Abstract
Tularemia in humans is caused mainly by two subspecies of the Gram-negative facultative anaerobe Francisella tularensis: F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B). The current serological test for tularemia is based on agglutination of whole organisms, and the reactive antigens are not well understood. Previously, we profiled the antibody responses in type A and B tularemia cases in the United States using a proteome microarray of 1,741 different proteins derived from the type A strain Schu S4. Fifteen dominant antigens able to detect antibodies to both types of infection were identified, although these were not validated in a different immunoassay format. Since type A and B subspecies are closely related, we hypothesized that Schu S4 antigens would also have utility for diagnosing type B tularemia caused by strains from other geographic locations. To test this, we probed the Schu S4 array with sera from 241 type B tularemia cases in Spain. Despite there being no type A strains in Spain, we confirmed the responses against some of the same potential serodiagnostic antigens reported previously, as well as determined the responses against additional potential serodiagnostic antigens. Five potential serodiagnostic antigens were evaluated on immunostrips, and two of these (FTT1696/GroEL and FTT0975/conserved hypothetical protein) discriminated between the Spanish tularemia cases and healthy controls. We conclude that antigens from the type A strain Schu S4 are suitable for detection of antibodies from patients with type B F. tularensis infections and that these can be used for the diagnosis of tularemia in a deployable format, such as the immunostrip.
INTRODUCTION
Tularemia is a zoonotic disease caused by the Gram-negative facultative anaerobe Francisella tularensis. The organism infects a multitude of different animals, including mammals, birds, fish, and insects. Transmission to humans can occur by inhalation, although the most common natural route is likely to be direct contact with infected animals or via ticks and other biting arthropods that feed on infected mammals. Important animal reservoirs appear to be rodents, hares, and rabbits. For example, the 1997 outbreak in Castilla y León, Spain (1, 2), was transmitted by the handling of infected hares. The following year, another outbreak which was attributed to crayfish handling occurred in Castilla-La Mancha, Spain (3). Human-to-human transmission is rare.
Four closely related subspecies have been defined: F. tularensis subsp. tularensis (type A), F. tularensis subsp. holarctica (type B), F. tularensis subsp. novicida, and F. tularensis subsp. mediasiatica. Each subspecies possesses a different pathogenic potential in humans, as well as a distinctive geographic distribution, host preference, and route of transmission. Type A is found mainly in North America and is associated with severe tularemia that may be fatal, whereas type B is found throughout the Northern Hemisphere and is less virulent in humans (4–6). F. tularensis subsp. novicida is relatively nonvirulent in healthy humans but has been responsible for a few rare cases of tularemia in immunodeficient individuals in the United States (7). The virulence of F. tularensis subsp. mediasiatica, which is found mainly in central Asia, is not presently known.
Different F. tularensis subspecies show a high level of sequence identity to each other (8), and optimal resolution is best achieved from whole-genome sequencing (9–11). Comparison of the type B strain OSU18 with the laboratory type A strain Schu S4 revealed approximately 99% DNA sequence identity, although numerous rearrangements and pseudogenes that may underlie the differential pathogenicities were discovered (9). A recent genomic characterization of five strains belonging to F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, and F. tularensis subsp. novicida revealed a high level of overall similarity (>95%) between genomes (10). Phylogenetic analyses have revealed that the lineage of F. tularensis subsp. novicida is distinct and diverged from the ancestral line leading to the other subspecies, with F. tularensis subsp. holarctica being more distantly related to F. tularensis subsp. tularensis and F. tularensis subsp. mediasiatica (8). Improved molecular subtyping has recently revealed two clades of F. tularensis subsp. tularensis (type A.I and A.II), and these also have different patterns of pathogenicity, geography, and transmission (12). As the genome sequences of more subspecies become available, a clearer understanding of how limited sequence variation can become manifest in such a wide variety of phenotypes is likely to emerge.
Very few organisms are needed to cause disease in humans. For example, the infectious dose of the type A laboratory strain Schu S4 is 10 to 50 organisms by inhalation (13, 14). It is also relatively straightforward to aerosolize the organism. In October 2012, the Centers for Disease Control and Prevention (CDC) reviewed the select agent list. F. tularensis was among 13 biological select agents and toxins (BSATs) that were given tier 1 status. These present the greatest risk of deliberate misuse with the most significant potential for mass casualties or devastating effects to the economy, critical infrastructure, or public confidence (15). An attenuated live vaccine (LVS), derived from F. tularensis type B, is partially protective against pathogenic F. tularensis in humans (16).
Currently, serology is the most widely used method for the diagnosis of tularemia. The agglutination of F. tularensis by immune sera, first reported in 1926 by Francis and Evans (17), provides the basis for a useful quantitative antibody test for acute infection. An individual who presents with symptoms but who is seronegative by the conventional microagglutination assay (MA) is usually retested 2 weeks later to determine if the titer has increased. No increase in titer helps rule out the possibility of an acute infection. Other than adapting from a tube to a 96-well plate format, the MA has remained essentially unchanged since its inception (18, 19). The test utilizes whole formalin-fixed and stained F. tularensis cells. To determine the agglutinating titer, these cells are added to serial dilutions of patient sera in round-bottomed microtiter plates. The sensitivity of the MA is usually 100% by 1 week after infection. Uninfected controls usually show titers below 1/16, while exposed individuals show titers of 1/64 or above (18, 20). As there are reports of potential cross-reactivity with antibodies against other bacteria, such as Brucella, Yersinia, and Proteus spp. (21, 22), a positive result by the MA must be interpreted with caution. The reactive antigens in the MA are not well understood, and a test based on defined antigens might be preferable. When F. tularensis lipopolysaccharide is used in an enzyme-linked immunosorbent assay (ELISA) format, the specificity of the assay is comparable to that of the MA (20). Outer membrane antigens have also been used for detection of antibodies (23, 24). Tularemia can also be diagnosed by bacterial culture or PCR, both of which are more sensitive and specific than current serological tests. However, culture requires biosafety level 3 containment in the case of type A and is more time-consuming. PCR is rapid and specific, but it is effective only when organisms are present in the sample. Despite the problems of specificity and the availability of defined antigens as alternatives, the MA is cost-effective and sensitive, and it remains the assay of choice in most diagnostic laboratories worldwide.
In a previous study, we used a proteome microarray derived from the type A strain Schu S4 to identify potential serodiagnostic antigens recognized by type A and B tularemia cases in the United States (25). The utility of these antigens for detecting tularemia in other geographic locations is not known. Given the high degree of sequence identity between subspecies, we hypothesized that type A antigens would be able to detect antibodies to other, type B infections. To test this, we used the Schu S4 array to profile antibodies from a tularemia outbreak caused by F. tularensis subsp. holarctica (type B) in Castilla y León, Spain. We also produced immunostrips based on the immunodominant antigens identified and tested them with sera from tularemia cases and controls from Spain. Several of the same potential serodiagnostic antigens that we reported previously in type A and B cases in the United States were also identified here (25). Additional differentially recognized antigens not seen in the original study were also identified (25). We conclude that antigens from the type A strain Schu S4 can detect antibodies to type B subspecies in Spain and that type A antigens may have general utility for the diagnosis of tularemia caused by either subspecies in a deployable format, such as the immunostrip.
MATERIALS AND METHODS
Sera.
The sera used for this study were from the collections of the Laboratorio de Espiroquetas y Patógenos Especiales, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain (Table 1). These comprised sera from individuals who presented to hospitals throughout Spain with a clinical diagnosis of tularemia and whose sera were sent to the laboratory for testing (groups 1 to 4). A diagnosis of tularemia was made according to Red Nacional de Vigilancia Epidemiológica (RENAVE) guidelines. A case was confirmed when a clinically compatible picture together with a positive laboratory test result (seroconversion by microagglutination or isolation of Francisella tularensis from a clinical specimen) was present. A case was classified as presumptive when a single serum sample was positive by microagglutination or the F. tularensis genome was detected by PCR in the presence of a compatible clinical picture. All data were processed according to data protection legislation. The use of Spanish samples was approved by Comité de Bioética y Bienestar Animal, Instituto de Salud Carlos III (approval no. PI 33), in accordance with the Declaration of Helsinki and the Spanish legislation respecting the individual privacy of the patients (the entire study was conducted in Spain). All human samples were anonymized, eliminating the link between the patient’s identity and the sample, in accordance with the Spanish Law of Biomedicine Research (14/2007). Of the 236 Spanish samples, 208 were paired samples from 104 donors taken at two time points. Agglutinating antibodies against the whole F. tularensis live vaccine strain were determined as described previously (19, 26). Briefly, the stock antigen was obtained from F. tularensis strain LVS (ATCC 29684). The MA was performed by placing 25 μl of the serum sample in the first well of the row of a U-bottomed 96-well plate containing 25 μl of phosphate-buffered saline (PBS) and adding 25 μl of PBS to the remaining 11 wells in the row. The sera were then serially diluted and 25 μl of the antigen was added to each well. Therefore, the dilutions of antisera tested ranged from 1:2 and 1:8,192 in the first row. MA titers of ≥1:64 were considered seropositive. Sera were obtained from 24 individuals consisting of anonymized blood donors (Ramón y Cajal Hospital, Madrid, Spain) with no known history of tularemia (group 5) as well as 5 Spanish donors with culture-confirmed tularemia (group 6). Sera from 12 healthy U.S. adults, collected in Orange County, CA (under University of California, Irvine, Institutional Review Board protocol 2007-5896, Healthy control serum panel for protein microarray serology), were used as U.S. naive controls (group 7).
TABLE 1.
Summary of sera used in this study
| Group | Description of sera | No. of patients or donors (no. of samples) |
|---|---|---|
| 1 | Spanish samples from 2 time points, seronegative (titer < 64) to seropositive (titer ≥ 64) | 40 patients (80) |
| 2 | Spanish samples from 2 time points, seropositive (titer ≥ 64) to seropositive (≥2-fold increase in titer) | 41 patients (82) |
| 3 | Spanish samples from 2 time points, seropositive (titer ≥ 64) to seropositive (but <2-fold increase in titer) | 18 patients (36) |
| 4 | Spanish samples from 2 time points, seronegative (titer = 0) to seronegative (titer = 0) | 5 patients (10) |
| 5 | Samples from healthy Spanish blood donors | 23 donors (23) |
| 6 | Spanish samples from a single time point, confirmation of tularemia by bacterial culture | 5 patients (5) |
| 7 | Samples from healthy adult controls from Orange County, CA (an area of nonendemicity), with no known history of tularemia | 12 donors (12) |
Protein microarrays.
F. tularensis subsp. tularensis proteome microarrays displaying 1,741 different F. tularensis subsp. tularensis proteins were produced at Antigen Discovery, Inc. (Irvine, CA), from individual bacterial open reading frames by amplification from the Schu S4 strain and expression by in vitro transcription translation (IVTT) reactions as described previously (25, 27). Arrays were probed according to published methods (28) with serum diluted 1/100 in protein array blocking buffer (Maine Manufacturing, GVS North America, Sanford, ME). The blocking buffer was supplemented with Escherichia coli lysate (Antigen Discovery, Inc., Irvine, CA) at a final concentration of 10 mg/ml protein to block anti-E. coli antibodies.
Protein purification.
The same T7 expression plasmids carrying the genes for the F. tularensis subsp. tularensis proteins used for protein microarray fabrication were used for protein expression in E. coli cells in vivo. The plasmids were introduced into E. coli strain BL21 Star(DE3) cells, the cells were recloned after colony selection, and the sequence was verified. Master cell banks (MCBs) of a single clone that showed high levels of protein expression in vivo were then deposited as glycerol stocks. Expression was evaluated by SDS-PAGE and Western blot analysis using antibodies to the polyhistidine and hemagglutinin epitope tags engineered into the N and C termini of each protein, respectively (29). Pilot expression experiments were first conducted in small cultures to determine the expression levels and solubility characteristics of the proteins. For this, cells scraped from frozen MCBs were inoculated into 5 ml LB broth containing 50 μg/ml kanamycin and incubated at 37°C with vigorous shaking (300 rpm) and aeration. Protein expression was induced with a final concentration of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an optical density at 600 nm (OD600) of 0.4 to 0.6, and incubation was continued overnight until the culture reached stationary phase. Pelleted bacterial cells were lysed using the BugBuster protein extraction reagent (EMD Millipore, Billerica, MA) according to the manufacturer's instructions. After centrifugation, the pellet and supernatant fractions were examined by dot blotting to determine the content of the protein.
Scale-up cultures were performed as described above, except that 1-ml starter cultures were inoculated in terrific broth (TB) and added to 1 liter of TB containing 50 μg/ml kanamycin on the following day. Protein expression was induced with IPTG at a final concentration of 0.1 mM when the culture reached an OD600 of 0.4 to 0.6, and incubation was continued overnight until the culture reached stationary phase. Soluble proteins were recovered from the culture supernatant following centrifugation (16,000 × g, 20 min, 4°C), and purified by fast-protein liquid chromatography (FPLC; Äkta, GE Healthcare, Pittsburgh, PA) on nickel columns as described previously (30). Briefly, 5 ml HisTrap HP columns (GE Healthcare Life Sciences, Chalfont, United Kingdom) were equilibrated with PBS containing 20 mM imidazole, pH 7.4 (binding buffer). Sample lysate was loaded, and the column was washed with binding buffer at a flow rate of 1 ml/min. Elution of protein was performed using a step gradient of 35% (175 mM imidazole) to 75% (375 mM imidazole) elution buffer at a flow rate of 1 ml/min. The elution buffer was then exchanged with PBS, pH 7.4, by dialysis using Amicon Ultra centrifugal filters (Millipore, Billerica, MA). Insoluble proteins found as inclusion bodies (IBs) in the pellet fraction were extracted using the BugBuster reagent. IB pellets were solubilized in 20 mM Tris-HCl, pH 8.0, containing 7 M urea, 0.5 M NaCl, 5 mM imidazole, and 1 mM 2-mercaptoethanol, and protein was purified by FPLC. Solubilized proteins were then bound to HisTrap HP columns and eluted with 75% (375 mM imidazole) elution buffer (7 M urea, 20 mM Tris-HCl, 0.5 M NaCl, 0.5 M imidazole, pH 8.0) at a flow rate of 1 ml/min. Eluted proteins were refolded using a Slide-A-Lyzer dialysis unit (Thermo Fisher Scientific, Waltham, MA) with a 10-kDa cutoff, concentrated to ∼1 mg/ml by the Bradford assay using centrifugal filter units with a 10-kDa cutoff (Amicon Ultra-15; EMD Millipore), and stored at −80°C. The average amount of protein recovered from a 1- to 2-liter culture was 1 to 3 mg, and the protein had a purity of 80 to 90%, as estimated by 4 to 12% gradient SDS-PAGE.
Immunostrips.
Purified protein antigens were diluted in PBS to 100 μg/ml and printed onto a 0.45-μm-pore-size Whatman Optitran BA-S 85 nitrocellulose membrane (GE Healthcare) using a BioJet dispenser (BioDot Inc., Irvine, CA) at a rate of 1 μl/cm. Titrations of human IgG were printed as positive controls for the secondary antibody. The membranes were dried, cut into 3-mm strips, and stored at room temperature (RT) in sealed pouches containing desiccant. For probing of serum, strips were placed into 8-well strip mini-incubation trays (Bio-Rad, Hercules, CA) and rehydrated for 30 min at RT in a blocking buffer consisting of 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20 (T-TBS). Serum samples were diluted 1:250 in blocking buffer, supplemented with E. coli lysate (Antigen Discovery, Inc.) at a final concentration of 10 mg/ml protein, and incubated for 30 min at RT with gentle agitation. Pretreated sera were then applied to the strips at 500 μl per strip, and the strips were incubated for 1 h at RT with gentle rocking. After incubation, the strips were washed 3 times for 5 min each time in T-TBS and then incubated for 1 h in 500 μl of alkaline phosphatase-conjugated donkey anti-human IgG, Fcγ fragment-specific antibody (Jackson ImmunoResearch, West Grove, PA) diluted to 1:5,000 in blocking buffer solution. Finally, the strips were washed 3 times for 5 min each time in T-TBS, followed by 3 5-min washes in TBS, and the bands were visualized by applying 1-step nitroblue tetrazolium chloride–5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt (NBT/BCIP) substrate (Thermo Fisher Scientific) for 2 min at RT. The reaction was stopped with several washes with deionized water. The strips were air dried, aligned, and scanned at 2,400 dots per inch using a document scanner (Hewlett-Packard). The band intensities of each strip were quantified using NIH ImageJ software (downloaded from http://imagej.nih.gov/ij/) for further analysis.
Data analysis.
The arrays were scanned in a ScanArray Express HT array scanner (Perkin-Elmer, Waltham, MA), and image files were quantified using ScanArray software. Mean pixel intensities from which the local background was subtracted were imported into R, and a floor of 100 was set for the data. An antigen was considered seropositive if the signal was >2 times the median for the sample-specific IVTT reaction control spots in at least 10% of the samples for either the Spanish acute- or convalescent-phase samples. The data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median for the 77 IVTT reaction control spots printed throughout the chip (fold over control [FOC]) and then taking the base 2 logarithm of the ratio (log2 FOC). The normalized data provide a relative measure of specific antibody binding to nonspecific antibody binding to the IVTT reaction controls (i.e., the background). With the normalized data, a value of 0.0 means that the intensity is no different from the background intensity and a value of 1.0 indicates a doubling with respect to the background intensity. The base stats package in R was used to calculate Mann-Whitney P values and 2-sided t test P values and to correct P values for false discovery using the Benjamini-Hochberg method (31). The ROCR package in R was used to generate receiver operating characteristic (ROC) plots and to calculate the areas under the curves. Graphical representations were generated in Excel software (Microsoft, Redmond, WA).
RESULTS
Protein microarrays reveal broad serological reactivity.
In this study, we used a protein microarray displaying 1,741 different proteins from F. tularensis Schu S4 to profile the antibodies present in Spanish tularemia cases and healthy controls. This expands on our previous studies with this array, in which we used samples from U.S. donors only (25) or from mouse models (27, 32).
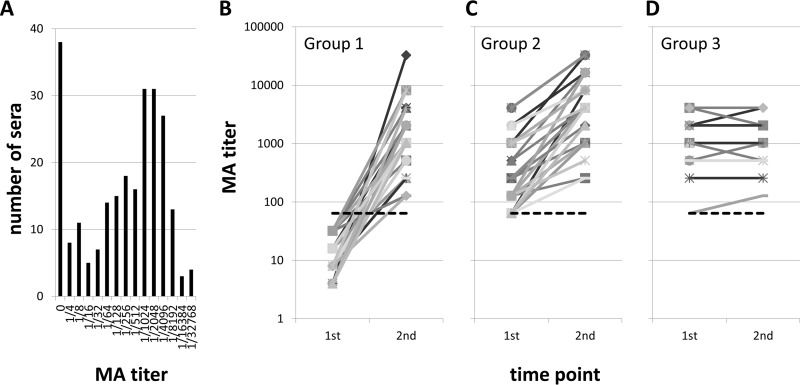
The sera used in the present study included samples collected in Spain during an outbreak of tularemia occurring in 2007 (Table 1, groups 1 to 4 and 6). Routine serological diagnosis was performed by microagglutination assay (MA) at the Centro Nacional de Microbiología, Madrid, Spain. A seronegative result (MA titer < 1/64) for a suspected case is tentatively diagnosed as an acute infection, whereas a seropositive result (MA titer ≥ 1/64) indicates current tularemia or a history of tularemia. A second sample is normally taken about 2 weeks later to help confirm the diagnosis. The distribution of MA titers in the Spanish sample collection ranged from 0 to 32,768, with a median titer of 512 and a mean titer of 2,110 (Fig. 1A). We classified the Spanish donors according to the change in MA titer between the first and second time points. Group 1 (Fig. 1B) comprised 40 patients, who were seronegative at the first time point and who seroconverted by the second time point (i.e., acute infections). Group 2 (Fig. 1C) comprised 41 patients with late acute infections who were seropositive at the first time point and who showed an increase in titer (≥2-fold) at the second time point. Group 3 (Fig. 1D) comprised 18 patients who were also seropositive at the first time point but who did not show a ≥2-fold increase in titer at the second time point and who were considered to be patients in the convalescent phase. Group 4 comprised 5 individuals who had an MA titer of 0 at both time points (not shown); these individuals were tentatively considered to be free of infection, although it is not known if they subsequently seroconverted. Data from group 6 (patients with culture-confirmed tularemia) are described in a later section.
FIG 1.
Classification by MA of sera used in this study. (A) Distribution of titers of all Spanish serum samples used. MA titers ranged from 0 to 32,768, with the median titer being 512. The cutoff for seroconversion is considered to be ≥1/64. (B to D) Changes in MA titers measured at two time points; the first is when the patient presents at the clinic, and the second is at the follow-up approximately 2 weeks later. Each line represents an individual patient. (B) Results for group 1 patients (n = 40). These patients had seroconverted, which normally confirms a diagnosis of acute infection. (C) Results for group 2 patients (n = 41). These patients were in the late acute phase of infection and were seropositive at both time points, with the second sample showing a ≥2-fold rise in MA titer. (D) Results for group 3 patients (n = 18). These patients were in the convalescent phase and seropositive at both time points, but the second sample did not show a ≥2-fold rise in MA titer. A fourth group (not shown) comprised 5 patients who had MA titers of 0 at both time points and who were diagnosed to be free of infection. Dashed line, cutoff for seroconversion (≥1/64).
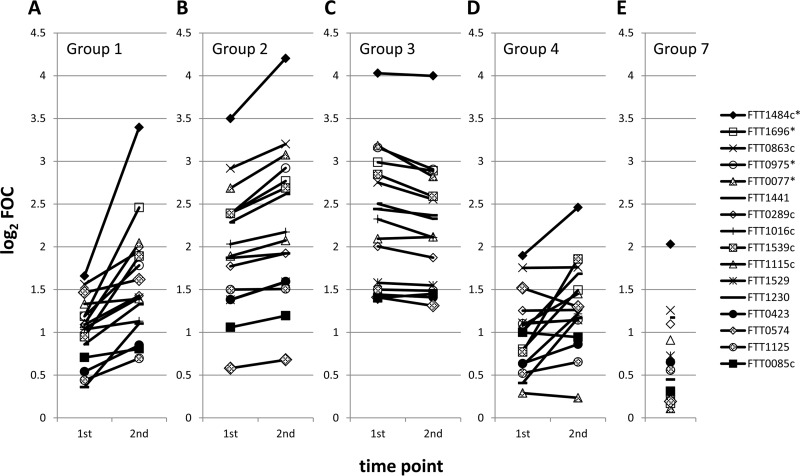
Overall, there was a good concordance in the dynamic trends of the antibody titers determined by MA and by use of the proteome microarray. Figure 2 shows the array signals for the top 16 most reactive antigens at both time points for groups 1 to 4 (Fig. 2A to D, respectively) and for the U.S. controls at a single time point (Fig. 2E). Individuals who seroconverted by MA (group 1) also showed an overall increase in array signals by the second time point, consistent with acute infection (Fig. 2A). Group 2 individuals, i.e., those who were already seropositive by MA and who showed an increased MA titer at the second time point, had a higher starting signal on the array than group 1 individuals and showed a more modest increase in the array signal, which was consistent with those individuals being at a later stage of infection (Fig. 2B). Those individuals who were seropositive at both time points but who showed no change in MA titer (group 3) also did not show an increase in signal by the array. Indeed, there was a slight overall decline in the signal, consistent with those individuals being in the convalescent phase of infection (Fig. 2C). Group 4 patients, i.e., those with an MA titer of 0 at both time points, showed the lowest array signals of the four groups (Fig. 2D), although several antigens gave signals higher than those for the U.S. controls (Fig. 2E). This indicates that these individuals had an infection that was detectable by the protein array but that was present at titers that were below the threshold for detection by the MA.
FIG 2.
Kinetic profiles revealed by the proteome microarray reflect those defined by conventional MA. The average array signal intensities of the top 16 most reactive antigens indicating exposure to tularemia in Spain are shown (the results are analyzed in detail in Fig. 3). Each line represents a different antigen. (A) Results for group 1 patients (n = 40). These patients had acute infection, seroconverted according to the MA titer, and showed an increase in signal intensity by array analysis. (B) Results for group 2 patients (n = 41). These patients were in the late acute phase of infection, were seropositive at both time points according to the MA titer, and showed a modest increase in signal intensity by array analysis. (C) Results for group 3 patients (n = 18). These patients were in the convalescent phase of infection and were seropositive by MA at both time points but did not show an increase in signal intensity by array analysis. (D) Results for group 4 patients (n = 5). These patients had MA titers of 0 at both time points and were tentatively considered to be free of F. tularensis infection. They showed the lowest signals of the Spanish samples. (E) Results for group 7, consisting of U.S. controls. Array data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median of the 77 IVTT reaction control spots printed throughout the chip to give the FOC and then taking the base 2 logarithm of the ratio (log2 FOC). *, antigens identified to be significant in the work of Sundaresh et al. (25).
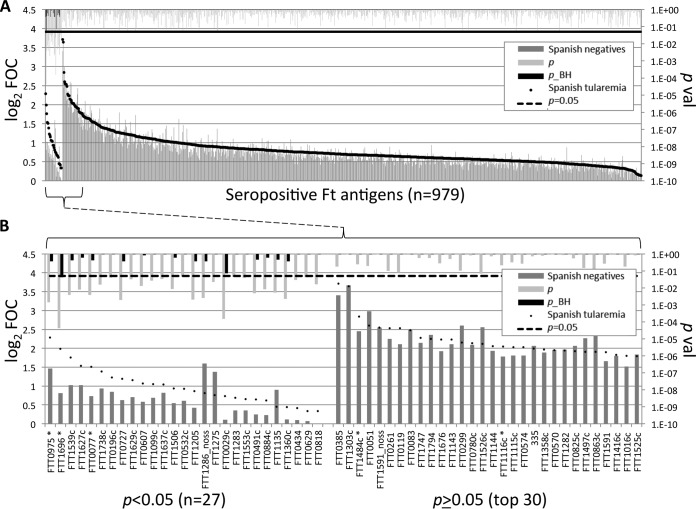
We first wished to identify antigens that were differentially recognized between Spanish cases and controls (Fig. 3). For this, we compared Spanish tularemia cases (samples from the second time points for individuals in groups 1 to 3) and Spanish seronegative individuals (samples from the second time point for individuals in group 4 and group 5). Overall, there were 979 seropositive antigens (defined by array signals greater than the median for the IVTT reaction control spots in 10% of the samples from either group), and 27 of these antigens were differentially recognized by sera from the two groups when the reactions were compared by the t test (P < 0.05). The remaining antigens (P ≥ 0.05) were nondifferentially recognized. Twelve of the discriminatory antigens gave signals 2-fold over the background (i.e., log2 FOC > 1) with tularemia patient sera, of which one (FTT1696, GroEL) withstood linear model correction for experimental effects (Table 2). The relative paucity of robust diagnostic antibodies is due to broadly similar antibody profiles in both the Spanish MA-seropositive and the Spanish MA-seronegative groups. In Fig. 3B, 5 of 15 antigens defined previously (25) were also identified to be among the top seroreactive antigens in the present study (indicated by asterisks), although the results for 2 antigens failed to reach significance.
FIG 3.
Identification of antigens associated with F. tularensis exposure using proteome microarrays. The microarrays were probed with sera from Spanish tularemia cases (sera obtained from groups 1 to 3 at the second time point) and Spanish individuals negative for tularemia (sera obtained from groups 4 and 5 at the second time point), and IgG was visualized by indirect immunofluorescence (see Materials and Methods). A serum sample was defined to be positive for an antigen if the array signal was >2 times the median for the IVTT reaction control spots in at least 10% of the samples from either group. (A) Antigens to which the serum samples were reactive (n = 979). Ft, F. tularensis. (B) Enlargement of the 27 discriminatory antigens and top 30 nondiscriminatory antigens from panel A. Array data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median for the 77 IVTT reaction control spots printed throughout the chip to give the FOC and then taking the base 2 logarithm of the ratio (log2 FOC). *, antigens identified to be significant in the work of Sundaresh et al. (25); noss, the signal sequence was deleted; p_BH, P values corrected for false discovery using the Benjamini-Hochberg method (31).
TABLE 2.
Reactive antigens identified using the Spanish tularemia seraa
| Antigena | Gene | Product | Avg signal intensity (log2 FOC) of Spanish samples: |
P value | P_BHb | |
|---|---|---|---|---|---|---|
| Negative for tularemia | Positive for tularemia | |||||
| FTT0975c | Conserved hypothetical protein | 1.468 | 2.283 | 0.0014 | 0.3622 | |
| FTT1696c | mopA | Chaperone protein GroEL | 0.810 | 1.985 | 0.0000 | 0.0418 |
| FTT1539c | Conserved hypothetical protein | 1.025 | 1.759 | 0.0038 | 0.4156 | |
| FTT1627c | Hypothetical protein | 1.022 | 1.531 | 0.0080 | 0.6150 | |
| FTT0077c | sucB | Dihydrolipoamide succinyltransferase component | 0.732 | 1.509 | 0.0038 | 0.4156 |
| FTT1738c | kdpB | Potassium-transporting ATPase B chain | 0.932 | 1.386 | 0.0155 | 0.9500 |
| FTT0196c | glnA | Glutamine synthetase | 0.844 | 1.229 | 0.0399 | 0.9873 |
| FTT0727 | Conserved hypothetical protein | 0.626 | 1.196 | 0.0019 | 0.3622 | |
| FTT1629c | Hypothetical membrane protein | 0.706 | 1.154 | 0.0325 | 0.9873 | |
| FTT0607 | ispG | 1-Hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate | 0.580 | 1.067 | 0.0129 | 0.8396 |
| FTT1099c | Hypothetical protein | 0.690 | 1.054 | 0.0266 | 0.9873 | |
| FTT1637c | Hypothetical protein | 0.818 | 1.039 | 0.0336 | 0.9873 | |
Listed are the top 12 of 27 discriminatory antigens shown in Fig. 3B, i.e., antigens to which sera were significantly reactive when sera from Spanish tularemia cases were compared with sera from Spanish controls by t test (P < 0.05) and were recognized by sera from the tularemia cases with a signal (log2 FOC) of >1.
P_BH, P values corrected for false discovery using the Benjamini-Hochberg method (31).
Proteins identified to be significant by Sundaresh et al. (25).
Since a diagnosis of tularemia in individuals in groups 1 to 3 was made on the basis of the MA titer without an additional confirmatory diagnostic test, it was of interest to compare the array data with the MA titers for culture-confirmed cases of tularemia to evaluate the relative sensitivities of the two assays. Figure 4 shows a comparison of the MA titers and reactivity to antigen FTT1696 (GroEL; identified in Fig. 3 to be an antigen strongly discriminating Spanish tularemia cases and controls) for five culture-confirmed tularemia cases (group 6). The data show that while two of the five cases were only borderline seropositive by MA (≥1/64), four of the five cases were strongly positive for FTT1696 reactivity.
FIG 4.
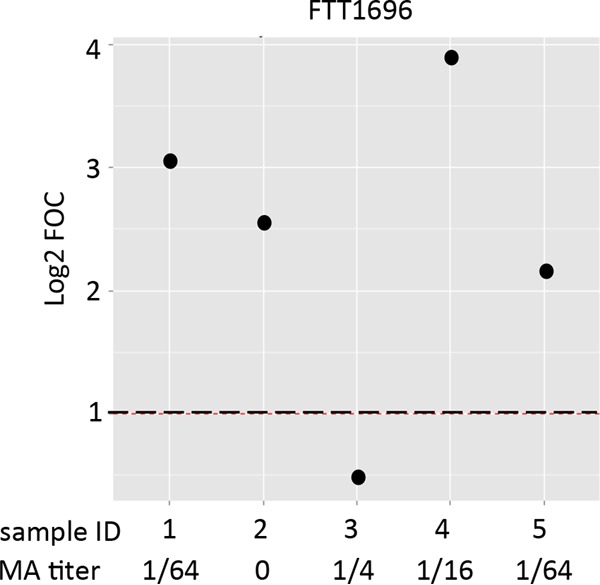
Relative sensitivity of MA titer and GroEL (FTT1696) on the protein array for detection of tularemia in 5 culture-confirmed tularemia cases. Sera from 5 culture-confirmed Spanish tularemia cases (group 6) were assayed by MA and on the protein microarrays for reactivity to the GroEL (FTT1696) antigen. MA titers are shown beneath the plot of the array values (cutoff, ≥1/64). The GroEL antigen was identified in Fig. 3 to discriminate Spanish tularemia cases and controls. Array data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median for the 77 IVTT reaction control spots printed throughout the chip to give the FOC and then taking the base 2 logarithm of the ratio (log2 FOC). Dashed line, log2 FOC of 1 (2-fold over the background intensity) for the array.
Antigens that are differentially recognized by different patient groups.
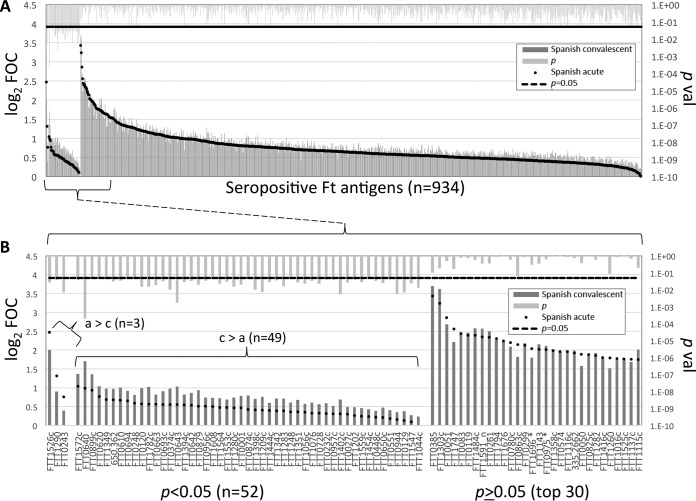
A desirable property for any improved serodiagnostic test would be the ability to discriminate between acute- and convalescent-stage infections. Establishment of the disease stage using the conventional MA requires that two serum samples be taken 2 weeks apart to assess the change in titer. Using the microarray approach, we hypothesized that it may be possible to identify a particular antigen(s) or antigen combinations that are differentially recognized at the acute and convalescent stages of disease. Therefore, we performed a paired analysis of Spanish tularemia patients where there was a MA titer increase (Fig. 5). This analysis identified 52 antigens that were differentially recognized (P < 0.05), and 3 of these gave stronger signals with the acute-phase samples and 49 gave stronger signals with the convalescent-phase samples. Of the three antigens that gave stronger signals with the acute-phase samples, the titers of antibodies to two of them were at least 2-fold above the IVTT reaction background titer (log2 FOC = 1). These were FTT1526c (Idh, isocitrate dehydrogenase) and FTT1290 (MetG, methionyl-tRNA synthetase).
FIG 5.
Identification of antigens that discriminate between Spanish acute- and convalescent-stage tularemia. (A) The IgG profiles of Spanish tularemia patients with low to high MA titers were compared and segregated by P value. (B) Enlargement of the 52 differentially reactive antigens (P < 0.05) and top 30 nondifferentially reactive antigens (P ≥ 0.05). a, acute phase; c, convalescent phase. Array data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median for the 77 IVTT reaction control spots printed throughout the chip to give the FOC and then taking the base 2 logarithm of the ratio (log2 FOC). *, antigens identified to be significant in the work of Sundaresh et al. (25); noss, the signal sequence was deleted.
Immunoassay development.
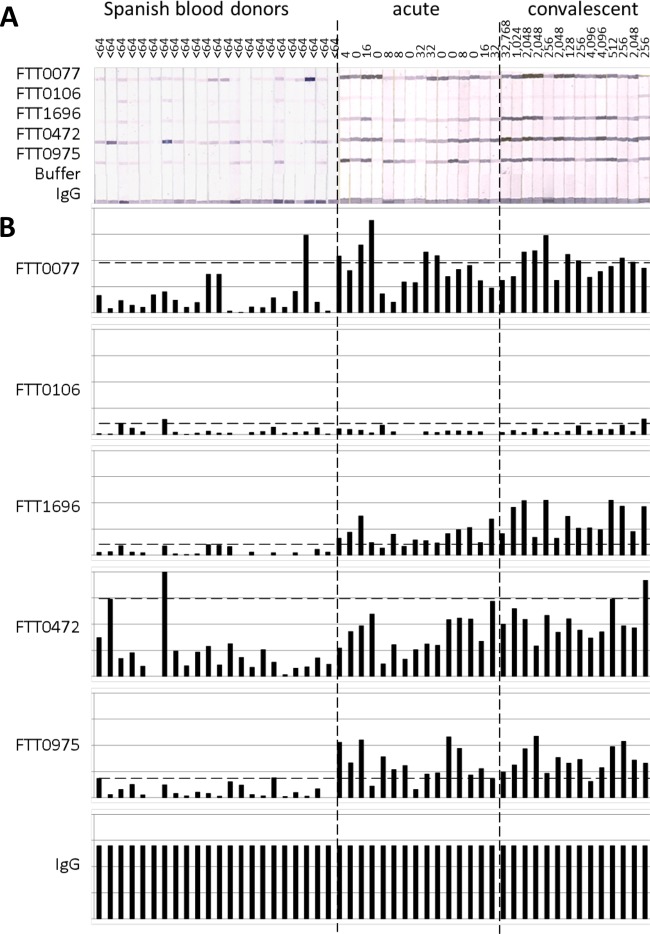
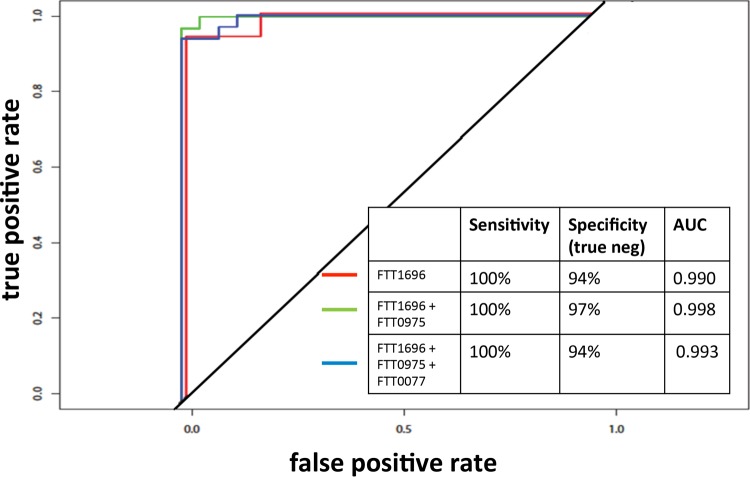
In parallel with the expanded protein array study reported here, we also aimed to purify potentially serodiagnostic antigens that were identified in our previous study (25) and evaluate their use in a more deployable immunoassay format. Of the 15 antigens reported previously, 5 were successfully purified by nickel chelate affinity chromatography in quantities sufficient for further development. These were FTT0077 (SucB), FTT0106, FTT1696 (GroEL), FTT0472 (AccB), and FTT0975. Purified proteins were printed onto nitrocellulose membranes, and the membranes were cut into strips and then probed with paired serum samples from Spanish tularemia cases obtained before and after seroconversion (group 1 in Table 1) and Spanish blood donors with no known history of tularemia (group 5 in Table 1). Bound IgG was then detected colorimetrically (Fig. 6A). Overall, the sera from the tularemia patients had a higher reactivity than the sera from the blood donors, as revealed by visual estimation. Moreover, the reactivities of sera from patients in both the acute and convalescent stages of infection were very similar, even though the sera from patients in the acute stage of infection were negative by MA. This finding is consistent with the array data for group 1 Spanish sera (Fig. 2A), which revealed several antigens with reactivities at least 2-fold above the IVTT reaction background (log2 FOC = 1) at the first time point. This indicates that the detection of IgG by the use of purified recombinant antigens may be more sensitive than detection by the conventional MA. The band intensities on the immunostrips were quantified and normalized by median scaling against the intensities of the IgG control bands (Fig. 6B). Using a cutoff based on the mean + 2 standard deviations (SDs) for the control population (n = 22), two of the antigens gave acceptable results with the tularemia samples (n = 30). These were FTT1696 and FTT0975, with 28 (93%) and 26 (87%) of the samples having reactivities above the cutoff, respectively. The use of both antigens combined provided 100% coverage of the tularemia patients. A third antigen, FTT0077, may also have diagnostic utility, but the result for this antigen was marred by a single outlier control serum sample. If the result for this outlier was removed from the calculation of the cutoff, 26 of the 30 tularemia serum samples were seropositive. The results for FTT0472 were similarly marred by two strong signals for the control group. Although the FTT0106 antigen was significant in our previous study (25), it was not recognized either on the array or by the ELISA format in the present study. ROC analyses (Fig. 7) revealed that antigen FTT1696 alone provided the best discrimination between Spanish cases and controls, although the specificity was improved slightly by combining it with FTT0975.
FIG 6.
Immunostrips with 5 purified F. tularensis subsp. tularensis antigens probed with sera from Spanish tularemia cases and controls. (A) Scan of immunostrips. The MA titers are shown above each strip. (B) Histograms of the corresponding band intensities after quantification and normalization by median scaling against the IgG printed on the strip. Horizontal dashed lines, cutoff generated by using the mean + 2 SDs for the blood donors (n = 23).
FIG 7.
ROC analysis of the immunostrip data shown in Fig. 6. Antigen FTT1696 alone provides the best discrimination between Spanish cases and controls, although the specificity was improved slightly by combining FTT1696 with FTT0975. neg, negative; AUC, area under the curve.
We hypothesized that the sporadic reactivity of sera from the Spanish blood donor controls with FTT0472 (and, to a lesser extent, FTT0077) may have been because the control samples were from an area where type B tularemia is endemic. We therefore probed a second batch of immunostrips with sera from Spanish tularemia cases, Spanish blood donor controls, and donors from a region of nonendemicity in the United States. Data were normalized by median scaling against the human IgG band, as before (Fig. 8). Antigen FTT0472, which give a high reactivity with sera from the Spanish control population, as before (n = 14 [100%] samples with values above the cutoff), showed negligible reactivity with U.S. control sera (n = 2 [4.5%] positive samples). Antigen FTT0077 reacted at equally low levels with U.S. control sera (n = 3 [6.5%] positive samples), whereas it had higher reactivity with sera from Spanish donors (n = 2 [14.3%] positive samples). These data support the notion that reactivity to both antigens by sera from controls is higher in areas of endemicity.
FIG 8.
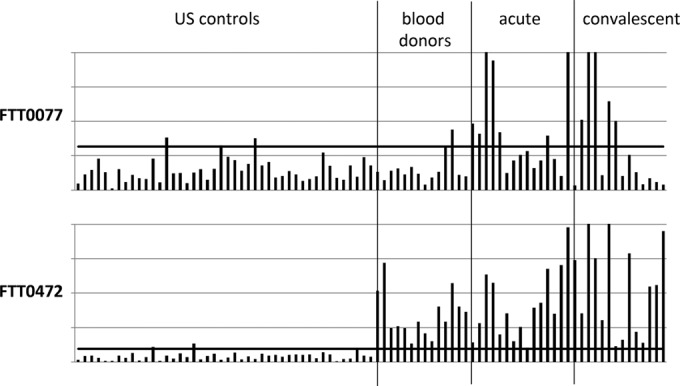
Immunostrips of 2 purified F. tularensis subsp. tularensis antigens probed with sera from Spanish tularemia cases and controls and U.S. controls. The histograms are described in the legend to Fig. 6. Solid horizontal line, cutoff generated by using the mean + 2 SDs for the U.S. controls (n = 44).
DISCUSSION
Despite the availability of ELISAs using recombinant antigens, MA remains the “gold standard” for the serodiagnosis of tularemia in many diagnostic laboratories. A negative result in the MA (titer < 1/64) may indicate an acute infection in individuals with a clinical diagnosis. However, a negative result per se is unsatisfactory for diagnosis, and an acute infection can be confirmed only if the patient seroconverts at a later time point. Elimination of problems of cross-reactivity of the MA with other bacteria, such as Brucella, Yersinia, and Proteus, requires the discovery of individual antigens able to bind Francisella antibodies specifically. The use of recombinant antigens discovered by proteomic screens and configured into ELISA or immunostrip assay formats promises to overcome many of the shortcomings of the traditional MA.
Comparative genomic studies have revealed a high degree of sequence conservation between subspecies of F. tularensis (8, 11). However, the influence of the genetic diversity of F. tularensis on the host immune response and serodiagnosis, in particular, is not well understood. Different subspecies are not discriminated by the conventional MA (8), although it has long been recognized that subspecies can be defined serologically using immune sera (33). While the targets of subspecies-specific antibodies presumably correspond to variable gene products, the DNA sequences of the various subspecies, which share extensive nucleotide sequence identity, likely encode mostly shared antigens. The core gene set is a large component of the genome. The first comparative genomic study of F. tularensis was of strains Schu S4 (type A) and OSU18 (type B), which revealed 97.6% nucleotide sequence identity (9). A more recent analysis of 5 type A and B isolates with a range of pathogenicities revealed over 95% sequence conservation (10).
Because type A and B subspecies of F. tularensis are virulent in humans, a primary diagnostic test should be able to detect both types. Here, we used a heterologous array approach, in which serum samples from individuals involved in a type B tularemia outbreak in the Castilla y León region of Spain in 2007 (1) were used to probe a F. tularensis Schu S4 (type A) proteome array. In this way, the influence of variation was likely to be minimized and allowed antigens shared by both subspecies to be identified more readily. A genomic analysis of cultured isolates from the same outbreak and another outbreak from the same region that took place a decade earlier revealed that both outbreaks were caused by strain with the highly conserved genotype B.Br:FTNF002-00 (1), which is a subclade of F. tularensis subsp. holarctica (type B) found throughout central and western Europe (34–36). The results of the study described here indicate that antigens of the type A strain Schu S4 are able to detect serological responses in type B infections, presumably owing to the high degree of sequence identity that permits antibody cross-reactivity.
Using the protein microarray approach, we consistently detected reactivity with antibodies in sera classified as seronegative by MA and even in sera with MA titers of 0 (for example, Fig. 2A, first time point, or Fig. 2D). We speculate that one or more antigens displayed on the array may be more sensitive than the MA for the detection of specific antibodies. This notion remains speculative in the absence of a confirmatory test for tularemia (e.g., by bacterial culture or PCR) in these individuals. However, of the 5 cases with culture-confirmed tularemia (group 6 in Table 1), a comparison of the array data for the major differentially recognized antigen FTT1696 (GroEL) and the MA titer (Fig. 4) showed that while 2 of 5 cases were seropositive by MA, 4 of 5 cases were positive for GroEL reactivity. While we cannot draw firm conclusions from a sample size this small, the results of the analysis are consistent with the notion that tests with certain protein antigens might be more sensitive than the MA.
We also noted elevated reactivity of sera from the Spanish MA-negative individuals (blood donors) relative to that of sera from the U.S. controls (Fig. 2), consistent with an elevated background serology in Spain compared to that in the United States. It is not clear if this is due to elevated exposure to F. tularensis, related Francisella species, or other antigenically similar organisms. Unexpectedly, serological reactivity was not completely absent in individuals from the United States with no known exposure to F. tularensis but instead was characterized by broad, low-level reactivity to numerous antigens. Such reactivity in samples from an area of nonendemicity again points toward environmental exposure to nonpathogenic related Francisella species or other antigenically related organisms (37, 38). Any such background reactivity that is static will not yield significant antigens when different groups, such as Spanish tularemia cases and controls (Fig. 3) or individuals with acute versus convalescent stages of infection (Fig. 5), are compared. Such antigens will not, therefore, become lead candidates for further development of a diagnostic test.
In this study, we have begun to address the utility of the use of individual protein antigens for serodiagnosis. These were FTT0077 (SucB), FTT0106, FTT1696 (GroEL), FTT0472 (AccB), and FTT0975. Of these, FTT1696 (GroEL) was of particular interest, as it gave a sensitivity and a specificity of 100% and 94%, respectively, for discriminating between Spanish tularemia cases and Spanish blood donors by use of the immunostrip. GroEL, a member of the heat shock family of proteins, is well conserved among other bacteria, and studies are needed to evaluate whether an assay based on this antigen would cross-react with antibodies to other bacteria. Another antigen of interest is FTT0975, a hypothetical protein of unknown function.
We also aimed to identify antigens that could discriminate between acute- and convalescent-stage infections. Ideally, antibodies to such an antigen would be strongly reactive during the acute phase of tularemia, but their reactivities would rapidly wane thereafter. A similar strategy has been employed in toxoplasmosis with some degree of success (39) and more recently in malaria, where a number of Plasmodium falciparum antigens that predicted recent exposure were identified by a proteome array (40). Examination of the data in Fig. 5 reveals three potential biomarkers of acute F. tularensis infection, although further studies are needed to evaluate such candidates.
ACKNOWLEDGMENTS
This study was supported by an PSW RCE New Opportunities Grant (grant AI065359 to D.H.D. and D.A.), grant AI078213 (to P.L.F.), and Ministerio de Economía y Competitividad grant CGL2015-66962-C2-2-R (to R.E. and P.A.).
D.H.D. and P.L.F. declare patent applications related to protein microarray fabrication and have stock positions with Antigen Discovery, Inc.
REFERENCES
- 1.Ariza-Miguel J, Johansson A, Fernández-Natal MI, Martínez-Nistal C, Orduña A, Rodríguez-Ferri EF, Hernández M, Rodríguez-Lázaro D. 2014. Molecular investigation of tularemia outbreaks, Spain, 1997-2008. Emerg Infect Dis 20:754–761. doi: 10.3201/eid2005.130654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centro Nacional de Epidemiología Instituto de Salud Carlos III. 1997. Brote de tularemia en Castilla y León. Bol Epidemiol Sem 5:249–252. [Google Scholar]
- 3.Anda P, Segura del Pozo J, Díaz García JM, Escudero R, García Peña FJ, López Velasco MC, Sellek RE, Jiménez Chillarón MR, Sánchez Serrano LP, Martínez Navarro JF. 2001. Waterborne outbreak of tularemia associated with crayfish fishing. Emerg Infect Dis 7(3 Suppl):575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjostedt A. 2003. Virulence determinants and protective antigens of Francisella tularensis. Curr Opin Microbiol 6:66–71. doi: 10.1016/S1369-5274(03)00002-X. [DOI] [PubMed] [Google Scholar]
- 5.McLendon MK, Apicella MA, Allen LA. 2006. Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol 60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K, Working Group on Civilian Biodefense. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 7.Kingry LC, Petersen JM. 2014. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol 4:35. doi: 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keim P, Johansson A, Wagner DM. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci 1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 9.Petrosino JF, Xiang Q, Karpathy SE, Jiang H, Yerrapragada S, Liu Y, Gioia J, Hemphill L, Gonzalez A, Raghavan TM, Uzman A, Fox GE, Highlander S, Reichard M, Morton RJ, Clinkenbeard KD, Weinstock GM. 2006. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J Bacteriol 188:6977–6985. doi: 10.1128/JB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, Borowsky M, Young S, Koehrsen M, Engels R, Pearson M, Howarth C, Larson L, White J, Alvarado L, Forsman M, Bearden SW, Sjostedt A, Titball R, Michell SL, Birren B, Galagan J. 2009. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog 5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, Keim P, Johansson A. 2009. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog 5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staples JE, Kubota KA, Chalcraft LG, Mead PS, Petersen JM. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg Infect Dis 12:1113–1118. doi: 10.3201/eid1207.051504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch Intern Med 107:702–714. [DOI] [PubMed] [Google Scholar]
- 14.Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin Microbiol Rev 15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention, U.S. Department of Health and Human Services. 2012. Possession, use, and transfer of select agents and toxins; biennial review. Final rule. Fed Regist 77:61083–61115. [PubMed] [Google Scholar]
- 16.Isherwood KE, Titball RW, Davies DH, Felgner PL, Morrow WJ. 2005. Vaccination strategies for Francisella tularensis. Adv Drug Deliv Rev 57:1403–1414. doi: 10.1016/j.addr.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 17.Francis E, Evans AC. 1926. Agglutination, crossagglutination, and agglutinin absorption in tularaemia. Public Health Rep 41:1273–1295. doi: 10.2307/4577917.19315040 [DOI] [Google Scholar]
- 18.Sato T, Fujita H, Ohara Y, Homma M. 1990. Microagglutination test for early and specific serodiagnosis of tularemia. J Clin Microbiol 28:2372–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massey ED, Mangiafico JA. 1974. Microagglutination test for detecting and measuring serum agglutinins of Francisella tularensis. Appl Microbiol 27:25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porsch-Ozcurumez M, Kischel N, Priebe H, Splettstosser W, Finke EJ, Grunow R. 2004. Comparison of enzyme-linked immunosorbent assay, Western blotting, microagglutination, indirect immunofluorescence assay, and flow cytometry for serological diagnosis of tularemia. Clin Diagn Lab Immunol 11:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behan KA, Klein GC. 1982. Reduction of Brucella species and Francisella tularensis cross-reacting agglutinins by dithiothreitol. J Clin Microbiol 16:756–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koskela P, Herva E. 1982. Immunity against Francisella tularensis in northern Finland. Scand J Infect Dis 14:195–199. doi: 10.3109/inf.1982.14.issue-3.07. [DOI] [PubMed] [Google Scholar]
- 23.Bevanger L, Maeland JA, Kvan AI. 1994. Comparative analysis of antibodies to Francisella tularensis antigens during the acute phase of tularemia and eight years later. Clin Diagn Lab Immunol 1:238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bevanger L, Maeland JA, Naess AI. 1989. Competitive enzyme immunoassay for antibodies to a 43,000-molecular-weight Francisella tularensis outer membrane protein for the diagnosis of tularemia. J Clin Microbiol 27:922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundaresh S, Randall A, Unal B, Petersen JM, Belisle JT, Hartley MG, Duffield M, Titball RW, Davies DH, Felgner PL, Baldi P. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 26.Brown SL, McKinney FT, Klein GC, Jones WL. 1980. Evaluation of a safranin-O-stained antigen microagglutination test for Francisella tularensis antibodies. J Clin Microbiol 11:146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, Randall A, Mu Y, Hirst S, Molina DM, Davies DH, Milne T, Griffin KF, Baldi P, Titball RW, Felgner PL. 2007. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 28.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, Crotty S, Karem KL, Damon IK, Ahmed R, Villarreal L, Felgner PL. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 29.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A 102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun 76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 32.Nuti DE, Crump RB, Dwi Handayani F, Chantratita N, Peacock SJ, Bowen R, Felgner PL, Davies DH, Wu T, Lyons CR, Brett PJ, Burtnick MN, Kozel TR, AuCoin DP. 2011. Identification of circulating bacterial antigens by in vivo microbial antigen discovery. mBio 2:e00136–11. doi: 10.1128/mBio.00136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen CR, Buker EO, Jellison WL, Lackman DB, Bell JF. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J Bacteriol 87:676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogler AJ, Birdsell DN, Lee J, Vaissaire J, Doujet CL, Lapalus M, Wagner DM, Keim P. 2011. Phylogeography of Francisella tularensis ssp. holarctica in France. Lett Appl Microbiol 52:177–180. doi: 10.1111/j.1472-765X.2010.02977.x. [DOI] [PubMed] [Google Scholar]
- 35.Chanturia G, Birdsell DN, Kekelidze M, Zhgenti E, Babuadze G, Tsertsvadze N, Tsanava S, Imnadze P, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Champion MD, Sinari S, Gyuranecz M, Farlow J, Pettus AH, Kaufman EL, Busch JD, Pearson T, Foster JT, Vogler AJ, Wagner DM, Keim P. 2011. Phylogeography of Francisella tularensis subspecies holarctica from the country of Georgia. BMC Microbiol 11:139. doi: 10.1186/1471-2180-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maraha B, Hajer G, Sjodin A, Forsman M, Paauw A, Roeselers G, Verspui E, Frenay I, Notermans D, de Vries M, Reubsaet F. 2013. Indigenous infection with Francisella tularensis holarctica in The Netherlands. Case Rep Infect Dis 2013:916985. doi: 10.1155/2013/916985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barns SM, Grow CC, Okinaka RT, Keim P, Kuske CR. 2005. Detection of diverse new Francisella-like bacteria in environmental samples. Appl Environ Microbiol 71:5494–5500. doi: 10.1128/AEM.71.9.5494-5500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berrada ZL, Telford SR III. 2010. Diversity of Francisella species in environmental samples from Martha's Vineyard, Massachusetts. Microb Ecol 59:277–283. doi: 10.1007/s00248-009-9568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felgner J, Juarez S, Hung C, Liang LI, Jain A, Doskaya M, Felgner PL, Caner A, Guruz Y, Davies DH. 2015. Identification of Toxoplasma gondii antigens associated with different types of infection by serum antibody profiling. Parasitology 142:827–838. doi: 10.1017/S0031182014001978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helb DA, Tetteh KK, Felgner PL, Skinner J, Hubbard A, Arinaitwe E, Mayanja-Kizza H, Ssewanyana I, Kamya MR, Beeson JG, Tappero J, Smith DL, Crompton PD, Rosenthal PJ, Dorsey G, Drakeley CJ, Greenhouse B. 2015. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci U S A 112:E4438–E4447. doi: 10.1073/pnas.1501705112. [DOI] [PMC free article] [PubMed] [Google Scholar]