Summary
We describe the impact of two propeptides and PedC on the production yield and the potency of recombinant pediocins produced in L actococcus lactis. On the one hand, the sequences encoding the propeptides SD or LEISSTCDA were inserted between the sequence encoding the signal peptide of Usp45 and the structural gene of the mature pediocin PA‐1. On the other hand, the putative thiol‐disulfide oxidoreductase PedC was coexpressed with pediocin. The concentration of recombinant pediocins produced in supernatants was determined by enzyme‐linked immunosorbent assay. The potency of recombinant pediocins was investigated by measuring the minimal inhibitory concentration by agar well diffusion assay. The results show that propeptides SD or LEISSTCDA lead to an improved secretion of recombinant pediocins with apparently no effect on the antibacterial potency and that PedC increases the potency of recombinant pediocin. To our knowledge, this study reveals for the first time that pediocin tolerates fusions at the N‐terminal end. Furthermore, it reveals that only expressing the pediocin structural gene in a heterologous host is not sufficient to get an optimal potency and requires the accessory protein PedC. In addition, it can be speculated that PedC catalyses the correct formation of disulfide bonds in pediocin.
Introduction
Bacteriocins from lactic acid bacteria are defined as antimicrobial proteinaceous compounds synthesized by ribosomes (Riley, 2009). They are used in food as bio‐preservatives and were lately suggested as drug candidates and probiotic promoting factors (Dobson et al., 2012). As bio‐preservatives, they can be added to food following three different strategies: in situ into fermented food by bacterial culture which constitutes the starter culture, directly in a purified or semi‐purified form (e.g. nisin A, nisaplin, Danisco) and as an ingredient based on a fermentation of a bacteriocin‐producing strain (pediocin PA‐1, ALTA 2431, Quest International) (Cotter et al., 2013). Utilization of bacteriocins as drugs in human or veterinary health has also been considered (Cotter et al., 2013; Van Heel et al., 2011). The industrial use of bacteriocins requires efficient downstream and upstream processes. However, low production rate by natural bacterial producers can dramatically impair further application of bacteriocins (Jack et al., 1996; Guyonnet et al., 2000; Jasniewski et al., 2008).
Heterologous expression is an efficient strategy to enhance bacteriocin production yields. The lactic acid bacterium Lactococcus lactis was successfully used to produce bacteriocins (Rodríguez et al., 2003) including pediocin PA‐1 (Horn et al., 1998; 1999; 2004; Reviriego et al., 2005, 2007a, 2007b; Martín et al., 2007a; Arqués et al., 2008), and could therefore be used as a host for large‐scale production. Besides, due to its generally recognized as safe status, L. lactis is an interesting bacterium for in vivo delivery of bacteriocins for probiotic purpose (Osmanagaoglu et al., 2010; Pontes et al., 2011; Dobson et al., 2012) or in fermented food (Renye and Somkuti, 2010). Another strategy, which aims at improving the secretion step and thereby the overall productivity, is to replace the wild‐type signal peptide of the recombinant protein by the Usp45 signal peptide (SPusp45), thus allowing secretion of pediocin PA‐1 through the secretory pathway (Li et al., 2011). However, although recombinant pediocin PA‐1 was successfully produced, little attention has been paid to the fact that the positive charges of pediocin PA‐1 might impair secretion through the secretory pathway. Indeed, the study of recombinant protein production in L. lactis showed that the insertion of the peptide LEISSTCDA between the signal peptide and the recombinant protein NucB allowed to increase production of NucB from 3 mg/L to 15 mg/L, highlighting the importance of the nature of the amino acids localized immediately after the cleavage site of the signal peptidase (Le Loir et al., 1998; Pontes et al., 2011). The class IIa bacteriocin divercin RV41 was successfully secreted through the secretory pathway with the remaining propeptide SD fused to the bacteriocin at the N‐terminus after the cleavage site. This propeptide SD results from the cloning procedure in the NsiI restriction site that allows translational fusion between SPusp45 and the coding sequence of interest (Bermúdez‐Humarán et al., 2007). As previously suggested (Morello et al., 2008), it could be expected that the negative charge of aspartate in the propeptide SD might enhance protein secretion. However, this point, as well as the impact of the propeptide SD on the divercin RV41 specific activity, has not been investigated.
Pediocin PA‐1 is a model class IIa bacteriocin produced by several strains from Pediococcus and Lactobacillus species. This bacteriocin exhibits antibacterial activity against a wide spectrum of food‐borne pathogens and food spoilage gram‐positive bacteria (Rodríguez et al., 2002). As other class IIa bacteriocins, it affects both membrane permeability and membrane potential (Rodríguez et al., 2002). Mature pediocin PA‐1 is a 44 amino acid peptide containing four cysteine residues involved in two disulfide bonds (Henderson et al., 1992). The pediocin PA‐1 operon consists of the four genes named pedA, pedB, pedC and pedD, which encode the prepediocin PA‐1, the pediocin PA‐1 immunity protein, an accessory export protein and an ABC transporter (involved in the excision of the signal peptide and the secretion of pediocin) respectively (Venema et al., 1995).
The aim of this work was to investigate the efficiency of heterologous production of pediocin in Lactococcus lactis at the quantitative and the qualitative levels with a special focus on protein secretion and post‐translational modification.
Results
Impact of propeptides on secretion efficiency
The three plasmids pSec::rpedA, pSec::s‐rpedA and pSec::l‐rpedA were constructed with the aim to produce three different recombinant pediocins (Fig. 1). The three plasmids contain chimeras where spusp45, which encodes the signal peptide of Usp45, was fused to the mature pediocin encoding sequence ΔsppedA. In the plasmid pSec::rpedA, spusp45 was directly fused to ΔsppedA which theoretically leads, after cleavage by the signal peptidase, to the release of a recombinant pediocin (Rpediocin) exhibiting the primary structure of the wild‐type mature pediocin PA‐1. In the plasmids pSec::s‐rped and pSec::l‐rped, the sequences encoding the propeptides SD and LEISSTCDA, respectively, were inserted between spusp45 and Δsp::pedA. These plasmids theoretically lead to the secretion of recombinant fusion pediocins PA‐1, named S‐Rpediocin and L‐Rpediocin, that would exhibit the peptide SD and LEISSTCDA at the N‐terminus respectively. The three plasmids were used to transform L. lactis, and the resulting strains were cultivated in order to produce the expected recombinant pediocins. The strain L. lactis NZ9000_pSec::nucB was used as control. This control strain produces an extracellular staphylococcal nuclease (NucB) with no known antibacterial property.
Figure 1.
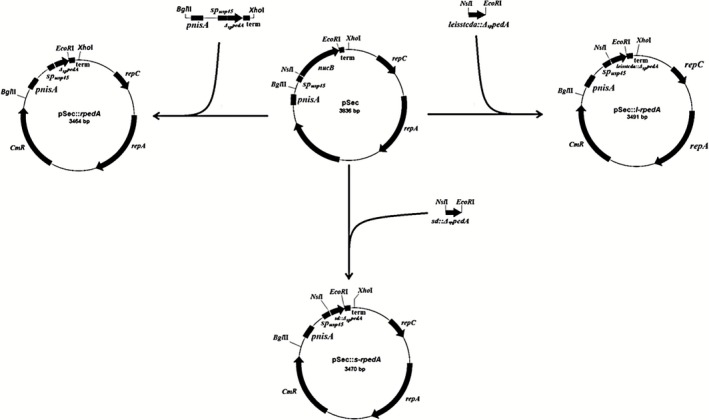
Construction of pSec derivatives. The restriction sites used for cloning purpose are indicated. The plasmid pSec and derivatives contain the nisin inducible promoter PnisA, the replication genes repA and repC, the gene conferring resistance to chloramphenicol and a transcriptional terminator term.
For each producer, a dry weight of 0.7 g/L was measured, indicating that all strains approximately produced the same biomass. Furthermore, the maximum specific growth rate (0.8 h−1) was not significantly different between strains. The concentration of each recombinant pediocin was measured by Enzyme‐Linked Immunosorbent Assay (ELISA), and the specific production was determined by normalizing pediocin PA‐1 concentration to dry weight bacteria (Table 1). The strain L. lactis NZ9000_pSec::rpedA produced approximately 2.3 ± 0.4 μmol/g of recombinant pediocin, while the wild‐type producer strain Lactobacillus plantarum LMAX exhibited a specific productivity below 0.1 μmol/g. This result shows that specific production of recombinant pediocin by L. lactis is approximately 20 times more efficient than production of pediocin by the wild‐type strain. Subsequently, ELISA experiments revealed that the strains L. lactis NZ9000_pSec::s‐rped and L. lactis NZ9000_pSec::l‐rped produced approximately 3.5 ± 0.2 and 3.2 ± 0.3 μmol/g of pediocin respectively. The results thus show that the SD and LEISSTCDA propeptides allow a 1.5‐fold increase in secreted recombinant pediocin production.
Table 1.
Specific production and antimicrobial activity
| Strain | Specific productiona , b | MICa , c |
|---|---|---|
| L. lactis NZ9000_pSec::rpedA | 2.3 ± 0.4 | 1.5 ± 0.3 |
| L. lactis NZ9000_pSec::s‐rpedA | 3.5 ± 0.2d | 1.2 ± 0.1 |
| L. lactis NZ9000_pSec::l‐rpedA | 3.2 ± 0,3d | 1.2 ± 0.2 |
| L. lactis NZ9000_pSec::s‐rpedA_pOri23 | 4.0 ± 0,7 | 1.3 ± 0.5 |
| L. lactis NZ9000_pSec::s‐rpedA_pOri::pedC | 1.4 ± 0.1e | 0.4 ± 0.1 |
Mean ± standard deviation.
Specific production was expressed as μmol pediocin PA‐1 equivalents/g of dry weight bacteria.
MIC values of culture supernatants were expressed in μM pediocin PA‐1 equivalents.
Indicate a significant difference (Student's t‐test, P‐value < 0,05) of specific production between the strains L. lactis NZ9000_pSec::s‐rpedA L. lactis NZ9000_pSec::l‐rpedA and the control strain L. lactis NZ9000_pSec::rpedA.
Indicate a significant difference (Student's t‐test, P‐value < 0,05) of specific production between the strains L. lactis NZ9000_pSec::s‐rpedA_pOri::pedC and the control strain L. lactis NZ9000_pSec::s‐rpedA_pOri::pedC.
Antibacterial activity
The antibacterial activity of supernatants was assessed by agar‐well diffusion assay using Carnobacterium maltaromaticum DSM20730_pSec::nucB_pOri23 as target strain. Inhibition areas were obtained from the culture supernatants of the three strains L. lactis NZ9000_pSec::rpedA, L. lactis NZ9000_pSec::s‐rpedA and L. lactis NZ9000_pSec::l‐rpedA (Fig. 2A, panels 2, 3 and 4). No inhibition was obtained with the culture supernatant of the control strain L. lactis NZ9000_ pSec::nucB (Fig. 2A, panel 1). Moreover, no inhibition was observed when the same experiment was carried out with an indicator strain previously transformed with pOri::pedB, which confers immunity to pediocin PA‐1 (Fig. 2B, panels 2, 3 and 4). These results show the three strains L. lactis NZ9000_pSec::s‐rpedA, L. lactis NZ9000_pSec::l‐rpedA and L. lactis NZ9000_pSec::rpedA produce active recombinant pediocins. The supernatants containing the recombinant pediocins Rpediocin, S‐Rpediocin and L‐Rpediocin exhibited minimal inhibitory concentration (MIC) values of 1.5 ± 0.3 μM, 1.2 ± 0.1 μM and 1.2 ± 0.2 μM (Table 1) respectively. These results show the propeptides SD and LEISSTCDA have no major impact on the MIC value of the supernatants containing recombinant pediocins.
Figure 2.
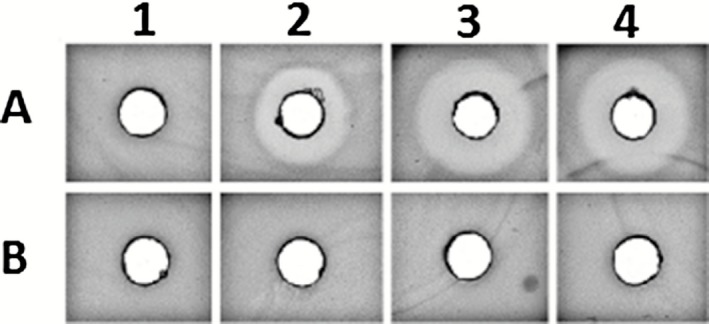
Antibacterial activity of recombinant pediocins. The antibacterial activity of recombinant pediocins was assessed on agar plate using the sensitive strain C . maltaromaticum DSM20730_pSec::nucB_pOri23 (line A) or the pediocin PA‐1 resistant strain C . maltaromaticum DSM20730_pSec::nucB_pOri::pedB (line B) as target strains. Pictures in columns 1, 2, 3, 4 show the inhibitory activity of the culture supernatants of the negative control (heterologous production of NucB from S taphylococcus aureus), Rpediocin (recombinant mature pediocin PA‐1), S‐Rpediocin (recombinant pediocins PA‐1 fused to the peptide SD at the N‐terminus) and L‐Rpediocin (recombinant pediocins PA‐1 fused to the peptide LEISSTCDA at the N‐terminus) respectively.
Co‐production of PedC with S‐Rpediocin
The potency of the recombinant pediocins produced in L. lactis was compared with the wild‐type pediocin produced by Pediococcus acidilactici. Surprisingly, the wild‐type pediocin PA‐1 produced from P. acidilactici exhibited a MIC value of 0.05 μM, and was thus approximately 30 times more active than the recombinant pediocins. This result suggests that the structure of recombinant pediocins is different from the wild‐type pediocin (Fig. 3). Although class IIa bacteriocins are described to be non‐post‐translationally modified, they all exhibit at least one disulfide bond. It was previously described that disulfide bonds are important for the antibacterial activity of pediocin PA‐1 (Fimland et al., 2000). Even if disulfide bond formation can be spontaneous, this is a slow and aspecific process that can require enzymatic catalysis in bacteria (Kadokura et al., 2003). It was hypothesized that such enzymes could allow correct formation of disulfide bonds in wild‐type pediocin PA‐1 bacterial producers (Fimland et al., 2000). Lately, it was shown that thiol‐disulfide oxidoreductases (TDORs), which belong to the thioredoxin superfamily and are characterized by a conserved CXXC motif, are involved in the post‐translational maturation of the bacteriocins sublancin 168 and BlpGst (Dorenbos et al., 2002; Fontaine and Hols, 2008). Interestingly, sequence analysis of the pediocin PA‐1 operon pedABCD revealed that pedC encodes a putative secreted protein with a CXXC motif and a thioredoxin‐like fold. It can be therefore hypothesized that PedC is a TDOR and allows native disulfide bond formation of pediocin PA‐1.
Figure 3.
Strategies used to quantitatively and qualitatively act on pediocin production in L lactis. Two propeptides were tested to improve secretion, and PedC was coexpressed with recombinant pediocin to improve its potency.
The encoding sequence of pedC was cloned into pOri23, and the resulting plasmid was used to transform L. lactis NZ9000_pSec::s‐rpedA. In parallel, the same recipient strain was transformed with pOri23, and the resulting strain was used as a control strain. Inhibition assays resulted in broader growth inhibition halos for the strain transformed with pOri::pedC (Fig. 4A) than for the strain transformed with pOri23 (Fig. 4C). Consistently, the MIC value of the recombinant pediocin decreased from 1.2 ± 0.1 μM to 0.4 ± 0.1 μM when pedC was co‐expressed with the recombinant pediocin (Table 1). Besides, the strain producing PedC exhibited a three times lower specific productivity compared with the control strain (Table 1). These results show that even if lower amounts of recombinant pediocin are produced, a higher activity was obtained with strain co‐producing the recombinant pediocin and PedC. When the same experiment was carried out with the indicator strain transformed with the pOri::pedB plasmid, which encodes the pediocin PA‐1 immunity factor, no inhibition was observed (Fig. 2B and D). Furthermore, no inhibition was observed when the control strain L. lactis NZ9000_pSec::nucB was transformed with pOri::pedC, indicating that PedC does not exhibit detectable antibacterial activity in our experimental conditions (data not shown). These results show that PedC allows increasing the antibacterial potency of the recombinant pediocin S‐Rpediocin in L. lactis.
Figure 4.
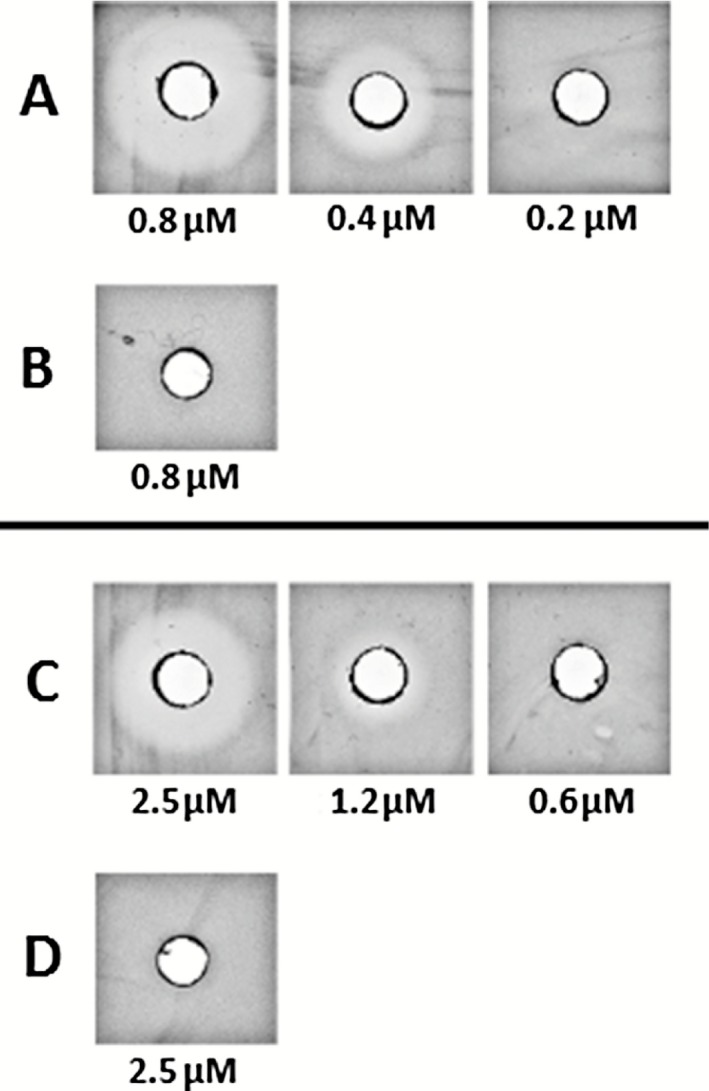
The antimicrobial activity of recombinant pediocin produced by L . lactis NZ9000_pSec::s‐rped_pOri::pedC (line A and line B) and L . lactis NZ9000_pSec::s‐rped_pOri23 (line C and line D) was assessed on agar plate using the sensitive strain C . maltaromaticum DSM20730_pSec::nucB_pOri23 (line A and C) or the pediocin PA‐1 resistant strain C . maltaromaticum DSM20730_pSec::nucB_pOri::pedB as target strains (line B and D). Picture in lines A and C show serial twofold dilutions of culture supernatants of strains L . lactis NZ9000_pSec::s‐ rpedA_pOri::pedC and L . lactis NZ9000_pSec::s‐ rpedA_pOri23 respectively. The concentrations below the pictures are expressed in μM equivalents of pediocin PA‐1.
PedC homologues as putative antibacterial activity promoting factors of bacteriocins
Previous studies already showed that the TDORs BlpGst and BdbA are involved in disulfide bond formation of the bacteriocin thermophilin 13 (Fontaine and Hols, 2008) and the bacteriocin sublancin 168 (Dorenbos et al., 2002) respectively. BLASTP analysis revealed that BlpGst shares 26% of identity (e‐value = 2,00E‐008) with PedC, and that BdbA shares 17% of identity (e‐value = 4,00E‐008) with PedC. Furthermore, BlpGst and PedC both exhibit a thioredoxin‐like fold (Interproscan accession number IPR012336), a CXXC motif and a bacteriocin transport accessory protein‐like fold (Interproscan accession number IPR005985). The proteins SkgC, PlaC, CoaC which are transport accessory proteins of the class IIa bacteriocins sakacin G, plantaricin 423 and coagulin A (Fig. 5), respectively, appears to have the same thioredoxin‐like fold and the same bacteriocin transport accessory protein‐like fold as PedC. Moreover, PedC shares 47% (e‐value = 1,00E‐056), 99% (e‐value = 1,00E‐132) and 99% (e‐value = 3,00E‐132) of identity with SkgC, PlaC and CoaC respectively. This suggests that SkgC, PlaC and CoaC might enhance the activity of the class IIa bacteriocins sakacin G, plantaricin 423 and coagulin A respectively.
Figure 5.
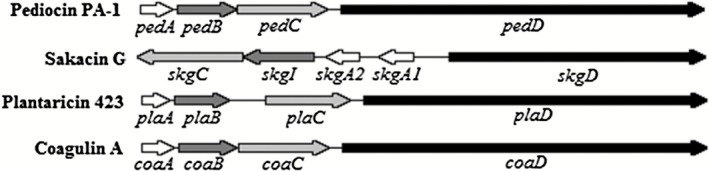
Genetic structure of gene clusters of class IIa bacteriocin containing a pedC homologue.
Discussion
The general secretion (Sec) system of L. lactis was widely used to improve secretion of recombinant proteins including bacteriocins (Herranz and Driessen, 2005; Bermúdez‐Humarán et al., 2007; Borrero et al., 2011a, 2011b; Jiménez et al., 2013) such as the bacteriocin pediocin PA‐1 (Martín et al., 2007a; Li et al., 2011). In order to allow secretion via the Sec system in L. lactis, the signal peptide of the Usp45 protein SPusp45 is commonly fused to the N‐terminus of recombinant proteins (Morello et al., 2008). Since it is thought that positive charges impair secretion through the secretory pathway, sequences encoding SD or LEISSTCDA propeptides were fused to the nucleotide sequence encoding the mature pediocin PA‐1 (ΔsppedA) at the 5′ end. Both propeptides allowed the improvement of the production of recombinant pediocins with no marked difference between propeptides SD and LEISSTCDA.
The effect of the signal peptide SPusp45 and the propeptide LEISSTCDA were previously described to improve the overall productivity in L. lactis (Le Loir et al., 1998; 2001; Langella and Le Loir, 1999; Freitas et al., 2005; Nouaille et al., 2005; Zhang et al., 2010). The replacement of the signal peptide of NucB by the signal peptide of the major secreted protein Usp45 (SPusp45) increased NucB secretion. It was hypothesized that enhancement of protein secretion resulted from a more efficient recognition of the SPusp45 by the secretory secretion machinery of L. lactis. Moreover, when the propeptide LEISSTCDA was inserted between SPusp45 and NucB, secretion was further improved. The underlying mechanism is still unknown, it is speculated that the negative charges of the propeptide LEISSTCDA would increase the cleavage efficiency by signal peptidase and/or would fold into a conformational state that would lead to optimal secretion efficiency. This optimal secretion efficiency would therefore allow the protein precursor to be less sensitive to intracellular degradation or would help to escape intracellular degradation thus increasing the production yield (Le Loir et al., 2005). A similar mechanism could explain the higher productivity of the recombinant pediocins fused to SD or LEISTCDA propeptides.
Propeptides SD and LEISSTCDA did not have any significant impact on the antibacterial specific activity of recombinant pediocin PA‐1. Thorough analysis of the impact of additional amino acid residues on specific activity of sakacin P revealed that an extra lysine residue at the C‐terminal end had no marked effect on antibacterial activity. However, a lysine residue added at the N‐terminus strongly reduced the potency of sakacin P (Kazazic et al., 2002). These results indicate that adding amino acid residues at the N‐terminal end of class IIa bacteriocins can have various impacts on their potency and peptide engineering implying peptide fusion at the N‐terminus would require checking the impact on antibacterial potency of pediocin‐like bacteriocins.
It was shown that all recombinant pediocins in the culture supernatants, including Rpediocin, are less active than the wild‐type pediocin PA‐1 produced by P. acidilactici. Similar results were already reported for pediocin PA‐1, enterocin A, divercin RV41 and sakacin A, and it was hypothesized that recombinant bacteriocins might be subjected to self‐aggregation, degradation or could be misfolded (Bermúdez‐Humarán et al., 2007; Martín et al., 2007a, 2007b; Borrero et al., 2011b; Jiménez et al., 2013). Another hypothesis would be linked to the fact that pediocin PA‐1 has two disulfide bonds. The disulfide bond at the N‐terminus is conserved in class IIa bacteriocins, whereas the disulfide bond at the C‐terminus is less conserved and is important for both pediocin PA‐1 spectrum and stabilization inside the bacterial membrane (Fimland et al., 1996; 2000; 2005). When heterologous production of pediocin PA‐1 is performed using Lactobacillus sakei, non‐native disulfide bonds are formed (Fimland et al., 2000). We hypothesized that similar cystein mispairing were formed in recombinant pediocin PA‐1 produced in L. lactis. Sequence analysis of the pedABCD operon revealed pedC would encode a protein with a putative thioredoxin fold and a CXXC catalytic site, suggesting a probable role in the status of the pediocin PA‐1 disulfide bonds. In this study, co‐production of PedC and the recombinant pediocin S‐Rpediocin led to the production of a bacteriocin with higher potency. Although PedC is described as an accessory transport protein, it was indeed concluded to be essential for secretion by the dedicated secretion system in both Pediococci and Escherichia coli (Venema et al., 1995). Nevertheless, since the amount of pediocin PA‐1 produced was not measured, it remains unclear whether the absence of activity recorded in this study was due to a decrease of secretion efficiency and/or of potency. In line with this, another study demonstrated that PedD was sufficient to allow secretion of pediocin PA‐1 in E. coli (Bukhtiyarova et al., 1994). Our results further support that PedC increases the potency of pediocin PA‐1. Sequence analyses showed that pedC homologous genes are genetically linked to genes encoding the class IIa bacteriocin sakacin G, plantaricin 423 and coagulin A suggesting that TDORs are critical functions for these bacteriocins. Two other studies revealed the role of TDORs in the potency of bacteriocins belonging to other classes: the class VI sublancin 168 (Dorenbos et al., 2002) and the class IIb thermophilin 9 (Fontaine and Hols, 2008), suggesting that the role of TDORs in the potency of bacteriocins would be not restricted to one particular class of bacteriocins.
When PedC was co‐expressed with recombinant pediocin S‐Rpediocin, a lower concentration of S‐Rpediocin was obtained in the culture supernatant. Sequence analysis of PedC suggested it contains a secretory‐dependent signal peptide. In addition, pedC is under the control of the strong constitutive promoter p23 (Van der Vossen et al., 1987). Therefore, it can be speculated that PedC competes with the recombinant pediocin for the recruitment by the secretory machinery for secretion.
This study revealed that the model class IIa bacteriocin pediocin PA‐1 is sufficiently flexible to tolerate fusions at the N‐terminal end. This property was used here to improve secretion efficiency and could also be used for other purposes such as genetic engineering of innovative antibacterial compounds as previously shown for other bacteriocins (Qiu et al., 2003; Acuña et al., 2012). Moreover, this work revealed that solely expressing the structural gene of class IIa pediocin PA‐1 in a heterologous host may not be sufficient to get an optimal antibacterial activity, and that PedC might be helpful to enhance the activity of recombinant bacteriocins, probably by increasing the concentration of correct disulfide bond containing peptides. Furthermore, these results suggest that the protein PedC is responsible for the formation of disulfide bonds in pediocin PA‐1 in the wild‐type context.
Experimental procedures
Bacterial strains, culture conditions and plasmids
Bacterial strains and plasmids used in this study are listed in Table 2. Lactococcus lactis NZ9000 was grown at 30°C in M17 (Difco, Sparks, USA) supplemented with 1% (w/v) glucose (GM17 medium). Carnobacterium maltaromaticum DSM20730 was grown in Trypton Salt Broth medium (TSB; Biomérieux, Marcy‐l'Étoile, France) supplemented with 6 g/L yeast extract (TSBYE) at 30°C. Lactobacillus plantarum LMAX, which was isolated from HOLDBAC Listeria (Danisco), was grown in Man, Rogosa and Sharpes medium (MRS, Biokar, Beauvais, France) at 30°C. Lactococcus lactis NZ9000 transformants were grown in GM17 containing 10 μg/ml chloramphenicol or/and 10 μg/ml erythromycin (Sigma‐Aldrich, Saint‐Louis, USA). Carnobacterium maltaromaticum DSM20730 transformants were grown in TSBYE containing 5 μg/ml chloramphenicol or/and 5 μg/ml of erythromycin.
Table 2.
Strains, plasmids and synthetic genes used in this study
| Plasmid, strain and synthetic genes | Description | Reference |
|---|---|---|
| Strains | ||
| L. lactis NZ9000 | MG1363 derivative; pepN::nisRK+ | (Kuipers et al., 1998) |
| C. maltaromaticum DSM20730a | Pediocin sensitive strain | (Hiu et al., 1984) |
| Lb plantarum LMAX | Natural pediocin producer, isolated from HOLDBAC Listeria | Danisco |
| L. lactis NZ9000_pSec::nucB | Carry the plasmid pSec which contains the nucB gene under the control of PnisA | This study |
| L. lactis NZ9000_pOri::pedB | Carry the plasmid pOri::pedB which contains the pedB gene under the control of P23 | This study |
| L. lactis NZ9000_pSec::rpedA | Carry the plasmid pSec::rpedA which contains the ΔsppedA gene under the control of PnisA. | This study |
| L. lactis NZ9000_pSec::s‐rpedA | Carry the plasmid pSec::s‐rpedA which contains the sd::ΔsppedA gene under the control of PnisA | This study |
| L. lactis NZ9000_pSec::l‐rpedA | Carry the plasmid pSec::l‐rpedA which contains the leisstcda::ΔsppedA gene under the control of PnisA | This study |
| L. lactis NZ9000_pSec_pOri::pedC | Carry the plasmid pSec which contains the nucB gene under the control of PnisA and pOri::pedC which contains the pedC gene under the control of P23. | This study |
| L. lactis NZ9000_pSec::s‐rpedA_pOri23 | Carry the plasmid pSec::s‐rpedA which contains the sd::ΔsppedA under the control of PnisA and the plasmid pOri23 | |
| L. lactis NZ9000_pSec::s‐rpedA_pOri::pedC | Carry the plasmid pSec::s‐rpedA which contains the sd::ΔsppedA under the control of PnisA and pOri::pedC which contains the pedC gene under the control of P23. | This study |
| C. maltaromaticum DSM20730_pSec::nucB_pOri23 | Pediocin sensitive strain resistant to erythromycin and chloramphenicol | This study |
| Plasmids | This study | |
| pSec::nucB | CmR, PnisA, spusp45, nucB, repA, repC | Bermúdez‐Humarán et al., 2007) |
| pOri23 | ErmR, p23, repD, repE | Que et al., 2000) |
| pOri::pedB | ErmR, p23, pOri23 derivative carrying pedB | This study |
| pSec::rpedA | CmR, pSec derivative carrying spusp45::rpedA and encoding secreted Rpediocin | This study |
| pSec::s‐rpedA | CmR, pSec: derivative carrying spusp45::s‐rpedA and encoding secreted S‐Rpediocin | This study |
| pSec::l‐rpedA | CmR, pSec derivative carrying spusp45::l‐rpedA and encoding secreted L‐Rpediocin | This study |
| pOri::pedC | ErmR, p23, pOri23 derivative carrying pedC | This study |
| Synthetic genes | Amino acid sequence | |
| sd::ΔsppedA | b tctgataaatattatggtaatggagttacttgtggaaaacattcatgttctgttgattggggtaaagctacaacttgtattattaataatggagctatggcatgggctactggtggacatcaaggtaatcataaatgttaa | Genscript |
| leisstcda::ΔsppedA | b ttagaaatttcatcaacatgtgatgctaatattatggtaatggagttacttgtggaaaacattcatgttctgttgattggggtaaagctacaacttgtattattaataatggagctatggcatgggctactggtggacatcaaggtaatcataaatgttaa | Genscript |
The strain was from Deutsche Sammlung von Mikroorganismen und Zellkulturen collection.
The nucleotide sequences encoding propeptides SD and LEISSTCDA fused to the N‐terminus of pediocin PA‐1 are in bold.
Electroporation
Electrocompetent L. lactis NZ9000 was obtained as previously described (Holo and Nes, 1989). For electrocompetent C. maltaromaticum, an overnight culture was diluted 10‐fold in fresh TSBYE medium and incubated at 30°C until an OD600 nm of 0.7 was reached. Then bacteria were harvested by centrifugation at 6000 g for 15 min. Following three washes in ice‐cold 10% glycerol solution, the pellet was re‐suspended in 1 ml of the same washing solution and stored at –80°C. The electric pulse was delivered with a Gene‐Pulser (Bio‐Rad Laboratories, Richmond, USA.) set up at 25 μF, 2.1 kV and 200 Ω. Then, the suspension was mixed with 4 ml of TSBYE and incubated 4 h at 30°C prior plating on selective medium containing 5 μg/ml chloramphenicol or/and 5 μg/ml erythromycin.
Plasmid construction
All the polymerase chain reaction (PCR) products used for cloning purpose were obtained with the oligonucleotide primers (Eurogentec, Seraing, Belgique) described in Table 3 by using the high‐fidelity thermostable polymerase Pfx50 (Invitrogen, Carlsbad, USA). The expression vectors pSec::s‐rpedA and pSec::l‐rpedA were constructed by using pSec::nucB as recipient vector. Polymerase chain reaction product PCRSD and PCRLEI (Table 2) were digested with NsiI and EcoRI (New England Biolabs, Massachusetts, Ipswich, USA) and were ligated into the pSec expression vector previously digested with the same enzymes to generate pSec::s‐rpedA and pSec::l‐rpedA expression vectors respectively. The plasmid pSec::rpedA was constructed by deleting the SD encoding sequence by crossover PCR. To do so, the PCR products named PEDA1 and PEDA2 which flank the SD encoding region, were obtained by using pSec::s‐rpedA as DNA matrix. The product named PEDA3 was obtained by crossover PCR by using PEDA1 and PEDA2 as DNA matrix and was subsequently digested with BglII and XhoI and cloned into the pSec expression vector previously digested with the same enzymes in order to generate the expression vector pSec::rpedA. Plasmid pOri::pedC was obtained by digesting the PCR product PEDC with BamHI and PstI (FastDigest, Thermo Fisher Scientific, Waltham, Massachusetts, USA). The plasmid pOri::pedB was constructed similarly by using the primers described in Table 3. All the recombinant plasmids were obtained by using the ligation mixes to electrotransform L. lactis NZ9000. The integrity of plasmid inserts was checked by DNA sequencing according to the method of Sanger (GATC‐BIOTECH, Konstanz, Germany).
Table 3.
Primers and PCR products
| PCR product name | Matrix | Primersa |
|---|---|---|
| PEDB | Lb. plantarum LMAX | Forward : ATCGGGATCCTAAAAAGGGAGGCCAAATATAATGAATAAGACTAAGTCGGAA |
| Reverse : ATCGCTGCAGCTATTGGCTAGGCCACGT | ||
| PCRSD | sd::ΔsppedA | Forward : CCCCCCCCATGCATCTGATAAATATTATGGTAATGGAGTTACTTGTGGA |
| Reverse : CCCCCCGAATTCACTAGTCCTTAACATTTATGATTACCTTG | ||
| PCRLEI | leisstcda::ΔsppedA | Forward : CCCCCCCCATGCATTAGAAATTTCATCAACATGTGATGCTAAATAT |
| Reverse : CCCCCCGAATTCACTAGTCCTTAACATTTATGATTACCTTG | ||
| PEDA1 | pSec::s‐rpedA | Forward : ACAGCTCCAAGATCTAGTCTT |
| Reverse : AGTAACTCCATTACCATAATATTTTGCATAAACACCTGACAACGG | ||
| PEDA2 | pSec::s‐rpedA | Forward : CCGTTGTCAGGTGTTTATGCAAAATATTATGGTAATGGAGTTACT |
| Reverse : ACATGCTGAAGAGCATCTCATT | ||
| PEDA3 | PEDA1 and PEDA2 | Forward : ACAGCTCCAAGATCTAGTCTT |
| Reverse : ACATGCTGAAGAGCATCTCATT | ||
| PEDC | Lb. plantarum LMAX | Forward : GGGGGGGGATCCTAAAAAGGGAGGCCAAATATAATGTCTAAGAAATTTTGGTCAAA |
| Reverse : GGGGGGCTGCAGCTACTGATTATTGTAATCAGC |
Restriction sites are in bold, Ribosome Binding Site sequences are italicized.
Heterologous expression
For strains containing pSec::nucB or pSec derivatives, a culture incubated for 17 h was used to inoculate 200 ml of GM17 supplemented with chloramphenicol (10 μg/ml) in a 250 ml flask at an initial OD600 nm of 0.02. Cultures were grown at 100 r.p.m. stirring (Amplitude: 20 mm; Sanyo, Osaka, Japan) and at 30°C. For strains containing both pSec::nucB or derivatives, and pOri23 or derivatives, a culture incubated for 17 h was used to inoculate 200 ml of GM17 medium supplemented with chloramphenicol (2 μg/ml) and erythromicyn (2 μg/ml) in a 250 ml flask at an initial OD600 nm of 0.1. For all strains, heterologous production was induced with 100 ng/ml of semi‐purified nisin (balance sodium chloride and denatured milk solids containing 2.5% of nisin from Sigma‐Aldrich, St Louis, USA) when OD600 nm reached 0.4–0.5. After 24 h of incubation, cell dry weights were determined gravimetrically. Then the supernatants were collected after centrifugation (5000 g; 15 min). Each supernatant was neutralized with 6M NaOH, sterilized by microfiltration (0.2 μm) and stored at −20°C. The experiments were independently performed three times.
ELISA
Recombinant pediocin concentrations were measured by ELISA. Rabbit serum containing anti‐pediocin PA‐1 polyclonal antibodies was purchased from Eurogentec (Seraing, Belgique). Goat anti‐rabbit IgG antibodies coupled with Horseradish peroxidase (HRP) were purchased from Sigma‐Aldrich (St Louis, USA). Between each step described below, the microtitre plate was washed eight times with washing buffer (phosphate buffer saline containing 0.05% of Tween 20). Microtitre plate wells (Corning, New york, USA) were coated overnight at 4°C with 200 μl of filtered culture supernatant containing recombinant pediocins previously diluted with phosphate buffer saline (PBS) when required. Wells were subsequently blocked with 300 μl of skimmed milk (10% w/v in PBS, Régilait, Saint‐Martin‐Belle‐Roche, France) for 1 h at room temperature. Wells were then incubated with 150 μl of anti‐pediocin polyclonal antibodies (1:1000 in PBS) for 1 h at room temperature, followed by incubation with 100 μl of HRP‐labelled goat anti‐rabbit IgG (1:2000 in PBS) for 1 h. Bound secondary antibodies were detected by adding 100 μl of 3,3′,5,5′‐tetramethylbenzidine (TMB, Thermo Fisher Scientific, Waltham, USA). The reaction was stopped with 100 μl of 2M H2SO4 and absorbance was read at 450 nm. Concentration of recombinant pediocins in the supernatants were extrapolated from a calibration curve constructed using purified pediocin PA‐1 from P. acidilactici (Sigma‐Aldrich, St Louis, USA) diluted with PBS and were expressed in μM equivalents of pediocin PA‐1.
Antimicrobial assay
Minimal inhibitory concentration of pediocin PA‐1 and recombinant pediocins were determined by well diffusion assay on agar plates (Mathieu et al., 1993) by using the strain C. maltaromaticum DSM20730_pSec_pOri23 as indicator. Minimal inhibitory concentration is defined as the lowest concentration exhibiting a distinct inhibition zone after 24 h at 30°C.
Sequence analysis
The nucleotide sequence of putative thiol disulfide oxidoreductases were analysed using blast (Altschul et al., 1997), SignalP (Nielsen et al., 1997), Conserved Domain Database CDD (Marchler‐Bauer and Bryant, 2004; Marchler‐Bauer et al., 2007; 2013) and Interproscan (Quevillon et al., 2005). The amino acid sequence of PedC (accession number: P37249), BdbA (accession number: P68569), SkgC (accession number: B2LS01 ), CoaC (accession number: Q9EZB1), PlaC (accession number: Q93FV5), BlpGst (accession number: Q03J39) used for analysis are from UniProt.
Conflict of interest
The authors declare no conflict of interests.
Acknowledgements
We thank John Renye Jr for his valuable help and Luis G. Bermúdez‐Humarán for the plasmid pSec::nucB and for helpful discussions. We thank Arnaud Khemisti, Myriam Michel, Sylvie Wolff, Laureen Hallez and Alexis Boutillas for technical assistance. We are grateful to Nadine Pavlov, Marianne Lasaulce, Claude Didierjean, Stéphane Delaunay, Raphaël Duval and Bertrand Aigle for helpful discussions during the course of this work.
Microbial Biotechnology (2016) 9(4), 466–477
Funding Information This study was supported by a PhD grant from the French ministry of education and research.
References
- Acuña, L. , Picariello, G. , Sesma, F. , Morero, R.D. , and Bellomio, A. (2012) A new hybrid bacteriocin, Ent35–MccV, displays antimicrobial activity against pathogenic Gram‐positive and Gram‐negative bacteria. FEBS Open Bio 2: 12–19. doi:10.1016/j.fob.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arqués, J.L. , Rodríguez, J.M. , Gasson, M.J. , and Horn, N. (2008) Immunity gene pedB enhances production of pediocin PA‐1 in naturally resistant Lactococcus lactis strains. J Dairy Sci 91: 2591–2594. [DOI] [PubMed] [Google Scholar]
- Bermúdez‐Humarán, L.G. , Rihakova, J. , Langella, P. , Demnerova, K. , Nazef, L. , Prévost, H. , and Drider, D. (2007) Antimicrobial activity of divercin RV41 produced and secreted by Lactococcus lactis . J Mol Microbiol Biotechnol 13: 259–263. [DOI] [PubMed] [Google Scholar]
- Borrero, J. , Jiménez, J.J. , Gútiez, L. , Herranz, C. , Cintas, L.M. , and Hernández, P.E. (2011a) Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis . Appl Microbiol Biotechnol 89: 131–143. [DOI] [PubMed] [Google Scholar]
- Borrero, J. , Jiménez, J.J. , Gútiez, L. , Herranz, C. , Cintas, L.M. , and Hernandez, P.E. (2011b) Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. J Biotechnol 156: 76–86. [DOI] [PubMed] [Google Scholar]
- Bukhtiyarova, M. , Yang, R. , and Ray, B. (1994) Analysis of the pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin‐negative Pediococcus acidilactici strain. Appl Environ Microbiol 60: 3405–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, P.D. , Ross, R.P. , and Hill, C. (2013) Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol 11: 95–105. doi:10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- Dobson, A. , Cotter, P.D. , Ross, R.P. , and Hill, C. (2012) Bacteriocin production: a probiotic trait? Appl Environ Microbiol 78: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorenbos, R. , Stein, T. , Kabel, J. , Bruand, C. , Bolhuis, A. , Bron, S. , et al (2002) Thiol‐disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem 277: 16682–16688. [DOI] [PubMed] [Google Scholar]
- Fimland, G. , Blingsmo, O.R. , Sletten, K. , Jung, G. , Nes, I.F. , and Nissen‐Meyer, J. (1996) New biologically active hybrid bacteriocins constructed by combining regions from various pediocin‐like bacteriocins: the C‐terminal region is important for determining specificity. Appl Environ Microbiol 62: 3313–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimland, G. , Johnsen, L. , Axelsson, L. , Brurberg, M.B. , Nes, I.F. , Eijsink, V.G. , and Nissen‐Meyer, J. (2000) A C‐terminal disulfide bridge in pediocin‐like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J Bacteriol 182: 2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimland, G. , Johnsen, L. , Dalhus, B. , and Nissen‐Meyer, J. (2005) Pediocin‐like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J Pept Sci 11: 688–696. [DOI] [PubMed] [Google Scholar]
- Fontaine, L. , and Hols, P. (2008) The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD‐9 depends on the production of multiple peptides and the activity of BlpG(St), a thiol‐disulfide oxidase. Appl Environ Microbiol 74: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas, D.A. , Leclerc, S. , Miyoshi, A. , Oliveira, S.C. , Sommer, P.S.M. , Rodrigues, L. , et al (2005) Secretion of Streptomyces tendae antifungal protein 1 by Lactococcus lactis . Braz J Med Biol Res 38: 1585–1592. doi:10.1590/S0100‐879X2005001100004. [DOI] [PubMed] [Google Scholar]
- Guyonnet, D. , Fremaux, C. , Cenatiempo, Y. , and Berjeaud, J.M. (2000) Method for rapid purification of class IIa bacteriocins and comparison of their activities. Appl Environ Microbiol 66: 1744–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, J.T. , Chopko, A.L. , and van Wassenaar, P.D. (1992) Purification and primary structure of pediocin PA‐1 produced by Pediococcus acidilactici PAC‐1.0. Arch Biochem Biophys 295: 5–12. doi:10.1016/0003‐9861(92)90480‐K. [DOI] [PubMed] [Google Scholar]
- Herranz, C. , and Driessen, A.J.M. (2005) Sec‐mediated secretion of bacteriocin enterocin P by Lactococcus lactis . Appl Environ Microbiol 71: 1959–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiu, S.F. , Holt, R.A. , Sriranganathan, N. , Seidler, R.J. , and Fryer, J.L. (1984) Lactobacillus piscicola, a new species from Salmonid fish. Int J Syst Bacteriol 34: 393–400. doi:10.1099/00207713‐34‐4‐393. [Google Scholar]
- Holo, H. , and Nes, I.F. (1989) High‐frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55: 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, N. , Martinez, M.I. , Martinez, J.M. , Hernandez, P.E. , Gasson, M.J. , Rodriguez, J.M. , and Dodd, H.M. (1998) Production of pediocin PA‐1 by Lactococcus lactis using the lactococcin a secretory apparatus. Appl Environ Microbiol 64: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, N. , Martinez, M.I. , Martinez, J.M. , Hernandez, P.E. , Gasson, M.J. , Rodriguez, J.M. , and Dodd, H.M. (1999) Enhanced production of pediocin PA‐1 and coproduction of nisin and pediocin PA‐1 by Lactococcus lactis . Appl Environ Microbiol 65: 4443–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, N. , Fernández, A. , Dodd, H.M. , Gasson, M.J. , and Rodríguez, J.M. (2004) Nisin‐controlled production of pediocin PA‐1 and colicin V in nisin‐ and non‐nisin‐producing Lactococcus lactis strains. Appl Environ Microbiol 70: 5030–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, R.W. , Wan, J. , Gordon, J. , Harmark, K. , Davidson, B.E. , Hillier, A.J. , et al (1996) Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126 . Appl Environ Microbiol 62: 2897–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasniewski, J. , Cailliez‐Grimal, C. , Gelhaye, E. , and Revol‐Junelles, A.‐M. (2008) Optimization of the production and purification processes of carnobacteriocins Cbn BM1 and Cbn B2 from Carnobacterium maltaromaticum CP5 by heterologous expression in Escherichia coli . J Microbiol Methods 73: 41–48. doi:10.1016/j.mimet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Jiménez, J.J. , Borrero, J. , Diep, D.B. , Gútiez, L. , Nes, I.F. , Herranz, C. , et al (2013) Cloning, production, and functional expression of the bacteriocin sakacin A (SakA) and two SakA‐derived chimeras in lactic acid bacteria (LAB) and the yeasts Pichia pastoris and Kluyveromyces lactis . J Ind Microbiol Biotechnol 40: 977–993. doi:10.1007/s10295‐013‐1302‐6. [DOI] [PubMed] [Google Scholar]
- Kadokura, H. , Katzen, F. , and Beckwith, J. (2003) Protein disulfide bond formation in prokaryotes. Annu Rev Biochem 72: 111–135. doi:10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- Kazazic, M. , Nissen‐Meyer, J. , and Fimland, G. (2002) Mutational analysis of the role of charged residues in target‐cell binding, potency and specificity of the pediocin‐like bacteriocin sakacin P. Microbiology 148: 2019–2027. [DOI] [PubMed] [Google Scholar]
- Kuipers, O.P. , de Ruyter, P.G.G. , Kleerebezem, M. , and de Vos, W.M. (1998) Quorum sensing‐controlled gene expression in lactic acid bacteria. J Biotechnol 64: 15–21. [Google Scholar]
- Langella, P. , and Le Loir, Y. (1999) Heterologous protein secretion in Lactococcus lactis: a novel antigen delivery system. Braz J Med Biol Res 32: 191–198. [DOI] [PubMed] [Google Scholar]
- Le Loir, Y. , Gruss, A. , Ehrlich, S.D. , and Langella, P. (1998) A nine‐residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis . J Bacteriol 180: 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir, Y. , Nouaille, S. , Commissaire, J. , Brétigny, L. , Gruss, A. , and Langella, P. (2001) Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis . Appl Environ Microbiol 67: 4119–4127. doi:10.1128/AEM.67.9.4119‐4127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir, Y. , Azevedo, V. , Oliveira, S.C. , Freitas, D.A. , Miyoshi, A. , Bermúdez‐Humarán, L.G. , et al (2005) Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb Cell Fact. 4: 2–15. doi:10.1186/1475‐2859‐4‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Takala, T.M. , Qiao, M. , Xu, H. , and Saris, P.E.J. (2011) Nisin‐selectable food‐grade secretion vector for Lactococcus lactis . Biotechnol Lett 33: 797–803. doi:10.1007/s10529‐010‐0503‐6. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , and Bryant, S.H. (2004) CD‐Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331. doi:10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Anderson, J.B. , Derbyshire, M.K. , DeWeese‐Scott, C. , Gonzales, N.R. , Gwadz, M. , et al (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35: D237–D240. doi:10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Zheng, C. , Chitsaz, F. , Derbyshire, M.K. , Geer, L.Y. , Geer, R.C. , et al (2013) CDD: conserved domains and protein three‐dimensional structure. Nucleic Acids Res 41: D348–D352. doi:10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín, M. , Gutiérrez, J. , Criado, R. , Herranz, C. , Cintas, L.M. , and Hernández, P.E. (2007a) Chimeras of mature pediocin PA‐1 fused to the signal peptide of enterocin P permits the cloning, production, and expression of pediocin PA‐1 in Lactococcus lactis . J Food Prot 70: 2792–2798. [DOI] [PubMed] [Google Scholar]
- Martín, M. , Gutiérrez, J. , Criado, R. , Herranz, C. , Cintas, L.M. , and Hernández, P.E. (2007b) Cloning, production and expression of the bacteriocin enterocin A produced by Enterococcus faecium PLBC21 in Lactococcus lactis . Appl Microbiol Biotechnol 76: 667–675. doi:10.1007/s00253‐007‐1044‐3. [DOI] [PubMed] [Google Scholar]
- Mathieu, F. , Michel, M. , and Lefebvre, G. (1993) Properties of a bacteriocin produced by Carnobacterium piscicola CP5. Biotechnol Lett 15: 587–590. doi:10.1007/BF00138545. [Google Scholar]
- Morello, E. , Bermúdez‐Humarán, L.G. , Llull, D. , Solé, V. , Miraglio, N. , Langella, P. , and Poquet, I. (2008) Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol 14: 48–58. doi:10.1159/000106082. [DOI] [PubMed] [Google Scholar]
- Nielsen, H. , Engelbrecht, J. , Brunak, S. , and von Heijne, G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Nouaille, S. , Bermúdez‐Humarán, L.G. , Adel‐Patient, K. , Commissaire, J. , Gruss, A. , Wal, J.M. , et al (2005) Improvement of bovine beta‐lactoglobulin production and secretion by Lactococcus lactis . Braz J Med Biol Res. 38: 353–359. doi:http://dx.doi.org/10.1590/S0100‐879X2005000300005. [DOI] [PubMed] [Google Scholar]
- Osmanagaoglu, O. , Kiran, F. , and Ataoglu, H. (2010) Evaluation of in vitro probiotic potential of Pediococcus pentosaceus OZF isolated from human breast milk. Probiotics Antimicrob Proteins 2: 162–174. doi:10.1007/s12602‐010‐9050‐7. [DOI] [PubMed] [Google Scholar]
- Pontes, D.S. , de Azevedo, M.S.P. , Chatel, J.‐M. , Langella, P. , Azevedo, V. , and Miyoshi, A. (2011) Lactococcus lactis as a live vector: heterologous protein production and DNA delivery systems. Protein Expr Purif 79: 165–175. doi:10.1016/j.pep.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Qiu, X.‐Q. , Wang, H. , Lu, X.‐F. , Zhang, J. , Li, S.‐F. , Cheng, G. , et al (2003) An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat Biotechnol 21: 1480–1485. doi:10.1038/nbt913. [DOI] [PubMed] [Google Scholar]
- Que, Y.A. , Haefliger, J.A. , Francioli, P. , and Moreillon, P. (2000) Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun 68: 3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon, E. , Silventoinen, V. , Pillai, S. , Harte, N. , Mulder, N. , Apweiler, R. , and Lopez, R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120. doi:10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renye, J.A., Jr , and Somkuti, G.A. (2010) Nisin‐induced expression of pediocin in dairy lactic acid bacteria. J Appl Microbiol 108: 2142–2151. doi:10.1111/j.1365‐2672.2009.04615.x. [DOI] [PubMed] [Google Scholar]
- Reviriego, C. , Fernández, A. , Horn, N. , Rodríguez, E. , Marín, M.L. , Fernández, L. , and Rodríguez, J.M. (2005) Production of pediocin PA‐1, and coproduction of nisin A and pediocin PA‐1, by wild Lactococcus lactis strains of dairy origin. Int Dairy J 15: 45–49. doi:10.1016/j.idairyj.2004.05.003. [Google Scholar]
- Reviriego, C. , Fernández, L. , Kuipers, O.P. , Kok, J. , and Rodríguez, J.M. (2007a) Enhanced production of pediocin PA‐1 in wild nisin‐ and non‐nisin‐producing Lactococcus lactis strains of dairy origin. Int Dairy J 17: 574–577. doi:10.1016/j.idairyj.2006.05.013. [Google Scholar]
- Reviriego, C. , Fernández, L. , and Rodríguez, J.M. (2007b) A food‐grade system for production of pediocin PA‐1 in nisin‐producing and non‐nisin‐producing Lactococcus lactis strains: application to inhibit Listeria growth in a cheese model system. J Food Prot 70: 2512–2517. [DOI] [PubMed] [Google Scholar]
- Riley, M.A. (2009) Bacteriocins, biology, ecology, and evolution In Encyclopedia of Microbiology, 3rd edn Schaechter M. (ed.). Oxford, UK: Academic Press, pp. 32–44. [Google Scholar]
- Rodríguez, J.M. , Martínez, M.I. , and Kok, J. (2002) Pediocin PA‐1, a wide‐spectrum bacteriocin from lactic acid bacteria. Crit Rev Food Sci Nutr 42: 91–121. doi:10.1080/10408690290825475. [DOI] [PubMed] [Google Scholar]
- Rodríguez, J.M. , Martínez, M.I. , Horn, N. , and Dodd, H.M. (2003) Heterologous production of bacteriocins by lactic acid bacteria. Int J Food Microbiol 80: 101–116. doi:10.1016/S0168‐1605(02)00153‐8. [DOI] [PubMed] [Google Scholar]
- Van der Vossen, J.M. , van der Lelie, D. , and Venema, G. (1987) Isolation and characterization of Streptococcus cremoris Wg2‐specific promoters. Appl Environ Microbiol 53: 2452–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel, A.J. , Montalban‐Lopez, M. , and Kuipers, O.P. (2011) Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol 7: 675–680. doi:10.1517/17425255.2011.573478. [DOI] [PubMed] [Google Scholar]
- Venema, K. , Kok, J. , Marugg, J.D. , Toonen, M.Y. , Ledeboer, A.M. , Venema, G. , and Chikindas, M.L. (1995) Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol 17: 515–522. doi:10.1111/j.1365‐2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Zhong, J. , Liang, X. , Liu, W. , and Huan, L. (2010) Improvement of human interferon alpha secretion by Lactococcus lactis . Biotechnol Lett 32: 1271–1277. doi:10.1007/s10529‐010‐0285‐x. [DOI] [PubMed] [Google Scholar]


