Abstract
Bacterial pathogens employ type IV secretion systems (T4SSs) for various purposes to aid in survival and proliferation in eukaryotic host. One large T4SS subfamily, the conjugation systems, confers a selective advantage to the invading pathogen in clinical settings through dissemination of antibiotic resistance genes and virulence traits. Besides their intrinsic importance as principle contributors to the emergence of multiply drug-resistant ‘superbugs’, detailed studies of these highly tractable systems have generated important new insights into the mode of action and architectures of paradigmatic T4SSs as a foundation for future efforts aimed at suppressing T4SS machine function. Over the past decade, extensive work on the second large T4SS subfamily, the effector translocators, has identified a myriad of mechanisms employed by pathogens to subvert, subdue, or bypass cellular processes and signaling pathways of the host cell. An overarching theme in the evolution of many effectors is that of molecular mimicry. These effectors carry domains similar to those of eukaryotic proteins and exert their effects through stealthy interdigitation of cellular pathways, often with the outcome not of inducing irreversible cell damage but rather of reversibly modulating cellular functions. This chapter summarizes the major developments for the actively studied pathogens with an emphasis on the structural and functional diversity of the T4SSs and the emerging common themes surrounding effector function in the human host.
INTRODUCTION
Bacterial type IV secretion systems (T4SSs) are widely distributed among Gram-negative and -positive bacteria. These systems contribute in various ways to infection processes among clinically-important pathogens, including Helicobacter pylori, Brucella and Bartonella species, Bordetella pertussis and Legionella pneumophila (1–3). The list of pathogens employing T4SSs to subvert host cellular pathways for establishment of a replication niche continues to expand, making these machines an important subject of study for defining critical features of disease progression and development of strategies aimed at suppressing T4SS function (4). Also of importance, studies of T4SSs and effector functions have coincidentally and appreciably augmented our understanding of basic cellular processes in the human host.
The T4SSs are a highly diverse translocation superfamily in terms of i) overall machine architecture, ii) the secretion substrates translocated, and iii) target cell types which can include bacteria, amoebae, fungal, plant, or human (5). Several classification schemes have emerged to describe the T4SSs, the most widely used being based on overall machine function (1). Accordingly, one subfamily existing within nearly all species of bacteria and even some Archaea are the conjugation systems (Fig. 1) (5). These systems mediate transfer of mobile genetic elements (MGEs) in the form of conjugative plasmids or chromsomally-located integrative and conjugative elements (ICEs) to other bacteria by a mechanism requiring direct cell-to-cell contact (6–8). These systems are highly important vehicles for the widespread and rapid transmission of antibiotic resistance genes and virulence traits among medically important pathogens. In a context of the recent emergence of multiply-resistant ‘superbugs’, the dissemination of MGEs via conjugation represents a huge threat to human health and an enormous financial burden to society (9).
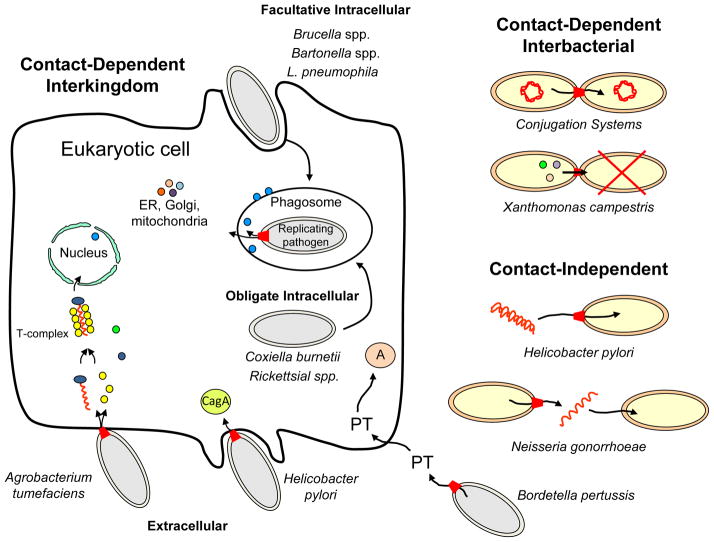
FIG. 1. Bacterial pathogens employing T4SSs for establishment within the human host, acquisition of DNA encoding virulence traits, or outcompetition of other bacteria for niche occupation.
Extracellular pathogens deliver substrates to human or plant cells by contact-dependent or -independent mechanisms. These pathogens deliver diverse substrates including oncogenic T-DNA, monomeric CagA, and multimeric PT toxin. Facultative intracellular pathogens enter the host cell from an environmental sample, whereas obligate intracellular pathogens enter directly from another host cell. The intracellular pathogens employ T4SSs to deliver a myriad of effectors whose collective function is to subvert host cellular processes principally for establishment of replicative niches. Shown are T4SSs on the bacterial cell envelope (red trapezoids), effectors (proteins: multicolor circles; T-DNA, red wavy line) and various target organelles/sites of effector action within the host cell. T4SSs also mediate interbacterial transfer by contact-dependent mechanisms for conjugative DNA transfer or to kill neighboring bacteria (red X), or by contact-independent mechanisms to exchange DNA with the environment.
The second subfamily, the ‘effector translocator’ systems, evolved from the conjugation systems but have acquired a different substrate repertoire composed mainly but not exclusively of proteins (1). Thus far, these systems have been identified only in Gram-negative pathogens, but recent work suggests a number of medically important Gram-positive species also rely on T4SSs for colonization through mechanisms not exclusively related to gene transfer (4, 10). The effector translocator systems deliver their cargoes into the eukaryotic cell cytosol usually by a cell-contact-dependent mechanism (Fig. 1). Upon translocation, the effector proteins target specific physiological pathways or biochemical processes with a variety of biological consequences that benefit survival, colonization, and transmission of the invading pathogens (3, 11, 12).
A third T4SS subfamily, the ‘DNA release and uptake systems’ presently is composed of a Neisseria gonorrhoeae T4SS functioning to deliver substrate DNA to the extracellular milieu and a H. pylori competence system used for DNA uptake by the bacterium (1). Both systems are closely functionally related to conjugation systems but adapted for DNA translocation in the absence of direct recipient cell contact (13, 14).
T4SSs alternatively have been designated as Type IVA, IVB or, recently, IVC (10, 15, 16). The IVA systems are composed of a dozen or so subunits homologous to components of the paradigmatic Agrobacterium tumefaciens VirB/VirD4 T4SS (17). This subfamily includes the well-characterized conjugation machines encoded by plasmids R388, pKM101, RP4, and F, as well as effector translocators employed by H. pylori, B. pertussis, Bartonella and Brucella spp., and Rickettsia spp (5). The IVB systems, exemplified by the L. pneumophila Dot/Icm system, bear little sequence relatedness to the IVA systems and are composed of over 25 subunits (16, 18). Other members of this family include the plasmid ColIb-P9 conjugation system and the Coxiella burnetii Dot/Icm effector translocator. The IVC systems, found almost exclusively in Gram-positive species, are composed of as few as five subunits and thus are also termed ‘minimized’ T4SSs (4, 10). Subunits of the IVC systems exhibit sequence similarities to a subset of the IVA components, and results of phylogenetic analyses suggest the IVC systems arose from the IVA systems (19).
An inherent difficulty in classifying T4SSs on the basis of function, structure, or phylogeny is that these are highly versatile and adaptive machines. A prominent example is the A. tumefaciens VirB/VirD4 T4SS, which functions both as a conjugation machine and an effector translocator and, while its target is plant cells in nature, it can also deliver substrates to other bacteria as well as various fungal and human cells (20). Many T4SSs also are highly mosaic in their subunit composition. For example, although the type IVA systems are built from homologs of VirB and VirD4 subunits, many systems have evolved specialized functions through loss of certain components or acquisition of others from unrelated ancestries (5, 19).
This chapter will summarize our recent progress in understanding the mechanism of action of T4SSs and their contributions to pathogenesis. We will focus the discussion mainly on contributions of the effector translocators to infection. To further highlight mechanistic themes and variations, we will group these systems according to the invasive mechanism, e.g., extracellular, facultative intracellular, obligate intracellular, of the pathogen, as illustrated in Figure 1. To further orient the reader, the pathogens discussed herein and some relevant properties relating to infection are presented in Table 1.
Table 1.
Type IV secretion systems and disease manifestations.
| Bacteria | T4SS | Diseases | Substrates | References |
|---|---|---|---|---|
| Interkingdom Transfer | ||||
| Extracellular Pathogens | ||||
| Agrobacterium tumefaciens | VirB/VirD4 | Crown Gall | Oncogenic T-DNA VirE2, VirE3, VirF | (20) |
| Bordetella pertussis | Ptl | Whooping cough | Pertussis Toxin | (47) |
| Helicobacter pylori | Cag | Gastritis, peptic ulcer, cancer | CagA | (65, 66, 90) |
| Facultative Intracellular | ||||
| Bartonella spp. | VirB/VirD4 | Cat-scratch, angiomatosis | BepA – BepG | (105) (95) |
| Trw | None | |||
| Brucella spp. | VirB | Brucellosis | ~14 | (95, 121, 222) |
| Legionella pneumophila | Dot/Icm | Legionnaire’s pneumonia | ~300 | (18, 127, 135, 137) |
| Obligate Intracellular | ||||
| Coxiella burnetti | Dot/Icm | Q fever | ~130 | (169, 223) |
| Anaplasma phagocytophilum | VirB/VirD4 | Granuloctyic anaplasmosis | AnkA, Ats-1, APH_0455 | (190, 224) |
| Anaplasma marginales | VirB/VirD4 | anaplasmosis | AM185, AM1141, AM470, AM705[AnkA] | (196) |
| Ehrlichia spp. | VirB/VirD4 | Ehrlichiosis | ECH0825 | (202) |
| Rickettsia spp. | VirB/VirD4 | Epidemic typhus, Mediterranean spotted fever | Unknown | (179, 188) |
| Wolbachia spp. | VirB/VirD4 | Endosymbiont of Filarial nematodes and arthropods | Unknown | (187) |
| Interbacterial Transfer | ||||
| Conjugation machines | Tra | Virulence and antibiotic resistance gene transfer Genome plasticity | Mobile elements | (5, 8, 9) |
| Agrobacterium tumefaciens | VirB/VirD4 | Conjugation | IncQ plasmid | (35) |
| Bartonella spp. | VirB/VirD4 | Conjugation | IncQ plasmid | (111) |
| Legionella pneumophila | Dot/Icm | Conjugation | IncQ plasmid | (129) |
| Neisseria gonorrhoeae | Tra | DNA release | Chromosomal DNA | (14) |
| Helicobacter pylori | Com | DNA uptake | Exogenous DNA | (13) |
| Xanthomonas campestris | Xac | Killing: Interbacterial competition | Xac toxins | (219) |
T4SS ARCHITECTURES AND ADAPTATIONS
By way of introduction to these fascinating and complex machines, we will first summarize recent exciting progress in structure - function studies of the paradigmatic A. tumefaciens VirB/VirD4 T4SS and related conjugation systems, and then briefly describe some of the structural adaptations acquired by T4SSs for specialized functions in pathogenic settings.
The paradigmatic A. tumefaciens VirB/VirD4 T4SS
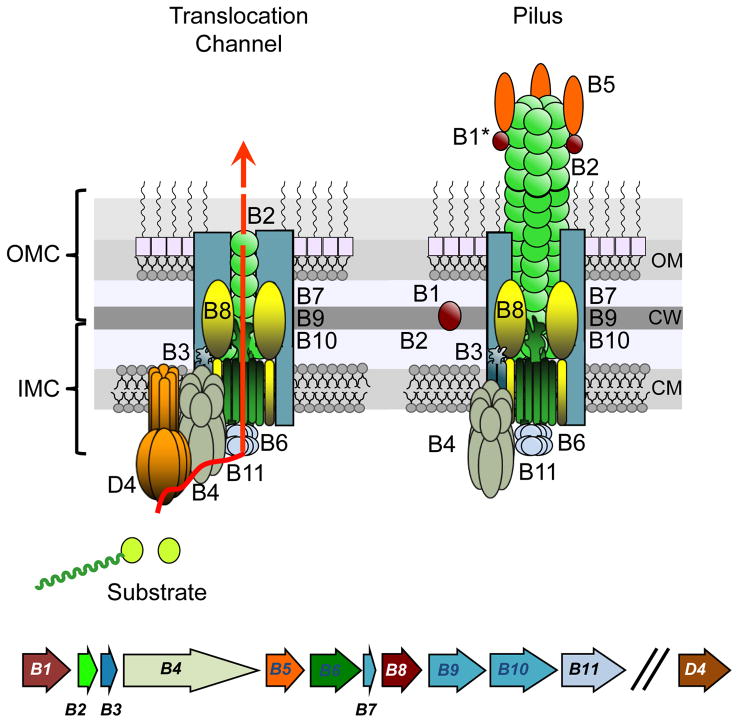
The T4SSs of Gram-negative bacteria are composed of four distinct machine subassemblies: i) the type IV coupling protein (T4CP), a hexameric ATPase related to the SpoIIIE/FtsK DNA translocases that recruits secretion substrates to the translocation machinery, ii) an inner membrane complex (IMC) responsible for substrate transfer across the inner membrane, iii) an envelope spanning outer membrane complex (OMC) required for substrate passage across the periplasm and outer membrane, and iv) the conjugative pilus, an extracellular organelle that initiates contact with potential recipient cells (Fig. 2) (8, 21, 22).
FIG. 2. Schematic of the Agrobacterium VirB/VirB T4SS.
Lower: virB genes are expressed from the same virB promoter and virD4 from a separate promoter (indicated by two slashes). Upper: The VirB and VirD4 subunits assemble as the translocation channel, which presents as two subcomplexes termed the outer membrane complex (OMC), composed of VirB7, VirB9, VirB10, and VirB2, and the inner membrane complex (IMC), composed of VirB3, VirB4, VirB6, VirB8, VirB11, and VirD4. VirB2, VirB5, and a proteolytic fragment of VirB1 (B1*) also assemble as the conjugative pilus without a requirement for VirD4. The physical and functional relationships between the translocation channel and the conjugative pilus are not yet known. OM, outer membrane; CW, cell wall; IM, CM, cytoplasmic membrane. See (24, 25, 237) for recent structures of related T4SSs.
A. tumefaciens is a phytopathogen that uses the VirB/VirD4 system to deliver oncogenic T-DNA and effector proteins to plants (Fig. 1) (20). A combination of structural and functional studies of the VirB/VirD4 T4SS and of closely related systems encoded by Escherichia coli conjugative plasmids, e.g., pKM101, R388, have generated for the first time a view of how the VirB/VirD4 T4SSs are architecturally arranged and function to convey secretion substrates across the Gram-negative cell envelope (Fig. 2). In studies carried out over a decade ago, the route of transfer of the oncogenic T-DNA substrate through the A. tumefaciens VirB/VirD4 T4SS was mapped with a formaldehyde (FA)-crosslinking assay termed transfer DNA immunoprecipitation (TrIP) (23). Results of the TrIP studies showed that the DNA substrate engages sequentially with VirD4, which then transfers it to the VirB11 ATPase. VirB11 in turn delivers the substrate to a putative translocation channel composed of VirB6 and VirB8 for delivery across the inner membrane. The substrate then passes through the periplasm and across the outer membrane via a channel minimally composed of VirB2 and VirB9.
More recent structural work has generated high-resolution structures of T4SS complexes, the latest of the near entire VirB/VirD4-like T4SS encoded by the conjugative plasmid R388 (24). Although the VirD4 and VirB11 ATPase subunits as well as the extracellular pilus were missing from this structure, the large ~3.2 MDa structure identified two VirB4 hexamers as part of a larger substructure the authors termed the IMC. Other IMC components include VirB3, VirB6 and VirB8. In the periplasm, the IMC is connected by a narrow stalk to the so-called OMC (also called the core complex). The OMC of the closely related E. coli pKM101-encoded T4SS is configured as a large barrel composed of 14 copies each of homologs of VirB7, VirB9, and VirB10 that extends across the entire cell envelope (25). Combining the results of the TrIP studies and the recent R388 structure, a cohesive model can thus be presented depicting the route(s) of substrate transfer across the Gram-negative bacterial cell envelope (Fig. 2) (22). Interestingly, in the pKM101 and R388 structures, the C-terminal regions of the VirB10-like scaffold proteins are predicted to form a channel across the outer membrane. While this might correspond to the pore through which substrates pass, it is likely this region of the channel is structurally more complex than currently depicted. This is because the structures solved to date lacked the pilin subunit and the conjugative pilus, which is also part of many T4SS structures (24, 25). In the TrIP studies, we did not detect DNA substrate crosslinking with A. tumefaciens VirB10, but rather contacts with the VirB2 pilin and VirB9. Conceivably, the distal portion of the translocation channel consists of a pilus-like structure that protrudes through the pore formed by the C terminus of VirB10 (Fig. 2) (17).
The A. tumefaciens VirB/VirD4 T4SS elaborates a conjugative pilus to mediate attachment to target cells. Assembly of the pilus does not require the VirD4 T4CP but does require the VirB1 lytic transglycosylase (17, 26). This pilus is composed of the pilin subunit VirB2 and the pilus-tip adhesin VirB5 (Fig. 2) (26). Its physical relationship to the VirB/VirD4 T4SS is not yet defined. However, in view of its role as an attachment organelle, it is interesting to speculate that the T4SS initially elaborates a pilus structure to establish contact with target cells. Then, once productive mating junctions are formed, the T4SS recruits the VirD4 T4CP for activation of the channel and subsequent substrate transfer. In such a model, the T4SS would function sequentially, first for elaboration of the pilus and second for biogenesis of the transfer channel.
Structure/function adaptations among T4SSs
While the R388 IMC/OMC complex can be viewed as a structural unit conserved among most if not all Gram-negative T4SSs, a large body of evidence now establishes that this structural unit has undergone extensive adaptation during establishment of pathogen - host relationships. Generally, the adaptations allow for i) substrate specific trafficking and spatiotemporal control of translocation during infection and ii) elaboration of novel and potentially antigenically variable pili or other surface structures. Examples of such adaptations are described briefly here and in Table 2, and discussed in more detail in subsequent sections.
Table 2.
T4SS machine adaptations enabling specialized functions.
| Bacteria | T4SS | T4SS Machine adaptation(s) | Specialized functions | References |
|---|---|---|---|---|
| A. tumefaciens | VirB/VirD4 | VirB1-VirB11, VirD4 | Transfer channel, Conjugative pilus Translocates DNA and protein substrates |
(35, 57) |
| B. pertussis | Ptl | No VirD4, VirB1, VirB5 homologs | Two-step translocation: PT subunits cross IM via GSP, PT crosses OM via Ptl T4SS No extracellular pilus |
(47) |
| H. pylori | Cag | VirB7-like CagT VirB10-like CagT have surface-exposed variable or repeat sequences | Phenotypic variation: Binding to different cell types, immune evasion | (40, 60, 69) |
| CagT | β1 integrin binding | |||
| VirB5-like CagL RGD motif | β1 integrin binding | |||
| CagA substrate at pilus tip | β1 integrin binding; translocation intermediate | |||
| Bartonella spp. | VirB/VirD4 | None | Translocates effectors & DNA substrate to human cells | (105)(110) |
| Trw | No VirD4 homolog | No substrate transfer | (42, 96) | |
| Multiple copies of VirB2 & VirB5 homologs | Variant forms of surface-exposed pilus for binding to different erythrocyte receptors? | |||
| Multiple copies of VirB6 & VirB7 homologs | Unknown | |||
| Brucella spp. | VirB | No VirD4 homolog | One-(?) and Two-step translocation via GSP and VirB systems | (114, 118, 225) |
| VirB12, orf13 | Serological marker, surface-exposed? | |||
|
A. phagocytophilum E. chaffeensis Rickettsia spp Wolbachia |
VirB/VirD4 | No VirB1, VirB5 homologs, but carries multiple copies of VirB2 paralogs and of ‘extended VirB6’ subunits | Variable pilus? Binding of different host cell/receptors? Immune modulation? | (41, 43, 187, 226,227) |
|
L. pneumophila C. burnetii |
Dot/Icm: Unrelated to VirB/VirD4 system | Translocates effectors & DNA substrate (to bacteria) | (18,44,130,131) | |
| VirB7 lipoprotein: N0 domain | Novel OMC structure | |||
| Fibrous surface mesh | Host cell binding? Immune modulation? | |||
| N. gonorrhoeae | Tra | No VirB11 homolog, Additional subunits related to F plasmid T4SS | DNA release to milieu | (228) |
| Variant forms of TraA pilin | Unknown | (14) | ||
| H. pylori | Com | No VirB, VirB5, VirB11 Com: DNA uptake across OM ComEC: DNA uptake across CM |
Natural transformation | (13) |
| X. campestris | Xac | VirB/VirD4 T4SS; VirB7 ortholog has N0 domain | Interbacterial delivery of Xac toxins for killing | (219) |
Substrate recruitment and translocation across the inner membrane
With a few notable exceptions, the T4SSs employ a T4CP receptor to recruit cognate substrates to the transfer channel. For docking with the T4CP, secretion substrates carry translocation signals located either C terminally and composed of clusters of positively-charged or hydrophobic residues, one or more internal signals of unspecified composition, or a combination of C-terminal and internal signals (27–31). Substrates also can have distinct translocation signals for docking with different T4SSs, providing another example of the functional versatility of these systems (30). In addition to these intrinsic translocation signals, T4SSs employ various chaperones or adaptor proteins for conferring substrate specificity and maintaining the substrate in a translocation competent form (32, 33). In the A. tumefaciens T4SS system, for example, translocation of the VirF effector proceeds independently of other known factors, whereas translocation of VirE2 requires cosynthesis of the VirE1 chaperone (34, 35). VirE2 is a single-stranded DNA binding protein that, upon transfer to plant cells interacts with the cotranslocated T-DNA to protect the DNA substrate and facilitate its delivery to the plant nucleus (Fig. 1). VirE1 shares several features of the specialized chaperones associated with type III secretion systems (T3SSs), including a small size, acidic pI, and an amphipathic helix. As discussed further below, the L. pneumophila Dot/Icm T4SS translocates hundreds of effectors to mammalian cells during infection. A subset of these is dependent on four accessory proteins residing in the cytoplasm or inner membrane. These include DotM, DotN, IcmS, and IcmW, which interact with each other in different combinations to mediate binding and recruitment of different substrates to the DotL T4CP (32). Thus, in L. pneumophila, a combination of C-terminal and internal translocation signals, together with a complex network of chaperone/adaptor/substrate interactions, regulates delivery of specific subsets of effector proteins through the Dot/Icm T4SS to host cells during L. pneumophila infection (32, 33).
T4SS structural and surface organelle variations
Many VirB/VirD4-like systems also have appropriated novel domains of functional importance (Table 2) (36). For example, some VirB6 subunits possess >30-kDa domains located at their C termini that have been shown or are proposed to localize at the cell surface (37, 38). Some VirB7-like lipoproteins and VirB10-like structural scaffolds also carry novel surface-localized motifs. Most notably, H. pylori CagT and CagY are classified as VirB7- and VirB10-like, respectively, but both subunits are considerably larger than their VirB counterparts and both localize extracellularly as a component of a large sheathed filament produced by the Cag T4SS (39). CagT and CagY also possess repeat regions that differ in size and composition among Cag systems of different H. pylori isolates (5, 40). Additionally, several systems including the Bartonella Trw system and VirB/VirD4 T4SSs carried by species in the order Rickettsiales carry multiple pilin genes in tandem array in their genomes, suggesting that variable pilus structures are elaborated through differential expression or intergenic recombination (41–43). Other T4SSs lack pilin genes or genes encoding the pilus tip protein VirB5, which is essential for elaboration of pili. These systems therefore probably do not elaborate pili but might still display surface structures, as exemplified by the L. pneumophila Dot/Icm, which encodes a fibrous structure on the cell surface (44). Collectively, the novel surface variable proteins or structures acquired by various T4SSs during evolution are thought to contribute to establishment of pathogen-host cell interactions, e.g., by mediating attachment to different host cells or host cell receptors or through modulation of the immune response.
T4SS EFFECTORS AND THEIR ROLES IN PATHOGENESIS
Besides appropriating novel structural motifs for specialized functions, the versatility of T4SSs is reflected in the diversity of substrates translocated to bacterial or eukaryotic target cells. Among the effector translocators employed by pathogens during infection, some translocate a single substrate, most deliver a restricted number of a half dozen or so, and a few highly promiscuous systems are estimated to translocate from fifty to several hundred effectors (Fig. 1, Table 1). In the following sections, we will review recent information about these systems, focusing mainly on the biochemical and cellular activities of effectors upon delivery to the host. Over the past decade, as studies have progressed on the T4SSs as well as the functionally (but not ancestrally) related T3SSs, it has become clear that common themes have emerged in the evolutionary design of many effectors. Most prominently, many effectors carry eukaryotic-like domains and thus exert their effects on various cellular processes or signaling pathways by mimicking activities of endogenous cellular proteins. For both the T4SSs and T3SSs, molecular mimicry has evolved predominantly to ‘fine-tune’ specific cellular functions to the advantage of the invading pathogen rather than irreversibly blocking cellular homeostasis. Indeed, recent work shows that both of these dedicated secretion pathways often target apoptotic pathways with the aim of inhibiting premature death of the host cell in which the pathogen is attempting to establish a replicative niche.
Extracellular pathogens
Bordetella pertussis is the causative agent of whooping cough, a respiratory illness spread mainly through aerosolization (45). Vaccination initiated in the 1940’s led to a decline in incidence, but since the 1990s the number of cases of whooping cough has increased worldwide (46). The reasons underlying the resurgence are not clear, but are generally attributed to waning of protective immunity from vaccination or natural infection, low vaccination coverage, transmission from individuals vaccinated with the currently used acellular vaccines, or genetic changes in B. pertussis to resistance against current vaccines (46). B. pertussis has several virulence factors of importance to infection and human-to-human spread, including pertussis toxin (PT) which is the sole substrate of the Ptl T4SS (47). PT is an ADP-ribosylating toxin composed of the catalytic A subunit and 5 B subunits required for translocation of the A subunit across eukaryotic cell membranes (48–51). Genes encoding PT subunits and the Ptl T4SS are genetically linked and coexpressed from the same promoter (52, 53).
Although the Ptl T4SS was recognized over 20 years ago to be closely related in overall gene order and subunit composition to the A. tumefaciens VirB/VirD4 T4SS and conjugation machines (54), it has several important distinctions that impart novel structural and functional features (Table 2). First, it lacks a homolog for the VirD4 substrate receptor, implying that an alternative mechanism has evolved for recruitment of the PT substrate to the transfer machine. In fact, the A and B subunits each carry a classical sec signal sequence and are translocated across the inner membrane via the general secretory pathway (GSP), completely independently of the Ptl system (55). Once in the periplasm, the subunits associate with the outer membrane and then assemble as the holotoxin, a process requiring extensive folding and disulfide bond formation. Mature holotoxin no longer associates with the outer membrane, but instead is released to periplasm, and becomes available for recruitment to the Ptl T4SS (47). The signal(s) mediating toxin recruitment to the T4SS is presently unspecified, although a domain of the A subunit may comprise part of it. Upon recruitment, the Ptl T4SS delivers PT across the outer membrane and to the extracellular milieu (56). Thus, two distinct machines, the Sec translocase and the Ptl T4SS, export PT in a two-step translocation reaction, in contrast to the one-step translocation route envisioned for the T4CP-dependent T4SSs (Fig. 2) (57).
Also in contrast to other T4SSs, which translocate partially or completely unfolded substrates, the Ptl system exports PT as a large, tightly folded multimer. The most recent structures obtained for VirB/VirD4 conjugation systems show no obvious portals of entry or outer membrane channels of a size sufficient to accommodate PT (24, 25, 58). This suggests that the Ptl T4SS must have acquired a novel outer membrane architecture for exporting this AB5-type toxin. Finally, the Ptl system lacks homologs for VirB1 and VirB5, two subunits required for biogenesis of pili (54). Accordingly, the Ptl system releases its cargo to the milieu, dispensing with a need for pilus or pilus-dependent contacts with the host cell.
Once PT is released to the milieu, it can interact with various mammalian cells, predominantly respiratory epithelial cells (Fig. 3) (45). The B oligomer of the toxin mediates binding to cell surface receptors, predominantly glycosylated proteins or lipids, which can induce host cellular responses such as T cell mitogenicity and hemagglutination independently of the biological activities attributable to the catalytic function of the A subunit within the host cell (59). Entry of PT into the host cell occurs by receptor-mediated endocytosis. PT trafficking follows the retrograde transport system sequentially through the endosomal compartment, Golgi apparatus, and endoplasmic reticulum (ER). Independently of the B moiety, the A moiety translocates through the ER into the cytosol to its target protein, membrane-associated trimeric G proteins. Upon binding, the A subunit catalyzes ADP-ribosylation of Cys residues in the C-terminal part of the α subunit of trimeric G proteins. ADP-ribosylation of Giα locks the protein in an inactive state, resulting in a loss of inhibition of adenylate cyclase activity and an increase in intracellular concentration of cAMP. Increased cAMP levels disrupts many cellular processes, accounting for the majority of pathological consequences of B. pertussis infection. Common effects are increased insulin secretion, sensitization to histamine, and inhibition of immune cell recruitment (Fig. 3) (45, 47).
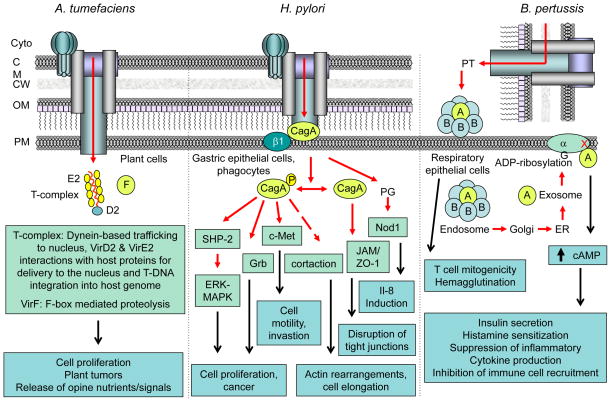
FIG. 3. T4SS effectors and cellular consequences of translocation by extracellular pathogens.
A. tumefaciens and H. pylori deliver substrates through direct cell-cell contact, and B. pertussis by a contact-independent mechanism. Pathogens target the host cell types listed. Translocated effectors interact (red arrows, dash denotes indirect interaction) with host cell proteins (light green boxes) to modulate various cellular processes and signaling pathways. Cellular consequences of the effector - host protein interactions (black arrows) are listed (aqua boxes).
Overall, the B. pertussis Ptl system represents a fascinating and, to date, novel example of a T4SS that evolved through loss of conserved subunits from an ancestral machine as a two-step translocation pathway capable of translocating a folded multimeric toxin to eukaryotic cells independently of direct cell-cell contact.
Helicobacter pylori is an extracellular pathogen that colonizes the gastric epithelium. It is the principal cause of chronic active gastritis and peptic ulcer disease and a risk factor for the development of gastric carcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma (Table 1) (60, 61). Virulent strains of H. pylori associated with the enhanced risk of developing peptic ulcers or adenocarcinoma harbor a 37-kilobase (kb) pathogenicity island (PAI) encoding the cytotoxin-associated gene (Cag) T4SS (62, 63). Also present on this PAI is a gene encoding CagA, the only known protein substrate of this T4SS (64). CagA is a ~120–145 kDa protein that shows no significant homology with other known proteins. Its size variation is due to structural diversity in its C-terminal region (65). In contrast to the B. pertussis Ptl system, the Cag T4SS recruits CagA from the bacterial cytoplasm via the Cagβ substrate receptor for transfer in one step across the H. pylori cell envelope and into mammalian target cells (Fig. 3).
The Cag T4SS is related to the A. tumefaciens VirB/VirD4 system but is also considerably more complex, as evidenced by the appropriation of more than a half dozen subunits and subdomains from unknown ancestries (66). As mentioned earlier, CagT and CagY are respectively classified as VirB7- and VirB10-like, yet both subunits are considerably larger than their VirB counterparts. These subunits comprise part of an OMC that is probably similar in overall structure to those solved for conjugation machines, while their additional domains localize extracellularly as a component of an extracellular pilus or large sheathed filament associated with the Cag T4SS (Table 2) (39). Importantly, these domains are composed of repeat regions that differ in size and composition among the various H. pylori strains (67, 68). Functions of these repeat regions are not yet defined, but are postulated to affect binding of H. pylori cells to mammalian cells through modulation of host β1 integrin interactions (69). There is also evidence that passage of H. pylori in mouse models results in variant forms of VirB10-like CagY, originally prompting the suggestion that CagY undergoes antigenic variation through homologous recombination within its repeat regions to evade the host immune response while maintaining T4SS function (67, 68). However, very recent work showed that immune driven DNA rearrangements in CagY serve not so much to evade the host immune response altogether, but rather to fine tune the response so as to establish the optimal homeostatic conditions of inflammation for persistent infection (40).
The Cag T4SS reportedly elaborates two types of surface structures, a large sheathed structure and pili, both implicated in interactions with host cells (39, 70, 71). The sheathed structures are 70 nm in diameter and are composed of pilin-like proteins CagC and CagL, as well as extracellular domains of CagY, CagT, and CagX (39). CagY and CagL bind β1 integrins, consistent with a role for this structure in attachment to host cells (69). More recent work has focused on the much narrower (~13 nm wide) pilus-like structures (71, 72). These pili are detected only when H. pylori cells are cocultured with gastric epithelial cells, in an abundance of 3 to 4 pili per adherent H. pylori cell. Most of the Cag T4SS subunits are required for pilus assembly, although strikingly not the VirB2 pilin-like CagC subunit or VirB10-like CagY, even though both of these subunits are required for T4SS function as monitored by induction of IL-8 production and CagA translocation (72). Together, these observations raise the interesting possibility that the Cag T4SS elaborates distinct organelles for different purposes in relation to the infection process. Although specific functions of these T4SS structures awaits further definition, it is noteworthy that CagL, a homolog of the VirB5 pilus tip protein in A. tumefaciens, clearly plays a role in mediating interaction with host cell β1 integrins via its RGD motif (73). Through its RGD-dependent integrin binding activities, purified CagL has been shown to elicit several responses in human cells, including cell adhesion, cell spreading, activation of host cell tyrosine kinases, and secretion of IL-8 (74, 75).
The importance of Cag T4SS-encoded pili to the H. pylori infection process is further underscored by the finding that strains that fail to produce pili are defective in Cag T4SS-dependent IL-8 induction in gastric epithelial cells (71, 76). Furthermore, changes in environmental conditions, such as reduced iron concentration, result in correlative increases in pilus production and Cag T4SS-associated activities (77). Also of interest, the sole protein substrate, CagA, of this T4SS is detectable on the H. pylori cell surface, particularly at the tip of pili (Fig. 3) (40, 69, 73). Both the surface display and pilus association of CagA are unique features among other known T4SS secretion substrates, and at least two lines of evidence support the notion that CagA surface accessibility is a biologically relevant entity. First, CagA binds phosphatidylserine at the outer leaflet of the host cell cytoplasmic membrane, which in turn induces its uptake into the cell (78). Second, as noted above, CagA translocation depends on the presence of β1 integrins as receptors for binding by the T4SS components CagL, CagY, CagI. However, CagA itself also binds strongly to β1 integrins, suggesting that the pilus-tip bound form is a translocation intermediate (69).
Several recent studies have explored the underlying mechanisms responsible for recruitment and passage of CagA through the Cag T4SS. Docking of CagA with the T4SS requires three proteins, a chaperone CagF, the VirD4 homolog Cagβ, and CagZ (79, 80). CagA is highly labile in H. pylori cells unless it is coproduced and forms a complex in a 1:1 stoichiometry with the CagF chaperone. CagF differs from other T4SS or T3SS secretion chaperones, e.g., A. tumefaciens VirE1. Besides being much larger (32-kDa) than the other (< 5-kDa) chaperones, CagF is highly hydrophobic and recently was shown to bind all five domains of CagA, presumably forming a broad interaction surface that protects the entire protein from degradation (81). CagF binding is also thought to prevent CagA intramolecular interactions of importance for protein function upon translocation to mammalian cells but might block CagA docking with the Cagβ receptor in H. pylori. While the nature of the CagA - Cagβ docking reaction remains to be defined, it is evident that the Cag T4SS has appropriated a novel CagF accessory factor to promote stabilization of the large, multidomain CagA substrate and docking with the Cagβ receptor (81).
Upon delivery into gastric epithelial cells, CagA is localized to the inner leaflet of the plasma membrane where it undergoes tyrosine phosphorylation (Fig. 3). The phosphorylated protein interacts with SH2-domain-containing protein tyrosine phosphatase (SHP2) (82). CagA resembles the Gab family of scaffold proteins, which also form a complex with SHP2 in a tyrosine phosphorylation dependent manner in juxtaposition to the plasma membrane. Thus, partly through Gab mimicry, CagA exerts its many effects in host cells through promiscuous binding of a variety of human proteins by both phosphorylation-dependent and -independent mechanisms (Table 3) (83).
Table 3.
Molecular Mimicry: Eukaryotic protein domains carried by T4SS effectors.
| Motif | Biochemical function | T4SS Effector* | References |
|---|---|---|---|
| EPIYA | Tyrosine phosphorylation |
Hp CagA Bh BepD, BepE, BepF Ap AnkA |
(87–89, 229) |
| Ank | Protein-protein interaction |
Ap, Ec, Rr AnkA Ec p200 Lp AnkB, AnkX, AnkH, AnkJ, 7 others Cb AnkA,B, D*,F,G Wolbachia wPip: 60 Ank proteins Ot: 50 Ank proteins Rf: 22 Ank proteins |
(198, 199, 230, 231) |
| F box | Interaction with SCF ubiquitination complexes |
At VirF Lp AnkB, LicA, LegU1, PpgA, Lpg2525 Cb AnkD, CBU0814, CBU0014 Ot Anklu9 |
(158, 159, 232, 233) |
| Farnesylation/Prenylation | Post-translational modification for protein - protein, protein - membrane interactions |
Lp AnkB Lp Lpg2525, LegU1? |
(234, 235) |
| FIC | AMPylation, phosphorylation, UMPylation, phosphocholination |
Lp AnkX Bh BepA, BepB, BepC |
(96, 102) |
| Gab | Family of scaffold protein | Hp CagA | (83) |
| ARF-, Ras-GEF | Guanine nucleotide exchange | Lp RalF, LegG2 | (152, 236) |
| SREBPs | Sterol regulatory element binding proteins | Am APH_0455 | (197) |
Abbreviations: Hp, Helicobacter pylori; Bh, Bartonella henselae; Ap, Anaplasma phagocytophilum; Ec, Ehrlichia chafeensis; Lp, Legionella pneumophila; Cb, Coxiella burnetii; Ot, Orientia tsutsugamushi; At, Agrobacterium tumefaciens; Am, Anaplasma marginales
CagA is phosphorylated by Src family kinases (SFKs) at Tyr residues in the Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs that resemble c-Src consensus phosphorylation sites and are located in variable numbers in the C-terminal region of the protein (Table 3) (84). Binding of phosphorylated CagA (CagA-P) to SHP2 causes aberrant activation of SHP2 and consequently of the ERK-MAPK (mitogen-activated protein kinase) pathway, which reportedly contributes to carcinogenesis by inducing mitogenic responses (Fig. 3) (65). CagA-activated SHP2 also dephosphorylates focal adhesion kinase, causing impaired focal adhesions that are associated with an elongated cell shape known as the hummingbird phenotype (85). Interestingly, East Asian CagA and Western CagA are respectively characterized by the presence of an EPIYA-D segment and an EPIYA-C segment to which SHP2 binds in a phosphorylation dependent manner. The EPIYA-D segment binds SHP2 more strongly than the -C segment, and it is speculated that H. pylori carrying the biologically more active East Asian CagA variant are responsible for the propensity of these strains to induce severe gastric atrophy and gastric cancers in infected patients (86).
Since the discovery of the CagA EPIYA motif, at least nine other T4SS or T3SS effectors were shown to carry such eukaryotic-like motifs with similar biochemical activities upon translocation to host cells. Among the T4SSs discussed in more detail below, the Bartonella henselae VirB/VirD4 T4SS translocates three effectors (BepD, PepE, and BepF) each with one or more EPIYA or EPIYA-like motifs (87). The Anaplasma phagocytophilum AnkA effector also bears multiple EPIYA motifs that display variations in number and subtypes among different bacterial strains (88). Other pathogens employing T3SSs to translocate EPIYA-bearing effectors during infection include Chlamydia trachomatis (the Tarp effector, induces cytoskeletal rearrangements) and enteropathogenic E. coli (Tir, triggers actin pedestal formation). Haemophilus dureyi also delivers two EPIYA-containing effectors, LspA1 and LspA2, via a two-partner secretion system to eukaryotic cells whereupon inhibition of Src family kinase (SFK) functions blocks Fcγ-mediated triggering of phagocytosis (Table 3) (89).
In addition to the SHP2 interaction, an array of other CagA-binding partners has been described, including carboxy-terminal Src kinase (Csk), growth factor receptor-bound protein 2 (Grb2), tight junction protein zonula occludens 1 (ZO-1), scatter factor receptor c-Met and phospholipase C-γ. This plethora of CagA phosphorylation-dependent and –independent interactions modulates pathways involved in the innate immune response, cell shape regulation, and signal transduction (Fig. 3) (see (90, 91)). There is also compelling evidence that the Cag T4S machine, through receptor dependent activation or translocation of fragments of peptidoglycan or another unidentified effector(s), operates independently of CagA and of VirD4-like Cagβ to elicit stress-response pathways that result in induction of IL-8 secretion (Fig. 3) (92).
Facultative intracellular pathogens
Bartonella spp. reside in erythrocytes and cause long-lasting bacteremic infections in diverse mammalian hosts. They are transmitted by blood-sucking arthropods most often through dermal inoculation. Upon reaching the blood stream, Bartonella spp. invade erythrocytes and persist intracellularly for the lifetime of the red blood cell. Adaptation to the reservoir host causes no or mild disease symptoms, whereas infection of incidental hosts can cause a spectrum of diseases including cat-scratch disease, Carrion’s disease, chronic lymphadenopathy, trench fever, chronic bacteraemia, endocarditis, bacillary angiomatosis, peliosis hepatis and neurological disorder. Without treatment, Bartonella infections are associated with high mortality and the potential for relapse due to the existence of an intraerythrocytic phase that may provide a protective niche for the bacteria (93–95).
Bartonella spp. employ two T4SSs for different purposes relating to the infection process (Table 2) (96). The Trw T4SS shares strong sequence similarities with the Trw system encoded by the IncW plasmid R388, with the exception that it lacks a homolog for the VirD4-like substrate receptor (97). The Trw homologs of the two systems share 20 to 80% sequence identities, and interestingly the Bartonella tribocorum Trw genes encoding VirB5-like TrwH, VirB10-like TrwE, and VirB11-like TrwD substitute for their homologs in the TrwR388 system (97, 98). This finding underscores the conservation of machine architecture between two T4SS that have evolved for completely different functions. However, since the Bartonella Trw system lacks a VirD4-like T4CP, it is postulated to function not as a translocation system but rather to mediate attachment to host cells. In line with such a function, the Trw locus possesses tandem copies of genes encoding homologs of the VirB2 and VirB5 pilin proteins, suggesting that this system is specifically adapted for the production of antigenically variable pili through intergenic recombination (97). The adaptation of a T4SS for elaboration of a variable surface structure aligns this system with the H. pylori Cag system, which elaborates variant forms of surface-exposed channel subunits, as well as Rickettsial systems discussed later in this chapter.
Studies have shown that the Trw system is required for establishing intra-erythrocytic infection in a B. tribocorum - rat model (93, 97). A signature-tagged mutagenesis (STM) screen further identified mutations in the trw locus that severely impaired adhesion of Bartonella birtlesii to erythrocytes (99). Based on the postulated surface location of the Trw components, this T4SS might directly interact with the erythrocyte surface thereby restricting the host range of the erythrocyte infection. A role for this system in host-specificity was confirmed through a demonstration that swapping of Trw from rat-specific B. tribocorum for that of cat-specific Bartonella henselae and human-specific Bartonella quintana shifted the host ranges of these latter species for erythrocyte infection towards rats (42). The further observation that the Trw pilin subunits exist in multiple copies suggests the trw locus encodes variant forms of surface-exposed pilus which might facilitate the specific interaction with polymorphic erythrocyte receptors either within the reservoir host population or among different reservoir hosts (42).
The second T4SS, designated VirB/VirD4, is composed of a complete set of homologs of the A. tumefaciens VirB and VirD4 subunits (100, 101). It contributes to pathogenicity by delivering up to seven effector proteins termed Beps (Bartonella effector proteins) into nucleated cells (Fig. 4). The Beps were identified originally by the presence of a ~140-residue domain near their C termini that was termed BID (Bep Intracellular Delivery). This domain and a positively charged C-terminal tail sequence correspond to a bipartite translocation signal required for delivery of several of the Beps through the VirB/VirD4 T4SS (28). For a few Beps, the BID domain is not required for translocation but rather contributes to effector function in the host cell.
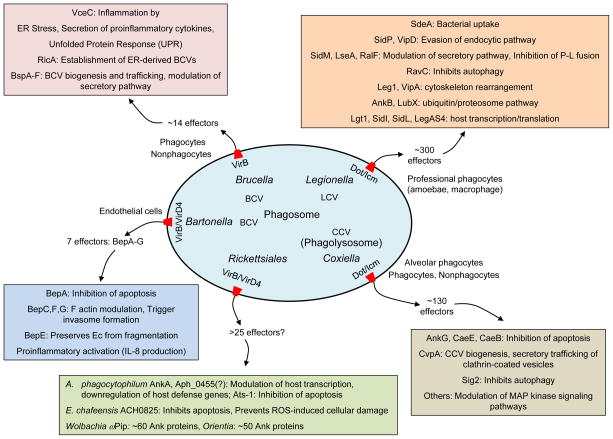
FIG. 4. T4SS effectors and cellular consequences of translocation by intracellular pathogens.
The pathogens listed deliver effector proteins to establish a replicative niche within phagosomal compartments. Associated with each organism is the name of the T4SS, the replicative niche (e.g., LCV for Legionella-containing vacuole), the host cell type(s) targeted, and the current number of known or estimated effectors. Also for each organism is a list of representative effectors and cellular consequences of their activity within the host cell (colored boxes).
Once inside the host cells, Beps target host components and modulate cellular processes to the benefit of the bacteria. A deletion of all seven bep genes (bepA-bepG) abolishes all VirB/VirD4-dependent cellular phenotypes, including cell invasion, pro-inflammatory activation, and the inhibition of apoptosis (Fig. 4) (28). The Beps have a modular architecture that includes one or more BID domains. BepA, BepB, and BepC also carry a FIC (filamentation induced by cAMP) domain, which is present in many bacteria and eukaryotes including mammals (Table 3) (96). FIC enzymes posttranslationally modify proteins through AMPylation, UMPylation, phosphorylation, or phosphocholination. All FIC enzymes catalyze the transfer of a part of a metabolite upon cleavage of a pyrophosphate-bond (102). Besides their presence in Bartonella Beps, FIC domains have been identified in effectors translocated by the L. pneumophila Dot/Icm system (AnkX) as well as T3SSs employed by Pseudomonas syringae (AvrB), Vibrio parahaemolyticus (VopS), and Xanthomonas campestris (AvrAC) (102). At this point, however, the contributions of Bep FIC domains to Bartonella infection processes have not been defined. As mentioned earlier, some Beps also carry EPIYA motifs serving as sites of tyrosine phosphorylation upon delivery into the mammalian host (Table 3) (28, 103).
Considerable progress has been made toward defining specific contributions of the Bep proteins to stages of the infection cycle (Fig. 4). In brief, upon deposition on the skin by a blood sucking arthropod, one or a few Bartonella cells are taken up by endocytosis into a vacuole called Bartonella-containing vacuole (BCV) (104, 105). The process of endocytosis is arrested by Beps (BepC, BepF, BepG) delivered into the host cytoplasm by the VirB/VirD4 T4SS (106, 107). Bacterial aggregates form at the cell surface, and these bacteria then invade the endothelial cells via F-actin-dependent invasome-mediated internalization (105). Bartonella then colonize the ‘dermal niche’ most likely within dendritic cells, which are considered important for dissemination to the ‘blood-seeding niche’. BepE is essential for invasion of cells in the dermal niche and for subsequent spread to the blood stream. Recently, it was reported that the BID domains of BepE contribute to endothelial cell migration and prevent cell fragmentation through interference of the RhoA signaling pathway (108). Furthermore, BepA mediates protection of endothelial cells from apoptosis through binding of a BID domain to host cell adenylyl cyclase with a consequence of elevated cAMP production and inhibition of apoptosis (109).
In sum, investigations to date point to essential contributions of the VirB/VirD4 T4SS and Bep effectors to the primary stages of infection. All Bep-dependent phenotypes have been shown to rely on the different BID domains, which collectively exert their effects on different physiological or signaling pathways in the human host and also serve as translocation signals in the bacterium presumably for productive binding with the VirD4 receptor. At this time, nothing is known of how the FIC and EPIYA domains contribute to the Bep-dependent phenotypes, despite evidence for tyrosine-phosphorylation of BepE’s EPIYA motif and binding of phosphorylated BepE to SHP2 and Csk, two well-characterized binding targets of H. pylori CagA. Of final note, the B. henselae VirB/VirD4 T4SS was shown to be capable of delivering DNA into vascular endothelial cells (110). This T4SS is closely related in subunit composition to conjugation systems, underscoring both the common ancestry and functional similarities of effector translocator and conjugation systems (111). Whether DNA transfer is a natural substrate that Bartonella delivers to eukaryotic cells during the infection process is an intriguing question for further study.
Brucella spp. cause abortion in their natural animal hosts (e.g., swine, goats/sheep, cattle, bison) and are responsible for human brucellosis. Brucella infect and replicate in phagocytic and non-phagocytic cells. The bacteria can persist for years in the reticuloendothelial system (RES) of their natural hosts as well as incidental human hosts (112). Human brucellosis is characterized by undulant fevers and constitutional symptoms including malaise, mayalgia, and enlarged spleens and livers. Chronic infection can progress to severe complications including arthritis and endocarditis. Factors mediating uptake are not completely defined, but once endocytosed, Brucella reside initially in the Brucella-containing vacuole (BCV). The BCV interacts sequentially with early and late endosomes and lysosomes, finally establishing an intracellular replicative niche derived from the ER. Subsequent to replication, Brucella exit host cells to reinfect adjacent cells (113).
The VirB T4SS of Brucella plays a critical role in establishment of the ER-derived replicative BCVs, and is an essential virulence factor in a mouse model of infection (114–116). It is also essential for persistence of the bacteria in a mouse model and goat host (115, 117). While the Brucella T4SS gene cluster contains homologs of all of the A. tumefaciens VirB genes, there is no VirD4-like receptor and the operon contains two additional genes, virB12 and orf13 (Table 2) (114). The absence of a VirD4-like receptor suggests either that an alternative routing mechanism exists for substrate transfer across the inner membrane (as shown for B. pertussis PT translocation) or that this machine does not translocate substrates but provides another function, e.g., attachment, of importance for infection (as proposed for the Bartonella Trw system). Confirming the former model, several Brucella effectors are now known to be translocated by a VirB-dependent mechanism to host cells (see below). Very intriguingly, only a subset of the effectors carry a classical sec signal suggestive of a two-step translocation route reminiscent of the PT toxin export system (118). The mechanism(s) by which other Brucella effectors engage with the VirB machine, and in which cellular compartment (e.g., cytoplasm, periplasm), remains to be determined.
Acidification of the phagosome is required for induction of the virB operon encoding the Brucella VirB T4SS during infection of macrophages (119). Production of the T4SS and effector translocation allows for evasion of lysosomal fusion with the BCV and association with the ER-derived intracellular compartment (120). Effector candidates were identified by bioinformatics screens for eukaryotic-like domains, genes that are coregulated with the virB operon, and genes whose products interact with human ER exit site (ERES)-associated proteins. At this time, 14 effectors have been identified, although in most cases their functions during infection are not yet established (Fig. 4). Interestingly, an additional 6 candidate effectors also appear to be translocated to human cells independently of the T4SS (121).
The first identified effectors were VceA and VceE, and VceE is currently the best characterized among all Brucella effectors (118). VceC has been shown to induce ER stress and secretion of proinflammatory cytokines (Fig. 4). Ectopically expressed VceC binds the ER in HeLa cells via an interaction with the ER chaperone BiP/Grp78, and VceC production is correlated with reorganization of ER structures (122). Furthermore, in macrophages, VceC triggers the unfolded protein response (UPR), resulting in the induction of inflammation by Brucella abortus (122). Whether VceC exerts its activity on the UPR through binding of BiP remains to be determined. VceC is not thought to be involved in establishment of the BCV, but rather contributes to long-term colonization and overall fitness of the infecting Brucella. However, another effector termed RicA (Rab2 interacting conserved protein A) binds the GDP-bound form of Rab2, a host GTPase shown to be essential for Brucella intracellular replication through binding of BCVs. Correspondingly, ricA mutants show a decrease in Rab2 recruitment to BCVs (123). These findings led the authors to postulate that T4SS-mediated translocation of RicA is important for establishment of the ER-derived replicative BCVs by enabling precise interactions with secretory pathway organelles (Fig. 4).
Two effectors, BrpA and BtpB, share a conserved TIR (Toll/Interleukin-1 receptor)-containing domain and both are essential for virulence (124, 125). BtpA exerts its effects through inhibition of NF-kB and the toll-like receptor (TLR) signaling pathway, whereas BtpB activates NF-kB and the TLR pathway. TIR-containing adaptors are widely present in mammals where the associated proteins regulate the signaling cascades of innate immune recognition. However, many bacteria besides Brucella employ TIR domain-containing proteins for modulation of the TLR signaling cascade, thus providing another example of molecular mimicry by pathogens for subversion of host cellular processes.
Finally, by use of a reporter assay for translocation, 5 candidate effectors designated BspA-F were identified whose delivery to human cells was dependent on the VirB T4SS (121). Importantly, several of these effectors were shown to localize to compartments of the secretory pathway when ectopically expressed in HeLa cells. Furthermore, ectopically expressed BsbA, BspB, and BspF were found to specifically inhibit host protein secretion and membrane trafficking along the secretory pathway, in line with observations that such perturbations are also observed during Brucella infection. Additionally, evidence was presented that these Bsp’s contribute to the biogenesis of BCVs and to bacterial replication in macrophages, as well as to long-term persistence in the liver of chronically infected mice. These new findings establish a possible link between BCV trafficking and VirB T4SS-dependent modulation of the secretory pathway (Fig. 4) (121).
Legionella pneumophila is ubiquitous in the environment, preferentially thriving in water systems. They colonize a variety of ecological niches, including various amoebae and other protozoa in which they can grow and replicate (126). Legionella enters the human lung via inhalation of Legionella-containing aerosols. The bacteria replicate in alveolar macrophages and can cause a potentially fatal pneumonia termed Legionnaires’ disease. The L. pneumophila infection cycle involves host-cell entry by phagocytosis, creation of a specialized vacuole termed the ‘Legionella-containing vacuole’ (LCV) for replication, macrophage lysis, and infection of neighboring cells (127). Creation of the LCV requires the Dot/Icm T4SS, which has the amazing capacity to translocate an estimated 300 effector proteins, approximately 10% of this organism’s proteome, into target cells (18, 128). Like the Bartonella spp. and A. tumefaciens VirB/VirD4 systems, the Dot/Icm T4SS has retained a functional vestige of its ancestral conjugation system in being able to mobilize transfer of IncQ plasmids to bacterial recipients, although the functional significance of DNA transfer during the infection process is not known (33, 129).
As mentioned earlier, the Dot/Icm systems are only weakly related to the VirB/VirD4 systems and thus were termed type IVB systems (15). The Dot/Icm systems are composed of at least 25–27 proteins, many of which localize to the cytoplasm or inner membrane (32). A number of these are chaperones or adaptors that fulfill the complex task of delivering the multitude of effectors to the host cell at appropriate times during the infection cycle. Recent structural work has shown that the cell envelope-spanning channel consists in part of a ring-shaped OMC similar to that of the VirB/VirD4 T4SSs (130). A notable variation from the latter systems is that DotD, a lipoprotein thought to provide a stabilizing function similar to that of VirB7, possesses a large domain structurally similar to N0 domains (131). Such domains are also found associated with secretins in the types II and III secretion systems, and might serve as a structural scaffold or for recruitment of substrates in the periplasm (see (131)). The Dot/Icm T4SS is thought to translocate substrates in one step across the cell envelope, but this model has not been rigorously tested, so a role for the N0 domain of DotD in substrate binding remains a possibility. Another difference with the VirB/VirD4 T4SSs is that the Dot/Icm system does not elaborate extracellular pili but rather a fibrous mesh on the bacterial cell surface composed at least in part of the DotH and DotO proteins. (44)
Most of the candidate effectors were identified through a combination of bioinformatics screens and assays for translocation using reporter protein fusions (132). Efforts to establish the importance of given effectors to the infection process, however, have been hampered by the fact that many effectors have redundant functions (133). In some cases, mutant strains lacking a single effector show intracellular growth defects upon depletion of specific host factors or are outcompeted by wild-type bacteria in competition assays (127, 134). Functions have been assigned for about 50 of the candidate effectors, which can be grouped based on their major cellular target pathway(s) during the infection cycle (135). A full description of the biochemical activities of the Dot/Icm T4SS effectors is beyond the scope of this chapter, and the interested reader is referred to several excellent reviews (127, 135–137). Here, we will identify and describe activities of a subset of the effectors shown to affect specific secretory pathways and cellular functions relevant to the infection process.
L. pneumophila replicates in ‘professional’ phagocytes such as amoebae and macrophages (133, 138). It employs the Dot/Icm T4SS to deliver effector proteins as soon as it contacts the cell (Fig. 4). One effector, SdeA, is implicated as being important for bacterial adherence and uptake, although further work is needed to confirm this function (139). However, the internalization process activates T4SS function, resulting in the translocation of presynthesized effector molecules (140, 141). Effector translocation enables L. pneumophila to evade endocytic (lysosome) fusion and intercept ER-to-Golgi vesicular traffic to remodel its phagosome into an ER-derived vacuole that serves as a proliferation niche (133). Internalization progresses to formation of the LCV, which communicates extensively with the endosomal trafficking route largely through acquisition of host molecules and effectors on the LCV membrane. For example, the LCV acquires two types of molecules, PI lipid phosphatidylinositol-3-phosphate [Ptdlns(3)P] and Rab GTPases, that are key regulators of the early endocytic pathway (135). Several Dot/Icm substrates including LidA, LptD, RidL, and SetA have been shown to bind Ptdlns(3)P possibly as an LCV membrane anchor (142–145). The effector SidP is a PI 3-phosphatase that hydrolyses Ptdlns(3)P and thus might contribute to evasion of the endocytic pathway (146). A number of Rab GTPases (Rab5A, Rab7, Rab14, Rab21) are recruited to the LCV, and several effectors interact with these GTPases. One of these, VipD, is activated by Rab5 and one of its functions is to remove Ptdlns(3)P from the LCV membrane (147). Thus, like SidP, VipD is thought to promote LCV evasion of the endocytic pathway. Other effectors acting on the endocytic pathway include SidK, which binds one of the subunits of V-type H+-ATPase (a late endosomal marker that also binds the LCV) and inhibits ATP hydrolysis, proton translocation and LCV acidification (148).
The LCV compartment also modulates the secretory pathway by intercepting early secretory vesicles released from ER exit sites towards the Golgi apparatus. The Dot/Icm effectors SidM and LseA have been shown to modulate this secretory pathway by promoting the fusion of LCVs with ER-derived vesicles through effects on SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex formation (149, 150). SNARES are responsible for the docking and fusion of many different vesicle-mediated transport events. Another PI, Ptdlns(4)P, as well as the GTPase Rab1, are major regulators of Golgi-bound, secretory vesicle trafficking. Ptdlns(4)P is formed on the LCV membrane through the actions of SidF (151) and other effectors. Several Dot/Icm effectors bind the LCV via Ptdlns(4)P or Rab1. These effectors collectively modulate the secretory pathway through their various biochemical activities including AMPylation, deAMPylation, ubiquitination, phosphocholination, and guanine nucleotide exchange (see (135)). For example, RalF, the first identified Dot/Icm effector, functions as a guanine nucleotide exchange factor (GEF) that mediates LCV fusion with ER-derived secretory vesicles (ESVs) and the ER by recruiting and activating Arf1, another small GTPase involved in vesicle trafficking from the ER to Golgi (152).
Besides altering the endocytic and secretory pathways, L. pneumophila uses the Dot/Icm T4SS to translocate numerous other effectors that act on a plethora of other host cellular processes to benefit survival in the host cell (Fig. 4). In brief, effectors such as RavZ are known to inhibit autophagy (153) and RidL to modulate the retrograde pathway (145). Others such as Legs2 associate with mitochondria possibly disrupting host cell bioenergetics (154), VipA with actin to effect cytoskeleton remodeling (155, 156), or AnkB with the ubiquitin/proteasome pathway (157–159). Others modulate programmed cell death in several ways that include modulation of host transcription and translation (see (135)).
Studies over the past decade have generated an extensive literature about the L. pneumophila T4SS and contributions of effectors to the infection process. Equally importantly, these studies have identified novel features of fundamental biochemical and cellular processes in the host cell. Given that only a small subset of the candidate effectors have been characterized so far, future studies will assuredly continue to unveil new mechanistic insights at many levels into the fascinating L. pneumophila - host cell relationship.
Obligate intracellular pathogens
Coxiella burnetii is an intracellular bacterium that causes the zoonotic disease Q fever. It is transmitted by inhalation of contaminated aerosols, and can cause life threatening illness with vague symptoms similar to those described above for Rickettsial infections that can progress to pneumonia, hepatitis, myocarditis or central nervous system complications (160). C. burnetii can also cause long-term chronic disease that can manifest as endocarditis years after the initial infection (161). Ruminants are the natural reservoir for Coxiella, and infections of these animals can cause abortion and subsequent contamination of the environment. As for the Rickettsial species, studies of C. burnetii pathogenesis have been hampered by the obligate intracellular lifestyle of the bacterium. However, an axenic culture condition was described in 2009, which has revolutionized studies of this pathogen that have supplied important new information about the bacterial factors, particularly its Dot/Icm T4SS and translocated effectors, to virulence (162).
Coxiella has the capacity to enter both phagocytic and non-phagocytic cells, where it replicates to high numbers inside a specialized vacuole termed the Coxiella-containing vacuole (CCV) (163). The CCV undergoes normal endocytic trafficking through early and late endosomes to a lysosome. In contrast to the Legionella-containing vacuole that subverts the endocytic trafficking to avoid lysosome fusion, however, the CCV fuses with the lysosome. Within this acidic environment, Coxiella then differentiates from an environmentally stable small cell variant (SCV) into a replicating large cell variant (LCV). The CCV then undergoes expansion by recruitment of cellular vesicles that fuse with the vacuole membrane, during which time Coxiella replicates to large numbers (164).
The Dot/Icm system was discovered by genome sequencing and was shown to be essential for virulence through construction and analysis of dot/icm transposon insertion or deletion mutations (165–168). As discussed above, the L. pneumophila T4SS assembles prior to host cell uptake and delivers effectors upon initial contact to subvert the endocytic pathway and promote fusion with secretory vesicles. By contrast, the Coxiella T4SS translocates effectors only several hours after infection, following CCV-lysosome fusion, with the overall objective of remodeling this vacuole to facilitate replication.
To date, over 130 candidate effectors have been identified for this system, through bioinformatics screens to identify proteins with eukaryotic-like motifs and experimental verification by use of reporter protein translocation assays (see (169)). Interestingly, a large number of the candidate effectors were classified as pseudogenes in some strains that can block synthesis or translocation, or alter the protein’s activity in the host through loss of one or more domains. Such genetic alterations have been postulated to contribute to the evolution of different Coxiella variants (170).
Although biochemical characterization of Coxiella effectors is still in its infancy, a variety of screens and approaches have identified effectors that contribute to CCV biogenesis and intracellular growth, and that modulate different cellular processes and pathways (Fig. 4). The host cell must remain viable to support Coxiella replication, and several effectors have been shown to inhibit the host cell death pathway. AnkG, for example, blocks apoptosis by interacting first with the host mitochondrial protein p32 and subsequently with the nucleus where it exerts its anti-apoptotic effects by an unknown mechanism (171, 172). Other anti-apoptic effectors include CaeA and CaeB, which bind respectively to the mitochondria and nucleus where they are thought to interfere with apoptotic signaling (173). A number of other effectors, identified through large-scale transposon mutagenesis screens, have been shown to be important for intracellular replication. This list now includes CirA-E (Coxiella effector for intracellular replication) as well as Cig57, CoxCC8, CBU1754, and CvpA-CvpE (174, 175). Identification of CvpA was of interest in view of evidence that clathrin-mediated vesicular trafficking is important for CCV biogenesis. Clathrin was shown to be present on the CCV membrane, and reduction in the level of CCV-bound clathrin was correlated with diminished Coxiella replication. Further studies showed that CvpA interacts with the clathrin-adaptor complex AP2, leading to a proposal that this effector contributes to CCV biogenesis and intracellular replication through recruitment of host cell clathrin transport machinery and possibly other factors to the CCV (176). Another effector, Cig2, was shown to contribute to establishment of the replicative niche by blocking autophagy, a process used by the host cell to remove misfolded proteins, damaged organelles and intracellular pathogens through targeting to the lysosome (175). Finally, several effectors have been shown to modulate MAP kinase signaling pathways in yeast, possibly through disruption of the phosphorylation state of pro-apoptotic proteins (166, 177, 178).
At this time, studies of effector function have identified important contributions to establishment of the CCV and intracellular replication through effects on autophagy, secretory trafficking of clathrin-coated vesicles, apoptosis, and signal transduction. The challenges for the future are to define the specific biochemical functions of these effectors. Another important goal is to define the contributions of the Dot/Icm system and other virulence factors to the unique capacity of this bacterium to survive and replicate in the acidic, proteolytic and oxidative environment of the lysosome-derived CCV.
Rickettsial spp. are obligate intracellular pathogens or endosymbionts. The major genera include Anaplasma, Ehrlichia, Orientia, Rickettsia and Wolbachia (179). The Rickettsiales associate with diverse eukaryotic hosts in pathogenic or symbiotic relationships. Rickettsial species are responsible for major human diseases including Rocky Mountain Spotted Fever (Rickettsia rickettsii), epidemic typhus (Rickettsia prowazekii), and zoonotic diseases of granulocytic anaplasmosis (Anaplasma phagocytophilum) and monocytic ehrlichiosis (Ehrlichia chaffeensis) (179, 180). Orientia tsutsugamushi is the causative agent of scrub typhus (181). Clinical signs of these diseases are similar and include fever, headache, myalgia, anorexia, and chills, frequently accompanied by leukopenia. Diseases can progress to meningitis and disseminated intravascular coagulation, which if left untreated can progress to multiple organ failure. Transmission is generally by a tick, mite or other insect vector from animal reservoirs (179, 180).
Wolbachia are endosymbionts or parasites associated with numerous invertebrates including arthropods and nematodes. In filarial nematodes, Wolbachia are essential for survival and replication of their host (182). In arthropods, Wolbachia are mostly parasites that affect reproduction of their hosts in ways that enhance their own transmission through maternal inheritance from infected females to progeny (183). Strikingly, there is evidence for widespread horizontal gene transfer of Wolbachia chromosomal fragments into many species of arthropods and nematodes, and in the latter for expression of the integrated genes (184). These findings raise the intriguing possibility that Wolbachia might employ their T4SSs to translocate chromosomal DNA for establishment of symbiotic or parasitic relationships with their hosts.
All characterized members of the Rickettsiales carry genes for T4SSs, although the genes typically are not clustered in a single operon but rather in two or three operons scattered around the genome (43, 185–188). O. tsutsugamushi offers the most extreme known example of the fragmentation of T4SS genes around the chromosome (189). The chromosomes of these bacteria are themselves highly fragmented, presumably as a result of extensive recombination at the 4,197 identical repeat sequences shown to be distributed around the genome. The O. tsutsugamushi genome also carries over 1,100 mobile genetic elements, which is especially intriguing in view of the fact that mobile elements are only rarely present in the genomes of other Rickettsial species (189). These findings provide compelling evidence that conjugative transfer has played an important role in the shaping of the O. tsutsugamushi genome throughout evolution.
Besides the unusual distribution of T4SS genes around the chromosome, the Rickettsial T4SSs possess a number of variations from the archetypal VirB/VirD4 T4SSs (Table 2). First, these systems lack pilus-associated proteins (VirB1, VirB5) thought to be involved in intercellular attachment processes, possibly reflecting the intracellular lifestyle of these bacteria (41, 43, 190). Another novel adaptation is the presence of multiple copies of genes encoding several of the VirB homologs. The genomes typically carry genes in single copy for the structural subunits VirB8, VirB9, and VirB10, as well as the ATPases VirB11 and VirD4. However, multiple copies exist for genes encoding inner membrane channel subunits VirB3 and the VirB4 ATPase as well as polytopic VirB6 (43, 190). Most noteworthy, the multiple VirB6 subunits display considerable variability in length and sequence composition, particularly in their hydrophilic C-terminal regions (5). There is evidence for surface exposure of VirB6 homologs in E. chaffeensis and Wolbachia, leading to the suggestion that these variable proteins promote survival in the host through binding of host cell receptors or evasion of host immune defenses (37, 191–193). Additionally, multiple copies exist of genes encoding sequence variable VirB2 pilin-like subunits, reminiscent of the Bartonella Trw T4SS (Table 2) (41). How these T4SS-encoded surface structures contribute to the infection processes of these intracellular pathogens is an intriguing question for further study.
Identification of T4SS effectors functioning in Rickettsial species has been challenging given a lack of genetic systems for mutant strain constructions and the inability to culture these bacteria axenically. However, a number of candidate or confirmed effectors have been identified through a combination of bioinformatics screens, two-hybrid assays for protein interactions with the VirD4-like substrate receptors, and use of surrogate T4SSs, e.g., the A. tumefaciens VirB/VirD4 and Legionella Dot/Icm systems, to assay for translocation (194–197). In bioinformatics screens, the ankryin repeat (Ank) is a particularly prominent eukaryotic-like domain identified in the genomes of various members of the Rickettsiales. Wolbachia strain ΩPip and O. tsutsugamushi respectively carry an astonishing 60 and 50 ank genes, the most identified to date for any bacterial species (198).
The Ank repeat is a 33-residue motif, often in tandem arrays, that cooperatively folds into structures that mediate molecular recognition via protein-protein interactions (Table 2). These are widespread interaction motifs in eukaryotes, and they are also features of T4SS as well as T3SS effectors, and there is evidence that such proteins play important roles in pathogenesis by mimicking or interfering with host cell functions (199). Indeed, the first T4SS effector identified among the Rickettsiales was AnkA shown to be a secretion substrate of the A. phagocytophilum T4SS (Fig. 4) (194). AnkA contains 11 Ank repeat motifs and a positively-charged C-terminal tail similar to those carried by secretion substrates of the A. tumefaciens VirB/VirD4 T4SS. Accordingly, AnkA was shown to be translocated through the A. tumefaciens VirB/VirD4 T4SS to plant cells (194). In its natural mammalian host cell, AnkA is phosphorylated by the Abl-1 and Src kinases, and phospho-AnkA then binds the host protein Src homology phosphatase-1 (SHP-1) (88). Interestingly, AnkA binds nuclear proteins and forms complexes with AT-rich DNA sequences, resulting in modulation of host gene transcription (200). Recently, AnkA was found to downregulate expression of multiple host defense genes, including the NADH oxidase component, CYBB. AnkA also binds the histone deacetylase 1 (HDAC1), which is critical for AnkA-mediated CYBB repression. The consequence of CYBB downregulation is a decrease in superoxide anion production by the NADH oxidase, which is a key mechanism of pathogen killing for neutrophils (201). Given the observation that AnkA binds to numerous sites in the host genome and histone deacetylation is frequently observed at host promoters of host defense genes, it is likely that AnkA broadly impacts the host transcriptional response to A. phagocytophilum infection (Fig. 4).
A second confirmed A. phagocytophilum effector, termed Ats-1 (Anaplasma translocated substrate) was identified through a two-hybrid screen for binding partners of the VirD4 receptor (195). Ats-1 also has a positively charged C terminus but was not translocated through the A. tumefaciens VirB/VirD4 T4SS. Nevertheless, it is a bona fide substrate of the A. phagocytophilum T4SS, as shown by its abundant secretion into mammalian cells where a large proportion localizes to the A. phagocytophilum inclusion. Ats-1 lacks known protein motifs, however, it does carry an N-terminal mitochondria-targeting presequence. This translocation signal directs Ats-1 into the mitochondrial matrix of infected human neutrophils, HeLa cells, and even yeast cells (195). Consequently, Ats-1 passes through a total of 5 membranes - bacterial inner and outer membranes, the inclusion membrane, and the mitochondrial membranes - enroute to its postulated site of action. One of the hallmarks of this infection process is the inhibition of spontaneous induced apoptosis of human neutrophils. Although several cellular mechanisms have been described by which A. phagocytophilum inhibits apoptosis (inhibition of the loss of mitochondrial membrane potential, inhibition of Bax translocation to the mitochondria, inhibition of activation of caspase 3), the role of Ats-1 in one or more of these processes is not yet known (Fig. 4).
Several candidate effectors of the E. chaffeensis T4SS also have been identified. The best-characterized, ECH0825, carries a positively-charged C terminus and interacts with the VirD4Ec receptor. ECH0825 is highly upregulated during infection, translocated to the host cell cytoplasm and, reminiscent of Ats-1, localizes to the mitochondria (202). Ectopically expressed ECH0825 inhibits apoptosis in response to etoposide treatment, and also inhibits Bax-induced apoptosis. ECH0825 production also was correlated with upregulation of mitochondrial manganese superoxide dismutase (MnSOD) and reduced reactive oxygen species (ROS). By preventing ROS-induced cellular damage and apoptosis, ECH0825 thus might promote intracellular infection (Fig. 4) (202).
Although no other effectors have been confirmed for Rickettsial T4SSs, a bioinformatics screen identified 21 possible effectors for A. marginales of which four (AM185, AM1141, AM470, AM705[AnkA]) were shown to be translocated through the surrogate L. pneumophila Dot/Icm T4SS (196). More recently, a study seeking to identify potential A. phagocytophilum-derived nuclear-transported proteins that could impact host cell transcription identified 50 candidate proteins through a combination of bioinformatics and iTRAQ protein profiling (197). Nuclear localization was confirmed for six of these candidate effectors through ectopic expression, but only one (APH_0455) was translocated through a surrogate C. burnetii Dot/Icm T4SS at detectable levels. APH_0455 is of interest because it carries domains resembling those found in sterol regulatory element binding proteins (SREBPs), which in humans can modulate chromatin structure and gene transcription (Table 3, Fig. 4) (197).
INTERBACTERIAL SYSTEMS
The effector translocator systems clearly play important and varied roles in establishment of the replicative niches of many extracellular and intracellular pathogens. As mentioned earlier, conjugation systems also play important roles in promoting growth of bacterial pathogens in clinical settings. It is becoming increasingly evident, however, that bacteria have adapted the T4SSs in ways not limited to conjugative DNA transfer to enhance their growth potential. Below, we describe T4SSs functioning in novel ways to facilitate colonization and proliferation of invading pathogens.
Conjugation machines
It is well established that conjugation provides a selective advantage for growth and colonization of invading pathogens through rapid intra- and interspecies dissemination of antibiotic resistance genes and virulence determinants in clinical settings (9). Even without gene transfer, however, the conjugation machines mediate functions of importance for virulence. Most notably, they promote bacterial attachment to biotic and abiotic surfaces, interbacterial aggregation, and development of biofilms in the human host (203–205). Among the Gram-negative species, these processes are mediated largely by conjugative pili (203, 204). The Gram-positive bacteria lack such pili, but their conjugation systems typically encode one or more surface adhesins as functional substitutes (5). The Enterococcus faecalis pCF10 conjugation system, for example, codes for three surface adhesins of which one, termed aggregation substance (AS), was shown to play an important role in intercellular aggregation, formation of robust biofilms, and bacterial attachment to heart tissues and endocarditis development in animal models (205, 206). The Gram-positive conjugation systems also code for multi-domain cell wall hydrolases possibly involved in autolysis and release of eDNA or other matrix components, as well as release of cell wall fragments, all of which could serve as potentiators of inflammatory responses in the human host (38). Conjugation machines are widely distributed among the medically-important pathogens, and their multifaceted contributions to the infection process cannot be overstated.
Additionally, although the Gram-positive bacterial T4SSs currently are known to function only as conjugation machines, there is accumulating evidence that T4SSs might also be employed for effector translocation. It was recently shown that clusters of T4SS genes are much more widely distributed among the genomes of Gram-positive species than previously thought, largely because gene functions were misannotated during genomic sequence analyses (4). Typically, the T4SS gene clusters code for homologs of subunits that in the Gram-negative systems comprise the inner membrane translocase, e.g., VirB3, VirB4, VirB6, VirB8, and VirD4, as well as a VirB1-like cell wall hydrolase. These Gram-positive T4SSs lack genes encoding the OMCs and pili of the Gram-negative systems, which is consistent with the lack of an outer membrane or evidence for elaboration of conjugative pili. The Gram-positive T4SSs were designated as type IVC or ‘mimimized’ T4SSs, the latter reflecting the fact that these are the simplest known T4SSs in terms of subunit composition (4, 10). The Gram-positive T4SSs are therefore excellent subjects for detailed structure-function studies of the cytoplasmic membrane translocase stripped of other T4SS adaptations acquired over evolutionary time.
Whether these ‘minimized’ systems translocate effector proteins during the course of infection is still not known. However, there is some experimental support for an effector function by a T4SS encoded by the 89 K pathogenicity island of Streptococcus suis strain 05ZYH33, which was the cause of a recent outbreak of streptococcal toxic shock in China (207–209). Mutations introduced into the virB4- or virD4-like genes abolished virulence of strain 05ZYH33, and the mutant strains also failed to trigger a host immune response in a mouse infection model (10). Based on these findings, it was proposed that this pathogen utilizes the 89 K T4SS to deliver an unknown effector protein(s) to the cell surface or into eukaryotic target cells. Whether Gram-positive T4SSs function in effector translocation and immune modulation remains an intriguing question for future investigation.
H. pylori Competence system
H. pylori carries a second T4SS adapted for the novel purpose of importing DNA from the extracellular milieu (210, 211). While not directly related to virulence, the ability to acquire foreign DNA imparts plasticity to the genome and genetic diversity of potential benefit for the invading pathogen in changing environmental settings. Natural transformation or competence (Com), for example, allows for recombinogenic alteration of surface antigens enabling both attachment to different cell types and immune evasion. The H. pylori Com system consists of homologs of most VirB/VirD4 proteins with the exception of the pilus assembly factors VirB1 and VirB5, VirD4 T4CP, and VirB11 ATPase (211, 212). Additionally, the VirB machinery functionally interfaces with an inner membrane channel protein ComEC (13). Thus, in a unique two-step translocation reaction, the VirB complex is postulated to take up double-stranded DNA from the milieu into the periplasm and then deliver the substrate to ComEC, which degrades one strand while importing the second across the inner membrane (13). The importance of the H. pylori Com system to infection is underscored by a recent study showing that mutations of the Com machine resulted in reduced persistence in a mouse model, suggesting that DNA exchange between genetically heterogeneous H. pylori contributes to establishment of chronic infection (213).
Neisseria gonorrhoeae DNA Release System
N. gonorrhoeae is the causative agent of gonorrhoeae. It is an exclusively human pathogen transmitted by sexual contact. Approximately 80% of gonococcal strains carry a 57-kb gonococcal genetic island (GGI), which is thought to be disseminated through release of genomic DNA to the milieu followed by acquisition by gonococci in the vicinity via a natural competence system (214). Gonococci naturally undergo autolysis, which represents one source for DNA uptake by transformation. Additionally, the GGI encodes a T4SS with the novel capacity to deliver DNA to the milieu (215). This DNA release system strikingly contrasts with the mechanism of conjugation, which requires a signal(s) transduced by recipient cell contact to activate the process of intercellular DNA transfer (216).
The GGI-encoded DNA release system is closely related in subunit composition and, probably architecture, to the E. coli F plasmid conjugation system (214). Both of these systems are assembled from homologs of the twelve VirB/VirD4 subunits, plus another subset of proteins unique to F-like T4SSs. One striking difference between the GGI- and F-encoded T4SSs, however, is that, while the TraA pilin is required for F plasmid transfer, the GGI system carries variant alleles of the TraA pilin subunits. Indeed, TraA is completely dispensable for DNA release by the GGI T4SS, as shown through mutational analyses (14). Conjugative pili are thought to initiate contacts with recipient cells, and the F-encoded pilus dynamically extends and retracts to bring donors and recipients into close apposition. However, N. gonorrhoeae do not require a target cell contact for GGI-dependent DNA release, thus dispensing with the need for a conjugative pilus. Besides their roles in attachment processes, conjugative pili or pilin subunits might contribute to recipient-stimulated DNA transfer through conformational effects on the OMC channel, thus ensuring substrate transfer only upon establishment of productive mating junctions. In N. gonorrhoeae, the lack of a TraA pilin requirement suggests the possibility that this F-like system evolved as a nonspecific DNA release system through loss of a critical gating activity. While the GGI T4SS is the only system known to mediate DNA release, as mentioned above there is some evidence that T4SSs of Gram-positive bacteria contribute directly or indirectly to the release of extracellular DNA (eDNA), thereby stimulating colonization, biofilm formation and infection (205).
Xanthomonas: A T4SS killing machine
Xanthomonas citri is a gammaproteobacterial phytopathogen that causes citrus canker, a disease that affects all citrus plants. Genome sequencing identified a virB/virD4 locus closely resembling the A. tumefaciens locus with the exception that the virB7 homolog encodes a much larger (139 residues) lipoprotein than A. tumefaciens VirB7 (4.5 kDa) (217). An X-ray structure revealed a N0 structural fold, which as described above is also present in DotD of the L. pneumophila Dot/Icm system and other outer membrane secretins (218). This domain is postulated to form an extra ring in the OMC, which might be a functionally important motif in view of recent work showing that the Xanthomonas T4SS functions not as a conjugation system or an effector translocator - at least in the classical sense (219). This system does translocate proteins, but to bacterial recipients with the goal of killing them! The effectors are toxins that were originally identified in a two-hybrid screen for VirD4-interacting proteins (220). All of the thirteen candidate effectors carry a conserved motif (XVIPCD) near their C termini that is implicated as a potential translocation signal and VirD4 interaction domain. These proteins also carry toxin motifs with peptidoglycan binding or hydrolase, lipase, or nuclease activities. Recent work confirmed that Xanthomonas employs its T4SS to kill neighboring bacteria, and that donors are themselves immune to killing by virtue of a coproduced cognate antitoxin (219). This killing activity increases competitiveness of Xanthomonas over coresident bacteria, presumably yielding a growth advantage through access to limiting nutrients in the environment. This striking new finding functionally aligns the T4SSs with the type VI secretion systems (T6SSs), which also have been exploited by bacteria for the specialized purpose of killing their neighbors (221).
Summary and Future Directions
In the last decade, there has been striking progress in our understanding of the structural diversity and functional versatility of the type IV secretion systems. In this chapter, we have highlighted the principal ways pathogens have adapted T4SSs for specialized purposes geared toward establishment of replicative niches, proliferation, and spread in the human host. Studies of the paradigmatic T4SSs have generated detailed views of how T4SS are architecturally arranged and how they recruit and mediate the transfer of DNA and protein substrates across the bacterial cell envelope. Corresponding work on the T4SSs employed by many medically important pathogens has further generated important insights into how the ancestral conjugation systems were adapted through appropriation of novel structural features. These structural modifications have endowed invading pathogen with the capacity to translocate a specific substrate repertoire - ranging from 1 to many hundreds of effectors - to host cells during the infection process. Structural adaptations also arm the pathogen with the ability to attach to specific host cells or, alternatively through intra- or intergenic recombination of variant T4SS genes, a range of different host cell types. The display of surface-variable antigens also potentially cloaks the invading pathogen, allowing evasion of the host immune response.
The explosion of interest in T4SS effectors in recent years has yielded an extensive body of new information about the infection processes of many medically important pathogens. The known extracellular pathogens employ T4SSs primarily for delivery of one or a few effectors that target major cellular pathways with a variety of physiological consequences. The intracellular pathogens, on the other hand, appear to use their T4SSs for delivery of a number of effectors ranging from a half dozen to many hundreds, some acting on specific biochemical functions and others more generally on a number of cellular targets. Several themes have been identified in the evolutionary design of the T4SS effectors. Most widespread is the concept of molecular mimicry, which enables the pathogen essentially to fly below the radar of host cellular and immune responses. Also, in line with the overarching aim of a pathogen to survive and replicate within an established niche, effectors often exert subtle and modulatory effects on cellular processes as opposed to blocking cellular functions and activating cell death pathways. Two other themes are evident among intracellular pathogens such as L. pneumophila that employ their T4SSs to deliver many hundreds of effectors to the host. First, many effectors target similar pathways and carry out redundant activities which, for interested scientists, has unfortunately thwarted efforts to assign their functional importance. From the pathogen’s perspective, however, functional redundancy can serve as a useful strategy for maintaining a broad host range of infection and for minimizing the number of effectors that are absolutely required for growth of the pathogen within the human host (127). Second, it is increasingly recognized that effectors often work in concert with other effectors to modulate complex biochemical pathways. With respect to the T4SS, this requires precise delivery in space and time of coordinately acting effectors and, hence, the evolution of recruitment mechanisms, e.g., chaperones/adaptors, to achieve such spatiotemporal control (11).
Finally, although the list of pathogens shown to employ T4SSs for delivery of effector proteins during infection continues to expand, it is still small compared to the number of bacteria that carry mobile genetic elements capable of elaborating conjugation systems. Most if not all members of the human microbiota carry such elements, many of which can become serious opportunistic pathogens in immunocompromised hosts or individuals suffering microbial dysbiosis as a result of antibiotic therapy or various illnesses. In such settings, conjugation systems can contribute significantly to establishment of infection not just through genetic exchange, but also by the elaboration of surface pili or adhesins capable of promoting attachment, colonization, and biofilm formation. Also of considerable interest is the recent discovery that a T4SS was adapted to deliver toxins interbacterially. This expanded feature of the T4SS arsenal enables invading bacteria to kill members of the normal flora, thus gaining access to space and resources for establishment of a replicative niche.
Clearly, there are many exciting avenues for further exploration toward the ultimate goal of gaining a comprehensive understanding of how T4SSs contribute to infection processes. While considerable headway has been made for the paradigmatic conjugation machines, fundamental questions remain about the architectures and mechanisms of action of T4SSs employed by pathogens. In particular, it is imperative to determine the structures of T4SS surface organelles and proteins, identify the basis for surface variability, and understand how these surface components mediate host cell binding and immune evasion. Studies of the effectors will assuredly continue to add detail to our understanding of various infection processes and of the mechanistic themes and variations associated with T4SS-mediated effector translocation. In particular, studies aimed at defining the T4SS effectors translocated by the obligate intracellular Rickettsial species appear poised to make significant progress over the next few years. Finally, as evident from the recent work and explosion of interest, we can anticipate that studies of specific pathogen - host cell interactions will not only define the functions of new T4SS effectors but also yield a broader understanding of basic biological processes of human host cells.
Acknowledgments
We thank members of the Christie laboratory for helpful discussions. We also thank reviewers of this chapter for insightful comments. Studies in the Christie laboratory were supported by NIH R01GM48476 and R21AI105454. C. G.-R. was supported in part by NRSA fellowship F32 AI114182.
Contributor Information
Christian Gonzalez-Rivera, Email: Christian.Gonzalez@uth.tmc.edu.
Minny Bhatty, Email: Minny.Bhatty@uth.tmc.edu.
Peter J. Christie, Email: Peter.J.Christie@uth.tmc.edu.
References
- 1.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Asrat S, Davis KM, Isberg RR. Modulation of the host innate immune and inflammatory response by translocated bacterial proteins. Cell Microbiol. 2015;17:785–795. doi: 10.1111/cmi.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Micro. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2014;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 9.Juhas M. Horizontal gene transfer in human pathogens. Crit Rev Microbiol. 2015;41:101–108. doi: 10.3109/1040841X.2013.804031. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Rong C, Chen C, Gao GF. Type-IVC secretion system: A novel subclass of type IV secretion system (T4SS) common existing in Gram-positive genus Streptococcus. PloS ONE. 2012;7:e46390. doi: 10.1371/journal.pone.0046390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llosa M, Roy C, Dehio C. Bacterial type IV secretion systems in human disease. Mol Microbiol. 2009;73:141–51. doi: 10.1111/j.1365-2958.2009.06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Nat Acad Sci USA. 2010;107:1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey ME, Woodhams KL, Dillard JP. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front Microbiol. 2011;2:61. doi: 10.3389/fmicb.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- 17.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai H, Kubori T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2012;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzfira T, Citovsky V. Agrobacterium: From Biology to Biotechnology. Springer; New York, NY: 2008. [Google Scholar]
- 21.Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 22.Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol. 2014;27:16–23. doi: 10.1016/j.sbi.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiol. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- 27.Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci USA. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohlfeld S, Pattis I, Puls J, Plano GV, Haas R, Fischer W. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- 30.Alperi A, Larrea D, Fernandez-Gonzalez E, Dehio C, Zechner EL, Llosa M. A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system. J Bacteriol. 2013;195:4999–5006. doi: 10.1128/JB.00367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol. 2013;89:324–333. doi: 10.1111/mmi.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong KC, Sutherland MC, Vogel JP. Novel export control of a Legionella Dot/Icm substrate is mediated by dual, independent signal sequences. Mol Microbiol. 2015;96:175–188. doi: 10.1111/mmi.12928. [DOI] [PubMed] [Google Scholar]
- 34.Sundberg CD, Ream W. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol. 2009;191:278–286. doi: 10.1128/JB.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laverde Gomez JA, Bhatty M, Christie PJ. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J Bacteriol. 2014;196:527–539. doi: 10.1128/JB.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 40.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, Cariaga TA, Suarez G, Peek RM, Jr, Cover TL, Solnick JV. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9:e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2009;4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, Danchin A, Marignac G, Lenaour E, Boulouis HJ, Mavris M, Arnaud L, Yang H, Wang J, Quebatte M, Engel P, Saenz H, Dehio C. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 2010;6:e1000946. doi: 10.1371/journal.ppat.1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Khedery B, Lundgren AM, Stuen S, Granquist EG, Munderloh UG, Nelson CM, Alleman AR, Mahan SM, Barbet AF. Structure of the type IV secretion system in different strains of Anaplasma phagocytophilum. BMC Genomics. 2012;13:678. doi: 10.1186/1471-2164-13-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watarai M, Andrews HL, Isberg R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol Microbiol. 2000;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 45.Melvin JA, Scheller EV, Miller JF, Cotter PA. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol. 2014;12:274–288. doi: 10.1038/nrmicro3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Althouse BM, Scarpino SV. Asymptomatic transmission and the resurgence of Bordetella pertussis. BMC Med. 2015;13:146. doi: 10.1186/s12916-015-0382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278:4668–82. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 48.Katada T, Ui M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc Natl Acad Sci USA. 1982;79:3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura M, Nogimori K, Murai S, Yajima M, Ito K, Katada T, Ui M, Ishii S. Subunitstructure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry. 1982;21:5516–5522. doi: 10.1021/bi00265a021. [DOI] [PubMed] [Google Scholar]
- 50.Burns DL, Hewlett EL, Moss J, Vaughan M. Pertussis toxin inhibits enkephalin stimulation of GTPase of NG108-15 cells. J Biol Chem. 1983;258:1435–1438. [PubMed] [Google Scholar]
- 51.Stein PE, Boodhoo A, Armstrong GD, Cockle SA, Klein MH, Read RJ. The crystal structure of pertussis toxin. Structure. 1994;2:45–57. doi: 10.1016/s0969-2126(00)00007-1. [DOI] [PubMed] [Google Scholar]
- 52.Weiss AA, Johnson FD, Burns DL. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 54.Winans SC, Burns DL, Christie PJ. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicosia A, Perugini M, Franzini C, Casagli MC, Borri MG, Antoni G, Almoni M, Neri P, Ratti G, Rappuoli R. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc Natl Acad Sci USA. 1986;83:4631–4635. doi: 10.1073/pnas.83.13.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farizo KM, Huang T, Burns DL. Importance of holotoxin assembly in Ptl-mediated secretion of pertussis toxin from Bordetella pertussis. Infect Immun. 2000;68:4049–4054. doi: 10.1128/iai.68.7.4049-4054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christie PJ. Bacterial type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochem Biophys Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivera-Calzada A, Fronzes R, Savva CG, Chandran V, Lian PW, Laeremans T, Pardon E, Steyaert J, Remaut H, Waksman G, Orlova EV. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013;32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witvliet MH, Burns DL, Brennan MJ, Poolman JT, Manclark CR. Binding of pertussis toxin to eucaryotic cells and glycoproteins. Infect Immun. 1989;57:3324–3330. doi: 10.1128/iai.57.11.3324-3330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 61.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 62.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1997;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 64.Backert S, Ziska E, Brinkmann V, Zimny-Arndt U, Fauconnier A, Jungblut PR, Naumann M, Meyer TF. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol. 2000;2:155–164. doi: 10.1046/j.1462-5822.2000.00043.x. [DOI] [PubMed] [Google Scholar]
- 65.Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15:306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Fischer W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 2011;278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 67.Aras RA, Fischer W, Perez-Perez GI, Crosatti M, Ando T, Haas R, Blaser MJ. Plasticity of repetitive DNA sequences within a bacterial (Type IV) secretion system component. J Exp Med. 2003;198:1349–1360. doi: 10.1084/jem.20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delahay RM, Balkwill GD, Bunting KA, Edwards W, Atherton JC, Searle MS. The highly repetitive region of the Helicobacter pylori CagY protein comprises tandem arrays of an alpha-helical repeat module. J Mol Biol. 2008;377:956–971. doi: 10.1016/j.jmb.2008.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5:e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 2003;5:395–404. doi: 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 71.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011;7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82:3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 74.Tegtmeyer N, Hartig R, Delahay RM, Rohde M, Brandt S, Conradi J, Takahashi S, Smolka AJ, Sewald N, Backert S. A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J Biol Chem. 2010;285:23515–23526. doi: 10.1074/jbc.M109.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorrell RJ, Guan J, Xin Y, Tafreshi MA, Hutton ML, McGuckin MA, Ferrero RL, Kwok T. A novel NOD1- and CagA-independent pathway of interleukin-8 induction mediated by the Helicobacter pylori type IV secretion system. Cell Microbiol. 2013;15:554–570. doi: 10.1111/cmi.12055. [DOI] [PubMed] [Google Scholar]
- 76.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 77.Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, Tan S, Morgan DR, Wilson KT, Bravo LE, Correa P, Cover TL, Amieva MR, Peek RM., Jr Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479–492. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe. 2010;7:399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 79.Couturier MR, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jurik A, Hausser E, Kutter S, Pattis I, Prassl S, Weiss E, Fischer W. The coupling protein Cagbeta and its interaction partner CagZ are required for type IV secretion of the Helicobacter pylori CagA protein. Infect Immun. 2010;78:5244–5251. doi: 10.1128/IAI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonsor DA, Weiss E, Iosub-Amir A, Reingewertz TH, Chen TW, Haas R, Friedler A, Fischer W, Sundberg EJ. Characterization of the translocation-competent complex between the Helicobacter pylori oncogenic protein CagA and the accessory protein CagF. J Biol Chem. 2013;288:32897–32909. doi: 10.1074/jbc.M113.507657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 83.Hatakeyama M. Helicobacter pylori CagA--a potential bacterial oncoprotein that functionally mimics the mammalian Gab family of adaptor proteins. Microbes Infect. 2003;5:143–150. doi: 10.1016/s1286-4579(02)00085-0. [DOI] [PubMed] [Google Scholar]
- 84.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 85.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277:6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 86.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Selbach M, Paul FE, Brandt S, Guye P, Daumke O, Backert S, Dehio C, Mann M. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5:397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 88.JWIJ, Carlson AC, Kennedy EL. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell Microbiol. 2007;9:1284–1296. doi: 10.1111/j.1462-5822.2006.00871.x. [DOI] [PubMed] [Google Scholar]
- 89.Hayashi T, Morohashi H, Hatakeyama M. Bacterial EPIYA effectors--where do they come from? What are they? Where are they going? Cell Microbiol. 2013;15:377–385. doi: 10.1111/cmi.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki N, Murata-Kamiya N, Yanagiya K, Suda W, Hattori M, Kanda H, Bingo A, Fujii Y, Maeda S, Koike K, Hatakeyama M. Mutual reinforcement of inflammation and carcinogenesis by the Helicobacter pylori CagA oncoprotein. Sci Reports. 2015;5:10024. doi: 10.1038/srep10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 93.Dehio C. Molecular and cellular basis of Bartonella pathogenesis. Annu Rev Microbiol. 2004;58:365–390. doi: 10.1146/annurev.micro.58.030603.123700. [DOI] [PubMed] [Google Scholar]
- 94.Pulliainen AT, Dehio C. Bartonella henselae: subversion of vascular endothelial cell functions by translocated bacterial effector proteins. Int J Biochem Cell Biol. 2009;41:507–510. doi: 10.1016/j.biocel.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 95.Ben-Tekaya H, Gorvel JP, Dehio C. Bartonella and Brucella--weapons and strategies for stealth attack. Cold Spring Harbor Perspect Med. 2013;3 doi: 10.1101/cshperspect.a010231. pii: a010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dehio C. Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell Microbiol. 2008;10:1591–1598. doi: 10.1111/j.1462-5822.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seubert A, Hiestand R, de la Cruz F, Dehio C. A bacterial conjugation machinery recruited for pathogenesis. Mol Microbiol. 2003;49:1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x. [DOI] [PubMed] [Google Scholar]
- 98.de Paz HD, Sangari FJ, Bolland S, Garcia-Lobo JM, Dehio C, de la Cruz F, Llosa M. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiol. 2005;151:3505–3516. doi: 10.1099/mic.0.28410-0. [DOI] [PubMed] [Google Scholar]
- 99.Saenz HL, Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr Opin Microbiol. 2005;8:612–619. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 100.Padmalayam I, Karem K, Baumstark B, Massung R. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 2000;19:377–382. doi: 10.1089/10445490050043344. [DOI] [PubMed] [Google Scholar]
- 101.Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC, Dehio C. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet. 2007;39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- 102.Garcia-Pino A, Zenkin N, Loris R. The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem Sci. 2014;39:121–129. doi: 10.1016/j.tibs.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 103.Backert S, Selbach M. Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 2005;13:476–484. doi: 10.1016/j.tim.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 104.Schulein R, Seubert A, Gille C, Lanz C, Hansmann Y, Piemont Y, Dehio C. Invasion and persistent intracellular colonization of erythrocytes. A unique parasitic strategy of the emerging pathogen Bartonella. J Exp Med. 2001;193:1077–1086. doi: 10.1084/jem.193.9.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siamer S, Dehio C. New insights into the role of Bartonella effector proteins in pathogenesis. Curr Opin Microbiol. 2015;23:80–85. doi: 10.1016/j.mib.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 106.Eicher SC, Dehio C. Bartonella entry mechanisms into mammalian host cells. Cell Microbiol. 2012;14:1166–1173. doi: 10.1111/j.1462-5822.2012.01806.x. [DOI] [PubMed] [Google Scholar]
- 107.Truttmann MC, Rhomberg TA, Dehio C. Combined action of the type IV secretion effector proteins BepC and BepF promotes invasome formation of Bartonella henselae on endothelial and epithelial cells. Cell Microbiol. 2011;13:284–299. doi: 10.1111/j.1462-5822.2010.01535.x. [DOI] [PubMed] [Google Scholar]
- 108.Okujava R, Guye P, Lu YY, Mistl C, Polus F, Vayssier-Taussat M, Halin C, Rolink AG, Dehio C. A translocated effector required for Bartonella dissemination from derma to blood safeguards migratory host cells from damage by co-translocated effectors. PLoS Pathog. 2014;10:e1004187. doi: 10.1371/journal.ppat.1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pulliainen AT, Pieles K, Brand CS, Hauert B, Bohm A, Quebatte M, Wepf A, Gstaiger M, Aebersold R, Dessauer CW, Dehio C. Bacterial effector binds host cell adenylyl cyclase to potentiate Galphas-dependent cAMP production. Proc Natl Acad Sci USA. 2012;109:9581–9586. doi: 10.1073/pnas.1117651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schroder G, Schuelein R, Quebatte M, Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA. 2011;108:14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Llosa M, Schroder G, Dehio C. New perspectives into bacterial DNA transfer to human cells. Trends Microbiol. 2012;20:355–359. doi: 10.1016/j.tim.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 112.von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- 113.Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol. 2011;65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- 114.O’Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 115.Hong PC, Tsolis RM, Ficht TA. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun. 2000;68:4102–4107. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med. 2003;198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kahl-McDonagh MM, Elzer PH, Hagius SD, Walker JV, Perry QL, Seabury CM, den Hartigh AB, Tsolis RM, Adams LG, Davis DS, Ficht TA. Evaluation of novel Brucella melitensis unmarked deletion mutants for safety and efficacy in the goat model of brucellosis. Vaccine. 2006;24:5169–5177. doi: 10.1016/j.vaccine.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 118.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O’Callaghan D. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci USA. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 121.Myeni S, Child R, Ng TW, Kupko JJ, 3rd, Wehrly TD, Porcella SF, Knodler LA, Celli J. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog. 2013;9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.de Jong MF, Starr T, Winter MG, den Hartigh AB, Child R, Knodler LA, van Dijl JM, Celli J, Tsolis RM. Sensing of bacterial type IV secretion via the unfolded protein response. MBio. 2013;4:e00418–00412. doi: 10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, Muller A, Twizere JC, Nkengfac B, Vandenhaute J, Hill DE, Salcedo SP, Gorvel JP, Letesson JJ, De Bolle X. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- 124.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, Comerci DJ, Ugalde RA, Pierre P, Gorvel JP. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salcedo SP, Marchesini MI, Degos C, Terwagne M, Von Bargen K, Lepidi H, Herrmann CK, Santos Lacerda TL, Imbert PR, Pierre P, Alexopoulou L, Letesson JJ, Comerci DJ, Gorvel JP. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol. 2013;3:28. doi: 10.3389/fcimb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barbaree JM, Fields BS, Feeley JC, Gorman GW, Martin WT. Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl Environ Microbiol. 1986;51:422–424. doi: 10.1128/aem.51.2.422-424.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Isaac DT, Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 2014;9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 129.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 130.Kubori T, Koike M, Bui XT, Higaki S, Aizawa S, Nagai H. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci USA. 2014;111:11804–11809. doi: 10.1073/pnas.1404506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakano N, Kubori T, Kinoshita M, Imada K, Nagai H. Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 2010;6:e1001129. doi: 10.1371/journal.ppat.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O’Connor TJ, Boyd D, Dorer MS, Isberg RR. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Finsel I, Hilbi H. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol. 2015;17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 136.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol. 2011;2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prashar A, Terebiznik MR. Legionella pneumophila: homeward bound away from the phagosome. Curr Opin Microbiol. 2015;23:86–93. doi: 10.1016/j.mib.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 138.Hoffmann C, Harrison CF, Hilbi H. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol. 2014;16:15–26. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 139.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 140.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen J, Reyes M, Clarke M, Shuman HA. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell Microbiol. 2007;9:1660–1671. doi: 10.1111/j.1462-5822.2007.00899.x. [DOI] [PubMed] [Google Scholar]
- 142.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hilbi H, Weber S, Finsel I. Anchors for effectors: subversion of phosphoinositide lipids by legionella. Front Microbiol. 2011;2:91. doi: 10.3389/fmicb.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jank T, Bohmer KE, Tzivelekidis T, Schwan C, Belyi Y, Aktories K. Domain organization of Legionella effector SetA. Cell Microbiol. 2012;14:852–868. doi: 10.1111/j.1462-5822.2012.01761.x. [DOI] [PubMed] [Google Scholar]
- 145.Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe. 2013;14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 146.Toulabi L, Wu X, Cheng Y, Mao Y. Identification and structural characterization of a Legionella phosphoinositide phosphatase. J Biol Chem. 2013;288:24518–24527. doi: 10.1074/jbc.M113.474239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci USA. 2014;111:4560–4565. doi: 10.1073/pnas.1316376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arasaki K, Toomre DK, Roy CR. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 2012;11:46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.King NP, Newton P, Schuelein R, Brown DL, Petru M, Zarsky V, Dolezal P, Luo L, Bugarcic A, Stanley AC, Murray RZ, Collins BM, Teasdale RD, Hartland EL, Stow JL. Soluble NSF attachment protein receptor molecular mimicry by a Legionella pneumophila Dot/Icm effector. Cell Microbiol. 2015;17:767–784. doi: 10.1111/cmi.12405. [DOI] [PubMed] [Google Scholar]
- 151.Hsu F, Zhu W, Brennan L, Tao L, Luo ZQ, Mao Y. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci USA. 2012;109:13567–13572. doi: 10.1073/pnas.1207903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 153.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Degtyar E, Zusman T, Ehrlich M, Segal G. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 2009;11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 155.Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Franco IS, Shohdy N, Shuman HA. The Legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog. 2012;8:e1002546. doi: 10.1371/journal.ppat.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol. 2008;70:908–923. doi: 10.1111/j.1365-2958.2008.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, Buchrieser C. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 160.Raoult D, Marrie T, Mege J. Natural history and pathophysiology of Q fever. Lancet Infect Dis. 2005;5:219–226. doi: 10.1016/S1473-3099(05)70052-9. [DOI] [PubMed] [Google Scholar]
- 161.Mazokopakis EE, Karefilakis CM, Starakis IK. Q fever endocarditis. Infect Disord Drug Targets. 2010;10:27–31. doi: 10.2174/187152610790410918. [DOI] [PubMed] [Google Scholar]
- 162.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 164.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol. 2004;186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio. 2011;2:e00175–00111. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 2011;7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol. 2011;2:97. doi: 10.3389/fmicb.2011.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol. 2012;78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell Microbiol. 2015;17:621–631. doi: 10.1111/cmi.12432. [DOI] [PubMed] [Google Scholar]
- 170.Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, Williams KP, Sobral BW, Kupko JJ, 3rd, Porcella SF, Samuel JE, Heinzen RA. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. 2009;77:642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Luhrmann A, Nogueira CV, Carey KL, Roy CR. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci USA. 2010;107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, Berens C, Luhrmann A. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun. 2014;82:2763–2771. doi: 10.1128/IAI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol. 2013;15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 174.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog. 2014;10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 2014;10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Larson CL, Beare PA, Howe D, Heinzen RA. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci USA. 2013;110:E4770–4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol. 2013;195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lifshitz Z, Burstein D, Schwartz K, Shuman HA, Pupko T, Segal G. Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect Immun. 2014;82:3740–3752. doi: 10.1128/IAI.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Renvoise A, Merhej V, Georgiades K, Raoult D. Intracellular Rickettsiales: Insights into manipulators of eukaryotic cells. Trends Mol Med. 2011;17:573–583. doi: 10.1016/j.molmed.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 180.Rikihisa Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol. 2010;8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 181.Ge Y, Rikihisa Y. Subversion of host cell signaling by Orientia tsutsugamushi. Microbes Infect. 2011;13:638–648. doi: 10.1016/j.micinf.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 182.Bandi C, McCall JW, Genchi C, Corona S, Venco L, Sacchi L. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int J Parasitol. 1999;29:357–364. doi: 10.1016/s0020-7519(98)00200-8. [DOI] [PubMed] [Google Scholar]
- 183.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 184.Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, Ingram J, Nene RV, Shepard J, Tomkins J, Richards S, Spiro DJ, Ghedin E, Slatko BE, Tettelin H, Werren JH. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 185.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 186.Ohashi N, Zhi N, Lin Q, Rikihisa Y. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect Immun. 2002;70:2128–2138. doi: 10.1128/IAI.70.4.2128-2138.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pichon S, Bouchon D, Cordaux R, Chen L, Garrett RA, Greve P. Conservation of the type IV secretion system throughout Wolbachia evolution. Biochem Biophys Res Commun. 2009;385:557–562. doi: 10.1016/j.bbrc.2009.05.118. [DOI] [PubMed] [Google Scholar]
- 188.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2015;39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Kim SY, Darby AC, Fuxelius HH, Yin J, Kim JH, Kim J, Lee SJ, Koh YS, Jang WJ, Park KH, Andersson SG, Choi MS, Kim IS. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rikihisa Y, Lin M, Niu H. Type IV secretion in the obligatory intracellular bacterium Anaplasma phagocytophilum. Cell Microbiol. 2010;12:1213–1221. doi: 10.1111/j.1462-5822.2010.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Niu H, Rikihisa Y, Yamaguchi M, Ohashi N. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell Microbiol. 2006;8:523–534. doi: 10.1111/j.1462-5822.2005.00643.x. [DOI] [PubMed] [Google Scholar]
- 192.Ge Y, Rikihisa Y. Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun. 2007;75:3833–3841. doi: 10.1128/IAI.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ge Y, Rikihisa Y. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J Bacteriol. 2007;189:7819–7828. doi: 10.1128/JB.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 195.Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 2010;6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS ONE. 2011;6:e27724. doi: 10.1371/journal.pone.0027724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sinclair SH, Garcia-Garcia JC, Dumler JS. Bioinformatic and mass spectrometry identification of Anaplasma phagocytophilum proteins translocated into host cell nuclei. Front Microbiol. 2015;6:55. doi: 10.3389/fmicb.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Voth DE. ThANKs for the repeat: Intracellular pathogens exploit a common eukaryotic domain. Cell Logist. 2011;1:128–132. doi: 10.4161/cl.1.4.18738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun. 2009;77:2385–2391. doi: 10.1128/IAI.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rennoll-Bankert KE, Garcia-Garcia JC, Sinclair SH, Dumler JS. Chromatin-bound bacterial effector ankyrin A recruits histone deacetylase 1 and modifies host gene expression. Cell Microbiol. 2015 doi: 10.1111/cmi.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu H, Bao W, Lin M, Niu H, Rikihisa Y. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 2012;14:1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 204.Reisner A, Holler BM, Molin S, Zechner EL. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J Bacteriol. 2006;188:3582–3588. doi: 10.1128/JB.188.10.3582-3588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bhatty M, Cruz MR, Frank KL, Gomez JA, Andrade F, Garsin DA, Dunny GM, Kaplan HB, Christie PJ. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 2015;95:660–677. doi: 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schlievert PM, Gahr PJ, Assimacopoulos AP, Dinges MM, Stoehr JA, Harmala JW, Hirt H, Dunny GM. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PloS ONE. 2007;2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang A, Yang M, Hu P, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics. 2011;12:523. doi: 10.1186/1471-2164-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol. 2011;79:1670–1683. doi: 10.1111/j.1365-2958.2011.07553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hofreuter D, Odenbreit S, Puls J, Schwan D, Haas R. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res Microbiol. 2000;151:487–491. doi: 10.1016/s0923-2508(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 211.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 212.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188:882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. Natural competence promotes Helicobacter pylori chronic infection. Infect Immun. 2013;81:209–215. doi: 10.1128/IAI.01042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 215.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 216.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol. 2011;82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, do Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CF, Miyaki CY, Moon DH, Moreira LM, Novo MT, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 218.Souza DP, Andrade MO, Alvarez-Martinez CE, Arantes GM, Farah CS, Salinas RK. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011;7:e1002031. doi: 10.1371/journal.ppat.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, Guzzo CR, Farah CS. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 220.Alegria MC, Docena C, Khater L, Ramos CHI, da Silva ACR, Farah CS. Identification of new protein-protein interactions involving products of the chromosome - and plasmid - encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J Bacteriol. 2004;187:2315–2325. doi: 10.1128/JB.187.7.2315-2325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lacerda TL, Salcedo SP, Gorvel JP. Brucella T4SS: the VIP pass inside host cells. Curr Opin Microbiol. 2013;16:45–51. doi: 10.1016/j.mib.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 223.Voth DE, Heinzen RA. Coxiella type IV secretion and cellular microbiology. Curr Opin Microbiol. 2009;12:74–80. doi: 10.1016/j.mib.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol. 2010;13:59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rolan HG, den Hartigh AB, Kahl-McDonagh M, Ficht T, Adams LG, Tsolis RM. VirB12 is a serological marker of Brucella infection in experimental and natural hosts. Clin Vacc Immun. 2008;15:208–214. doi: 10.1128/CVI.00374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rances E, Voronin D, Tran-Van V, Mavingui P. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol. 2008;190:5020–5030. doi: 10.1128/JB.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, Azad AF, Sobral BW, Caler E. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 2012;194:376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pachulec E, Siewering K, Bender T, Heller EM, Salgado-Pabon W, Schmoller SK, Woodhams KL, Dillard JP, van der Does C. Functional analysis of the gonococcal genetic island of Neisseria gonorrhoeae. PLoS ONE. 2014;9:e109613. doi: 10.1371/journal.pone.0109613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 230.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino KG, Heinzen RA, Richards AL, Carlyon JA. Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol. 2014;4:186. doi: 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 233.Hubber A, Kubori T, Nagai H. Modulation of the ubiquitination machinery by Legionella. Curr Top Microbiol Immunol. 2013;376:227–247. doi: 10.1007/82_2013_343. [DOI] [PubMed] [Google Scholar]
- 234.Price CT, Jones SC, Amundson KE, Kwaik YA. Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of Legionella pneumophila. Front Microbiol. 2010;1:131. doi: 10.3389/fmicb.2010.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]