobilka B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 22.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong S.S., Abrol R., Goddard W.A., 3rd The predicted ensemble of low-energy conformations of human somatostatin receptor subtype 5 and the binding of antagonists. ChemMedChem. 2015;10:650–661. doi: 10.1002/cmdc.201500023. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y., Zhou X.E., Xu H.E. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manglik A., Kim T.H., Kobilka B.K. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin R.E., Mohr P., Christ A.D. Benzoxazole piperidines as selective and potent somatostatin receptor subtype 5 antagonists. Bioorg. Med. Chem. Lett. 2009;19:6106–6113. doi: 10.1016/j.bmcl.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Feytens D., De Vlaeminck M., Reubi J.C. Highly potent 4-amino-indolo[2,3-c]azepin-3-one-containing somatostatin mimetics with a range of sst receptor selectivities. J. Med. Chem. 2009;52:95–104. doi: 10.1021/jm801205x. [DOI] [PubMed] [Google Scholar]
- 28.Ozenberger B.A., Hadcock J.R. A single amino acid substitution in somatostatin receptor subtype 5 increases affinity for somatostatin-14. Mol. Pharmacol. 1995;47:82–87. [PubMed] [Google Scholar]
- 29.Warne T., Moukhametzianov R., Tate C.G. The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebon G., Warne T., Tate C.G. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wacker D., Wang C., Stevens R.C. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., Jiang Y., Xu H.E. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340:610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White J.F., Noinaj N., Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D., Flock T., Veprintsev D.B. Probing Gαi1 protein activation at single-amino acid resolution. Nat. Struct. Mol. Biol. 2015;22:686–694. doi: 10.1038/nsmb.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong H., Raynor K., Reisine T. Mutation of an aspartate at residue 89 in somatostatin receptor subtype 2 prevents Na+ regulation of agonist binding but does not alter receptor-G protein association. Mol. Pharmacol. 1993;44:380–384. [PubMed] [Google Scholar]
- 36.Roche J.P., Bounds S., Mackie K. A mutation in the second transmembrane region of the CB1 receptor selectively disrupts G protein signaling and prevents receptor internalization. Mol. Pharmacol. 1999;56:611–618. doi: 10.1124/mol.56.3.611. [DOI] [PubMed] [Google Scholar]
- 37.Schrodinger, LLC . Schrodinger, LLC; New York: 2009. MacroModel, version 9.7. [Google Scholar]
- 38.Schrodinger, LLC . Schrodinger, LLC; New York: 2010. Maestro, version 9.1. [Google Scholar]
- 39.Chang G., Guida W.C., Still W.C. An internal-coordinate Monte Carlo method for searching conformational space. J. Am. Chem. Soc. 1989;111:4379–4386. [Google Scholar]
- 40.Saunders M., Houk K.N., Guida W.C. Conformations of cycloheptadecane. A comparison of methods for conformational searching. J. Am. Chem. Soc. 1990;112:1419–1427. [Google Scholar]
- 41.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 42.Schrodinger, LLC . Schrodinger, LLC; New York: 2009. Jaguar, version 7.6. [Google Scholar]
- 43.Manglik A., Kruse A.C., Granier S. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choe H.-W., Kim Y.J., Ernst O.P. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 45.Okada T., Sugihara M., Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 46.Jaakola V.-P., Griffith M.T., Stevens R.C. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrodinger, LLC . Schrödinger, LLC; New York: 2012. Maestro, version 9.3. [Google Scholar]
- 48.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 49.Case D.A., Darden T., Merz K. University of California; San Francisco: 2010. Amber 11. [Google Scholar]
- 50.Lim K.-T., Brunett S., Goddard W.A. Molecular dynamics for very large systems on massively parallel computers: the MPSim program. J. Comput. Chem. 1997;18:501–521. [Google Scholar]
- 51.Schwede T., Kopp J., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 53.Flock T., Ravarani C.N., Babu M.M. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.