Abstract
Disease or injury to articular cartilage results in loss of extracellular matrix components which can lead to the development of osteoarthritis (OA). To better understand the process of disease development, there is a need for evaluation of changes in cartilage composition without the requirement of extensive sample preparation. Near infrared (NIR) spectroscopy is a chemical investigative technique based on molecular vibrations that is increasingly used as an assessment tool for studying cartilage composition. However, the assignment of specific molecular vibrations to absorbance bands in the NIR spectrum of cartilage, which arise from overtones and combinations of primary absorbances in the mid infrared (MIR) spectral region, has been challenging. In contrast, MIR spectroscopic assessment of cartilage is well-established, with many studies validating the assignment of specific bands present in MIR spectra to specific molecular vibrations. In the current study, NIR imaging spectroscopic data were obtained for compositional analysis of tissues that served as an in vitro model of OA. MIR spectroscopic data obtained from the identical tissue regions were used as the gold-standard for collagen and proteoglycan (PG) content. MIR spectroscopy in transmittance mode typically requires a much shorter pathlength through the sample (≤10 microns thick) compared to NIR spectroscopy (millimeters). Thus, this study first addressed the linearity of small absorbance bands in the MIR region with increasing tissue thickness, suitable for obtaining a signal in both the MIR and NIR regions. It was found that the linearity of specific, small MIR absorbance bands attributable to the collagen and PG components of cartilage (at 1336 and 856 cm−1, respectively) are maintained through a thickness of 60 μm, which was also suitable for NIR data collection. MIR and NIR spectral data were then collected from 60 μm thick samples of cartilage degraded with chondroitinase ABC as a model of OA. Partial least squares (PLS) regression using NIR spectra as input predicted the MIR-determined compositional parameters of PG/collagen within 6% of actual values. These results indicate that NIR spectral data can be used to assess molecular changes that occur with cartilage degradation, and further, the data provide a foundation for future clinical studies where NIR fiber optic probes can be used to assess the progression of cartilage degradation.
Keywords: Near infrared spectroscopy, Mid infrared spectroscopy, Articular cartilage, Collagen, Proteoglycan, Water, Partial least squares regression
Graphical Abstract
1. Introduction
The function of articular cartilage is to provide a near frictionless, load bearing surface for smooth articulation of joints [1]. Cartilage matrix, comprised primarily of type II collagen and proteoglycans (PGs), is prone to degeneration due to age, disease and or trauma, and the degeneration of matrix leads to the development of a painful condition called osteoarthritis (OA). Current clinical practice for identification of cartilage degeneration and severity of OA involves identification of symptoms related to OA progression, including pain and loss of motion. Subsequently, plain radiographs, and at times arthroscopic investigations, are used to confirm the diagnosis [2]. However, these techniques only offer a macroscopic or external view of the articular surface and are not useful in assessing cartilage molecular composition [3,4]. To monitor the progression of disease, advanced imaging techniques such as computer tomography (CT), ultrasound and magnetic resonance imaging (MRI) are used, but these techniques offer low specificity and sensitivity to macromolecular changes in cartilage degeneration [5–11]. Histological and biochemical assessment of biopsies offer insights into changing tissue composition during OA, but are not typically performed clinically [12,13]. In vitro studies have used mechanical testing as a tool for assessing OA progression, and studies have shown correlations with biochemical and histological findings [14]. However, mechanical testing only provides bulk properties of tissue and, as in the case of histological and biochemical assessments, involves destruction of samples. Thus, there is a pressing need for a technique that can reliably assess changes in the molecular composition of articular cartilage during OA progression with improved specificity and sensitivity to macromolecular content. Previous studies have shown that the degradation of proteoglycan is a hallmark of OA and cartilage degeneration [15,16]. Naturally, there is a turnover of PG in cartilage tissue, but in early stages of OA, PG degradation rate exceeds the regeneration of the molecules, causing tissue degradation. There is a significant inter-individual variation in changes in PG content in early stages of OA [16]. However, it is likely that clinical evaluation of changes in composition within joint tissue from an individual patient could identify early disease.
Fourier transform infrared imaging spectroscopy (FT-IRIS) has been used extensively to evaluate the composition of native and degraded cartilage, including semi-quantitative changes in collagen and PG concentration [17–23]. As each molecule has a unique absorption profile across the infrared spectral range, quantification of specific molecules is possible using the Beer–Lambert law, which relates the concentration of a chemical substance in a sample to the amount of infrared radiation absorbed [24]. Although there is an abundance of literature on the use of FT-IRIS in the mid infrared (MIR) range for cartilage assessment, there are far fewer studies that utilize the NIR spectral region, primarily due to the fact that the NIR spectral characteristics of tissues are not as distinct as those observed in the MIR region. The assignment of specific molecular vibrations to absorbance bands in the NIR spectrum of cartilage, which arise from overtones and combinations of primary absorbances in the MIR spectral region, has been challenging, and many absorbances attributable to matrix components are not unique to either collagen or PG [25]. However, the depth of penetration of NIR radiation is greater than MIR radiation (on the order of millimeters to centimeters, compared to microns) which potentially makes it more useful for investigations of intact tissue involving regions deeper than the superficial zone of cartilage [26–28]. Another advantage of studies in the NIR spectral range is that little to no sample preparation is required. This is in contrast to the use of FT-IRIS in the MIR range, which requires extensive sample preparation, typically embedding and thin sectioning of tissues.
NIR spectra of cartilage are dominated by water absorbances at 5200 cm−1 (bound and free water) [29,30], with smaller absorbances from matrix peaks present in the spectral range of interest between 4000 and 6000 cm−1 [25,30]. Changes in these absorbance bands have been shown to occur with tissue degradation [26,31], which can be very heterogeneous. To date, NIR evaluation of cartilage has been performed primarily with fiber optic probes on tissues with thicknesses in the millimeter range, with few studies directly comparing spectral data to gold standard biochemical measurements [25,32,33]. In contrast to NIR fiber optic data, collection of NIR spectral imaging data provides the opportunity to obtain NIR spectra from tissues at high spatial resolution, and enables direct correlation of those spectra with compositional information from MIR spectral data from the same tissues. In the current study, we obtained NIR spectral imaging data from an in vitro model of OA. Concomitantly, we also collected MIR spectral imaging data from the same locations, and used those data as the gold standard for molecular composition. Here, MIR data provides a faster and more quantitative tool to validate NIR measurements compared to traditional gold standard methods of biochemical or histological evaluations. Collection of MIR spectral data in transmittance mode typically requires a much shorter path length through the sample (≤10 microns thick) compared to NIR spectroscopy (mms). Thus, we first addressed the linearity of small absorbance bands in the MIR region with increasing tissue thickness to identify a suitable thickness for simultaneously obtaining a signal in the MIR and NIR regions. The optimal thickness was then used for sequential collection of mid and near infrared spectral imaging data. This study provides data that can serve as a foundation for eventual in vivo NIR spectroscopic fiber optic applications for monitoring cartilage tissue compositional changes for clinical applications.
2. Methods
2.1. Cartilage tissue preparation
Juvenile bovine knee joints (Research 87, Boylston, MA) and nasal snouts (Green Village Packing, Green Village, NJ) were obtained within 24 h of slaughter. Bovine nasal cartilage (BNC) and bovine articular cartilage (BAC) plugs (both hyaline cartilage) were harvested using a 5 mm biopsy punch (Ted Pella, Redding, CA) from nasal septa, and from femoral-patellar grooves and medial condyles, respectively. The plugs were immersed in a 1 × phosphate buffered saline (PBS) solution containing a cocktail of protease inhibitors (Sigma Aldrich, St. Louis, MO) and stored at −20 °C. BNC plugs were thawed at room temperature, rinsed twice in 1 × PBS, embedded in OCT compound (Sakura Finetek, Torrance, CA) and flash frozen. The BNC plugs were utilized for thickness studies, due to their greater homogeneity of matrix composition compared to articular cartilage. They were embedded such that three fourths of the length of the plug was left free standing, thereby ensuring stability of the base for grasping the tissue in the cryostat while ensuring that the cut tissue sections were free of OCT. Serial sections of nasal cartilage at thicknesses of 10, 20, 40, 60 and 80 μm were obtained using a HM 525 cryotome (Thermo Scientific, Dreieich, Germany). The cryosections were deposited on low-e slides (Kevley Technologies, Chesterland, OH) and silicon (Si) wafers (Fuzere Manufacturing Co., Sunnyvale, CA). The cryosections were air dried for 2 h prior to being imaged using FT-IRIS.
2.2. Enzymatic degradation of articular cartilage (in vitro model of OA)
BAC plugs were allowed to thaw at room temperature before being rinsed twice in PBS (Invitrogen Life Technologies, Gaithersburg, MD). The plugs were subjected to an enzyme treatment procedure to degrade PGs, which produces tissue matrix changes that model OA [34]. Tissue from a control group (n = 7 plugs) were incubated in buffer solution (0.05 M Tris–HCl, 0.06 M sodium acetate, 2 mM PMSF, 2 mM EDTA, 5 mM benzamidine HCl and 10 mM N-ethylmaleimide (Sigma Aldrich, St. Louis, MO), pH 8.0) and tissue from the treatment group (n = 8 plugs) were incubated in buffer solution containing 0.125 units of chondroitinase ABC per mL for 24 h at 25 °C with gentle agitation. The enzymatic degradation was stopped by washing the plugs in PBS for 15 min twice. Five plugs from the enzymatic degradation group and 4 control group plugs were immersed in PBS containing protease inhibitors for mechanical testing. The remaining plugs (n = 3) from each group were embedded in OCT (Tissue-Tek, Sakura Finetek, Torrance, CA) and flash frozen. The plugs were embedded in OCT such that three fourth of the plug was left free standing, and the tissues cryosectioned at 60 μm thickness onto Si wafers for spectral imaging, and at 20 μm thickness onto electrostatically charged plus slides (Fisher Scientific, Fair Lawn, NJ) for histology. Similar to the BNC samples, these tissues were only partially embedded in the OCT to facilitate grasping the sample for sectioning, and there was not any OCT in the actual thin tissue sections. The cryosections on Si wafers were air dried for 2 h prior to being used for FT-IRIS.
2.3. Infrared spectral imaging data collection
The tissues on Si wafers were imaged in transmittance mode and the tissues on low-e slides were imaged in transflectance mode (two common infrared data collection modes) on a Spotlight 400 imaging spectrometer system with a Cassegrain condenser and objective (Perkin Elmer, Waltham, MA). The radiation was focused onto the surface of the tissue for data collection. Three sections per sample thickness were imaged in the spectral range of 4000–780 cm−1 at 8 cm−1 spectral resolution, 25 μm spatial resolution and 8 co-added scans per pixel. Immediately after MIR spectral data acquisition for each section was complete, NIR spectral data were collected from a region of the tissue section. Only a portion of the tissue was imaged in the NIR due to increased data collection time, as a result of increased co-added scans required to obtain a reasonable signal to noise ratio. For NIR spectral images, a spectral range of 6000–4000 cm−1 was used at 8 cm−1 spectral resolution, 25 μm spatial resolution and 32 co-added scans per pixel. Background spectra were collected at 8 cm−1 spectral resolution, 25 μm spatial resolution and 15 co-added scans for the MIR region and 8 cm−1 spectral resolution, 25 μm spatial resolution and 60 co-added scans for the NIR region. The sample images were ratioed to the background spectra for the MIR and NIR regions, respectively. Total spectral image acquisition time was approximately 10 min for the MIR spectral range, and 40 min for the NIR spectral range.
2.4. Histological staining of articular cartilage
Alcian blue staining was used to visualize PG depletion in articular cartilage cryosections [35]. Stained sections were imaged on an inverted light microscope (Olympus, Center Valley, PA) with a 4× objective.
2.5. Mechanical testing of articular cartilage
Mechanical testing of the control and degraded articular cartilage samples was performed to validate the damage to cartilage matrix due to enzymatic degradation. Uniaxial compression tests were performed on an Instron mechanical testing machine (Instron Mini 55, Instron, Norwood, MA) to assess equilibrium and dynamic compressive moduli, respectively. Samples were pre-loaded to 2 N at a rate of 1 mm/min and allowed to relax for 5 min before testing began. For equilibrium modulus, serial stress relaxation analysis was performed at compressive strains of 5, 10, 12.5 and 15% (time per step: 1 min for loading, 9 min for stress relaxation). The linear relationship of equilibrium stresses at each strain step determines equilibrium stiffness. Subsequently, the loading rate dependent dynamic modulus, calculated as the ratio of stress amplitude per strain amplitude, was interpreted from a 1% peak–peak compressive strain oscillation (mean 15%) applied at several frequencies.
Matlab's (Mathworks Inc, Natick, MA) curve fitting toolbox was used to determine (1) the equilibrium stress from each stress relaxation segment through a stretched exponential decay model [36] equilibrium modulus as a first order polynomial as a function of equilibrium stress at each strain step, and (3) oscillatory stress and strain amplitudes from Fourier transforms to calculate dynamic moduli.
2.6. Mid and near infrared spectral data processing
MIR and NIR spectral data were analyzed using Isys 5.0 software (Malvern Instruments, Worcestershire, UK). Univariate data analysis of second derivative peak heights and integrated areas of the MIR 1336 and 856 cm−1 absorbances were used to semi-quantitatively assess the amount of collagen and proteoglycan present in the samples, respectively, and the linearity of the MIR data with increasing tissue thickness. Pre-processing the spectra using second derivatives helped reveal the smaller matrix peaks that were not easily visible in the raw spectra due to the dominance of the water absorbances. An average mean value of all image pixels is reported for all samples groups. The area under the small absorbance peak near 1336 cm−1, that arises from the CH2 side chain vibrations, correlates to collagen present in cartilage [37]. Similarly, the area under the small absorbance peak at 856 cm−1 that arises due to C–O–S stretching linearly increases with aggrecan content [17]. The second derivative peak heights were multiplied by negative one to make the peaks positive. Regions in the spectral images from the control and enzymatically degraded cartilage that displayed a wide range of values in the collagen and PG absorbances were chosen and the corresponding NIR and MIR spectra extracted for analysis using Unscrambler X statistical software (CAMO Software, Oslo, Norway). The scattering effect in NIR images due to different particle sizes was minimized by applying an extended multiplicative scatter correction to the raw spectral data [38]. The scatter corrected spectra were then area normalized and converted to second derivative spectra using a Savitzky Golay differentiation window of 164 cm−1 and 3rd order polynomial. An algorithm was applied to the NIR spectra to filter noisy pixels (outliers) based on ± 2 standard deviations from the mean spectrum of each sample. A total of 672 spectra from the control group and 613 spectra from the chondroitinase ABC-treated group were analyzed. The second derivative peak heights at 5200 cm−1 were assessed to reflect the relative amount of water in the tissues [30]. As the tissues had been air dried for 2 h prior to imaging, and then exposed to the atmosphere for an additional 50 min while data were collected, the majority of the free water had evaporated from the sample, and only bound water remained [30].
Partial least square (PLS) regression, a multivariate variable reduction method, was used to predict the MIR parameters from the NIR scatter-corrected second derivative spectral data [39]. PLS models were validated using cross validation that included spectra from combined control and chondroitinase groups (n = 922 spectra). A random column of spectra (4 spectra per column) were kept out of the calibration model for each iteration of the cross validation. An independent test set comprising of a total of 363 spectra from both control and treatment groups was used to test the performance of the model. The quality of the prediction model was evaluated based on the root mean square error of prediction (RMSEP) and the R2 of actual vs. predicted values.
3. Results and discussion
3.1. Linearity of MIR parameters with increasing thickness
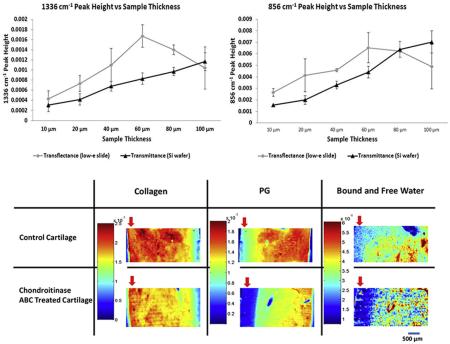
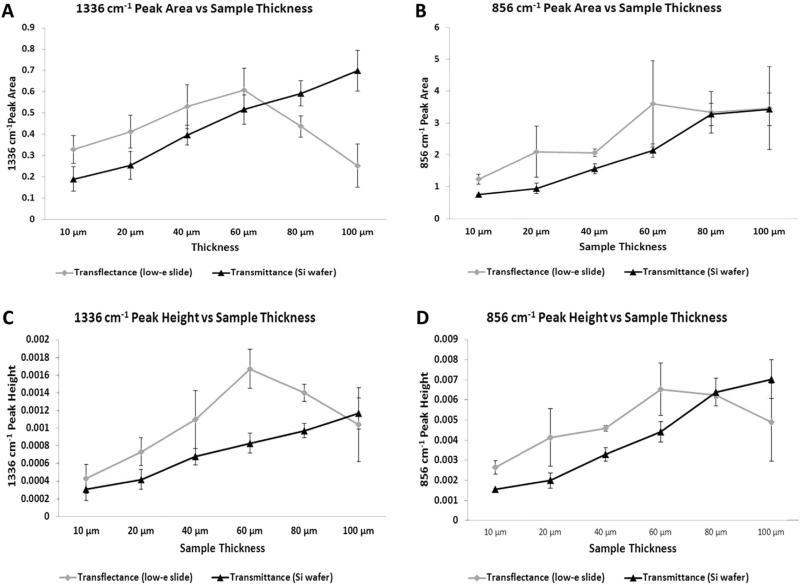
As sample thickness (pathlength) increases, the MIR absorbances with a high absorptivity coefficient can approach saturation. To address this issue in thicker tissue sections, we hypothesized that the intensity of absorbance bands with smaller absorptivity coefficients, such as those at 1336 and 856 cm−1, corresponding to collagen and PG respectively, would remain linear with increasing tissue thickness. In transflectance mode, there was a general increase in integrated peak areas and second derivative peak heights of both the 1336 and 856 cm−1 absorbances from 10 to 60 microns, although large standard deviations were present across multiple sections from the same sample (Fig. 1).
Fig. 1.
Average peak areas (A,B) and peak heights (C,D) of MIR parameters at 1336 cm−1 and 856 cm−1 with varying sample thicknesses and data collection modes (transflectance and transmittance).
Interestingly, in transflectance, the peak areas and peak heights decreased with tissue thickness greater than 60 μm, suggesting a breakdown of linearity in Beer–Lambert's law [24]. A contributor to this is likely the effect of radiation going through a “double pass” through the sample in transflectance mode [40]. In contrast, the peak areas and peak heights from MIR data of samples imaged on silicon wafers in transmittance exhibit increased absorbance with increasing thickness for the full range of thickness investigated through 100 μm (Fig. 1). Although the standard deviations at each thickness from multiple sections are low initially, the values are generally higher when section thickness is greater than 60 μm.
The peak areas and peak heights obtained from tissues on Si wafers in transmittance mode exhibit a more uniform trend with increasing sample thickness as compared to transflectance data from low-e slides. Nonetheless, the change in absorbance is not exactly linear. As BNC is not a completely homogeneous mixture, this non-linearity might be due to changes in concentrations of collagen and PG with increasing thickness. Increased scattering with thicker tissue sections leads to non-linear changes in absorbance, which might also contribute to the observed non-linearity. The process of microsectioning using a cryotome also can result in small deviations in sample thickness with increasing thickness. Together, these data supported the use of transmittance sampling in the thickness range of 20–60 μm, which demonstrated increases in absorbances and peak heights.
3.2. Optimization of sample thickness for MIR and NIR data acquisition in transmittance
The ratio of changes in peak areas of the 1336 and 856 cm−1 absorbances with increasing sample thickness were similar to those obtained with peak height ratios, supporting the use of either analysis (Table 1). Nonetheless, it is interesting to note that although thickness increased three-fold from 20 to 60 microns, the absorbances and peak heights only increased ~ two-fold. This likely indicates a loss of radiation due to scattering.
Table 1.
Average ratio of MIR parameter values obtained from 60 to 20 micron thick tissues imaged in transmittance on silicon wafers (N = 6).
| MIR data Parameters | Parameter ratio ± SD (value at 60 μm/value at 20 μm) |
|---|---|
| 1336 cm−1 peak area | 2.03 ± 0.200 |
| 1336 cm−1 second derivative peak height | 1.98 ± 0.200 |
| 856 cm−1 peak area | 2.25 ± 0.116 |
| 856 cm−1 second derivative peak height | 2.23 ± 0.131 |
To select the appropriate thickness for NIR spectral imaging, sample thicknesses of 20, 40, 60 and 80 μm were imaged and the signal to noise ratios (SNR) were evaluated (Table 2). The NIR SNR from the 60 μm tissue sections was 2 times greater than the SNR of the 40 μm sections with better peak resolution (data not shown). The 60 μm sections had lower SNR than 80 μm sections, but the resolution of the peak absorbances were comparable to that at 80 μm. Since use of 60 μm thick slices would result in a smaller amount of tissue being consumed for cryosectioning compared to 80 μm thick slices, a 60 μm sample thickness was chosen for further analysis, and for comparison of second derivative peak heights at 1336 and 856 cm−1 obtained from tissue sections imaged on Si wafers in transmittance mode. Data from samples sectioned at 60 μm are used to show the compositional and spectral changes in the figures.
Table 2.
Signal-to-noise (SNR) ratios of near infrared spectral images collected in transmittance at varying thicknesses (N = 3 images per thickness, 1000 pixels each image).
| Sample thickness | Average SNR | Standard deviation |
|---|---|---|
| 20 μm | 400 | ±136 |
| 40 μm | 700 | ±148 |
| 60 μm | 1400 | ±211 |
| 80 μm | 2200 | ±301 |
3.3. Histological and mechanical evaluation of enzymatically degraded articular cartilage
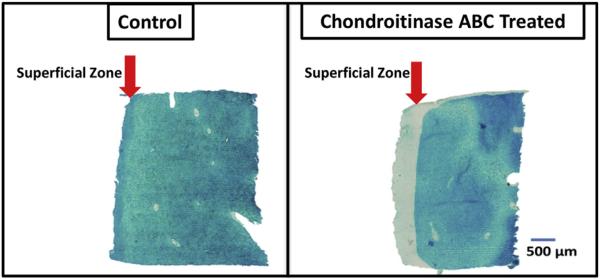
Many studies have employed enzymatic cartilage degradation, including the use of chondroitinases, to simulate osteoarthritic conditions in vitro [14,41–47]. Chondroitinases, a family of endoglycosidases that act on glycoproteins, specifically proteoglycans, have been extensively used in articular cartilage studies to degrade proteoglycans [48–53]. In this study we treated articular cartilage plugs with chondroitinase ABC to leach PGs and create regions of low PG content in tissue. Histological staining with alcian blue, which stains mucopolysaccharides, showed depletion of PGs from the superficial surface and edges of the treated samples (Fig. 2). The surface of the enzyme treated samples also showed severe fibrillation due to loss of structural integrity. Some minor fibrillations, attributed to cutting artifacts, were also present on the control surface. In previous studies where cartilage enzymatic degradation was used to model OA, the decrease in PG content with enzymatic degradation ranged from 50% [41] to between 5 and 18% [46,47]. Although the specific amount of degradation was not quantified here, the histological images show depleted PG through approximately 20% of the tissue, which is in the range of other studies.
Fig. 2.
Representative light microscopy images of alcian blue stained control (left) and chondroitinase ABC treated (right) articular cartilage samples. The arrow indicates the superficial zone. As the fluid diffuses into the sample from the outer edges, digestion removes the proteoglycan from the superficial zone to the greatest extent. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
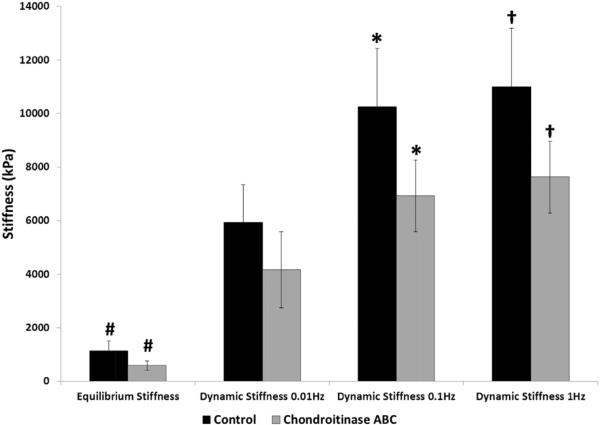
Mechanical testing revealed a significant decrease in mechanical properties of the enzymatically-degraded cartilage plugs compared to controls (Fig. 3). The chondroitinase ABC treated samples exhibited significantly lower equilibrium stiffness values, as well as dynamic stiffness values, at 1 and 0.1 Hz. Compressive equilibrium and dynamic stiffness of a viscoelastic material explain the mechanical behavior of a material under extended periods of loading and repeated loading, respectively. The loading forces used in the mechanical testing are analogous to standing for a period of time in the case of equilibrium loading, or motion at various rates in the case of dynamic loading. The lower values of equilibrium and dynamic stiffness in enzyme treated samples can be attributed to significant leaching of the PGs from the surfaces. Proteoglycans are negatively charged and entrap water in the collagen mesh network, enabling the cartilage to resist compressive forces. Loss of PGs results in loss of bound water from the cartilage matrix, decreasing the ability of cartilage matrix to effectively counter the loading force. Taken together, the histological and mechanical testing results suggest that there was a significant difference in the composition and function of enzymatically degraded vs. control cartilage, and confirms that a clinical level of degradation exists in these samples.
Fig. 3.
Mechanical properties of enzymatically treated articular cartilage samples of control (N = 4) and chondroitinase ABC digested (N = 5) sample groups. *, #, † indicate significant differences between groups with p < 0.05.
3.4. Mid and near infrared spectral imaging analysis of degraded articular cartilage
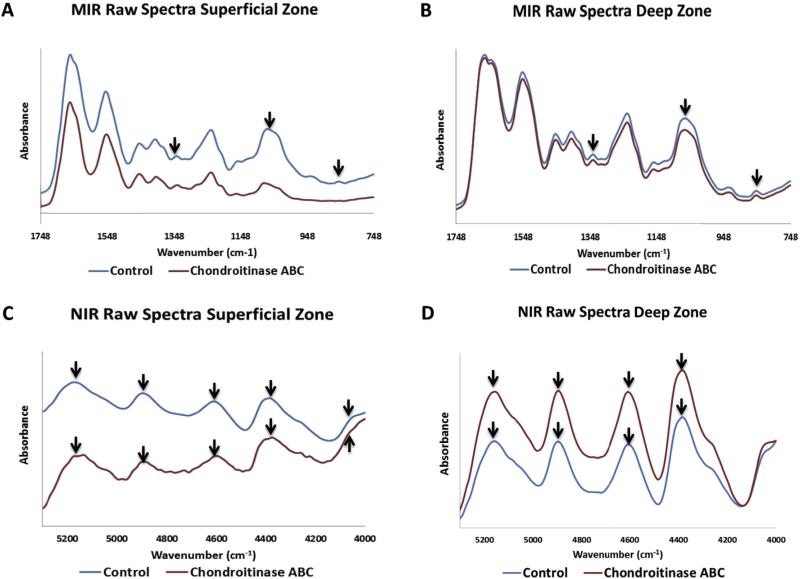
As expected from the contrast in the histology images, MIR spectra of control and enzyme treated samples show differences in the proteoglycan content, in particular in the superficial zone (Fig. 4A). The overall contour of the MIR spectra from the deep zone is similar for control and chondroitinase-treated samples (4B). The NIR spectral contours are also similar, but with slight differences in baseline offsets (Fig. 4C, D). The distribution of PG (856 cm−1 second derivative peak heights) in the control samples increased through the middle and deep zone (Fig. 5), in accordance with literature reports of matrix distribution in articular cartilage [1], and in agreement with the alcian blue histology for PG (Fig. 2). The distribution of collagen (1336 cm−1) followed the opposite trend, with higher content in the superficial zone and middle zone compared to the deep zone of cartilage (Fig. 5).
Fig. 4.
Average MIR (A,B) and NIR (C,D) spectra from the superficial and deep zones of control and chondroitinase ABC-treated samples. Arrows indicate peaks of interest, for MIR (collagen, 1336 cm−1, and PG, 856 cm−1) and NIR (water, 5200 cm−1 and matrix peaks, 4060, 4390, 4600 and 4890 cm−1). PG depletion is evident in the chondroitinase ABC-treated superficial zone MIR spectra compared to control. Imaging spectroscopy was done in transmittance mode from tissues sectioned on Si wafer substrates.
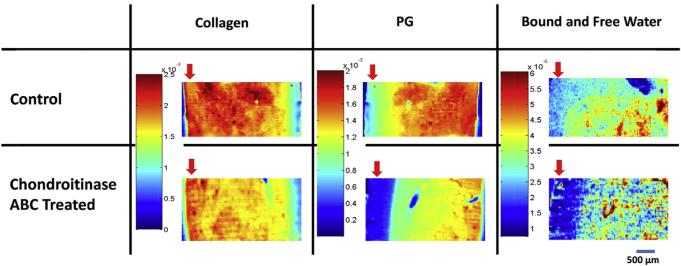
Fig. 5.
Representative 2nd derivative peak height spectral images based on the MIR collagen (assessed at 1336 cm−1) and PG absorbance (assessed at 856 cm−1), and NIR water content (assessed at 5200 cm−1). Component distribution is shown in control and chondroitinase-treated ABC treated samples. The arrow indicates the superficial zone. Imaging spectroscopy was done in transmittance mode from tissues sectioned on Si wafer substrates.
Compared to control samples, the chondroitinase ABC treated samples showed a marked decrease in the peak height values of 856 cm−1 (PG content), especially in the superficial zone (Fig. 5). This PG depletion pattern is due to the fact that the enzyme solution diffuses into the sample from the outer edges. The NIR imaging spectra displayed broad matrix peaks or shoulders at 4060, 4390, 4600 and 4890, and a water peak at 5200 cm−1 (Fig. 4C and D). Regional differences in water content were evident between control and chondroitinase ABC treated samples, primarily in the superficial zone (Fig. 5). As expected, the water distribution generally follows the PG distribution in the MIR and NIR spectral images [54].
3.5. Validation of NIR spectral imaging data with MIR spectral imaging data– PLS analysis
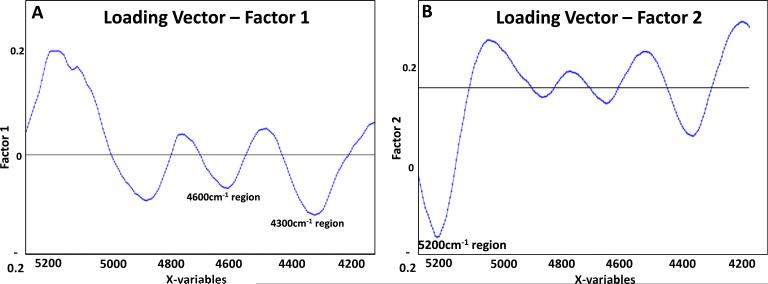
Multivariate analyses (e.g. PLS s) are powerful techniques for investigation of spectral data when overlapping absorbances are present, or if absorbances cannot be assigned to specific components, as is often the case for NIR spectral data. Although the water absorbances in cartilage have been assigned, many of the absorbances attributable to matrix components are not unique to either collagen or PG, and so cannot be utilized as a surrogate for concentration. However, based on the MIR-determined peak heights, we can predict the relative amounts of collagen and PG, as well as the ratio of collagen to PG, from NIR spectra using the entire 4000–6000 cm−1 spectral range and PLS modeling (Table 3). The best prediction was for the ratio of PG/collagen, with a percent error of 6%, while the prediction errors for either component separately was ~10%. The best prediction error presented in this work was in the same range, but slightly improved, as previously reported for our PLS models from other studies, where articular cartilage composition was predicted based on mixtures of pure components of collagen and chondroitin sulfate [25], and engineered cartilage composition was predicted based on biochemical evaluations [55]. In the current data set, spectra from control and enzymatically degraded tissues were used both to create the model and for the independent prediction set. This reflects the most physiologically relevant case, where tissues being investigated would likely have some regions that are “normal”, and some regions that are degraded or otherwise diseased. In PLS multivariate analysis, original data (infrared spectra in this study) are reduced based on a new coordinate system with orthogonal components (factors) that are able to explain the maximum amount of variation among samples. Loading vectors reflect these factors, and can be correlated to different regions of the original dataset. PLS loading vectors in the current study were evaluated for all three models presented in Table 3 to see whether there are specific regions of the infrared spectra that best explain the differences among samples, and underlie the correlation between the MIR and NIR spectral data. Loading vectors associated with Factors 1 and 2 were able to explain at least 63% of variation in the models (ranging from 63% to 87%), where Factor 1 has large contributions at 4300 cm−1 and 4600 cm−1, regions that are correlated to matrix components proteoglycan and collagen, and Factor 2 is dominated by the water peak at 5200 cm−1 (Fig. 6). Notwithstanding these encouraging results, it is likely that the error in the PLS model predictions could be minimized by using separate training and prediction sets from a wider range of tissue composition, or alternatively by the use of non-linear regression techniques, which were not investigated in this study.
Table 3.
PLS models used for predicting MIR parameters from NIR scatter-corrected, second derivative spectral data. Models were created based on a training set of 488 spectra from two control samples and 434 spectra of two enzymatically-digested samples. Prediction was conducted on a test set of 184 spectra of one control sample and 179 spectra of one enzymatically-digested sample.
| Peak height or peak height ratio | RMSEP | Range (peak height or ratio) | RMSEP (percent of range) | R-squared (prediction vs. Experiment) |
|---|---|---|---|---|
| 1336 cm−1 | 3.6 × 10−4 | 5.3 × 10−4 – 4.5 × 10−4 | 9% | 0.55 |
| 856 cm−1 | 3.1 × 10−4 | 9.7 × 10−5 – 2.8 × 10−3 | 11% | 0.54 |
| 856/1336 cm−1 | 2.2 × 10−1 | 3.6 × 10−2 – 3.4 | 6% | 0.46 |
Fig. 6.
Loading vectors associated with Factors 1 (A) and 2 (B) explained 27% and 36% of the variation in the model, respectively, where Factor 1 has a greater contribution from the matrix peaks, and Factor 2 is dominated by the water peak at 5200 cm−1.
In summary, the minimum tissue thickness required for acquisition of MIR and NIR spectral data from the same sample were established. By analyzing the MIR spectral imaging data of BNC tissues of varying thickness, the linear range of small absorbances peaks attributable to collagen and PG was demonstrated. MIR spectroscopy faces a penetration depth limitation in clinical applications. However, MIR measurements of cartilage matrix components are well defined from previous studies, and provide a robust tool for validation of NIR measurements. Thus, NIR imaging spectroscopy analysis was validated using MIR spectral data in this study to provide a foundation for eventual clinical applications using a fiber optic probe modality of NIR spectroscopy during arthroscopic procedures, as have recently been described. Future fiber optic studies will have several challenges not addressed in the current study, including evaluation of tissues in a physiological environment where the water content is much greater, and understanding the natural variability of cartilage composition due to age, gender, and anatomic location. Nevertheless, the results of the current study are encouraging, and support further NIR spectral studies of pathologic cartilage changes [26,56–58].
HIGHLIGHTS.
Near infrared (NIR) and mid infrared (MIR) spectral imaging were used to assess articular cartilage composition from the identical tissue region.
An optimal thickness was determined for obtaining NIR and MIR spectral data simultaneously from the same tissue.
NIR spectra in 4000–6000 cm−1 range were used to build a PLS model to predict MIR-derived cartilage compositional parameters within 6% of actual values.
Results support use of NIR spectroscopy for assessment of cartilage composition.
Acknowledgments
This work was supported by NIH R01AR056145 and EB000744 grants.
References
- 1.Fox AJS, Bedi A, Rodeo SA. The basic science of articular cartilage: structure, composition, and function. Sports Health A Multidiscip. Approach. 2009;1(6):461–468. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377(9783):2115–2126. doi: 10.1016/S0140-6736(11)60243-2. [DOI] [PubMed] [Google Scholar]
- 3.Oakley SP, et al. Arthroscopy – a potential “gold standard” for the diagnosis of the chondropathy of early osteoarthritis. Osteoarthr. Cartil. 2005;13(5):368–378. doi: 10.1016/j.joca.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Bekkers J, et al. Diagnostic modalities for diseased articular cartilage—from defect to degeneration a review. Cartilage. 2010;1(3):157–164. doi: 10.1177/1947603510364539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal PN, et al. Contrast enhanced computed tomography can predict the glycosaminoglycan content and biomechanical properties of articular cartilage. Osteoarthr. Cartil. 2010;18((2):184–191. doi: 10.1016/j.joca.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Keen HI, Conaghan PG. Ultrasonography in osteoarthritis. Radiol. Clin. North Am. 2009;47(4):581–594. doi: 10.1016/j.rcl.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Eckstein F, Burstein D, Link TM. Quantitative MRI of cartilage and bone: degenerative changes in osteoarthritis. NMR Biomed. 2006;19(7):822–854. doi: 10.1002/nbm.1063. [DOI] [PubMed] [Google Scholar]
- 8.Yusuf E, et al. Do knee abnormalities visualised on MRI explain knee pain in knee osteoarthritis? A systematic review. Ann. Rheum. Dis. 2011;70(1):60–67. doi: 10.1136/ard.2010.131904. [DOI] [PubMed] [Google Scholar]
- 9.Hunter DJ, et al. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score) Osteoarthr. Cartil. 2011;19((8):990–1002. doi: 10.1016/j.joca.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutter R, et al. Hip MRI: how useful is intraarticular contrast material for evaluating surgically proven lesions of the labrum and articular cartilage? AJR Am. J. Roentgenol. 2014;202(1):160–169. doi: 10.2214/AJR.12.10266. [DOI] [PubMed] [Google Scholar]
- 11.Siebelt M, et al. Clinically applied CT arthrography to measure the sulphated glycosaminoglycan content of cartilage. Osteoarthr. Cartil. 2011;19(10):1183–1189. doi: 10.1016/j.joca.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Pritzker KP, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthr. Cartil. 2006;14((1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Hoemann CD. Cartilage and Osteoarthritis. Springer; 2004. Molecular and biochemical assays of cartilage components; pp. 127–156. [DOI] [PubMed] [Google Scholar]
- 14.Knecht S, Vanwanseele B, Stussi E. A review on the mechanical quality of articular cartilage - implications for the diagnosis of osteoarthritis. Clin. Biomech. (Bristol, Avon) 2006;21(10):999–1012. doi: 10.1016/j.clinbiomech.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Brown CP, et al. Characterization of early stage cartilage degradation using diffuse reflectance near infrared spectroscopy. Phys. Med. Biol. 2011;56(7):2299–2307. doi: 10.1088/0031-9155/56/7/024. [DOI] [PubMed] [Google Scholar]
- 16.Lohmander S. Proteoglycans of joint cartilage. Structure, function, turnover and role as markers of joint disease. Baillieres Clin. Rheumatol. 1988;2(1):37–62. doi: 10.1016/s0950-3579(88)80004-9. [DOI] [PubMed] [Google Scholar]
- 17.Camacho NP, et al. FTIR microscopic imaging of collagen and proteoglycan in bovine cartilage. Biopolymers. 2001;62(1):1–8. doi: 10.1002/1097-0282(2001)62:1<1::AID-BIP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.West PA, et al. Fourier transform infrared spectral analysis of degenerative cartilage: an infrared fiber optic probe and imaging study. Appl. Spectrosc. 2004;58(4):376–381. doi: 10.1366/000370204773580194. [DOI] [PubMed] [Google Scholar]
- 19.Bi X, et al. Fourier transform infrared imaging and MR microscopy studies detect compositional and structural changes in cartilage in a rabbit model of osteoarthritis. Anal. Bioanal. Chem. 2007;387(5):1601–1612. doi: 10.1007/s00216-006-0910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi X, et al. Fourier transform infrared imaging spectroscopy investigations in the pathogenesis and repair of cartilage. Biochim. Biophys. Acta. 2006;1758(7):934–941. doi: 10.1016/j.bbamem.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Boskey A, Pleshko Camacho N. FT-IR imaging of native and tissue-engineered bone and cartilage. Biomaterials. 2007;28(15):2465–2478. doi: 10.1016/j.biomaterials.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spahn G, et al. Evaluation of Cartilage Degeneration by Near Infrared Spectroscopy (NIRS): Methodical Description and Systematic Literature Review. Zeitschrift fur Orthopadie und Unfallchirurgie. 2013 doi: 10.1055/s-0032-1327873. [DOI] [PubMed] [Google Scholar]
- 23.Hanifi A, et al. Infrared fiber optic probe evaluation of degenerative cartilage correlates to histological grading. Am. J. Sports Med. 2012;40(12):2853–2861. doi: 10.1177/0363546512462009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith BC. Fundamentals of Fourier Transform Infrared Spectroscopy. CRC press; 2011. [Google Scholar]
- 25.Palukuru UP, McGoverin CM, Pleshko N. Assessment of hyaline cartilage matrix composition using near infrared spectroscopy. Matrix Biol. 2014;38:3–11. doi: 10.1016/j.matbio.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Afara I, et al. Non-destructive evaluation of articular cartilage defects using near-infrared (NIR) spectroscopy in osteoarthritic rat models and its direct relation to Mankin score. Osteoarthr. Cartil. 2012;20(11):1367–1373. doi: 10.1016/j.joca.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Afara I, Singh S, Oloyede A. Application of near infrared (NIR) spectroscopy for determining the thickness of articular cartilage. Med. Eng. Phys. 2012 doi: 10.1016/j.medengphy.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Brown CP, et al. Acoustic, mechanical and near-infrared profiling of osteoarthritic progression in bovine joints. Phys. Med. Biol. 2012;57(2):547–559. doi: 10.1088/0031-9155/57/2/547. [DOI] [PubMed] [Google Scholar]
- 29.Luck WAP. Proceedings of the International Symposium Held at Marburg in July 1973. Verlag Chemie; 1974. Structure of water and aqueous solutions. [Google Scholar]
- 30.Padalkar MV, Spencer RG, Pleshko N. Near Infrared Spectroscopic Evaluation of Water in Hyaline Cartilage. Ann. Biomed. Eng. 2013;41(11):2426–2436. doi: 10.1007/s10439-013-0844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown CP, et al. Diffuse reflectance near infrared spectroscopy can distinguish normal from enzymatically digested cartilage. Phys. Med. Biol. 2009;54(18):5579–5594. doi: 10.1088/0031-9155/54/18/015. [DOI] [PubMed] [Google Scholar]
- 32.Spahn G, et al. Evaluation of cartilage defects with near-infrared spectroscopy (NIR): an ex vivo study. Med. Eng. Phys. 2008;30(3):285–292. doi: 10.1016/j.medengphy.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 33.McGoverin CM, et al. Non-destructive Assessment of Engineered Cartilage Composition by Near Infrared Spectroscopy. Ann. Biomed. Eng. 2016 doi: 10.1007/s10439-015-1536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt MB, et al. Effects of proteoglycan extraction on the tensile behavior of articular cartilage. J. Orthop. Res. 1990;8(3):353–363. doi: 10.1002/jor.1100080307. [DOI] [PubMed] [Google Scholar]
- 35.Potter K, et al. Imaging of collagen and proteoglycan in cartilage sections using Fourier transform infrared spectral imaging. Arthritis Rheum. 2001;44(4):846–855. doi: 10.1002/1529-0131(200104)44:4<846::AID-ANR141>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.June RK, et al. Cartilage stress-relaxation is affected by both the charge concentration and valence of solution cations. Osteoarthr. Cartil. 2009;17(5):669–676. doi: 10.1016/j.joca.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West PA, et al. Fourier transform infrared imaging spectroscopy analysis of collagenase-induced cartilage degradation. J. Biomed. Opt. 2005;10(1):14015. doi: 10.1117/1.1854131. [DOI] [PubMed] [Google Scholar]
- 38.Rinnan Å, Berg F.v.d, Engelsen SB. Review of the most common preprocessing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009;28(10):1201–1222. [Google Scholar]
- 39.Esbensen K, et al. Multivariate Data Analysis-in Practice: an Introduction to Multivariate Data Analysis and Experimental Design. Multivariate Data Analysis. 2002 [Google Scholar]
- 40.Hanifi A, et al. Differences in infrared spectroscopic data of connective tissues in transflectance and transmittance modes. Anal. Chim. Acta. 2013;779:41–49. doi: 10.1016/j.aca.2013.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grenier S, Bhargava MM, Torzilli PA. An in vitro model for the pathological degradation of articular cartilage in osteoarthritis. J. Biomech. 2014;47(3):645–652. doi: 10.1016/j.jbiomech.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katta J, et al. Biotribology of articular cartilage–a review of the recent advances. Med. Eng. Phys. 2008;30(10):1349–1363. doi: 10.1016/j.medengphy.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Ernst S, et al. Enzymatic Degradation of Glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 1995;30(5):387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 44.Tortorella MD, et al. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthr. Cartil. 2001;9(6):539–552. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- 45.Toyras J, et al. Characterization of enzymatically induced degradation of articular cartilage using high frequency ultrasound. Phys. Med. Biol. 1999;44(11):2723–2733. doi: 10.1088/0031-9155/44/11/303. [DOI] [PubMed] [Google Scholar]
- 46.Wheaton AJ, et al. Detection of changes in articular cartilage proteoglycan by T(1rho) magnetic resonance imaging. J. Orthop. Res. 2005;23(1):102–108. doi: 10.1016/j.orthres.2004.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borthakur A, et al. Sensitivity of MRI to proteoglycan depletion in cartilage: comparison of sodium and proton MRI, Osteoarthr. Cartil. 2000;8(4):288–293. doi: 10.1053/joca.1999.0303. [DOI] [PubMed] [Google Scholar]
- 48.Yamagata T, et al. Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 1968;243(7):1523–1535. [PubMed] [Google Scholar]
- 49.Frank EH, et al. Streaming potentials: a sensitive index of enzymatic degradation in articular cartilage. J. Orthop. Res. 1987;5(4):497–508. doi: 10.1002/jor.1100050405. [DOI] [PubMed] [Google Scholar]
- 50.Lyyra T, et al. Experimental validation of arthroscopic cartilage stiffness measurement using enzymatically degraded cartilage samples. Phys. Med. Biol. 1999;44(2):525–535. doi: 10.1088/0031-9155/44/2/017. [DOI] [PubMed] [Google Scholar]
- 51.Korhonen RK, et al. Fibril reinforced poroelastic model predicts specifically mechanical behavior of normal, proteoglycan depleted and collagen degraded articular cartilage. J. Biomech. 2003;36(9):1373–1379. doi: 10.1016/s0021-9290(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 52.Rieppo J, et al. Structure-function relationships in enzymatically modified articular cartilage. Cells Tissues Organs. 2003;175(3):121–132. doi: 10.1159/000074628. [DOI] [PubMed] [Google Scholar]
- 53.Keiser HD, Hatcher VB. Comparison of Bovine Nasal Cartilage Proteoglycan Core Protein Produced by Chondroitinase and Hyaluronidase e Possible Role of Protease Contaminants. Connect. Tissue Res. 1977;5(3):147–155. doi: 10.3109/03008207709152265. [DOI] [PubMed] [Google Scholar]
- 54.Choi JA, Gold GE. MR imaging of articular cartilage physiology. Magn. Reson Imaging Clin. N. Am. 2011;19(2):249–282. doi: 10.1016/j.mric.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baykal D, et al. Nondestructive Assessment of Engineered Cartilage Constructs Using Near-Infrared Spectroscopy. Appl. Spectrosc. 2010;64(10):1160–1166. doi: 10.1366/000370210792973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofmann GO, et al. Detection and evaluation of initial cartilage pathology in man: A comparison between MRT, arthroscopy and near-infrared spectroscopy (NIR) in their relation to initial knee pain. Pathophysiology. 2010;17(1):1–8. doi: 10.1016/j.pathophys.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Spahn G, et al. Near-infrared (NIR) spectroscopy. A new method for arthroscopic evaluation of low grade degenerated cartilage lesions, Results a pilot study. BMC Musculoskelet Disord. 2007;8:47. doi: 10.1186/1471-2474-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw RA, et al. Arthritis diagnosis based upon the near-infrared spectrum of synovial fluid. Rheumatol. Int. 1995;15:159–165. doi: 10.1007/BF00301774. [DOI] [PubMed] [Google Scholar]