Abstract
The majority of species within the genus Malassezia are lipophilic yeasts that colonize the skin of warm-blooded animals. Two species, Malassezia globosa and Malassezia restricta, are implicated in the causation of seborrheic dermatitis/dandruff (SD/D). During our survey of SD/D cases, we isolated several species of Malassezia and noticed vast variations within a few lipid-dependent species. Variations observed in the phenotypic characteristics (colony morphology, absence of catalase activity, growth at 37°C, and precipitation surrounding wells containing Tween 20 or Cremophor EL) suggested the possible presence of a novel species. Sequence divergence observed in the internal transcribed spacer (ITS) region, the D1/D2 domain, and the intergenic spacer 1 (IGS1) region of rDNA and the TEF1 gene, PCR-restriction fragment length polymorphism (RFLP) analysis of the ITS2 region, and fluorescent amplified fragment length polymorphism analysis support the existence of a novel species. Based on phenotypic and molecular characterization of these strains, we propose a new species, namely, M. arunalokei sp. nov., and we designate NCCPF 127130 (= MTCC 12054 = CBS 13387) as the type strain.
INTRODUCTION
Malassezia spp. are lipophilic, unipolar budding, commensal yeasts that inhabit body surfaces rich in sebaceous glands in warm-blooded animals. In humans, these microorganisms have been implicated in a variety of skin diseases, including pityriasis versicolor (PV), seborrheic dermatitis/dandruff (SD/D), pityrosporum folliculitis (PF), and some subsets of psoriasis and atopic dermatitis (AD) (1). Weather conditions (such as temperature and humidity), the cutaneous microenvironment, or host defense mechanisms may predispose individuals to Malassezia infection or colonization. After a thorough analysis of morphological and biochemical characteristics, Gueho et al. described seven species (Malassezia furfur, Malassezia obtusa, Malassezia globosa, Malassezia slooffiae, Malassezia sympodialis, Malassezia pachydermatis, and Malassezia restricta) (2). Seven more Malassezia spp. (Malassezia dermatis, Malassezia equina, Malassezia japonica, Malassezia nana, Malassezia yamatoensis, Malassezia caprae, and Malassezia cuniculi) were described later by different groups (3–9). Of these, M. caprae, M. equina, M. nana, and M. cuniculi have been isolated only from domestic animals; the host specificity of these species prompted other workers to look for new species in the genus (10).
In human infections, certain species possibly play important roles in pathogenesis. M. globosa is commonly associated with SD/D (11). Other species, including M. restricta, M. furfur, M. sympodialis, M. obtusa, and M. slooffiae, are occasionally associated with SD/D at different geographical locations (12). SD/D is widely prevalent in India, and M. globosa and M. restricta are commonly associated with this condition (13, 14). While conducting a detailed survey of SD/D cases, however, we noticed vast variations within a few lipid-dependent strains of Malassezia, which are characterized and described here as a new species, Malassezia arunalokei sp. nov.
MATERIALS AND METHODS
Subjects.
The prospective case-control study was conducted in villages in provinces in northwestern India (Punjab) (Badashapur, Karial, Salimgarh, Nial, Brass, and Bhunderpeni) and northern India (Haryana) (Firozpur, Lalpur, Taparian, Galodi, UjjarMajri, and Mirjapur and Rathouli) in December 2011 through February 2013. The area was selected on the basis of convenience for performing a field study. The subjects included 124 patients with SD/D and 55 healthy control subjects. Patients and controls with a history of application of topical antifungals and/or steroid preparations to the scalp during the month preceding sampling were excluded from the study. Convenience sampling was carried out by random screening for SD/D cases in the selected villages. The study protocol was approved by the Institute Ethics Committee of the Postgraduate Institute of Medical Education and Research (Chandigarh, India). The samples were collected after informed consent was obtained from the subjects.
Clinical specimen collection and processing.
Flakes or scales were collected from SD/D patients by partitioning the hair with a sterile comb and scraping an ∼1-inch area using a blunt scalpel. SD/D severity was graded as mild, moderate, or severe on the basis of the size, color, and surface of the flakes or scales (13). In healthy individuals, the scrapings were collected from the vertex and temporal regions. Samples were also collected from the nasolabial folds of both patients and controls, using cotton swabs moistened with phosphate-buffered saline (pH 7.2), and were inoculated on modified Dixon's agar plates. The plates were incubated in a humid chamber at 30°C for 4 weeks (13).
Standard strains.
M. furfur (MTCC 1374) was provided by the Microbial Type Culture Collection and Gene Bank (MTCC), Council of Scientific and Industrial Research-Institute of Microbial Technology (Chandigarh, India). Type/standard strains of other Malassezia spp. were obtained from the CBS-KNAW Fungal Biodiversity Centre (Utrecht, The Netherlands) and are included in the study for comparison.
Identification of Malassezia spp.
Colonies resembling Malassezia were identified on the basis of phenotypic (morphological, biochemical, and physiological) characteristics, and the identity of the isolates was confirmed by molecular techniques (DNA sequencing of the D1/D2 region of 26S rDNA and PCR-restriction fragment length polymorphism [RFLP] analysis of the internal transcribed spacer 2 [ITS2] region of rDNA) (15). Catalase-negative Malassezia isolates were subjected to detailed phenotypic and molecular characterization.
Phenotypic characterization.
For morphological studies, Malassezia cells were stained with methylene blue (0.1%) and observed under ×1,000 magnification with dynamic measuring software (DS-Fi1C-U3 digital sight color camera system; Nikon, Tokyo, Japan). The areas and lengths of ∼100 cells were measured, and the arithmetic mean values were determined. Micromorphological characteristics were studied by scanning electron microscopy (AG-EVO 40 series; Carl Zeiss, Jena, Germany). Physiological properties such as catalase activity, ability to grow at different temperatures (32°C, 37°C, and 40°C), assimilation of Tween compounds (Tween 20, 40, 60, and 80 and Cremophor EL), and hydrolysis of esculin (β-glucosidase activity) were recorded (2, 16).
Molecular characterization.
Genomic DNA extraction was carried out by harvesting yeast colonies from the surface of Leeming-Notman agar (LNA) (2). Total genomic DNA was extracted using a phenol-chloroform extraction method (6) and was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). DNA was stored at −20°C until use.
Amplification and sequencing of the D1/D2 region of 26S rDNA, the ITS and intergenic spacer 1 (IGS1) regions of rDNA, and the exon 1 region of translation elongation factor 1 alpha (TEF1) were performed with the primer pairs NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′) (17), ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (18), 26SF (5′-ATCCTTTGCAGACGACTTGA-3′) and 5SR (5′-AGCTTGACTTCGCAGATCGG-3′) (19), and EF1-983F (5′-GCYCCYGGHCAYCGTGAYTTYAT-3′) and EF1-2218R (5′-ATGACACCRACRGCRACRGTYTG-3′) (20), respectively, in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany). Purification of amplified PCR products were performed using a gel extraction kit (QIAquick; Qiagen, Bangalore, India). Sequencing was performed using the aforementioned primers and a BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems, Foster City, CA). Sequencing reactions were analyzed on an ABI 3130 genetic analyzer (Applied Biosystems). Consensus sequences obtained using Bionumerics software (version 7.1; Applied Maths, Ghent, Belgium) were compared with the GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) and CBS-KNAW Fungal Biodiversity Centre (http://www.cbs.knaw.nl) databases.
Evolutionary relationships were inferred using the neighbor-joining method (21). A bootstrap analysis was conducted with 1,000 pseudoreplicates (22), to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates were collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) were shown next to the branches (22). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using three different models (Kimura 2-parameter, maximum composite likelihood, and Tamura-Nei models) and were presented in the units of the number of base substitutions per site (23). All positions containing gaps and missing data were excluded from the analysis. Evolutionary analyses were conducted using MEGA5 software (24).
PCR-RFLP analysis of the ITS2 region was performed by following the standard method described earlier, with minor modifications (25). In addition to the three enzymes (Alu, BanI, and MSPA1) used in the previous protocol, we included BglI (Thermo Scientific, Vilnius, Lithuania) after analyzing the sequences with the software Clone Manager 6 (version 6.00; Sci-Ed, Denver, CO). Amplification, restriction, and electrophoresis followed a protocol described earlier (25).
Molecular typing of the isolates was performed using a fluorescent amplified fragment length polymorphism (FAFLP) technique as described earlier, with minor modifications (26). EcoRI and HindIII restriction enzymes (New England BioLabs, Ipswich, MA) and two adapters were used. Amplification was performed using the preselective primers EcoRI (5′-GACTGCGTACCAATTC-3′) and HindIII (5′-GACTGCGTACCAGCTT-3′). A HindIII primer with one selective nucleotide (5′-GACTGCGTACCAGCTTT-3′) and an EcoRI primer with two selective nucleotides (5′-GACTGCGTACCAATTCAC-3′; 6-carboxyfluorescein [6-FAM] labeled) were used. Capillary electrophoresis of the amplified products (labeled with 6-FAM) with a standard marker (LIZ 500; Applied Biosystems, Foster City, CA) was performed using an ABI 3130 genetic analyzer. Typing data were imported to Bionumerics software, and the similarity coefficient was determined by Pearson correlation analysis, with negative similarities clipped to zero. Cluster analysis was performed using the unweighted pair group method with arithmetic means, in Bionumerics software. Comparison of the distributions of Malassezia spp. between SD/D patients and healthy individuals was performed with the chi-square test (Epi Info software, version 7.1.0.6; Centers for Disease Control and Prevention, Atlanta, GA).
Accession numbers.
The following are the GenBank accession numbers of the sequences determined in this study: D1/D2 region of 26S rDNA, KM235687 to KM235689, KJ847247 to KJ847249, and KJ847253 to KJ847258; ITS region, KJ698644, KJ847244 to KJ847246, KJ847259 to KJ847264, KM235690, and KM235691; IGS1 region, KM235692 to KM235694, KJ847250 to KJ847252, and KJ847265 to KJ847269; TEF1 gene, KU761997, KU761998, KU894649 to KU894652, KU852491, and KU867871 to KU867875. Data for Malassezia arunalokei were deposited in MycoBank under accession no. MB 808048.
RESULTS
Species distribution.
The distribution of Malassezia spp. among seborrheic dermatitis patients and healthy individuals is given in Table 1. M. globosa was isolated significantly more often from the scalps of controls than from SD/D patients (P = 0.03), whereas M. restricta was recovered significantly more often from the scalps of SD/D patients than from controls (P = 0.02). We isolated a new Malassezia sp. from 11 patients (scalp, n = 7; nasolabial fold, n = 4) and 3 healthy individuals (scalp, n = 2; nasolabial fold, n = 1), and we named it M. arunalokei sp. nov.
TABLE 1.
Distribution of Malassezia spp. among SD/D patients and healthy individuals
| Species yielding growth | No. (%)a |
|||
|---|---|---|---|---|
| SD/D patients (n = 124) |
Controls (n = 55) |
|||
| Scalp (n = 87 [70%]) | Nasolabial fold (n = 31 [25%]) | Scalp (n = 35 [63.6%]) | Nasolabial fold (n = 12 [21.6%]) | |
| M. globosa | 31 (25) | 6 (4.8) | 18 (32.7) | 4 (7.2) |
| M. furfur | 18 (14.5) | 17 (13.8) | 5 (9.1) | 4 (7.2) |
| M. globosa + M. restricta | 10 (8) | 0 | 6 (10.9) | 0 |
| M. restricta | 21 (16.9) | 4 (3.2) | 4 (7.3) | 3 (5.4) |
| M. arunalokei sp. nov. | 7 (5.6) | 4 (3.2) | 2 (3.6) | 1 (1.8) |
| No growth | 37 (30) | 93 (75) | 20 (36.4) | 43 (78.4) |
Malassezia spp. yielding growth from samples of patients (n = 124) and controls (n = 55).
Phenotypic characteristics.
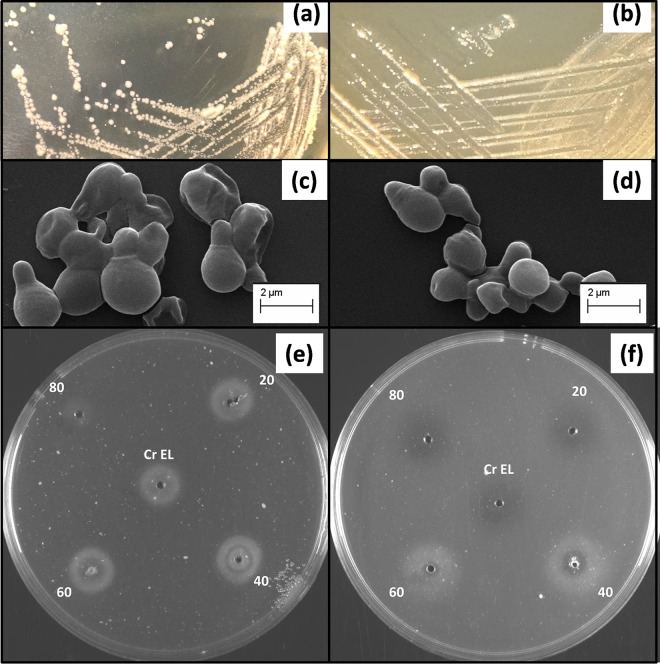
Among the catalase-negative Malassezia spp. isolated from the scalps of SD/D patients, colonies of 14 isolates were morphologically distinct from those of M. restricta, the only known catalase-negative Malassezia spp. Compared with colonies of M. restricta (flat to somewhat raised, dull, hard and brittle, pale yellowish brown, and somewhat ridged near the edge, with a lobate margin) (Fig. 1b), M. arunalokei colonies were flat, glossy, smooth, whitish to cream in color, and moderately convex, with an entire margin (Fig. 1a). On microscopic examination, yeast cells were ovoid to globose and larger (mean length, 4.0 μm; mean area, 5.5 μm2) than M. restricta cells (mean length, 2.6 μm; mean area, 3.2 μm2). Hyphae and pseudohyphae were not produced. Buds formed monopolarly on a narrow base. The scanning electron microscopic features of M. arunalokei and M. restricta are shown in Fig. 1c and d, respectively.
FIG 1.
Phenotypic characteristics differentiating M. arunalokei sp. nov. from the closely related species M. restricta. (a and b) Colonies of M. arunalokei sp. nov. (CBS 13387T) (a) and M. restricta (CBS 7877T) (b) cultured on modified Dixon's agar at 34°C. (c and d) Scanning electron microscopic images of M. arunalokei sp. nov. (CBS 13387T) (c) and M. restricta (CBS 7877T) (d) cells. (e and f) Tween 20, 40, 60, and 80 and Cremophor EL (Cr EL) assimilation by M. arunalokei sp. nov. (CBS 13387T) (e) and M. restricta (CBS 7877T) (f).
M. arunalokei isolates did not grow at or above 40°C but grew well at 37°C on modified Dixon's agar. There was no growth with media containing Tween 20, 40, 60, or 80 or Cremophor EL for M. arunalokei, but a precipitate appeared as a ring surrounding the wells containing Tween 20, 40, or 60 or Cremophor EL (Fig. 1e). The different Tween assimilation pattern of M. restricta is shown in Fig. 1f. The catalase and β-glucosidase activities of M. arunalokei were negative. Characteristics that differentiated the novel species from the 14 known Malassezia spp. are summarized in Table 2.
TABLE 2.
Salient phenotypic characteristics of Malassezia spp.a
| Species | Cell morphology | Growth on mDAb | Lipid dependency | Utilization |
Activity |
Growth |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tween 20 | Tween 40 | Tween 60 | Tween 80 | Cremophor EL | Catalase | β-Glucosidase | At 37°C | At 40°C | ||||
| M. furfur | Globose, ellipsoidal, cylindrical | + | + | + | + | + | + | + | + (−) | − (w) | + | + |
| M. pachydermatis | Ellipsoidal | + | − (w) | + | + | + | + | + | + (w) | + (−) | + | + |
| M. sympodialis | Ellipsoidal | + | + | −, w | + | + | + | − (w) | + | + | + | + |
| M. globosa | Globose | + | + | − | −, ppt | −, ppt | − | − | + | − | − (w) | − |
| M. obtusa | Ellipsoidal, cylindrical | + | + | − | − | − | − | − | + | + | − (w) | − |
| M. slooffiae | Ellipsoidal, cylindrical | + | + | +, w | + | + | − (w) | − | + | − | + | + |
| M. dermatis | Ellipsoidal, globose | + | + | + | + | + | + | w (+) | + | ? | + | + |
| M. japonica | Globose, ellipsoidal | + | + | − | w | + | − | ? | + | ? | + | − |
| M. nana | Ellipsoidal | + | + | v | + | + | w | − | + | − | + | v |
| M. yamatoensis | Ellipsoidal | + | + | + | + | + | + | ? | + | ? | + | − |
| M. caprae | Globose, ellipsoidal | + | + | − | + | + | + (−) | − | + | + (−) | − (w) | − |
| M. equina | Ellipsoidal | + | + | w | + | + | + | − | + | − | w | − |
| M. cuniculi | Globose | − (w) | + | − | − | − | − | − | + | + | + | + |
| M. restricta | Globose, ellipsoidal | + | + | − | −, ppt | −, ppt | − | − | − | − | v | − |
| M. arunalokei sp. nov. | Ovoid, globose | + | + | −, ppt | −, ppt | −, ppt | v | −, ppt | − | − | + | − |
mDA, modified Dixon's agar; +, positive; −, negative; v, variable; ?, not included in the description of the species; w, weak; ppt, appearance of precipitate or opaque zone without any growth of yeast. Parentheses indicate rare deviations from the main pattern.
Molecular analysis.
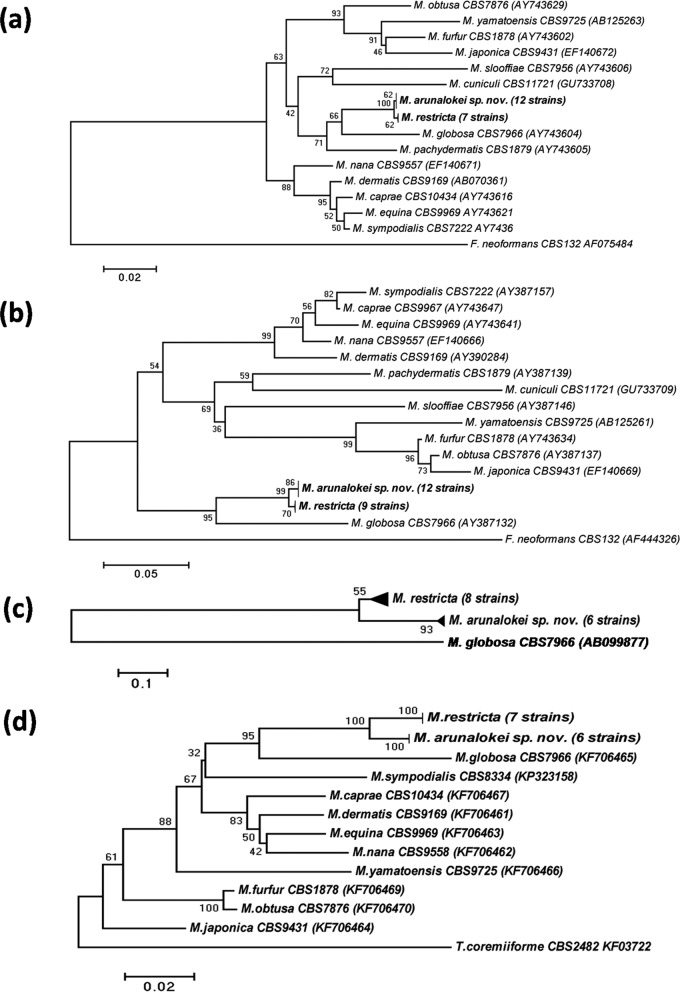
Various molecular techniques were used to confirm the novelty of the 14 isolates. In the phylogenetic analysis (D1/D2 sequences), the novel isolates clustered separately from M. restricta, with 100% bootstrap support (∼5 substitution variations) (Fig. 2a). The ITS sequence of M. arunalokei differed by 6.4% (3.4% variation in the 218-bp ITS1 region and 8.8% variation in the 273-bp ITS2 region) from the ITS sequence of M. restricta CBS 7877T and other strains of this species (99% bootstrap support) (Fig. 2b). Even in the phylogram constructed using IGS1 sequences, the novel strains clustered together and were distinctly different from M. restricta CBS 7877T and other strains of this species (Fig. 2c). Comparison of the IGS1 sequences of M. restricta and the novel species showed that they differed by 18 to 24.6%. IGS1 sequence variation ranged from 0.9 to 8.3% among the strains of M. restricta but only from 0.7 to 2.4% among the strains of M. arunalokei. Similarly, the sequences of the novel species showed 3.2% variation (885/914 bp) in the TEF1 gene, compared to M. restricta (CBS 7877T and other strains of this species), with 100% bootstrap support (Fig. 2d). Phylogenetic evolutionary distances inferred from the 26S, ITS, and IGS1 regions and the TEF1 gene with three different models (Kimura 2-parameter, maximum composite likelihood, and Tamura-Nei models) clustered M. arunalokei sp. nov. with high bootstrap values (99 to 100%). PCR-RFLP analysis using the BglI restriction enzyme digested the ITS2 region of M. restricta into two fragments, measuring 381 bp and 90 bp, whereas the novel isolates showed a single band, indicating the absence of a BglI site in the ITS2 region.
FIG 2.
Phylogenetic relationships of M. arunalokei sp. nov. and 14 recognized Malassezia spp. The phylograms of the presently known Malassezia spp. were inferred using the neighbor-joining method. The maximum composite likelihood model was used to compute evolutionary distances. The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic trees (1,000 bootstrap replicates). (a and b) Phylograms of Malassezia spp. based on the D1/D2 regions of 26S rDNA (a) and ITS-5.8S rDNA (b), using Cryptococcus neoformans CBS 132T as an outgroup. (c) Phylogram of M. arunalokei sp. nov. and M. restricta strains based on the IGS1 region, using M. globosa CBS 7996T as an outgroup. (d) Phylograms of Malassezia spp. based on the TEF1 gene, using Trichosporon coremiiforme CBS 2482 as an outgroup.
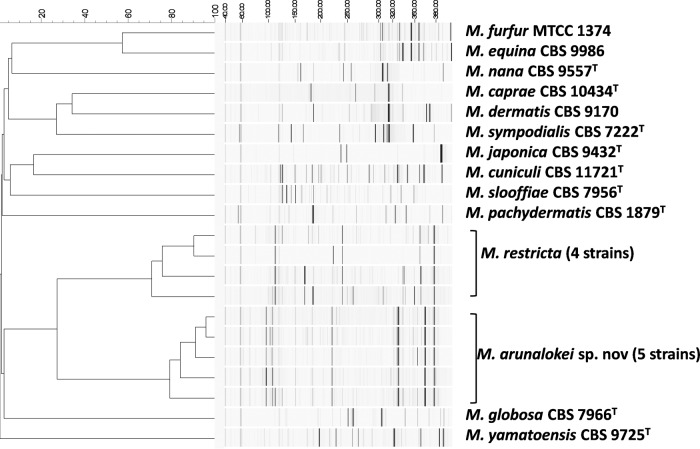
FAFLP analysis was used in the present study to differentiate the strains of M. arunalokei from M. restricta and other Malassezia spp. The FAFLP profiles of all known Malassezia spp. and five representative strains of M. arunalokei were analyzed. Fragments in the range of 40 to 350 bp were included, and approximately 100 fragments were analyzed. M. arunalokei sp. nov. isolates showed only 25% similarity to M. restricta (Fig. 3). The similarity coefficient for M. arunalokei sp. nov. isolates was 80%, and that for M. restricta isolates was 70%.
FIG 3.
FAFLP banding patterns of five representative M. arunalokei sp. nov. strains and presently known Malassezia spp. Fragments in the range of 40 to 350 bp were included in the analysis. Bars, percentages of similarity. The strains of M. arunalokei sp. nov. showed 80% similarity among themselves, but the M. restricta isolates showed only 25% similarity to the M. arunalokei sp. nov. isolates.
Taxonomy.
Malassezia arunalokei Honnavar, Rudramurthy, and Prasad, sp. nov. MycoBank accession no. MB 808048. Etymology: Malassezia arunalokei, the species epithet arunalokei (a.ru.na.lo'ke.i. N.L. masc. gen. n. arunalokei, of Arunalokei) in honor of Arunaloke Chakrabarti, for his numerous contributions to the development of medical mycology in India and Asia. Diagnosis: Malassezia arunalokei, the new proposed species in Malassezia, is closely related to M. restricta. Asexual reproduction of this yeast occurs by budding. Hyphae and pseudohyphae are not produced. After 7 days of incubation on modified Dixon's agar at 34°C, colonies are small, 2 to 3 mm in diameter on average, flat, glossy, smooth, whitish to cream in color, and moderately convex, with an entire margin. Cells of M. arunalokei are ovoid to globose, 3 to 6 by 2.5 to 4 μm (mean area, 5.5 μm2). Buds are formed monopolarly, on a narrow base. No growth on Sabouraud's dextrose agar was observed. Catalase and β-glucosidase activities are negative. This species, which is the most fastidious of the genus, did not grow at 40°C but grew at 37°C. A teleomorph is unknown. A precipitate formed in a white disk or a ring, without any growth of the fungus, surrounding the wells with Tween 20, 40, or 60 or Cremophor EL. The molecular techniques used in this study confirmed that the novel strains are distinct and differed from all other described species of the genus Malassezia. On the basis of the micromorphological features, phenotypic characteristics, modified PCR-RFLP analysis of the ITS2 region, sequence analysis of the D1/D2 region of 26S rDNA, the ITS-5.8S, IGS1 region of rDNA, and the TEF1 gene, and FAFLP data presented above, the 14 isolates are proposed as strains of a novel species in the genus Malassezia. Holotype: M. arunalokei NCCPF 127130 (National Culture Collection of Pathogenic Fungi [NCCPF], Chandigarh, India) was designated the type strain of the novel species. It was isolated from the scalp of a seborrheic dermatitis patient in Chandigarh, India, in December 2012. This strain also has been deposited in the Centraalbureau voor Schimmelcultures (CBS) (Utrecht, The Netherlands) (CBS 13387T) and the Microbial Type Culture Collection and Gene Bank (MTCC) (Chandigarh, India) (MTCC 12054T). Other representative strains of M. arunalokei include NCCPF 127125, 127126, 127127, 127128, 127129, 127131, and 127132, which are deposited at the NCCPF and at the MTCC (i.e., MTCC 12055 [NCCPF 127125] and MTCC 12056 [NCCPF 127127]).
DISCUSSION
The taxonomy of Malassezia is evolving. Unlike other yeast species, few phenotypic tests are available to differentiate Malassezia spp. Many of the phenotypic characteristics described for differentiating the 14 known species overlap, making it difficult to identify species. However, the novel species M. arunalokei could be unambiguously differentiated from the closest species, M. restricta, by better growth at 37°C on modified Dixon's agar, larger cell size (3 to 6 by 2.5 to 4 μm; mean area, 5.5 μm2), flat glossy-smooth colonies (∼2 to 3 mm) with an entire margin, and the ability to form precipitate surrounding the wells with Tween 20 and Cremophor EL and occasionally surrounding the wells with Tween 80 (Table 2).
Due to the limitations in identification by phenotypic characteristics alone, molecular methods are essential to confirm the species (27). The ITS sequences of M. arunalokei sp. nov. and M. restricta differed from each other by 6.4%, indicating that these are distinctly different species. Minor variation (<1%) in the ITS region was observed among the strains of M. arunalokei, suggesting that these are conspecific strains. Sequences of the D1/D2 region of 26S rDNA are reported to have less variation (<1%) among newly described species in Malassezia (M. caprae, M. equina, and M. dermatis) and M. sympodialis. In addition, these species share similar phenotypic characteristics (28). Ascomycetous and basidiomycetous yeasts, which differ in their ITS and D1/D2 sequences by more than 1 to 2% from the existing species, are considered novel species (28–33). Recently, the ITS region has been accepted as the universal target, superior to the large subunit (LSU) region for identification of fungi due to the presence of a clearly defined barcode gap (34). Although a previous publication reported high levels of variation of the ITS sequences within M. pachydermatis, M. globosa, and M. restricta (35), analysis of those sequences (data not shown) showed that the variation was due to the presence of a large number of indels in the ITS1 region, with few (<1%) substitutions in the ITS2 region. As the rates and patterns of indel formation are not well understood, they are generally considered “missing data” in the construction of phylograms (36). Within the ITS region, ITS2 is more reliable for the differentiation of closely related species than is ITS1 (37). Muller et al. (38) analyzed ITS sequences of >1,300 closely related species and confirmed this finding. They also suggested that ITS2 secondary structural differences between the two species are associated with sexual incompatibility (38). Recently, Amado et al. (39) showed that ITS2 secondary structures can be used for the precise identification of Malassezia spp., with good resolution due to high bootstrap support values. They predicted that the ITS2 region could accurately classify unknown isolates (39). M. arunalokei and M. restricta differed by more than 8.8% in the ITS2 region (273 bp), with 8 deletions and 16 substitutions. In addition to the transcribed regions (ITS1 and ITS2 regions), nontranscribed spacer regions (IGS regions) have been successfully used to identify fungal species and to discriminate between closely related species (40). Based on variations in the IGS1 region, Sugita et al. (41) divided M. restricta into two subgroups. In the present study, the large variations (18 to 24%) observed among the IGS1 sequences of M. arunalokei and M. restricta suggest that these are different species. Analysis of the IGS1 sequences of M. restricta strains obtained from nucleotide databanks and five of our M. restricta strains showed that all were closely related, forming a single cluster with CBS strain 7991. Divergence in exon 1 of the TEF1 gene also differentiated M. arunalokei and M. restricta. This gene has been used for the first time for Malassezia systematic analysis. The high bootstrap support values among Malassezia spp. probably show that this gene can be used as a molecular marker to identify new species and to assess diversity. The combination of four loci, i.e., three rRNA genes (D1/D2 26S rRNA, ITS-5.8S rRNA, and IGS1 regions) and a protein-encoding gene (TEF1), and three different phylogenetic approaches, i.e., Kimura 2-parameter, maximum composite likelihood, and Tamura-Nei models, provided robust support in clustering M. arunalokei sp. nov. with a high bootstrap value. PCR-RFLP analysis of ITS2 is a standard cost-effective method recommended for identification of all Malassezia spp. of human origin (25). In the present study, the standard PCR-RFLP technique failed to differentiate the novel isolates from M. restricta. The standard protocol was slightly modified to include the BglI restriction enzyme, which could cut the amplified ITS2 region of M. restricta, but not M. arunalokei sp. nov., into two fragments.
FAFLP analysis is considered one of the most discriminative molecular techniques for phylogenetic studies and genotyping (42). FAFLP analysis has been shown to differentiate closely related Ustilaginomycetes isolates that could not be discriminated by comparisons of ITS sequences (43). Compared to other random amplification techniques, FAFLP analysis has higher sensitivity, reproducibility, and resolution. As this technique screens the whole genome, it has been successfully used to differentiate closely related species such as M. sympodialis, M. caprae, M. equina, and M. dermatis, which differ by only a few substitutions in the D1/D2 domain (4). This technique has not been exploited much to type Malassezia spp. (4, 44, 45). In the present study, we showed that the FAFLP fingerprints obtained from different species of Malassezia distinctly differentiated all of the species, including distinguishing M. arunalokei sp. nov. from the phenotypically closely related species M. restricta. These fingerprint data also confirm that the M. arunalokei isolates are distinct from the other species in the genus.
Malassezia spp. implicated in SD/D vary depending on the geographical location and the methods used for detection (12). M. globosa and M. restricta are the predominant agents associated with SD/D, and M. furfur and five other species are occasionally involved (46). In our recent study (13) and in the present study, M. restricta and M. globosa were found to be the predominant species isolated from the scalps of SD/D patients and controls, both as single agents and together. In some culture-dependent studies from Canada, Sweden, and Japan, however, M. restricta was not recovered from the scalps of SD/D patients (47–49). Interestingly, a mixture of M. globosa and M. restricta was found to be predominant when DNA-based techniques were employed for detection of Malassezia spp. (28). M. restricta is a fastidious species, which can easily be masked by other fast-growing species in culture; therefore, its clinical importance might have been underestimated (46). Thorough inspection and processing of different morphological forms of the colonies from the culture plates helped us to isolate several M. restricta strains and related strains. Further examination of these isolates using various phenotypic and molecular techniques revealed the presence of a novel species related to M. restricta.
M. arunalokei was isolated from patients with either mild or moderate SD/D and from healthy controls, from both the scalp and nasolabial folds. Except for one patient, all subjects were female in the age range of 13 to 18 years. The subjects were not related, and no geographical clustering of M. arunalokei was noted. Antifungal susceptibility testing of novel species was not carried out because a standard technique for Malassezia spp. is not available. The novel species was not isolated from patients with psoriasis (15) or pityriasis versicolor (S. M. Rudramurthy, P. Honnavar, S. Dogra, S. Handa, and A. Chakrabarti, unpublished data). Interestingly, a search of GenBank revealed 99% identity of the ITS rDNA of M. arunalokei sp. nov. with that of an uncultured basidiomycete from indoor dust in Finland (GenBank accession no. FR682278), an uncultured fungus from deep sea sediments of the central Indian basin in India (GenBank accession no. GU370752), an uncultured eukaryote from sea cruise samples in south China (GenBank accession no. GU942302), and an uncultured Malasseziales clone from plant tissue cultures in Germany (GenBank accession no. EU812484). This points to the possibility that M. arunalokei occupies various ecological niches and possesses a wide geographical distribution that warrants further study. In support of our observation, Malassezia was recently described as “ecologically hyper-diverse,” suggesting that Malassezia-like fungi are among the most widespread fungi on the planet (50).
ACKNOWLEDGMENTS
We acknowledge A. S. Shamanth for his assistance in sample collection.
There are no conflicts of interest to declare for any of the authors in the study.
REFERENCES
- 1.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. 2012. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25:106–141. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gueho E, Midgley G, Guillot J. 1996. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek 69:337–355. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 3.Batra R, Boekhout T, Gueho E, Cabanes FJ, Dawson TL Jr, Gupta AK. 2005. Malassezia Baillon, emerging clinical yeasts. FEMS Yeast Res 5:1101–1113. doi: 10.1016/j.femsyr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Cabanes FJ, Theelen B, Castella G, Boekhout T. 2007. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res 7:1064–1076. doi: 10.1111/j.1567-1364.2007.00217.x. [DOI] [PubMed] [Google Scholar]
- 5.Cabanes FJ, Vega S, Castella G. 2011. Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Med Mycol 49:40–48. doi: 10.3109/13693786.2010.493562. [DOI] [PubMed] [Google Scholar]
- 6.Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance MA, Yamaguchi H, Hasegawa A. 2004. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol 54:623–627. doi: 10.1099/ijs.0.02776-0. [DOI] [PubMed] [Google Scholar]
- 7.Sugita T, Tajima M, Takashima M, Amaya M, Saito M, Tsuboi R, Nishikawa A. 2004. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol Immunol 48:579–583. doi: 10.1111/j.1348-0421.2004.tb03554.x. [DOI] [PubMed] [Google Scholar]
- 8.Sugita T, Takashima M, Kodama M, Tsuboi R, Nishikawa A. 2003. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J Clin Microbiol 41:4695–4699. doi: 10.1128/JCM.41.10.4695-4699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, Ogawa H, Nishikawa A. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol 40:1363–1367. doi: 10.1128/JCM.40.4.1363-1367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabanes FJ. 2014. Malassezia yeasts: how many species infect humans and animals? PLoS Pathog 10:e1003892. doi: 10.1371/journal.ppat.1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson TL., Jr 2007. Malassezia globosa and restricta: breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J Investig Dermatol Symp Proc 12:15–19. doi: 10.1038/sj.jidsymp.5650049. [DOI] [PubMed] [Google Scholar]
- 12.Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. 2004. Skin diseases associated with Malassezia species. J Am Acad Dermatol 51:785–798. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 13.Rudramurthy SM, Honnavar P, Dogra S, Yegneswaran PP, Handa S, Chakrabarti A. 2014. Association of Malassezia species with dandruff. Indian J Med Res 139:431–437. [PMC free article] [PubMed] [Google Scholar]
- 14.Manuel F, Ranganathan S. 2011. A new postulate on two stages of dandruff: a clinical perspective. Int J Trichology 3:3–6. doi: 10.4103/0974-7753.82117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudramurthy SM, Honnavar P, Chakrabarti A, Dogra S, Singh P, Handa S. 2014. Association of Malassezia species with psoriatic lesions. Mycoses 57:483–488. doi: 10.1111/myc.12186. [DOI] [PubMed] [Google Scholar]
- 16.Gueho E, Boekhout T, Ashbee HR, Guillot J, Van Belkum A, Faergemann J. 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med Mycol 36(Suppl 1):220–229. [PubMed] [Google Scholar]
- 17.Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 19.Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T. 2002. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J Clin Microbiol 40:1826–1830. doi: 10.1128/JCM.40.5.1826-1830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T. 2014. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33:41–47. doi: 10.3767/003158514X682313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaitanis G, Robert V, Velegraki A. 2006. Verifiable single nucleotide polymorphisms of the internal transcribed spacer 2 region for the identification of 11 Malassezia species. J Dermatol Sci 43:214–217. doi: 10.1016/j.jdermsci.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti A, Shivaprakash MR, Curfs-Breuker I, Baghela A, Klaassen CH, Meis JF. 2010. Apophysomyces elegans: epidemiology, amplified fragment length polymorphism typing, and in vitro antifungal susceptibility pattern. J Clin Microbiol 48:4580–4585. doi: 10.1128/JCM.01420-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaitanis G, Velegraki A, Mayser P, Bassukas ID. 2013. Skin diseases associated with Malassezia yeasts: facts and controversies. Clin Dermatol 31:455–463. doi: 10.1016/j.clindermatol.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Sugita T, Velegraki A. 2010. Epidemiology of Malassezia-related skin diseases, p 65–120. In Boekhout T, Gueho E, Mayser P, Velegraki A (ed), Malassezia and the skin. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 29.Kurtzman CP. 2014. Use of gene sequence analyses and genome comparisons for yeast systematics. Int J Syst Evol Microbiol 64:325–332. doi: 10.1099/ijs.0.054197-0. [DOI] [PubMed] [Google Scholar]
- 30.Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 31.Peterson SW, Kurtzman CP. 1991. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst Appl Microbiol 14:124–129. doi: 10.1016/S0723-2020(11)80289-4. [DOI] [Google Scholar]
- 32.Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res 2:495–517. doi: 10.1016/S1567-1356(02)00128-9. [DOI] [PubMed] [Google Scholar]
- 33.Sugita T, Nishikawa A, Ikeda R, Shinoda T. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol 37:1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. 2000. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol 49:29–35. doi: 10.1099/0022-1317-49-1-29. [DOI] [PubMed] [Google Scholar]
- 36.Saitou N, Ueda S. 1994. Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol Biol Evol 11:504–512. [DOI] [PubMed] [Google Scholar]
- 37.Leaw SN, Chang HC, Sun HF, Barton R, Bouchara JP, Chang TC. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microbiol 44:693–699. doi: 10.1128/JCM.44.3.693-699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller T, Philippi N, Dandekar T, Schultz J, Wolf M. 2007. Distinguishing species. RNA 13:1469–1472. doi: 10.1261/rna.617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amado Y, Patino-Uzcategui A, Cepero de Garcia MC, Tabima J, Motta A, Cardenas M, Bernal A, Restrepo S, Celis A. 2013. Seborrheic dermatitis: predisposing factors and ITS2 secondary structure for Malassezia phylogenic analysis. Med Mycol 51:868–875. doi: 10.3109/13693786.2013.820001. [DOI] [PubMed] [Google Scholar]
- 40.Harrington TC, Rizzo DM. 1999. Defining species in fungi, p 43–71. In Worrall JJ. (ed), Structure and dynamics of fungal populations. Kluwer Press, Dordrecht, The Netherlands. [Google Scholar]
- 41.Sugita T, Tajima M, Amaya M, Tsuboi R, Nishikawa A. 2004. Genotype analysis of Malassezia restricta as the major cutaneous flora in patients with atopic dermatitis and healthy subjects. Microbiol Immunol 48:755–759. doi: 10.1111/j.1348-0421.2004.tb03601.x. [DOI] [PubMed] [Google Scholar]
- 42.Klaassen CH, Osherov N. 2007. Aspergillus strain typing in the genomics era. Stud Mycol 59:47–51. doi: 10.3114/sim.2007.59.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakkeren G, Kronstad JW, Levesque CA. 2000. Comparison of AFLP fingerprints and ITS sequences as phylogenetic markers in Ustilagomycetes. Mycologia 92:510–521. doi: 10.2307/3761510. [DOI] [Google Scholar]
- 44.Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. 2004. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol 42:4253–4260. doi: 10.1128/JCM.42.9.4253-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theelen B, Silvestri M, Gueho E, van Belkum A, Boekhout T. 2001. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res 1:79–86. doi: 10.1111/j.1567-1364.2001.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 46.Kellermann EBR, Boekhout T. 2011. Malassezia Baillon, p 1807–1832. In Kurtzman C, Fell JW, Boekhout T (ed), The yeasts: a taxonomic study, 5th ed Elsevier, London, United Kingdom. [Google Scholar]
- 47.Gupta AK, Kohli Y, Summerbell RC, Faergemann J. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med Mycol 39:243–251. doi: 10.1080/mmy.39.3.243.251. [DOI] [PubMed] [Google Scholar]
- 48.Nakabayashi A, Sei Y, Guillot J. 2000. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol 38:337–341. doi: 10.1080/mmy.38.5.337.341. [DOI] [PubMed] [Google Scholar]
- 49.Sandstrom Falk MH, Tengvall Linder M, Johansson C, Bartosik J, Back O, Sarnhult T, Wahlgren CF, Scheynius A, Faergemann J. 2005. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm Venereol 85:17–23. doi: 10.1080/00015550410022276. [DOI] [PubMed] [Google Scholar]
- 50.Amend A. 2014. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog 10:e1004277. doi: 10.1371/journal.ppat.1004277. [DOI] [PMC free article] [PubMed] [Google Scholar]