Summary
Coordination of transcription in bacteria occurs at supra-operonic scales, but the extent, specificity, and mechanisms of such regulation are poorly understood. Here, we tackle this problem by profiling the transcriptome of the model organism Mycoplasma pneumoniae across 115 growth conditions. We identify three qualitatively different levels of co-expression corresponding to distinct relative orientations and intergenic properties of adjacent genes. We reveal that the degree of co-expression between co-directional adjacent operons, and more generally between genes, is tightly related to their capacity to be transcribed en bloc into the same mRNA. We further show that this genome-wide pervasive transcription of adjacent genes and operons is specifically repressed by DNA regions preferentially bound by RNA polymerases, by intrinsic terminators, and by large intergenic distances. Taken together, our findings suggest that the basal coordination of transcription is mediated by the physical entities and mechanical properties of the transcription process itself, and that operon-like behaviors may strongly vary from condition to condition.
Graphical Abstract
Highlights
-
•
Basal coordination of transcription is driven by pervasive transcription
-
•
It is repressed by terminators, long intergenic regions, and stalled RNA polymerases
-
•
Operon-like behaviors may strongly vary from condition to condition
By analyzing multiple transcriptomes and structural features of a minimal bacterial genome, Junier et al. show that the basal coordination of transcription in bacteria relies on the capacity of RNA polymerases to transcribe consecutive genes and operons in one go. Stalled RNA polymerases, large intergenic regions, and intrinsic terminators delineate genomic domains of this coordination, for which operon-like behaviors may strongly vary from condition to condition.
Introduction
Transcriptional regulatory mechanisms can be broadly categorized into two classes. On one hand, response mechanisms can convert environmental cues into specific transcriptional responses. This occurs mostly through the action of dedicated transcription factors (TFs), as in the well-known case of the lac operon (Jacob et al., 1960). On the other hand, gene expression is continuously adjusted to adapt to varying environmental conditions, leading to quantitative relationships between the molecular content of cells and their growth rates (Scott and Hwa, 2011). “Physiological factors” (Berthoumieux et al., 2013), such as the concentration of RNA polymerases (RNAPs) (Klumpp and Hwa, 2008), the regulation of RNA degradation (Chen et al., 2015), and topological properties of DNA (Dorman, 1995, Hatfield and Benham, 2002, Travers and Muskhelishvili, 2005), are thus known to continuously affect, at a system level, the transcriptional activity of genes. The specificity, if any, of each of these mechanisms with respect to the set of co-regulated genes, nevertheless remains to be understood.
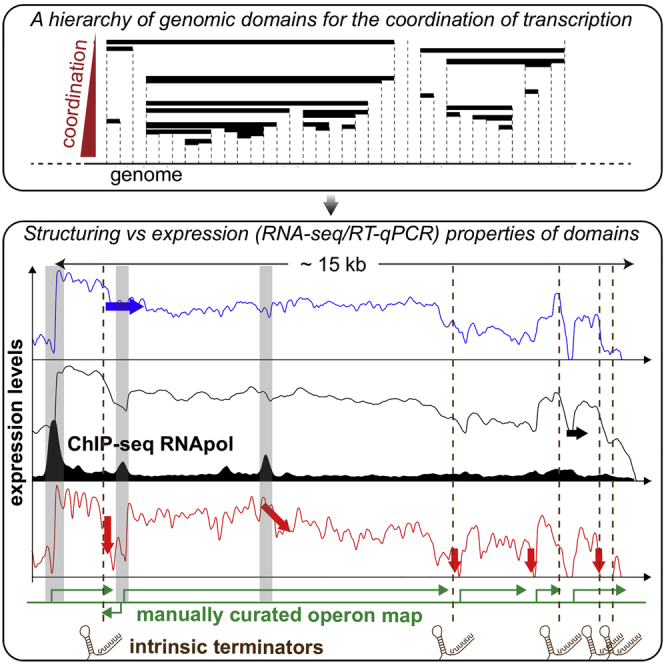
In bacteria, the coordination of transcription is strongly related to the linear organization of genomes (Képès et al., 2012). At the smallest scale, many co-regulated genes are thus found within operons so that they can be co-transcribed into the same mRNAs. Despite their apparent simplicity, the operons have nevertheless raised important questions, not only about their determination but also about their definition (Okuda et al., 2007, Güell et al., 2011, Mazin et al., 2014) and utility (de Lorenzo and Danchin, 2008, Junier, 2014). For instance, although certain operons are easily recognizable by their functional homogeneity (as, e.g., for the lac operon), many of them are composed of genes whose function appears unrelated (de Lorenzo and Danchin, 2008). Soon after the seminal work of Jacob and Monod, studies on operons such as trp also revealed the possibility to have specific internal regulation of termination (Yanofsky, 2000): mRNA-based intrinsic terminators may abort transcription midway, whereas competing mRNA secondary structures (anti-terminators) may attenuate this effect (Merino and Yanofsky, 2005, Santangelo and Artsimovitch, 2011). Together with the observation that the majority of operons actually contain alternative transcription start sites (TSSs) (Sharma et al., 2010, Cho et al., 2014), high-throughput data have thus revealed the presence, inside operons, of differential initiation and termination points (Okuda et al., 2007, Güell et al., 2011, Nicolas et al., 2012, Mazin et al., 2014). Yet the impact of these internal elements on the genome-wide coordination of transcription has remained unexplored.
Model systems such as the bacteriophage further revealed that operons may be part of larger functional genomic units with, in particular, the possibility of having subsequent operons transcribed in one go (Gottesman et al., 1980). RNAP can indeed override termination, which is called “transcriptional read-through” (TRT). TRT has actually been shown to be frequent and regulated by dedicated proteins (Stülke, 2002, Nudler and Gottesman, 2002), with the so-called factor playing a major role in many bacteria (Richardson, 2002). Transcriptional co-expression has thus been shown to extend beyond operons (Jeong et al., 2004, Carpentier et al., 2005, Nicolas et al., 2012). Yet the systematic identification of supra-operonic units and of their regulatory mechanisms remains an open problem.
A system-level understanding of transcriptional coordination thus requires abandoning, at least in the first stage, our preconception of the potential units that may come at play. A promising avenue along this line consists in analyzing in detail the genomic properties of proximal genes as a function of their degree of co-expression and to question the structural and regulatory properties that might be associated to the observed patterns (Ma et al., 2013). Here, we perform such analysis in M. pneumoniae, a model organism with a reduced genome (∼820 kb) that offers ideal properties to address questions about the fundamental mechanisms that govern bacterial cell physiology (Güell et al., 2009, Kühner et al., 2009, Yus et al., 2009). In particular, although M. pneumoniae has two sigma factors (Torres-Puig et al., 2015), a tiny TF repertoire (Table S1) and no factor (Himmelreich et al., 1996), it shows genome-wide complex specific regulatory patterns in response to different external perturbations (Güell et al., 2009). This suggests the existence of fundamental mechanisms ensuring coordination of transcription, different from TFs and from the factor.
To test this hypothesis and to identify associated regulatory mechanisms, we analyzed RNA sequencing (RNA-seq) data obtained from M. pneumoniae under 115 different conditions (Figure 1A). To this end, we built a co-expression measure particularly well poised to highlight basal co-expression. Using a hierarchical clustering framework that is constrained to respect the 1D organization of the genome, we then reveal the existence of three qualitatively distinct levels of co-expression associated to different organizations of adjacent genes and to different properties of intergenic regions. We next show that the degree of co-expression between co-directional genes and operons is tightly related to the capacity of the RNAP to transcribe them as if they belonged to the same operon. We then reveal that such TRT is both ubiquitous and condition dependent and that it is repressed by DNA-bound RNAPs, strong intrinsic terminators, and large intergenic distances.
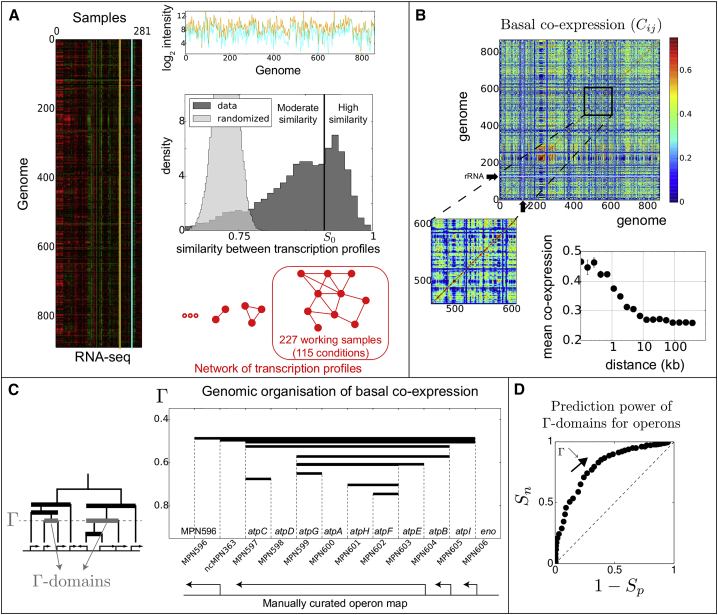
Figure 1.
A Hierarchical Genomic Analysis of RNA-Seq Data across More Than 100 Conditions
(A) Given the initial set of RNA-seq samples (2 of which are shown at the top in cyan and orange), we computed all possible pairwise similarities (Pearson coefficient). These were in general high (dark gray distribution), even after shuffling, for each gene separately, the expression values between the conditions (light gray distribution). Given the bimodal shape of the resulting distribution, we defined a threshold (=0.91, vertical black line) above which profiles with larger similarities were connected to form a network, as schematically represented in red. The largest component of the network contained 227 samples (115 conditions), which we used to compute the basal co-expression.
(B) Top: heatmap of the basal co-expression for which the genes are sorted according to their genomic position. The black arrows indicate the position of the rRNAs, which were used to normalize the data and, hence, whose co-expression values were discarded (thin white lines). Bottom left: zoom in. Bottom right: average co-expression level between pairs of genes as a function of their genomic distance.
(C) Using a hierarchical clustering constrained to respect the linear organization of the genome, we built a dendrogram (bottom left) by fusing genes on the basis of their co-expression level. -domains are maximal segments of the genome inside which all pairs of adjacent genes have a co-expression larger than (gray thick lines; all thick lines correspond to a specific -domain but for various values of ). They thus correspond to the clades of the dendrogram at the level . As shown on the right panel for the F-ATPase genes, although different, -domains share similarities with operons.
(D) Receiver-operating characteristic analysis to evaluate the predictive power of -domains for operons (AUC = 0.76). and respectively indicate the sensitivity and specificity of the resulting domains.
Results
Profiling Basal Gene Expression across Conditions
We measured the transcriptional activity of 869 M. pneumoniae genes, of which 701 encode proteins (Lluch-Senar et al., 2015), across 141 conditions (282 samples). To this end, M. pneumoniae M129 (passage 34, NC_000912 reference genome in the National Center for Biotechnology Information [NCBI]) was grown in modified Hayflick medium and transformed by electroporation (Yus et al., 2009). RNA-seq data were then collected at various stages of the cell growth, after various perturbations and overexpression of different regulators (Table S2). The resulting transcription profiles were generally highly similar, even after shuffling the expression values between the conditions for each gene separately (light gray distribution in Figure 1A), showing that genes have mostly stable expression. To focus specifically on basal co-expression, we discarded “aberrant” transcription profiles using a network approach (Figure 1A). We identified a large set of 227 highly similar samples corresponding to 115 conditions (112 for which two technical replicates are present), the 29 remaining conditions being characterized by a particularly low level of transcription (Table S2).
A Hierarchical Genomic Analysis of Basal Co-expression
To analyze the basal coordination of transcription between all pairs of genes, we built a specific measure of transcriptional co-expression, , hereafter referred to as “basal correlation.” quantifies the tendency of the expression of the two genes to systematically vary in parallel (Figure S1), here among the 227 working samples. Specifically, it is equal to the difference between the number of pairs of samples for which the two genes co-vary and the number for which they vary in the opposite direction, normalized by the total number of possible pairs of samples. Compared with other correlation measures to which it is related, such as the Pearson correlation, is well suited to highlight the basal coordination of genes (Figure S1). In particular, indicates that the expression of the two genes varies as many times in the same direction as in the opposite direction. In contrast, indicates that they always vary in the same direction, whereas indicates a systematic tendency to vary in the opposite direction.
Figure 1B shows the resulting heatmap of , for which the genes are sorted according to their genomic position, and each pixel indicates the co-expression level between two genes. One can distinguish the presence of specific contiguous clusters of highly co-expressed genes with, on average, a genomic extension that typically extends up to 10 kbp (Figure 1B, bottom). This 10 kbp length scale is larger than the typical length scale of operons, corroborating that the co-expression of proximal genes extend beyond operons. It is actually similar to that found in E. coli and B. subtilis when performing a similar analysis of co-expression (Jeong et al., 2004, Carpentier et al., 2005, Junier and Rivoire, 2016).
To delineate in a more precise way the relationship between basal coordination of transcription and the established organization of M. pneumoniae into operons (see Experimental Procedures for their definition), we analyzed in detail the genomic organization of co-expression. To this end, we developed a hierarchical description of co-expression constrained to respect the linear organization of the genome. Briefly, we built a dendrogram in which pairs of adjacent genes were hierarchically fused on the basis of their co-expression (Figure 1C). Using this dendrogram, we defined domains of the genome, which we call -domains, as the contiguous domains of genes for which all the adjacent genes have a co-expression larger than (Figure 1C).
An analysis of -domains for all possible values of reveals that although these domains may coincide with operons at certain values of , they are generally different. This can be qualitatively appreciated for specific clusters of genes, such as that of the F-ATPase machinery (Figure 1C). More quantitatively, we evaluated the capacity of -domains to predict operons using our most recent manual annotation of operons as the ground truth (Table S1). To this end, we made vary from 1 to −1 and we assessed both the specificity and the sensitivity of predictions. The resulting area under the receiver operating curve (AUC), which summarizes the balance between specificity and sensitivity by a single value, was equal to 0.76 (Figure 1D). -domains are thus not perfect predictors (in which case the AUC would have been equal to 1), corroborating the necessity to analyze co-expression properties independently of our knowledge of operons, at least in the first stage.
Three Qualitatively Different Levels of Basal Co-expression
To better understand the hierarchical properties of co-expression, we analyzed the relative orientations of adjacent genes as a function of their co-expression (846 pairs analyzed for which expression values were available for the two genes) (Figure 2A). We also analyzed the tendency of co-directional genes to overlap (Figure 2B), as well as the intergenic distances between the non-overlapping ones (Figure 2C).
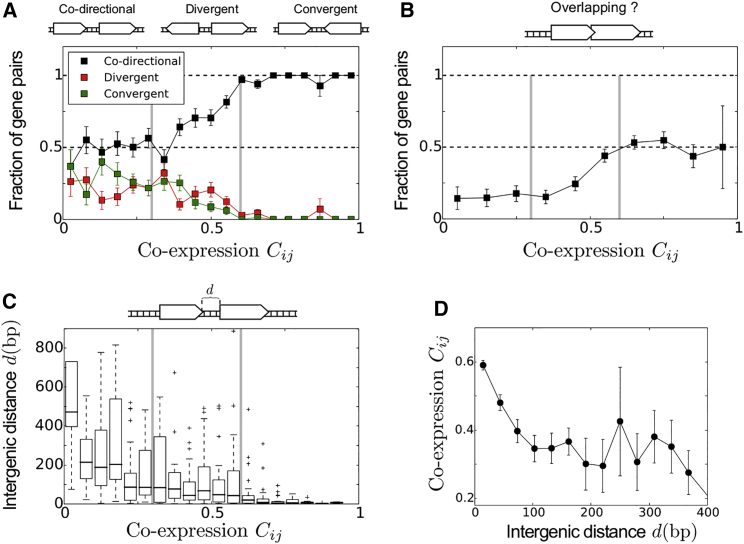
Figure 2.
Evidence for the Existence of a Three-Level Organization in the Basal Coordination of Transcription
(A) The distribution of relative orientations of adjacent genes as a function of their co-expression reveals the existence of three qualitatively different levels of co-expression, with threshold occurring at ∼0.3 and ∼0.6 (vertical gray lines).
(B and C) A similar three-level organization can be distinguished both from the fraction of overlapping pairs (B) and from the distribution of the intergenic distances that separate co-directional genes (C).
(D) Mean co-expression as a function of the distance separating co-directional adjacent genes, revealing a characteristic length scale of 100 bp below which co-expression is all the higher that the distance is small. Error bars correspond to SEM.
Notably, the three properties (relative orientation, overlapping, and distance) suggest a similar three-level organization of co-expression. Specifically, for co-expression > 0.6 (strong co-expression), 227 of 234 pairs of genes are co-directional, with a high proportion (∼52%) of overlapping cases (, hypergeometric test). For co-expression between 0.3 and 0.6 (moderate co-expression, 375 pairs), genes significantly tend to be co-directional and to overlap , all the more that co-expression is large. At this level, although intergenic distances between non-overlapping genes are larger than those with strong co-expression, they remain relatively small. Plotting the co-expression level of all pairs of non-overlapping co-directional genes as a function of their intergenic distance actually reveals the existence of a 100 bp length scale below which typical co-expression is larger than 0.3 and above which co-expression is low and statistically insensitive to distances (Figure 2D). Finally, below 0.3 (low co-expression, 237 pairs), there is no enrichment for a specific relative orientation of genes ( 1, binomial test of the hypothesis that the probability of co-directionality is equal to 0.5). Moreover, intergenic distances between co-directional genes are large, exceeding 100 bp and reaching typically 400 bp at very low co-expression (Figure 2C).
Performing the same analyses using Pearson correlation led qualitatively to the same findings (Figure S2). The sharp delineation of the three different regimes as well as their correspondence between the different properties (relative orientation, overlapping, and distance) is less clear, though, than those obtained using the basal correlation.
Transcription En Bloc Coordinates Co-directional Genes and Operons
Because a significant number of operons have moderate co-expression levels and because TRT pervades the transcriptomes of bacteria (Wade and Grainger, 2014), we wondered whether TRT could explain the co-expression levels of co-directional genes belonging to distinct operons. We thus investigated the tendency of all adjacent co-directional genes belonging to two different operons to be transcribed as a single transcript (268 pairs analyzed). We examined the variation of expression in their intergenic region as a function of the variation of expression of the downstream gene (Figure 3A). Our rationale was that TRT, if present, should leave a trace on the expression of the intergenic region that precedes the downstream gene. We thus compared the co-expression between the downstream gene and the sense (5′→ 3′) intergenic region (co-expression CS in Figure 3A) with that between the two genes (co-expression C); as a control, we considered the anti-sense (3′→ 5′) region (Figure 3A, right) (co-expression CA). To prevent any bias arising from the transcription of the UTRs inside operons, for this analysis, we defined intergenic regions as the sequences that separate the transcription termination site (TTS) of the upstream gene and the TSS of the downstream gene.
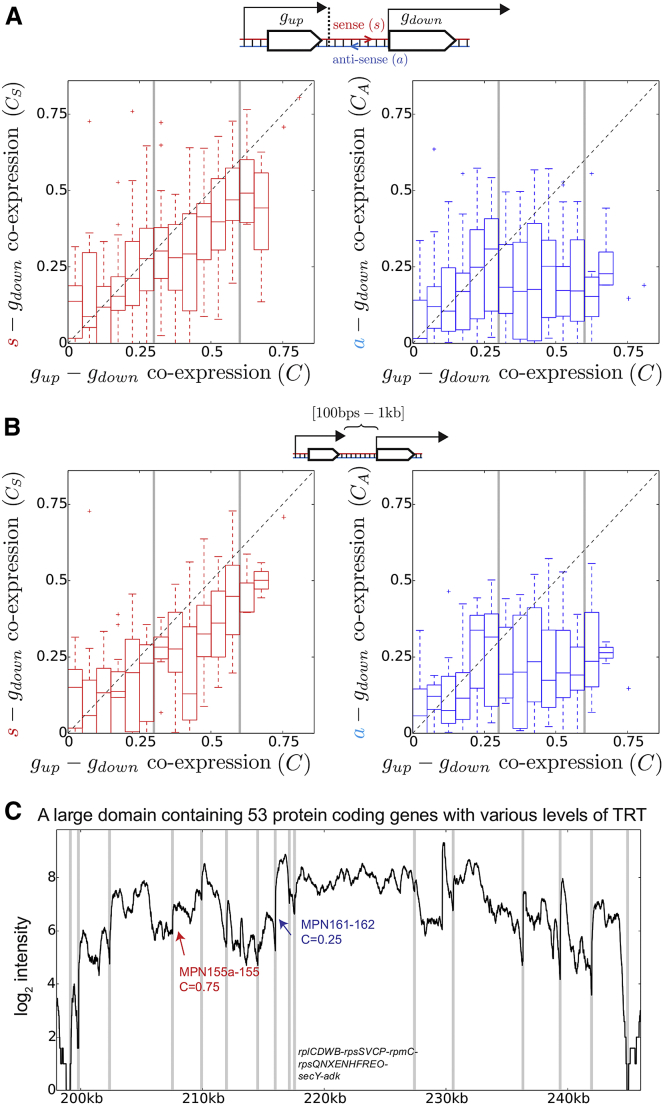
Figure 3.
TRT at the Core of the Basal Coordination of Transcription
(A) Left: for pairs of co-directional adjacent genes belonging to different operons, we compare the co-expression, CS, between the downstream gene and the sense (5′→ 3′) intergenic region with the co-expression, C, between the two genes. Right: as a control, we consider the anti-sense (3′→ 5′) region (co-expression CA) instead of the sense region. Results show that for , CS and C are strongly correlated, while CA and C are not. Correlations for C < 0.3 might be explained by local concentration effects and the presence of pervasive transcription (Wade and Grainger, 2014).
(B) Same as in (A) but keeping only pairs of operons that are separated by more than 100 bp; distances are measured from the TTS of the upstream operon to the TSS of the downstream operon.
(C) Example of a large domain with a high-level background expression surrounding the ribosomal protein genes and containing 53 genes (15 operons) and for which 46 of the 52 pairs show a significant basal co-expression (> 0.3); for clarity, we indicate the composition of only the largest operon. Although the TSSs of most operons (vertical gray lines) can be distinguished by a steep fold change of the expression, real-time qPCR analysis confirms that TRT occurs between strongly co-expressed operons, as indicated in red for the pair MPN155a-MPN155. In contrast, TRT does not seem to occur at a significant level for low co-expression as in the case indicated in blue (see Figure S3 for details). The RNA-seq profile was obtained at 24 hr (late exponential) of the growth curve.
For co-expression levels larger than ∼0.3, we observed that the degree of co-expression between co-directional adjacent operons was higher when there was co-expression with the sense intergenic region (Figure 3A); in contrast, it did not show any dependency with the anti-sense expression (Figure 3A, right). The same analyses, but considering genes that are separated by more than 100 bp (Figure 3B), or using the Pearson correlation (Figure S2E), led to the same conclusions.
Next, to explicitly demonstrate the role of TRT in basal coordination of transcription, we first studied the efficiency, , of TRT extending between two co-directional adjacent operons, say, X and Y with X preceding Y (Figure S3A). was defined as the ratio between the RNA-seq expression levels measured at the TSS of Y and at the stop codon of the last cistron of X (independently whether this was associated to a well-defined terminator). We thus assumed that the RNA-seq level just preceding Y would result from the TRT of X and would be representative of the basal level of Y. According to this model, for which we provide below an experimental validation, the overall expression of Y is thus equal to the sum of its basal level coming from TRT, plus some contribution from its own TSS (Figure S3A).
We analyzed seven pairs of genes with various degrees of correlations and distances: two pairs with strong correlation, including one overlapping case (MPN155a-MPN155); four pairs with moderate correlation and distances larger than 100 bp, including two pairs with an intermediate gene located on the opposite strand of their intergenic region; and one pair with low co-expression. As shown in Figure S3B, was close to 1 and varied little for pairs with strong correlation, but also for the pair MPN160-MPN161 (highest moderate correlation, ) except during heat shock. For the other pairs, was both smaller and more variable. In particular, for the pair MPN161-MPN162 (low correlation), we observed a 2-fold variation during cold shock that poorly correlated to the expression variation of the downstream gene.
We then tested the validity of our TRT-based model of transcriptional coordination by confronting predictions of the model (using RNA-seq data) to direct measurements of transcripts using real-time quantitative PCR (qPCR). Specifically, for the aforementioned seven pairs, we measured the level of transcripts extending between the genes during cold shock, heat shock, and exponential growth (control). As shown in Figure S3C, the variations of extended TRT measured by real-time qPCR were qualitatively (quantitatively in most cases) similar to those predicted by the model. Notably, this was true for the cases with an intermediate gene on the opposite strand.
We thus conclude that TRT is ubiquitous and can explain, in principle, many of the significant co-expression levels of co-directional adjacent operons. These results also suggest that large pieces of genomes that extend beyond operons may be transcribed en bloc. An instructive example concerns the ribosome-encoding genes: these genes are surrounded by transcription-related genes and other biological pathways, apparently forming altogether a large domain containing more than 50 genes (corresponding to 15 manually annotated operons) with a high level of background expression (Figure 3C). Although some of this background is expected to result from the strong tendency of these promoters to initiate transcription, as demonstrated by our real-time qPCR analysis (Figure S3) it also results from TRT extending between operons. In this context, it is important to recognize that large domains of coordinated expression, in which several genes and operons may be transcribed in a row, remain compatible with the very presence of operons. This can be seen by the decrease of expression at the end of certain operons or by the presence of steep fold changes at their promoter (vertical gray lines in Figure 3C). The pair MPN155a-MPN155 (strong basal co-expression) provides a good example of this effect as it shows extended TRT between the two corresponding operons (Figures S3B and S3C) but also a sharp TSS at the downstream gene at late exponential phase (in red in Figure 3C) and at stationary phase (Figure S3B).
TRT Variations and Its Regulation
RNA-seq data and real-time qPCR show that TRT may vary not only along the genome but also among conditions (Figure S3). The ten-gene (four-operon) domain containing the heat shock gene (grpE) provides an insightful example of such variations (Figure 5C), with the operon containing grpE differentiating into two sub-operons during heat shock and distinct operons becoming transcribed as a single operon during cold shock (Figures S3B and S3C). Notably, both our RNA-seq analysis and our real-time qPCR measurements further suggest that TRT is globally enhanced during cold shock (Table S3; Figure S3), in accord with reports in E. coli and B. subtilis of the anti-terminator role of CspA cold-shock proteins (Bae et al., 2000, Stülke, 2002).
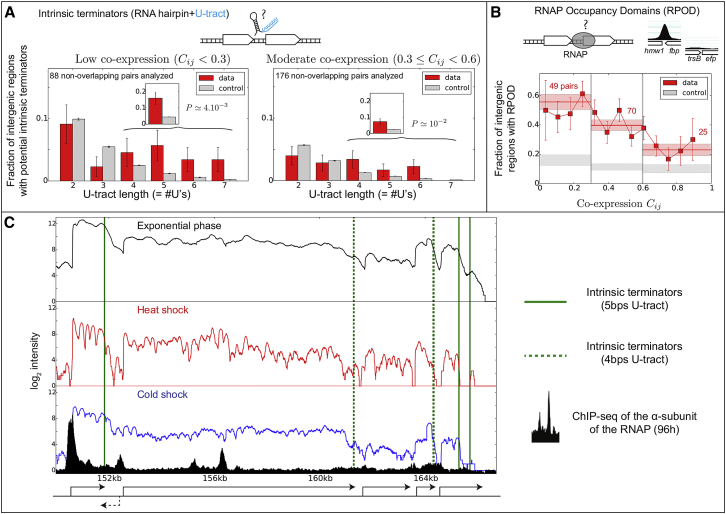
Figure 5.
Intergenic Properties of Co-directional Genes Relevant to Delineate Domains of Transcription En Bloc
(A) Fraction of intergenic regions containing a potential intrinsic terminator for low co-expression levels (left) and for moderate co-expression levels (right). Potential terminators were defined as RNA hairpins immediately followed by a U tract. Several lengths of the U tracts were analyzed (x axis of the bar plots). As a null model, we considered intergenic regions that were shifted by various amounts of base pairs (gray bars; Supplemental Experimental Procedures), allowing us to evaluate the statistical significance of the results (error bars indicate SEM). Insets show the results by cumulating the cases in which , revealing an enrichment that is absent with shorter U tracts .
(B) Fraction of intergenic regions containing a RPOD as a function of the basal coordination of transcription. The red bands indicate the SEM computed over the whole region; the red numbers indicate the number of corresponding pairs among the 386 pairs of non-overlapping genes analyzed. The gray bands indicate the same values but for data for which the positions of the intergenic regions were globally shifted by an arbitrary amount of base pairs.
(C) RNA-seq profiles of a large ten-gene (four-operon) domain around the heat shock gene (grpE) showing condition-dependent TRT; one additional gene (dashed arrow) is present on the opposite strand. Bottom, in black: ChIP-seq profile of the α-subunit of the RNAP (data obtained at 96 hr), revealing in particular the presence of a large RPOD at the start of the domain. Vertical green lines, positions of strong intrinsic terminators as identified in (A).
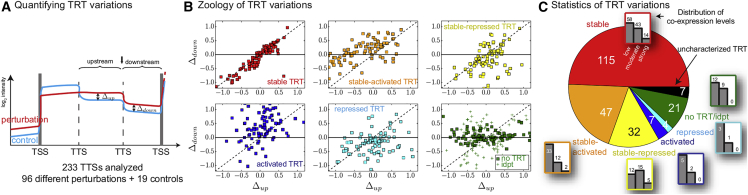
To systematically quantify TRT variations among conditions, we analyzed the behavior of the TTSs internal to pairs of co-directional genes belonging to different operons (233 TTSs analyzed), independently of whether a well-defined terminator was associated to the TTS. For each TTS, we computed the variation of its downstream expression as a function of the variation of its upstream expression in response to perturbations (96 perturbations tested with respect to 19 controls; Figure 4A). Using this approach, we could identify at least six types of TTSs (Figure 4B). The three first types concern TTSs for which a statistically significant positive correlation exists between the values of and that are computed over the different perturbations (see legend of Figure 4 for details of the statistical analyses). They are respectively defined by (stable TRT, in red in Figures 4B and 4C), (stable TRT plus some activation, in orange), and (stable TRT plus some repression, in yellow). For these TSSs, TRT thus tends to be maintained at a similar level, irrespective of the conditions. Notably, these correspond to ∼85% of the total amount of TTSs (∼50% if only considering ) and appear to account for all strong co-expression levels (Figure 4C). The two next types correspond to activation only, with a majority of (in blue), and to repression only, with a majority of (in cyan), irrespective of the value of . Together with the TTSs having independent or no apparent statistically significant variations of (in green), these three last types contribute mainly to low co-expression levels.
Figure 4.
Quantification of TRT Variations
(A) For each pair of adjacent operons, we analyzed at the TTS of the last gene of the upstream operon (black arrow) the behavior of the downstream variation of expression as a function of the upstream variation of expression ; the corresponding regions were defined by the closest TTS or TSS on each side of the TTS of interest.
(B) We identified six types of TTS, for which an example of each type is shown in every panel; the 96 color points inside every panel correspond to the resulting behavior of the corresponding TTS for the 96 perturbations. To this end, we used two p values, and , respectively associated to the null hypotheses that and are not linearly correlated and that on average, is equal to , and considered for significance thresholds a multiple hypothesis correction procedure (Supplemental Experimental Procedures). Stable TRT was then defined by a significant and a non-significant , stable-activated (repressed) TRT by significant values of both and with , activated (repressed) TRT by a non-significant and a significant with , and the set “no TRT or independent TRT” by non-significant values of both and .
(C) Distribution of the TTS types as identified in (B). Uncharacterized types (in black) correspond to those that did not fit the criteria of the p values. For each type, we show in addition the distribution of basal co-expression (low, moderate, or strong, indicated by the gray bars), revealing that only stable TRTs contribute to strong basal co-expression.
Altogether, these results thus corroborate both the ubiquity of TRT and its major role in basal co-expression. The observation of pairs having both low co-expression and stable TRTs also suggest that TRT does not systematically extend to the next operon.
Finally, to apprehend whether TRT is “stochastic” or specifically regulated, we analyzed the behavior of the TTSs upon each perturbation. We found three interesting results (see Table S3 for details). First, in accord with our real-time qPCR experiments (Figure S3), we observed that the variations of TRT (activation or repression) for a given perturbation tend to be the same for all TTSs, suggesting that TRT is specifically regulated at a genome-wide level. Second, we found a larger number of perturbations with TRT activation. These include cold shock, osmotic shock, and novobiocin treatments, whereas low pH and heat stresses tend to repress TRT. Finally, by identifying the TTSs and the corresponding perturbations for which the variation of TRT was extreme (Supplemental Experimental Procedures), we found that conditions for which a large number (≥12) of TTSs had an extreme behavior were strongly enriched in novobiocin (gyrase inhibitor) perturbations (, hypergeometric test) and strongly depleted in single-gene perturbations (Table S3). Because novobiocin targets topoisomerases and, hence, modify DNA supercoiling, these results suggest that the mechanical properties of DNA and its interaction with RNAPs might play a crucial role in TRT variations (see the following discussion for further details).
The Role of Genome Compactness and Intrinsic Terminators
Our observations of a TRT that depends strongly on conditions, with operons that can be transcribed uniformly, en bloc (super-operons), or differentially (sub-operons), raise at least two fundamental questions: what mechanisms are responsible (1) for promoting an operon-like transcription of adjacent genes and (2) for preventing it?
In answer to the first issue, using co-expression levels as a proxy of transcription en bloc, the results in Figures 2C and 2D suggest that compactness, with a distance between open reading frames smaller than 100 bp, may be required for efficient operon-like co-expression. Notably this length scale corresponds to the typical distance that is usually considered for operon prediction (McClure et al., 2013). We note, nevertheless, that pairs of genes with intergenic regions larger than 100 bp can have a high level of TRT (Figures 3 and S3). Our analysis also shows that compactness alone is not sufficient, because a substantial number (52) of pairs of co-directional genes with low co-expression levels are separated by less than 100 bp (among which 17 pairs concern overlapping genes).
In answer to the second question, let us first mention that although compactness properties call for an important role of distances on the capacity of the RNAP to transcribe multiple genes in a row, co-expression does not depend primarily on distances when these exceed 100 bp (Figure 2D). To better understand the differences between intermediate co-expression levels and low co-expression levels, we thus investigated the possible impact of -independent intrinsic terminators found within mRNA sequences ( -dependent termination is absent in M. pneumonia). Canonical intrinsic terminators consist of an RNA hairpin followed by a U tract, a combination that is believed to favor the disruption of the mRNA-DNA template hybridization necessary for the RNAP to process transcription (Peters et al., 2011). We thus evaluated the presence, in the intergenic regions of all pairs of co-directional genes, of RNA hairpins that were immediately followed by U tracts of various lengths ; as a control, we considered intergenic regions that were translated by an arbitrary amount of base pairs, which allowed us handling distance effects of intergenic regions, a longer sequence being more likely to contain an RNA hairpin (Figures S4A and S4B), independently whether the latter plays a functional role.
We found that more than 15% of the gene pairs with low co-expression contained an RNA hairpin with (Figure 5A), a proportion that was highly significant with respect to the control (, two-sided t test with unequal variances). Note here that the absence of an enrichment of terminators with shorter U tracts corroborates previous observations that long U tracts are needed to have efficient termination (Chen et al., 2013). Similar trends were observed for intermediate co-expression levels, although involving a lower fraction (typically half) of gene pairs, in accord with the fact that at this level, TRT is expected to occur in a larger subset of conditions.
Correlation with RNAP Occupancy Domains and Transcript Half-Lives
According to the above analysis, more than 80% of the co-directional gene pairs with low co-expression do not contain any strong intrinsic terminator in their intergenic region. Non-perfect U tracts or more complex termination signals that are yet to be identified might explain part of this low co-expression. The action of nucleoid-associated proteins (NAPs) such as H-NS in E. coli (Singh et al., 2014) could also be invoked. Data from our lab nevertheless show that M. pneumoniae contains only one NAP, IHF (gene MPN529), with a low copy number (<100).
To better apprehend the mechanisms related to the repression of TRT, we thus performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis of the α-subunit of the RNAP. Consistent with results obtained in E. coli (Mooney et al., 2009), ChIP-seq profiles from cells in the stationary phase revealed the presence of well-defined peaks corresponding to preferentially RNAP occupancy domains (RPOD) (Figures 5B and 5C; Table S4). Notably, we found that the majority of gene pairs with low co-expression contain RPODs in their intergenic regions (Figure 5B). For intermediate co-expression, RPODs are less present but remain over-compared to strong co-expression. Note that in contrast to hairpins, the presence of RPODs does not depend on the intergenic distances (Figure S4C), meaning that larger intergenic distances cannot simply explain these results.
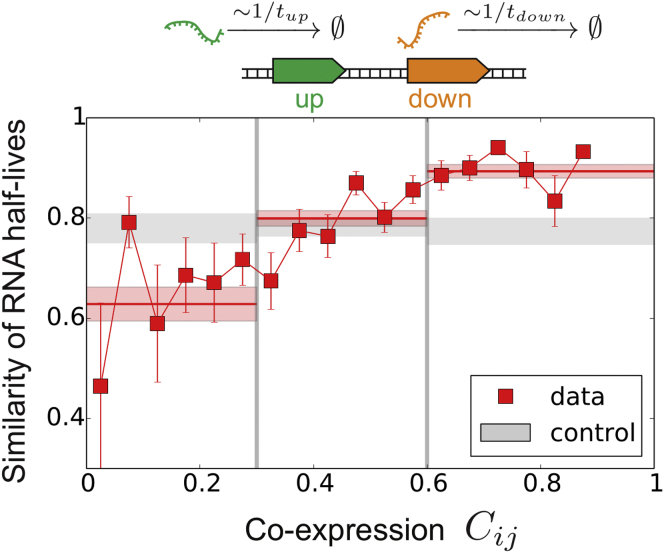
Because the average level of transcription strongly depends on RNA degradation, we eventually compared the half-lives of transcripts between adjacent genes. We found a remarkable correlation between the degree of transcriptional coordination and the similarity of half-lives (Figure 6).
Figure 6.
Relative Stability of Transcripts of Adjacent Co-directional Genes
The relative stability is defined as , with and the transcript half-lives of the upstream and downstream genes, respectively; this parameter is therefore close to 1 for similar half-lives and close to 0 for very different ones. The red bands indicate the SEM computed over the corresponding region of co-expression. The gray bands indicate the same values but for a random set of pairs of genes.
Discussion
From Operonic Transcription to Stochastic Condition-Dependent Transcription En Bloc
Using a correlation measure well poised to quantify basal co-expression and applying it to RNA-seq data obtained in more than 100 different conditions, we have revealed the existence in M. pneumoniae of three distinct levels of basal coordination of transcription (strong, moderate, and low), corresponding to three qualitatively different properties for the relative orientations and intergenic regions of adjacent genes. In accord with the major role of operons in the coordination of gene expression, we have found that strong basal co-expression requires adjacent genes to be co-directional. We have also found the existence of a 100 bp length scale, below which an operon-like behavior appears to be quasi-systematic and above which co-expression depends strongly on the sequence and structural properties of the intergenic region. In particular, although pairs of adjacent genes with low basal co-expression do not show any preferential relative orientations, ∼70% of the intergenic region of the co-directional pairs either contain a domain preferentially occupied by RNAPs (RPODs) or strong terminators (∼55% and ∼15% of the cases, respectively).
By focusing specifically on co-directional adjacent genes, we have further revealed that the coordination of transcription is tightly related to the tendency of proximal genes to be transcribed en bloc, even though these genes may have not been categorized to belong to the same operon. Three extreme scenarios can then be considered (Figure S5A), which is in accord with our observation of the three qualitatively distinct co-expression levels. The transcription en bloc may be systematic (i.e., it occurs with probability close to 1 in all conditions), in which case the genes behave as canonical operons (green light in Figure S5A). Or it occurs from time to time, meaning that it can take place in specific conditions and be absent in others. Variations may also occur in a given condition, because transcription may terminate with a certain probability due, for example, to the presence of an intrinsic terminator. In this case, gene expression may present staircase-like patterns (Güell et al., 2009) (orange light). Finally, transcription en bloc can “never” occur, in which case genes must be considered to belong to different transcriptional units (red light).
In accord with the rich zoology of operon-related structures that have been described over the past decade (Okuda et al., 2007, Güell et al., 2009, Cho et al., 2009, Nicolas et al., 2012, Mazin et al., 2014) and with the ubiquitous presence of pervasive transcription (Wade and Grainger, 2014), our findings thus indicate that operon-like behaviors are often stochastic and condition dependent, with frequencies of occurrence that depend on intergenic sequences. In particular, transcriptional initiation may often occur on top of a background level of continuous expression. In this context, we surmise that one of the most fundamental mechanism for the coordination of transcription relies on a high probability to have specific large domains of genes that are transcribed in a row, independently of the fact that these domains may contain several internal entry points and exit points for the RNAPs (see Figure S5B for a schematic representation of this model). These internal landmarks might then be used by the bacterium to adapt to a wide range of conditions (see, e.g., Figure 5C). They might also contribute to the activation of a given domain (see the following discussion).
Minimal Prescriptions for Generating Specific Domains of Transcriptional Coordination
Our scenario implies, on one hand, the existence of two mechanisms internal to the domains, which are a priori necessary to maintain a proper balance between transcripts. First, there should exist a mechanism that enhances the transcription of upstream genes whenever transcription is initiated within the domain, in order to avoid a gradient of transcripts along the domain (with downstream genes in larger quantity than upstream genes). Although at this stage we have no direct evidence of such phenomenon, this prediction suits the proposal, in bacteria, of a control of gene expression by DNA supercoiling (Dorman, 1995, Hatfield and Benham, 2002, Travers and Muskhelishvili, 2005). It is also in accord with our observation of a strong impact of novobiocin (a gyrase inhibitor) treatment on TRT properties (Table S3). The negative supercoiling that is generated upstream of the transcribing RNAPs might indeed enhance the initiation of the upstream genes (Meyer and Beslon, 2014). Considering that these effects can propagate all the way up to the borders of the domain because of the long-range nature of the transmission of supercoiling constraints (Krasilnikov et al., 1999), an internal initiation event should in principle be able to activate the expression of the whole domain (Figure S5B), in particular without the additional action of TFs. Second, produced transcripts should have similar degradation rates, which we confirmed by analyzing RNA half-lives (Figure 6).
Well-defined domains of basal co-expression require, on the other hand, the ability, upstream, to prevent the activation of genes and, downstream, to terminate the transcription process. Supposing that the upstream activation is mainly the result of supercoiling transmission, stalled RNAPs, as suggested by the presence of RPODs (Reppas et al., 2006, Mooney et al., 2009), could act as topological barriers (Higgins, 2014) (see Figure 5C for a suggestive example). Downstream, in addition to the possibility of RPOD roadblocks, strong intrinsic terminators are expected to play an important role in terminating transcription (Figure 5). Other mechanisms can be contemplated, such as anti-sense transcription (Lybecker et al., 2014) or the action of small RNAs, although recent work from our lab shows that the latter have little impact on gene expression (Lloréns-Rico et al., 2016). Here, and more particularly in the absence of the factor and of NAPs, which have been shown to prevent pervasive transcription (Singh et al., 2014), the mechanisms at the core of the basal coordination of transcription in M. pneumoniae thus appear to rely solely on the physical entities (RNAP and mRNA) and mechanical properties of the transcription process itself.
Local Concentration Effects
Although a strong terminator can efficiently prevent TRT, it may prevent co-expression only partially. This can be seen for instance by the adjacent genes 5S rRNA (Mpnr03) and MPN095, which is the unique pair showing both strong co-expression and the presence of a strong intergenic terminator. Although some specific processing of rRNA might occur, overriding of the terminal signal, as suggested by the high level of co-expression with the sense intergenic region , might explain the strong co-expression. Local concentration effects of RNAPs might also contribute, more particularly because of the high expression level of the 5S rRNA. In such situations, intergenic distances might play a crucial role in the isolation of adjacent genes. Specifically, compared to the 20–30 nm size of the RNAP, the 130 bp that separate the TTS of the 5S rRNA from the TSS of MPN095 correspond to a maximal spatial distance of ∼45 nm; 400 bp, the typical distance for pairs of genes with co-expression close to 0 (Figure 2C), correspond to ∼135 nm.
Conclusions
Our scenario reckons with the intrinsic stochastic nature of transcriptional initiation, with the capacity of the RNAP to transcribe multiple operons in one go (Santangelo and Artsimovitch, 2011), and with the possible role of supercoiling to transmit regulatory properties, especially in a bacterium that is depleted in TFs (Zhang and Baseman, 2011, Dorman, 2011). It also opens new roads to understand the existence of preferential regions and promoters for the binding of RNAPs (Reppas et al., 2006, Mooney et al., 2009) and suggests that a large part of the specific basal coordination of transcription might rely exclusively on the interplay among RNAP, DNA, and mRNA.
Importantly, our findings appear to hold in a wide range of bacterial species. A similar three-level organization of co-expression, with the same properties of relative orientations and of intergenic distances (including the existence of a ∼100 bp length scale), is indeed observed both in E. coli and in B. subtilis, (Figure S6). Domains of proximal genes that are conserved in phylogenetically distant bacteria have also been shown to correspond, both in E. coli and in B. subtilis, to domains of highly co-expressed genes and operons where TRT is particularly enhanced (Junier and Rivoire, 2016). Finally, we note that -independent terminators, as well as attenuators of these terminators through, for example, the action of riboswitches, are often conserved among distant bacteria (Vitreschak et al., 2004, Merino and Yanofsky, 2005). Together with the dynamical interplay between DNA and RNAPs, they may thus correspond to ancestral mechanisms upon which the basal functioning of bacteria has been tinkered. In particular, TFs and other types of gene control such as the invertible DNA switches of Bacteroides (Kuwahara et al., 2004) may represent evolutionary solutions dedicated to specific needs related to the lifestyle of each bacterium.
Experimental Procedures
RNA-seq and ChIP-seq Data
RNA isolation was performed using miRNeasy kits from Qiagen, and an in-column DNase treatment was included. RNA was measured using a Nanodrop (Thermo), and integrity was confirmed in a 6000 Nano chip Bioanalyzer (Agilent). We then used the TruSeq Stranded mRNA Sample Prep Kit v2 (Illumina) to obtain a paired-end strand-specific RNA-seq library. See Table S2 for further details of conditions.
ChIP-seq of RNAP (TAP-tagged; see Kühner et al., 2009) was performed as previously described (Yus et al., 2012).
TSSs and Manual Annotation of Operons and Sub-operons
We identified all mRNA TSSs from their associated tssRNAs (Yus et al., 2012). We distinguished productive promoters from short tssRNAs as explained previously (Lloréns-Rico et al., 2015). Regarding 3′ sites, we used strand-specific deep sequencing and tiling array data to define approximately their positions (Güell et al., 2009). We then used these data to refine our previously published operon map (Güell et al., 2009) (updated map in Table S1).
Real-Time qPCR of Regions Encompassing Distinct Operons
Cells were collected in the indicated conditions, and RNA was purified as described above. Retrotranscription and real-time qPCR of ∼800 base long regions were done in one step with the GoTaq 1-Step RT-qPCR System (Promega). Oligos (Table S5) were used at 0.15 μM, and 25 ng total RNA was used as a template. mRNA of the stable gene MPN517 was used as control and reference.
Intrinsic Terminators and RPODs
Potential intrinsic terminators were defined as a RNA hairpin immediately followed by a U tract. RNA hairpins were identified as described previously (Mathews et al., 1999).
RPODs were identified by the presence of significant peaks (see Supplemental Experimental Procedures) in the ChIP-seq data of the RNAP α-subunit (gene MPN191) at 6 and 96 hr.
RNA Half-Lives
RNA half-lives were determined using a DNA gyrase inhibitor (novobiocin), which alters the chromosomal supercoiling releasing the RNAP, thus stopping transcription (Dorman, 2011). After novobiocin treatment, RNA was extracted at different time points, and RNA-seq was performed to determine transcript levels. Half-lives were estimated by fitting RNA decays using an exponential function.
Author Contributions
I.J., E.Y., and L.S. conceived the analysis. E.Y. performed the experiments. I.J., E.B.U., E.Y., V.L.-R., and L.S. analyzed the data. All authors participated in writing the manuscript.
Acknowledgments
I.J. is supported by an ATIP-Avenir grant (Centre National de la Recherche Scientifique). E.B.U. was co-funded by Marie Curie Actions. This work was supported by Fundación Marcelino Botin and the Spanish Ministerio de Economía y Competitividad (BIO2007-61762). This project was financed by Instituto de Salud Carlos III and co-financed by Federación Española de Enfermedades Raras under grant agreement PI10/01702 and the European Research Council and European Union’s Horizon 2020 research and innovation program under grant agreements 634942 (MycoSynVac) and 670216 (MYCOCHASSIS). The Centre for Genomic Regulation acknowledges the support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013-2017,” SEV-2012-0208.
Published: May 26, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.04.015.
Contributor Information
Ivan Junier, Email: ivan.junier@univ-grenoble-alpes.fr.
Luis Serrano, Email: luis.serrano@crg.eu.
Accession Numbers
The accession numbers for sequencing data for RNA-seq and ChIP-seq have been deposited in the EMBL-EBI ArrayExpress Archive: E-MTAB-3771, E-MTAB-3772, E-MTAB-3773, and E-MTAB-3783.
Supplemental Information
Sheet 1: List of known or putative transcriptional regulators in M. pneumoniae. The last column indicates the name of the strains in which the TF is perturbed (see Table S2)
Sheet 2: Manual operon and sub-operon annotation of the M. pneumoniae genome. The table indicates the following information for each of the manually annotated transcriptional units (operons and sub-operons): operon number, sub-operon ID, genes belonging to each sub-operon, TSS of the sub-operon, TTS of the sub-operon and strand.
Sheet 1: List of RNAseq experiments used in this work. For each sample, we indicate the strain (wt, M129 or mutant), transgene (indicates the gene that was overexpressed or mutated), timeOfGrowth_experimentPerformedAt in h (time of growth after inoculum), medium used, treatment (type of drug/perturbation), perturbant (drug, condition…), finalConcentration_perturbant (working dilution), durationOfPerturbation in min, Filtered? (in case it was left out of the analysis, see main Materials and Methods)
Sheet 2: list of samples discarded for the co-expression analysis. Sheet 3: Corresponding list of conditions effectively used in the analysis of basal co-expression and of TRT variations. The last column indicates whether the condition was analyzed for TRT variation or if it corresponded to a control. The red names indicate that a single gene was perturbed, in contrast to more global perturbation (various stress shocks, Novobiocin treatments, etc…). The yellow boxes indicate that the perturbed gene is a putative TF (see Table S1).
Sheet 1: Leftmost list: conditions leading to an overall repression of TRT, that is, showing a tendency for having . The average value of Δdown − Δup (third column) is computed over all the TTSs. The list is sorted according to the p values of the bias of the distribution of Δdown − Δup (second column, one sample t test value). The horizontal dashed and full lines respectively indicate the values where the false discovery rate (FDR) is equal to 0.005 and 0.05 (Benjamini–Hochberg procedure). Rightmost list: same thing but for conditions leading to an overall activation of TRT, that is, showing a tendency for having
Sheet 2: Leftmost list: perturbations for which no pair of adjacent genes shows an extreme variation of TRT. Rightmost list: perturbations for which at least 12 pairs of adjacent genes show an extreme variation of TRT. The color codes are those of Table S2.
ChIP-seq peaks associated to RNAP (see Methods and Materials and Supp. Methods text for experimental procedures and for the identification of peaks). For each of the peaks, the following information is displayed: peak position (in bps); peak height (in arbitrary units); peak width (in bps covered); peak score, based on the confidence in the intra-peak distance (see Supplementary Methods); associated TSS(s), if any, otherwise is “NONE”; associated TSS strand(s), if any, otherwise is “NONE”; and time point of the corresponding experiment (6h or 96h).
Oligos used for the RT-qPCR.
References
- Bae W., Xia B., Inouye M., Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U S A. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoumieux S., de Jong H., Baptist G., Pinel C., Ranquet C., Ropers D., Geiselmann J. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol. Syst. Biol. 2013;9:634. doi: 10.1038/msb.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier A.-S., Torrésani B., Grossmann A., Hénaut A. Decoding the nucleoid organisation of Bacillus subtilis and Escherichia coli through gene expression data. BMC Genomics. 2005;6:84. doi: 10.1186/1471-2164-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shiroguchi K., Ge H., Xie X.S. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 2015;11:781. doi: 10.15252/msb.20145794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J., Liu P., Nielsen A.A., Brophy J.A., Clancy K., Peterson T., Voigt C.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- Cho B.-K., Zengler K., Qiu Y., Park Y.S., Knight E.M., Barrett C.L., Gao Y., Palsson B.Ø. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B.-K., Kim D., Knight E.M., Zengler K., Palsson B.Ø. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014;12:4. doi: 10.1186/1741-7007-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Danchin A. Synthetic biology: discovering new worlds and new words. EMBO Rep. 2008;9:822–827. doi: 10.1038/embor.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C.J. 1995 Flemming Lecture. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology (Reading, England) 1995;141:1271–1280. doi: 10.1099/13500872-141-6-1271. [DOI] [PubMed] [Google Scholar]
- Dorman C.J. Regulation of transcription by DNA supercoiling in Mycoplasma genitalium: global control in the smallest known self-replicating genome. Mol. Microbiol. 2011;81:302–304. doi: 10.1111/j.1365-2958.2011.07718.x. [DOI] [PubMed] [Google Scholar]
- Gottesman M.E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J. Mol. Biol. 1980;140:57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Güell M., van Noort V., Yus E., Chen W.-H., Leigh-Bell J., Michalodimitrakis K., Yamada T., Arumugam M., Doerks T., Kühner S. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- Güell M., Yus E., Lluch-Senar M., Serrano L. Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat. Rev. Microbiol. 2011;9:658–669. doi: 10.1038/nrmicro2620. [DOI] [PubMed] [Google Scholar]
- Hatfield G.W., Benham C.J. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- Higgins N.P. RNA polymerase: chromosome domain boundary maker and regulator of supercoil density. Curr. Opin. Microbiol. 2014;22:138–143. doi: 10.1016/j.mib.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R., Hilbert H., Plagens H., Pirkl E., Li B.C., Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Perrin D., Sánchez C., Monod J. L’opéron: groupe de gènes à expression coordonnée par un opérateur. CR Acad. Sci. Paris. 1960;250:1727–1729. [Google Scholar]
- Jeong K.S., Ahn J., Khodursky A.B. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier I. Conserved patterns in bacterial genomes: A conundrum physically tailored by evolutionary tinkering. Comput. Biol. Chem. 2014;53:125–133. doi: 10.1016/j.compbiolchem.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Junier I., Rivoire O. Conserved Units of Co-Expression in Bacterial Genomes: An Evolutionary Insight into Transcriptional Regulation. PLOS One. 2016 doi: 10.1371/journal.pone.0155740. Published online May 19, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Képès F., Jester B.C., Lepage T., Rafiei N., Rosu B., Junier I. The layout of a bacterial genome. FEBS Lett. 2012;586:2043–2048. doi: 10.1016/j.febslet.2012.03.051. [DOI] [PubMed] [Google Scholar]
- Klumpp S., Hwa T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl. Acad. Sci. U S A. 2008;105:20245–20250. doi: 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov A.S., Podtelezhnikov A., Vologodskii A., Mirkin S.M. Large-scale effects of transcriptional DNA supercoiling in vivo. J. Mol. Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- Kühner S., van Noort V., Betts M.J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Yamashita A., Hirakawa H., Nakayama H., Toh H., Okada N., Kuhara S., Hattori M., Hayashi T., Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U S A. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloréns-Rico V., Lluch-Senar M., Serrano L. Distinguishing between productive and abortive promoters using a random forest classifier in Mycoplasma pneumoniae. Nucleic Acids Res. 2015;43:3442–3453. doi: 10.1093/nar/gkv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloréns-Rico V., Cano J., Kamminga T., Gil R., Latorre A., Chen W.-H., Bork P., Glass J.I., Serrano L., Lluch-Senar M. Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2016;2:e1501363. doi: 10.1126/sciadv.1501363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M., Delgado J., Chen W.-H., Lloréns-Rico V., O’Reilly F.J., Wodke J.A., Unal E.B., Yus E., Martínez S., Nichols R.J. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol. Syst. Biol. 2015;11:780. doi: 10.15252/msb.20145558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker M., Bilusic I., Raghavan R. Pervasive transcription: detecting functional RNAs in bacteria. Transcription. 2014;5:e944039. doi: 10.4161/21541272.2014.944039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Yin Y., Schell M.A., Zhang H., Li G., Xu Y. Computational analyses of transcriptomic data reveal the dynamic organization of the Escherichia coli chromosome under different conditions. Nucleic Acids Res. 2013;41:5594–5603. doi: 10.1093/nar/gkt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mazin P.V., Fisunov G.Y., Gorbachev A.Y., Kapitskaya K.Y., Altukhov I.A., Semashko T.A., Alexeev D.G., Govorun V.M. Transcriptome analysis reveals novel regulatory mechanisms in a genome-reduced bacterium. Nucleic Acids Res. 2014;42:13254–13268. doi: 10.1093/nar/gku976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C.A., Vanderpool C.K., Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E., Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Meyer S., Beslon G. Torsion-mediated interaction between adjacent genes. PLoS Comput. Biol. 2014;10:e1003785. doi: 10.1371/journal.pcbi.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R.A., Davis S.E., Peters J.M., Rowland J.L., Ansari A.Z., Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Nudler E., Gottesman M.E. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Okuda S., Kawashima S., Kobayashi K., Ogasawara N., Kanehisa M., Goto S. Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli. BMC Genomics. 2007;8:48. doi: 10.1186/1471-2164-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Vangeloff A.D., Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J. Mol. Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppas N.B., Wade J.T., Church G.M., Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- Santangelo T.J., Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011;9:319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M., Hwa T. Bacterial growth laws and their applications. Curr. Opin. Biotechnol. 2011;22:559–565. doi: 10.1016/j.copbio.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Singh S.S., Singh N., Bonocora R.P., Fitzgerald D.M., Wade J.T., Grainger D.C. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014;28:214–219. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol. 2002;177:433–440. doi: 10.1007/s00203-002-0407-5. [DOI] [PubMed] [Google Scholar]
- Torres-Puig S., Broto A., Querol E., Piñol J., Pich O.Q. A novel sigma factor reveals a unique regulon controlling cell-specific recombination in Mycoplasma genitalium. Nucleic Acids Res. 2015;43:4923–4936. doi: 10.1093/nar/gkv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A., Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Wade J.T., Grainger D.C. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014;12:647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yus E., Maier T., Michalodimitrakis K., van Noort V., Yamada T., Chen W.-H., Wodke J.A., Güell M., Martínez S., Bourgeois R. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- Yus E., Güell M., Vivancos A.P., Chen W.-H., Lluch-Senar M., Delgado J., Gavin A.C., Bork P., Serrano L. Transcription start site associated RNAs in bacteria. Mol. Syst. Biol. 2012;8:585. doi: 10.1038/msb.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Baseman J.B. Transcriptional response of Mycoplasma genitalium to osmotic stress. Microbiology. 2011;157:548–556. doi: 10.1099/mic.0.043984-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1: List of known or putative transcriptional regulators in M. pneumoniae. The last column indicates the name of the strains in which the TF is perturbed (see Table S2)
Sheet 2: Manual operon and sub-operon annotation of the M. pneumoniae genome. The table indicates the following information for each of the manually annotated transcriptional units (operons and sub-operons): operon number, sub-operon ID, genes belonging to each sub-operon, TSS of the sub-operon, TTS of the sub-operon and strand.
Sheet 1: List of RNAseq experiments used in this work. For each sample, we indicate the strain (wt, M129 or mutant), transgene (indicates the gene that was overexpressed or mutated), timeOfGrowth_experimentPerformedAt in h (time of growth after inoculum), medium used, treatment (type of drug/perturbation), perturbant (drug, condition…), finalConcentration_perturbant (working dilution), durationOfPerturbation in min, Filtered? (in case it was left out of the analysis, see main Materials and Methods)
Sheet 2: list of samples discarded for the co-expression analysis. Sheet 3: Corresponding list of conditions effectively used in the analysis of basal co-expression and of TRT variations. The last column indicates whether the condition was analyzed for TRT variation or if it corresponded to a control. The red names indicate that a single gene was perturbed, in contrast to more global perturbation (various stress shocks, Novobiocin treatments, etc…). The yellow boxes indicate that the perturbed gene is a putative TF (see Table S1).
Sheet 1: Leftmost list: conditions leading to an overall repression of TRT, that is, showing a tendency for having . The average value of Δdown − Δup (third column) is computed over all the TTSs. The list is sorted according to the p values of the bias of the distribution of Δdown − Δup (second column, one sample t test value). The horizontal dashed and full lines respectively indicate the values where the false discovery rate (FDR) is equal to 0.005 and 0.05 (Benjamini–Hochberg procedure). Rightmost list: same thing but for conditions leading to an overall activation of TRT, that is, showing a tendency for having
Sheet 2: Leftmost list: perturbations for which no pair of adjacent genes shows an extreme variation of TRT. Rightmost list: perturbations for which at least 12 pairs of adjacent genes show an extreme variation of TRT. The color codes are those of Table S2.
ChIP-seq peaks associated to RNAP (see Methods and Materials and Supp. Methods text for experimental procedures and for the identification of peaks). For each of the peaks, the following information is displayed: peak position (in bps); peak height (in arbitrary units); peak width (in bps covered); peak score, based on the confidence in the intra-peak distance (see Supplementary Methods); associated TSS(s), if any, otherwise is “NONE”; associated TSS strand(s), if any, otherwise is “NONE”; and time point of the corresponding experiment (6h or 96h).
Oligos used for the RT-qPCR.