Abstract
Aims: Carbon monoxide is a respiratory poison and gaseous signaling molecule. Although CO-releasing molecules (CORMs) deliver CO with temporal and spatial specificity in mammals, and are proven antimicrobial agents, we do not understand the modes of CO toxicity. Our aim was to explore the impact of CO gas per se, without intervention of CORMs, on bacterial physiology and gene expression. Results: We used tightly controlled chemostat conditions and integrated transcriptomic datasets with statistical modeling to reveal the global effects of CO. CO is known to inhibit bacterial respiration, and we found expression of genes encoding energy-transducing pathways to be significantly affected via the global regulators, Fnr, Arc, and PdhR. Aerobically, ArcA—the response regulator—is transiently phosphorylated and pyruvate accumulates, mimicking anaerobiosis. Genes implicated in iron acquisition, and the metabolism of sulfur amino acids and arginine, are all perturbed. The global iron-related changes, confirmed by modulation of activity of the transcription factor Fur, may underlie enhanced siderophore excretion, diminished intracellular iron pools, and the sensitivity of CO-challenged bacteria to metal chelators. Although CO gas (unlike H2S and NO) offers little protection from antibiotics, a ruthenium CORM is a potent adjuvant of antibiotic activity. Innovation: This is the first detailed exploration of global bacterial responses to CO, revealing unexpected targets with implications for employing CORMs therapeutically. Conclusion: This work reveals the complexity of bacterial responses to CO and provides a basis for understanding the impacts of CO from CORMs, heme oxygenase activity, or environmental sources. Antioxid. Redox Signal. 24, 1013–1028.
Introduction
CO is a notoriously toxic gas, binding primarily to ferrous oxygen-reactive heme proteins (21). Nevertheless, CO is generated endogenously in mammals by heme oxygenase (HO)-catalyzed breakdown of heme (62), and, although relatively inert from a chemical standpoint, it is a gasotransmitter or small gaseous signaling molecule with critical biological activities, including vasodilation and anti-inflammatory mediation (47).
The role of CO in pathogenesis and host defense against microbial infection is obscure, with conflicting and unexpected roles reported [reviewed in Wareham et al. (70)]. For example, Wegiel et al. recently hypothesized that bacteria exposed to CO release ATP, which activates inflammatory pathways (71). Although CO may be toxic toward microorganisms [being used to preserve meat (52)], many bacteria are relatively resistant, in part, because they possess CO-insensitive oxidases, such as cytochrome bd (32). Indeed, airborne bacteria survive high urban CO concentrations (39), and bacterial cultures may be bubbled with CO with little toxicity (71).
Innovation.
The beneficial effects of CO-releasing compounds (CORMs) in physiological and antimicrobial therapies are generally attributed to CO, yet bacteria tolerate this gas. This is the first analysis of the global impact of CO (without a CORM) on bacterial growth, gene expression, and responses to stress, thus underpinning interpretation of studies that employ CORMs. Tightly controlled chemostat growth and statistical modeling show that not only global transcriptional responses occur in energy metabolism but also iron transport and thus metal chelator sensitivity and the metabolism of arginine and sulfur amino acids. Unlike other gasotransmitters (H2S and NO), CO provides negligible protection against antibiotics.
CO-releasing molecules (CORMs) were developed for temporal and spatial CO delivery in therapy without intoxication. CORMs are generally metal carbonyls with one or more labile CO groups, which are released by ligand exchange reactions, enzymatic activation, or photoactivation [references in Wareham et al. (70)]. Many CORMs have potent antimicrobial effects, but the mechanism of toxicity has been debated. Even the significance of CO release is unclear, although CORM-derived CO does target oxidases in vivo and CORMs elicit multiple transcriptomic changes in respiratory gene expression (41). Since the actions of CORMs are distinct from antibiotics (70), they are promising replacements for, or alternatively adjuvants to, conventional overused antibiotics in fighting antibiotic-resistant strains. The antibiotic-potentiating effects of certain CORMs (65) have not been reported for CO gas, even though NO and H2S confer some defense against antibiotics (24, 59).
In this study, we present the first systematic multilevel analysis of the bacterial effects of CO gas. Transcription factor (TF) measurements and modeling reveal that gene expression is highly perturbed with major consequences for energy metabolism, iron homeostasis, and amino acid metabolism. Interestingly, a CORM, but not CO gas, is an effective adjuvant to antibiotics, highlighting the importance of the metal ion in bacterial toxicity.
Results
Growth of Escherichia coli aerobically and anoxically in the presence of CO gas
Apart from carboxydobacteria, which oxidize CO to CO2, little is known about the effects of CO on growth of bacteria. To establish a sublethal concentration of CO for analyses, E. coli cells were grown in a batch bioreactor in Evans medium (41) with glucose. In the mid-log phase, the gas mix was switched to 50% CO (by volume, 100 ml·min−1). CO only slightly inhibited growth aerobically (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/ars): growth was linear (not exponential) and the doubling time at the point of CO addition was about 1.6 h, increasing to 2.2 h with CO. Anoxically, however (Supplementary Fig. S1B), CO inhibited growth and a lower biomass was attained.
Overview of transcriptomic responses to CO
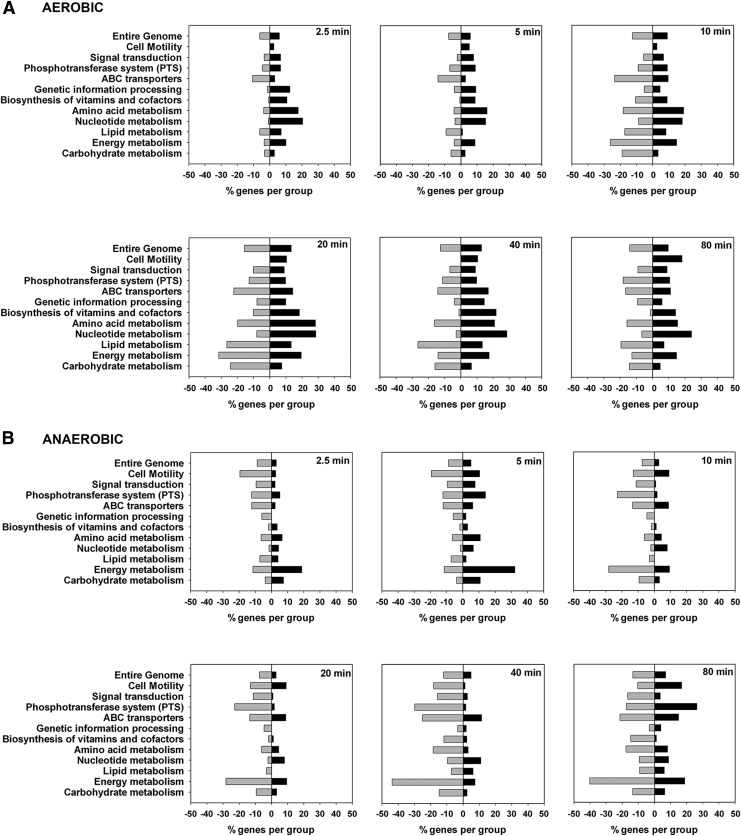
To determine how cells respond globally to CO, we conducted transcriptomic analyses on samples taken immediately before CO gas addition and at regular timed intervals thereafter. CO altered expression of numerous genes, defined as ≥2-fold up or ≥2-fold down (the latter representing a change of ≥0.5 of the control transcript level). Aerobically, exposure to CO gas for 20 min led to 29% of the genome significantly changing, decreasing to 24% after 80 min. Anoxically, most significant gene changes were seen only after 80 min of CO exposure, where 20% of the genome changed significantly in expression (Fig. 1).
FIG. 1.
Functional categories of genes affected by CO gas addition in aerobic and anaerobic conditions. Transcriptomic analyses were performed at 2.5, 5, 10, 20, 40, and 80 min after the flow of CO gas was initiated in both aerobic (A) and anaerobic (B) conditions as described in the Materials and Methods section. Genes are grouped according to functional categories. The percentages of genes in each panel that showed elevated expression (black bars, right) or reduced expression (gray bars, left) are shown.
Aerobic and anaerobic treatments with CO perturb central energy metabolism
CO competes with oxygen for oxidases, inhibiting aerobic respiration. Classically, a CO:O2 ratio of 4:1 (11) is used to measure CO inhibition of respiration and its photochemical relief; thus, the transcriptomic responses to CO of genes involved in energy metabolism were of special interest. In facultative bacteria, the tricarboxylic acid (TCA; Krebs) cycle and glycolysis generate reducing equivalents that feed into multiple oxygen (or other oxidant)-terminated respiratory chains that conserve energy as the protonmotive force for ATP synthesis and membrane transport. Maximal transcript changes occurred after 20 min aerobically [52% of 149 genes in this category; 32% of genes were downregulated and 20% were upregulated (Fig. 1A)].
Figure 2 illustrates pathways of aerobic and anaerobic central metabolism and the CO-induced changes in expression of genes that encode each step. Each color block (separated into six vertical strips) represents a single gene; the strips from left to right in each block denote the transcript level, by reference to the heat scales, at each time increment in the experiment, that is, 2.5, 5, 10, 20, 40, and 80 min after CO treatment (as in Fig. 1). For example, the conversion of acetyl-CoA to citrate is mediated by gltA that encodes citrate synthase (Fig. 2A, left). The adjacent color block indicates that aerobically it is transiently downregulated (blue).
FIG. 2.
CO-induced transcriptomic changes of genes involved in the TCA (Krebs) cycle, glycolytic pathways, and O2-dependent electron transport pathways in aerobic and anaerobic conditions. (A) The major routes of carbon flow through the Embden–Meyerhof pathway and Krebs cycle are shown. (B) Reducing equivalents from NADH are fed via two NADH dehydrogenases (Ndh, Nuo) to a quinone pool (Q) and thence to one of three terminal oxidases (cytochromes bd-I, bd-II, and bo′). Each block of color strips indicates a single gene involved in the reaction step shown and, within each block, the vertical strips show (from left to right) changes in gene expression at the sampling points (2.5, 5, 10, 20, 40, and 80 min after introducing CO gas). Changes in gene expression are illustrated by the heat map (right): blue color indicates a gene that is downregulated, red color indicates upregulation, and yellow color indicates no change in transcriptomic level. TCA, tricarboxylic acid. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Aerobically, CO caused little change in genes involved in glycolysis, but transient downregulation of most Krebs cycle genes, including aconitase (catalyzing citrate to isocitrate via cis-aconitate; acnAB); α-oxoglutarate dehydrogenase; succinyl-CoA synthetase (α-oxoglutarate to succinyl-CoA; sucAB); lipoamide dehydrogenase [the lpd gene product, E3 component of three multicomponent enzyme complexes (16)]; succinate dehydrogenase (succinate to fumarate; sdhABCD); and fumarase (fumarate to malate; fumABC) (Supplementary Fig. S2A for fold changes and details of relevant TFs). The downregulation of Krebs cycle genes in aerobic conditions may reflect a backlog of reducing equivalents on respiratory inhibition by CO.
In contrast (Fig. 2A, right and Supplementary Fig. S2A), anaerobically, CO upregulated certain genes of the Krebs cycle, in particular genes encoding pyruvate dehydrogenase (PDH; aceEF, lpd), aconitase (acnAB), and succinate dehydrogenase (sdh) (Supplementary Fig. S2A), with smaller changes in glycolysis. The anaerobic response was immediate: after 5 min of CO bubbling, aceEF and lpd (together encoding the PDH complex) were upregulated 7-fold and 5-fold, respectively. PDH generates acetyl CoA and acts as a gateway for flux of metabolites into the Krebs cycle. Most highly upregulated after 2.5 min, by up to 13-fold, were the succinate dehydrogenase genes (sdhABCD), followed by a 10-fold downregulation of sdhD after 40 min.
Consequences of CO exposure for components of the respiratory chain
CO inhibits aerobic respiration by binding to all three terminal oxidases (32). Oxidase structural genes are under complex transcriptional control, primarily by oxygen (55), but there are no reports of CO effects. However, aerobically (Fig. 2B, left), there was a 5- to 10-fold decrease in expression of the cyo operon (cyoABCD) that encodes the cytochrome bo′ (bo3) oxidase, but an increase in cydAB expression, which encodes cytochrome bd-I (4-fold upregulation after 20 min) (Supplementary Fig. S2B shows fold changes and relevant TFs). This is consistent with the properties of the oxidases: cytochrome bd-I has high oxygen affinity (19) and a stoichiometry of 1 H+/electron, whereas cytochrome bo′ has lower oxygen affinity and a stoichiometry of 2 H+/electrons (18, 57). Interestingly, cytochrome bd is also less sensitive than cytochrome bo′ to other respiratory inhibitors such as cyanide (3), azide (50), and NO (40). Aerobically, changes in expression of the enigmatic cytochrome bd-II encoded by appBC were slight. In the presence of CO, electron flow into the aerobic transport chain is facilitated in both aerobic and anoxic conditions by upregulation of NADH dehydrogenase (ndh), whereas succinate dehydrogenase (sdhABCD) transcripts diminished (Fig. 2B, left and Supplementary Fig. S2A).
Major changes in terminal oxidase genes were not expected anaerobically due to the absence of oxygen or alternative respiratory electron acceptors. However, the cyoABCD genes responded dramatically to CO by upregulation of the entire operon by 20- to 30-fold after only 5 min of CO exposure (Fig. 2B, right and Supplementary Fig. S2B). The response dampened after 40 min. The effects of CO on ndh and cydAB echoed the response aerobically (Fig. 2B, left and right), but the responses anoxically were greater, with ndh maximally expressed at 80 min; however, in contrast to aerobic conditions, the nuo genes were uniformly downregulated (2- to 5-fold) after 40 min with CO (Fig. 2B, right).
We also assayed the levels of the various quinone species (labeled Q in Fig. 2B) that act as membrane-bound electron and proton carriers from dehydrogenases to the oxidases. Although E. coli responds to reduced oxygen availability on a timescale of seconds, with synthesis (under post-translational control) of quinones involved in anaerobic respiration (7), we found no significant changes in the pool sizes of ubiquinone (UQ), menaquinone (MK), or demethylmenaquinone (DMK) on CO challenge (Supplementary Fig. S3).
Finally, certain CORMs cause accumulation of ROS (66). However, inspection of the complete data sets (which can be consulted at GEO, see the Materials and Methods section) reveals that oxidative stress genes (sodA, sodB, sodC, katE, katG, oxyR, oxyS) were not upregulated by CO gas, either aerobically or anaerobically. This suggests that CO gas does not induce oxidative stress.
Modeling of the transcriptomic data by TFInfer
The abundance of TFs that underlie multiple fluctuations in transcript levels frustrates data interpretation. In this study, we used the TFInfer approach (4, 37), a Bayesian statistical method that integrates gene expression data with regulon information (culled from Regulon DB or EcoCyc) to identify TF activity profiles that aid in understanding of the raw transcriptional changes. To compare results from TFInfer on different data sets (CO gas under aerobic and anoxic conditions), we used intuitive coherence plots that highlight differences in the magnitude and kinetics of the response to the two stimuli.
Figure 3 summarizes the differences between two sets of TFInfer data (growth with CO either aerobically or anoxically) in a scatter plot. The x-coordinate of each point (labeled with the TF identity) represents the profile difference between the two conditions, computed as 1 minus the absolute Pearson correlation coefficient between the two profiles, while the y-coordinate represents the change in magnitude of the response (computed as the absolute difference of the norms of the two profiles). Hence, TFs whose response is similar both in magnitude and kinetics are located near the origin of the coherence plot, while TFs in the top right corner of the plot respond differently in both kinetics and amplitude. In quadrants A, C, and D, one representative TF is shown to illustrate the value of the plots. Error bars (shown only in Supplementary Fig. S4) represent 95% confidence intervals; to reduce clutter, we have omitted TFs where the error bars for the absolute Pearson correlation were greater than ±0.15.
FIG. 3.
TFInfer correlation profile of TF activities in CO aerobic conditions versus anoxic conditions. Profile differences are plotted on the abscissa and differences in magnitude on the ordinate. In quadrants (A, C, D), the dynamics of one representative TF are shown both aerobically and anaerobically. For example, the response profiles for the TFs, FNR and CysB, are similar in both conditions, in both the magnitude of the response and their correlation (appearing in the lower left quadrant), while GadX and PdhR (upper left) have a similar response in terms of the shape of the profile, while the magnitude of the response is greater in anaerobic conditions than aerobic conditions. On the other hand, the response of the TF MetJ is different both in magnitude and in the shape of the profile, lying near the interface of the upper and lower right quadrants. ArgR and HNS have a similar response in terms of the magnitude of the profile, while the shape of the activity curve of the response is greater in aerobic conditions than anaerobic conditions. Absolute Pearson correlations in the middle indicate only weak similarity between profiles. FNR, fumarate nitrate reduction regulator; TF, transcription factor.
For example, under acid conditions, GadX (with GadE) positively regulates gadA and gadBC, which encode components of the glutamate-dependent acid response (68). GadX is inducible by NO (9), but was not known to be regulated by CO. While the magnitude of the GadX response is different aerobically and anaerobically (quadrant A), that of CysB is similar in both conditions, reflected in its location in quadrant C of Figure 3. Its role in cysteine metabolism is discussed below. Finally, ArgR in quadrant D shows marked profile differences aerobically and anaerobically; its role in arginine metabolism is also explained below.
The inferred role of ArcA in the response of E. coli to CO
The TFInfer coherence plot reveals three TFs that collectively regulate genes in central metabolism: PdhR (Fig. 3A), fumarate nitrate reduction regulator (FNR) (Fig. 3C), and ArcA (Fig. 3D). Each TF responds distinctively to CO. ArcA responded differently under each condition (Figs. 3D and 4). ArcAB, a two-component system, indirectly senses oxygen via the redox state of the quinone pool, fermentation products, and perhaps other factors (2). Under anoxic or microaerobic conditions, ArcB autophosphorylates, then transphosphorylates ArcA through a phosphorelay, thereby increasing the affinity of ArcA for its DNA targets (31). Phosphorylated active ArcA (ArcA-P) then represses expression of genes involved in respiration (e.g., electron transport enzymes, cytochrome bo′, and the Krebs cycle enzymes) and activates genes involved in fermentative metabolism and cytochrome bd. Thus, in a mutant lacking two of the three oxidases, the aerobic expression of ArcA-P-activatable genes, such as cydAB, is elevated, but that of ArcA-P-repressible genes, such as cyoABCDE, is lowered (30) because the quinone pool is trapped in a reduced form and unable to inhibit the autokinase activity of ArcB. CO would be expected to mimic such an absence of oxidase activity by inhibiting electron transfer to oxygen.
FIG. 4.
Inferred activity of the TFs, ArcA, FNR, and PdhR, in response to CO. Response profiles for ArcA (top, circles), FNR (middle, squares), and PdhR (bottom, triangles) are shown as predicted by TFInfer. TF activity is plotted for each time point using TFInfer data modeled on gene changes in response to CO at each time point. Aerobic profiles are shown with closed symbols (left) and anoxic profiles with open symbols (right).
The modeled changes in ArcA activity (Fig. 4) are consistent with this: aerobically, ArcA activity increases rapidly, in part, due to quinone over-reduction. After 20 min, declining ArcA reflects partial reversal of the effects of CO. This pattern may relate to a transient overshoot of the UQH2/UQ ratio observed in transitions from aerobic to sustained anaerobic conditions [(7); JWA van Beilen and KJ Hellingwerf, unpublished]. The increased activity of ArcA represses Krebs cycle gene expression in the first 20 min of CO exposure (Fig. 2A), but enhances cydAB expression (Fig. 2A, B). Thus cells respond, not directly to CO, but to diminished oxygen availability due to competition by CO.
Anoxically, the falling ArcA(-P) activity correlates with the increase in expression of Krebs cycle and cyo genes (Fig. 2A, right) (55). This may perhaps be attributed to inhibition by fermentation products of ArcB phosphatase activity (29, 31) rather than the respiratory quinone pools since these would not be expected to display redox changes under anoxia in the absence of alternative electron acceptors (6, 55). Anoxic CO exposure increased cytochrome bd-1 gene expression (Fig. 2B), as expected from the initial rise in ArcA activity.
Phosphorylation of ArcA and pyruvate accumulation in the response to CO
The role of Arc in the CO response was confirmed by direct assay of the phosphorylation state of ArcA (Fig. 5A–C). To measure the proportion of phosphorylated ArcA as a percentage of the total protein, Phos-tag™-acrylamide gel electrophoresis was used, followed by Western blotting. Within 5 min of aerobic CO exposure, and persisting for >20 min, there was a marked increase in ArcA-P, consistent with the inferred ArcA activity profile (Fig. 4). Anaerobically, significant levels of ArcA-P were present at 5–80 min, irrespective of the presence of CO (Fig. 5C), also consistent with ArcA activity (Fig. 4). Precise temporal correlation is impractical given the limitations of sampling.
FIG. 5.
The phosphorylation state of ArcA in the absence and presence of CO and corresponding pyruvate levels in cells. (A, B) Typical Western blots developed using ArcA polyclonal antiserum for Phos-tag™ gels and cultures grown with/without CO at the time points shown, both aerobically and anaerobically. Controls with purified ArcA are shown: the lane labeled “+” shows ArcA-P (25 ng purified ArcA phosphorylated in vitro using carbamoyl phosphate), and the lane labeled “−” shows 25 ng unphosphorylated ArcA. (C) The percentage of Arc-P in each sample was calculated from band intensities; results show ArcA-P expressed as a percentage of total ArcA/ArcA-P in each sample.(D) Pyruvate levels were assayed in culture supernatants collected over time (in each condition: solid bars, t = 0; hashed bars, t = 5; spotted bars, t = 20; checkered bars, t = 80).
The effect of fermentation products, such as lactate and pyruvate, on ArcB is obscure, but D-lactate, pyruvate, and NADH enhance the autophosphorylation of ArcB (29). We therefore assayed extracellular pyruvate and showed (Fig. 5D) that (over 80 min) CO markedly increases aerobic accumulation of pyruvate (67). Anaerobically, as expected, pyruvate accumulated to higher levels, irrespective of the presence of CO. Interestingly, PdhR activity (Fig. 4) declined during aerobic accumulation of pyruvate (Fig. 5).
The role of FNR in the response of E. coli to CO
The ligand-reactive FeS clusters of FNR make it a direct sensor of oxygen availability. Under microaerobic/anaerobic conditions, it negatively regulates Krebs cycle (e.g., fumA, sdhABCD, sucABCD) and oxidase genes (cydAB and cyo operons) (14). TFInfer (Fig. 4) predicted that FNR activity increases in the presence or absence of oxygen after CO treatment. Nevertheless, the cydAB operon was also upregulated after CO exposure in both conditions (Fig. 2B and Supplementary Fig. S2B), implicating additional factors. Indeed, the cydAB operon is under complex regulation, being repressed by FNR and the nucleoid DNA-binding protein H-NS (22) and induced by ArcA and FruR (53). In addition, the expression of aerobic respiratory pathways is influenced by iron availability (13) (Figs. 6, 7). Whatever the mechanism, the upregulation of the cydAB operon by CO suggests that CO induces a transition to a more anoxic-like state (20).
FIG. 6.
Predicted TF activity of Fur, a transcriptional repressor of Fe-regulated genes, and corresponding gene changes. TFInfer data over time for Fur are shown under (A) aerobic and (B) anaerobic conditions in the presence of CO. (C) and (D) map changes in iron-regulated genes aerobically and anaerobically, respectively. Yellow indicates genes that remain unchanged in the presence of CO, red indicates genes upregulated, and blue indicates genes downregulated, according to the heat map (right). The intensity of the color is indicative of the trust placed on that measured level of regulation after three biological repeats. Feature extraction from the scanned arrays and subsequent data analysis used GeneSpring GX v7.3. The clustering at the left is a GeneSpring gene tree and shows genes organized by their similarity in response to the imposed conditions. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 7.
Cells exposed to CO produce higher levels of siderophores as free intracellular iron pools are depleted. Stationary Escherichia coli cultures were grown and 10 μl aliquots of culture were spotted onto 25-ml CAS agar plates. Cells were incubated in aerobic (air)/aerobic + CO (air +50% CO) or anaerobic (N2)/anaerobic + CO (N2/50% CO) conditions for 24 h. The diameter of the cells plus halo and the diameter of the cells alone were measured and the difference plotted in (A). The presence of CO increased siderophore production in aerobic and anaerobic conditions; results are plotted as mean ± SD of two biological repeats each with four technical replicates. A paired t-test shows *p of 0.03 and **p of 0.008. (B) The ferric iron in rhombic coordination in whole cells as measured by electron paramagnetic resonance (EPR) spectroscopy shows that the iron levels decreased both aerobically and anaerobically on exposure to CO gas. Results are plotted as mean ± SD of two biological repeats each with two technical replicates. A paired t-test shows *p of 0.05 and **p of 0.005. CAS, Chrome Azurol S.
In contrast, the aerobic downregulation of cyo expression is explicable by rising FNR activity alone. Anoxically, cyo expression is elevated, but this operon too is under complex regulation, being repressed by ArcA, Fur, and PdhR, as well as FNR. GadE (28) and CRP (75) upregulate cyo expression.
PdhR and other regulators
The pdhR gene is the proximal gene of the pdhR-aceE-aceF-lpd operon encoding the PDH complex. PdhR functions with other central regulators as an additional master switch (46), negatively regulating expression of the pdh promoter, with pyruvate serving as an inducing coeffector (51) and repressing the cyo and ndh operons (46). The structural genes of the PDH operon (Fig. 2A) as well as cyoABCD and ndh were all upregulated by CO under anoxic conditions (Fig. 2B and Supplementary Fig. S2), as was the cyo operon, implying loss of the repressing activity of PdhR, perhaps due to pyruvate (46). TFInfer analysis (Fig. 4) also shows that aerobically PdhR activity declines, consistent with a CO-induced block of respiration and pyruvate accumulation (Fig. 5D).
Electron flow into the respiratory chains when CO is present is facilitated both aerobically and anoxically (Fig. 2B) by immediate and marked upregulation of ndh, encoding NADH:ubiquinone oxidoreductase II, a single-subunit nonproton-translocating enzyme. FNR represses ndh expression anoxically and expression of an ndh-lacZ fusion in an fnr mutant is enhanced by anaerobiosis (23). Therefore, the inferred increase in FNR activity (Fig. 4) and ndh upregulation are not consistent with simple repression of ndh by FNR. Other regulators may be implicated, such as Fis, which enhances ndh expression during rapid growth (31). Expression of Ndh is associated with growth where protonmotive force generation is not favored (10), consistent with the presence of CO. In contrast to ndh, the nuo genes encoding a multisubunit, proton-pumping NADH dehydrogenase were downregulated (2- to 5 fold) in response to CO in anoxic conditions (Supplementary Fig. S2B). These results together with downregulation of the cyo operon (Fig. 2) suggest that CO promotes assembly of a respiratory chain that is not optimized for energy-efficient growth, but for the disposal of reducing equivalents (i.e., NAD(P)H) (10).
CO exposure perturbs genes involved in amino acid biosynthesis
Global responses to CO included gross alterations in amino acid metabolism (Fig. 1). First, many genes in the cys and tau operons were upregulated, suggesting a shortage of sulfur in response to CO (Supplementary Fig. S5A). The major regulator is CysB (35), which lies adjacent to FNR in the coherence profile (Fig. 3; bottom left quadrant), indicating that the response was similar in magnitude and kinetics both aerobically and anaerobically. In Salmonella, CysB, activated by N-acetylserine in the absence of cysteine (35), forms a DNA-binding tetramer. CysB activates the cys regulon for sulfur utilization and uptake, the tau operon for taurine utilization, the cbl gene encoding a regulator of the sulfate starvation response, and, in coordination with the Cbl protein, the sulfonate utilization genes (ssuABCDE) (69). Many of these genes are upregulated in response to CO (Supplementary Fig. S5A) after 10 min of CO exposure, especially cysB, cysC, cysD, cysH, and cysP, as well as most tau genes.
Second, many genes involved in methionine biosynthesis were perturbed by CO (Supplementary Fig. S5B), but with complex patterns of expression. Aerobically, a requirement for methionine appears immediate with many met biosynthetic genes upregulated by CO, especially metA, metB, metF, metK, metN, and metR. MetJ, which represses the expression of genes involved in biosynthesis and transport of methionine (5), was elevated in the coherence plot (Fig. 3D), indicating differences in the aerobic and anoxic responses (Supplementary Fig. S5B).
Third, ArgR responded differently to CO aerobically and anoxically (Figs. 3 and Supplementary Fig. S6A). Most arginine biosynthesis genes were highly upregulated aerobically as well as carA encoding carbamoyl synthetase. Additionally, arginine transport genes (artJ-Q) were upregulated aerobically with artJ, encoding the periplasmic protein of the L-arginine transporter, upregulated by 20-fold after 10 min. However, anaerobically, these genes were predominantly downregulated (Supplementary Fig. S6A). Thus, CO appears to lead to arginine deprivation under aerobic conditions. Interestingly, peroxynitrite also increases arginine biosynthesis transcript levels in E. coli due to nitration of the ArgR repressor (42). However, CO is relatively inert (CO dehydrogenase being the only documented biological redox activity) and it is unclear how it could modify or degrade ArgR (see the Discussion section).
CO leads to altered expression of iron acquisition genes
The location of Fur in the upper left quadrant of the coherence plot (Fig. 3A) indicates that it exhibited significantly different responses under aerobic and anoxic conditions. Since the primary biological targets of CO are heme- and [Fe-S] cluster-containing proteins (8), we investigated the role of Fur, the principal TF involved in detecting iron levels and repressing genes for iron acquisition (56). Aerobically (Fig. 6A), the inferred Fur activity initially rose, consistent with downregulation of ent genes involved in enterochelin biosynthesis (Figs. 6C and Supplementary Fig. S6B) and the fep and fhu genes involved in ferric enterobactin transport and hydroxamate-dependent iron uptake, respectively. Anoxically, there was extensive and sustained derepression of iron-regulated genes (Figs. 6D and Supplementary Fig. S6B) and inferred Fur activity fell sharply (Fig. 6B). For example, entE (encoding a component of enterobactin synthase) was 30-fold upregulated anaerobically after 10 min; genes involved in enterobactin (fepABCDG) and ferrichrome transport (fhuA-F) were also upregulated. Overall, the data point to CO-induced iron deprivation, particularly anoxically. The changes in respiratory genes (Fig. 2B and Supplementary Fig. S2B) are also consistent with modulation by iron. Upregulation of cydAB expression, particularly anoxically, is consistent with elevated cydAB expression by the iron chelator, 2,2′-dipyridyl (13).
Exposure of cells to CO leads to increased siderophore production and a reduction in the levels of intracellular free iron
Given the changes in ent gene expression, it was important to determine whether cell physiology reflected transcript levels. Therefore, cultures growing on Chrome Azurol S (CAS) agar plates were incubated in CO atmospheres. Fe(III) reacts with CAS to produce an intense blue color, but siderophores compete with the dye for Fe(III), causing a color change (blue to orange). Siderophore production on CAS plates caused a clear orange halo around the bacterial growth (not shown). Both aerobically and anaerobically, CO increased halo diameter (corrected for the spot size, representing siderophore production; Fig. 7A), supporting the transcriptomic data, and suggesting that CO leads to iron limitation.
To test whether free intracellular iron pools were depleted upon exposure to CO, EPR analysis of ferric ions in rhombic coordination was performed (34). To measure total free iron, the cell-permeable iron chelator, desferrioxamine (which promotes the oxidation of Fe(II) to Fe(III) and chelates all Fe(III)), was used. Importantly, desferrioxamine does not chelate protein-bound iron. Fe(III) in rhombic coordination gives an EPR signal with g = 4.3, which was measured on whole cells (Fig. 7B). Total iron levels decreased by about 50% on exposure to CO both aerobically and anaerobically. Aerobically, free intracellular iron levels are ∼40 μM in the absence of CO, falling to 15 μM with CO (Fig. 7B). As hypothesized, free iron levels decreased anaerobically from ∼200 to ∼100 μM with CO. Thus, CO depletes free cellular iron, consistent with patterns of gene expression and elevated siderophore synthesis.
CO renders cultures hypersensitive to external chelators
Given the changes in iron-related genes and iron pools, we hypothesized that iron chelators would inhibit growth in CO since intracellular iron limitation would be exacerbated by exogenous chelating activity. Bacteria were therefore grown with 8-hydroxyquinoline or citric acid in CO atmospheres (Fig. 8A, B). For each, the aerobic (left) and anoxic data (right) show the effects of increasing concentrations (as a percentage of the no-chelator control in the same gas atmosphere). The effects of CO (25% by volume of the incubation jar) are compared with the same atmospheres, but in which 25% nitrogen was used as control. CO alone slightly inhibited aerobic growth. 8-Hydroxyquinoline inhibited aerobic growth, relative to the nonchelator control, by up to 62% with CO, but only 15% without (Fig. 8A). Anaerobically, the values were 27% and 18%, respectively. 8-Hydroxyquinoline has a high affinity for Fe(III) (log stability constant = 26.3) and, to a lesser extent, Fe(II) (log stability constant = 15.0) (49), so other chelators with lower specificity were also tested. Cells grown with citric acid (log stability constant with Fe(III) = 11.4) experienced inhibition in growth; anoxically, CO gave up to 65% inhibition of growth relative to the nonchelator control aerobically and up to 62% anaerobically (Fig. 8B). We also performed assays using Biolog phenotype arrays and found that 1,10-phenanthroline (log stability constant with Fe(III) = 14.1) gave similar results to citrate; it inhibited aerobic growth, relative to the nonchelator control, by 78%–88% with CO, but only 37%–81% without. Anaerobically, the values were 54%–87% and 19%–74%, respectively. These data are not shown as the chelator concentrations in proprietary Biolog plates are unknown. We did not use desferrioxamine in these experiments designed to test extracellular retention of iron since this chelator is cell permeable. In conclusion, CO exacerbates inhibition of E. coli growth by chelating agents.
FIG. 8.
Effects of CO gas on bacterial sensitivity to metal chelators and antibiotics. Aliquots of E. coli in defined medium were dispensed into the wells of a 96-well plate and supplemented with the following compounds at the concentrations shown: (A) 8-hydroxyquinoline, (B) citric acid, (C) the antibiotics doxycycline (DC), trimethoprim (TR), and cefotaxime (CT). Percentage inhibition was calculated as 100 − (X/Y × 100), where X denotes growth observed in a CO atmosphere (25% CO +75% air or 25% CO +75% N2) and Y the observed growth in the equivalent atmosphere without CO. Data are representative of two biological repeats and each of four technical repeats, expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (Paired t-test).
CO and a CORM: antimicrobial agents or protectants against antibiotics?
With the advent of antibiotic resistance, combinatorial approaches may reduce dependency on the use of single antibiotics (45). Both NO and H2S protect bacteria from a broad range of antibiotics (24, 59), but CO effects have not been reported. Without antibiotics, CO reduced aerobic growth yield by 36%; anoxically, however, CO stimulated growth by 33% (Fig. 8C). This is shown also in the no-chelator plots in Figure 8A and B, but is currently unexplained. CO only marginally affected growth in the presence of the antibiotics, doxycycline (DC), trimethoprim (TR), or cefotaxime (CT). For example, CT (Fig. 8C) inhibited the no-CO control cells by 42%–44%, but cells grown in a CO-supplemented atmosphere by 31%–33%. The results were similar, irrespective of the presence of oxygen.
Since CO gas is not a potent antimicrobial agent (Supplementary Fig. S1), yet CORMs generally are (70), we compared the effects of CO gas with an extensively used ruthenium CORM, CORM-2. The toxicity of this compound allowed us to use it in agar plates where its effects on the minimal inhibitory concentrations (MICs) of three well-established antibiotics (TR, DC, and CT) were determined. In each case, 30 μM CORM-2 dramatically reduced the MICs, whereas the inactive form of the control compound iCORM-2 (Ru(II)Cl2(DMSO)4) and the solvent DMSO were without significant effect on the MICs (Supplementary Fig. S7).
Discussion
CO delivery to key sites offers novel pharmacological and therapeutic approaches, but we know too little of how CO mediates its multiple biological effects. Although CO's affinity for heme proteins is undisputed, its actions in vivo are remarkably complex (54), having further roles in NO release, reactive oxygen species formation, and ion channels (54). It has been tacitly assumed that the biological impacts of CORMs are attributable largely to CO delivery to hemes, but microbial (70) and mammalian (54) studies point to greater complexity. Indeed, bacteria are often resistant to CO gas (71), but not to certain CORMs, and demonstrate multiple transcriptomic changes that cannot be understood in terms of known CO biochemistry [e.g., Wilson et al. (72)]. Furthermore, even iCORMs, frequently assumed to be CO depleted, can exert toxicity in bacterial systems (41). Most tellingly, cells lacking all hemes are also inhibited by CORM-3 and reveal multiple transcriptomic changes (73). Finally, other compounds of Ru are taken up and have antimicrobial properties, even though they are not CORMs [e.g., Li et al. (38)].
We therefore characterized the effects of CO gas per se, without mediation of a CORM, on bacterial growth, gene expression, and physiology. HO-derived CO induces the dormancy regulon in Mycobacterium tuberculosis (60). The CO-oxidizing Archaeon Archaeoglobus fulgidus responds to CO with only limited transcriptional effects (26): of 52 genes observed to alter, inorganic ion transport was the most prominent category, along with cooF, encoding a CO dehydrogenase Fe-S enzyme. Bacterial mechanisms for resisting CO are unclear. Zacharia et al. (74) used a 2% CO atmosphere to screen for M. tuberculosis mutants susceptible to CO, but the resistance test reported for the cor gene identified used CORM-2, not CO gas.
We demonstrate here for the first time that genes encoding energy transduction are significantly affected by CO via the master regulators, ArcA and FNR. In the case of Arc, CO inhibition of respiration promotes pyruvate accumulation (Fig. 5D) and may elicit changes in the redox state of the quinone pool (which is not significantly altered in its composition; Supplementary Fig. S3). The effect of CO on UQ reduction levels could be studied in a UQ-only mutant. These changes indirectly result in modulation of Arc activity (6, 29, 31) (Fig. 9). In contrast, FNR is primarily a direct sensor of oxygen, although the 4Fe-4S cluster of the transcriptionally active factor is also sensitive to NO, rendering it inactive as a repressor (15). It is not known whether the Fe-S cluster of FNR is reactive with CO, but CO can bind to other nonheme sites (36, 63).
FIG. 9.
Schematic diagram of the global impacts of CO gas on E. coli. Inhibition of the electron transfer chain results in stimuli that cause ArcA phosphorylation, namely over-reduction of the quinone pool and formation of the fermentation product, pyruvate, in concentrations that can be assayed extracellularly. Transcriptomic responses are consistent with direct or indirect modulation by CO of Fnr and Fur activities.
A striking consequence of CO exposure was enhanced expression of iron acquisition genes and changes in Fur activity. The reason for the loss of total free iron in cells is unclear, but CO may result in more iron being protein bound, perhaps in inactive states. Bacteria thus respond by producing more iron-containing proteins, further draining iron resources. The change in Fur activity may also be due to a direct effect of CO on the protein since the active form of Fur contains a nonheme ferrous iron site with oxygen and nitrogen donor ligands (1). NO reacts with Fe(II) in Fur, inhibiting Fur repressor activity and Fur DNA binding (17), so interaction with other ligands cannot be discounted. Significantly, Fur in Anabaena interacts with and binds to heme (61), affecting DNA binding (25). The binding of heme to Fur exhibits the same physicochemical features as heme sensor proteins and the Fe(II) heme-Fur binds CO (48); His is the probable axial ligand trans to CO. Binding of CO to Fur-associated heme would be more likely anoxically, consistent with the upregulation of Fe-related genes under anoxic conditions.
The upregulation of genes involved in amino acid metabolism was unexpected (Supplementary Figs. S5 and S6), although a transcriptomic study with CORM-3 showed similar results, with sulfur metabolism being a prime target (41). The introduction of carbonyl groups into proteins by oxidation of amino acid side chains or by oxidative cleavage of proteins is also possible. Protein carbonylation is metal-catalyzed oxidation, which occurs when reduced metal ions (e.g., Fe(II) or Cu(I)) react with H2O2 in Fenton chemistry to produce highly reactive hydroxyl radicals. This in turn oxidizes amino acid side chains or causes protein backbone cleavage, both resulting in the formation of carbonyl groups. In bacteria, metal-catalyzed oxidations appear to be the predominant source of carbonylated proteins (44). Arginine is one of the four most commonly carbonylated amino acids. Interestingly, the present data point to iron deprivation on CO treatment and could reflect loss of iron due to carbonylation.
Contrary to recent findings with NO (24) and H2S (59), we found no evidence that CO significantly protected bacteria against antibiotics, nor did it potentiate their effect. However, CORM-2, as reported previously for Helicobacter pylori (65), markedly reduced the MIC for three commonly used antibiotics. The results imply that the effects of CORMs are not attributed to CO alone and support a role for the metal in CORM toxicity. The mechanisms underlying these effects require urgent investigation to facilitate future applications of combination therapy.
Materials and Methods
E. coli strains and growth conditions
E. coli K12 derivative MG1655 was used throughout. For transcriptomic studies, cells were grown in Evans medium (41) with glucose as the carbon source. In continuous culture, cells were grown in an Infors Multifors bioreactor (working volume 200 ml) adapted to fit a Labfors-3 fermenter base unit. Temperature was maintained at 37°C with continuous stirring at 200 rpm, and pH was maintained at 7.0. Mass flow controllers maintained gas mixes for aerobic and anaerobic conditions by continuous bubbling with air (aerobic) or with N2 gas (anaerobic) at 200 ml·min−1; CO was bubbled at a rate of 100 ml·min−1 with the appropriate gas mix for continuous culture conditions.
Transcriptomic analysis
Experiments were performed and analyzed as before (41) on samples from chemostat cultures immediately before CO gas addition and at time intervals thereafter. RNA was isolated using phenol–ethanol extraction, and cDNA was synthesized and labeled with Cy-3 or Cy-5-dCTP (41). Arbitrary values of ≥2-fold or ≤0.5-fold changes were chosen to show genes with significantly altered expression. Functional category gene lists were created using KEGG (Kyoto Encyclopedia of Genes and Genomes) (33). Regulatory proteins for each gene were identified using EcoCyc. Modeling of TF activities was carried out using TFInfer (4, 37). Data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE50713 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50713).
Arc phosphorylation assays
For measurements of ArcA phosphorylation, and quinone and pyruvate levels, batch cultures were grown in 200 ml Evans medium (supplemented with 5% LB broth [LB; Miller] for anaerobic growth) in Duran bottles with modified lids allowing gas bubbling into the culture. Cultures were inoculated (5%) with prewashed LB-grown cells and incubated at 37°C in a static water bath with a gas supply of either air (aerobic) or 95% N2 with 5% CO2 (anaerobic) until an OD600 of 0.5–0.6 was reached. Mass flow controllers as above controlled gas mixing and delivery. At time zero, the gas supply to the culture was switched, where appropriate, to a mix of 50% CO and 50% of the original gas. Anaerobic samples were removed using a sterile syringe and needle through a tube without the need to remove the lid.
For measuring ArcA phosphorylation, samples (1 ml) were taken at t = 0 and at 5, 20, 40, and 80 min thereafter and added to a microfuge tube containing 200 μl formic acid (6 M). After centrifugation, the pellet was resuspended in 50 μl formic acid (1 M), centrifuged again, and the final pellet frozen in liquid nitrogen, then stored at −20°C. ArcA phosphorylation levels were measured using Phos-tag™-acrylamide gel electrophoresis (NARD Institute, Ltd.) and subsequent Western immunoblotting (55). ArcA was immunodetected using rabbit ArcA polyclonal antiserum and the ECL system (GE Healthcare). ArcA protein was purified as before (6). Values were corrected for the OD600 of the culture at the point of harvest. ArcA phosphorylation of samples in each of two independent cultures (biological repeats) was analyzed in duplicate and the results of one experiment are shown in Figure 5.
Quinone extraction and analysis
Culture samples (3 ml) taken at t = 0 and t = 40 min, and immediately frozen in liquid nitrogen, were stored at −20°C. Quinone extraction and measurement methods were adapted from (58). During evaporation of the petroleum ether, 100 μl hexanol was added and the remaining material, dissolved in hexanol, was transferred to HPLC vials and stored at −20°C until analyzed. Standards for UQ, DMK, and MK were prepared from isolated quinone extracts from E. coli MG1655. Concentrations for each quinone type were determined using extinction coefficients (27, 43). The quinone content of two independent cultures was analyzed in duplicate.
Quantification of pyruvate
Cultures were sampled as shown in Figure 5D and the supernatants frozen in liquid nitrogen, then stored at −20°C until the assay was performed. A Pyruvate Assay Kit (MAK071; Sigma Aldrich) was used according to the manufacturer's protocol. Concentrations were corrected for the OD600 of the culture at harvest. Pyruvate content of two independent cultures was analyzed in duplicate.
Siderophore assays
Siderophore production was assayed on CAS agar plates (12) supplemented with a trace element solution that (at 100 × strength) contained (L−1) 8 ml concentrated HCl, 0.41 g ZnO, 2 g MnCl2.4H2O, 0.17 g CuCl2.2H2O, 0.48 g CoCl2.6H2O, 64 mg H3BO3, and 4 mg Na2MoO4.2H2O. Batch cultures were grown in LB medium to an OD600 of ∼0.5 and 10 μl portions spotted on 25-ml plates that were incubated for 48 h aerobically (100% air) or in anaerobic jars (HP0011A, Oxoid). A vacuum was first used to withdraw air and the vacuum gauge was read; the first gas (e.g., CO) was then admitted via a valve to restore 50% of atmospheric pressure, and then the second gas (e.g., air) was admitted to restore normal atmospheric pressure. Siderophore production was determined by measuring halo diameters (Fig. 7).
Whole cell electron paramagnetic resonance analysis
The method was derived from (34, 64). Cells were grown in LB to an OD of ∼0.2 before centrifugation at 7800 g at room temperature and concentrated 200- to 300-fold before incubation at 37°C for 15 min in 20 mM desferrioxamine. Cells were again centrifuged, washed with cold 20 mM Tris (pH 7.4), and resuspended in a final volume of ∼0.5 ml Tris/10% glycerol. Aliquots (0.3 ml) were placed in EPR tubes, which were flash-frozen in methanol kept on dry ice. EPR measurements were performed at 10 K on a Bruker EMX EPR spectrometer equipped with a spherical high-quality Bruker resonator SP9703 and an Oxford Instruments liquid helium system for low temperature measurements. To measure intensities of the g = 4.3 EPR signal from rhombic ferric iron, the procedure of spectral subtraction with a variable coefficient was used (64).
Growth of E. coli in anaerobic jars to test chelators
Aliquots of a cell suspension (OD 0.05) were dispensed into the wells of a 96-well plate and supplemented with the chelators. Plates were incubated both aerobically (25% CO +75% air or 25% N2: 75% air) and anaerobically (25% CO: 75% N2 or pure N2) in anaerobic jars, as above, at 37°C for 24 h. Preliminary testing of the combined effects of CO and antimicrobial compounds was done using Biolog Phenotype Microarrays.
Effect of CO gas and CORM-2 on bacterial growth in conjunction with chelators and antibiotics
Aliquots of a cell suspension (OD 0.05) were dispensed into a 96-well plate and supplemented with the following compounds at the concentrations shown in Figures 8 and S7: 8-hydroxyquinoline, citric acid, or the antibiotics DC, TR, and CT. For aerobic experiments, growth in 25% CO +75% air was compared with growth in 25% N2 + 75% air. For anaerobic experiments, growth in 25% CO +75% N2 was compared with growth in N2 alone. Optical density was measured after 24 h of incubation at 37°C. Percentage inhibition was calculated as 100 − (X/Y × 100), where X denotes growth observed in a CO atmosphere (25% CO +75% air or 25% CO +75% N2) and Y the observed growth in the equivalent atmosphere without CO.
Determination of MIC values for antibiotics
Cell suspension (100 μl) was added to 4 ml of 0.5% agar and the mixture poured onto defined minimal agar plates. Plates were left to dry for ∼10 min before antibiotic strips (E-test, bioMérieux) were placed on the surface of the plates, which were then incubated at 37°C for 24 h.
Supplementary Material
Abbreviations Used
- ABC
ATP-binding cassette (in ABC transporters)
- CAS
Chrome Azurol S
- CORM
carbon monoxide-releasing molecule
- CORM-2
tricarbonyldichlororuthenium(II) dimer, [Ru(CO)3Cl2]2
- CORM-3
Ru(CO)3Cl(glycinate)
- CT
cefotaxime
- DC
doxycycline
- DMK
demethylmenaquinone
- DMSO
dimethyl sulfoxide
- EPR
electron paramagnetic resonance
- Fe-S
iron-sulfur (as in cluster)
- FNR
fumarate nitrate reduction regulator)
- HO
heme oxygenase
- iCORM-2
inactivated CORM-2
- MIC
minimal inhibitory concentration
- MK
menaquinone
- OD
optical density
- PDH
pyruvate dehydrogenase
- TCA
tricarboxylic acid
- TF
transcription factor
- TR
trimethoprim
- UQ
ubiquinone
Acknowledgments
This work was supported, in part, by the Biotechnology and Biological Sciences Research Council (UK, BBSRC) and the Kurdistan Regional Government (KRG, Iraq). G.S. acknowledges support from the European Research Council under grant MLCS306999.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Adrait A, Jacquamet L, LePape L, dePeredo AG, Aberdam D, Hazemann JL, Latour JM, and Michaud-Soret I. Spectroscopic and saturation magnetization properties of the manganese- and cobalt-substituted Fur (Ferric uptake regulation) protein from Escherichia coli. Biochemistry 38: 6248–6260, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Alvarez AF. and Georgellis D. In vitro and in vivo analysis of the ArcB/A redox signaling pathway. Methods Enzymol 471: 205–228, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft JR. and Haddock BA. Synthesis of alternative membrane-bound redox carriers during aerobic growth of Escherichia coli in the presence of cyanide. Biochem J 148: 349–352, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asif HM, Rolfe MD, Green J, Lawrence ND, Rattray M, and Sanguinetti G. TFInfer: a tool for probabilistic inference of transcription factor activities. Bioinformatics 26: 2635–2636, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Augustus AM. and Spicer LD. The MetJ regulon in gammaproteobacteria determined by comparative genomics methods. BMC Genomics 12: 558, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bekker M, Alexeeva S, Laan W, Sawers G, Teixeira de Mattos J, and Hellingwerf K. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J Bacteriol 192: 746–754, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, and de Mattos MJT. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153: 1974–1980, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Boczkowski J, Poderoso JJ, and Motterlini R. CO-metal interaction: vital signaling from a lethal gas. Trends Biochem Sci 31: 614–621, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Branchu P, Matrat S, Vareille M, Garrivier A, Durand A, Crepin S, Harel J, Jubelin G, and Gobert AP. NsrR, GadE, and GadX interplay in repressing expression of the Escherichia coli O157:H7 LEE pathogenicity island in response to nitric oxide. PLoS Pathog 10: e1003874, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calhoun MW, Oden KL, Gennis RB, Demattos MJT, and Neijssel OM. Energetic efficiency of Escherichia coli—effects of mutations in components of the aerobic respiratory chain. J Bacteriol 175: 3020–3025, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castor LN. and Chance B. Photochemical action spectra of carbon monoxide-inhibited respiration. J Biol Chem 217: 453–465, 1955 [PubMed] [Google Scholar]
- 12.Cook GM, Loder C, Soballe B, Stafford GP, Membrillo-Hernandez J, and Poole RK. A factor produced by Escherichia coli K-12 inhibits the growth of E. coli mutants defective in the cytochrome bd quinol oxidase complex: enterochelin rediscovered. Microbiology 144: 3297–3308, 1998 [DOI] [PubMed] [Google Scholar]
- 13.Cotter PA, Darie S, and Gunsalus RP. The effect of iron limitation on expression of the aerobic and anaerobic electron transport pathway genes in Escherichia coli. FEMS Microbiol Lett 100: 227–232, 1992 [DOI] [PubMed] [Google Scholar]
- 14.Crack JC, Green J, Hutchings MI, Thomson AJ, and Le Brun NE. Bacterial iron-sulfur regulatory proteins as biological sensor-switches. Antioxid Redox Signal 17: 1215–1231, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, and Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J 21: 3235–3244, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham L, Georgellis D, Green J, and Guest JR. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol Lett 169: 403–408, 1998 [DOI] [PubMed] [Google Scholar]
- 17.D'Autreaux B, Touati D, Bersch B, Latour JM, and Michaud-Soret I. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci U S A 99: 16619–16624, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'mello R, Hill S, and Poole RK. The oxygen affinity of cytochrome bo′ in Escherichia coli determined by the deoxygenation of oxyleghemoglobin and oxymyoglobin: Km values for oxygen are in the submicromolar range. J Bacteriol 177: 867–870, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'mello R, Hill S, and Poole RK. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142: 755–763, 1996 [DOI] [PubMed] [Google Scholar]
- 20.Davidge KS, Sanguinetti G, Yee CH, Cox AG, McLeod CW, Monk CE, Mann BE, Motterlini R, and Poole RK. Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. J Biol Chem 284: 4516–4524, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Engel RR, Rodkey FL, O'Neal JD, and Collison HA. Relative affinity of human fetal hemoglobin for carbon monoxide and oxygen. Blood 33: 37–45, 1969 [PubMed] [Google Scholar]
- 22.Govantes F, Orjalo AV, and Gunsalus RP. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (CydAB) operon. Mol Microbiol 38: 1061–1073, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Green J. and Guest JR. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol Microbiol 12: 433–444, 1994 [DOI] [PubMed] [Google Scholar]
- 24.Gusarov I, Shatalin K, Starodubtseva M, and Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325: 1380–1384, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez JA, Lopez-Gomollon S, Bes MT, Fillat MF, and Peleato ML. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol Lett 236: 275–282, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Hocking WP, Roalkvam I, Magnussen C, Stokke R, and Steen IH. Assessment of the carbon monoxide metabolism of the hyperthermophilic sulfate-reducing Archaeon Archaeoglobus fulgidus VC-16 by comparative transcriptome analyses. Archaea 2015: 235384, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollander R. Correlation of the function of demethylmenaquinone in bacterial electron transport with its redox potential. FEBS Lett 72: 98–100, 1976 [DOI] [PubMed] [Google Scholar]
- 28.Hommais F, Krin E, Coppee JY, Lacroix C, Yeramian E, Danchin A, and Bertin P. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150: 61–72, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Iuchi S. Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem 268: 23972–23980, 1993 [PubMed] [Google Scholar]
- 30.Iuchi S, Chepuri V, Fu HA, Gennis RB, and Lin ECC. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol 172: 6020–6025, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iuchi S. and Lin ECC. Adaptation of Escherichia coli to redox environments by gene expression. Mol Microbiol 9: 9–15, 1993 [DOI] [PubMed] [Google Scholar]
- 32.Jesse HE, Nye TL, McLean S, Green J, Mann BE, and Poole RK. The terminal oxidase cytochrome bd-I in Escherichia coli has lower susceptibility than cytochromes bd-II or bo′ to inhibition by the carbon monoxide-releasing molecule, CORM-3: N-acetylcysteine reduces CO-RM uptake and inhibition of respiration. Biochim Biophys Acta 1834: 1693–1703, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Furumichi M, and Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keyer K. and Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A 93: 13635–13640, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kredich NM. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol Microbiol 6: 2747–2753, 1992 [DOI] [PubMed] [Google Scholar]
- 36.Kung Y, Doukov TI, Seravalli J, Ragsdale SW, and Drennan CL. Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry 48: 7432–7440, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence ND, Sanguinetti G, and Rattray M. Modelling transcriptional regulation using Gaussian processes. In: Advances in Neural Information Processing Systems (NIPS). B Scholkopf, J. Platt and T. Hofmann (eds), MIT Press, Cambridge, MA, pp. 785–792, 2006 [Google Scholar]
- 38.Li F, Collins JG, and Keene FR. Ruthenium complexes as antimicrobial agents. Chem Soc Rev 44: 2529–2542, 2015 [DOI] [PubMed] [Google Scholar]
- 39.Lighthart B. Survival of airborne bacteria in a high urban concentration of carbon monoxide. Appl Microbiol 25: 86–91, 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, and Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol 5: 94–96, 2009 [DOI] [PubMed] [Google Scholar]
- 41.McLean S, Begg R, Jesse HE, Mann BE, Sanguinetti G, and Poole RK. Analysis of the bacterial response to Ru(CO)3Cl(glycinate) (CORM-3) and the inactivated compound identifies the role played by the ruthenium compound and reveals sulfur-containing species as a major target of CORM-3 action. Antiox Redox Signal 19: 1999–2012, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean S, Bowman LAH, Sanguinetti G, Read RC, and Poole RK. Peroxynitrite toxicity in Escherichia coli K12 elicits expression of oxidative stress responses and protein nitration and nitrosylation. J Biol Chem 285: 20724–20731, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMillan DG, Marritt SJ, Butt JN, and Jeuken LJ. Menaquinone-7 is specific cofactor in tetraheme quinol dehydrogenase CymA. J Biol Chem 287: 14215–14225, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moller IM, Rogowska-Wrzesinska A, and Rao RS. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J Proteomics 74: 2228–2242, 2011 [DOI] [PubMed] [Google Scholar]
- 45.Nightingale CH, Ambrose PG, Drusano GL, and Murakawa T. (Eds). Antimicrobial Pharmacodynamics in Theory and Clinical Practice, Second Edition (Infectious Disease and Therapy). New York, London, Informa Healthcare, 2007 [Google Scholar]
- 46.Ogasawara H, Ishida Y, Yamada K, Yamamoto K, and Ishihama A. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J Bacteriol 189: 5534–5541, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onyiah JC, Sheikh SZ, Maharshak N, Steinbach EC, Russo SM, Kobayashi T, Mackey LC, Hansen JJ, Moeser AJ, Rawls JF, Borst LB, Otterbein LE, and Plevy SE. Carbon monoxide and heme oxygenase-1 prevent intestinal inflammation in mice by promoting bacterial clearance. Gastroenterology 144: 789–798, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellicer S, Gonzalez A, Peleato ML, Martinez JI, Fillat MF, and Bes MT. Site-directed mutagenesis and spectral studies suggest a putative role of FurA from Anabaena sp. PCC 7120 as a heme sensor protein. FEBS J 279: 2231–2246, 2012 [DOI] [PubMed] [Google Scholar]
- 49.Pirt SJ. Principles of Microbe and Cell Cultivation. Oxford: Blackwell Scientific Publications, 1985, p. 274 [Google Scholar]
- 50.Poole RK, Williams HD, Downie JA, and Gibson F. Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: Identification and mapping of a fourth locus, cydD. J Gen Microbiol 135: 1865–1874, 1989 [DOI] [PubMed] [Google Scholar]
- 51.Quail MA. and Guest JR. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol Microbiol 15: 519–529, 1995 [DOI] [PubMed] [Google Scholar]
- 52.Ramamoorthi L, Toshkov S, and Brewer MS. Effects of carbon monoxide-modified atmosphere packaging and irradiation on E. coli K12 survival and raw beef quality. Meat Sci 83: 358–365, 2009 [DOI] [PubMed] [Google Scholar]
- 53.Ramseier TM, Chien SY, and Saier MH. Cooperative interaction between Cra and Fnr in the regulation of the cydAB operon of Escherichia coli. Curr Microbiol 33: 270–274, 1996 [DOI] [PubMed] [Google Scholar]
- 54.Roderique JD, Josef CS, Feldman MJ, and Spiess BD. A modern literature review of carbon monoxide poisoning theories, therapies, and potential targets for therapy advancement. Toxicology 334: 45–58, 2015 [DOI] [PubMed] [Google Scholar]
- 55.Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HM, Sanguinetti G, de Mattos JT, Poole RK, and Green J. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286: 10147–10154, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, and Palsson BO. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5: 4910, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma P, Hellingwerf KJ, de Mattos MJT, and Bekker M. Uncoupling of substrate-level phosphorylation in Escherichia coli during glucose-limited growth. Appl Environ Microbiol 78: 6908–6913, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma P, Stagge S, Bekker M, Bettenbrock K, and Hellingwerf KJ. Kinase activity of ArcB from Escherichia coli is subject to regulation by both ubiquinone and demethylmenaquinone. PLoS One 8: e75412, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shatalin K, Shatalina E, Mironov A, and Nudler E. H2S: a universal defense against antibiotics in bacteria. Science 334: 986–990, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Shiloh MU, Manzanillo P, and Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microb 3: 323–330, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith A, Hooper NI, Shipulina N, and Morgan WT. Heme binding by a bacterial repressor protein, the gene product of the ferric uptake regulation (fur) gene of Escherichia coli. J Protein Chem 15: 575–583, 1996 [DOI] [PubMed] [Google Scholar]
- 62.Soares MP. and Bach FH. Heme oxygenase-1: from biology to therapeutic potential. Trends Mol Med 15: 50–58, 2009 [DOI] [PubMed] [Google Scholar]
- 63.Stripp ST, Goldet G, Brandmayr C, Sanganas O, Vincent KA, Haumann M, Armstrong FA, and Happe T. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc Natl Acad Sci U S A 106: 17331–17336, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svistunenko DA. Reaction of haem containing proteins and enzymes with hydroperoxides: the radical view. Biochim Biophys Acta 1707: 127–155, 2005 [DOI] [PubMed] [Google Scholar]
- 65.Tavares AF, Parente MR, Justino MC, Oleastro M, Nobre LS, and Saraiva LM. The bactericidal activity of carbon monoxide-releasing molecules against Helicobacter pylori. PLoS One 8: e83157, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tavares AF, Teixeira M, Romao CC, Seixas JD, Nobre LS, and Saraiva LM. Reactive oxygen species mediate bactericidal killing elicited by carbon monoxide-releasing molecules. J Biol Chem 286: 26708–26717, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trotter EW, Rolfe MD, Hounslow AM, Craven CJ, Williamson MP, Sanguinetti G, Poole RK, and Green J. Reprogramming of Escherichia coli K-12 metabolism during the initial phase of transition from an anaerobic to a micro-aerobic environment. PLoS One 6: e25501, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, and Conway T. Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol 185: 3190–3201, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Ploeg JR, Eichhorn E, and Leisinger T. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch Microbiol 176: 1–8, 2001 [DOI] [PubMed] [Google Scholar]
- 70.Wareham LK, Poole RK, and Tinajero-Trejo M. CO-releasing metal carbonyl compounds as antimicrobial agents in the post-antibiotic era. J Biol Chem 290: 18999–19007, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wegiel B, Larsen R, Gallo D, Chin BY, Harris C, Mannam P, Kaczmarek E, Lee PJ, Zuckerbraun BS, Flavell R, Soares MP, and Otterbein LE. Macrophages sense and kill bacteria through carbon monoxide-dependent inflammasome activation. J Clin Invest 124: 4926–4940, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson JL, Jesse HE, Hughes BM, Lund V, Naylor K, Davidge KS, Cook GM, Mann BE, and Poole RK. Ru(CO)3Cl(glycinate) (CORM-3): a CO-releasing molecule with broad-spectrum antimicrobial and photosensitive activities against respiration and cation transport in Escherichia coli. Antioxid Redox Signal 19: 497–509, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson JL, Wareham LK, McLean S, Begg R, Greaves S, Mann BE, Sanguinetti G, and Poole RK. CO-Releasing molecules have nonheme targets in bacteria: transcriptomic, mathematical modeling and biochemical analyses of CORM-3 [Ru(CO)Cl(glycinate)] Actions on a heme-deficient mutant of Escherichia coli. Antioxid Redox Signal 23: 148–162, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zacharia VM, Manzanillo PS, Nair VR, Marciano DK, Kinch LN, Grishin NV, Cox JS, and Shiloh MU. cor, a Novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis. mBio 4: e00721-13, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng DL, Constantinidou C, Hobman JL, and Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res 32: 5874–5893, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.