ABSTRACT
The secretion of immune-mediators is a critical step in the host innate immune response to pathogen invasion, and Rab GTPases have an important role in the regulation of this process. Rab4/Rab11 recycling endosomes are involved in the sorting of immune-mediators into specialist Rab11 vesicles that can traffic this cargo to the plasma membrane; however, how this sequential delivery process is regulated has yet to be fully defined. Here, we report that Drosophila Pkaap, an orthologue of the human dual-specific A-kinase-anchoring protein 2 or D-AKAP2 (also called AKAP10), appeared to have a nucleotide-dependent localisation to Rab4 and Rab11 endosomes. RNAi silencing of pkaap altered Rab4/Rab11 recycling endosome morphology, suggesting that Pkaap functions in cargo sorting and delivery in the secretory pathway. The depletion of pkaap also had a direct effect on Rab11 vesicle exocytosis and the secretion of the antimicrobial peptide Drosomycin at the plasma membrane. We propose that Pkaap has a dual role in antimicrobial peptide traffic and exocytosis, making it an essential component for the secretion of inflammatory mediators and the defence of the host against pathogens.
KEY WORDS: Drosophila, Innate immunity, Endosomes, Rab4, Rab11, Pkaap, Antimicrobial peptide Drosomycin
Summary: Pkaap has a dual role in antimicrobial peptide trafficking and exocytosis, making it an essential component for the secretion of inflammatory mediators and the defence of the host against pathogens.
INTRODUCTION
The innate immune system enables a coordinated host response to microorganism invasion, resulting in the secretion of immune-mediators, which can either induce inflammation or kill pathogens (Beisswenger and Bals, 2005; Cederlund et al., 2011; Jenssen et al., 2006; Lai and Gallo, 2009; Stow and Murray, 2013). However, excessive or uncontrolled secretion of immune-mediators can be associated with important pathological states, including rheumatoid arthritis, diabetes, Alzheimer's, multiple sclerosis, cancer and cardiovascular disease (Charo and Ransohoff, 2006; Compston and Coles, 2002; Grateau, 2006; Hansson and Libby, 2006; Hotamisligil, 2006; McInnes and Schett, 2007). The critical role of innate immunity in host defence and inflammation has warranted studies on the molecular mechanisms involved in generating an effective immune response and for the secretion of specific immune cargo.
The Toll-like receptors recognise pathogens and initiate an innate immune response (Barton and Kagan, 2009). This pathogen recognition system at the cell surface of immune cells can trigger different intracellular signalling cascades, including the Toll-interleukin 1 pathway or the immune-deficiency-tumour necrosis factor alpha (TNFα) pathway, which results in the synthesis of immune-mediators (Lemaitre, 2004). Cytokines and antimicrobial peptides that are synthesised in response to this immune stimulation are subsequently delivered to the endosomal system for processing and packaging into specialist exocytic vesicles (Bonifacino and Glick, 2004). Small Rab GTPases are critically involved in the regulation of this sequential transport process, with cargo sorting in Rab4/Rab11 recycling endosomes (Ang et al., 2004), and traffic to the plasma membrane in Rab11 vesicles (Chen et al., 1998; Damiani et al., 2004; Li et al., 2008; Mohrmann et al., 2002; Shandala et al., 2011). Rab11-mediated exocytosis has been reported for various cytokines, including TNFα (Murray et al., 2005; Reefman et al., 2010), interferon gamma (Reefman et al., 2010), interleukin 10 (Stanley et al., 2012) in mammals, and the antimicrobial peptide Drosomycin in Drosophila (Shandala et al., 2011). While the immune signalling cascades as well as the synthesis and delivery of cytokines and antimicrobial peptides have been well characterised, the regulatory mechanisms for Rab11-mediated exocytosis and release of innate immune-mediators have yet to be fully defined.
GTPase activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) are known to regulate Rab GTPase activity, but these regulatory elements have yet to be defined for innate immune cargo delivery and secretion at the plasma membrane (Bos et al., 2007; Csépányi-Kömi et al., 2012; Liao et al., 2008). GAPs inactivate Rab GTPases by stimulating the hydrolysis of guanosine triphosphate (GTP) to guanosine diphosphate (GDP; Scheffzek and Ahmadian, 2005; Zurita et al., 2010), whereas GEFs facilitate their activation by exchanging GDP for GTP (Bos et al., 2007; Liao et al., 2008). Multiple GAPs and GEFs have been reported to be involved in Rab11 nucleotide exchange. For example, TBC1D15 exhibits GAP activity against Rab11, but can also affect the activity of Rab7 (Zhang et al., 2005). Similarly, TBC1D11/GAPCenA exhibits GAP activity to both Rab4 and Rab11 GTPases (Fuchs et al., 2007). The Drosophila protein Evi5 acts as a GAP for Rab11 to regulate the migration of border cells (Dabbeekeh et al., 2007; Laflamme et al., 2012). In contrast, the vesicular transport of Rhodospin to the rhabdomere membrane involves Rab11 activation by Calmodulin-binding protein related to a Rab3 GDP/GTP exchange protein (also called Crag) that acts as a GEF in Drosophila photoreceptors (Xiong et al., 2012). The dual-specific A-kinase-anchoring protein 2 (also called D-AKAP2 or AKAP10) appears to have GEF activity towards Rab11 in humans (Eggers et al., 2009). Two highly conserved regulatory G protein signalling (RGS) domains in D-AKAP2, referred to as RGS1 and RGS2, are sufficient to bind simultaneously to both Rab4 and Rab11 (Fukuda et al., 2008), and this is used in the regulation of transferrin receptor recycling in HeLa cells (Eggers et al., 2009). D-AKAP2 preferentially binds to inactive GDP-locked Rab11S25N and active GTP-locked Rab4Q67L, and therefore D-AKAP2 may exhibit GEF activity towards Rab11 and GAP activity towards Rab4 (Eggers et al., 2009). Consequently, proteins involved in the regulation of Rab11 can be very selective and function preferentially on this GTPase, while others are promiscuous and regulate multiple Rab proteins, making it difficult to define the precise mechanism controlling innate immune secretion.
Little is known about the GEF and GAP proteins that are involved in regulating Rab11 function during an immune response and how this impacts on the secretion of immune-mediators. Here, we have investigated Drosophila Pkaap, an orthologue of human D-AKAP2, as a candidate for the regulation of Rab4/Rab11 sorting and Rab11 exocytosis of innate immune cargo. The response to pkaapRNAi was studied in live mode in the fat body, due to the large cell size (high DNA ploidy) and proportionally enlarged endosome/exocytic compartments, which facilitated the visualisation of molecular events during an immune response. The specific focus on Drosophila Pkaap was based on previous evidence for D-AKAP2 appearing to have a role in exocytosis (Eggers et al., 2009) and the immune response (Kim et al., 2011).
RESULTS
pkaap depletion increased Drosophila susceptibility to acute bacterial infection
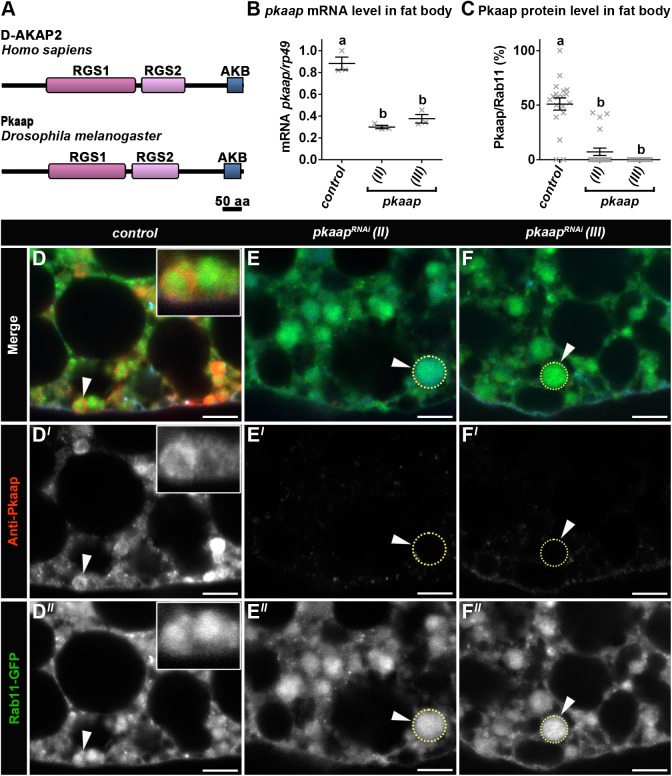
The pkaap gene (CG4132) encodes a 662 amino acid protein that is 27% identical (152/571) to human D-AKAP2. Pkaap and D-AKAP2 share two tandem conserved regulatory G protein signalling domains and a protein kinase A (PKA)-binding domain (Fig. 1A). Pkaap RGS1 and RGS2 domains displayed respectively 22% and 34% sequence identity with those of D-AKAP2. RNAi silencing of pkaap significantly reduced mRNA levels of pkaap (P<0.0001; Fig. 1B) and the amount of Pkaap protein detected in Drosophila fat body tissue (Fig. 1C,E,E′,F,F′), when compared to control larval tissue (Fig. 1D,D′). There was a significant reduction in the survival of pkaapRNAi Drosophila larvae, when compared to control third instar larvae (P<0.05). The viability was 91.7±1.5% for control larvae, compared to 82.0±1.9% for pkaapRNAi chromosome II and 83.1±1.9% for pkaapRNAi chromosome III transgenic larvae. When Drosophila early third instar larvae were infected with a combination of Gram-positive Micrococcus luteus and Gram-negative Escherichia coli, there was significantly reduced survival for pkaapRNAi transgenic larvae compared to control larvae (P<0.05). The viability was 84.6±2.3% for control larvae, compared to 80.0±2.3% for pkaapRNAi chromosome II and 63.3±5.4% for pkaapRNAi chromosome III transgenic larvae. The reduced survival of pkaapRNAi transgenic larvae compared to control larvae, either without or with bacterial challenge, might be attributed to a potential problem in immune function.
Fig. 1.
Expression of CG-GAL4>UAS-pkaap in fat body tissue. (A) A schematic diagram showing the domain structure of D-AKAP2 and Pkaap proteins. D-AKAP2 and Pkaap proteins contain regulator of G protein signalling (RGS) domains and PKA-binding (AKB) domain. (B) The expression of pkaap mRNA was characterised by quantitative real-time PCR in fat body tissue from the following genotypes: control, pkaapRNAi chromosome II and pkaapRNAi chromosome III. mRNA levels were normalised against rp49 mRNA levels. Three independent sets of samples were analysed from late third larval instar (−4 h puparium formation). One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m. (C) Percentage of Pkaap colocation with Rab11 vesicles in the fat body cells from the following genotypes: control, pkaapRNAi chromosome II and pkaapRNAi chromosome III. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). (D-F) Confocal micrographs showing localisation of Pkaap detected with an anti-Pkaap antibody (red in D-F; greyscale in D′-F′) in relation to Rab11-GFP vesicles (green in D-F; greyscale in D″-F″) in (D) control, (E) pkaapRNAi chromosome II and (F) pkaapRNAi chromosome III fat bodies. The plasma membrane was detected with Alexa Fluor® 568 Phalloidin (cyan in D-F). Data is representative of at least ten independent replicates. Scale bar: 5 μm.
pkaap depletion resulted in altered Rab11 vesicle morphology and exocytosis
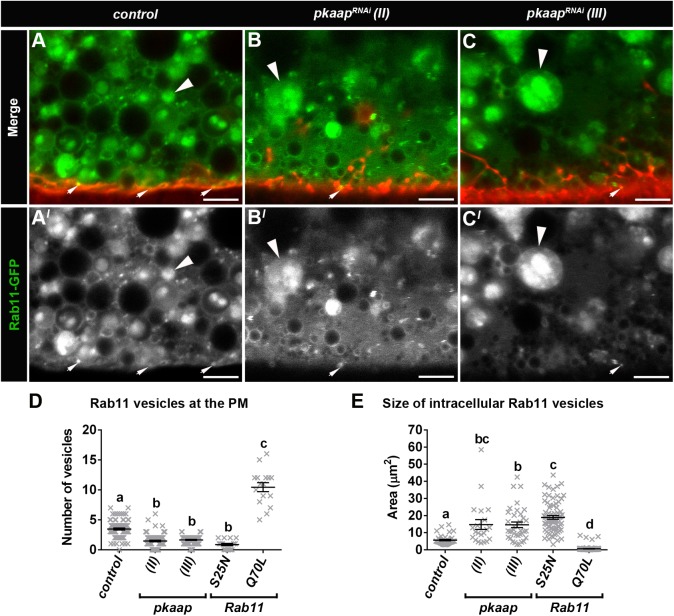
In control Drosophila fat body tissue from third larval instars, Pkaap protein colocated with Rab11 vesicles in close proximity to the plasma membrane (Fig. 1D-D″). The amount of Pkaap protein colocating with Rab11 vesicles was significantly reduced in pkaapRNAi larval fat body cells, when compared to control (P<0.0001; Fig. 1C). In addition, the morphology of Rab11 vesicles was altered, with the appearance of larger endosomes in the fat body tissue from pkaapRNAi (Fig. 1E,E″,F,F″), when compared to control larvae that exhibited smaller Rab11 vesicles (Fig. 1D,D″). The distribution and morphology of Rab11 vesicles was examined in relation to the plasma membrane, which was stained with CellMask™ Deep Red in live tissues. There were significantly more small ≤1 μm2 Rab11 vesicles (P<0.0001) near the plasma membrane of fat body cells in control larvae (Fig. 2A,A′) than for pkaapRNAi transgenic larvae (Fig. 2B,B′,C,C′,D). Moreover, there was a concomitant appearance of large Rab11 endosomes with intraluminal vesicles in the intracellular regions of fat body cells from pkaapRNAi transgenic larvae (Fig. 2B,B′,C,C′), when compared to control larvae (Fig. 2A,A′; Fig. S1A,A′). The size of these intracellular Rab11 endosomes was significantly increased in pkaapRNAi, when compared to control fat body cells (Fig. 2E). The Rab11 vesicle phenotype caused by pkaapRNAi (Fig. 2B,B′,C,C′) was similar to that observed for the GDP-bound dominant negative form of Rab11, Rab11S25N (Fig. 2D,E; Fig. S1C,C′), but not the GTP-bound constitutively active form of Rab11, Rab11Q70L (Fig. 2D,E; Fig. S1B,B′).
Fig. 2.
pkaap depletion resulted in a decreased number of Rab11 vesicles at the plasma membrane and changed the morphology of intracellular Rab11 endosomes. (A-C) Confocal micrographs of cross-section through the fat body cells showing Rab11-GFP vesicles (green in A-C; greyscale in A′-C′) with the plasma membrane outlined by CellMask™ Deep Red (red in A-C). Arrows depict small ≤1 μm2 Rab11 vesicles at the plasma membrane. Arrowheads depict large Rab11-GFP endosomes with area ≥3 μm2. Representative larval fat body cells were from the following genotypes: (A) control, (B) pkaapRNAi chromosome II and (C) pkaapRNAi chromosome III. Scale bar: 5 μm. Histograms showing (D) comparative analysis of the number of small ≤1 μm2 Rab11 vesicles at the plasma membrane, and (E) size of intracellular Rab11 endosomes with area ≥3 μm2. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m. PM, plasma membrane.
The GTPase activities of Rab4 and Rab11 affected the intracellular distribution of Pkaap
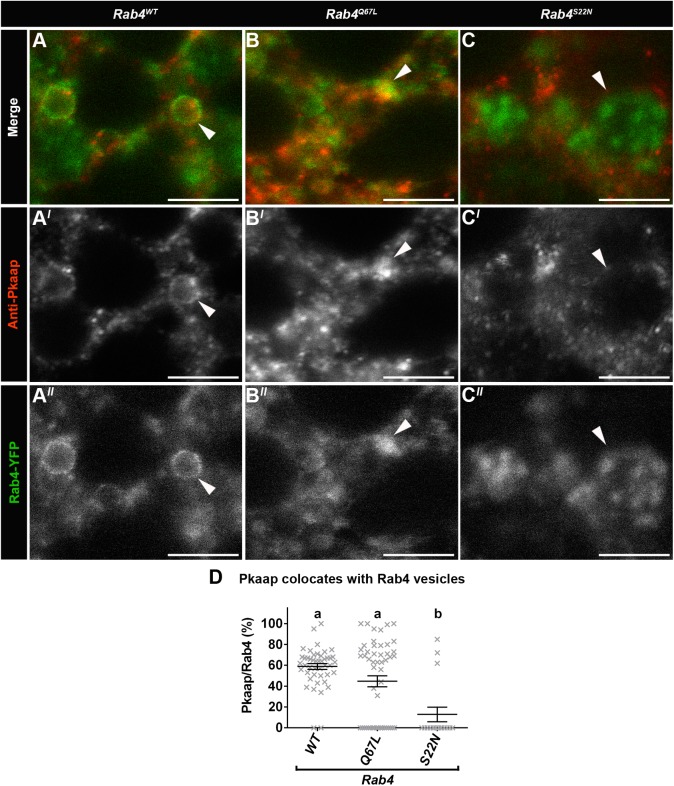
In fat body cells from larvae expressing wild-type Rab4 (Rab4WT), Pkaap was detected on Rab4-YFP vesicles (Fig. 3A-A″) and in the cytosol (as previously observed for D-AKAP2; Eggers et al., 2009). In fat body cells expressing constitutively active Rab4Q67L, while the size of the Rab4 vesicles was reduced, Pkaap was still detected colocating with Rab4 vesicles (Fig. 3B-B″). In contrast, in fat body cells expressing dominant negative Rab4S22N, the size of the Rab4 vesicles was increased and Pkaap showed no colocation with these vesicles, but rather was only detected in the same proximity as these Rab4 endosomes (Fig. 3C-C″). Quantitative analysis showed that the expression of Rab4S22N significantly reduced Pkaap association with Rab4 vesicles, when compared to Rab4WT and Rab4Q67L (P<0.0001; Fig. 3D).
Fig. 3.
Pkaap colocates with Rab4 endosomes. (A-C) Confocal micrographs showing localisation of Pkaap detected with an anti-Pkaap antibody (red in A-C; greyscale in A′-C′) in relation to Rab4-YFP endosomes (green in A-C; greyscale in A″-C″) in fat body cells. Representative larval fat body cells were from the following genotypes: (A-A″; Rab4WT) wild-type control of Rab4, (B-B″; Rab4Q67L) GTP-bound constitutively active form of Rab4 and (C-C″; Rab4S22N) GDP-bound dominant negative form of Rab4. Scale bar: 5 μm. (D) Percentage of Pkaap colocation with Rab4-YFP vesicles in the fat body cells from Rab4WT, Rab4Q67L and Rab4S22N. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m.
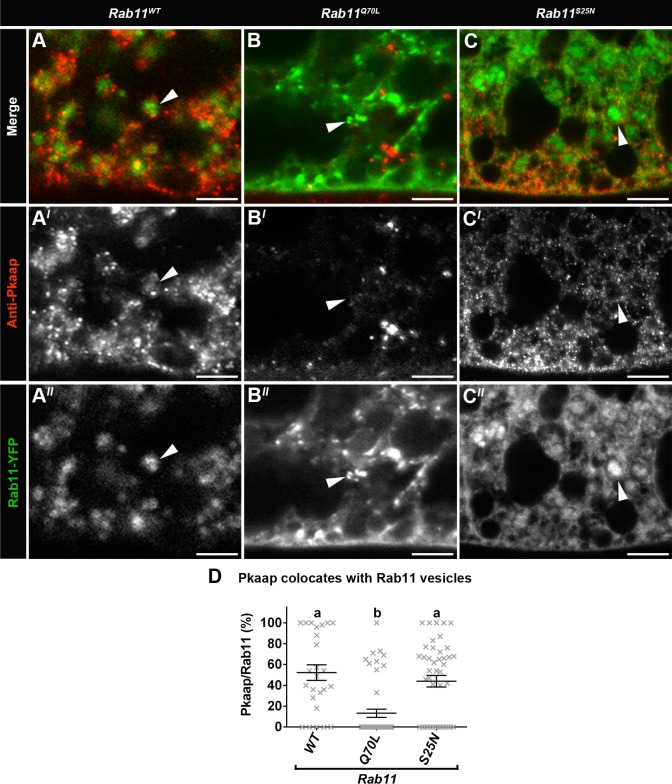
In fat body cells from larvae expressing wild-type Rab11 (Rab11WT), Pkaap was detected in association with Rab11 endosomes (Fig. 4A-A″), and also showed diffused cytosolic localisation (as previously observed for D-AKAP2; Eggers et al., 2009). However, there was little or no Pkaap detected on Rab11 vesicles in fat body cells expressing constitutively active Rab11Q70L (Fig. 4B-B″). In fat body cells expressing dominant negative Rab11S25N, Pkaap showed significant interaction with Rab11 endosomes (Fig. 4C-C″), particularly in the vicinity of the plasma membrane. Quantification confirmed that Rab11WT and Rab11S25N resulted in Pkaap association with Rab11 endosomes, while there was significantly less Pkaap colocation with Rab11 endosomes for Rab11Q70L (P<0.0001; Fig. 4D).
Fig. 4.
Pkaap colocates with Rab11 endosomes. (A-C) Confocal micrographs showing localisation of Pkaap detected with anti-Pkaap antibody (red in A-C; greyscale in A′-C′) in relation to Rab11-YFP endosomes (green in A-C; greyscale in A″-C″) in fat body cells. Representative larval fat body cells were from the following genotypes: (A-A″; Rab11WT) wild-type contol of Rab11, (B-B″; Rab11Q70L) GTP-bound constitutively active form of Rab11 and (C-C″; Rab11S25N) GDP-bound dominant negative form of Rab11. Scale bar: 5 μm. (D) Percentage of Pkaap colocation with Rab11-YFP vesicles in the fat body cells from Rab11WT, Rab11Q70L, and Rab11S25N. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m.
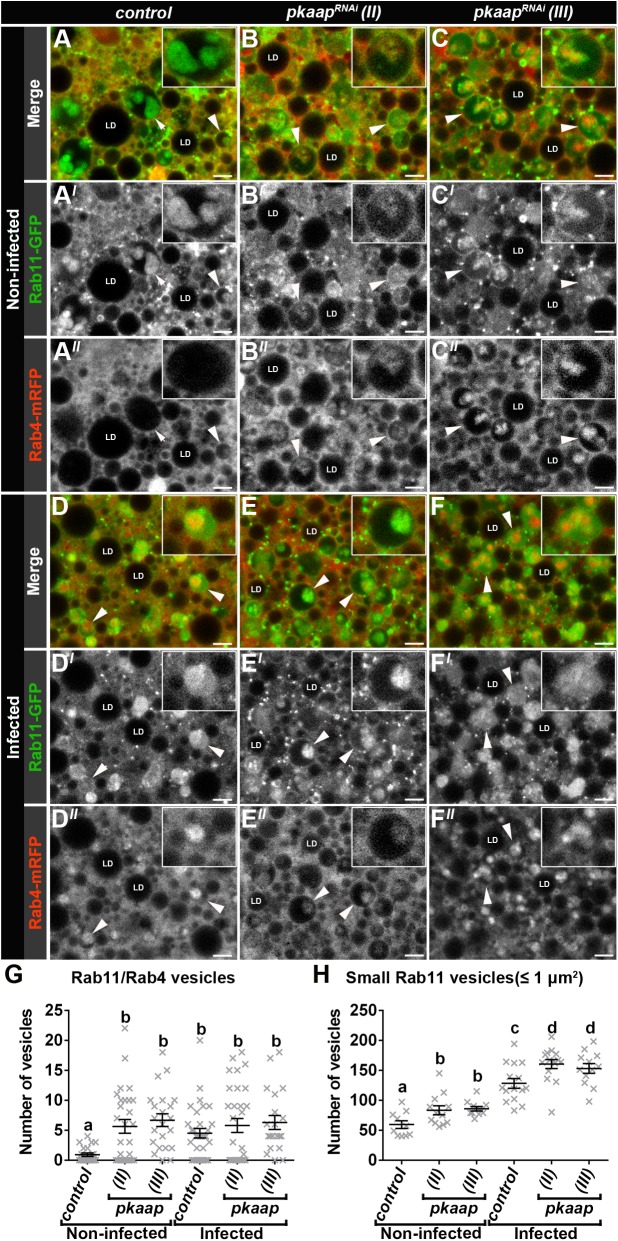
pkaap depletion resulted in altered Rab4/Rab11 recycling endosome morphology
In fat body cells from control larvae, there were large Rab11 endosomes that contained Rab11 intraluminal vesicles; as well as other smaller Rab11 vesicles (Fig. 5A). Recycling endosomes with Rab4 and Rab11 colocalisation were identified, but the larger Rab11 multivesicular endosomes did not appear to contain appreciable amounts of Rab4 (Fig. 5A-A″). In pkaapRNAi fat body cells, there were Rab4 compartments that contained little or no Rab11 (Fig. 5B-C″), and the larger Rab11 endosomes had intraluminal vesicles with both Rab4 and Rab11 (Fig. 5B-C″); with intraluminal Rab4/Rab11 vesicles in 64.6±8.3% for pkaapRNAi chromosome II and 70.9±8.9% for pkaapRNAi chromosome III compared to 13.1±7.2% in the controls. There was also an increase in the number of small Rab11 vesicles at the periphery of the larger multivesicular endosomes (Fig. 5B,B′,C,C′), particularly for pkaapRNAi chromosome III (Fig. 5C,C′).
Fig. 5.
pkaap depletion altered the morphology of Rab4/Rab11 endosomes. (A-F) Confocal micrographs of cross-section through the fat body cells showing Rab11-GFP (green in A-F; greyscale in A′-F′) and Rab4-mRFP vesicles (red in A-F; greyscale in A″-F″) in close proximity to the perinuclear region. Fat body cells were visualised from (A-C) non-infected larvae and (D-F) larvae infected orally with Micrococcus luteus for 4 h. Representative larval fat body cells were from the following genotypes: (A,D) control, (B,E) pkaapRNAi chromosome II and (C,F) pkaapRNAi chromosome III. Arrow depicts Rab11-GFP endosome (A). Arrowheads depict Rab4/Rab11 endosomes. LD, lipid droplets. Scale bar: 5 μm. (G) Histogram showing comparative analysis of the number of Rab4/Rab11 endosomes. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P=0.0025). (H) Histogram showing comparative analysis of the number of small ≤1 μm2 intracellular Rab11 vesicles. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means for the genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m.
The formation of Rab4/Rab11 multivesicular endosomes was also investigated under conditions of bacterial challenge to help define the role of Pkaap during an innate immune response. At four hours post-infection with Micrococcus luteus (Fig. 5D-F), there were variable amounts of Rab4 (e.g. Fig. 5E,E″), while there were similar numbers of Rab4/Rab11 multivesicular endosomes in the fat body cells from control and pkaapRNAi larvae; with 65.5±7.9% in control and 70.4±8.4% for pkaapRNAi chromosome II, 77.3±6.7% for pkaapRNAi chromosome III. Consequently, while there was a significant increase in the number of Rab4/Rab11 multivesicular endosomes in infected control cells, there was not a significant difference between the number of Rab4/Rab11 multivesicular endosomes before and after infection in pkaapRNAi fat body cells (P=0.0025; Fig. 5G). In control cells there were also significantly more small Rab11 vesicles (≤1 μm2) after infection (P<0.0001; Fig. 5D,D′,H), and more small Rab11 vesicles in pkaapRNAi fat body cells (P<0.0001; Fig. 5E,E′,F,F′,H).
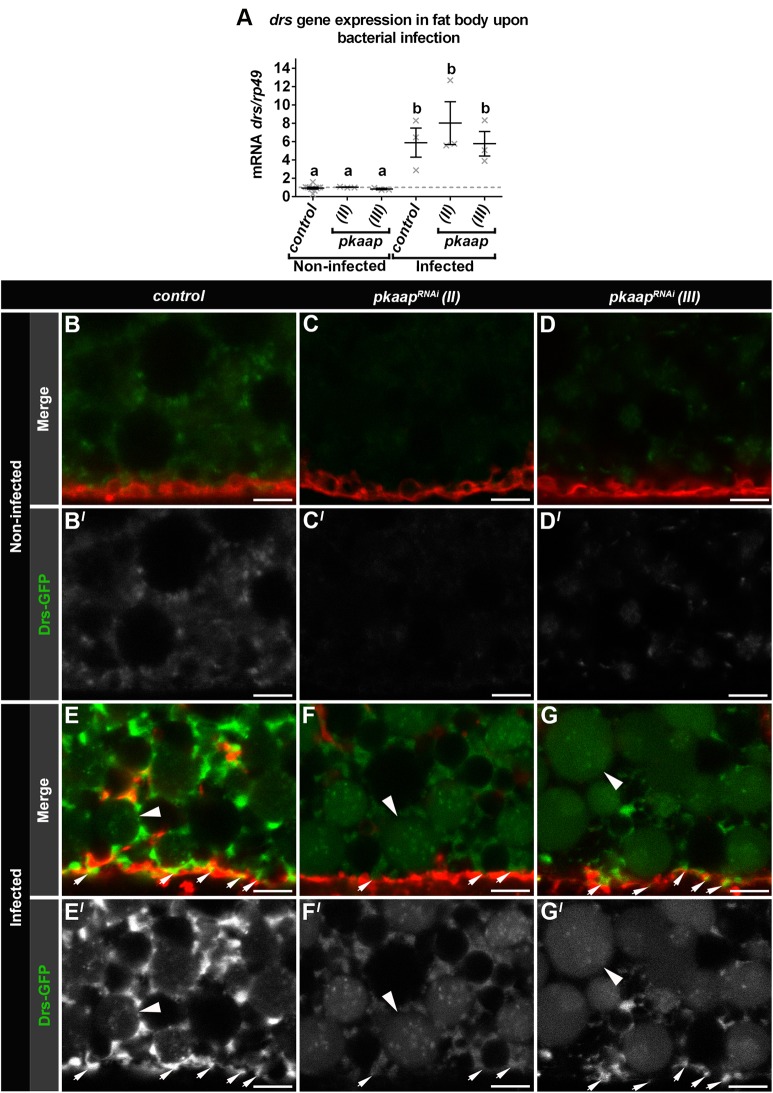
pkaap depletion resulted in reduced antimicrobial peptide Drosomycin delivery to the plasma membrane
To determine the effect of pkaapRNAi on innate immune cargo delivery, we examined by quantitative real-time PCR the expression of the antimicrobial peptide Drosomycin in fat body cells, either before or after infection. In non-infected control larvae, only minimal Drosomycin was detected prior to infection (Fig. 6A for mRNA and protein visualised in Fig. 6B,B′), but there was a marked increase in Drosomycin expression four hours after infection (Fig. 6A for mRNA and protein visualised in Fig. 6E,E′). In addition, the Drosophila NF-κB homolog Dorsal, which normally activates Drosomycin transcription in response to infection (Manfruelli et al., 1999), was translocated from the cytoplasm to the nucleus in pkaapRNAi transgenic larvae, and there were no differences in Dorsal subcellular localisation for control and pkaapRNAi [i.e. Dorsal was cytoplasmic in non-infected larvae (Fig. S2A-C) and predominantly nuclear within 30-60 min after infection (Fig. S2D-I)]. In Micrococcus luteus-infected control larvae (Fig. 6E,E′), the Drosomycin-GFP cargo was detected in small ∼1 µm2 vesicles either in close proximity to larger vesicles that contained less concentrated cargo or near the cell surface colocating with CellMask™ Deep Red. In contrast, for pkaapRNAi larvae there was little or no detectable Drosomycin-GFP protein detected prior to infection (Fig. 6C-D′), even though the level of mRNA was similar to that detected in control fat body tissue (Fig. 6A). While there was an increase in mRNA expression in response to infection in pkaapRNAi fat body cells (Fig. 6A), there was only a relatively small amount of Drosomycin-GFP detected, when compared to control larvae (Fig. 6F-G′). This Drosomycin-GFP, in pkaapRNAi fat body cells, was detected as a diffuse signal in large vesicular structures that contained intraluminal vesicles with only minimal Drosomycin-GFP (Fig. 6F-G′). In addition, there was a reduced amount of this Drosomycin-GFP cargo near the cell surface (Fig. 6F-G′), when compared to controls (Fig. 6E,E′).
Fig. 6.
pkaap depletion reduced the amount of Drosomycin at the plasma membrane. (A) The expression of drosomycin (drs) mRNA was characterised by quantitative real-time PCR. mRNA levels were normalised against rp49 mRNA levels. Three independent sets of samples were collected from late third larval instar (−4 h puparium formation) from non-infected larvae and larvae infected orally with Micrococcus luteus and Escherichia coli. One-way ANOVA and Tukey's multiple comparison test showed significant differences between the means in genotypes (depicted by different letters on the bars, P<0.0001). Data presented as mean±s.e.m. (B-G) Confocal micrographs showing the intracellular distribution of Drosomycin-GFP in fat body cells (green in A-G; greyscale in A′-G′) with the plasma membrane outlined by CellMask™ Deep Red (red in A-G). Fat body cells were visualised from (B-D) non-infected larvae and (E-G) larvae infected orally with Micrococcus luteus for 4 h. Arrows depict Drosomycin-GFP at the plasma membrane. Arrowheads depict large Drosomycin-GFP compartments. Representative larval fat body cells were from the following genotypes: (B,E) control, (C,F) pkaapRNAi chromosome II and (D,G) pkaapRNAi chromosome III. Scale bar: 5 μm.
DISCUSSION
Following host invasion, cells of the innate immune system detect microbial pathogens via pattern recognition receptors and peptidoglycan-recognition receptors (Ferrandon et al., 2007; Royet et al., 2005). These receptors can trigger the activation of immune cells by two distinct intracellular cascades; the Toll-interleukin 1 pathway and the immune deficiency-TNFα pathway (Lemaitre, 2004). This innate immune signalling initiates a sequence of events that leads to the transcription of pro-inflammatory cytokine and antimicrobial peptide genes (Beisswenger and Bals, 2005; Cederlund et al., 2011; Jenssen et al., 2006; Lai and Gallo, 2009; Medzhitov, 2008; Stow and Murray, 2013). Newly synthesised immune-mediators are transported through the endoplasmic reticulum–Golgi network and delivered to endosomes for final processing and packaging into Rab11 vesicles; ready for exocytosis at the plasma membrane. Endosomal trafficking is regulated by a complex set of vesicular machinery including, for example A-kinase anchoring proteins, and here, we have investigated the role of the Drosophila A-kinase anchoring protein, Pkaap, in the control of innate immune secretion.
Relatively little is known about how A-kinase anchoring proteins regulate immune defence mechanisms, although they have been previously implicated in modulating the transcriptional activation of cytokines in immune cells (Kim et al., 2011; Shibolet et al., 2007; Wall et al., 2009; Williams, 2002). For instance, AKAP13 has a role in the nuclear translocation of transcription factor NF-κβ, and therefore the TLR2-dependent activation of pro-inflammatory cytokine secretion (Shibolet et al., 2007). In contrast, AKAP95 was required for targeting of type II regulatory subunit of PKA to supress the early expression of TNFα in lipopolysaccharide-stimulated RAW264.7 macrophages (Wall et al., 2009). D-AKAP2-anchored type I regulatory subunit of PKA is involved in prostaglandin E2 potentiation of lipopolysaccharide-induced nitric oxide synthesis downstream of Toll-like receptors and the expression of interleukin 6 and interleukin 10 in alveolar macrophages (Kim et al., 2011). Our analysis of pkaap depleted larvae showed a normal activation of immune response pathways, resulting in Dorsal nuclear translocation and antimicrobial peptide drosomycin gene expression, suggesting that Pkaap did not have a role in modulating antimicrobial peptide transcriptional regulation. In future studies the effect of pkaapRNAi on phagocytosis and morphology of Rab4/Rab11 endosomes in haemocytes might be determined as the expression of transgenes was driven by fat body- and haemocyte-specific CG-GAL4 driver. Given that D-AKAP2 is involved in the regulation of transferrin receptor recycling via Rab4/Rab11 endosomes (Eggers et al., 2009) and there were no difference in Rab11-GFP fluorescence in control and pkaapRNAi fat body cells, it was postulated that Pkaap might be involved in antimicrobial peptide intracellular traffic.
In Drosophila, antimicrobial peptide targeting to the plasma membrane is known to involve Rab4 and Rab11 GTPases (Shandala et al., 2011). GAPs and GEFs control membrane trafficking by modulating Rab protein activities (Bos et al., 2007; Csépányi-Kömi et al., 2012; Liao et al., 2008); and the nucleotide-dependent location of Pkaap to endosomes suggested that it could have a role as both a GAP and GEF. We found that Pkaap colocated with wild-type and GTP-bound constitutively active Rab4, but not the GDP-bound dominant negative form of Rab4, which was consistent with it acting as a GAP for Rab4. We also showed that Pkaap colocated with wild-type and GDP-bound dominant negative form of Rab11, but not GTP-bound constitutively active Rab11, suggesting that it may also be acting as a GEF for Rab11. This was consistent with observations of D-AKAP2 co-immunoprecipitation studies for both Rab4 and Rab11 GTPases in HEK293 cells (Eggers et al., 2009; Fukuda et al., 2008) and that some AKAPs have been shown to act as molecular switches in regulating GTPase activity. For example, AKAP-Lbc binds to the GDP-bound or nucleotide-free forms of RhoA and possesses Rho-selective GEF activity in HeLa and HEK-293 cells (Diviani et al., 2004, 2001). Thus, by regulating the activity of Rab4 and Rab11, Pkaap could modulate two critical steps during the vesicular traffic of cargo along the immune secretory pathway, cargo sorting in recycling endosomes and exocytosis at the plasma membrane.
The depletion of pkaap had a direct effect on Rab11 vesicle traffic and antimicrobial peptide immune cargo delivery to the plasma membrane and resulted in the accumulation of enlarged Rab11 vesicles at the cellular periphery. The GDP-bound dominant negative form of Rab11 also reduced the number of small Rab11 vesicles in close proximity to the cell surface and resulted in the accumulation of enlarged Rab11 multivesicular endosomes, suggesting that pkaap depletion trapped Rab11 in a GDP-bound form. These enlarged Rab11 compartments with intraluminal vesicles were identified as multivesicular endosomes by definition. To further explore this question, future work may investigate fat body tissues expressing Rab11-GFP with antibodies detecting other multivesicular endosome markers, such as Hrs or Vps16. Constitutively active Rab11 produced a different vesicular phenotype, with enhanced delivery of small Rab11 vesicles to the plasma membrane. The results also revealed low Pkaap levels in fat body cells expressing constitutively active form of Rab11, suggesting a negative feedback mechanism by which the intracellular level of GEF proteins is regulated, but this is yet to be determined. Immune cargo delivery is dependent on Rab11 vesicles (Shandala et al., 2011) and pkaap depletion abrogated the intracellular traffic and delivery of the antimicrobial peptide Drosomycin to the plasma membrane upon bacterial challenge, but did not cause changes in the size of the Rab4/Rab11 endosomes. Concomitantly, pkaap depletion caused Drosomycin to accumulate in Rab4/Rab11 endosomes and this reduction in antimicrobial peptide delivery coupled with reduced secretion into the haemolymph could explain reduced larval viability following bacterial challenge. Due to limitations in the availability of transgenic stocks that could be used to study secreted proteins in live mode, we were limited to using Drosomycin-GFP and ideally the findings should be confirmed with other immune mediators. It would also be interesting to determine whether treatment of pkaapRNAi transgenic larvae with beta-lactam antibiotics (e.g. tetracycline) can improve survival rates after infection with Micrococcus luteus.
The structural changes to Rab4/Rab11 endosomes caused by pkaap depletion could affect the sorting and compartmentalisation of immune cargo in recycling endosomes. This would be consistent with the effects that pkaap depletion had on both of the GTPases Rab4 and Rab11 and the morphology of Rab4/Rab11 endosomes. Instead of correct antimicrobial peptide cargo sorting in Rab4/Rab11 recycling endosomes and packaging into Rab11 vesicles, a large amount of Drosomycin appeared to be degraded. For example, after immune challenge, while drosomycin mRNA expression was not reduced in response to pkaap depletion there was less Drosomycin protein, suggesting that a lysosomal degradative pathway may have been evoked in response to failed sorting in Rab4/Rab11 recycling endosomes; therapeutic agents, such as bafilomycin A and chloroquine, could be used in future studies. The specific effects of pkaap depletion on the dynamics and morphology of different endosomes suggested that pkaapRNAi transgenic larvae could have multiple impairments in the antimicrobial peptide trafficking and secretion pathway that contributed to immune dysfunction.
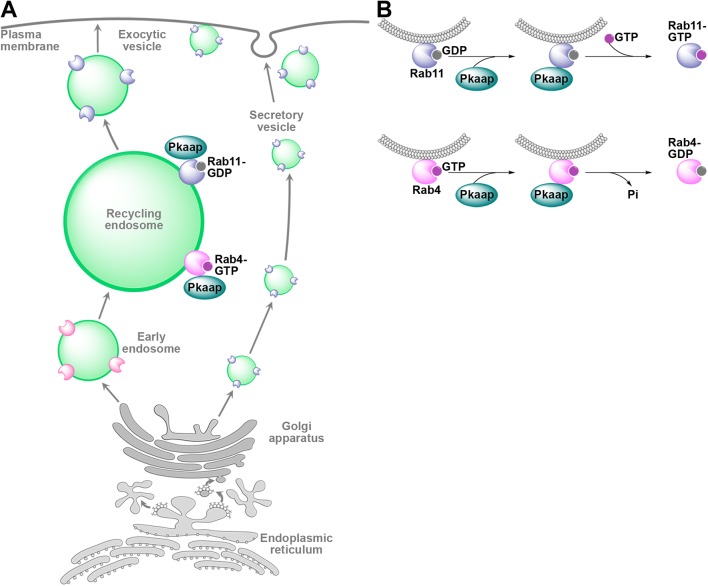
We concluded that Pkaap has a critical role in effecting an innate immune response and is important for Drosophila viability. The colocation of Pkaap with Rab11 endosomes suggested that Pkaap might be acting as a regulator of exocytosis and the effect of pkaap depletion on Rab11 vesicle and antimicrobial peptide cargo delivery would support this hypothesis (Fig. 7A,B). However, Pkaap also appeared to have an important role in Rab4/Rab11 recycling endosome morphology and function suggesting that it is also involved in cargo sorting and delivery earlier in the secretory pathway. It appears that Pkaap has a dual role in antimicrobial peptide trafficking and exocytosis making it an essential component for the secretion of inflammatory mediators and the defence of the host against pathogens. In future studies, the role of Pkaap might be investigated in other secretory tissues, such as the salivary glands, to ascertain if a similar molecular mechanism is used to control non-immune secretion.
Fig. 7.
The role of Pkaap in the regulation of antimicrobial peptide exocytosis in fat body cells. (A) Schematic representation of the nucleotide-dependent location of Pkaap to recycling endosomes. Pkaap binds to GTP-bound constitutively active form of Rab4 and GDP-bound dominant negative form of Rab11. (B) Schematic diagram showing the role of Pkaap in the regulation of membrane trafficking by modulating Rab4 and Rab11 activities. GTP, guanosine triphosphate; GDP, guanosine diphosphate.
MATERIALS AND METHODS
Fly stocks
Fly stocks were maintained in standard medium at 25°C. The yeast GAL4-UAS two component system was used for fat body-specific expression of genes of interest (Brand and Perrimon, 1993; Schmid et al., 2014). Expression of transgenes from the UAS was driven by CG-GAL4 (Asha et al., 2003). RNA interference (RNAi) silencing stocks of pkaap, UAS-pkaapRNAi chromosome II and UAS-pkaapRNAi chromosome III, were obtained from the Vienna Drosophila RNAi Centre (Vienna, Austria; Dietzl et al., 2007). Transgenic stocks UAS-Rab4WT-YFP, UAS-Rab4Q67L-YFP, UAS-Rab4S22N-YFP, UAS-Rab11WT-YFP, UAS-Rab11Q70L-YFP and UAS-Rab11S25N-YFP were obtained from the Bloomington Drosophila Stock Center (Indiana University, IN, USA; Zhang et al., 2007). Transgenic stocks UAS-Rab4-mRFP and UAS-Rab11-GFP were obtained from Markos González-Gaitán (University of Geneva, Geneva, Switzerland; Entchev et al., 2000; Wucherpfennig et al., 2003) and Donald F. Ready (Purdue University, West Lafayette, IN; Satoh et al., 2005). Drosomycin-GFP was a kind gift from Dominique Ferrandon (Equipe Fondation Recherche Médicale, Strasbourg, France; Ferrandon, 2007).
Natural bacterial infection
Drosophila early third larval instars were infected by the oral route with Micrococcus luteus and Escherichia coli for four hours at 25°C (to avoid temperature stress; Shandala et al., 2011). Fat body tissues were collected and stored at −80°C until required for antimicrobial peptide drosomycin gene expression analysis by quantitative real-time PCR. Secretion of the antimicrobial peptide Drosomycin into the haemolymph was not assessed because insect hemolymph had variable degrees of clotting (and therefore caused issues for Drosomycin recovery), making this task technically impossible. To assess survival rates (Shandala et al., 2011), pupae were scored as either live or dead within one week following infection and for each group (control, pkaapRNAi chromosome II and pkaapRNAi chromosome III) n=1200-1400 individual larvae were scored. The genotypes used for these experiments were CG-CAL4>+/+; +/+ (control), CG-CAL4>UAS-pkaapRNAi/+; +/+ (pkaapRNAi chromosome II) and CG-CAL4>+/+; UAS-pkaapRNAi/+ (pkaapRNAi chromosome III).
Gene expression
For quantitative real-time PCR analysis, RNA was isolated from the fat body tissue of 30 larvae using an RNAqueous® kit according to the manufacturer's protocol (Ambion, USA). cDNA was synthesised using a High Capacity RNA-to-cDNA kit (Applied Biosystems, USA). Quantitative real-time PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems, USA) using Fast SYBR® Green Master Mix kit (Applied Biosystems, USA). Three independent biological samples were analysed for each genotype. The mRNA expression of genes was normalised against the endogenous control gene rp49, using the ΔΔCT method. PCR primers were obtained from GeneWorks (Adelaide, Australia). The primers used for the quantitative real-time PCR were: pkaap (CG4132) forward, 5′-CTCCGATGGCATCAGTCTCG-3′, and reverse, 5′-CAGGCAGTTCGGATCGTTGA-3′; drosomycin (CG10810) forward, 5′-GTACTTGTTCGCCCTCTTCG-3′, and reverse, 5′-ATTTAGCATCCTTCGCACCA-3′; and rp49 (CG7939, used as an endogenous control) forward, 5′-CGAGTTGAACTGCCTTCAAGATGACCA-3′, reverse 5′-GCTTGGTGCGCTTCTTCACGATCT-3′.
Live cell imaging
For ex vivo live cell imaging, the fat body tissues were dissected from late third larval instars (−4 h before puparium formation) and incubated with CellMask™ Deep Red Plasma Membrane Stain (Applied Biosystems, USA) for 2 min at room temperature. Fat body tissues were attached to a coverslip using Carbomer 940-based gel (Snowdrift Farm, USA) and analysed for a maximum of 30 min; as a slower motility for endosome vesicles was observed after this time period (Sorvina et al., 2013).
Immunostaining and confocal microscopy
Fat body tissues expressing simultaneously Rab4-mRFP/Rab4-YFP and Rab11-GFP/Rab11-YFP were imaged in order to show the similarity in the distribution of the endosomes in these transgenic lines (Fig. S3A,B). Drosophila fat body tissues were fixed and stained as previously described (Shandala et al., 2011). Antibodies used for immunofluorescence were rabbit polyclonal anti-Pkaap (GeneScript, USA) and mouse monoclonal anti-Dorsal 7A4 (DSHB, USA). Anti-Pkaap antibody was pre-absorbed against fixed fat body tissues in 5% bovine serum albumin for 45 min at room temperature. This custom-made anti-Pkaap antibody only worked for immunofluorescence and did not work for western blot analysis or co-immunoprecipitation, and therefore protein-protein interaction studies were not technically possible due to problems with current reagents. Secondary anti-IgG antibody conjugates with Cy3 and Cy5 labels were obtained from Jackson Immuno Research Laboratories (Shandala et al., 2011). The Alexa Fluor® 488 Phalloidin, Alexa Fluor® 568 Phalloidin and Hoechst 33258 DNA stain were obtained from Invitrogen, USA. Imaging was performed using a Zeiss LSM710 NLO confocal microscope equipped with argon-gas, 543 nm and 633 nm solid-state lasers (Zeiss, Germany) and a two-photon Mai-Tai®, tunable Ti:Sapphire femtosecond pulse laser (Spectra-Physics, USA). All images were acquired using a Plan-APOCHROMAT 63×/NA 1.4 oil immersion objective to enable detailed visualisation of endosome/exocytic compartments. Each confocal micrograph represented 1.5 μm thin optical sections and the data was representative of at least ten independent replicates.
Image processing
The final preparation of the figures was conducted with Adobe Photoshop CS6 (Adobe Systems Inc.).
Measurements and statistical analysis
The number of small ≤1 µm2 Rab11 vesicles at the plasma membrane (22 µm sections), the size of intracellular Rab11 endosomes within a total cells area, and the number of Rab4/Rab11 recycling endosomes within 60 µm2 regions of interest was defined using Volocity 6.2.1 software.
The difference between group means was assessed by one-way analysis of variance (ANOVA), with individual group variance assessed by a Bartlett's test. Where the level of significance was P<0.05, post-hoc tests were performed using a Tukey's multiple comparison test (using GraphPad Prism version 6.00 for Windows, GraphPad Software, San Diego, CA USA). Data was presented as the mean±s.e.m.
Acknowledgements
We thank M. González-Gaitán, D. Ready and D. Ferrandon for kindly providing us with the fly stocks. We also thank Dr Emma Parkinson-Lawrence and Dr Christie A. Bader for the helpful comments on this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
A.S. performed experiments, analysed and interpreted the data and wrote the manuscript. T.S. designed experiments and interpreted the data. D.A.B. designed experiments, interpreted the data and edited the manuscript.
Funding
This work was supported by funding from the National Health and Medical Research Council of Australia [NHMRC 631915].
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.016642/-/DC1
References
- Ang A. L., Taguchi T., Francis S., Fölsch H., Murrells L. J., Pypaert M., Warren G. and Mellman I. (2004). Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167, 531-543. 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I. and Dearolf C. R. (2003). Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G. M. and Kagan J. C. (2009). A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 9, 535-542. 10.1038/nri2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswenger C. and Bals R. (2005). Functions of antimicrobial peptides in host defense and immunity. Curr. Protein Pept. Sci. 6, 255-264. 10.2174/1389203054065428 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S. and Glick B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153-166. 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H. and Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865-877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Cederlund A., Gudmundsson G. H. and Agerberth B. (2011). Antimicrobial peptides important in innate immunity. FEBS J. 278, 3942-3951. 10.1111/j.1742-4658.2011.08302.x [DOI] [PubMed] [Google Scholar]
- Charo I. F. and Ransohoff R. M. (2006). The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 354, 610-621. 10.1056/NEJMra052723 [DOI] [PubMed] [Google Scholar]
- Chen W., Feng Y., Chen D. and Wandinger-Ness A. (1998). Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9, 3241-3257. 10.1091/mbc.9.11.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compston A. and Coles A. (2002). Multiple sclerosis. Lancet 359, 1221-1231. 10.1016/S0140-6736(02)08220-X [DOI] [PubMed] [Google Scholar]
- Csépányi-Kömi R., Lévay M. and Ligeti E. (2012). Small G proteins and their regulators in cellular signalling. Mol. Cell. Endocrinol. 353, 10-20. 10.1016/j.mce.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Dabbeekeh J. T. S., Faitar S. L., Dufresne C. P. and Cowell J. K. (2007). The EVI5 TBC domain provides the GTPase-activating protein motif for RAB11. Oncogene 26, 2804-2808. 10.1038/sj.onc.1210081 [DOI] [PubMed] [Google Scholar]
- Damiani M. T., Pavarotti M., Leiva N., Lindsay A. J., McCaffrey M. W. and Colombo M. I. (2004). Rab coupling protein associates with phagosomes and regulates recycling from the phagosomal compartment. Traffic 5, 785-797. 10.1111/j.1600-0854.2004.00220.x [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Diviani D., Soderling J. and Scott J. D. (2001). AKAP-Lbc anchors protein kinase A and nucleates Galpha 12-selective Rho-mediated stress fiber formation. J. Biol. Chem. 276, 44247-44257. 10.1074/jbc.M106629200 [DOI] [PubMed] [Google Scholar]
- Diviani D., Abuin L., Cotecchia S. and Pansier L. (2004). Anchoring of both PKA and 14-3-3 inhibits the Rho-GEF activity of the AKAP-Lbc signaling complex. EMBO J. 23, 2811-2820. 10.1038/sj.emboj.7600287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers C. T., Schafer J. C., Goldenring J. R. and Taylor S. S. (2009). D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J. Biol. Chem. 284, 32869-32880. 10.1074/jbc.M109.022582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev E. V., Schwabedissen A. and Gonzalez-Gaitan M. (2000). Gradient formation of the TGF-beta homolog Dpp. Cell 103, 981-992. 10.1016/S0092-8674(00)00200-2 [DOI] [PubMed] [Google Scholar]
- Ferrandon D. (2007). Ubiquitin-proteasome: pallbearer carries the deceased to the grave. Immunity 27, 541-544. 10.1016/j.immuni.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Imler J.-L., Hetru C. and Hoffmann J. A. (2007). The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862-874. 10.1038/nri2194 [DOI] [PubMed] [Google Scholar]
- Fuchs E., Haas A. K., Spooner R. A., Yoshimura S.-I., Lord J. M. and Barr F. A. (2007). Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177, 1133-1143. 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Kanno E., Ishibashi K. and Itoh T. (2008). Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031-1042. 10.1074/mcp.M700569-MCP200 [DOI] [PubMed] [Google Scholar]
- Grateau G. (2006). Autoinflammatory diseases. Acta Clin. Belg. 61, 264-269. 10.1179/acb.2006.045 [DOI] [PubMed] [Google Scholar]
- Hansson G. K. and Libby P. (2006). The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 6, 508-519. 10.1038/nri1882 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2006). Inflammation and metabolic disorders. Nature 444, 860-867. 10.1038/nature05485 [DOI] [PubMed] [Google Scholar]
- Jenssen H., Hamill P. and Hancock R. E. W. (2006). Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491-511. 10.1128/CMR.00056-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Serezani C. H., Okunishi K., Zaslona Z., Aronoff D. M. and Peters-Golden M. (2011). Distinct protein kinase A anchoring proteins direct prostaglandin E2 modulation of Toll-like receptor signaling in alveolar macrophages. J. Biol. Chem. 286, 8875-8883. 10.1074/jbc.M110.187815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme C., Assaker G., Ramel D., Dorn J. F., She D., Maddox P. S. and Emery G. (2012). Evi5 promotes collective cell migration through its Rab-GAP activity. J. Cell Biol. 198, 57-67. 10.1083/jcb.201112114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. and Gallo R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131-141. 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B. (2004). The road to Toll. Nat. Rev. Immunol. 4, 521-527. 10.1038/nri1390 [DOI] [PubMed] [Google Scholar]
- Li H., Li H.-F., Felder R. A., Periasamy A. and Jose P. A. (2008). Rab4 and Rab11 coordinately regulate the recycling of angiotensin II type I receptor as demonstrated by fluorescence resonance energy transfer microscopy. J. Biomed. Opt. 13, 031206 10.1117/1.2943286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Shima F., Araki M., Ye M., Muraoka S., Sugimoto T., Kawamura M., Yamamoto N., Tamura A. and Kataoka T. (2008). Two conformational states of Ras GTPase exhibit differential GTP-binding kinetics. Biochem. Biophys. Res. Commun. 369, 327-332. 10.1016/j.bbrc.2008.01.169 [DOI] [PubMed] [Google Scholar]
- Manfruelli P., Reichhart J.-M., Steward R., Hoffmann J. A. and Lemaitre B. (1999). A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18, 3380-3391. 10.1093/emboj/18.12.3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes I. B. and Schett G. (2007). Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol. 7, 429-442. 10.1038/nri2094 [DOI] [PubMed] [Google Scholar]
- Medzhitov R. (2008). Origin and physiological roles of inflammation. Nature 454, 428-435. 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- Mohrmann K., Leijendekker R., Gerez L. and van Der Sluijs P. (2002). rab4 regulates transport to the apical plasma membrane in Madin-Darby canine kidney cells. J. Biol. Chem. 277, 10474-10481. 10.1074/jbc.M111237200 [DOI] [PubMed] [Google Scholar]
- Murray R. Z., Kay J. G., Sangermani D. G. and Stow J. L. (2005). A role for the phagosome in cytokine secretion. Science 310, 1492-1495. 10.1126/science.1120225 [DOI] [PubMed] [Google Scholar]
- Reefman E., Kay J. G., Wood S. M., Offenhauser C., Brown D. L., Roy S., Stanley A. C., Low P. C., Manderson A. P. and Stow J. L. (2010). Cytokine secretion is distinct from secretion of cytotoxic granules in NK cells. J. Immunol. 184, 4852-4862. 10.4049/jimmunol.0803954 [DOI] [PubMed] [Google Scholar]
- Royet J., Reichhart J.-M. and Hoffmann J. A. (2005). Sensing and signaling during infection in Drosophila. Curr. Opin. Immunol. 17, 11-17. 10.1016/j.coi.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Satoh A. K., O'Tousa J. E., Ozaki K. and Ready D. F. (2005). Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487-1497. 10.1242/dev.01704 [DOI] [PubMed] [Google Scholar]
- Scheffzek K. and Ahmadian M. R. (2005). GTPase activating proteins: structural and functional insights 18 years after discovery. Cell. Mol. Life Sci. 62, 3014-3038. 10.1007/s00018-005-5136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. R., Anderl I., Vesala L., Vanha-aho L.-M., Deng X.-J., Rämet M. and Hultmark D. (2014). Control of Drosophila blood cell activation via Toll signaling in the fat body. PLoS ONE 9, e102568 10.1371/journal.pone.0102568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandala T., Woodcock J. M., Ng Y., Biggs L., Skoulakis E. M. C., Brooks D. A. and Lopez A. F. (2011). Drosophila 14-3-3 epsilon has a crucial role in anti-microbial peptide secretion and innate immunity. J. Cell. Sci. 124, 2165-2174. 10.1242/jcs.080598 [DOI] [PubMed] [Google Scholar]
- Shibolet O., Giallourakis C., Rosenberg I., Mueller T., Xavier R. J. and Podolsky D. K. (2007). AKAP13, a RhoA GTPase-specific guanine exchange factor, is a novel regulator of TLR2 signaling. J. Biol. Chem. 282, 35308-35317. 10.1074/jbc.M704426200 [DOI] [PubMed] [Google Scholar]
- Sorvina A., Brooks D. A., Ng Y. S., Bader C. A., Weigert R. and Shandala T. (2013). Bacterial challenge initiates endosome-lysosome response in Drosophila immune tissues. IntraVital 2, e23889 10.4161/intv.23889 [DOI] [Google Scholar]
- Stanley A. C., Lieu Z. Z., Wall A. A., Venturato J., Khromykh T., Hamilton N. A., Gleeson P. A. and Stow J. L. (2012). Recycling endosome-dependent and -independent mechanisms for IL-10 secretion in LPS-activated macrophages. J. Leukoc. Biol. 92, 1227-1239. 10.1189/jlb.0412191 [DOI] [PubMed] [Google Scholar]
- Stow J. L. and Murray R. Z. (2013). Intracellular trafficking and secretion of inflammatory cytokines. Cytokine Growth Factor Rev. 24, 227-239. 10.1016/j.cytogfr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Wall E. A., Zavzavadjian J. R., Chang M. S., Randhawa B., Zhu X., Hsueh R. C., Liu J., Driver A., Bao X. R., Sternweis P. C. et al. (2009). Suppression of LPS-induced TNF-alpha production in macrophages by cAMP is mediated by PKA-AKAP95-p105. Sci. Signal. 2, ra28 10.1126/scisignal.2000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. O. (2002). Cutting edge: A-kinase anchor proteins are involved in maintaining resting T cells in an inactive state. J. Immunol. 168, 5392-5396. 10.4049/jimmunol.168.11.5392 [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T., Wilsch-Bräuninger M. and González-Gaitán M. (2003). Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J. Cell Biol. 161, 609-624. 10.1083/jcb.200211087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong B., Bayat V., Jaiswal M., Zhang K., Sandoval H., Charng W.-L., Li T., David G., Duraine L., Lin Y.-Q. et al. (2012). Crag is a GEF for Rab11 required for rhodopsin trafficking and maintenance of adult photoreceptor cells. PLoS Biol. 10, e1001438 10.1371/journal.pbio.1001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-M., Walsh B., Mitchell C. A. and Rowe T. (2005). TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem. Biophys. Res. Commun. 335, 154-161. 10.1016/j.bbrc.2005.07.070 [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., Fish M., Acar M., Hoskins R. A., Bellen H. J. and Scott M. P. (2007). Thirty-one flavors of Drosophila rab proteins. Genetics 176, 1307-1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita A., Zhang Y., Pedersen L., Darden T. and Birnbaumer L. (2010). Obligatory role in GTP hydrolysis for the amide carbonyl oxygen of the Mg(2+)-coordinating Thr of regulatory GTPases. Proc. Natl. Acad. Sci. USA 107, 9596-9601. 10.1073/pnas.1004803107 [DOI] [PMC free article] [PubMed] [Google Scholar]